Abstract
Sustained activity of mammalian methionine synthase (MS) requires MS reductase (MSR), but there have been few studies of the interactions between these two proteins. In this study, recombinant human MS (hMS) and MSR (hMSR) were expressed in baculovirus-infected insect cells and purified to homogeneity. hMSR maintained hMS activity at a 1:1 stoichiometric ratio with a Kact value of 71 nM. Escherichia coli MS, however, was not activated by hMSR. Moreover, hMS was not significantly active in the presence of E. coli flavodoxin and flavodoxin reductase, which maintain the activity of E. coli MS. These results indicate that recognition of MS by their reductive partners is very strict, despite the high homology between MS from different species. The effects of hMSR on the formation of hMS holoenzyme also were examined by using crude extracts of baculovirus-infected insect cells containing hMS apoenzyme (apoMS). In the presence of MSR and NADPH, holoenzyme formation from apoMS and methylcobalamin was significantly enhanced. The observed stimulation is shown to be due to stabilization of human apoMS in the presence of MSR. Apoenzyme alone is quite unstable at 37°C. MSR also is able to reduce aquacobalamin to cob(II)alamin in the presence of NADPH, and this reduction leads to stimulation of the conversion of apoMS and aquacobalamin to MS holoenzyme. Based on these findings, we propose that MSR serves as a special chaperone for hMS and as an aquacobalamin reductase, rather than acting solely in the reductive activation of MS.
Keywords: aquacobalamin reductase, cobalamin, holoenzyme formation
Bioinformatic methods permit us to forecast protein structure and function from deduced amino acid sequences predicted by DNA sequences. Nevertheless, it is important to confirm the predicted functions of novel proteins to ascertain actual functions and to verify the scope of the predictions made based on the similarity of conserved domains in evolutionarily distant proteins. The current study concerns the in vivo function of human methionine synthase reductase (hMSR), identified as the reactivating enzyme of human methionine synthase (hMS) on genetic and biochemical grounds and by sequence and domain similarity to two Escherichia coli proteins, flavodoxin and ferredoxin (flavodoxin)-NADP+ oxidoreductase (FNR), the reactivating enzymes of bacterial MS.
The importance of MS to human metabolism relates to the MS-catalyzed conversion of homocysteine to methionine (Met) by transfer of a methyl group from methyltetrahydrofolate (CH3-H4folate) as shown by
Homocysteine is generated by the transfer of the methyl group of Met, via an S-adenosylmethionine (AdoMet) intermediate to hundreds of methyl acceptors that contribute to all facets of metabolism and homeostasis in cells. This transfer of methyl groups derived from Met and the regeneration of Met by methylation of homocysteine constitute the Met cycle in eukaryotic cells. It is intimately involved with folate metabolism via the CH3-H4folate substrate and critically dependent on B12 [cobalamin (Cbl)] as the methylcobalamin (MeCbl) cofactor of MS. An elevated level of homocysteine in blood has garnered much attention as a putative risk factor for cardiovascular disease (1, 2) and neural tube defects (3). Given its critical participation in the remethylation of homocysteine, low folate status leads to elevated plasma homocysteine (1), and dietary supplementation with folate can significantly lower homocysteine levels (4).
E. coli MS (MetH) is the most characterized Cbl-dependent MS. The bacterial enzyme shares high homology (55% identity in deduced amino acid sequences) with hMS (5). The Cbl cofactor acts as an intermediate in the methyl transfer catalyzed by MS, accepting methyl groups from CH3-H4folate and donating them to homocysteine to form Met. Demethylation of MeCbl leaves the Cbl cofactor as cob(I)alamin, which is very sensitive to oxidation. Under the microaerophilic conditions typical of mammalian cells, the cofactor is oxidized about once in every 2,000 turnovers (6), resulting in the accumulation of inactive enzyme. The inactivated MS needs reductive reactivation of the Cbl to recover its catalytic function, requiring AdoMet and an electron donor to return the cofactor to MeCbl (8, 9, ‖). For in vitro assays, DTT with aquacobalamin (aqCbl), reduced flavin mononucleotide (9), or Ti(III) citrate (10) are commonly used as electron donors. E. coli MS also can be reactivated enzymatically in vitro by using flavodoxin, FNR, and NADPH (11). Flavodoxin is a small flavin mononucleotide-containing, electron-transfer protein that is not present in mammalian cells. For mammalian MS, MSR has been shown to be the reductive partner (12, 13). hMSR shares homology with E. coli flavodoxin and FNR at its N and C termini, respectively, a structural configuration that places hMSR in the P450 reductase family. As expected, recombinant hMSR expressed in E. coli cells is a dual flavoprotein-containing flavin mononucleotide and FAD (13).
hMSR was cloned in 1998 (12), and the enzyme was shown to activate porcine MS in vitro (13, 14). Extensive studies of the interactions of purified hMSR with NADPH have been performed (15–17). However, overexpression of an active recombinant hMS has heretofore not been achieved, preventing biochemical studies of the interactions between hMS and hMSR. In this paper, we describe the successful overexpression of hMS apoenzyme in a baculovirus–insect cell expression system and its purification to homogeneity on a Cbl-affinity column. We show that purified hMSR, in addition to catalyzing the reductive reactivation of MS, specifically stabilizes the apoenzyme as well and serves as an independent aqCbl reductase in the generation of a suitable cofactor substrate for the formation of MS holoenzyme (holoMS).
Results
Expression and Purification of hMS and hMSR.
Recombinant hMS expressed in insect cells was almost entirely apoenzyme as judged by the lack of enzyme activity in the absence of added MeCbl. This finding is consistent with our previous report that rat MS is expressed as an apoenzyme in baculovirus-infected insect cell cultures (18). hMS was purified by using Cbl-affinity chromatography. This procedure allowed us to obtain nearly homogenous protein in a single step and to form holoenzyme at the same time. After the affinity purification, the eluate was treated with chitin beads, because the only contamination at this point was a viral chitinase that was identified by tryptic fragment mass fingerprinting analysis (data not shown). By this method, purified holoMS could be obtained in a simple two-step procedure (Table 1).
Table 1.
Purification of hMS
| Total protein, mg | Total activity, nmol/min | Specific activity, nmol·min−1·mg−1 | Yield, % | Purification, n-fold | |
|---|---|---|---|---|---|
| Crude extract | 366 | 2,530 | 6.9 | 100 | 1 |
| B12-affinity | 0.47 | 823 | 1,750 | 32 | 253 |
| Chitin beads | 0.35 | 678 | 1,940 | 27 | 279 |
Six grams of wet Sf9 cells infected with recombinant baculovirus was used. MS activity was measured by using the aqCbl/DTT reducing system, as described in Materials and Methods. The activity in the crude insect cell extract, where MS is present as apoenzyme, was measured after incubation of the apoMS with 50 μM MeCbl for 15 min at 37°C to reconstitute holoMS.
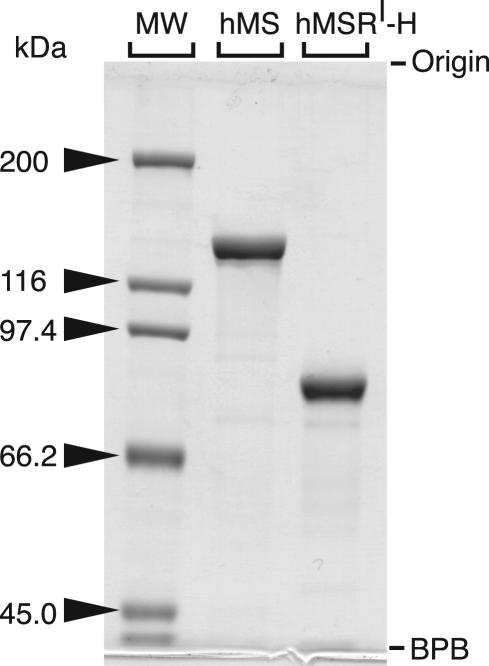
hMSR has been expressed in E. coli as a glutatione S-transferase fusion protein (13). We have achieved hMSR expression by using baculovirus-infected insect cells to produce the protein containing an N-terminal His tag. The smaller tag was expected to facilitate studies of the interaction of MS with MSR. The protein is purified by a simple two-step purification procedure using DE-52 followed by Ni-affinity chromatography. We found that the flavin mononucleotide cofactor readily dissociates from hMSR upon dilution (K.Y., C. L. Elmore, and R.G.M., unpublished data). Hence, it is important that the concentration of MSR be kept high whenever possible during purification. The temperature was maintained at 4°C throughout the procedure. An SDS/PAGE analysis of purified hMS and hMSR is shown in Fig. 1.
Fig. 1.
SDS/PAGE of purified hMS and hMSR. Purity was analyzed with 7.5% acrylamide gel electrophoresis under denaturing conditions. The gel was stained with Coomassie brilliant blue R-250 (Sigma). MW, molecular marker (Bio-Rad); hMSRI-H, Ile-22 variant of His-tagged hMSR; BPB, bromophenol blue.
Reductive Activation of hMS by hMSR.
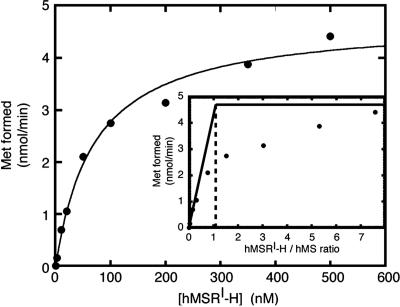
The primary function of MSR is expected to be the reductive activation of MS. Because of our purification method, the MS isolated was holoenzyme, which, after its release from the Cbl-affinity column by photolysis, contained bound a mixture of cob(II)alamin and aqCbl. This enzyme is inactive, and MSR would be required to catalyze the initial reduction of the bound cofactor before the priming methylation using the methyl group of AdoMet. Fig. 2 shows the dependence of MS activity on the concentration of MSR. MS activities were determined in the presence of various concentrations of MSR using purified proteins. When Eq. 2 was applied to the data, the Kact value was estimated to be 71 nM.
A titration to determine the stoichiometry is shown in Fig. 2 Inset. The initial slope of the titration indicates that a 1:1 ratio of MSR to MS leads to maximal activation.
Fig. 2.
Activation of hMS in the presence of hMSR. Purified MS (2.3 μg), whose specific activity was 2.2 μmol·min−1·mg−1 protein as measured by using the aqCbl/DTT reducing system, and the indicated amount of purified MSR were used for each reaction. MS activity was measured under aerobic conditions as described in Materials and Methods, with DTT present to allow nonenzymatic reduction of aqCbl to cob(II)alamin. The reactions were initiated by addition of AdoMet after a 3-min preincubation and quenched after 1 min. Shown is Met formation as a function of MSR concentration. (Inset) The stoichiometry of the reaction.
Spectral Properties of Recombinant hMS.
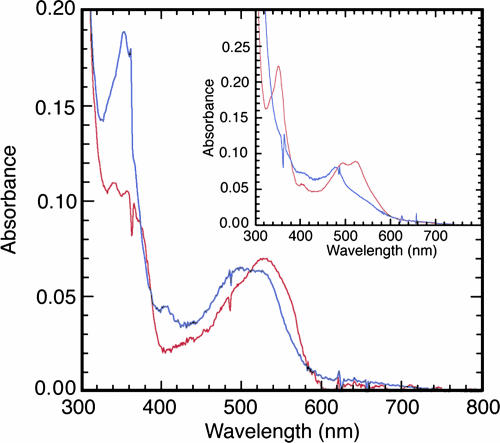
The spectrum of purified hMS as obtained after release from the Cbl-affinity column is shown in Fig. 3Inset and resembles that of aqCbl. The spectrum is changed to one typical of cob(II)alamin after incubation with DTT (Fig. 3 Inset), suggesting that the enzyme-bound cofactor can be reduced by excess DTT. To assess the spectrum of MS after treatment with MSR, a spectrophotometric assay of MSR-dependent reductive methylation of hMS was then performed as described in Materials and Methods. Spectra of hMS as isolated, incubated in the presence or absence of MSR but in the absence of DTT, and then purified by removal of the MSR, are shown in Fig. 3. After incubation with MSR, AdoMet, and NADPH, the spectrum of hMS resembles that of MeCbl, indicating that the Cbl cofactor of hMS was successfully methylated in an MSR-dependent manner.
Fig. 3.
Spectral properties of recombinant hMS. hMS was incubated with AdoMet and NADPH in the presence or absence of equimolar MSR and in the absence of DTT, and then the MSR was removed as described in Materials and Methods. The resulting spectra are shown by red and blue lines, indicating the presence and the absence of MSR, respectively. (Inset) The spectrum of purified hMS in 50 mM potassium phosphate buffer, pH 7.2, before (red line) and after (blue line) incubation with DTT.
Reductive Partners.
The effects of various reducing systems on the activity of purified hMS and E. coli MS were examined (Table 2). MS activities, measured as the formation of Met from CH3-H4folate and homocysteine, were determined as described in Materials and Methods. hMS was as active in the presence of MSR and NADPH as it was when the chemical aqCbl/DTT reducing system was used. The E. coli reducing system of flavodoxin and FNR was not effective for hMS, giving negligible Met formation when it was used in place of the chemical reducing system. Species restriction was also observed for the E. coli reducing system. E. coli flavodoxin and FNR activated E. coli MS, but human MSR did not activate the bacterial enzyme, even in the presence of a 10-fold higher (1.2 μM) concentration of MSR (data not shown).
Table 2.
Effects of reducing systems on Met formation
| Reducing system | Met formation, nmol/min |
|
|---|---|---|
| hMS | E. coli MS | |
| hMSRI-H, NADPH, DTT | 0.25 | <0.01 |
| E. coli Fld and FNR, NADPH, DTT | 0.02 | 0.20 |
| aqCbl, DTT | 0.26 | 0.50 |
| DTT | 0.03 | <0.01 |
Met formation was measured by using physiological MS activity assays as described in Materials and Methods. These assays were performed under semianaerobic conditions, in which the assay solution was bubbled with argon and the reaction was initiated by addition of the proteins and CH3-H4 folate. All proteins used in this assay were purified to apparent homogeneity. Concentrations of protein components were as follows: 0.12 μM hMSRI-H, 1 μM E. coli Fld (flavodoxin), 1μM E. coli FNR, 3 nM E. coli MS, and 50 nM hMS.
MS Apoenzyme (apoMS) Protection by MSR.
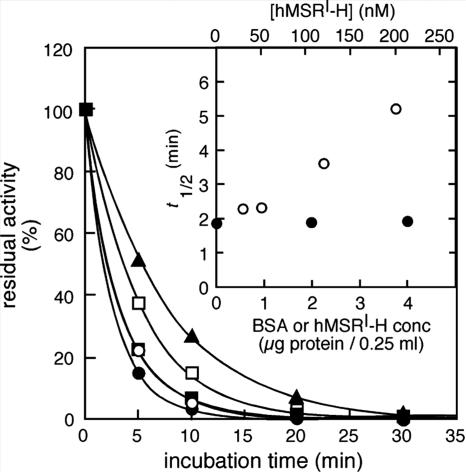
Previous studies had indicated that rat liver apoMS was unstable at 37°C (18). We were therefore interested in determining whether human apoMS was similarly unstable, and whether MSR might affect its stability. The stability of apoMS was determined at 37°C by preincubating the protein in the presence of various concentrations of MSR (Fig. 4). Apoenzyme preincubated in the absence of MSR almost completely lost the ability to form holoenzyme after the subsequent addition of MeCbl and incubation at 37°C for 10 min. Reconstitution of MS with MeCbl does not require reduction and was therefore expected to be independent of the presence of MSR. However, in the presence of MSR, dramatically increased holoenzyme formation was seen, and the amount of holoenzyme depended on the concentration of added MSR. BSA at the same concentration did not increase holoenzyme formation during the incubation. These results indicate that apoMS is specifically stabilized in the presence of MSR. During the time course, band intensities of hMS on an SDS polyacrylamide gel were not changed (data not shown). Thus, the reduction in enzyme activity with time of incubation is probably explained by denaturation of apoenzyme rather than by protein degradation. In addition, denatured apoMS was not reactivated by MSR, given the fact that addition of MSR after preincubation at 37°C without MSR could not restore holoenzyme formation to initial levels.
Fig. 4.
Protection of apoMS from denaturation in the presence of hMSR. A crude extract of Sf21 cells containing human apoMS was incubated with various concentrations of purified MSR: 0 (●), 30 (○), 50 (■), 120 (□), and 200 (▴) nM at 37°C. At the indicated time points, aliquots of the mixture were used to measure holoenzyme formation in the presence of MeCbl to estimate residual active apoenzyme as described in Materials and Methods. Residual activity is plotted against incubation time. (Inset) The t1/2 values for apoenzyme denaturation are plotted as a function of the concentration of MSR in the preincubation (○). In a control experiment, the crude extract was incubated with equivalent amounts of BSA (●).
Activity of hMSR as holoMS Synthase.
In early studies, it was shown that a two-component system, presumably flavodoxin and FNR, can function as a holoMS synthase in E. coli by promoting the addition of a Cbl cofactor to apoMS (19, 20). We have explored the possibility that hMSR can function in a similar fashion, exhibiting holoMS synthase activity. Because of the extreme instability of apoMS, apoenzyme in crude extracts of Sf cells was treated with a gel filtration column to remove small molecules and used immediately for the assay. Apoenzyme in crude extract was incubated at 37°C for 15 min with or without MSR and various forms of Cbl. Formation of holoenzyme was evaluated by measuring MS activity using aqCbl and DTT (Table 3). apoMS that failed to form holoenzyme was inactivated during the incubation (see last line of Table 3 showing precincubation without additions), and thus the amount of holoMS formed could be estimated from the resulting enzyme activity. When MeCbl was used as a prosthetic group in the absence of MSR and NADPH, MS bound the cofactor with high efficiency, generating significant MS activity. In the presence of MSR and NADPH, holoenzyme formation with MeCbl was more than double that of MS incubated with MeCbl alone. aqCbl alone was not effective in the generation of active holoenzyme. In the presence of MSR, however, holoenzyme formation from aqCbl is increased >10-fold. Holoenzyme formation in the presence of MSR was further enhanced in the presence of NADPH, although holoenzyme formation was not stimulated by NADPH alone. The effects of MSR and NADPH on holoMS formation in the presence of aqCbl were observed more than five times in independent experiments. Thus MSR apparently functions as a holoMS synthase.
Table 3.
Holoenzyme synthase activity of hMSR
| apoMS treatment | MS activity, nmol/min |
|---|---|
| MeCbl | |
| w/hMSRI-H and NADPH | 0.83 |
| w/hMSRI-H | 0.62 |
| w/NADPH | 0.39 |
| w/o hMSRI-H and NADPH | 0.35 |
| aqCbl | |
| w/hMSRI-H and NADPH | 0.24 |
| w/hMSRI-H | 0.11 |
| w/NADPH | 0.02 |
| w/o hMSRI-H and NADPH | 0.01 |
| No Cbl | |
| w/hMSRI-H and NADPH | 0.06 |
| w/o hMSRI-H and NADPH | <0.01 |
A crude extract from Sf9 cells infected with recombinant baculovirus was used as the source of human apoMS. A mixture (total volume was 200 μl) that contained 150 μl of crude extract (2.2 mg/ml protein) and the components indicated (w/, with; w/o, without) was preincubated for 15 min at 37°C prior to measurement of MS activity by the aqCbl/DTT assay. The concentrations of the components were 50 μM MeCbl or aqCbl, 200 nM hMSRI-H, and 200 μM NADPH.
The holoMS synthase activity of hMSR could result from stabilization of the apoMS in the presence of MSR during the preincubation and/or from NADPH-dependent reduction of exogenous aqCbl to cob(II)alamin, which is more readily incorporated into MS (see Discussion). The latter activity would depend on the inclusion of NADPH in the preincubations.
Reduction of Free aqCbl by Human MSR.
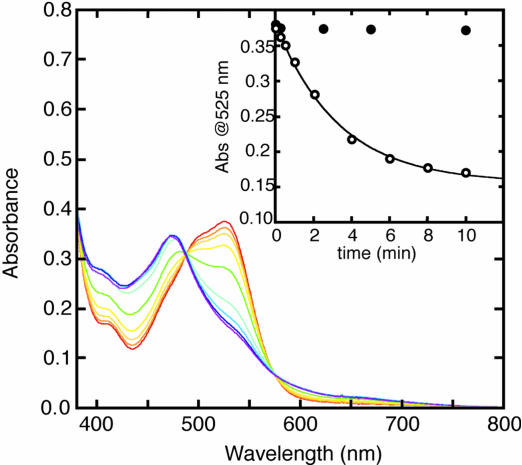
As shown in Fig. 5, MSR acts as an aqCbl reductase at 37°C. The turnover number was estimated to be 220 min−1 by using the differential molecular extinction coefficient of aqCbl to cob(II)alamin of 5.5 × 103 M−1·cm−1 at 525 nm (21). The Km value for aqCbl is ≈3.7 μM (data not shown). Reduction was not observed at 22°C (data not shown). This unanticipated property of hMSR probably explains the pronounced stimulation of holoenzyme formation from aqCbl shown in Table 3.
Fig. 5.
Reduction of aqCbl by hMSR. The reduction of aqCbl was monitored in a mixture containing 54 nM hMSR, 150 μM NADPH, and 35 μM aqCbl in aerobic 50 mM potassium phosphate buffer, pH 7.2, at 37°C. The initial spectrum is shown by the red line, and the final spectrum, which resembles the spectrum of cob(II)alamin, is shown by the purple line. (Inset) The spectral changes at 525 nm in the presence (○) or absence (●) of MSR.
Discussion
Although some of the properties of hMSR revealed by these studies were in good agreement with predictions from earlier biochemical and bioinformatic analyses of mammalian MSR, unexpected properties of hMSR became evident. The lack of crossreactivity between the E. coli and mammalian activation systems and the instability of apoMS at 37°C may have been responsible for the failure to overexpress hMS in E. coli and suggests that coexpression with MSR may be a plausible strategy for successful overexpression.
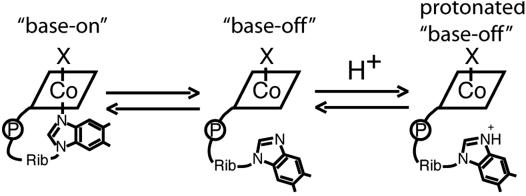
Perhaps most importantly, our studies have revealed a role for MSR in the stabilization of apoMS and the incorporation of Cbl into the enzyme. A fundamental problem in the successful conversion of apo- to holoMS is the requirement that the dimethylbenzimidazole base of Cbl be dissociated from the free cofactor and substituted by coordination of a His residue from the protein (22). The propensity for dissociation of dimethylbenzimidazole is strongly influenced by the nature of the cobalt substituent in the upper axial position. When the upper axial ligand is strongly electron-donating, the dissociation of dimethylbenzimidazole is more favorable than when the ligand is weakly electron-donating. As shown in Scheme 1, measurement of the apparent pKa of the dimethylbenzimidazole base of Cbl allows an estimation of the effect of the upper axial substituent on the equilibrium between base-off and base-on species. Our earlier studies on holoMS formation in the rat, conducted in the absence of MSR, had established that the apparent pKa of the dimethylbenzimidazole base of Cbl was critical for holoMS formation (18). MeCbl has an apparent pKa of +2.7 (23), whereas that of aqCbl is −2.4 (24), indicating an ≈100,000-fold increase in the proportion of the free cofactor in the base-off form in neutral solution. Thus, incorporation of MeCbl into apoMS is greatly favored over incorporation of aqCbl.
Scheme 1.
Equilibria defining the pKa values observed for Cbl derivatives. Shown are the equilibria for free Cbl derivatives, in which X is the upper axial ligand [HOH for aqCbl, CH3 for MeCbl, or an electron for cob(II)alamin]. The first step is the dissociation of the dimethylbenzimidazole ribonucleotide from the cobalt, and the second step is the protonation of the dissociated dimethylbenzimidazole ribonucleotide.
The stimulation of holoMS formation from apoMS in the presence of NADPH and MSR may be explained in part by the newly demonstrated aqCbl reductase activity of MSR. The apparent pKa for Cbl defined by Scheme 1 is dramatically changed by reduction; in aqCbl, where the cobalt is formally +3, the pKa = −2.4; whereas, in cob(II)alamin, the pKa = +2.5 (25). The pKa value of cob(II)alamin is comparable with that of MeCbl. Thus, one way in which MSR stimulates holoMS formation may be by catalyzing the reduction of aqCbl to cob(II)alamin, therefore providing a cofactor that is more readily incorporated into apoMS because its dimethylbenzimidazole base dissociates more readily from the cobalt.
Before this study, the proteins responsible for aqCbl reductase activity in the cell had not been identified. Watanabe et al. (21) described the purification of NADH- and NADPH-dependent aqCbl reductases in cells, although the genes corresponding to these proteins were not identified. Genetic complementation has been used to identify genes that potentially might be involved in Cbl reduction because they lead to defective synthesis of MeCbl and adenosylcobalamin (cblC, cblD, and cblF complementation groups) (reviewed in ref. 26). Deficiency in cblF results in defective lysosomal Cbl transport (27), and the cblC gene was recently shown to be a homologue of TonB, a bacterial protein involved in Cbl transport (28). We have now demonstrated an aqCbl reductase function for MSR. MSR deficiency, identified in the cblE complementation group (12), results in normal formation of adenosylcobalamin, but MeCbl synthesis is defective. Our results suggest that Cbl reductases may be specific for either MeCbl or adenosylcobalamin generation, perhaps because they will be associated closely with MS or methylmalonyl CoA mutase (the only adenosylcobalamin-containing enzyme in mammalian cells). Although an ATP:Cbl adenosyltransferase also is required for the biosynthesis of adenosylcobalamin (29, 30), a comparable enzyme catalyzing formation of MeCbl has not been identified, and we believe that MS itself is the only source of MeCbl in mammalian cells.
We have also shown that free cob(II)alamin can be converted to enzyme-bound MeCbl by the combined action of MSR and MS. The ability of MSR to stabilize apoMS is likely to be important in vivo. The tissue concentration of Cbl is very low, so that the stabilizing function would be needed to allow time for Cbl to encounter apoMS. We note that this property of MSR was not expected from the genetic and bioinformatic analysis of MSR, the MTRR gene product, but could only be revealed by biochemical characterization of the purified enzyme.
Materials and Methods
Reagents.
d,l-homocysteine was obtained from Aldrich. CH3-H4folate and radiolabeled 14CH3-H4folate were purchased from Eprova and Amersham Pharmacia, respectively. MeCbl, aqCbl (which is a tautomer of hydroxocobalamin and the dominant species at pH 7.2), and AdoMet (chloride salt) were obtained from Sigma.
Expression of hMS and hMSR.
hMS and hMSR were expressed by using a baculovirus–insect cell expression system. The cDNA for hMS (5) was provided by Barry Shane (University of California, Berkeley). The MS cDNA was inserted into a pFastBac 1 donor vector (Invitrogen) by using recognition sites for BamHI and SalI endonucleases and was designated pFB1(hMSwt). The full-length hMSR cDNA (12) was amplified by PCR using a human fibroblast cDNA library with oligonucleotides 5′-GgggatccATGA-GGAGGTTTCTGTTACTATA-3′ and 5′-gggtcgacTTATGACCAAATATCCTGAA-GG-3′ as sense and antisense primers, respectively, and then subcloned into a pQE-30 vector (Qiagen, Valencia, CA). The clone had two missense mutations at 525T → C and 769T → A by comparison with the original MSR cDNA reported by LeClerc et al. (12). To fix these missense mutations and to change the 66G to A for MSRIle-22 (MSRI) expression, primer–PCR-based site-directed mutagenesis was done. The cDNA was subcloned into a donor vector pFastBacHTb (Invitrogen) by using BamHI and SalI sites and designated pFBHT(hMSRI). The DNA sequence of the His-tagged hMSR is shown in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Recombinant baculovirus for human protein expression were generated by using the Bac-to-Bac expression system (Invitrogen). This procedure allows expression of untagged hMS or N-terminally His-tagged hMSR in insect cells. Both donor vector cDNA sequences were confirmed by DNA sequencing. Sf9 and Sf21 cells (Invitrogen) were used for amplification of baculovirus stocks and overexpression of these proteins with suspension or monolayer culture, depending on the required amount of baculovirus stocks and cells. Sf cells were maintained at 27°C by using Sf900-II SFM (Invitrogen) in the presence of gentamycin.
hMS Purification.
For hMS purification, Cbl-affinity chromatography (31) was used as the first step. Cbl-agarose was prepared according to Sato et al. (31) with minor modification. Commercially available ω-aminohexylagarose (Sigma) was used instead of synthesizing aminohexylagarose from cyanogen bromide-activated resin. Our preparations of Cbl-agarose contained >70 nmol of immobilized Cbl per milliliter of resin. Sf cells (≈6 g of wet weight from an ≈1.2-liter suspension culture) infected with recombinant baculovirus carrying hMS cDNA were suspended in 50 mM potassium phosphate buffer and sonicated. After centrifugation twice at 10,000 × g for 10 min at 4°C, supernatant was collected as a crude extract. The crude extract was incubated with Cbl-agarose (2 ml) at 22°C for 15 min. The resin was washed with 50 mM potassium phosphate buffer, followed by 50 mM phosphate buffer containing 1 M NaCl. After reequilibrating the resin with 10 mM Tris·HCl buffer (pH 7.2), the resin, as a 50% slurry in the same buffer, was put in a 10-ml beaker and exposed to light (500-W tungsten lamp) for 1 min at 0°C. The released holoMS was eluted with 10 mM Tris·HCl buffer containing 0.5 M NaCl and then dialyzed against 50 mM potassium phosphate buffer at 4°C overnight. The dialyzate was passed through a 1-ml bed volume of chitin beads (New England Biolabs) to remove a viral chitinase that was the major remaining impurity (see Results). Throughout the purification, protein concentrations were determined with a Bio-Rad protein assay kit. Purified hMS was concentrated, and the protein was stored at −80°C.
hMSR Purification.
Sf cells (≈20 g of wet weight from an ≈2.5-liter suspension culture) infected with recombinant baculovirus derived from the pFBHT(hMSRI) donor vector were suspended in 50 mM potassium phosphate buffer (pH 7.2) containing 1 mM phenyl methyl sulfonyl fluoride and sonicated. Supernatant from centrifugation at 10,000 × g for 30 min at 4°C was used as a crude extract. The crude extract was mixed with 20 g of DE-52 (DEAE-cellulose; Whatman) at 4°C for 30 min, and the slurry was packed into an empty 2.5-cm column. The column was washed with 50 mM potassium phosphate buffer, and then the protein was eluted with 50 mM potassium phosphate buffer containing 0.3 M NaCl. Yellow fractions were combined and applied to a 5-ml Ni-affinity column (Hi-Trap column; Amersham Pharmacia). The column was washed with 50 mM potassium phosphate buffer containing 0.3 M NaCl and 0.02 M imidazole and then with 50 mM potassium phosphate buffer containing 0.3 M NaCl and 0.1 M imidazole. hMSR was eluted with 50 mM potassium phosphate buffer containing 0.3 M NaCl and 0.2 M imidazole. The eluate was collected and dialyzed overnight against 25 mM potassium phosphate buffer containing 0.5 mM EDTA at 4°C. The dialyzate was concentrated, the protein concentration was determined based on the visible absorbance of the flavin cofactors, and the protein was stored at −80°C.
Purified E. coli MS, Flavodoxin, and FNR.
Recombinant wild-type E. coli MS, flavodoxin, and His-tagged FNR were prepared according to the methods of Jarrett et al. (32) and Hall et al. (7).
MS Assay.
An assay of MS activity using aqCbl and DTT was performed as reported in ref. 18 with minor modifications. An assay mixture (250 μl total) consisted of 1 mM homocysteine, 250 μM14CH3-H4folate (1,200 dpm/nmol), 100 μM AdoMet, 25 mM DTT, 50 μM aqCbl, and MS in 0.2 M potassium phosphate buffer, pH 7.2. The enzymatic reaction was performed at 37°C for 10 min or as otherwise noted and quenched at 90°C for 3 min. After cooling down the mixture, a 200-μl portion was loaded onto an AG 1×8 column (Bio-Rad), and the column was washed with 1.5 ml of water. The radioactivity of the combined pass-through and wash fractions was quantitated by scintillation counting. For assays using physiological reducing systems, the same assay mixture was used except that aqCbl was replaced with NADPH (100 μM) and MSR or flavodoxin and FNR.
MSR-Mediated Methylation of hMS.
The spectrum of 32 μM enzyme in 50 mM potassium phosphate buffer, pH 7.2, was recorded, and then the enzyme was incubated with 1.2 mM AdoMet and 300 μM NADPH for 5 min at room temperature in the presence or absence of equimolar MSR. The mixture was loaded onto a 100-μl Ni-NTA spin column (Qiagen) that had been equilibrated with 50 mM potassium phosphate buffer containing 20 mM imidazole. The resin was washed with 50 μl of the equilibrating buffer. The eluted fractions were combined and passed through a Bio-Spin column (Bio-Rad) equilibrated with 50 mM potassium phosphate buffer, pH 7.2.
Measurement of holoMS Synthase Activity of MSR.
holoMS synthase activity was measured with a crude extract of Sf21 cells containing human apoMS after the removal of small molecules by gel filtration. The extract, with additions as noted, was incubated at 37°C for 15 min. holoMS activity was then measured by using aqCbl and DTT, as described in MS Assay. For experiments in which the ability of MSR to stabilize apoMS was measured, aliquots of the preincubation mixture were withdrawn at specified time points and incubated for 5 min at 37°C with 50 μM MeCbl/200 μM NADPH/0.2 μM MSR to generate holoMS from residual active apoMS, and the holoMS activity was measured by using the aqCbl/DTT assay.
Supplementary Material
Acknowledgments
We thank Prof. Barry Shane for sharing hMS cDNA and Vahe Bandarian (University of Arizona, Tucson), Diane Hall (University of Michigan, Ann Arbor), and Angela Fleischacker (University of Michigan, Ann Arbor) for providing purified bacterial proteins. This work was supported in part by National Institutes of Health Grants GM24908 (to R.G.M.) and HL58955 (to R.A.G. and R.G.M.).
Abbreviations
- MS
methionine synthase
- hMS
human MS
- MSR
MS reductase
- hMSR
human MSR
- hMSR
I-H, Ile-22 variant of His-tagged hMSR
- apoMS
MS apoenzyme
- holoMS
MS holoenzyme
- FNR
ferredoxin (flavodoxin)-NADP+ oxidoreductase
- Met
methionine
- AdoMet
S-adenosylmethionine
- Cbl
cobalamin
- MeCbl
methylcobalamin
- aqCbl
aquacobalamin.
Footnotes
Conflict of interest statement: No conflicts declared.
‖Mangum, J. H. & Scrimgeour, K. G. (1962) Fed. Proc. 21, 242 (abstr.).
References
- 1.Boushey C. J., Beresford S. A. A., Omenn G. S., Motulsky A. G. J. Am. Med. Assn. 1995;274:1049–1057. doi: 10.1001/jama.1995.03530130055028. [DOI] [PubMed] [Google Scholar]
- 2.Graham I. M., Daly L. E., Refsum H. M., Robinson K., Brattstrom L. E., Ueland P. M., Palma-Reis J., Boers G. H. J., Sheahan R. G., Israelsson B., et al. J. Am. Med. Assn. 1997;277:1775–1781. doi: 10.1001/jama.1997.03540460039030. [DOI] [PubMed] [Google Scholar]
- 3.Shields D. C., Kirke P. N., Milols J. L., Ramsbottom D., Molloy, A. M. B. H., Weir D. G., Scott J. M., Whitehead A. S. Am. J. Hum. Genet. 1999;64:1045–1055. doi: 10.1086/302310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Homocysteine Lowering Trialists’ Collaboration. Am. J. Clin. Nutr. 2005;82:806–812. doi: 10.1093/ajcn/82.4.806. [DOI] [PubMed] [Google Scholar]
- 5.Chen L. H., Liu M.-L., Hwang H.-Y., Chen L.-S., Korenberg J., Shane B. J. Biol. Chem. 1997;272:3628–3634. [PubMed] [Google Scholar]
- 6.Drummond J. T., Huang S., Blumenthal R. M., Matthews R. G. Biochemistry. 1993;32:9290–9295. doi: 10.1021/bi00087a005. [DOI] [PubMed] [Google Scholar]
- 7.Hall D. E., Jordan-Starck T. C., Loo R. O., Ludwig M. L., Matthews R. G. Biochemisty. 2000;39:10711–10719. doi: 10.1021/bi001096c. [DOI] [PubMed] [Google Scholar]
- 8.Loughlin R. E., Elford H. L., Buchanan J. M. J. Biol. Chem. 1964;239:2888–2895. [PubMed] [Google Scholar]
- 9.Taylor R. T., Weissbach H. In: The Enzymes. Boyer P. D., editor. Vol. 1. New York: Academic; 1973. pp. 121–165. [Google Scholar]
- 10.Chen Z., Chakraborty S., Banerjee R. J. Biol. Chem. 1995;270:19246–19249. doi: 10.1074/jbc.270.33.19246. [DOI] [PubMed] [Google Scholar]
- 11.Fujii K., Huennekens F. M. J. Biol. Chem. 1974;249:6745–6753. [PubMed] [Google Scholar]
- 12.LeClerc D., Wilson A., Dumas R., Gafuik C., Song D., Watkins D., Heng H. H. Q., Rommens J. M., Scherer S. W., Rosenblatt D. S., Gravel R. A. Proc. Natl. Acad. Sci. USA. 1998;95:3059–3064. doi: 10.1073/pnas.95.6.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olteanu H., Banerjee R. J. Biol. Chem. 2001;276:35558–35563. doi: 10.1074/jbc.M103707200. [DOI] [PubMed] [Google Scholar]
- 14.Olteanu H., Banerjee R. J. Biol. Chem. 2003;278:38310–38314. doi: 10.1074/jbc.M306282200. [DOI] [PubMed] [Google Scholar]
- 15.Wolthers K. R., Basran J., Munro A. W., Scrutton N. S. Biochemistry. 2003;42:3911–3920. doi: 10.1021/bi027290b. [DOI] [PubMed] [Google Scholar]
- 16.Olteanu H., Wolthers K. R., Munro A. W., Scrutton N. S., Banerjee R. Biochemistry. 2004;43:1988–1997. doi: 10.1021/bi035910i. [DOI] [PubMed] [Google Scholar]
- 17.Wolthers K. R., Scrutton N. S. Biochemistry. 2004;43:490–500. doi: 10.1021/bi0356303. [DOI] [PubMed] [Google Scholar]
- 18.Yamada K., Yamada S., Tobimatsu T., Toraya T. J. Biol. Chem. 1999;274:35571–35576. doi: 10.1074/jbc.274.50.35571. [DOI] [PubMed] [Google Scholar]
- 19.Rosales F., Ritari S. J., Sakami W. Biochem. Biophys. Res. Commun. 1970;40:271–276. doi: 10.1016/0006-291x(70)91005-3. [DOI] [PubMed] [Google Scholar]
- 20.Brot N., Weissbach H. J. Biol. Chem. 1966;241:2024–2028. [PubMed] [Google Scholar]
- 21.Watanabe F., Nakano Y. Methods Enzymol. 1997;281:295–305. doi: 10.1016/s0076-6879(97)81036-1. [DOI] [PubMed] [Google Scholar]
- 22.Drennan C. L., Huang S., Drummond J. T., Matthews R. G., Ludwig M. L. Science. 1994;266:1669–1674. doi: 10.1126/science.7992050. [DOI] [PubMed] [Google Scholar]
- 23.Hogenkamp H. P. C., Rush J. E., Swenson C. A. J. Biol. Chem. 1965;240:3641–3644. [PubMed] [Google Scholar]
- 24.Haywood G. C., Hill H. A. O., Pratt J. M., Vanston N. J., Williams R. J. P. J. Chem. Soc. 1965:6485–6493. [PubMed] [Google Scholar]
- 25.Hill H. A. O., Pratt J. M., Williams R. J. P. Proc. R. Soc. London Ser. A; 1965. pp. 352–358. [Google Scholar]
- 26.Rosenblatt D. S., Fenton W. A. In: The Metabolic and Molecular Bases of Inherited Disease. 8th Ed. Scriver C. R., Beaudet A. L., Sly W. S., Valle D., editors. Vol. 3. New York: McGraw–Hill; 2001. pp. 3897–3933. [Google Scholar]
- 27.Watkins D., Rosenblatt D. S. Am. J. Hum. Genet. 1986;39:404–408. [PMC free article] [PubMed] [Google Scholar]
- 28.Lerner-Ellis J. P., Tirone J. C., Pawelek P. D., Doré C., Atkinson J. L., Watkins D., Morel C. F., Fujiwara T. M., Moras E., Hosack A. R., et al. Nat. Genet. 2006;38:93–100. doi: 10.1038/ng1683. [DOI] [PubMed] [Google Scholar]
- 29.Lerner-Ellis J. P., Gradinger A. B., Watkins D., Tirone J. C., Villeneuve A., Dobson C. M., Montpetit A., Lepage P., Gravel R. A., Rosenblatt D. S. Mol. Genet. Metab. 2006;87:219–225. doi: 10.1016/j.ymgme.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 30.Zhang J., Dobson C. M., Wu S., Lerner-Ellis J., Rosenblatt D. S., Gravel R. A. Mol. Genet. Metab. 2006;87:315–322. doi: 10.1016/j.ymgme.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 31.Sato K., Hiei E., Shimizu S., Abeles R. FEBS Lett. 1978;85:73–76. doi: 10.1016/0014-5793(78)81251-4. [DOI] [PubMed] [Google Scholar]
- 32.Jarrett J. T., Goulding C. W., Fluhr K., Huang S., Matthews R. G. Methods Enzymol. 1997;281:196–213. doi: 10.1016/s0076-6879(97)81026-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.