Abstract
The mammalian retina contains an endogenous circadian pacemaker that broadly regulates retinal physiology and function, yet the cellular origin and organization of the mammalian retinal circadian clock remains unclear. Circadian clock neurons generate daily rhythms via cell-autonomous autoregulatory clock gene networks, and, thus, to localize circadian clock neurons within the mammalian retina, we have studied the cell type-specific expression of six core circadian clock genes in individual, identified mouse retinal neurons, as well as characterized the clock gene expression rhythms in photoreceptor degenerate rd mouse retinas. Individual photoreceptors, horizontal, bipolar, dopaminergic (DA) amacrines, catecholaminergic (CA) amacrines, and ganglion neurons were identified either by morphology or by a tyrosine hydroxylase (TH) promoter-driven red fluorescent protein (RFP) fluorescent reporter. Cells were collected, and their transcriptomes were subjected to multiplex single-cell RT-PCR for the core clock genes Period (Per) 1 and 2, Cryptochrome (Cry) 1 and 2, Clock, and Bmal1. Individual horizontal, bipolar, DA, CA, and ganglion neurons, but not photoreceptors, were found to coordinately express all six core clock genes, with the lowest proportion of putative clock cells in photoreceptors (0%) and the highest proportion in DA neurons (30%). In addition, clock gene rhythms were found to persist for >25 days in isolated, cultured rd mouse retinas in which photoreceptors had degenerated. Our results indicate that multiple types of retinal neurons are potential circadian clock neurons that express key elements of the circadian autoregulatory gene network and that the inner nuclear and ganglion cell layers of the mammalian retina contain functionally autonomous circadian clocks.
Keywords: circadian clock, clock gene, mouse retina, photoreceptor, real-time PCR
The mammalian retinal circadian clock exerts extensive control over retinal physiology and function, regulating a wide variety of retinal circadian rhythms, including rod disk shedding (1–3), melatonin release (4–6), dopamine synthesis (7, 8), electroretinogram (ERG) b-wave amplitude (9), extracellular pH (10), visual sensitivity (11, 12), and intraocular pressure (13, 14). The retinal circadian clock and its dopamine- and melatonin-signaling molecules also influence pathological processes in the eye, including the susceptibility of photoreceptors to degeneration from light damage (15, 16), photoreceptor survival in animal models of retinal degeneration (17), and the degree of refractive errors in primate models of myopia (18). Despite its widespread influence, the cellular origin and organization of the circadian clock in the mammalian retina remain unclear.
Neural circadian clocks generate endogenous circadian rhythms through cell-autonomous autoregulatory transcription-translation feedback loops comprised of a defined set of “clock genes” in subsets of circadian pacemaker neurons. Gene targeting has demonstrated that, in mammals, the genes Period (Per) 1 and 2 and Cryptochrome (Cry) 1 and 2, as well as Clock and Bmal1, are core molecular components of circadian clocks and that their expression is necessary for circadian rhythmicity (19). The prevailing model for circadian organization of vertebrate retinas, based on molecular and physiological data from amphibian and avian retinas, holds that photoreceptors contain self-sustained circadian clocks that rhythmically secrete melatonin as an output signal (20–23). Although mammalian retinas also exhibit circadian melatonin rhythms (4–6), in situ hybridization studies of the regional distribution of clock genes in the mammalian retina found them to be expressed predominantly in the inner nuclear and ganglion cell layers (24–28), suggesting the potential for an alternate circadian organization.
In the current study, we have examined the circadian organization of the mouse retina using cell morphology, as well as genetic and retrograde labeling, to identify individual retinal neurons; single-cell real-time RT-PCR; PER2::LUC fusion protein real-time reporting of molecular circadian rhythms (29); and quantitative real-time RT-PCR (qPCR) to investigate the cell type-specific expression and endogenous rhythmicity of core circadian clock genes. Our results indicate that there are neurons with the requisite molecular components to be circadian pacemakers among all of the major classes of retinal neurons, with the exception of photoreceptors, and that the ability to generate endogenous circadian rhythms resides in neurons of the inner nuclear and ganglion cell layers, independent of photoreceptors and of the central biological clock of the brain.
Results
Coordinate Expression of Core Clock Genes Occurs Preferentially in Neurons of the Inner Nuclear and Ganglion Cell Layers.
To localize potential clock neurons within the mouse retina, we used single-cell real-time RT-PCR to simultaneously detect all six core clock genes within single, identified retinal neurons. Six specific retinal neuron types were identified for collection either from acutely dissociated retinal cell cultures or from retinal whole mounts. Rods and rod bipolar cells were distinguished from other cells after dissociation based on their unique morphological characteristics (Fig. 1 a and b). Horizontal cells were harvested from whole-mount retinas of rd mice, in which the degeneration of photoreceptors permits direct access to horizontal cell somata (Fig. 1c; D.-Q.Z. and D.G.M., unpublished data). Two types of amacrine cells, dopaminergic (DA) amacrines and type 2 catecholamine (CA) amacrines, were collected by dissociating retinas from mice harboring a tyrosine hydroxylase (TH) promoter-driven red fluorescent protein (RFP) transgene and visualized by RFP fluorescence (30). Both DA and type 2 CA cells express the TH::RFP reporter but are readily distinguished by soma size (30), with DA cells having the relatively larger somas (Fig. 1 d and e). Ganglion cells were retrogradely labeled with 1,1’-dioctadecyl-3,3,3’,3’-tetramethylindocarbocyanine (DiI) by injection into the lateral geniculate nuclei and were visualized by fluorescence after retinal dissociation (Fig. 1f).
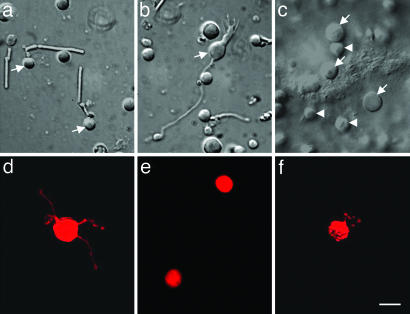
Fig. 1.
Six types of mouse retinal cells were identified and harvested for RT-PCR. Rods (a; arrows) and rod bipolar cells (b; arrow) were identified in dispersed cultures based on morphology. Horizontal cells (c; arrows) were identified and harvested from whole-mount rd mouse retinas. The relatively small surface cells (arrow heads) are either bipolar cells or remaining photoreceptors. DA cells (d) and type 2 CA cells (e) were identified in dispersed cultures of TH::RFP transgenic mouse retinas by RFP fluorescence. Ganglion cells (f) were identified in dispersed cultures by DiI fluorescence after injection and retrograde transport from the lateral geniculate nuclei. (Scale bar: 10 μm.)
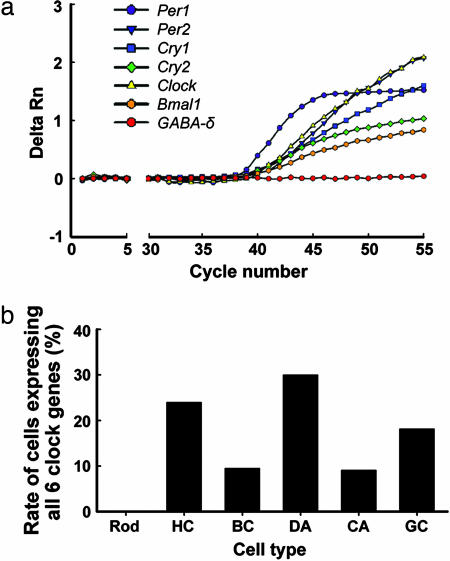
Cellular contents of identified cells were collected by suction with a patch-type pipette (Fig. 5, which is published as supporting information on the PNAS web site) and subjected to RT-PCR for the six core clock genes. Fig. 2a shows the real-time PCR amplification plot for a representative DA cell. Although all clock gene transcripts were detected in this cell, the transcript for the GABA-δ receptor subunit, known to be absent in DA cells (31), was not detected. We initially examined core clock gene expression in 31 rods, 25 horizontal cells, 21 rod bipolar cells, 30 DA cells, 22 type 2 CA cells, and 22 ganglion cells. Simultaneous detection of the six core clock genes in individual identified neurons yielded 0 rods (0%), 6 horizontal cells (24%), 2 rod bipolar cells (10%), 9 DA cells (30%), 2 type 2 CA cells (9%), and 4 ganglion cells (18%) that expressed all six clock genes (Fig. 2b). The remaining cells of each type expressed one to five clock genes, except for 10 rods (32%) and 2 horizontal cells (8%) that did not express any core clock genes. In addition, 8 cells identified as cone-like by morphology were collected, and the PCR results for them were similar to those obtained for rods. The percentage expression of each clock gene in each neuron type is shown in Fig. 6, which is published as supporting information on the PNAS web site. A number of steps were taken to minimize false positives and negatives in the PCR (see Supporting Materials and Methods, which is published as supporting information on the PNAS web site).
Fig. 2.
Simultaneous detection of all six core clock genes in individual retinal neurons. (a) Real-time PCR amplification plot of a representative DA cell in which all six clock gene transcripts, but not the GABA-δ receptor subunit transcript (a negative control gene in DA cells), were detected. Data are presented as the average of duplicate amplifications for each gene. Detection of specific PCR products was monitored as the relative fluorescence (delta Rn) in the intensity of reporter dye. (b) The percentage of cells expressing all six core clock genes in each cell type is shown. Rod, rods; HC, horizontal cells; BC, rod bipolar cells; DA, dopaminergic amacrine cells; CA, type 2 catecholamine amacrine cells; GC, ganglion cells.
The cells reported in Fig. 2 were collected at approximately Zeitgeber time (ZT) 7–8 during the day phase (4–5 h before lights off in the mouse colony). To test whether time of day has an effect on the detection rate of clock genes, an additional 31 rods and 16 DA cells were individually harvested at ZT 19–20 during the night, and core clock gene expression was examined as described above. At this point during the night phase, the Per and Cry gene transcripts are expressed at lower levels in the whole retina, whereas Bmal1 is expressed at higher levels (G-X.R. and D.G.M., unpublished data). The detection rates of the core clock genes at ZT 19–20 were similar to those at ZT 7–8 in both neuron types, with 0% of rods and 25% of DA neurons exhibiting expression of all six core clock genes (Fig. 7, which is published as supporting information on the PNAS web site). Thus, the circadian phase does not significantly affect the detection rates of clock genes in retinal neurons by using our methods. Cell-to-cell variability in absolute levels of the transcripts precluded using this single-cell RT-PCR approach to quantify potential clock gene rhythms in specific neuron populations. Taken together, our single-cell real-time PCR data demonstrate the presence of potential circadian oscillator neurons among all of the major cell types of the inner nuclear and ganglion cell layers of the retina.
rd Mouse Retinas Exhibit Robust Circadian Clock Gene Rhythms.
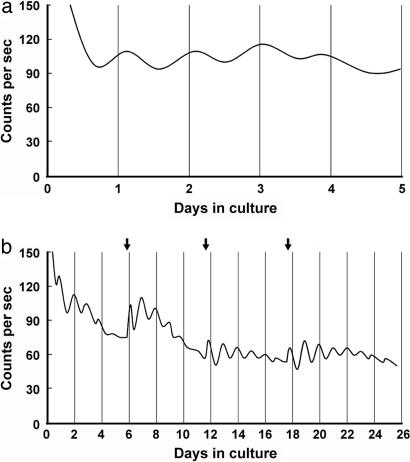
To test whether the clock gene-expressing neurons of the inner nuclear and ganglion cell layers can generate endogenous circadian rhythms, we isolated and cultured retinas from mPer2Luc knockin mice, which had been crossed with photoreceptor degenerate C3H mice, to introduce the rd photoreceptor degeneration gene. We used retinas from mPer2Luc mice that were homozygous for rd and were 85 ± 2 days old. At this age, virtually all photoreceptors have degenerated in the retinas of mice homozygous for rd (5, 32–34). Retinas were explanted onto membranes, and luminescence from the PER2::LUC fusion protein was continuously measured in real-time in constant darkness at 36.8°C. PER2::LUC expression in vitro was robustly rhythmic with the peak of PER2::LUC expression occurring at hour 14.28 ± 0.56 of projected Zeitgeber time from the previous light/dark (LD) cycles (Fig. 3a; n = 6 retinas from three mice), phase delayed by 2 h relative to the suprachiasmatic nucleus (SCN) (29). The period of the retinal rhythms was 22.70 ± 0.20 h, which is shorter than the period previously reported for the SCN (23.5 h; ref. 29), but is slightly longer than that of the cornea (22.2 h; 29). Most explants were maintained for only 4–5 days, but one culture studied over a longer term exhibited robust circadian rhythms for >25 days (Fig. 3b). As with the cultured SCN (29), fresh medium with fresh luciferin substrate, exchanged every 6–7 days, tended to increase the amplitude of the ongoing luminescence rhythms (Fig. 3b). In contrast to the results with photoreceptor degenerate retinas, PER2::LUC expression in retinas cultured from mPer2Luc knockin mice that were heterozygous for rd (i.e., that retained their photoreceptors) exhibited only 1–2 circadian cycles and then damped out (data now shown). This lack of rhythmicity might be due to a suppressive effect of the photoreceptors on gene expression in inner retinal neurons, as has been found in the Royal College of Surgeons (RCS) rat (35), or it might be due to nonoptimal culture conditions for photoreceptor-retaining retinas.
Fig. 3.
Real-time monitoring of PER2::LUC expression in isolated rd mouse retinas. (a) Representative record showing retinal rhythms during the initial 5 days of culture. Retinas were dissected and explanted just before lights off (ZT 12). Vertical lines indicate the projected time of lights off on each day in DD (projected ZT 12). Photon counts per second are plotted against days in culture. (b) Long-term culture of an rd mouse retina showing persistent circadian rhythms in PER2::LUC expression. Arrows indicate times of medium changes.
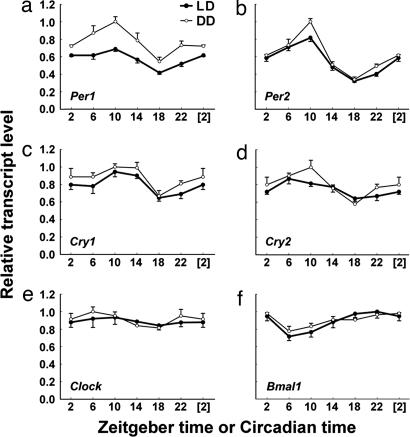
To further define the temporal expression patterns of all of the core clock genes in rd mouse retinas, we quantified the rhythmic expression of core clock genes in whole-retina RNA using qPCR. Mice 84–112 days of age were maintained in 12:12 LD cycles or in constant darkness (DD), and the temporal pattern of clock gene expression in their retinas was sampled at 4-h intervals. All core clock genes except Clock exhibited statistically significant variations in expression level over the 24-h sampling period consistent with ongoing circadian rhythms in both LD and DD conditions (Fig. 4). It is noteworthy that these retinal clock gene profiles were statistically significant but were generally lower in amplitude than those previously reported in the SCN and liver (36, 37), similar to retinal clock gene rhythms reported for the rat (26, 38). Peak-trough amplitudes ranged from 2.95-fold for Per2 in DD to 1.26-fold for Bmal1 in DD (Fig. 4). Per and Cry genes peaked in their expression at or near ZT/circadian time (CT) 10, whereas Bmal1 peaked in antiphase at or near ZT/CT 22. Peak Per2 transcript level occurred ≈4 h earlier than the peak PER2 protein level (Fig. 3; projected ZT 14). We also compared mean clock gene transcript levels between LD and DD conditions. Surprisingly, in rd retinas, the expression of Per1 was significantly higher in DD vs. LD (F = 52.5; P < 0.001; two-way ANOVA) (Fig. 4a), indicating a suppressive effect of light cycles on the overall retinal expression of Per1, even when the outer retinal photoreceptors had degenerated. Taken together, our whole-retina qPCR data further demonstrate the existence of the molecular circadian clockworks in the inner nuclear and ganglion cell layers.
Fig. 4.
Temporal expression profiles of Per1 (a), Per2 (b), Cry1 (c), Cry2 (d), Clock (e), and Bmal1 (f) in rd mouse retinas under LD (●) and DD (○) conditions. Data are presented as means ± SEM from five mice and expressed as a percentage of the maximum mean expression. Points at ZT/CT [2] are duplicates of ZT/CT 2 replotted to show 24-h trends. One-way ANOVA revealed P < 0.001 for Per1 and Per2 under both LD and DD conditions, P < 0.01 for Cry1 and Bmal1 under both LD and DD conditions, P < 0.05 for Cry2/LD, and P < 0.001 for Cry2/DD. For Per1, Per2, Cry1, Cry2, and Bmal1 expression, the peak-trough amplitudes under LD conditions were 1.65-, 2.54-, 1.46-, 1.35-, and 1.39-fold, respectively, whereas, under DD conditions, they were 1.84-, 2.95-, 1.50-, 1.73-, and 1.26-fold, respectively.
Discussion
Our examination of the circadian organization of the mouse retina using cell type-specific and temporal gene expression profiling of the core circadian clock genes, as well as real-time gene expression reporting, resulted in two main findings. First, coordinate expression of the six core circadian clock genes occurs in individual neurons from all of the major cell types of the inner nuclear and ganglion cell layers of the retina, but not in photoreceptors, and, second, these clock gene-expressing neurons of the inner retinal layers constitute a self-sustained circadian clock that can generate rhythmicity independently of the master SCN clock and of photoreceptors. These results support a paradigm for circadian organization of the retina in which retinal neurons other than rod or cone photoreceptors are a primary locus of endogenous circadian rhythms generation.
Circadian Clocks in the Inner Nuclear and Ganglion Cell Layers.
Individual horizontal, rod bipolar, DA amacrine, CA amacrine, and ganglion cells were found to coordinately express the core clock genes at varying rates according to cell type. Thus, retinal neurons harboring the molecular genetic basis for endogenous rhythms generation are widely distributed among the inner nuclear and ganglion cell layers of the mammalian retina. Of note is that the proportion of putative clock neurons expressing all six core clock genes in each of the retinal neuron populations tested was a minority, ranging from 9% in CA amacrines to 30% in DA amacrines. However, we consider these to be minimum estimates of the proportion of putative clock neurons. The combinatorial nature of our criteria (i.e., simultaneous detection of six genes) means that false negatives have a multiplicative effect and even the low false negative rates we have demonstrated will significantly reduce the apparent number of putative clock neurons. Thus, it is likely that the actual proportion of retinal cells expressing sufficient molecular elements to support circadian rhythms generation is somewhat higher than our current results indicate, although the expression rate for Bmal1 is limiting and effectively caps the maximum possible proportion of putative clock neurons in all cell types in a range of 40–60% (see Fig. 6). It should be noted that, although we have examined selected subpopulations of all of the major retinal neuron types, we do not have morphological or genetic markers for all subtypes, and, thus, there may be additional neuronal subpopulations in the inner nuclear and ganglion cell layers that have circadian pacemaker capability. In addition, it must be recognized that, at this point, coordinate clock gene expression in horizontal cells has been demonstrated only in rd mouse retinas wherein there is the potential for functional reorganization.
Can a minority of pacemaker cells be sufficient to generate rhythms within retinal neuron populations? Recent findings suggest that the SCN circadian clock and the retinal circadian clock may be organized in a similar manner, with minority populations of pacemaker neurons driving high amplitude circadian rhythms throughout the entire neural ensemble. Selected subpopulations of SCN neurons do not express endogenous rhythms in clock genes (39), and disruption of neuropeptide communication between SCN neurons blunts rhythmicity in many cells, revealing a pacemaker neuron population of 25–30% of SCN neurons in which self-sustained rhythmicity persists (40). Thus, by this criterion, the proportion of putative pacemaker neurons in the master SCN clock is similar to the proportions of DA cells, horizontal cells, and ganglion cells that we have identified as putative pacemaker neurons by molecular means. It is likely that these specific subpopulations engender endogenous rhythmicity to the inner retinal layers.
Real-time bioluminescence gene expression reporting in isolated mPer2Luc knockin rd mouse retinas demonstrated that the neurons of the inner nuclear and ganglion cell layers constitute a functional circadian clock in isolation from the SCN master clock and in the absence of virtually all rods and cones. These results also demonstrate that inner retinal circadian molecular rhythms persist in the absence of melatonin rhythms because rd mouse retinas exhibit low, arrhythmic melatonin levels (5). Because a few cones may have remained in the rd retinas we tested (32–34), we cannot rigorously conclude, based on this experiment alone, that the observed PER2 expression rhythms were completely endogenous to the inner nuclear and ganglion cell layers. However, previous in situ hybridization experiments have demonstrated low levels of clock gene expression in the mammalian photoreceptor layer (24–28), and our single-cell RT-PCR studies failed to find a single photoreceptor cell out of 70 tested that expressed the full complement of core clock genes. Thus, mammalian photoreceptors show a relative lack of molecular clock elements, and they are, on this basis, unlikely to be a locus of endogenous rhythms generation. In this regard, a recent study found temporal variations in the expression level of mRNA for retinal arylalkylamine N-acetyltransferase (AANAT), a key enzyme in melatonin synthesis, consistent with ongoing circadian rhythms after lesion of the inner retina, suggesting the possibility of photoreceptor clock function (41). However, neural mechanisms for SCN influence of mammalian retinal rhythms have been described (42), and these in vivo sampling studies, limited to one 24-h interval, did not address whether the AANAT variations were indeed endogenous rhythms that persist independently of the influence of the master SCN clock. Our data do not completely exclude the possibility of photoreceptor clock function, because it is possible that there is a small number of photoreceptors expressing the full complement of clock genes that escaped our detection. However, when taken together, the lack of demonstrated coordinate expression of clock genes in photoreceptors, the lack of evidence for endogenous photoreceptor rhythmicity independent of both the inner retina and the SCN, along with the coordinate clock gene expression in more inner retinal neurons, as well as the persistent in vitro circadian rhythmicity from inner retinal neurons in rd retinas, form convergent lines of evidence suggesting that neurons of the inner nuclear and ganglion cell layers constitute an endogenous circadian pacemaker within the mammalian retina.
Clock Genes in Photoreceptors.
Our results do not provide support for the proposition that mammalian photoreceptors contain endogenous circadian clocks, but they do demonstrate a limited expression of clock genes in these cells. Each of the six clock genes was indeed detected within the population of photoreceptor cells we sampled, and the genes exhibited a range in detection rate, with Per2 detected in ≈10% of photoreceptors and Bmal1 detected in ≈35% of photoreceptors (Fig. 6). Despite the demonstrated presence of clock genes in photoreceptors, consistent with previous in situ hybridization studies (24–28), coordinate expression of all six core clock genes was never detected. The great majority of photoreceptor cells sampled was morphologically identifiable as rods, but, based on a more limited sample, cones, like rods, showed similar low-frequency expression of clock genes and none showed coordinate expression of all six core clock genes. We must be more cautious in interpreting the cone results because the morphological identification of mouse cones is less certain. It is likely that the clock genes expressed in photoreceptors serve non-clock functions. For example, Clock and Bmal1, which are expressed together in ≈10% of rods, encode CLOCK and BMAL1 transcription factors that form heterodimers for activating the transcription of E-box-containing genes, such as AANAT. However, because the full complement of required negative elements of the circadian clockworks (Per and Cry genes) is missing from these cells, it is unlikely that they can form a viable, cell-autonomous circadian feedback loop. Thus, our data are consistent with the notion that clock genes expressed in mouse photoreceptors may play a role in regulation of gene expression, but that circadian rhythmicity in these cells is likely to be driven by exogenous signals.
Circadian Organization of Vertebrate Retinas.
Our results support a model for circadian organization of mammalian retinas distinct from the current prevailing paradigm based primarily upon data from amphibian and avian retinas. In amphibian and avian retinas, the photoreceptors are endogenous circadian oscillators that mediate rhythmic melatonin release (20–23) and express high levels of the core clock genes (43–47), whereas, in the mouse retina, clock genes and clock function have become more localized to the inner retinal layers. However, melatonin secreted by the photoreceptors may still serve as an important circadian output signal. Although a previous study (5) demonstrated a lack of circadian rhythmicity of melatonin synthesis in isolated C3H rd mouse retinas, our results show that endogenous molecular rhythms generation in retinal neurons is intact after photoreceptor degeneration. In addition, cultured Royal College of Surgeons (RCS) rat retinas with photoreceptor degeneration exhibit circadian rhythms of melatonin release mediated by the inner retina (35), and dopamine content varies between subjective night and subjective day even in retinas sampled from populations of photoreceptor-degenerate RCS rats (8) and melatonin-less mice (48), which are consistent with a photoreceptor-independent retinal circadian oscillator in mammals.
There is extensive influence upon photoreceptor function and gene expression of diffusible signals from inner retinal neurons. Basic photoreceptor functions that are under circadian control, such as disk shedding, melatonin secretion, and AANAT gene expression, as well as Na+/K+ ATPase function, are regulated by the inner retinal circadian signal dopamine (49–53), acting on mouse cones through D4 dopamine receptors (54). Dopamine even influences photoreceptor survival in rd mouse retinas (17). Dopamine is secreted rhythmically in the retina, and DA amacrine neurons, as a population, have previously been shown to exhibit circadian rhythms in the transcription of the clock gene Per1 (27), as well as to contain the clock gene products PER1, CRY1, CRY2, CLOCK, and BMAL1 (55). Our results specifically colocalize the core components of the molecular circadian clockworks within individual DA neurons, defining their potential for endogenous circadian oscillation and further strengthening the inference that they are clock neurons. In addition, nitric oxide (NO) and GABA are potential circadian neurotransmitters within the retina, based on clock gene rhythmicity in inner retinal neuron subpopulations. Nitric oxide synthase-expressing amacrine cells rhythmically express the clock gene Per1 under LD and DD conditions (56) and thus are also candidate circadian pacemaker neurons. Both DA and NO amacrines are also GABAergic. Whether NO and GABA, like dopamine and melatonin, are rhythmically secreted in the retina remains to be determined.
In summary, putative circadian clock neurons that express the core clock genes necessary for circadian rhythms generation are found in all of the major neuron populations of the mammalian retina, except in photoreceptors. In addition, after photoreceptor degeneration, retinal neuron populations continue to express endogenously generated molecular circadian rhythms. These findings provide an expanded basis for the study of mammalian retinal rhythms by localizing putative pacemaker neurons to the inner nuclear and ganglion cell neuronal layers, suggesting that there are likely many local pacemaker neurons embedded within the full spectrum of retinal circuits that could influence retinal physiology and function, photoreceptor survival, and eye growth, through endogenous rhythms generation and neurotransmitter secretion. Cell communication is important for rhythms production in the neural circadian pacemakers of the mammalian and fly brains (40, 57). The highly ordered and well characterized nature of retinal circuitry will likely facilitate elucidation of the general principles of neuronal communication in circadian pacemaking.
Materials and Methods
Harvesting of Individual Retinal Neurons.
Individual photoreceptors, rod bipolar cells, DA cells, type 2 CA cells, and ganglion cells were collected from acute retinal cell cultures. Ganglion cell labeling was performed as described (27). Two- to four-month-old C57BL/6J mice, hemizygous transgenic B6C3H mice harboring a TH::RFP reporter (30), and DiI-injected C57BL/6J mice were killed by cervical dislocation at ZT 6. Retinas were isolated in Leibovitz's L-15 medium (GIBCO), digested with papain (20 units/ml; Worthington), gently triturated in L-15 medium, and finally distributed into culture dishes at low density. Individual horizontal cells were harvested from whole-mount rd mouse retinas in which the photoreceptors had degenerated, allowing direct physical access to the horizontal cells. Three- to four-month-old C3H rd mice were killed at ZT 6. The retinas were isolated and transferred to a perfusion chamber, stabilized by using a nylon mesh with ganglion cell layer down, mounted on an upright microscope (Zeiss), and perfused for 1 h with 95% O2-5% CO2 bubbled Ames' medium (supplemented with 25 mM NaHCO3 and 10 mM glucose) before harvesting. Superficial horizontal cells with relatively large cell bodies were harvested.
Explant Cultures.
mPer2Luc knockin mice, which initially were on a 129SvEv × C57BL/6J genetic background (29, a gift from J. Takahashi to S. Yamazaki), were maintained as a continuous backcross to C57BL/6J for 10 generations. The resulting mPer2Luc mice were crossed with C3H rd mice twice to produce mPer2Luc knockin mice that are homozygous for rd gene. Approximately 30 min before lights off, mPer2Luc knockin rd mice were killed by cervical dislocation. Eyes were enucleated and placed in HBSS (Invitrogen). Retinas were isolated from the rest of the eyecup with as little disruption and manipulation as possible and then gently placed ganglion cell layer up on Millicell culture membranes (Millipore) in 35-mm culture dishes with 1.0 ml of Medium 199 (without l-glutamine, sodium bicarbonate, and phenol red; Sigma), supplemented with 0.7 mM l-glutamine, 4 mM sodium bicarbonate, 10 mM Hepes, 2% B27, 0.1 mM beetle luciferin (Promega), 25 units/ml penicillin, and 25 μg/ml streptomycin (Invitrogen). Bioluminescence was measured with a LumiCycle (Actimetrics, Wilmette, IL). Cultures were maintained in a light-tight incubator at 36.8°C. Data were analyzed by using lumicycle data analysis software (Actimetrics) and clocklab software (Actimetrics). The timing of the peak of the circadian oscillation was determined during the interval of 12 and 36 h in culture.
Animal Sampling for Whole-Retina qPCR.
Sixty 10- to 14-week-old male C3H rd mice were purchased from The Jackson Laboratory. The mice were either maintained in LD cycles or, before being used, transferred into constant darkness at the light-dark transition and kept there for 36–48 h. Five animals were sampled for each time point examined at ZT/CT 2, 6, 10, 14, 18, and 22. For dissections during the dark phase, night-vision goggles (B. E. Meyers, Redmond, WA) were used under infrared illumination so that no visible light was necessary for the retina isolation procedure. The two retinas from each mouse were pooled as one sample and immediately homogenized in TRIzol reagent (Invitrogen) under dim red light, frozen on dry ice, and stored at −80°C until RNA extraction.
TaqMan Real-Time PCR.
The primers and probes were designed by using Applied Biosystems primerexpress software (see Supporting Materials and Methods for sequences). Immediately after first-strand cDNA synthesis, real-time PCR was performed by using an Applied Biosystems Prism 7000. The reaction was first incubated at 50°C for 2 min, then at 95°C for 10 min, followed by 55 cycles (single-cell) or 40 cycles (whole retina) of 95°C for 15 s and 60°C for 1 min. Samples were assayed in duplicate for single-cell PCR and in triplicate for whole-retina qPCR. Each particular transcript was regarded as present in individual single-cell samples when its delta Rn exceeded a threshold value of 0.2 in <50 cycles of real-time PCR.
Data Analysis.
All statistical analyses were conducted with spss 12.0. For single-cell PCR, a χ2 test was applied to compare percentages of clock gene expression between two paired cell types, as well as expression percentages between clock gene pairs. For sample sizes <5, Fisher's exact test was performed instead of a χ2 test. For whole-retina qPCR, the normalization method used was based on Dijk et al. (58) with some modifications. In brief, the cycle threshold (Ct) was converted to absolute amount of transcript (E−Ct, where E is amplification efficiency) and presented as E−Ct/E−Ct(Max). E−Ct(Max) corresponds to the maximum E−Ct in six ZTs and six CTs. Data were analyzed by using one-way ANOVA for time differences and two-way ANOVA for light cycle differences.
Supplementary Material
Acknowledgments
We thank Dr. Gilbert Pitts for assistance with RT-PCR and Drs. Karen Gamble and Bashar Shakhtour for assistance with statistical analysis. This work was supported by National Institutes of Health Grant EY015815 (to D.G.M.).
Abbreviations
- DA
dopaminergic
- CA
catecholamine
- Per
Period
- Cry
Cryptochrome
- SCN
suprachiasmatic nucleus
- ZT
Zeitgeber time
- CT
circadian time
- LD
light/dark
- DD
constant darkness
- qPCR
quantitative real-time RT-PCR
- RFP
red fluorescent protein
- DiI
1,1’-dioctadecyl-3,3,3’,3’-tetramethylindocarbocyanine
- AANAT
arylalkylamine N-acetyltransferase
Note Added in Proof.
While this paper was under review, DeBruyne et al. (59) reported evidence that expression of the Clock gene is not necessary for the generation of SCN molecular rhythms or behavioral rhythms in mice. Although the significance of these results for retinal rhythmicity is not yet clear, they call into question the role of Clock as a core circadian clock gene and suggest the possibility that neurons lacking Clock could be endogenously rhythmic. This possibility would not appreciably alter our conclusions regarding the presence and prevalence of putative circadian clock neurons in the retinal neuron subtypes we tested. For all neuron types except photoreceptors, Bmal1 generally exhibited a lower expression rate than other clock genes and was the principal limiting gene, whereas photoreceptors exhibited very low expression rates of all clock genes (Fig. 6).
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.LaVail M. M. Science. 1976;194:1071–1074. doi: 10.1126/science.982063. [DOI] [PubMed] [Google Scholar]
- 2.Terman J. S., Reme C. E., Terman M. Brain Res. 1993;605:256–264. doi: 10.1016/0006-8993(93)91748-h. [DOI] [PubMed] [Google Scholar]
- 3.Grace M. S., Wang L. M., Pickard G. E., Besharse J. C., Menaker M. Brain Res. 1996;735:93–100. doi: 10.1016/0006-8993(96)00600-2. [DOI] [PubMed] [Google Scholar]
- 4.Tosini G., Menaker M. Science. 1996;272:419–421. doi: 10.1126/science.272.5260.419. [DOI] [PubMed] [Google Scholar]
- 5.Tosini G., Menaker M. Brain Res. 1998;789:221–228. doi: 10.1016/s0006-8993(97)01446-7. [DOI] [PubMed] [Google Scholar]
- 6.Niki T., Hamada T., Ohtomi M., Sakamoto K., Suzuki S., Kako K., Hosoya Y., Horikawa K., Ishida N. Biochem. Biophys. Res. Commun. 1998;248:115–120. doi: 10.1006/bbrc.1998.8916. [DOI] [PubMed] [Google Scholar]
- 7.Doyle S. E., Grace M. S., McIvor W., Menaker M. Visual Neurosci. 2002;19:593–601. doi: 10.1017/s0952523802195058. [DOI] [PubMed] [Google Scholar]
- 8.Doyle S. E., McIvor W. E., Menaker M. J. Neurochem. 2002;83:211–219. doi: 10.1046/j.1471-4159.2002.01149.x. [DOI] [PubMed] [Google Scholar]
- 9.Brandenburg J., Bobbert A. C., Eggelmeyer F. Behav. Brain Res. 1983;7:113–123. doi: 10.1016/0166-4328(83)90008-6. [DOI] [PubMed] [Google Scholar]
- 10.Dmitriev A. V., Mangel S. C. J. Neurosci. 2001;21:2897–2902. doi: 10.1523/JNEUROSCI.21-08-02897.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenwasser A. M., Raibert M., Terman J. S., Terman M. Physiol. Behav. 1979;23:17–21. [Google Scholar]
- 12.Bassi C. J., Powers M. K. Physiol. Behav. 1986;38:871–877. doi: 10.1016/0031-9384(86)90056-9. [DOI] [PubMed] [Google Scholar]
- 13.Boyd T. A., McLeod L. E. Ann. N.Y. Acad. Sci. 1964;117:597–613. doi: 10.1111/j.1749-6632.1964.tb48212.x. [DOI] [PubMed] [Google Scholar]
- 14.Rowland J. M., Potter D. E., Reiter R. J. Curr. Eye Res. 1981;1:169–173. doi: 10.3109/02713688109001822. [DOI] [PubMed] [Google Scholar]
- 15.Organisciak D. T., Darrow R. M., Barsalou L., Kutty R. K., Wiggert B. Invest. Ophthalmol. Visual Sci. 2000;41:3694–3701. [PubMed] [Google Scholar]
- 16.Sugawara T., Sieving P. A., Iuvone P. M., Bush R. A. Invest. Ophthalmol. Visual Sci. 1998;39:2458–2465. [PubMed] [Google Scholar]
- 17.Ogilvie J. M., Speck J. D. Neurobiol. Dis. 2002;10:33–40. doi: 10.1006/nbdi.2002.0489. [DOI] [PubMed] [Google Scholar]
- 18.Iuvone P. M., Tigges M., Stone R. A., Lambert S., Laties A. M. Invest. Ophthalmol. Visual Sci. 1991;32:1674–1677. [PubMed] [Google Scholar]
- 19.Reppert S. M., Weaver D. R. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 20.Cahill G. M., Besharse J. C. Neuron. 1993;10:573–577. doi: 10.1016/0896-6273(93)90160-s. [DOI] [PubMed] [Google Scholar]
- 21.Pierce M. E., Sheshberadaran H., Zhang Z., Fox L. E., Applebury M. L., Takahashi J. S. Neuron. 1993;10:579–584. doi: 10.1016/0896-6273(93)90161-j. [DOI] [PubMed] [Google Scholar]
- 22.Hayasaka N., LaRue S. I., Green C. B. J. Neurosci. 2002;22:1600–1607. doi: 10.1523/JNEUROSCI.22-05-01600.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivanova T. N., Iuvone P. M. Brain Res. 2003;973:56–63. doi: 10.1016/s0006-8993(03)02540-x. [DOI] [PubMed] [Google Scholar]
- 24.Gekakis N., Staknis D., Nguyen H. B., Davis F. C., Wilsbacher L. D., King D. P., Takahashi J. S., Weitz C. J. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 25.Miyamoto Y., Sancar A. Proc. Natl. Acad. Sci. USA. 1998;95:6097–6102. doi: 10.1073/pnas.95.11.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Namihira M., Honma S., Abe H., Masubuchi S., Ikeda M., Honmaca K. NeuroReport. 2001;12:471–475. doi: 10.1097/00001756-200103050-00010. [DOI] [PubMed] [Google Scholar]
- 27.Witkovsky P., Veisenberger E., LeSauter J., Yan L., Johnson M., Zhang D. Q., McMahon D., Silver R. J. Neurosci. 2003;23:7670–7676. doi: 10.1523/JNEUROSCI.23-20-07670.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson C. L., Rickman C. B., Shaw S. J., Ebright J. N., Kelly U., Sancar A., Rickman D. W. Invest. Ophthalmol. Visual Sci. 2003;44:4515–4521. doi: 10.1167/iovs.03-0303. [DOI] [PubMed] [Google Scholar]
- 29.Yoo S. H., Yamazaki S., Lowrey P. L., Shimomura K., Ko C. H., Buhr E. D., Siepka S. M., Hong H. K., Oh W. J., Yoo O. J., et al. Proc. Natl. Acad. Sci. USA. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang D. Q., Stone J. F., Zhou T., Ohta H., McMahon D. G. NeuroReport. 2004;15:1761–1765. doi: 10.1097/01.wnr.0000135699.75775.41. [DOI] [PubMed] [Google Scholar]
- 31.Gustincich S., Feigenspan A., Sieghart W., Raviola E. J. Neurosci. 1999;19:7812–7822. doi: 10.1523/JNEUROSCI.19-18-07812.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carter-Dawson L. D., LaVail M. M., Sidman R. L. Invest. Ophthalmol. Visual Sci. 1978;17:489–498. [PubMed] [Google Scholar]
- 33.Jimenez A. J., Garcia-Fernandez J. M., Gonzalez B., Foster R. G. Cell Tissue Res. 1996;284:193–202. doi: 10.1007/s004410050579. [DOI] [PubMed] [Google Scholar]
- 34.LaVail M. M., Matthes M. T., Yasumura D., Steinberg R. H. Exp. Eye Res. 1997;65:45–50. doi: 10.1006/exer.1997.0308. [DOI] [PubMed] [Google Scholar]
- 35.Sakamoto K., Liu C., Tosini G. J. Neurochem. 2004;90:1019–1024. doi: 10.1111/j.1471-4159.2004.02571.x. [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto T., Nakahata Y., Soma H., Akashi M., Mamine T., Takumi T. BMC Mol. Biol. 2004;5:18. doi: 10.1186/1471-2199-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ueda H. R., Hayashi S., Chen W., Sano M., Machida M., Shigeyoshi Y., Iino M., Hashimoto S. Nat. Genet. 2005;37:187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 38.Kamphuis W., Cailotto C., Dijk F., Bergen A., Buijs R. M. Biochem. Biophys. Res. Commun. 2005;330:18–26. doi: 10.1016/j.bbrc.2005.02.118. [DOI] [PubMed] [Google Scholar]
- 39.Silver R., Schwartz W. J. Methods Enzymol. 2005;393:451–465. doi: 10.1016/S0076-6879(05)93022-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aton S. J., Colwell C. S., Harmar A. J., Waschek J., Herzog E. D. Nat. Neurosci. 2005;8:476–483. doi: 10.1038/nn1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sakamoto K., Liu C., Kasamatsu M., Iuvone P. M., Tosini G. Mol. Vis. 2006;12:117–124. [PubMed] [Google Scholar]
- 42.Brandenburg J., Bobbert A. C., Eggelmeyer F. Int. J. Chronobiol. 1981;8:13–29. [PubMed] [Google Scholar]
- 43.Zhu H., LaRue S., Whiteley A., Steeves T. D., Takahashi J. S., Green C. B. Mol. Brain Res. 2000;75:303–308. doi: 10.1016/s0169-328x(99)00309-5. [DOI] [PubMed] [Google Scholar]
- 44.Zhuang M., Wang Y., Steenhard B. M., Besharse J. C. Mol. Brain Res. 2000;82:52–64. doi: 10.1016/s0169-328x(00)00177-7. [DOI] [PubMed] [Google Scholar]
- 45.Zhu H., Green C. B. Mol. Vis. 2001;7:210–215. [PubMed] [Google Scholar]
- 46.Bailey M. J., Chong N. W., Xiong J., Cassone V. M. FEBS Lett. 2002;513:169–174. doi: 10.1016/s0014-5793(02)02276-7. [DOI] [PubMed] [Google Scholar]
- 47.Haque R., Chaurasia S. S., Wessel J. H., III, Iuvone P. M. NeuroReport. 2002;13:2247–2251. doi: 10.1097/00001756-200212030-00016. [DOI] [PubMed] [Google Scholar]
- 48.Nir I., Haque R., Iuvone P. M. Brain Res. 2000;870:118–125. doi: 10.1016/s0006-8993(00)02409-4. [DOI] [PubMed] [Google Scholar]
- 49.Nguyen-Legros J., Hicks D. Int. Rev. Cytol. 2000;196:245–313. doi: 10.1016/s0074-7696(00)96006-6. [DOI] [PubMed] [Google Scholar]
- 50.Nguyen-Legros J., Chanut E., Versaux-Botteri C., Simon A., Trouvin J. H. J. Neurochem. 1996;67:2514–2520. doi: 10.1046/j.1471-4159.1996.67062514.x. [DOI] [PubMed] [Google Scholar]
- 51.Tosini G., Dirden J. C. Neurosci. Lett. 2000;286:119–122. doi: 10.1016/s0304-3940(00)01117-4. [DOI] [PubMed] [Google Scholar]
- 52.Iuvone P. M., Tosini G., Pozdeyev N., Haque R., Klein D. C., Chaurasia S. S. Prog. Retin. Eye Res. 2005;24:433–456. doi: 10.1016/j.preteyeres.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 53.Shulman L. M., Fox D. A. Proc. Natl. Acad. Sci. USA. 1996;93:8034–8039. doi: 10.1073/pnas.93.15.8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cohen A. I., Todd R. D., Harmon S., O’Malley K. L. Proc. Natl. Acad. Sci. USA. 1992;89:12093–12097. doi: 10.1073/pnas.89.24.12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gustincich S., Contini M., Gariboldi M., Puopolo M., Kadota K., Bono H., LeMieux J., Walsh P., Carninci P., Hayashizaki Y., et al. Proc. Natl. Acad. Sci. USA. 2004;101:5069–5074. doi: 10.1073/pnas.0400913101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang D. Q., Zhou T., Ruan G. X., McMahon D. G. Brain Res. 2005;1050:101–109. doi: 10.1016/j.brainres.2005.05.042. [DOI] [PubMed] [Google Scholar]
- 57.Nitabach M. N., Blau J., Holmes T. C. Cell. 2002;109:485–495. doi: 10.1016/s0092-8674(02)00737-7. [DOI] [PubMed] [Google Scholar]
- 58.Dijk F., Kraal-Muller E., Kamphuis W. Invest. Ophthalmol. Visual Sci. 2004;45:330–341. doi: 10.1167/iovs.03-0285. [DOI] [PubMed] [Google Scholar]
- 59.DeBruyne J. P., Nuton E., Lambert C. M., Maywood E. S., Weaver D. R., Reppert S. M. Neuron. 2006;50:465–477. doi: 10.1016/j.neuron.2006.03.041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.