Abstract
Since they are equipped with several strategies by which they evade the antimicrobial defense of host macrophages, it is surprising that members of the genus Haemophilus appear to be deficient in common antioxidant systems that are well established to protect prokaryotes against oxidative stress. Among others, no genetic evidence for glutathione (γ-Glu-Cys-Gly) (GSH) biosynthesis or for alkyl hydroperoxide reduction (e.g., the Ahp system characteristic or enteric bacteria) is apparent from the Haemophilus influenzae Rd genome sequence, suggesting that the organism relies on alternative systems to maintain redox homeostasis or to reduce small alkyl hydroperoxides. In this report we address this apparent paradox for the nontypeable H. influenzae type strain NCTC 8143. Instead of biosynthesis, we could show that this strain acquires GSH by importing the thiol tripeptide from the growth medium. Although such GSH accumulation had no effect on growth rates, the presence of cellular GSH protected against methylglyoxal, tert-butyl hydroperoxide (t-BuOOH), and S-nitrosoglutathione toxicity and regulated the activity of certain antioxidant enzymes. H. influenzae NCTC 8143 extracts were shown to contain GSH-dependent peroxidase activity with t-BuOOH as the peroxide substrate. The GSH-mediated protection against t-BuOOH stress is most probably catalyzed by the product of open reading frame HI0572 (Prx/Grx), which we isolated from a genomic DNA fragment that confers wild-type resistance to t-BuOOH toxicity in the Ahp-negative Escherichia coli strain TA4315 and that introduces GSH-dependent alkyl hydroperoxide reductase activity into naturally GSH peroxidase-negative E. coli. Finally, we demonstrated that cysteine is an essential amino acid for growth and that cystine, GSH, glutathione amide, and cysteinylglycine can be catabolized in order to complement cysteine deficiency.
Glutathione (GSH) is the most abundant nonprotein thiol compound present in nearly all eukaryotic cells and in almost all members of the purple bacteria and the cyanobacteria (14). In contrast, the thiol tripeptide is absent from gram-positive bacteria, with the exception of some members of the Bacillus-Clostridium group (8, 36, 68). For that group however, it was shown that the GSH present in these cells was most often not the result of intrinsic biosynthesis but was acquired by import of a reducible form of GSH from the growth medium. Sherrill and Fahey (59) showed in a recent report that Streptococcus mutans imports GSH in an energy-dependent fashion with very high affinity and that this transport is not discriminatory for symmetrical or various mixed disulfides resulting from the GSH thiol chemistry in an oxidative environment, such as the growth medium. However, the imported GSH appears to be dispensable for growth under defined laboratory conditions, as has been established for Escherichia coli by monitoring the growth of mutant GSH-deficient and wild-type E. coli (2, 27). This observation was rather unexpected, because it is well established that GSH plays numerous important roles in the cellular cytoplasm, including control of redox potential, protection against oxidative stress, detoxification of endogenously and exogenously derived toxins, protein folding, and storage and transport of organic sulfur (reviewed in reference 54).
The antioxidant properties of GSH thus appear to be redundant in order to protect aerobic bacteria against the reactive oxygen intermediates (ROI) produced by the incomplete reduction of molecular oxygen during respiration (2). Moreover, log-phase GSH-deficient E. coli mutants are unaffected in their resistance to compounds causing oxidative damage, including hydrogen peroxide (H2O2), cumene hydroperoxide, superoxide anion (O2.−), and gamma radiation (9, 27), but compared to their wild-type counterparts, they show an increased adaptive response to these triggers of oxidative stress (27, 55). This result is an illustration of the fact that a decrease in the cellular thiol/disulfide ratio, as can be caused by mutations that affect the expression of components of the thioredoxin-thioredoxin reductase and/or GSH-glutaredoxin (Grx)-glutathione reductase (GR) systems (the two most important systems maintaining redox homeostasis in living cells), evokes an oxidative stress-like response (55). In E. coli, key regulators of the adaptive response to H2O2 and O2.− are the OxyR and SoxR transcription factors, respectively (reviewed in reference 64). Among the OxyR-regulated genes are those encoding hydroperoxidase I (HP I), GR, Grx1, and the alkyl hydroperoxide reductase system (Ahp) (ahpC and ahpF). Among the SoxR-regulated genes are those encoding Mn-cofactored superoxide dismutase (MnSOD) and glucose-6-phosphate dehydrogenase (G6PD) (zwf). G6PD catalyzes the first rate-limiting step in the pentose phosphate cycle and is accepted to be an antioxidant enzyme because it fuels thioredoxin and GSH redox action by providing reducing equivalents in the form of NADPH (37).
In contrast to the case for bacteria, GSH is an essential reductant during normal metabolic processes in yeast (53), and, moreover, GSH-deficient yeast mutants are hypersensitive to a large collection of oxidative stress-inducing agents (26, 61). The constrained distribution among living organisms of the selenocysteine-containing GSH peroxidases, which are accepted to be universally present only in eukaryotes, is believed to account for the observed differences in sensitivity to oxidative stress of GSH-deficient mutant E. coli and yeast compared to their wild-type counterparts. Moreover, the finding that GSH-deficient yeast strains are hypersensitive to H2O2 suggests that detoxification of H2O2 is exerted primarily by GSH-based reactions rather than by catalase-mediated removal (26). It appears that GSH-producing bacteria lack significant GSH-dependent peroxidase activity (66). Biochemical evidence for such activity has been obtained for Enterococcus faecalis (52), but no gene encoding a GSH-dependent peroxidase has been cloned so far, although recently, genetic evidence has been provided for a peroxidase in the purple bacterium Chromatium gracile, designated Prx/Grx, fueled by the redox cycle of a GSH derivative (i.e., glutathione amide [GASH]) (72).
Haemophilus influenzae is a strictly human commensal capable of invasive infections, including meningitis, pneumonia, and epiglottitis. A complete scan of the entire genome sequence of H. influenzae Rd (21) suggests that this organism, compared to E. coli, possesses an abridged antioxidant armament to deal with ROI. For example, no homologs encoding γ-glutamylcysteine synthase and glutathione synthase, the GSH biosynthetic enzymes, or homologs encoding the components of the Ahp system are apparent from the H. influenzae Rd genome. In this report, we provide evidence that H. influenzae is indeed unable to produce GSH but acquires a complete and functional GSH metabolism by importing GSH from the environment. Furthermore, we ascertain the importance of the imported GSH in the supply of organic sulfur and in the maintenance of intracellular redox homeostasis. Finally, we present biochemical and genetic data showing that the H. influenzae GSH metabolism fuels a C. gracile GASH peroxidase (Prx/Grx) homolog which can complement the tert-butyl hydroperoxide (t-BuOOH) sensitivity of an Ahp negative E. coli strain and the H2O2 sensitivity of a totally catalase-deficient (acatalasemic) E. coli mutant.
MATERIALS AND METHODS
Materials.
All reagents and commercial enzymes were purchased from Sigma-Aldrich (St. Louis, Mo.), unless indicated otherwise. Bacterial culture material was from Difco Laboratories (Detroit, Mich.), unless indicated otherwise. GASH was prepared as described previously (72).
Media.
Mueller-Hinton (MH) broth was prepared from a dehydrate (Fluka at Sigma-Aldrich) and autoclaved. MHs broth was MH broth supplemented with V factor (NAD) and X factor (hemin), both at 10 μg/ml. GSH-depleted MHs broth (dpMHs) was prepared enzymatically by treating MH broth with γ-glutamyl transpeptidase (1 U/ml) for 4 h at 37°C before autoclaving. GSH-replete MHs broth (rpMHs) was prepared from dpMHs broth by adding glutathione disulfide (GSSG) from a sterile concentrated stock solution (final concentration, 20 μM). By use of the total GSH (GSX) (i.e., reducible symmetrical or mixed disulfide forms of GSH) quantification method described by Tietze (69), MHs and dpMHs media were determined to contain approximately 5 and less than 0.1 μM GSX, respectively.
H. influenzae specific minimal medium (MIc) was prepared essentially as described by Herriott et al. (30). The only modification was that cysteine was added from a sterile stock solution (final concentration, 100 μM) in place of cystine (Cys-Cys). GSH-replete MIc medium (rpMIc) was prepared by adding GSSG from a sterile concentrated stock solution (final concentration, 20 μM) to MIc medium. MIc and rpMIc agar plates were prepared as described previously (30).
Luria-Bertani medium (LB) broth and LB agar plates were supplemented with carbenicillin (100 μg/ml) and t-BuOOH (final concentration, 50 to 500 μM) when necessary.
Plasmids, bacterial strains, and growth conditions.
The nonencapsulated, nontypeable H. influenzae type strain NCTC 8143 and the H. influenzae Rd strain were purchased from the American Type Culture Collection (Manassas, Va.). For maintenance, H. influenzae NCTC 8143 cells were routinely cultivated in a candle extinction jar in dpMHs broth at 37°C for 24 h without rotary shaking. For comparison of the abilities of H. influenzae NCTC 8143 to grow in cysteine-deficient broth supplemented with dithiothreitol (DTT), hydrogen sulfide (H2S), or organic cysteine sources, a 20-μl portion of an overnight preculture was transferred into 1.9 ml of fresh MIc broth devoid of cysteine and supplemented with the sulfur source into capped culture tubes (flat bottom; outer diameter, 13 mm; height, 130 mm) (MLS, Menen, Belgium); these tubes fit tightly in the cuvette chamber of a Shimadzu (Duisburg, Germany) 1240 mini-single-beam UV-visible spectrophotometer and allow cell density measurements without the need to transfer culture samples into regular cuvettes. The resulting cultures were cultivated aerobically at 37°C with rotary shaking (190 rpm), and growth was monitored by measuring the turbidity at 600 nm for 9 h. Sulfur donor compounds were used at the following final concentrations: cysteine and DTT, 100 μM; Cys-Cys, 50 μM; and H2S, GSH, GSSG, GASH, γ-glutamylcysteine (γ-Glu-Cys), and cysteinylglycine (Cys-Gly), 20 μM.
E. coli HB101 (Bio-Rad, Hercules, Calif.) was used for propagation of the plasmid vector pUC18 (Gibco-BRL, Rockville, Md.) as well as derivatives of this plasmid. Complementation experiments were performed with E. coli strain K-12 (wild type), UM2 (catalase-negative) (39), and TA4315 (ahpΔ5) (65), all obtained from the E. coli Genetic Stock Center (New Haven, Conn.). E. coli cells were routinely cultivated in LB broth at 37°C with rotary shaking (190 rpm).
Growth inhibition assays of log-phase H. influenzae NCTC 8143.
A 1:20 dilution of an overnight H. influenzae NCTC 8143 preculture in fresh dpMHs or rpMHs broth (MIc or rpMIc medium, respectively, for the S-nitrosoglutathione [GSNO] growth inhibition assay) was aseptically divided in 2-ml portions into the capped culture tubes described above. These cultures were grown aerobically at 37°C with vigorous shaking (190 rpm) until they reached an A600 of approximately 0.11 (early exponential growth phase). One hundred microliters of stress-inducing test chemicals dissolved in phosphate-buffered saline (PBS), at three or four increasing final concentrations, or 100 μl of PBS alone (negative controls) was added to the culture tubes, and cell density was recorded every hour for at least 5 h. The percentage of growth of stressed cultures relative to the negative controls was assessed after 2 h (3 h for the GSNO growth inhibition assay) and expressed as the mean ± standard error of the mean (SEM) from three independent experiments (the SEM was <5%).
Agar disk diffusion susceptibility assays.
A 1:20 dilution of an overnight H. influenzae NCTC 8143 preculture in fresh MIc or rpMIc medium (20 ml in a 250-ml sidearm culture flask) was grown aerobically at 37°C with vigorous shaking (190 rpm) until the A600 reached 0.85 (early stationary phase). By using a sterile cotton swab, cells of the MIc and rpMIc cultures were inoculated onto the entire surfaces of MIc and rpMIc agar plates, respectively, as described previously (47). Five microliters of 3% H2O2, 500 mM t-BuOOH, 500 mM GSNO, or 100 mM sodium hypochlorite (NaOCl) was placed on 3MM paper (Whatman, Maidstone, United Kingdom) disks (diameter, 5.2 mm), and these disks were individually placed with sterile forceps onto the agar within 15 min after inoculation. After incubation at 37°C in a candle extinction jar for 48 h, the diameter of the zone of complete inhibition was recorded in millimeters.
Agar disk diffusion susceptibility assays with H2O2 and t-BuOOH for E. coli strains K12, UM2, and TA4315 transformed with plasmids pUC18 and pUC-HI0572 were carried out essentially as described above for H. influenzae NCTC 8143. Liquid cultures were grown in carbenicillin-LB broth, and paper disks were placed on the surface of carbenicillin-LB agar plates. The results were interpreted after an incubation period of 24 h at 37°C for the assay with H2O2 and after 62 h at 37°C for the assay with t-BuOOH.
Determination of GSX.
The GSX content of H. influenzae NCTC 8143 cells grown in different media was measured by the method of Tietze (69). Samples were prepared as described previously (60) from early-stationary-phase H. influenzae NCTC 8143 cultures (A600 = 1.0) grown aerobically at 37°C in MIc, rpMIc, dpMHs, or rpMHs medium (25 ml in a 250-ml sidearm culture flask). GSX content is given in micromoles per gram of cell residual dry weight.
Enzyme assays.
A Uvikon 943 double-beam UV-visible spectrophotometer (Kontron Instruments, Watford, United Kingdom) was used for the spectroscopic measurements. All enzyme assays were performed at room temperature in 0.5-ml quartz-Sephacil cuvettes on cell extracts prepared as follows. A 1:20 dilution of an overnight H. influenzae NCTC 8143 preculture in fresh MIc or rpMIc medium (150 ml in a 1,000-ml sidearm culture flask) was grown aerobically at 37°C with vigorous shaking (190 rpm) until the A600 reached approximately 0.17 (early exponential phase). The cultures were then aseptically divided in two, with one half treated with 100 μM H2O2 and the other half left untreated (i.e., an equal amount of PBS was added). Next, after a prolonged incubation for exactly 1 h, the cultures were placed on ice for 10 min. Cells were collected by centrifugation (from here on, bacterial pellets were kept on ice) and then washed twice with PBS (pH 7.3) containing 2 mM EDTA (reaction buffer). Subsequently, the bacterial pellets were suspended in 1 ml of reaction buffer and the cells were broken by sonication at 0°C, using a 15-s pulse for four cycles. The resulting crude suspensions were cleared by centrifugation (15,000 × g) for 10 min at 4°C, and the resulting supernatants were immediately used in the enzyme assays. Total protein concentration was determined by the Bradford dye binding assay (6), using the Bio-Rad 500-0006 kit with bovine serum albumin as a standard.
Catalase activity was assayed by monitoring the extract-dependent decomposition of H2O2 (final concentration, 10 mM) at 240 nm (ɛ = 40.0 M−1 cm−1) (28). GR activity (52) and Grx activity (73) were determined as described previously. G6PD, the first and rate-limiting enzyme of the pentose phosphate pathway, couples the oxidation of glucose-6-phosphate to the reduction of NADP+, generating NADPH and eventually (via chemical decomposition or enzymatic interference) 6-phosphogluconate. Because the latter product is the substrate of another enzyme of the pentose phosphate pathway, 6-phosphogluconate dehydrogenase (6PGD), which also generates NADPH, both 6PGD and total dehydrogenase activity (6PGD plus G6PD) were measured at 340 nm. In order to obtain accurate G6PD activity data, the activity of 6PGD was then subtracted from total dehydrogenase activity. For total dehydrogenase and 6PGD activity determinations, the blank and assay reaction mixture contained 50 μl of extract and 400 μM NADP+. The reactions were initiated by adding 1 mM glucose-6-phosphate plus 1 mM 6-phosphogluconate (total dehydrogenase activity) or 1 mM 6-phosphogluconate (6PGD activity) to the assay reaction mixture. The GSH-peroxidase assay was performed as described by Patel et al. (52), with substitution of t-BuOOH (final concentration, 400 μM) for H2O2.
DNA manipulations and sequence analysis.
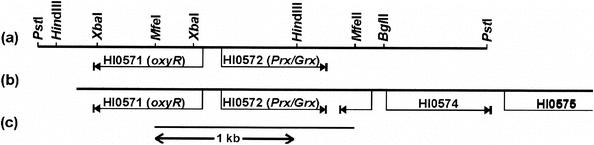
Restriction enzymes were from New England Biolabs (Herefordshire, United Kingdom), and the Expand Long Template PCR System and T4 DNA ligase were from Roche Diagnostics (Mannheim, Germany). H. influenzae Rd genomic DNA was prepared as described previously (3). A ∼4-kb chromosomal fragment containing open reading frames (ORFs) HI0571 to HI0574 (Fig. 1) was generated by PCR with the forward primer 5′-TGCCTGAACTTTCGCGTAATA-3′ and the reverse primer 5′-TGTTTGATTTGGCGGATGTA-3′. The PCR product was digested with MfeI, and the resulting 1,427-bp fragment was ligated to pUC18 linearized with EcoRI (not protected for religation via calf intestinal phosphatase treatment) to yield pUC18-HI0572. Selection of positive clones is described in Results. Determination of the nucleotide sequence of the insert of the pUC-HI0572 construct was performed with an ABI PRISM 377 automated sequencer (Applied Biosystems, Foster City, Calif.). Database searches were performed with the BLAST program (1) provided by the National Center for Biotechnology Information. Partially sequenced genomes were assessed and searched either from the National Center for Biotechnology Information genome database or from individual databases listed in and linked to The Institute for Genome Research website at http://www.tigr.org.
FIG. 1.
(a) Partial restriction enzyme map of the nontypeable H. influenzae 3.7-kb genomic DNA fragment cloned by Maciver and Hansen (41), containing ORF HI0571 and ORF HI0572, which expresses transferrin-binding activity in E. coli. The arrows indicate the direction of transcription. (b and c) A 3.9-kb H. influenzae Rd DNA fragment (b) was generated by PCR in order to clone the 1,427-bp MfeI-fragment (c) into EcoRI-linearized pUC18 plasmid DNA (pUC-HI0572).
RESULTS
Imported GSX is involved in cysteine metabolism.
In order to verify whether GSH import actually is responsible for GSH acquisition in H. influenzae NCTC 8143, we cultivated H. influenzae NCTC 8143 cells in minimal medium either deficient (MIc) or rich (rpMIc) in GSSG or in complex medium either deficient in (dpMHs) or physiologically loaded with (rpMHs) GSX. Cultures were cultivated until early stationary phase (MIc and rpMIc, A600 = 0.85; dpMHs and rpMHs, A600 = 0.60) before GSX determination was performed. Cells grown in broth supplemented with GSX accumulated the thiol tripeptide (2.5 ± 0.5 μmol/g [residual dry weight]) independent of whether the medium was minimal or rich, while cells grown in medium devoid of GSX were completely GSH deficient.
By comparing the generation times of H. influenzae NCTC 8143 cultures grown in MIc medium devoid of cysteine and in identical medium enriched with DTT, H2S, or organic cysteine sources, we examined whether the imported GSH is involved in the cysteine metabolism of H. influenzae NCTC 8143 (Table 1). No growth was found in MIc medium devoid of cysteine (note that MIc medium contains K2SO4 at the prescribed final concentration of 5.74 mM [30]). Comparable minimum generation times were observed for cultures grown in the presence of cysteine, Cys-Cys, GSSG, GSH, GASH, and Cys-Gly, while no growth was found in the presence of DTT or H2S. With γ-Glu-Cys as the sole sulfur source, the doubling time observed was 1 order of magnitude higher and stationary phase was already initiated at an A600 of approximately 0.05. Finally, the data in Table 1 show that when cysteine is present in the growth medium, GSH is not necessary for normal growth under standard laboratory conditions.
TABLE 1.
Minimum generation times of H. influenzae NCTC 8143 cells grown with different sulfur sourcesa
| Sulfur source (concn, μM) | Generation time (min)b |
|---|---|
| Cysteine (100) | 68 |
| Cys-Cys (50) | 68 |
| GSSG (20) | 68 |
| GSH (20) | 68 |
| Cys-Gly (20) | 65 |
| γ-Glu-Cys (20) | 556 |
| GASH (20) | 67 |
| DTT (100) | No growth |
| Cysteine + DTT | 70 |
| H2S (20) | No growth |
| Cysteine + H2S | 70 |
| No sulfur source added (i.e., SO42−) | No growth |
Cells were grown in cysteine-depleted MIc medium supplemented as indicated.
Derived from turbidimetric measurements at 600 nm on log-phase cultures; data are expressed as the means from four independent experiments.
Cellular GSH is involved in the detoxification of certain electrophilic and phagocyte-derived antimicrobial compounds.
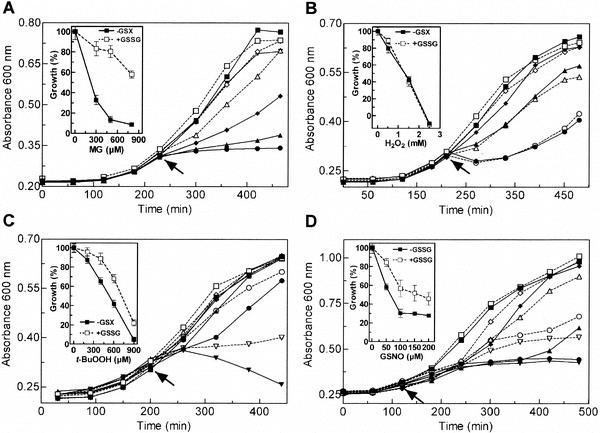
In order to establish whether the accumulated GSH not only functions as a nutrient source but also serves protective functions, we challenged early-exponential-phase H. influenzae NCTC 8143 cultures grown in dpMHs and rpMHs broth with increasing methylglyoxal (MG) concentrations (Fig. 2A). MG is a known endogenous and environmental mutagen that can modify both DNA and proteins (46, 51). While the growth of log-phase H. influenzae NCTC 8143 cultures devoid of GSSG was almost completely inhibited in the presence of 450 μM MG, this treatment was only modestly bacteriostatic (approximately 20% growth reduction) for cultures grown in the presence of GSSG.
FIG. 2.
Effects of sublethal and lethal concentrations of MG (A), H2O2 (B), and t-BuOOH (C) on actively growing cells of H. influenzae NCTC 8143 in culture medium dpMHs (−GSX) (closed symbols) or in culture medium rpMHs (+GSSG) (open symbols) and of GSNO (D) on actively growing cells of H. influenzae NCTC 8143 in culture medium MIc (−GSSG) (closed symbols) or in culture medium rpMIc (+GSSG) (open symbols). MG concentrations (in micromolar): 0 (squares), 300 (diamonds), 500 (triangles), and 800 (circles); H2O2 concentrations (in millimolar): 0 (squares), 0.5 (diamonds), 1.5 (triangles), and 2.5 (circles); t-BuOOH concentrations (in micromolar): 0 (squares), 200 (diamonds), 400 (triangles), 600 (circles), and 900 (inverted triangles); GSNO concentrations (in micromolar): 0 (squares), 50 (diamonds), 100 (triangles), 150 (circles), and 200 (inverted triangles). Growth was periodically monitored by measuring the absorbance at 600 nm, and stress induction is indicated by an arrow. The data are averages from three different experiments (error bars are not shown for clarity). Insets, percentage of growth of stressed cultures relative to the negative controls; data are expressed as the means ± SEMs from three independent experiments (the SEMs were <5%).
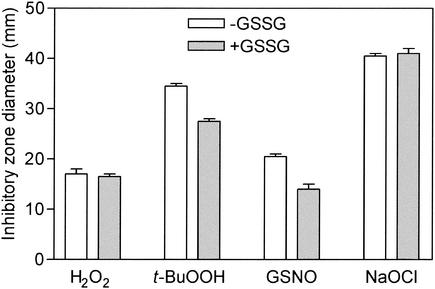
To investigate whether GSH is involved in the susceptibility of H. influenzae NCTC 8143 cells to ROI and to reactive nitrogen intermediates (RNI) generated by phagocytes to eliminate unwanted microorganisms, we also determined the susceptibility to H2O2, t-BuOOH, and GSNO by the zone of inhibition in agar disk diffusion susceptibility assays, in addition to growth curve measurements. The results of the two methods were found to correlate with each other. Growth characteristics (Fig. 2B) and inhibition zone diameters (Fig. 3) were identical for H2O2-challenged H. influenzae NCTC 8143 cultures grown in the presence or absence of GSSG. On the other hand, a significantly enhanced susceptibility to the small alkyl hydroperoxide t-BuOOH and to GSNO was demonstrated for H. influenzae NCTC 8143 cultures devoid of GSSG compared to GSSG-replete cultures (Fig. 2C and D and 3).
FIG. 3.
Susceptibility of H. influenzae NCTC 8143 to oxidative stress-generating compounds. Susceptibility was determined by a disk diffusion method with MIc agar plates (−GSSG) or rpMIc agar plates (+GSSG).
Finally, we could not determine inhibition zone variation for GSH-replete and GSH-deficient H. influenzae NCTC 8143 resulting from NaOCl-impregnated disks, i.e., hypochlorous acid (HOCl) stress (Fig. 3).
Extracts of H. influenzae NCTC 8143 display GSH-dependent peroxidase activity.
The foregoing results concerning the role of GSH in the vulnerability of H. influenzae NCTC 8143 to H2O2 and t-BuOOH suggest that the established protection of endogenous GSH against t-BuOOH relies on presently unknown enzymatic grounds and not on the participation of GSH in the nonenzymatic destruction of t-BuOOH. Therefore, we examined cell-free H. influenzae NCTC 8143 extracts for the presence of GSH-dependent alkyl hydroperoxide reductase activity. The assay confirmed the presence of GSH-dependent peroxidase activity in H. influenzae NCTC 8143 extracts (Table 2). In addition, linearity in plots of activity versus extract volume (data not shown) was observed. Since the peroxidase activity was almost totally inactivated in the presence of 4 mM iodoacetamide (data not shown), we reasoned that cysteine residues are involved in catalysis.
TABLE 2.
Effect of H2O2 treatment on antioxidant enzyme activities monitored in H. influenzae NCTC 8143 extracts derived from cultures grown in rpMIc medium or in thiol-disulfide stress-inducing MIc mediuma
| Growth medium and treatment | Catalase (HktE)b | G6PDc | GRd | Grxd | GSH-dependent peroxidased |
|---|---|---|---|---|---|
| rpMIc | 36 ± 7 | 15 ± 3 | 335 ± 5 | 32.1 ± 10 | 1.0 ± 0.2 |
| rpMIc + H2O2 | 89 ± 5 | 28 ± 1 | 376 ± 5 | 32.0 ± 9 | 0.9 ± 0.2 |
| MIc | 76 ± 13 | 31 ± 0.4 | 437 ± 50 | 34.5 ± 6 | 1.0 ± 0.1 |
| MIc + H2O2 | 315 ± 7 | 37 ± 1 | 531 ± 14 | 38.6 ± 8 | 1.0 ± 0.2 |
All results are means and standard deviations of three determinations. See Materials and Methods for experimental details.
Results are expressed as micromoles mol of H2O2 decomposed per minute per milligram of protein.
Results are expressed as nanomoles of NADP+ reduced per minute per milligram of protein.
Results are expressed as nanomoles of NADPH oxidized per minute per milligram of protein.
Cloning of an H. influenzae Rd genomic DNA fragment that confers resistance to peroxides.
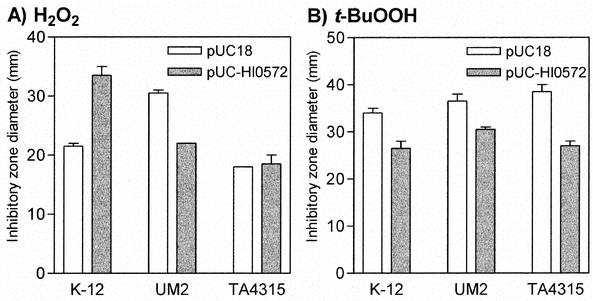
The H. influenzae Rd genome carries a homolog (ORF HI0572) of the gene encoding the C. gracile Prx/Grx peroxidase (64% identity at the amino acid sequence level). In order to establish whether the HI0572 gene product could be responsible for the observed GSH-dependent protection of H. influenzae NCTC 8143 cells against t-BuOOH stress, we cloned an MfeI fragment, containing ORF HI0572 and a large part of ORF HI0571 (Fig. 1), derived from a PCR-generated H. influenzae Rd DNA fragment into the pUC18 vector (linearized with EcoRI but not subsequently dephosphorylated). Clones containing the MfeI insert were selected by complementation of the t-BuOOH sensitivity of E. coli strain TA4315. Thus, strain TA4315 was transformed with the ligate and plated on LB-ampicillin plates containing 400 μM t-BuOOH, which allowed the growth of E. coli strain K-12 but not that of strain TA4315. Twenty colonies that grew on this selective medium were tested, and they all contained the proper construct, while only 1 out of 40 colonies that grew on LB-ampicillin plates without t-BuOOH selection were positive. Furthermore, agar disk diffusion susceptibility assays revealed that the HI0572 gene product is able to complement, in addition to the t-BuOOH sensitivity of strain TA4315, the H2O2 sensitivity of the acatalasemic E. coli strain UM2 (Fig. 4). Surprisingly, however, pUC-HI0572-carrying wild-type E. coli strain K-12 displays UM2-like sensitivity to H2O2 (Fig. 4).
FIG. 4.
Effect of the introduction of the pUC-HI0572 plasmid on the susceptibilities of wild-type (K-12), acatalasemic (UM2), and Ahp-negative (TA4315) E. coli strains to H2O2 (A) and t-BuOOH (B).
To determine whether the cloned H. influenzae NCTC 8143 DNA actually encodes GSH-dependent peroxidase activity, we assessed pUC-HI0572-transformed log-phase E. coli culture lysates for this activity. Table 3 shows that every E. coli strain that carries the pUC-HI0572 plasmid indeed acquired considerable GSH-dependent alkyl hydroperoxide reductase activity. Notably, the activity determined in log-phase acatalasemic E. coli extracts was shown to be almost twofold the activity measured in extracts derived from the wild-type or Ahp-negative lysates.
TABLE 3.
GSH-dependent peroxidase and catalase activities monitored in log-phase lysates derived from wild-type and mutant E. coli strains transformed with the pUC18 (control) or pUC-HI0572 plasmida
| E. coli strain (plasmid) | GSH-dependent peroxidaseb | Catalasec |
|---|---|---|
| Wild-type K-12 (pUC18) | 0 | 1,042 ± 32 |
| Acatalasemic UM2 (pUC18) | 0 | <10 |
| Ahp-negative TA4315 (pUC18) | 0 | 980 ± 24 |
| Wild-type K-12 (pUC-HI0572) | 1,326 ± 33 | 2,295 ± 81 |
| Acatalasemic UM2 (pUC-HI0572) | 2,248 ± 51 | <10 |
| Ahp-negative TA4315 (pUC-HI0572) | 1,150 ± 27 | 3,170 ± 52 |
Transformed cells were grown in carbenicillin-LB broth until the A600 reached 0.45, and then cell lysates were prepared essentially as described in Material and Methods for the preparation of H. influenzae NCTC8143 crude extracts. The total protein concentration was determined by the Bradford dye binding assay (6) with bovine serum albumin as a standard. All results are means and standard deviations of three determinations.
Results are expressed as nanomoles of NADPH oxidized per minute per milligram of protein.
Results are expressed as nanomoles of H2O2 decomposed per minute per milligram of protein.
Cellular GSH is involved in the regulation of particular antioxidant activities.
Since there is no soxR- or soxS-homologous sequence apparent from the H. influenzae Rd genome sequence and because of our finding that H. influenzae possesses an NADPH-driven GSH-dependent peroxidase system instead of the well-known NADH-driven Ahp-system of enteric bacteria (65), we evaluated whether the expression of the H. influenzae NCTC 8143 zwf gene positively responds to H2O2- and thiol-disulfide-mediated stress (induced by the lack of GSSG in the growth medium), as is the case for eukaryotic cells (35). Concurrently, we monitored the activities of the sole catalase, HktE (5), and the sole Grx (as is apparent from the H. influenzae Rd genome sequence [ORF HI1532]), along with GR and GSH-dependent peroxidase activity, in response to H2O2 and/or thiol-disulfide-mediated stress (Table 2). We found that the activities of HktE, GR, and G6PD in H. influenzae NCTC 8143 extracts derived from log-phase cultures grown in MIc medium are enhanced compared to the corresponding activities in the extracts derived from rpMIc-grown GSH-replete cultures, being induced approximately 2.1-fold for HktE and G6PD activity and 1.3-fold for GR activity. In response to H2O2 exposure, HktE, GR, and G6PD activities in extracts derived from MIc medium cultures were induced 3.5-, 1.4-, and 1.3-fold, respectively, compared to the corresponding activities monitored in rpMIc-derived extracts. Moreover, it appears that H2O2 stress and thiol-disulfide-mediated stress exert a cumulative positive effect on the activities of these antioxidant enzymes. Neither H2O2-induced stress nor thiol-disulfide-mediated stress appeared to affect GSH-dependent peroxidase or Grx activity.
DISCUSSION
The small-genome parasitic organism H. influenzae exhibits a nutritional requirement for organic sulfur.
Although biochemical evidence for GSH uptake has been reported for gram-positive bacteria (59), yeasts (44), plants (22), and mammalian cells (31), the import of GSX from the growth medium as the exclusive source of GSH for H. influenzae NCTC 8143 cells reported here is rather surprising because H. influenzae belongs to the aerobic gram-negative bacteria which are thought to endogenously synthesize the thiol tripeptide (15). While the uptake of GSX appears to be redundant under standard laboratory conditions for log-phase H. influenzae NCTC 8143 growing in complex medium, GSH accumulation is essential for growth in minimal broth lacking cysteine, indicating that GSH can be catabolized to meet the requirement of H. influenzae NCTC 8143 for cysteine.
Cysteine is a nonessential amino acid for eukaryotes and most prokaryotes but appears to be essential for H. influenzae NCTC 8143. Indeed, we established that H. influenzae NCTC 8143 is not able to use either environmental sulfate or sulfide as a source of sulfur in cysteine biosynthesis (Table 1), despite the fact that a cysK homolog (ORF HI1103, with 56% identity at the amino acid sequence level) is apparent from the genomic sequence (in enteric bacteria, the cysK gene product cysteine synthase A catalyzes cysteine biosynthesis from H2S and O-acetyl-l-serine). Taken together, the facts that H. influenzae NCTC 8143 is not able to grow with sulfate as the sole sulfur source and, in agreement with this result, that the H. influenzae Rd genome sequence gives no genetic evidence for either a single component of the sulfate assimilation pathway or a sulfite reductase protein raise the question (assuming that the cysK homolog of H. influenzae NCTC 8143 encodes a functional cysteine synthase A protein) of how this strain acquires the necessary sulfide to drive cysteine synthase A activity.
Another interesting finding is the observation that in addition to cysteine, Cys-Cys, GSH, and GSSG, and also Cys-Gly, but not γ-Glu-Cys, can be used as organic cysteine sources by H. influenzae NCTC 8143 cells. The only report of a GSH transport system in bacteria is a biochemical study of the import and metabolism of GSH by S. mutans (59). In that report, the authors showed that the glutamyl residue appears to be critical for the transport of GSH, and they thus established import of γ-Glu-Cys, but not of Cys-Gly, from the environment into the cytoplasm. Whether, in the case of H. influenzae NCTC 8143, γ-Glu-Cys is defective as a cysteine source because of the inability of the GSH transport system to recognize the dipeptide or because of the inability of the cytoplasm to catabolize the dipeptide remains to be answered. γ-Glu-Cys stock solutions remain clear, so precipitation of oxidized compound cannot be the reason for inadequate γ-Glu-Cys uptake.
Cellular GSH protects H. influenzae NCTC 8143 against MG and GSNO toxicity.
The production of the electrophile MG, catalyzed by the action of MG synthase, allows bacterial cells to control the rate of carbon flux and therefore enhances cell survival and the chance of colonizing new environments (20). Nevertheless, MG accumulation is lethal for all living cells, and therefore specialized detoxification pathways and protective mechanisms have evolved. In E. coli, the GSH-dependent glyoxalase I-II MG detoxification pathway (70), in conjunction with the GSH-mediated KefB and KefC ion channel systems (19), were shown to be vital for surviving MG stress. Because these two systems rely on GSH for proper activity, E. coli double mutants defective in the two protective systems and GSH-deficient mutants display equally increased sensitivity to MG stress (42, 56). From the H. influenzae Rd genome sequence, it has become apparent that this organism possesses homologs encoding, in addition to an MG synthase (ORF HI1234), a complete glyoxalase I-II system (ORF HI1274 and ORF HI0323) and a single Kef protein, which, at the sequence level, is intermediate between KefB and KefC and has been designated KefX (ORF HI0911) (48). The growth analysis assays described here for MG-stressed GSH-deficient and GSH-replete H. influenzae NCTC 8143 show that MG detoxification to a great extent depends on cellular GSH and that log-phase H. influenzae NCTC 8143 and E. coli display comparable sensitivities to MG (17, 18).
GSNO is a main side product of phagocyte nitric oxide synthase activity and is formed in close vicinity to and within microorganisms subjected to a nitrosative challenge (43, 50). GSNO formation in microorganisms was recognized at first to be the result of the RNI scavenging function of GSH, but S-nitrosothiols in general, and GSNO in particular, were later found to be bactericidal. Mutation of the gene encoding glutathione synthase, which, as mentioned before, controls GSH biosynthesis, renders Salmonella enterica serovar Typhimurium hypersusceptible to RNI (12). In agreement with this result, we here clearly show via growth analysis experiments and disk diffusion assays that GSH is involved in H. influenzae NCTC 8143 detoxification of GSNO as well. Whether the observed GSH-mediated protection is the result of pure scavenging of GSNO toxicity or of cofactor functioning for a GSNO reductase (e.g., GSH-dependent formaldehyde dehydrogenase [GS-FDH] [see below]) remains to be answered.
Metabolism of peroxides in H. influenzae.
Nonencapsulated, nontypeable H. influenzae strains, the major cause of both upper and lower respiratory tract infections in humans, are able to survive in macrophages for prolonged periods (10). Hence, these H. influenzae strains are skilled at circumventing the antimicrobial assault of mammalian phagocytes and therefore ought to possess protective systems against the nonspecific oxidative effects of ROI and RNI. However, a scan of the entire genome sequence suggests that the antioxidant armament of H. influenzae Rd is much more condensed than those of the enteric bacteria E. coli and S. enterica serovar Typhimurium. At present it is recognized that the latter organisms exploit the actions of HP I, HP II, Ahp, MnSOD, FeSOD, Cu-ZnSOD, G6PD, bacterioferritin comigratory protein (Bcp), and thiol peroxidase (Tpx) to directly neutralize ROI; of the Ahp (7), Cu-ZnSOD (11), G6PD (40), peptide methionine sulfoxide reductase (MsrA) (63), GS-FDH (38), and flavohemoglobin (62) antioxidant enzymes to directly neutralize RNI; and of the OxyR, SoxRS, MarA, Rob, RpoS, ArcA, Fnr, and Fur transcriptional regulators to respond to oxidative stress. Of these antioxidant enzymes and stress response regulators, only HP II (hktE gene product) (5), GS-FDH (29), MnSOD (13), and OxyR (41) are identified biochemically in H. influenzae strains, and G6PD, Bcp, Tpx, MsrA, ArcA, Fnr, and Fur are apparent from the H. influenzae Rd genome sequence. Haemophilus somnus, a pathogen of cattle that survives in bovine neutrophiles, displays an even further reduced antioxidant armament, being totally catalase negative (57). In agreement with this observation, it was reported that HktE-negative H. influenzae mutants were as viable and as virulent as wild-type controls (4). Therefore, we speculated that H. influenzae should exploit an unidentified alternative strategy to destroy peroxides.
We recently published the nucleotide sequence of a gene from the phototrophic purple sulfur bacterium C. gracile that translates into a 27.5-kDa enzyme displaying GASH-dependent peroxidase activity, designated Prx/Grx. This Prx/Grx peroxidase is a hybrid enzyme, with an N-terminal peroxiredoxin (Prx)-like domain and a C-terminal Grx-like domain (72). The H. influenzae Rd Prx/Grx homologous ORF HI0572 has been cloned by Maciver and Hansen (41) as part of a 3.7-kb subgenomic fragment derived from a nontypeable H. influenzae DNA library (Fig. 1). According to those authors, this 3.7-kb fragment contained two ORFs (ORF HI0571 and ORF HI0572) and was isolated because of its expression of transferrin-binding activity in E. coli, which the authors assigned to ORF HI0571, which almost certainly translates to a functional OxyR protein (41). In this report we argue that the 26.7-kDa HI0572 gene product, Prx/Grx, should represent an important role in the peroxide metabolism of H. influenzae because of the following: (i) ORFs HI0571 and HI0572 appear to be transcribed from divergent, overlapping promoters, which indicates that the regulation of HI0572 expression is somehow correlated with the titer of OxyR protein; (ii) the HI0572 gene product is a thiol peroxidase and thus inactive in the presence of iodoacetamide (GSH-dependent peroxidase activity is completely abolished in H. influenzae extracts due to the presence of iodoacetamide); (iii) when introduced in E. coli, the plasmid containing HI0572 is able to complement the t-BuOOH and H2O2 sensitivities of Ahp-negative and catalase-negative strains, respectively, and is able to deliver considerable alkyl hydroperoxide reductase activity to otherwise GSH peroxidase-negative E. coli; and (iv) recombinant H. influenzae Rd Prx/Grx produced in E. coli has been shown to be a high-affinity GSH-dependent reductase of H2O2 and small alkyl hydroperoxides (data not shown; the full characterization of the H. influenzae Rd Prx/Grx peroxidase will be published elsewhere).
Interestingly, while conferring greater resistance to t-BuOOH in all strains tested and to H2O2 in the acatalasemic E. coli strain UM2, plasmid pUC-HI0572 caused a substantially increased sensitivity to H2O2 in E. coli strain K-12 and a marginally increased sensitivity to H2O2 in E. coli strain TA4315. Storz et al. (65) reported an observation similar to that in our study for E. coli strain TA4315 carrying a cosmid clone that accommodates the complete Ahp locus. Those authors reasoned that multiple copies of the ahpC and ahpF genes may cause the OxyR regulator to be titrated away from other OxyR-regulated genes, such as the katG gene, which confer greater resistance to high levels of exogenous H2O2. This implies that the intergenic region between HI0571 and HI0572 should be able to accommodate the E. coli OxyR protein. To address this premise, we have monitored total catalase activity in extracts derived from pUC18- and pUC-HI0572-transformed K-12, UM2, or TA4315 cells. While it was expected that less catalase activity would be measured in pUC-HI0572-transformed E. coli than in the controls, we established an approximately twofold increase (Table 3). This may mean that the propagation of the multicopy plasmid pUC-HI0572 is associated with the generation of an aberrant quantity of HI0572 transcripts causing an increased oxidative (or general) stress condition. In combination with high H2O2 stress, this may make pUC-HI0572-transformed wild-type E. coli more vulnerable to high exogenous concentrations of H2O2 than the untransformed equivalent. The vast overproduction of Prx/Grx peroxidase, together with the endogenous AhpCF proteins, in E. coli strain K-12 may then inflict a more severe stress in the wild-type strain than in the Ahp-negative strain TA4315, which explains the different responses of the pUC-HI0572-transformed strains to H2O2.
The facts and considerations described above strongly indicate that the H. influenzae Prx/Grx system functionally replaces the well-known Ahp system of enteric bacteria, which was recently reported by Seaver and Imlay (58) to be the primary scavenger of endogenous H2O2 in E. coli. To address this premise, we are currently generating an H. influenzae HI0572 knockout strain.
Cellular GSH performs redox control.
GSH has long been implicated in the regulation of gene expression in E. coli. For example, GSH, in combination with Grx1, was found to be the main deactivator of OxyR activity (74), and hence, log-phase GSH-deficient E. coli displays higher activities of OxyR-regulated antioxidant enzymes than the proficient wild-type parent (24, 49). It has been previously reported that catalase activity of H. influenzae is most probably OxyR controlled in response to H2O2 stress (4, 5), and therefore one might expect that HktE activity would increase as a result of GSH deficiency and H2O2 stress. We established that thiol-disulfide and H2O2 stresses exert a cumulative effect, not only on catalase activity but also on GR and G6PD activities, in log-phase H. influenzae NCTC 8143, strongly suggesting that these three antioxidant enzymes are OxyR controlled. Unexpectedly, however, Grx activity monitored in H. influenzae NCTC 8143 extracts does not respond to either thiol-disulfide stress or H2O2 stress. This result is inconsistent with the Grx regulation described so far for E. coli (67) or yeast (25) and should thus be investigated in more detail.
In E. coli, zwf expression for G6PD is not regulated by the OxyR redox state but is positively controlled by the O2.−- and nitric oxide-responsive SoxRS regulon (16), and, accordingly, no increase in G6PD activity is detectable following H2O2 challenge. The peroxide and GSH redox-dependent control of H. influenzae NCTC 8143 zwf gene expression reported in this paper is also observed in yeast and animal cells (34, 71). In these cases, the zwf gene product has to provide sufficient NADPH for GSH-dependent peroxidases to remain effective H2O2 scavengers. In fact, these Sec-containing peroxidases are believed to be the predominant (in comparison with catalases) scavengers at moderate H2O2 concentrations (32, 33). Analogously, H2O2 induction of G6PD activity in H. influenzae NCTC 8143 may result from higher NADPH demands owing to increased Prx/Grx peroxidase activity. To our surprise, we did not observe variation in Prx/Grx peroxidase activity following peroxidatic stress.
The Prx/Grx peroxidase of H. influenzae is the first genetically and biochemically established GSH-dependent peroxidase in a prokaryotic system.
A database search of the presently available bacterial genome sequences for homologs of the mammalian Sec-containing GSH peroxidases produces numerous (∼50) significant alignments displaying up to 50% amino acid sequence identity and containing a conserved cysteine residue instead of the Sec responsible for peroxide attack. These aligned sequences are in most cases annotated as GSH peroxidase, even though this enzymatic activity was proven only once in a bacterial cell, i.e., Enterococcus faecalis (52), which appears (the genome sequence project is not yet completed) to be absent from the list of positive hits. Furthermore, animal GSH peroxidase homologs are present in bacteria which are incapable of synthesizing GSH, such as Streptomyces coelicolor, Bacillus subtilis, Clostridium spp., and Staphylococcus aureus. At present, one genetic approach (Neisseria meningitides [45]) and one biochemical approach (Synechocystis sp. strain PCC 6803 [23]) have been reported to establish the role or activity of these animal GSH peroxidase homologs in prokaryotes, and although both studies indicate a function in defense processes against oxidative stress, no evidence for GSH-dependent peroxidase activity has been provided. Given that it is universally present in prokaryotes, the question arises (especially since we now have shown that GSH in prokaryotes can function as a cofactor for a peroxidase) why this ancestral gene acquired GSH-dependent peroxidase activity only when adopted in a eukaryotic cell.
Acknowledgments
This work was supported by the Fund for Scientific Research-Flanders (grant 3G003601) and the Bijzonder Onderzoeksfonds of Ghent University (grant 12050198).
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apontoweil, P., and W. Berends. 1975. Isolation and initial characterization of glutathione-deficient mutants of Escherichia coli K 12. Biochim. Biophys. Acta 399:10-22. [DOI] [PubMed] [Google Scholar]
- 3.Barcak, G. J., M. S. Chandler, R. J. Redfield, and J. F. Tomb. 1991. Genetic systems in Haemophilus influenzae. Methods Enzymol. 204:321-342. [DOI] [PubMed] [Google Scholar]
- 4.Bishai, W. R., N. S. Howard, J. A. Winkelstein, and H. O. Smith. 1994. Characterization and virulence analysis of catalase mutants of Haemophilus influenzae. Infect. Immun. 62:4855-4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bishai, W. R., H. O. Smith, and G. J. Barcak. 1994. A peroxide/ascorbate-inducible catalase from Haemophilus influenzae is homologous to the Escherichia coli katE gene product. J. Bacteriol. 176:2914-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Bryk, R., P. Griffin, and C. Nathan. 2000. Peroxynitrite reductase activity of bacterial peroxiredoxins. Nature 407:211-215. [DOI] [PubMed] [Google Scholar]
- 8.Carlsson, J., J. T. Larsen, and M. B. Edlund. 1993. Peptostreptococcus micros has a uniquely high capacity to form hydrogen sulfide from glutathione. Oral Microbiol. Immunol. 8:42-45. [DOI] [PubMed] [Google Scholar]
- 9.Chesney, J. A., J. W. Eaton, and J. R. Mahoney, Jr. 1996. Bacterial glutathione: a sacrificial defense against chlorine compounds. J. Bacteriol. 178:2131-2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Craig, J. E., A. Cliffe, K. Garnett, and N. J. High. 2001. Survival of nontypeable Haemophilus influenzae in macrophages. FEMS Microbiol. Lett. 203:55-61. [DOI] [PubMed] [Google Scholar]
- 11.De Groote, M. A., U. A. Ochsner, M. U. Shiloh, C. Nathan, J. M. McCord, M. C. Dinauer, S. J. Libby, A. Vazquez-Torres, Y. Xu, and F. C. Fang. 1997. Periplasmic superoxide dismutase protects Salmonella from products of phagocyte NADPH-oxidase and nitric oxide synthase. Proc. Natl. Acad. Sci. USA 94:13997-14001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeGroote, M. A., and F. C. Fang. 1999. Nitric oxide and infection. Kluwer/Plenum, New York, N.Y.
- 13.D'Mello, R. A., P. R. Langford, and J. S. Kroll. 1997. Role of bacterial Mn-cofactored superoxide dismutase in oxidative stress responses, nasopharyngeal colonization, and sustained bacteremia caused by Haemophilus influenzae type b. Infect. Immun. 65:2700-2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fahey, R. C., W. C. Brown, W. B. Adams, and M. B. Worsham. 1978. Occurrence of glutathione in bacteria. J. Bacteriol. 133:1126-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fahey, R. C., and A. R. Sundquist. 1991. Evolution of glutathione metabolism. Adv. Enzymol. Relat. Areas Mol. Biol. 64:1-53. [DOI] [PubMed] [Google Scholar]
- 16.Fawcett, W. P., and R. E. Wolf, Jr. 1995. Genetic definition of the Escherichia coli zwf “soxbox,” the DNA binding site for SoxS-mediated induction of glucose 6-phosphate dehydrogenase in response to superoxide. J. Bacteriol. 177:1742-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferguson, G. P., A. D. Chacko, C. H. Lee, I. R. Booth, and C. Lee. 1996. The activity of the high-affinity K+ uptake system Kdp sensitizes cells of Escherichia coli to methylglyoxal. J. Bacteriol. 178:3957-3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferguson, G. P., D. McLaggan, and I. R. Booth. 1995. Potassium channel activation by glutathione-S-conjugates in Escherichia coli: protection against methylglyoxal is mediated by cytoplasmic acidification. Mol. Microbiol. 17:1025-1033. [DOI] [PubMed] [Google Scholar]
- 19.Ferguson, G. P., A. W. Munro, R. M. Douglas, D. McLaggan, and I. R. Booth. 1993. Activation of potassium channels during metabolite detoxification in Escherichia coli. Mol. Microbiol. 9:1297-1303. [DOI] [PubMed] [Google Scholar]
- 20.Ferguson, G. P., S. Totemeyer, M. J. MacLean, and I. R. Booth. 1998. Methylglyoxal production in bacteria: suicide or survival? Arch. Microbiol. 170:209-218. [DOI] [PubMed] [Google Scholar]
- 21.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, et al. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 22.Foyer, C. H., F. L. Theodoulou, and S. Delrot. 2001. The functions of inter- and intracellular glutathione transport systems in plants. Trends Plant Sci. 6:486-492. [DOI] [PubMed] [Google Scholar]
- 23.Gaber, A., M. Tamoi, T. Takeda, Y. Nakano, and S. Shigeoka. 2001. NADPH-dependent glutathione peroxidase-like proteins (Gpx-1, Gpx-2) reduce unsaturated fatty acid hydroperoxides in Synechocystis PCC 6803. FEBS Lett. 499:32-36. [DOI] [PubMed] [Google Scholar]
- 24.Gallardo-Madueno, R., J. F. Leal, G. Dorado, A. Holmgren, J. Lopez-Barea, and C. Pueyo. 1998. In vivo transcription of nrdAB operon and of grxA and fpg genes is triggered in Escherichia coli lacking both thioredoxin and glutaredoxin 1 or thioredoxin and glutathione, respectively. J. Biol. Chem. 273:18382-18388. [DOI] [PubMed] [Google Scholar]
- 25.Grant, C. M., S. Luikenhuis, A. Beckhouse, M. Soderbergh, and I. W. Dawes. 2000. Differential regulation of glutaredoxin gene expression in response to stress conditions in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1490:33-42. [DOI] [PubMed] [Google Scholar]
- 26.Grant, C. M., F. H. MacIver, and I. W. Dawes. 1996. Glutathione is an essential metabolite required for resistance to oxidative stress in the yeast Saccharomyces cerevisiae. Curr. Genet. 29:511-515. [DOI] [PubMed] [Google Scholar]
- 27.Greenberg, J. T., and B. Demple. 1986. Glutathione in Escherichia coli is dispensable for resistance to H2O2 and gamma radiation. J. Bacteriol. 168:1026-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenwald, R. A. 1985. Therapeutic benefits of oxygen radical scavenger treatments remain unproven. J. Free Radic. Biol. Med. 1:173-177. [DOI] [PubMed] [Google Scholar]
- 29.Gutheil, W. G., E. Kasimoglu, and P. C. Nicholson. 1997. Induction of glutathione-dependent formaldehyde dehydrogenase activity in Escherichia coli and Haemophilus influenzae. Biochem. Biophys. Res. Commun. 238:693-696. [DOI] [PubMed] [Google Scholar]
- 30.Herriott, R. M., E. Y. Meyer, M. Vogt, and M. Modan. 1970. Defined medium for growth of Haemophilus influenzae. J. Bacteriol. 101:513-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iantomasi, T., F. Favilli, P. Marraccini, T. Magaldi, P. Bruni, and M. T. Vincenzini. 1997. Glutathione transport system in human small intestine epithelial cells. Biochim. Biophys. Acta 1330:274-283. [DOI] [PubMed] [Google Scholar]
- 32.Inoue, Y., T. Matsuda, K. Sugiyama, S. Izawa, and A. Kimura. 1999. Genetic analysis of glutathione peroxidase in oxidative stress response of Saccharomyces cerevisiae. J. Biol. Chem. 274:27002-27009. [DOI] [PubMed] [Google Scholar]
- 33.Izawa, S., Y. Inoue, and A. Kimura. 1996. Importance of catalase in the adaptive response to hydrogen peroxide: analysis of acatalasaemic Saccharomyces cerevisiae. Biochem. J. 320:61-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Izawa, S., Y. Inoue, and A. Kimura. 1995. Oxidative stress response in yeast: effect of glutathione on adaptation to hydrogen peroxide stress in Saccharomyces cerevisiae. FEBS Lett. 368:73-76. [DOI] [PubMed] [Google Scholar]
- 35.Izawa, S., K. Maeda, T. Miki, J. Mano, Y. Inoue, and A. Kimura. 1998. Importance of glucose-6-phosphate dehydrogenase in the adaptive response to hydrogen peroxide in Saccharomyces cerevisiae. Biochem. J. 330:811-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumaresan, K. R., S. S. Springhorn, and S. A. Lacks. 1995. Lethal and mutagenic actions of N-methyl-N′-nitro-N-nitrosoguanidine potentiated by oxidized glutathione, a seemingly harmless substance in the cellular environment. J. Bacteriol. 177:3641-3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liochev, S. I., and I. Fridovich. 1991. Effects of overproduction of superoxide dismutase on the toxicity of paraquat toward Escherichia coli. J. Biol. Chem. 266:8747-8750. [PubMed] [Google Scholar]
- 38.Liu, L., A. Hausladen, M. Zeng, L. Que, J. Heitman, and J. S. Stamler. 2001. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature 410:490-494. [DOI] [PubMed] [Google Scholar]
- 39.Loewen, P. C., and B. L. Triggs. 1984. Genetic mapping of katF, a locus that with katE affects the synthesis of a second catalase species in Escherichia coli. J. Bacteriol. 160:668-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lundberg, B. E., R. E. Wolf, Jr., M. C. Dinauer, Y. Xu, and F. C. Fang. 1999. Glucose 6-phosphate dehydrogenase is required for Salmonella typhimurium virulence and resistance to reactive oxygen and nitrogen intermediates. Infect. Immun. 67:436-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maciver, I., and E. J. Hansen. 1996. Lack of expression of the global regulator OxyR in Haemophilus influenzae has a profound effect on growth phenotype. Infect. Immun. 64:4618-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.MacLean, M. J., L. S. Ness, G. P. Ferguson, and I. R. Booth. 1998. The role of glyoxalase I in the detoxification of methylglyoxal and in the activation of the KefB K+ efflux system in Escherichia coli. Mol. Microbiol. 27:563-571. [DOI] [PubMed] [Google Scholar]
- 43.Meyer, D. J., H. Kramer, N. Ozer, B. Coles, and B. Ketterer. 1994. Kinetics and equilibria of S-nitrosothiol-thiol exchange between glutathione, cysteine, penicillamines and serum albumin. FEBS Lett. 345:177-180. [DOI] [PubMed] [Google Scholar]
- 44.Miyake, T., T. Hazu, S. Yoshida, M. Kanayama, K. Tomochika, S. Shinoda, and B. Ono. 1998. Glutathione transport systems of the budding yeast Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 62:1858-1864. [DOI] [PubMed] [Google Scholar]
- 45.Moore, T. D., and P. F. Sparling. 1996. Interruption of the gpxA gene increases the sensitivity of Neisseria meningitidis to paraquat. J. Bacteriol. 178:4301-4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murata-Kamiya, N., H. Kaji, and H. Kasai. 1999. Deficient nucleotide excision repair increases base-pair substitutions but decreases TGGC frameshifts induced by methylglyoxal in Escherichia coli. Mutat. Res. 442:19-28. [DOI] [PubMed] [Google Scholar]
- 47.National Committee for Clinical Laboratory Standards. 1997. Performance standards for antimicrobial disk susceptibility tests, 6th ed. Approved standard M2-A6. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 48.Ness, L. S., and I. R. Booth. 1999. Different foci for the regulation of the activity of the KefB and KefC glutathione-gated K+ efflux systems. J. Biol. Chem. 274:9524-9530. [DOI] [PubMed] [Google Scholar]
- 49.Oktyabrsky, O. N., G. V. Smirnovam, and N. G. Muzyka. 2001. Role of glutathione in regulation of hydroperoxidase I in growing Escherichia coli. Free Radic. Biol. Med. 31:250-255. [DOI] [PubMed] [Google Scholar]
- 50.Padgett, C. M., and A. R. Whorton. 1995. S-nitrosoglutathione reversibly inhibits GAPDH by S-nitrosylation. Am. J. Physiol. 269:C739-C749. [DOI] [PubMed] [Google Scholar]
- 51.Papoulis, A., Y. al-Abed, and R. Bucala. 1995. Identification of N2-(1-carboxyethyl)guanine (CEG) as a guanine advanced glycosylation end product. Biochemistry 34:648-655. [DOI] [PubMed] [Google Scholar]
- 52.Patel, M. P., J. Marcinkeviciene, and J. S. Blanchard. 1998. Enterococcus faecalis glutathione reductase: purification, characterization and expression under normal and hyperbaric O2 conditions. FEMS Microbiol. Lett. 166:155-163. [DOI] [PubMed] [Google Scholar]
- 53.Penninckx, M. 2000. A short review on the role of glutathione in the response of yeasts to nutritional, environmental, and oxidative stresses. Enzyme Microb. Technol. 26:737-742. [DOI] [PubMed] [Google Scholar]
- 54.Penninckx, M. J., and M. T. Elskens. 1993. Metabolism and functions of glutathione in micro-organisms. Adv. Microb. Physiol. 34:239-301. [DOI] [PubMed] [Google Scholar]
- 55.Prieto-Alamo, M. J., J. Jurado, R. Gallardo-Madueno, F. Monje-Casas, A. Holmgren, and C. Pueyo. 2000. Transcriptional regulation of glutaredoxin and thioredoxin pathways and related enzymes in response to oxidative stress. J. Biol. Chem. 275:13398-13405. [DOI] [PubMed] [Google Scholar]
- 56.Riccillo, P. M., C. I. Muglia, F. J. de Bruijn, A. J. Roe, I. R. Booth, and O. M. Aguilar. 2000. Glutathione is involved in environmental stress responses in Rhizobium tropici, including acid tolerance. J. Bacteriol. 182:1748-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sample, A. K., and C. J. Czuprynski. 1991. Elimination of hydrogen peroxide by Haemophilus somnus, a catalase-negative pathogen of cattle. Infect. Immun. 59:2239-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seaver, L. C., and J. A. Imlay. 2001. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J. Bacteriol. 183:7173-7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sherrill, C., and R. C. Fahey. 1998. Import and metabolism of glutathione by Streptococcus mutans. J. Bacteriol. 180:1454-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smirnova, G. V., N. G. Muzyka, M. N. Glukhovchenko, and O. N. Oktyabrsky. 2000. Effects of menadione and hydrogen peroxide on glutathione status in growing Escherichia coli. Free Radic. Biol. Med. 28:1009-1016. [DOI] [PubMed] [Google Scholar]
- 61.Stephen, D. W., and D. J. Jamieson. 1996. Glutathione is an important antioxidant molecule in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Lett. 141:207-212. [DOI] [PubMed] [Google Scholar]
- 62.Stevanin, T. M., N. Ioannidis, C. E. Mills, S. O. Kim, M. N. Hughes, and R. K. Poole. 2000. Flavohemoglobin Hmp affords inducible protection for Escherichia coli respiration, catalyzed by cytochromes bo′ or bd, from nitric oxide. J. Biol. Chem. 275:35868-35875. [DOI] [PubMed] [Google Scholar]
- 63.St. John, G., N. Brot, J. Ruan, H. Erdjument-Bromage, P. Tempst, H. Weissbach, and C. Nathan. 2001. Peptide methionine sulfoxide reductase from Escherichia coli and Mycobacterium tuberculosis protects bacteria against oxidative damage from reactive nitrogen intermediates. Proc. Natl. Acad. Sci. USA 98:9901-9906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Storz, G., and J. A. Imlay. 1999. Oxidative stress. Curr. Opin. Microbiol. 2:188-194. [DOI] [PubMed] [Google Scholar]
- 65.Storz, G., F. S. Jacobson, L. A. Tartaglia, R. W. Morgan, L. A. Silveira, and B. N. Ames. 1989. An alkyl hydroperoxide reductase induced by oxidative stress in Salmonella typhimurium and Escherichia coli: genetic characterization and cloning of ahp. J. Bacteriol. 171:2049-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sundquist, A. R., and R. C. Fahey. 1989. Evolution of antioxidant mechanisms: thiol-dependent peroxidases and thioltransferase among procaryotes. J. Mol. Evol. 29:429-435. [DOI] [PubMed] [Google Scholar]
- 67.Tao, K. 1997. OxyR-dependent induction of Escherichia coli grx gene expression by peroxide stress. J. Bacteriol. 179:5967-5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thomas, E. L. 1984. Disulfide reduction and sulfhydryl uptake by Streptococcus mutans. J. Bacteriol. 157:240-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tietze, F. 1969. Enzymic method for quantitative determination of nanogram amounts of total and oxidized glutathione: applications to mammalian blood and other tissues. Anal. Biochem. 27:502-522. [DOI] [PubMed] [Google Scholar]
- 70.Uotila, L. 1973. Purification and characterization of S-2-hydroxyacylglutathione hydrolase (glyoxalase II) from human liver. Biochemistry 12:3944-3951. [DOI] [PubMed] [Google Scholar]
- 71.Ursini, M. V., A. Parrella, G. Rosa, S. Salzano, and G. Martini. 1997. Enhanced expression of glucose-6-phosphate dehydrogenase in human cells sustaining oxidative stress. Biochem. J. 323:801-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vergauwen, B., F. Pauwels, F. Jacquemotte, T. E. Meyer, M. A. Cusanovich, R. G. Bartsch, and J. J. Van Beeumen. 2001. Characterization of glutathione amide reductase from Chromatium gracile. Identification of a novel thiol peroxidase (Prx/Grx) fueled by glutathione amide redox cycling. J. Biol. Chem. 276:20890-20897. [DOI] [PubMed] [Google Scholar]
- 73.Vlamis-Gardikas, A., F. Aslund, G. Spyrou, T. Bergman, and A. Holmgren. 1997. Cloning, overexpression, and characterization of glutaredoxin 2, an atypical glutaredoxin from Escherichia coli. J. Biol. Chem. 272:11236-11243. [DOI] [PubMed] [Google Scholar]
- 74.Zheng, M., F. Aslund, and G. Storz. 1998. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 279:1718-1721. [DOI] [PubMed] [Google Scholar]