Abstract
We identified a response regulator in Mycobacterium smegmatis which plays an important role in adaptation to oxygen-starved stationary phase. The regulator exhibits strong sequence similarity to DevR/Rv3133c of M. tuberculosis. The structural gene is present on a multigene locus, which also encodes a sensor kinase. A devR mutant of M. smegmatis was adept at surviving growth arrest initiated by either carbon or nitrogen starvation. However, its culturability decreased several orders of magnitude below that of the wild type under oxygen-starved stationary-phase conditions. Two-dimensional gel analysis revealed that a number of oxygen starvation-inducible proteins were not expressed in the devR mutant. Three of these proteins are universal stress proteins, one of which is encoded directly upstream of devR. Another protein closely resembles a proposed nitroreductase, while a fifth protein corresponds to the α-crystallin (HspX) orthologue of M. smegmatis. None of the three universal stress proteins or nitroreductase, and a considerably lower amount of HspX was detected in carbon-starved wild-type cultures. A fusion of the hspX promoter to gfp demonstrated that DevR directs gene expression when M. smegmatis enters stationary phase brought about, in particular, by oxygen starvation. To our knowledge, this is the first time a role for a two-component response regulator in the control of universal stress protein expression has been shown. Notably, the devR mutant was 104-fold more sensitive than wild type to heat stress. We conclude that DevR is a stationary-phase regulator required for adaptation to oxygen starvation and resistance to heat stress in M. smegmatis.
Mycobacterium tuberculosis is a major cause of infectious disease in humans causing over two million deaths annually (41). Despite its high level of pathogenicity, few of the common virulence mechanisms found in other human pathogens have been described in M. tuberculosis (6). An important feature of the pathogenesis of tuberculosis is the prevalence of latent infection without disease (54). Although it has been estimated that one-third of the world's population harbor viable M. tuberculosis bacilli (4), the majority of carriers exhibit no clinical symptoms (44). It is believed that the immune system confines the organism to a latent state in which the organism can persist indefinitely (27). During latency, M. tuberculosis is thought to reside in pulmonary granulomatous lesions and possibly other host sites where it retains the ability to resume growth at a future stage and instigate disease (41). It has yet to be established whether the pathogen is maintained in a state of dormancy or in one of continuous but suppressed cell growth (41).
There is some evidence which suggests that M. tuberculosis persists in a state of nongrowth in the human host (36, 37). Growth arrest of a bacterial population can be caused by several conditions, including nutrient and oxygen depletion, environmental stress, and accumulation of toxic by-products. In many species, including mycobacteria, this transition is accompanied by changes in the composition and structure of cellular macromolecules and alterations in gene expression and RNA stability (30, 50). These physiological changes often coincide with the increased ability of stationary-phase bacteria to withstand starvation and stress (35, 50). A number of in vitro models, in particular oxygen and carbon starvation, have been used to study the stasis response of mycobacteria (50, 55). The use of these systems has enabled the identification of genes whose expression is induced at entry into stationary phase and whose products are important to mycobacterial survival and virulence (3, 32, 56).
The aim of our work is to investigate the genetic and physiological basis of mycobacterial persistence. To this end our approach has been to identify and characterize a regulator of mycobacterial stationary-phase survival and determine the function of the genes under its control. Due to its shorter generation time and the fewer constraints on its handling and manipulation, M. smegmatis is increasingly demonstrating its usefulness as a model for the study of mycobacterial physiology and gene regulation (20, 46, 59). We therefore sought to locate a regulatory system that could be involved in the adaptation and survival of M. smegmatis in stationary phase.
Upon starvation, the gram-positive bacterium Bacillus subtilis initiates a series of morphological changes that result in the formation of a dormant spore which is crucial for its survival under adverse conditions (12). One of the regulators controlling this process is a small 74-amino-acid protein, GerE, which directs the expression of cot genes encoding proteins that form the spore coat late in development (29). There are many bacterial transcriptional regulators that bind DNA through a so-called GerE domain. As with GerE, a number of these regulators are activated at the onset of growth arrest. An example of this is GacA, a regulator in gram-negative bacteria that controls the expression of secondary metabolites and extracellular proteins involved in pathogenicity to plants and animals, ecological fitness, and tolerance to stress (13, 15, 21, 23, 24, 28, 43, 45, 47, 57). Thus, we reasoned that regulatory proteins containing a GerE domain in other microbes, including mycobacteria, could be involved in the control of genes that function during the stationary phase.
In the present study, we commenced with a bioinformatic approach to locate a GerE-domain containing regulator in the genome sequence of M. smegmatis. After mutagenesis of the response regulator identified, we examined whether it controls gene expression upon growth arrest in M. smegmatis. In addition, we investigated whether this system confers a survival advantage upon M. smegmatis when exposed to nutrient and oxygen depletion and to a number of stress conditions.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Escherichia coli XL1-Blue [recA1 lac endA1 gyrA96 thi hsdR17 supE44 relA1 (F′ proAB lacIq lacZΔM15 Tn10)] was used as a host strain for all cloning experiments. The M. smegmatis strain used was mc2155 (51). A green fluorescent protein (GFP)-based expression plasmid was constructed from the mycobacterial vector pTKmx (34). The coding sequence of the gfpmut2 gene (7) was amplified by PCR with the primers gfpF1 (5′-GGGGGGATCCATGAGTAAAGGAGAAGAACTTTTC-3′) and gfpR1 (5′-GGGGGCATGCTTATTATTTGTATAGTTCATCCATGCC-3′) and the pKEN2gfpmut2 plasmid as a template (7). The incorporated restriction sites, BamHI and SphI, are highlighted in boldface. The gfpmut2 fragment generated was cloned into the pGEM-Te vector (Promega) and sequenced to verify the fidelity of the amplification. The xylE gene was excised from pTKmx by using the BamHI and SphI restriction enzymes and replaced with the gfpmut2 fragment. This generated the plasmid pOT71 which allows for translational fusions with the gfpmut2 gene. pSMT100 is a sacB-containing suicide vector used for mutagenesis in mycobacteria (52).
Media and growth conditions.
E. coli and M. smegmatis were routinely grown at 37°C in Trypticase soy broth (TSB; Becton Dickinson) or on TSA (TSB plus 1.5% agar). The nutrient limitation media used, Hartmans-de Bont minimal broth (HdeB), was prepared as previously described (50). Phosphate-buffered saline containing 0.05% (vol/vol) Tween 80 (PBST) was used for the washing and serial dilution of bacteria. The antibiotic concentrations were used as follows: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; and hygromycin, 50 μg/ml.
Nutrient starvation experiments.
M. smegmatis starter cultures were grown overnight in 5 ml of HdeB supplemented with 0.08% (vol/vol) glycerol. For the carbon starvation experiments, the starter cultures were subinoculated into 250-ml flasks containing 50 ml of HdeB supplemented with 0.08% (vol/vol) glycerol to an initial optical density at 600 nm (OD600) of 0.05. For the nitrogen starvation experiments, the (NH2)2SO4 concentration in the HdeB was reduced 100-fold to 0.15 mM, whereas glycerol was present at 0.2% (vol/vol). For the oxygen starvation experiments, cultures were grown in sealed (Suba seals; Scientific Laboratory Supplies) 250-ml flasks containing 150 ml of HdeB supplemented with 0.2% (vol/vol) glycerol, with a resultant headspace ratio (HSR) of 1.67. Experiments in which the HSR was varied inferred that with an HSR of 1.67, the cultures entered stationary phase due to oxygen starvation. For example, with 50 ml of medium giving an HSR of 5, the culture entered stationary phase with an OD600 of ca. 1.5, whereas with a HSR of 1.67 stationary phase was entered at an OD600 of ca. 0.6. To initiate experiments, the starter cultures were subinoculated into the experimental flask to an OD600 of 0.05. The OD600 of the cultures was monitored and when the cultures reached stationary phase, serial dilutions and viable plating were performed. For the oxygen starvation experiments, samples were taken by inverting the flask, inserting a fine 25-gauge needle (0.5 by 16 mm; Becton Dickinson) through the Suba seal and extracting culture by using a 1-ml syringe (Becton Dickinson). The viable counts of the cultures were assessed for up to 55 days. All nutrient limitation experiments were performed in triplicate.
Mutagenesis of the devR gene of M. smegmatis.
A 597-bp region was deleted from the coding sequence of the M. smegmatis devR gene and replaced with a hygromycin resistance cassette by homologous recombination. An upstream fragment of 1,502-bp was amplified by PCR with the primers Msm8F1 (5′-GGGGGGATCCCGCACCGACCGGCTGCCGATGTCG-3′) and Msm8R1 (5′-GGGGACTAGTCAGAAAAACCCTGATCATCTCCCG-3′) and M. smegmatis genomic DNA as the template. The product generated was cloned into pGEM-Te, sequenced, and subcloned into the BamHI and SpeI restriction sites of pSMT100 (52) upstream of the hygromycin resistance cassette, generating plasmid pDUB1. A downstream fragment of 1,501 bp was similarly generated by PCR with the primers Msm8F2 (5′-GGGGAGATCTAAACTGGACCGGCGCAACTAGGGC-3′) and Msm8R2 (5′-GGGGTCTAGACACGGCCTGCAGTTGCGCCACGGG-3′). The product generated was cloned into pGEM-Te, sequenced, and subcloned into the BglII and XbaI restriction sites of pDUB1 downstream of the hygromycin resistance cassette. The resulting knockout vector, pDUB2, contained a hygromycin cassette flanked by a fragment from upstream and a fragment from downstream of the intended deletion. Then, 5 μg of pDUB2 was used in an electroporation of M. smegmatis mc2155, and transformants were selected on TSA containing 50 μg of hygromycin/ml. To select for the second recombination event, colonies were grown overnight in TSB containing hygromycin and 5% (wt/vol) sucrose and subsequently plated onto TSA containing hygromycin and 5% (wt/vol) sucrose. Genomic DNA was prepared from sucrose-resistant colonies and screened by PCR for the presence of the hygromycin-resistance cassette with primers hygF2 (5′-GGCGAGAGCACCAACCCCGTAACTG-3′) and hygR2 (5′-GTCGCCCCGGAAGGCGTTGAGATG-3′) and for the presence of the deletion with primers internal to the M. smegmatis devR gene, namely, tcrRF2 (5′-CGGCTGCCCGACGGCAACGGC-3′) and tcrRR2 (5′-CACGTAGTTCTTGACGGTCTT-3′). Colonies indicating the presence of the hygromycin resistance cassette and a deletion in the devR gene were analyzed further by Southern hybridization by using the QuikHyb kit (Stratagene). The genomic DNA was treated with the restriction enzyme BstXI, which cuts outside of the regions used to construct the pDUB2 vector, and separated by gel electrophoresis. A probe to the hygromycin gene was generated by PCR with the primers hygF2 and hygR2 with plasmid pSMT100 as a template and labeled with [α-32P]dCTP by random oligonucleotide priming. A second probe was generated by PCR with the primers Msm8F1 and Msm8R1 with M. smegmatis genomic DNA as a template and labeled. The results of this analysis were consistent with the allelic exchange of an internal devR fragment for the hygromycin cassette, generating a devR mutant, OTL1.
Stress challenge experiments.
M. smegmatis strains were grown to stationary phase under carbon or oxygen starvation conditions as described above. After 24 h in stationary phase, serial dilutions and viable plating were performed. For the heat stress assay, the cultures were shifted to a shaking incubator held at 55°C. Incubation of the cultures was continued for a further 7.5 h with samples taken for viable plating at 90-min intervals. For the UV light sensitivity assay, serial 10-fold dilutions of the strains were plated on TSA agar. The plates were allowed to dry and were then exposed to 0, 25, and 50 mJ of UV light by using a UV Stratalinker 1800 (Stratagene). All stress challenge experiments were performed in triplicate.
Two-dimensional gel analysis.
M. smegmatis strains were grown to nutrient-starved stationary phase in HdeB medium, and cells were harvested 48 h after entry into stationary phase. Bacterial pellets were resuspended in 5 ml of sterile distilled water (dH2O) containing a cocktail of protease inhibitors (Complete; Roche), and cells were disrupted by passage three times through a One-Shot model cell disrupter (Constant Systems) at 17.5 MPa. A 100-μl sample was stored at −80°C for determination of protein concentration by using the Coomassie brilliant blue (CBB) protein reagent (Pierce). The remainder of the cell lysates was immediately added to a final concentration of 9 M urea, 2% Triton X-100, and 70 mM dithiothreitol (DTT). The solution was incubated for at least 1 h with gentle rocking at room temperature to fully denature the proteins before centrifugation at 12,000 × g. The supernatant was concentrated fivefold by using Ultrafree-four filter units with a molecular weight cutoff of 5 kDa (Millipore). Two-dimensional gel electrophoresis was performed by using the Multiphor II system from Amersham Biosciences according to the manufacturer's instructions. Briefly, 18-cm dehydrated isoelectric focusing strips with an immobilized pH gradient between pH 4 and 7, were rehydrated overnight in rehydration solution (12 g of urea, 0.13 ml of Triton X-100, 37 mg of DTT, and 0.25 ml of 10 mg of orange G/ml in 25 ml of dH2O). Then, 200 μg of protein was loaded on rehydrated gel strips, and proteins were focused at 20°C in two stages (5 h at 300 V, 2 mA, and 5 W followed by 23 h at 3,500 V, 2 mA, and 5 W) to a total of 70 to 80 kV · h. Focused gel strips were equilibrated for 10 min in equilibration solution A (per 100 ml: 0.05 M Tris HCl [pH 6.8], 30 ml of glycerol, 36 g of urea, 1 g of sodium dodecyl sulfate [SDS], 125 mg of DTT) and for another 10 min in 10 ml of equilibration solution B (per 100 ml: 0.05 M Tris HCl [pH 6.8], 30 ml of glycerol, 36 g of urea, 1 g of SDS, 2.25 g of iodoacetamide). For the second dimension we used precast polyacrylamide gels with a linear polyacrylamide gradient from 8 to 18% with the appropriate precast buffer strips (Amersham). Gels were run at 600 V, 20 mAmp, and 30 W for 25 min at 16°C, after which the first dimension strip gels were removed. The gel was then run at 600 V, 50 mAmp, and 30 W for a further 75 min at 16°C. Proteins were immediately transferred to a polyvinylidene difluoride membrane by electroblotting. The membranes were washed carefully (20 min in water, 20 min in 50% [vol/vol] methanol, and another 20 min in dH2O), rinsed briefly in 100% methanol, and stained in CBB staining solution (0.1% [wt/vol] CBB, 1% [vol/vol] acetic acid, and 40% [vol/vol] methanol) for 60 s. Membranes were destained in 50% (vol/vol) methanol, washed for 5 min in dH2O three times, and dried in a laminar flow cabinet. Proteins were excised from the membrane and N-terminal sequencing was performed by ProSeq, Inc. N-terminal sequences were used to identify the respective proteins from the genome sequence of M. smegmatis.
Measurement of promoter expression.
The promoter of one of the genes upregulated under oxygen limitation, hspX, was amplified by PCR. A fragment containing the first 6 codons of the hspX gene together with 501 bases of sequence upstream of the predicted start codon was generated with the primers hspXF1 (5′-GGGGGAATTCAACCGTGCGGCACGGGGAGATCTG-3′) and hspXR1 (5′-GGGGGGATCCTTCAGGAAGTTTGGTCATCGGTCCTCCTCA-3′) and M. smegmatis genomic DNA as the template. The PCR product was cloned into pGEM-Te, sequenced to verify fidelity, and then subcloned into the EcoRI and BamHI restriction sites of the reporter plasmid pOT71, i.e., upstream of and in frame with the gfpmut2 gene. The resulting plasmid, pOT73, was electroporated into the M. smegmatis wild-type and devR mutant strains, which were then grown under carbon and oxygen starvation conditions. The OD600 of the cultures were monitored over a 72-h period and, at 2-h intervals, 2-ml portions of the cultures were harvested by centrifugation, washed once, and resuspended in PBST to a final OD600 of 1.0. The green fluorescence was measured with a spectrofluorimeter (Shimadzu model RF-5301PC) set at an excitation wavelength of 480 nm and an emission detection wavelength of 510 nm. A negative control consisted of wild-type M. smegmatis containing pOT71 (promoterless gfpmut2). The background fluorescence readings for the negative control were between 2.0 and 3.0, and the results shown (Fig. 4) are the remainder of the fluorescence after subtraction of the background.
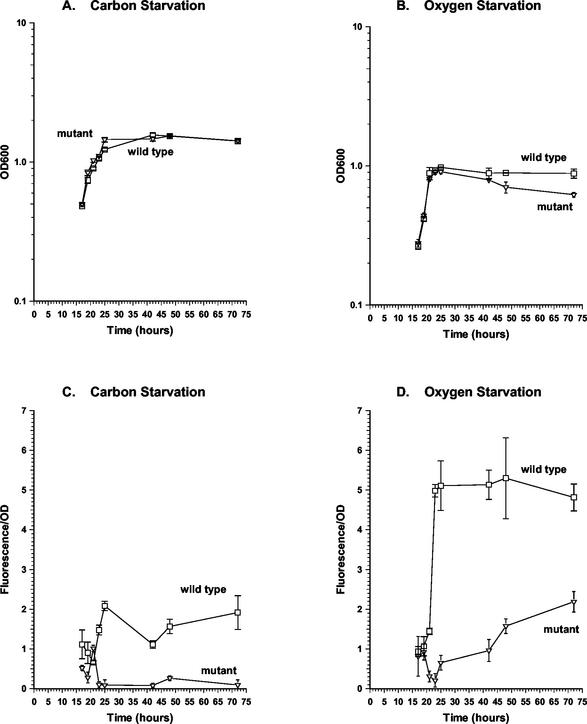
FIG. 4.
Effect of devR mutation on hspX expression. M. smegmatis wild type and devR mutant harboring a hspX-gfpmut2 fusion on the plasmid pOT73 were grown in HdeB medium to carbon-starved (A and C) or oxygen-starved (B and D) stationary phase. Growth rate (A and B) and hspX-gfpmut2 expression (C and D) were monitored. Symbols: □, wild-type strain mc2155; ▿, devR mutant OTL1. All experiments were performed by using three independent cultures and error bars represent standard deviation. Fluorescence is expressed as a function of OD. Under oxygen-starved conditions, the OD600 of the devR mutant culture began to decrease from 42 h onward. This feature was unique to this culture and was at least partially responsible for the observed increase in the relative fluorescence and OD values obtained for this strain at later time points.
RESULTS
Identification of a GerE-domain containing response regulator in the genome sequence of M. smegmatis.
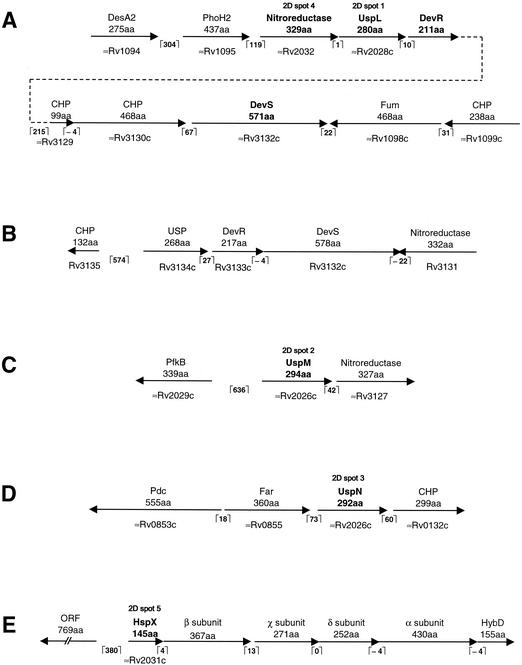
To identify a GerE-domain containing response regulator in M. smegmatis, the amino acid sequence of P. aeruginosa GacA was used in a BLAST search of the M. smegmatis genome at TIGR (http://www.tigr.org/tdb/mdb/mdbinprogress.html). One open reading frame (ORF) in particular encodes a potential 211-amino-acid protein which has the same domain organization as GacA in that it consists of an N-terminal CheY-domain (residues 2 to 123) and a C-terminal GerE DNA-binding domain (residues 147 to 211). Upon comparison with the M. tuberculosis H37Rv genome sequence (http://www.sanger.ac.uk/Projects/M_tuberculosis/blast_server.shtml), this potential protein exhibits strong similarity (85% identity over 209 residues) to the Rv3133c/DevR protein (9). The region surrounding this ORF was analyzed further and found to be flanked by several closely spaced ORFs (Fig. 1A). Three genes downstream of the response regulator gene an ORF, which potentially encodes a 571-amino-acid histidine kinase consisting of two N-terminal GAF-type domains (residues 64 to 201 and residues 232 to 363) and a C-terminal histidine kinase domain (residues 482 to 570), is located. The proximity of the response regulator and the downstream sensor kinase genes suggests that they may constitute a two-component system. Immediately upstream of the response regulator gene lies an ORF encoding a potential 280-amino-acid protein that contains a single N-terminal universal stress protein (USP) domain (residues 4 to 137). Upon comparing the layout of this locus with that of the devR locus in M. tuberculosis, it is apparent that a similar single USP domain-containing protein, Rv3134c (48), also lies directly upstream of the corresponding response regulator in M. tuberculosis (Fig. 1B). In addition, the histidine kinase encoded directly downstream of devR in M. tuberculosis, Rv3132c/DevS (9), also possesses two N-terminal GAF domains and exhibits 66% identity over 560 amino acids to the above-mentioned putative histidine kinase of M. smegmatis. Thus, it is apparent that the response regulator identified in M. smegmatis resembles DevR of M. tuberculosis both in terms of sequence conservation and gene linkage. Due to this similarity between the two systems, we will use the same nomenclature, i.e., DevS/DevR, in M. smegmatis. However, it is imperative to point out that it has still to be shown experimentally that DevS is the cognate sensor kinase of DevR in M. tuberculosis. Thus, we use the term DevS for the sense kinase encoded downstream of devR of M. smegmatis provisionally in this work. Directly downstream of the devR gene of M. smegmatis regulator lies an ORF, which encodes a 99-amino-acid protein which exhibits 63% identity to Rv3129 (110 residues) of M. tuberculosis (Fig. 1A). The stop codon of this ORF overlaps the start codon of another ORF which encodes a potential 468-amino-acid protein exhibiting 53% identity to Rv3130c of M. tuberculosis. Both Rv3129 and Rv3130c of M. tuberculosis encode conserved hypothetical proteins of an as-yet-unknown function. The devS coding region starts 68 bases downstream of the stop codon of the Rv3130c orthologue, whereas the gene located downstream of devS is transcribed in the opposite direction. This indicates that devS is the last gene in this gene cluster (Fig. 1A). Ending just two bases upstream of the start codon of the USP gene is an orthologue of the Rv2032 gene of M. tuberculosis. This gene has been proposed to encode a member of a classical family of nitroreductases (42).
FIG. 1.
(A) Schematic organization of the M. smegmatis multigene locus encoding the DevR response regulator. The positions of the genes encoding the proteins identified from the two-dimensional gel analysis, UspL (spot 1) and the putative nitroreductase (spot 4), are illustrated. The positions of the GAF domain-containing histidine kinase sensor gene, devS, and other neighboring genes are illustrated. (B) For comparison purposes, the devR locus in M. tuberculosis is also illustrated. (C) Schematic organization of the locus encoding the UspM protein (spot 2) identified from the two-dimensional analysis. (D) Schematic organization of the locus encoding the UspN protein (spot 3) identified from the two-dimensional analysis. (E) Schematic organization of the locus encoding the HspX protein (spot 5) identified from the two-dimensional analysis. The positions of the homologues of the α, β, χ, and δ subunits of the hydrogenase-sulfur reductase of C. tepidum and the hydrogenase maturation protease, HybD, of E. coli are shown. The identity of each protein spot was determined from its N-terminal sequence that was used in BLAST search analysis of the incomplete M. smegmatis genome sequence. The layout and gene spacing of each locus was established by analysis of the region for neighboring ORFs which encoded proteins showing significant similarity to known or hypothetical proteins. The annotation of the closest orthologue, where present, in M. tuberculosis is given. The spacing (number of nucleotides) between each coding sequence is shown in brackets. All figures are drawn to scale. CHP, conserved hypothetical protein.
Adaptation of the M. smegmatis devR mutant to growth-limiting conditions.
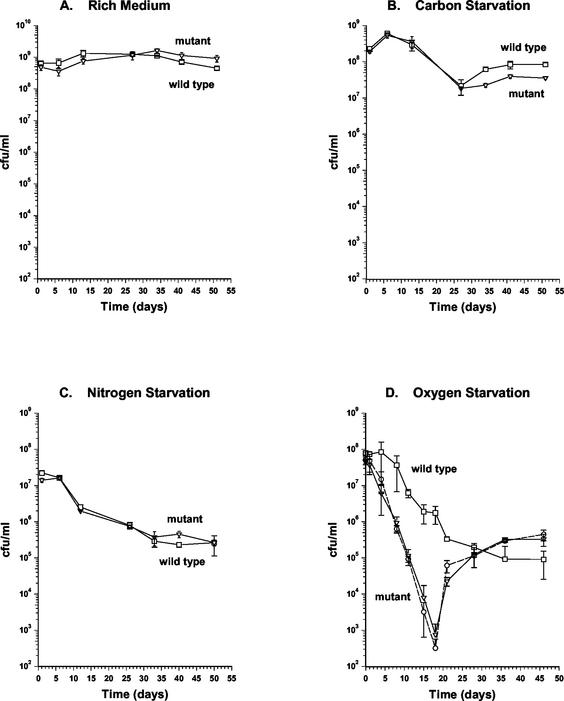
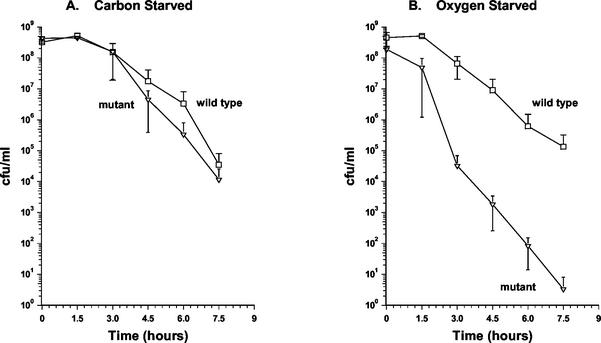
A 597-bp segment was deleted from the coding sequence of the devR gene. This corresponds to a removal of 199 of the potential 211 amino acid codons, whereas leaving behind the proximal 6 codons and the distal 7 codons, including the stop codon. The deleted region was replaced with a hygromycin resistance cassette by allelic exchange, which was confirmed by PCR and Southern hybridization analyses. The devR mutant was characterized for growth in both rich and minimal media and for survival in stationary phase in rich medium and under carbon-, nitrogen-, and oxygen-starved conditions. The exponential-phase growth rate and population density at entry into stationary phase in the mutant were found to be indistinguishable from that of the wild-type strain in all of the media tested (Fig. 2 and 4). The viable counts of the wild-type and mutant strains in stationary phase were then monitored for up to 55 days. In rich medium, the culturability of both strains remained constant (Fig. 2A). Under carbon and nitrogen starvation, the viable counts of both the wild-type and the mutant cultures diminished at similar rates (Fig. 2B and C). Under oxygen-starved conditions, the wild-type strain maintained its culturability for up to 4 days in stationary phase and then began to decrease gradually. In contrast, the viable counts of the mutant culture decreased at a considerably higher rate 24 h after entry into stationary phase (Fig. 2D). The mutant culture continued to lose culturability up to 18 days after entry into stationary phase at which point its viable counts were of the order of 104-fold lower than that of wild type (Fig. 2D). At later time points, the viable counts of the devR mutant began to increase and at 30 days after entry into stationary phase were restored to wild-type levels (Fig. 2D). This phenotype was highly reproducible. To examine whether the apparent recovery was due to isolation of a suppressor mutant that had regained culturability under oxygen starvation, cells were isolated from a devR mutant culture when its viable counts were returned to wild-type levels. When this isolate was grown to oxygen-starved stationary phase, its behavior was indistinguishable from that of the mutant in that it exhibited a similar drop in viable counts, followed by a return to wild-type levels (Fig. 2D). Therefore, the recovery in culturability of the devR mutant is not due to the accumulation of suppressor mutations.
FIG. 2.
Culturability of wild-type and devR mutant strains of M. smegmatis following entry into stationary phase. Strains were grown to stationary phase under the following conditions: rich medium (A), carbon starvation (B), nitrogen starvation (C), and oxygen starvation (D), after which their viability on plates was monitored for up to 55 days. Symbols: □, wild-type strain mc2155; ▿, devR mutant OTL1. Cells were isolated from the devR mutant at day 30 when culturability of this strain had recovered (panel D). This isolate was grown under oxygen starvation conditions, and its culturability on plates is represented by a dashed line and the symbol “○.” All experiments were performed by using three independent cultures, and error bars represent the standard deviation.
Protein expression in the devR mutant.
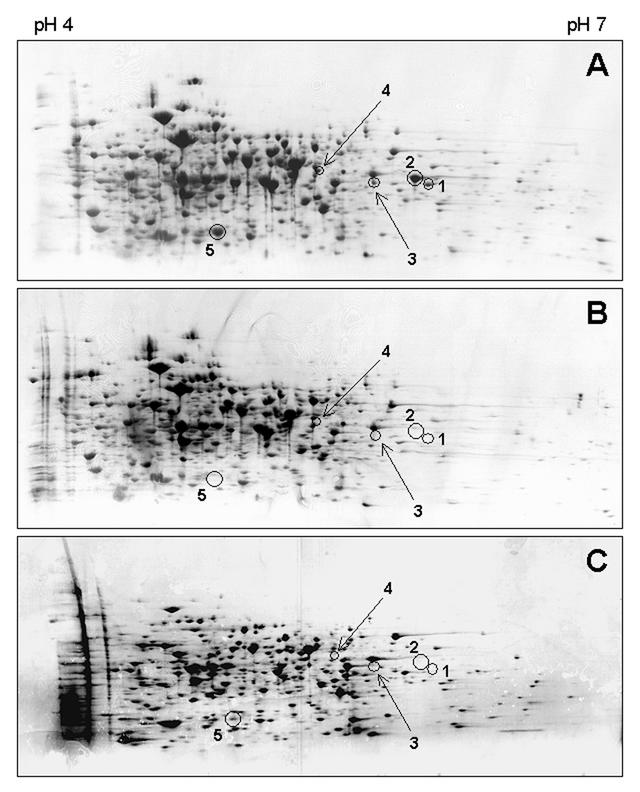
Given the devR mutant's deficiency at maintaining culturability under oxygen starvation conditions, we sought to determine whether the DevR response regulator controlled the expression of proteins under these conditions. Proteins were prepared from both the wild-type and mutant strains grown to oxygen-starved stationary phase and were analyzed by two-dimensional gel electrophoresis. We observed the absence of a number of protein spots in stationary-phase cultures of the mutant strain compared to wild type (Fig. 3A and B). Focusing on five of these protein spots, it was apparent that four were not detectable, whereas a fifth protein was present at considerably lower levels in stationary-phase cultures of the wild-type strain grown under carbon starvation conditions (Fig. 3C). This demonstrated that expression of the five proteins occurs in a DevR-dependent manner and is more pronounced under oxygen as opposed to carbon starvation. Subsequently, we determined the identity of these five DevR-dependent proteins by N-terminal sequencing of the protein spots and subsequent comparison of the sequences obtained with the M. smegmatis genome. The results of this analysis are shown in Table 1. Three of the proteins, spots 1, 2, and 3, are USPs which for ease of reference we have named UspL, UspM, and UspN, respectively. UspL, which contains a single USP domain, is encoded directly upstream of the devR gene (Fig. 1A). UspM consists of two USP domains and appears to be encoded at the start of an operon (Fig. 1C). Encoded 42 bases downstream is a protein that exhibits strong similarity to Rv3127 of M. tuberculosis, which is a proposed nitroreductase (42). The uspN gene appears to be the second gene in a three gene cluster (Fig. 1D). Located 60 bases downstream is an orthologue of Rv0132c of M. tuberculosis which encodes a putative oxidoreductase, while 73 bases upstream is an ORF with similarity to Rv0855, or far, which is predicted to encode a fatty acyl-coenzyme A racemase. The N-terminal sequence obtained from spot 4 corresponds to the protein encoded directly upstream of the uspL gene (Fig. 1A). This protein exhibits highest similarity to another proposed nitroreductase, Rv2032, of M. tuberculosis (42). Protein spot 5 corresponds to the HspX, or α-crystallin, orthologue of M. smegmatis. Like the hspX gene of M. tuberculosis, hspX of M. smegmatis is the first gene in a series of ORFs (Fig. 1E). However, the nature of its neighboring genes differs considerably. Downstream of the hspX gene of M. smegmatis are located four genes which encode proteins with similarity to the α, β, χ, and δ subunits of the hydrogenase/sulfur reductase of Chlorobium tepidum and a fifth gene which encodes an orthologue of the hydrogenase maturation protease, HybD, from E. coli (Fig. 1E). In contrast, the hspX gene of M. tuberculosis is present in a locus which includes two USPs, the carbohydrate kinase PfkB and a DevS-like sensor kinase, Rv2027c.
FIG. 3.
Two-dimensional gel analysis of total proteins isolated from stationary-phase M. smegmatis. (A) Wild-type strain mc2155 grown under oxygen-starvation conditions. (B) devR mutant OTL1 grown under oxygen starvation conditions. (C) Wild-type strain mc2155 grown under carbon starvation conditions. The positions of five DevR-dependent proteins are illustrated in each gel, and their identities were determined by N-terminal sequencing. Spot 1, UspL; spot 2, UspM, spot 3, UspN; spot 4, putative nitroreductase; spot 5, HspX. UspL, UspM, and UspN are members of the USP family. The nitroreductase shows strong similarity to Rv2032, a proposed nitroreductase in M. tuberculosis. HspX is the M. smegmatis homologue of the α-crystallin (HspX) protein of M. tuberculosis. Gels shown are representative of 3 independent experiments.
TABLE 1.
DevR-dependent oxygen starvation-inducible proteins of M. smegmatis identified by two-dimensional gel analysisa
| Two-dimensional gel spot no. | N-terminal sequence (aa range) | Protein description [aa range(s)] | Closest M. tuberculosis homologue (% identity) |
|---|---|---|---|
| 1 | SIARPVIVGI (2-11) | UspL; 280 aa; single USP domain (4-137) | Rv2028c (40% identity over 271 aa) |
| 2 | QSATEYGILV (3-12) | UspM; 294 aa; two USP domains (7-148; 157-292) | Rv2026c (60% identity over 293 aa) |
| 3 | STSTAPVVVGI (2-12) | UspN; 292 aa; two USP domains (4-145; 154-292) | Rv2026c (32% identity over 292 aa) |
| 4 | DTRLDVATLA (3-12) | Nitroreductase (329 aa) | Rv2032 (55% identity over 328 aa) |
| 5 | TKLPERSRAR (2-11) | HspX (145 aa) | Rv2031c (60% identity over 143 aa) |
N-terminal sequences were exact matches to the numbered amino acids from the N termini of the putative proteins identified from the M. smegmatis genome sequence. aa, amino acid.
Measurement of the dependence of hspX expression upon DevR.
The two-dimensional gel analysis showed that the expression of the UspL, UspM, UspN, HspX and nitroreductase proteins occurs in M. smegmatis grown under oxygen starvation conditions and that this induction is dependent upon the DevR regulator. To investigate the kinetics of this DevR-dependent expression further, the promoter region of one of the genes, hspX, was amplified by PCR and cloned in frame with the promoterless gfpmut2 gene in plasmid pOT71 generating a translational fusion. This construct, pOT73, was then introduced into both the wild-type and the mutant backgrounds; the resulting strains were grown under carbon and oxygen starvation conditions; and their growth and GFP fluorescence were monitored (Fig. 4). Under carbon starvation there was no significant hspX expression in logarithmic-phase cultures of either the wild-type or the mutant strains (Fig. 4C). At the point of entry into stationary phase (at 24 h of incubation) hspX expression became detectable in wild-type cultures, whereas in the mutant its expression remained low (Fig. 4C). Under oxygen limitation, expression of hspX was low in logarithmic-phase cultures of the wild-type and mutant strains (Fig. 4D). However, at the point of entry of the culture into stationary phase (at 24 h of incubation) there was a sudden induction of hspX expression in the wild-type cells (Fig. 4D). The magnitude of this hspX expression was considerably higher in oxygen-starved cultures compared to carbon-starved cultures. In contrast, low levels of hspX expression were detected in stationary-phase cells of the mutant strain (Fig. 4D). This confirms the dependence of expression of one of the proteins identified on the two-dimensional gels, namely, HspX, upon DevR and indicates that DevR activates gene expression after growth arrest due to oxygen starvation in particular.
Sensitivity of the devR mutant to stress challenge.
The dependence of USP proteins upon DevR suggested that this regulator might be required for the optimal survival of M. smegmatis under stress conditions in addition to oxygen starvation. To investigate this possibility, the wild type and devR mutant were exposed to a stress condition under which UspA of E. coli has been shown to be required for survival, i.e., UV light exposure (11, 22). The strains were also tested for their ability to resist heat stress, a condition which induces the expression of UspA in E. coli (39). Exposure of carbon-starved stationary-phase cultures to both UV light and heat shock revealed no difference in survival between the wild type and the devR mutant (Table 2 and Fig. 5A). We also tested the stress survival of the two strains when grown under oxygen starvation stationary-phase conditions. We observed no significant difference between the two strains with respect to resistance to UV light stress (Table 2). On the other hand, after exposure to heat stress, the viability of the mutant decreased more rapidly than that of wild type (Fig. 5B). At 3 h after the temperature shift to 55°C, there was 103-fold difference in survival between the two strains that increased to a 105-fold difference at 7.5 h (Fig. 5B). Control cultures kept at 37°C maintained their viability for the entire duration of the experiment (data not shown). This indicates that under oxygen limitation M. smegmatis DevR directs protein expression which is required for the resistance of stationary-phase cells to heat stress.
TABLE 2.
Survival of stationary-phase M. smegmatis after UV exposurea
| UV dose (mJ of UV exposure) | % Carbon starvation
|
% Oxygen starvation
|
||
|---|---|---|---|---|
| Wild type | devR | Wild type | devR | |
| 25 | 5.4 ± 2.2 | 2.9 ± 2.0 | 21.9 ± 6.9 | 28.6 ± 4.7 |
| 50 | <0.1 | <0.1 | <0.1 | <0.1 |
The strains were grown under either carbon- or oxygen-starvation conditions and at 24 hours after entry into stationary phase were spread on agar medium and exposed to UV light. Survival is expressed as a percentage of the viable counts obtained for untreated cells. Three independent experiments were perfromed, and the errors represent the standard error.
FIG. 5.
Heat stress resistance of the wild-type and devR mutant strains of M. smegmatis. The strains were grown under either carbon (A) or oxygen (B) starvation conditions and at 24 h after entry into stationary phase were shifted to 55°C at time zero. Samples were taken at 90-min intervals up to 7.5 h and plated on rich media. Symbols: □, wild-type strain mc2155; ▿, devR mutant OTL1. All experiments were performed by using three independent cultures, and the error bars represent the standard deviation.
DISCUSSION
We have identified a response regulator in M. smegmatis, DevR, with an important role in adaptation to growth arrest and survival under adverse environmental conditions. Encoded downstream of the response regulator is a histidine kinase sensor, DevS, whereas upstream a convergently transcribed gene, uspL, that encodes a protein with the signature domain of USPs is located (Fig. 1A). A very similar locus exists in M. tuberculosis in the form of the ORFs Rv3134c, Rv3133c, and Rv3132c (Fig. 1B), which are predicted to encode a USP, response regulator, and a histidine sensor kinase, respectively (9, 48). Mutation of M. smegmatis devR had no effect on culturability in stationary phase under carbon or nitrogen starvation or in rich medium but resulted in a clear effect under oxygen-starved conditions (Fig. 2). The mutant showed a severe loss of culturability after entry into stationary phase, which at its maximum was 104-fold less than that of the wild-type culture. This was followed by recovery of the viable counts to wild-type levels which was not due to the accumulation of suppressor mutations (Fig. 2D). This survival phenotype is similar to that of a M. smegmatis purF mutant, which we have previously shown to be defective for survival under oxygen- but not carbon-starved stationary phase (33). There are two possible explanations for the recovery in viable counts of the devR mutant. First, that loss of culturability results from a significant fraction of the population becoming dormant or active but nonculturable (2, 33) and that this is followed by resuscitation of the nonculturable cells. Recently, it has been shown that under certain conditions M. tuberculosis can enter and be resuscitated from a dormant state (49). A second possibility is that upon oxygen starvation, death of a large portion of the bacterial population occurs, followed by regrowth of surviving cells that are able to use the nutrients released by dead cells. This latter scenario would require that M. smegmatis can utilize an as-yet-unknown alternative terminal electron acceptor or fermentation mechanism. We are currently investigating the basis of the loss and subsequent recovery in the culturability of the oxygen-starved mycobacterial culture.
The devR locus was first described in M. tuberculosis by Dasgupta et al. (9) after a comparison of a virulent and an avirulent strain of M. tuberculosis for differentially expressed genes. Sherman et al. (48) found from microarray analysis that the devR gene is upregulated in response to hypoxia. They constructed a mutant in which expression of the Rv3133c/devR orthologue of M. bovis BCG was abolished due to a polar mutation in the upstream BCG Rv3134c gene. Importantly, they established that expression of the hypoxia-induced α-crystallin gene, hspX, was dependent upon DevR (48). They also found that 3 weeks into stationary phase a fivefold difference in CFU was observed between the polar Rv3134c mutant and wild type (48). These experiments were performed on roller cultures grown in a rich complex medium, and it is difficult to determine whether the bacteria entered stationary phase due to oxygen starvation or nutrient depletion. In our experiments on M. smegmatis, under defined carbon-starvation conditions, we observed an approximate fourfold difference in CFU per milliliter between the devR mutant and wild type after 52 days in stationary phase (Fig. 2B). Against the background of a 104-fold difference between wild type and devR mutant under oxygen starvation conditions, we do not regard the fourfold difference as being particularly significant. Notably, the rate of decrease in CFU per milliliter was the same for both strains under carbon starvation (Fig. 2B) in sharp contrast to oxygen starvation (Fig. 2D). Our data are the first direct demonstration of the role of a response regulator in preserving mycobacterial culturability under oxygen-starved conditions. Additional work would help establish the importance of DevR to the culturability of M. tuberculosis or M. bovis BCG under oxygen starvation conditions.
The phenotype of the M. smegmatis devR mutant under oxygen starvation conditions is significant given the proposed importance of oxygen deprivation during the transition of M. tuberculosis from active growth to the so-called nonreplicative persistent state found in latent tuberculosis (55, 56). This DevR-mediated response is likely to be assisted by its cognate histidine kinase sensor. The putative sensory region of the histidine kinase encoded downstream of the devR gene, DevS, consists of two N-terminal GAF domains. This type of domain is found in cGMP-specific phosphodiesterases, phytochromes, and the transcriptional activators NifA and FhlA (1, 18). The GAF domains of DevS lack the conserved cysteines of phytochromes and, thus, DevS is unlikely to be light stimulated. The cyclic nucleotides, cGMP and cAMP, which are known to affect a variety of fundamental physiological processes, have both been shown to be ligands of various GAF domains (26, 31) and, thus, either nucleotide could potentially represent a signal for DevS. Interestingly, the GAF domain is very similar in both structure and binding site location to the PAS domain (1) of sensors such as Aer, ArcB, FixL, and NifL which respond to changes in oxygen concentration (53, 60). In the case of ArcB, the PAS domain has been shown to mediate this response by sensing the changes in the electron transport system, in particular, changes in the redox state of quinones (19). Should DevS prove to be the cognate sensor kinase of DevR, it is likely that this signal transduction system responds to alterations in available oxygen by an indirect process involving its GAF domains. However, other possibilities exist. In the work by Sherman et al. (48), they showed that unlike in a DevR-deficient mutant, mutation of devS had no effect on the induction of the hspX gene under defined hypoxic conditions. They, as have other researchers (9), noted the presence of a second sensor encoded by Rv2027c which exhibits strong homology to DevS (>60% identity over its entire length) and raised the prospect that the presence of Rv2027c could be sufficient to activate DevR in a devS mutant. This is strengthened by our observations that Rv2027c, like DevS, possesses tandem N-terminal GAF domains. They also made the valid suggestion that DevS may activate DevR in response to signals other than hypoxia (48) from which one could postulate that a different type of sensor may be responsible for triggering DevR under hypoxic conditions.
We identified 5 proteins that are induced in response to oxygen starvation in a devR-dependent manner (Fig. 3). Three of these are USPs, which we have named UspL, UspM, and UspN. The UspA protein of E. coli is the primary USP to have its physiological role studied in detail (11, 38, 39, 40). It is a small cytoplasmic protein whose expression is enhanced several times when cell viability is challenged by a range of conditions that hinder growth such as nutrient starvation, heat shock, DNA damage, and oxidative and osmotic stress. UspA is an autophosphorylating serine/threonine phosphoprotein that becomes phosphorylated specifically in response to growth arrest (16, 17). uspA mutants are impaired in their ability to survive growth arrest, and both inactivation and overexpression of uspA result in global changes in the timing of gene expression. M. smegmatis UspL, which is encoded directly upstream of devR (Fig. 1), has a single USP domain, whereas both UspM and UspN contain two conserved USP domains. To our knowledge, this is the first time that the participation of a two-component response regulator in the expression of USPs has been described. Other researchers have also reported the upregulation of a USP (Rv2623) in M. bovis BCG and M. tuberculosis when grown under poorly aerated conditions (5, 14, 48) and it remains to be seen whether USPs are also subject to DevR-dependent regulation in these latter mycobacterial species. Given the recent demonstration that USPs of E. coli have a role in protection against DNA-damaging agents (11, 22), we looked at the sensitivity of the devR mutant to UV stress under stationary-phase conditions. We found no significant difference in the UV sensitivity of the devR mutant (Table 1), which indicates that neither DevR nor DevR-dependent USPs are required for providing resistance to UV-induced DNA damage in M. smegmatis. However, DevR clearly has an important role in protecting oxygen-starved cells against heat stress (Fig. 5), and we are currently examining whether this role extends to other stress protection mechanisms.
The fourth protein identified is encoded by the gene immediately upstream of uspL (Fig. 1A) and corresponds to an orthologue of the proposed nitroreductase encoded by Rv2032 in M. tuberculosis, which has been shown to be upregulated under oxygen starvation (42). These results demonstrate that DevR controls the expression of two genes, i.e., the putative nitroreductase gene and uspL, which are located directly upstream of its own structural gene. Given the close spacing of the nitroreductase, UspL, and DevR genes (Fig. 1) and the finding in M. tuberculosis that the uspL orthologue, Rv3134c, and devR are cotranscribed (9, 48), it is most likely that these genes form an operon and that devR is subject to autoregulation.
The N-terminal sequence of the fifth spot analyzed was derived from the alpha-crystallin or HspX orthologue of M. smegmatis. HspX is upregulated in M. tuberculosis in response to oxygen limitation (8, 10). A related member of the α-crystallin family in B. subtilis, CotM, is induced during spore development and has been shown to be essential for normal spore coat assembly (25). Thus, given the association of HspX with cell wall thickening (8) and its importance to the survival of the pathogen in macrophages (58), it is possible that HspX facilitates the rearrangement of the mycobacterial cell wall providing protection during adverse conditions. As noted above, HspX expression is dependent upon DevR in M. tuberculosis (48). That it should be regulated in a similar fashion in M. smegmatis indicates that aspects of the response to oxygen deprivation are common to both species. Our two-dimensional gel analysis and measurements of the activity of the hspX promoter indicate that some HspX expression also occurs in a DevR-dependent manner under carbon starvation conditions (Fig. 3C and 4C). Thus, a role for DevR under other conditions, in addition to oxygen starvation, cannot be ruled out at this stage.
In summary, we have determined that the response regulator DevR plays an important role in stationary-phase gene expression, adaptation to oxygen stavation-induced growth arrest, and resistance to environmental stress in M. smegmatis. We are currently investigating the importance and function of USPs in mycobacterial persistence and virulence.
Acknowledgments
We are very grateful to Douglas Young and Graham Stewart for advice on mutant construction and for providing plasmid pSMT100, to Gordon Churchward for the provision of plasmid pTKmx, and to Brendan Cormack for providing plasmid pKEN2gfpmut2.
This work was funded by the British Lung Foundation and the Biotechnology and Biological Sciences Research Council (United Kingdom).
REFERENCES
- 1.Aravind, L., and C. P. Ponting. 1997. The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem. Sci. 22:458-459. [DOI] [PubMed] [Google Scholar]
- 2.Barer, M. R., and C. R. Harwood. 1999. Bacterial viability and culturability. Adv. Microb. Physiol. 41:93-137. [DOI] [PubMed] [Google Scholar]
- 3.Betts, J. C., P. T. Lukey, L. C. Robb, R. A. McAdam, and K. Duncan. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717-731. [DOI] [PubMed] [Google Scholar]
- 4.Bloom, B. R., and C. J. Murray. 1992. Tuberculosis: commentary on a reemergent killer. Science 257:1055-1064. [DOI] [PubMed] [Google Scholar]
- 5.Boon, C., R. Li, R. Qi, and T. Dick. 2001. Proteins of Mycobacterium bovis BCG induced in the Wayne dormancy model. J. Bacteriol. 183:2672-2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et. al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 7.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 8.Cunningham, A. F., and C. L. Spreadbury. 1998. Mycobacterial stationary phase induced by low oxygen tension: cell wall thickening and localization of the 16-kilodalton alpha-crystallin homolog. J. Bacteriol. 180:801-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dasgupta, N., V. Kapur, K. K. Singh, T. K. Das, S. Sachdeva, K. Jyothisri, and J. S. Tyagi. 2000. Characterization of a two-component system, devR-devS, of Mycobacterium tuberculosis. Tuberc. Lung Dis. 80:141-159. [DOI] [PubMed] [Google Scholar]
- 10.Desjardin, L. E., L. G. Hayes, C. D. Sohaskey, L. G. Wayne, and K. D. Eisenach. 2001. Microaerophilic induction of the alpha-crystallin chaperone protein homologue (hspX) mRNA of Mycobacterium tuberculosis. J. Bacteriol. 183:5311-5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diez, A., N. Gustavsson, and T. Nystrom. 2000. The universal stress protein A of Escherichia coli is required for resistance to DNA damaging agents and is regulated by a RecA/FtsK-dependent regulatory pathway. Mol. Microbiol. 36:1494-1503. [DOI] [PubMed] [Google Scholar]
- 12.Driks, A. 1999. Bacillus subtilis spore coat. Microbiol. Mol. Biol. Rev. 63:1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eriksson, A. R., R. A. Andersson, M. Pirhonen, and E. T. Palva. 1998. Two-component regulators involved in the global control of virulence in Erwinia carotovora subsp. carotovora. Mol. Plant-Microbe Interact. 11:743-752. [DOI] [PubMed] [Google Scholar]
- 14.Florczyk, M. A., L. A. McCue, R. F. Stack, C. R. Hauer, and K. A. McDonough. 2001. Identification and characterization of mycobacterial proteins differentially expressed under standing and shaking culture conditions, including Rv2623 from a novel class of putative ATP-binding proteins. Infect. Immun. 69:5777-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frederick, R. D., J. Chiu, J. L. Bennetzen, and A. K. Handa. 1997. Identification of a pathogenicity locus, rpfA, in Erwinia carotovora subsp. carotovora subsp. carotovora that encodes a two-component sensor-regulator protein. Mol. Plant-Microbe Interact. 10:407-415. [DOI] [PubMed] [Google Scholar]
- 16.Freestone, P., T. Nystrom, M. Trinei, and V. Norris. 1997. The universal stress protein, UspA, of Escherichia coli is phosphorylated in response to stasis. J. Mol. Biol. 274:318-324. [DOI] [PubMed] [Google Scholar]
- 17.Freestone, P., M. Trinei, S. C. Clarke, T. Nystrom, and V. Norris. 1998. Tyrosine phosphorylation in Escherichia coli. J. Mol. Biol. 279:1045-1051. [DOI] [PubMed] [Google Scholar]
- 18.Galperin, M. Y., A. N. Nikolskaya, and E. V. Koonin. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203:11-21. [DOI] [PubMed] [Google Scholar]
- 19.Georgellis, D., O. Kwon, and E. C. Lin. 2001. Quinones as the redox signal for the arc two-component system of bacteria. Science 292:2314-2316. [DOI] [PubMed] [Google Scholar]
- 20.Gomez, M., L. Doukhan, G. Nair, and I. Smith. 1998. sigA is an essential gene in Mycobacterium smegmatis. Mol. Microbiol. 29:617-628. [DOI] [PubMed] [Google Scholar]
- 21.Goodier, R. I., and B. M. Ahmer. 2001. SirA orthologs affect both motility and virulence. J. Bacteriol. 183:2249-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gustavsson, N., A. Diez, and T. Nystrom. 2002. The universal stress protein paralogues of Escherichia coli are co-ordinately regulated and co-operate in the defence against DNA damage. Mol. Microbiol. 43:107-117. [DOI] [PubMed] [Google Scholar]
- 23.Hammer, B. K., E. S. Tateda, and M. S. Swanson. 2002. A two-component regulator induces the transmission phenotype of stationary-phase Legionella pneumophila. Mol. Microbiol. 44:107-118. [DOI] [PubMed] [Google Scholar]
- 24.Heeb, S., and D. Haas. 2001. Regulatory roles of the GacS/GacA two-component system in plant-associated and other gram-negative bacteria. Mol. Plant-Microbe Interact. 14:1351-1363. [DOI] [PubMed] [Google Scholar]
- 25.Henriques, A. O., B. W. Beall, and C. P. Moran, Jr. 1997. CotM of Bacillus subtilis, a member of the alpha-crystallin family of stress proteins, is induced during development and participates in spore outer coat formation. J. Bacteriol. 179:1887-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho, Y. S., L. M. Burden, and J. H. Hurley. 2000. Structure of the GAF domain, a ubiquitous signaling motif and a new class of cyclic GMP receptor. EMBO J. 19:5288-5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Honer zu Bentrup, K., and D. G. Russell. 2001. Mycobacterial persistence: adaptation to a changing environment. Trends Microbiol. 9:597-605. [DOI] [PubMed] [Google Scholar]
- 28.Hrabak, E. M., and D. K. Willis. 1992. The lemA gene required for pathogenicity of Pseudomonas syringae pv. syringae on bean is a member of a family of two-component regulators. J. Bacteriol. 174:3011-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ichikawa, H., and L. Kroos. 2000. Combined action of two transcription factors regulates genes encoding spore coat proteins of Bacillus subtilis. J. Biol. Chem. 275:13849-13855. [DOI] [PubMed] [Google Scholar]
- 30.Ishihama, A. 1997. Adaptation of gene expression in stationary phase bacteria. Curr. Opin. Genet. Dev. 7:582-588. [DOI] [PubMed] [Google Scholar]
- 31.Kanacher, T., A. Schultz, J. U. Linder, and J. E. Schultz. 2002. A GAF-domain-regulated adenylyl cyclase from Anabaena is a self-activating cAMP switch. EMBO J. 21:3672-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keer, J., M. J. Smeulders, K. M. Gray, and H. D. Williams. 2000. Mutants of Mycobacterium smegmatis impaired in stationary-phase survival. Microbiology 146(Pt. 9):2209-2217. [DOI] [PubMed] [Google Scholar]
- 33.Keer, J., M. J. Smeulders, and H. D. Williams. 2001. A purF mutant of Mycobacterium smegmatis has impaired survival during oxygen-starved stationary phase. Microbiology 147:473-481. [DOI] [PubMed] [Google Scholar]
- 34.Kenney, T. J., and G. Churchward. 1996. Genetic analysis of the Mycobacterium smegmatis rpsL promoter. J. Bacteriol. 178:3564-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kolter, R., D. A. Siegele, and A. Tormo. 1993. The stationary phase of the bacterial life cycle. Annu. Rev. Microbiol. 47:855-874. [DOI] [PubMed] [Google Scholar]
- 36.Nyka, W. 1967. Method for staining both acid-fast and chromophobic tubercle bacilli with carbolfuschsin. J. Bacteriol. 93:1458-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nyka, W. 1974. Studies on the effect of starvation on mycobacteria. Infect. Immun. 9:843-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nystrom, T., and F. C. Neidhardt. 1993. Isolation and properties of a mutant of Escherichia coli with an insertional inactivation of the uspA gene, which encodes a universal stress protein. J. Bacteriol. 175:3949-3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nystrom, T., and F. C. Neidhardt. 1994. Expression and role of the universal stress protein, UspA, of Escherichia coli during growth arrest. Mol. Microbiol. 11:537-544. [DOI] [PubMed] [Google Scholar]
- 40.Nystrom, T., and F. C. Neidhardt. 1996. Effects of overproducing the universal stress protein, UspA, in Escherichia coli K-12. J. Bacteriol. 178:927-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parrish, N. M., J. D. Dick, and W. R. Bishai. 1998. Mechanisms of latency in Mycobacterium tuberculosis. Trends Microbiol. 6:107-112. [DOI] [PubMed] [Google Scholar]
- 42.Purkayastha, A., L. A. McCue, and K. A. McDonough. 2002. Identification of a Mycobacterium tuberculosis putative classical nitroreductase gene whose expression is coregulated with that of the acr aene within macrophages, in standing versus shaking cultures, and under low oxygen conditions. Infect. Immun. 70:1518-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rahme, L. G., E. J. Stevens, S. F. Wolfort, J. Shao, R. G. Tompkins, and F. M. Ausubel. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899-1902. [DOI] [PubMed] [Google Scholar]
- 44.Raviglione, M. C., D. E. Snider, Jr., and A. Kochi. 1995. Global epidemiology of tuberculosis: morbidity and mortality of a worldwide epidemic. JAMA 273:220-226. [PubMed] [Google Scholar]
- 45.Reimmann, C., M. Beyeler, A. Latifi, H. Winteler, M. Foglino, A. Lazdunski, and D. Haas. 1997. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol. Microbiol. 24:309-319. [DOI] [PubMed] [Google Scholar]
- 46.Reyrat, J. M., and D. Kahn. 2001. Mycobacterium smegmatis: an absurd model for tuberculosis? Trends Microbiol. 9:472-474. [DOI] [PubMed] [Google Scholar]
- 47.Sacherer, P., G. Defago, and D. Haas. 1994. Extracellular protease and phospholipase C are controlled by the global regulatory gene gacA in the biocontrol strain Pseudomonas fluorescens CHA0. FEMS Microbiol. Lett. 116:155-160. [DOI] [PubMed] [Google Scholar]
- 48.Sherman, D. R., M. Voskuil, D. Schnappinger, R. Liao, M. I. Harrell, and G. K. Schoolnik. 2001. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha-crystallin. Proc. Natl. Acad. Sci. USA 98:7534-7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shleeva, M. O., K. Bagramyan, M. V. Telkov, G. V. Mukamolova, M. Young, D. B. Kell, and A. S. Kaprelyants. 2002. Formation and resuscitation of ′non-culturable' cells of Rhodococcus rhodochrous and Mycobacterium tuberculosis in prolonged stationary phase. Microbiology 148:1581-1591. [DOI] [PubMed] [Google Scholar]
- 50.Smeulders, M. J., J. Keer, R. A. Speight, and H. D. Williams. 1999. Adaptation of Mycobacterium smegmatis to stationary phase. J. Bacteriol. 181:270-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1919. [DOI] [PubMed] [Google Scholar]
- 52.Stewart, G. R., V. A. Snewin, G. Walzl, T. Hussell, P. Tormay, P. O'Gaora, M. Goyal, J. Betts, I. N. Brown, and D. B. Young. 2001. Overexpression of heat-shock proteins reduces survival of Mycobacterium tuberculosis in the chronic phase of infection. Nat. Med. 7:732-737. [DOI] [PubMed] [Google Scholar]
- 53.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wayne, L. G. 1994. Dormancy of Mycobacterium tuberculosis and latency of disease. Eur. J. Clin. Microbiol. Infect. Dis. 13:908-914. [DOI] [PubMed] [Google Scholar]
- 55.Wayne, L. G., and L. G. Hayes. 1996. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 64:2062-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wayne, L. G., and C. D. Sohaskey. 2001. Nonreplicating persistence of Mycobacterium tuberculosis. Annu. Rev. Microbiol. 55:139-163. [DOI] [PubMed] [Google Scholar]
- 57.Wong, S. M., P. A. Carroll, L. G. Rahme, F. M. Ausubel, and S. B. Calderwood. 1998. Modulation of expression of the ToxR regulon in Vibrio cholerae by a member of the two-component family of response regulators. Infect. Immun. 66:5854-5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yuan, Y., D. D. Crane, R. M. Simpson, Y. Q. Zhu, M. J. Hickey, D. R. Sherman, and C. E. Barry III. 1998. The 16-kDa alpha-crystallin (Acr) protein of Mycobacterium tuberculosis is required for growth in macrophages. Proc. Natl. Acad. Sci. USA 95:9578-9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zahrt, T. C., J. Song, J. Siple, and V. Deretic. 2001. Mycobacterial FurA is a negative regulator of catalase-peroxidase gene katG. Mol. Microbiol. 39:1174-1185. [DOI] [PubMed] [Google Scholar]
- 60.Zhulin, I. B., B. L. Taylor, and R. Dixon. 1997. PAS domain S-boxes in Archaea, Bacteria and sensors for oxygen and redox. Trends Biochem. Sci. 22:331-333. [DOI] [PubMed] [Google Scholar]