SUMMARY
The vertebrate retina contains seven major neuronal and glial cell types in an interconnected network that collects, processes and sends visual signals through the optic nerve to the brain. Retinal neuron differentiation is thought to require both intrinsic and extrinsic factors, yet few intrinsic gene products have been identified that direct this process. Math5 (Atoh7) encodes a basic helix-loop-helix (bHLH) transcription factor that is specifically expressed by mouse retinal progenitors. Math5 is highly homologous to atonal, which is critically required for R8 neuron formation during Drosophila eye development. Like R8 cells in the fly eye, retinal ganglion cells (RGCs) are the first neurons in the vertebrate eye. Here we show that Math5 mutant mice are fully viable, yet lack RGCs and optic nerves. Thus, two evolutionarily diverse eye types require atonal gene family function for the earliest stages of retinal neuron formation. At the same time, the abundance of cone photoreceptors is significantly increased in Math5−/− retinae, suggesting a binary change in cell fate from RGCs to cones. A small number of nascent RGCs are detected during embryogenesis, but these fail to develop further, suggesting that committed RGCs may also require Math5 function.
Keywords: Math5, Atoh7, Mouse, Retina, Optic nerves
INTRODUCTION
The vertebrate retina arises from an evagination of the anterior neural tube that forms a bilayered optic cup, comprising an inner neural retina and an outer pigmented epithelium. Mammalian retinal neurons and glia differentiate over an extended period, which in mice is from embryonic day 12 (E12) through postnatal day 21 (P21). In the initial stages, waves of dividing progenitor cells cease mitosis and migrate to the vitreal side of the optic cup, closest to the lens. This process of histogenesis continues through the ensuing 4 weeks and produces all retinal cell types. At later stages, as successive progenitors exit the cell cycle and become committed to particular cell fates, they migrate to fixed positions throughout the laminated retina, and establish synaptic connections to other neurons. [3H]thymidine-labeling experiments have demonstrated that retinal cell types are produced in sequence from multipotent progenitors (Sidman, 1961; Young, 1985). Each has a characteristic ‘birthdate’, defined by its terminal mitotic S phase, that strongly influences its identity. In the murine eye, the most likely birth order is retinal ganglion cells (RGCs), cone photoreceptors, amacrines and horizontal cells, followed by rod photoreceptors, bipolar cells and Müller glia, with significant overlap in the appearance of these different cell types (Cepko et al., 1996). Although phylogenetic differences in the birth order of retinal neurons have been observed, RGCs are the first neurons born in all vertebrate species examined (Altshuler et al., 1991).
RGCs are the only retinal neurons that extend axons outside the eye. As RGCs differentiate, their axons grow laterally toward the presumptive optic nerve head in response to molecular guidance cues (Birgbauer et al., 2000; Brittis and Silver, 1994; Deiner et al., 1997) and pass outward through the optic stalk (Hinds and Hinds, 1974; Silver and Sidman, 1980). These axons travel along the optic nerve to the chiasm, where they make characteristic pathway choices and project to six specific regions in the brain, including the lateral geniculate nucleus and superior colliculus (Rodieck, 1998). The optic nerve is largely composed of RGC axons, but also contains supporting glia and astrocytes, and the central retinal artery and vein. The physiology and histological characteristics of RGC subtypes are well known (Rodieck, 1998), yet no single intrinsic factor has been demonstrated that specifies RGC fate within the mammalian eye.
The cellular mechanisms of vertebrate and Drosophila retinogenesis are fundamentally different. In vertebrates, the continuous selection of committed neurons from a pool of multipotent progenitors leads to overlapping of neuronal birthdates, stochastic variation in clonal composition (Turner et al., 1990), and random spacing between cell bodies of different neuron types (Rockhill et al., 2000). In Drosophila, a strict spatial and temporal hierarchy of cell-cell inductive interactions creates a highly ordered and invariant array of ommatidia (Brennan and Moses, 2000; Rubin, 1989; Tomlinson, 1988). In addition, all fly photoreceptors synapse directly to neurons within the brain, while vertebrate photoreceptors connect to the brain via specialized interneurons and RGCs, which are the sole projection neurons for the eye.
bHLH transcription factors are central to retinal neurogenesis (Cepko, 1999; Kageyama et al., 1995). In Drosophila photoreceptor development, the proneural gene atonal (ato) specifies the founding R8 neuron in each ommatidium (Dokucu et al., 1996; Jarman et al., 1994; Jarman et al., 1995). In addition to R8 determination, atonal also controls neuronal subtype identity (Chien et al., 1996; Sun et al., 2000) and neurite arborization (Hassan et al., 2000) within the Drosophila peripheral nervous system and brain, respectively. Among murine bHLH genes, Math5 (also known as Atoh7) is the most closely related to atonal within the bHLH domain (Brown et al., 1998; Hassan and Bellen, 2000). This structural homology is consistent with the specific expression of Math5 in the developing mouse retina (Brown et al., 1998), Xath5 in the developing frog retina (Kanekar et al., 1997), Cath5 in the chick eye (Liu et al., 2001; Matter-Sadzinski et al., 2001) and Ath5 in zebrafish retinal progenitors (Masai et al., 2000). Ectopic expression of Xath5 during frog eye development biases progenitors to become RGCs at the expense of later-born neurons such as bipolars and Müller glia (Kanekar et al., 1997). Xath5 is therefore sufficient to specify RGC fate. However, ectopic expression of Math5 in the same assay promotes formation of bipolar cells rather than RGCs (Brown et al., 1998). Thus, despite highly conserved structure and expression patterns, the functional orthology between Math5, Xath5 and atonal is unclear.
In this report, we test the role of Math5 in RGC formation by removing its function in vivo. We show that mice homozygous for a targeted Math5 mutation have grossly normal eyes, but no optic nerves or chiasm. Histological and molecular analyses reveal an almost complete absence of RGCs in postnatal Math5−/− retinae and an increase in cone photoreceptors. We further demonstrate that loss of Math5 alters the early stages of RGC formation and conclude that Math5 acts as a proneural gene for mammalian RGC determination.
MATERIALS AND METHODS
Targeted deletion of Math5
The genomic locus was targeted by homologous recombination using a 6.5 kb EcoRI fragment isolated from a mouse J1 ES cell phage library. A 256 bp SmaI fragment encoding the bHLH domain was excised and replaced with a BamHI-linkered PGK-neo-bpA cassette. A cytoplasmic β-gal-pA cassette was then inserted in-frame after codon 18. The linearized construct was electroporated into R1 embryonic stem cells (Nagy et al., 1993) and colonies selected using G418. Homologous recombination was detected in 3% of the ES clones by long-range PCR, with one primer outside each arm and the other within the neo gene (Fig. 1A). These assays gave 2.3 kb (5′ arm) and 4 kb (3′ arm) PCR products only when recombination occurred correctly. Targeting was verified by the presence of a 17 kb BamHI fragment, instead of the normal 22 kb fragment, in genomic Southern blots hybridized with an external 5′ probe (Fig. 1B). F1 founders were generated by crossing chimeric males to C57BL/6J mice. Phenotypic analysis was performed on N2F2 mice derived by crossing F1 founders with strain 129S3/SvImJ and intercrossing the resulting progeny. For retinal cell counts and some β-gal staining experiments, albino adult mice and embryos were generated using an outbred CD-1 background. Internal primers were used for routine genotyping of mouse litters and further verification of homologous recombination (Fig. 1C).
Fig. 1.
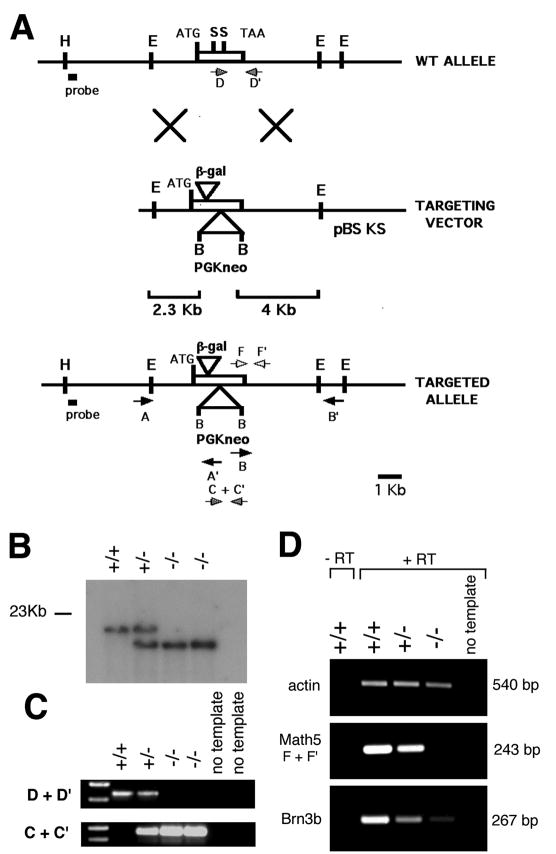
Targeted disruption of Math5. (A) Homologous recombination between 2.3 kb and 4 kb vector arms resulted in replacement of the bHLH domain (contained within the SmaI fragment) with a PGK-neo cassette and insertion of cytoplasmic β-gal near the N terminus of Math5. Positions of the single-copy probe and PCR primers are indicated. Primers A+A′ (5′ arm) and B+B′ (3′ arm) were used for long-range PCR. (B) BamHI Southern analysis of littermate tail DNA obtained from an F2 cross. (C) Internal PCR primers (D+D′ for wild-type, C+C′ for mutant) amplify allele-specific products from weanling tail DNA. (D) RT-PCR products (F+F′) from E15.5 total eye RNA show the absence of Math5 mRNA and a great reduction of Brn3b mRNA in the Math5−/− mutant, in comparison to β-actin. B, BamHI; E, EcoRI; H, HindIII; S, SmaI.
mRNA analysis
To assess Math5, β-actin (Actb) and Brn3b (Pou4f2) mRNA expression, parallel RT-PCR assays were performed on total RNA collected from E15.5 eyes and pretreated with DNase I. Primer pairs were located in the 3′ untranslated regions of Math5 (accession no. AFO71223) and Brn3b (accession no. S68377), and within the coding region of β-actin (accession no. M12481).
Histology, immunohistochemistry and in situ hybridization
Mice were quickly sacrificed and their eyes, brains, or embryos dissected in cold phosphate-buffered saline (PBS). Tissues were fixed in Bouins or buffered formalin for paraffin embedding, or 4% paraformaldehyde for β-gal histochemistry, or freshly frozen in OCT (Miles Scientific) for cryosectioning. Paraffin sections (10 μm) were either stained with Hematoxylin and Eosin, counterstained with Neutral Red following β-gal histochemistry, or processed for immunoperoxidase staining following antigen retrieval (Evers and Uylings, 1997). Cryosections (10 μm) were fixed in 4% paraformaldehyde PBS and used for Math5 digoxigenin in situ hybridization (Brown et al., 1998) or antibody labeling. Immunohistochemical and PNA lectin staining was performed according to published protocols (Rich et al., 1997; Sundin and Eichele, 1990) with the biotin-streptavidin system (ABC, Vector Labs), rabbit Pax6 antiserum (1:2000, Mastick and Andrews, 2001), rabbit S-cone opsin antiserum (1:20,000, Applebury et al., 2000), rabbit recoverin antiserum (1:500, Dizhoor et al., 1991), rabbit caspase-3 antisera (1:200, New England Biolabs), biotin-PNA (5 μg/ml, Vector Labs), and the following monoclonal antibodies: anti-neuron-specific β-tubulin (TUJ1, 1:1000, Babco), anti-protein kinase C (MC5, 1:400, Sigma), anti-vimentin (LN9, 1:200, Sigma), anti-neurofilament (NN18 for 160 kDa and NE14 for 200 kDa, 1:500, Sigma), anti-tyrosine hydroxylase (TH2, 1:1000, Sigma), anti-syntaxin (HPC1, 1:1000, Sigma), anti-calretinin (mAb1568, 1:500, Chemicon), anti-calbindin (CB955, 1:500, Sigma), VC1.1 (1:200, Sigma), and anti-rhodopsin (RET-P1, 1:1000, Sigma). For β-gal visualization, tissues were fixed and stained as described previously (Sanes et al., 1986).
Retinal disassociation and cell counts
Cone cell counts were performed on eyes from adult F2 littermates obtained by intercrossing CD-1 Math5+/− mice. At least two mice from each genotype were compared. Neural retinae were dissected free of other ocular tissues in HBSS-CMF (Ca2+/Mg2+-free Hanks buffered saline solution) and gently disassociated as described by Altshuler and Cepko with minor modifications (Altshuler and Cepko, 1992). Dissected retinae were first incubated for 15 minutes at 37°C in normal HBSS with 300 μg/ml hyaluronidase (Sigma, Type IV-S) and 1 mg/ml collagenase (Sigma, Type XI-S), gently washed, and then incubated for 10 minutes at 25°C in HBSS-CMF with 0.1% trypsin (Gibco). After trypsinization, 10% normal donkey serum (NDS) was added and the cells pelleted by low-speed centrifugation. The retinal cells from each mouse were then resuspended in 5 ml HBSS-CMF, 10% NDS, 10 mM Hepes pH 7.4, triturated gently to a single cell suspension, and plated on poly-d-lysine-coated glass slides for 90 minutes at 37°C in a humidified chamber. Slides were fixed in fresh ice-cold PBS with 4% PFA for 10 minutes, and processed for immunohistochemistry in TST (150 mM NaCl, 10 mM Tris pH 7.4, 0.1% Tween20) containing 2.5% NDS. S-cone opsin was detected using a 1:20,000 dilution of polyclonal rabbit antiserum (Applebury et al., 2000), a biotinylated secondary antibody (Jackson Immunoresearch), and streptavidin-DTAF (Molecular Probes). To visualize cell nuclei, RNaseA (200 μg/ml) and propidium iodide (50 μg/ml) were added to the secondary and tertiary reagents, respectively. Processed slides were viewed using a Nikon Eclipse E800 fluorescence microscope and SPOT camera. Two-color images (200×) were captured digitally from random fields selected using a TRITC filter, and the total and S-cone opsin-positive cells in each image counted manually. For each mouse, 10–25 images (800–3000 cells) were analyzed. Differences between wild-type and mutant values were evaluated for statistical significance using the nonparametric Wilcoxon two-sample test for n >10 measurements (Sokal and Rohlf, 1969).
RESULTS
Targeted deletion of the Math5 gene
To investigate the role of Math5 during in vivo retinal development, we deleted its bHLH domain by gene targeting in mouse embryonic stem (ES) cells (Fig. 1). Math5 contains a single exon, similar to most vertebrate atonal homologues. The bHLH domain was replaced with a PGK-neo-bpA cassette, thereby removing both DNA binding and dimerization functions and destroying the Math5 transcription unit. We also inserted a cytoplasmic β-galactosidase (β-gal) cassette near the N terminus. ES cell colonies with the desired homologous recombination events were identified by Southern and long-range PCR analyses (data not shown). One ES cell line heterozygous for the targeted Math5 mutation (4F1) gave rise to chimeric mice, which transmitted the mutant allele through the germline. Southern (Fig. 1B) and allele-specific PCR assays (Fig. 1C) were used to follow the mutant allele in descendants. Math5 homozygotes (Math5−/−) and heterozygotes (Math5+/−) are indistinguishable from wild-type (Math5+/+) littermates in their viability, growth rate and fertility. Consequently, the distribution of F2 genotypes was consistent with Mendelian inheritance: we observed 21 wild type (27%), 36 heterozygous (46%) and 21 homozygous (27%) mice at weaning. We confirmed the absence of Math5 mRNA in homozygous mutant embryos by reverse transcriptase (RT) PCR using RNA derived from E15.5 eyes (Fig. 1D).
Math5-βgal expression during retinogenesis
The targeted allele contains a histochemical reporter, Escherichia coli β-galactosidase, which is transcribed from the endogenous Math5 promoter (Fig. 1A). The β-gal reporter is an exquisitely sensitive tool for detecting gene expression (Lawson et al., 1999). Although Math5 encodes a nuclear protein, we used a cytoplasmic β-gal cassette to demarcate Math5 expression domains in the embryo and reveal the processes of differentiated neurons. In heterozygous E11.5 embryos, the pattern of β-gal expression in retinal progenitors was identical to Math5 mRNA (Fig. 2A,B) and coincides with the onset of histogenesis. After the initiation of RGC differentiation, we noted two layers of β-gal-expressing cells within the retinal neuroepithelium (Fig. 2C,D). One layer is located at the ventricular (sclerad) surface, closest to the pigmented epithelium (RPE), and corresponds to the zone where retinal progenitors undergo mitosis (Sidman, 1961). These retinal progenitors also express Math5 mRNA (compare Fig. 2D and E). The distribution of Math5 mRNA (Brown et al., 1998) and β-gal activity within this layer is not uniform, but instead suggests a radial columnar pattern. The second layer is located at the vitreal surface, closest to the lens, where terminally differentiating RGCs accumulate. These cells express β-gal within their somata and axons (Fig. 2D). We also noted β-gal expression within the optic nerve at E17.5 (Fig. 2F) and in cone photoreceptors at P21 (Fig. 2G). Consistent with the latter observation, we have noted a low level of Math5 mRNA expression in the photoreceptor cell layer at P0.5 (data not shown). β-gal expression in these differentiated cell types may reflect perdurance of this stable enzyme (Hall et al., 1983) or persistent, low level transcription of Math5 in some differentiated neurons, combined with the high sensitivity of this reporter system. No other retinal neuron expression domain was observed.
Fig. 2.
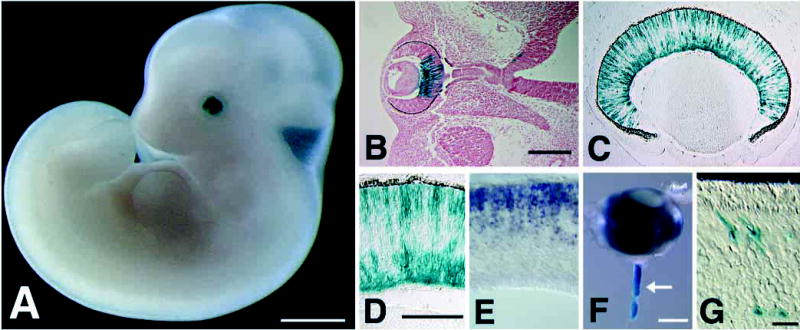
Embryonic β-gal expression in Math5+/− eyes. (A,B) E11.5 β-gal expression in the developing optic cup. B shows a horizontal section through the whole-mount embryo in A. The overall pattern is identical to endogenous Math5 mRNA (see Fig. 2 of Brown et al., 1998). β-gal staining begins in the dorsal central cup. A small number of positive cells are also visible in the diencephalon near the base of the optic stalk. (C,D) E15.5 retinal β-gal expression. β-gal is present in progenitors and differentiating RGCs. D shows a higher magnification. E. In situ hybridization at E15.5 showing Math5 mRNA expression solely in retinal progenitors. (F) β-gal expression in the optic nerve at day E17.5 (arrow). (G) β-gal-positive cells with characteristic cone morphology in the photoreceptor layer of a heterozygote at P21. Bars are 1 mm in A; 200 μm in B; 100 μm in D; 500 μm in F; and 20 μm in G.
Ocular phenotypes of adult Math5−/− mice
Math5−/− mice appear normal externally, and have average sized eyes with intact extraocular muscles and periocular tissues. However, they lack optic nerves and chiasmata (Fig. 3A,B). These structures are grossly normal in heterozygotes, and are indistinguishable from those of Math5+/+ littermates. Failure to form optic nerves strongly suggests that RGCs are missing or abnormal in Math5−/− mice.
Fig. 3.
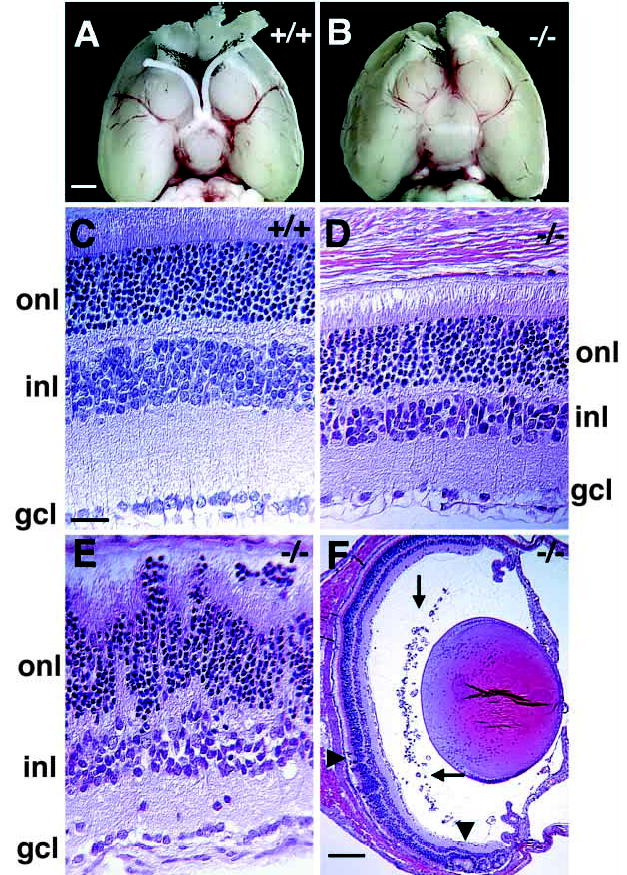
Ocular abnormalities in adult Math5−/− mice. (A,B) Ventral view of Math5+/+ and Math5−/− P21 brains, with rostral ends pointing upwards. Olfactory bulbs have been removed. Both optic nerves and the optic chiasm are absent in the Math5−/− mouse. (C–F) Hematoxylin and Eosin-stained transverse sections of P21 Math5+/+ (C) and Math5−/− (D–F) eyes. Mutant retinae have regions of normal (D,F) and disrupted laminar structure (E and arrowheads in F). However, even in areas with largely normal structure, cells and axons are missing from the INL, IPL and GCL (D,E). Ectopic blood vessels are present in the vitreal space of mutant eyes (arrows in F). gcl, ganglion cell layer; inl, inner nuclear layer; onl, outer nuclear layer. Bars are 1.5 mm in A; 20 μm in C; and 200 μm in F.
Histological analysis demonstrated overall structural similarity between postnatal Math5−/− and Math5+/+ eyes. However, mutant retinae have major abnormalities. First, they are 20–25% thinner than wild type, primarily due to decreased width of the inner plexiform (IPL) and nuclear (INL) layers (Fig. 3C,D). Second, we observed significantly fewer cells in the ganglion cell layer (GCL). In normal mice, only 41% of the cells in this layer are actually RGCs and contribute axons to the optic nerve; the remaining 59% are considered displaced amacrine cells (Drager and Olsen, 1981; Jeon et al., 1998; Perry, 1981). Third, the majority of mutant eyes have areas with disrupted cell layering (Fig. 3E), including rosettes (arrowheads in Fig. 3F). Fourth, we found no optic stalk remnant in serial sections of adult Math5−/− eyes, and the central artery and vein, which normally enter the eye via the optic nerve, were absent. Fifth, we observed ectopic blood vessels within the posterior chamber in many mutant animals (arrows in Fig. 3F) and less frequently within the GCL or IPL (not shown). These may reflect choroidal neovascularization or persistent hyaloid vessels.
Specific neuron classes are altered in Math5−/− retinae
To assess the abundance and patterning of the seven major cell types, we examined sections of postnatal Math5+/+, Math5+/− and Math5−/− eyes by light microscopy using a panel of 15 terminal differentiation markers. No significant difference was noted between Math5+/+ and Math5+/− eyes in the expression level or number of positive cells for any marker. Because laminar disruptions in Math5−/− eyes could differentially affect cell populations, we selected regions with relatively normal structure for comparison. Using both 160 kDa and 200 kDa neurofilament markers (Nixon et al., 1989), we discovered a total absence of RGCs and axon bundles in homozygous Math5 mutant retinae (Fig. 4A,B and data not shown). Immunostaining with the neuron-specific β-tubulin marker TUJ1 (Brittis and Silver, 1994; Macabe et al., 1999; Snow and Robson, 1994; Watanabe et al., 1991) further highlighted a complete absence of RGCs and axon bundles in P21 Math5−/− eyes (Fig. 4C,D).
Fig. 4.
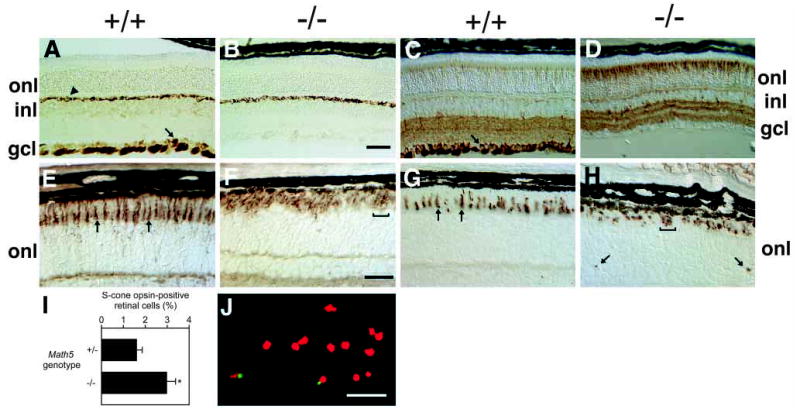
RGCs are absent and cone photoreceptors are increased in Math5−/− mice. (A,B) Neurofilament (160 kDa) is normally expressed by RGCs (arrow in A) and horizontal neurons (arrowhead in A). This marker highlights the loss of RGCs and their axon fibers in Math5 mutants (B). Horizontal cells are unaffected. (C,D) Anti-β-tubulin (TUJ1) labeling shows the absence of RGCs and an excess of cone photoreceptors in Math5−/− retinae. RGCs are strongly labeled in the wild-type section (arrow). (E,F) Peanut agglutinin (PNA) stains all cone photoreceptors. Individual cones are visible in the wild-type (arrows in E) but the density of cones is greatly increased in the Math5−/− mice (F). (G,H) The abundance of S-cones is also increased in Math5 mutants. The arrows in G indicate normal S-cone outer segments. Mutant cones are irregularly arranged in the outer retina (brackets in F and H). In some mutant sections, S-cone outer segments were observed within the ONL (arrows in H), OPL or INL. (I) Histogram comparing the abundance of S-cones in Math5+/− and Math5−/− mice as a percentage of total retinal cells. The bars show mean values and standard errors for these two genotypes. The increase in cones in Math5−/− mice is statistically significant (t s[∞] = 2.74, P <0.01). (J) Dissociated retinal cells labeled with S-cone opsin antisera (green) and propidium iodide (red). All retinal sections (A–H) are from P21 mice. gcl, ganglion cell layer; inl, inner nuclear layer; onl, outer nuclear layer. Bars are 100 μm in B; 30 μm in F; 50 μm in J.
Interestingly, we also discovered that the abundance of cone photoreceptors was substantially increased in Math5−/− retinae. We confirmed this increase using three different cone cell markers – peanut agglutinin (PNA), a lectin that binds to the matrix sheath surrounding cone but not rod photoreceptors (Blanks and Johnson, 1983; Ng et al., 2001; Rich et al., 1997; Fig. 4E,F); short wavelength (S) cone opsin, which is expressed in a ventral-to-dorsal gradient by approximately 70–80% of cones in normal mice (Applebury et al., 2000; Fig. 4G,H); and neuron-specific β-tubulin (TUJ1), which is present in cones at significant but lower levels than in RGCs (Fig. 4C,D). In mutant mice, these cones were often clustered or abnormally positioned in the outer segment layer (Fig. 4F,H). Similar cone cell findings were noted at P7 and P14 (data not shown). To determine the magnitude of the cone cell increase, we disassociated adult neural retinae and counted the proportion of cells that are S-cone opsin-positive (Fig. 4I,J). In normal laboratory mice, cones account for 2.8% of all photoreceptors (Carter-Dawson and LaVail, 1979a; Jeon et al., 1998), and thus approximately 2.5% of all retinal cells (Rodieck, 1998). We observed that 1.7±0.2% of disassociated retinal cells from Math5+/− mice were reactive with S-cone opsin antisera. These two values are similar, if one assumes that 70% of cones express S-opsin (Applebury et al., 2000). In Math5−/− littermates, the proportion of positive retinal cells was 3.0 ± 0.4%, which is significantly higher (Fig. 4I). The ratio of cones to RGCs in normal C57BL/6 mice is approximately four to one (roughly 180,000 to 45,000; Jeon et al., 1998). Since cones are increased by 75% in Math5−/− mice, this exceeds the loss of RGCs by two- to three-fold. Although this calculation ignores variations in the number of RGCs among normal inbred laboratory mouse strains (Jeon et al., 1998; Williams et al., 1998) and assumes that there is no significant decrease in the number of rod photoreceptors in the Math5 mutant (see below), the reciprocal changes in RGC and cone cell populations are comparable in magnitude.
Amacrine cells have highly overlapping birthdates with RGCs and cones. This class of cells, originally named because they have no axons (Ramón y Cajal, 1892), is extremely heterogeneous and consists of at least 22 different morphologically and molecularly distinct subtypes (MacNeil and Masland, 1998; Masland, 1988; Strettoi and Masland, 1996). Although the majority of amacrines reside in the INL, a significant proportion are located in the GCL, where they are termed displaced amacrines. We were therefore particularly interested in studying these cells in Math5−/− retinae. The protein syntaxin is present in the dendritic processes and perikarya of all amacrine cells (Barnstable et al., 1985). We observed similar patterns of immunostaining with this pan-amacrine marker in mutant and wild-type eyes (Fig. 5A,B), indicating that amacrine cells as a whole are grossly normal. The transcription factor Pax6 was also similarly expressed by INL and displaced amacrine cells in mutant and wild-type postnatal eyes (Fig. 5C,D). Although Pax6 protein is normally expressed by both amacrines and RGCs in postnatal retinae (Belecky-Adams et al., 1997; Davis and Reed, 1996; Hitchcock et al., 1996; Koroma et al., 1997), ganglion cells are absent in the Math5 mutants (Fig. 4B,D). On the basis of their relative density and expression profile (neurofilament- and TUJ1-negative, syntaxin- and Pax6-positive), we conclude that most, if not all, of cells remaining in the GCL of adult Math5−/− mice (Fig. 3D) are displaced amacrines.
Fig. 5.
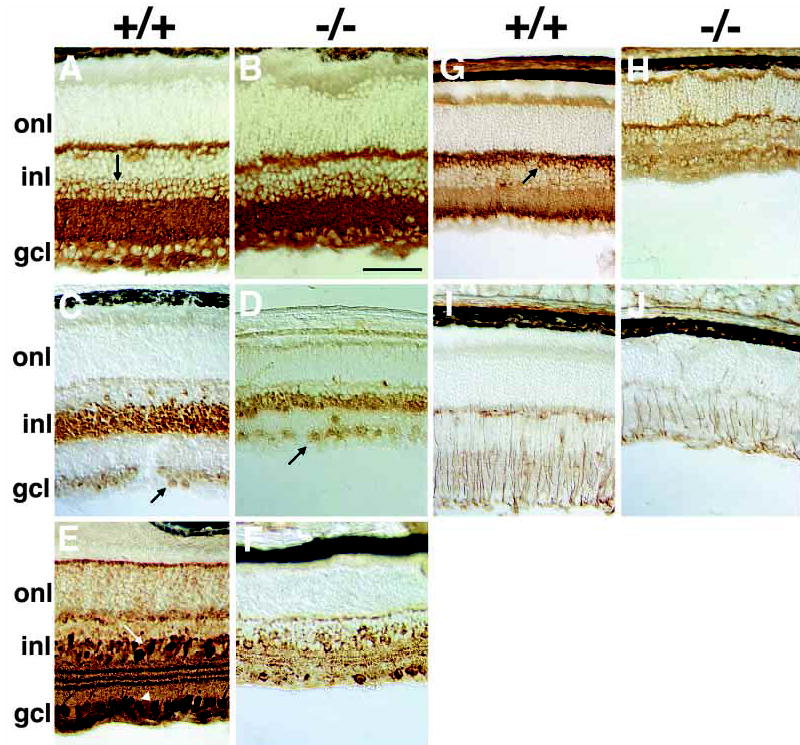
Other retinal neuron defects in Math5−/− mice. (A,B) Anti-syntaxin (HPC-1) immunostaining shows that amacrines in vitread INL (arrow) and displaced amacrines in the GCL are grossly unaffected. (C,D) Pax6 is similarly expressed by INL and displaced amacrine cells of wild-type (C) and Math5 mutant mice (D). (E,F) Calretinin-positive A2 amacrines in the INL (arrow) and GCL (arrowhead) are reduced in the Math5 mutant (F) compared to wild-type (E). (G,H) Rod bipolar cells labeled with anti-protein kinase C have cell bodies in the sclerad INL (arrow) and synapse with photoreceptors in the OPL and A2 amacrines in the IPL. These neurons are reduced in Math5−/− retinae and lack well organized synaptic termini in the IPL. (I,J) Anti-vimentin staining shows radial fibers of Müller glia extending across Math5+/+ retinal laminae. The abundance of Müller glia is reduced in Math5−/− eyes. Retinal sections in A–D are from P14 mice and those in E–J are from P21. gcl, ganglion cell layer; inl, inner nuclear layer; onl, outer nuclear layer. The bar is 100 μm in B.
To further characterize this diverse cell population, we examined two important amacrine subtypes. A2 cells are the single most common subtype of amacrines, accounting for 12.6% of all amacrines cells in the rabbit (MacNeil and Masland, 1998), and are selectively labeled with antibodies to the calcium-binding protein calretinin (Massey and Mills, 1999). We found that A2 amacrines are decreased in Math5 mutants (Fig. 5E,F) but that a substantial fraction of cells remaining in the GCL are calretinin-positive. These observations are consistent with our conclusion that the cells remaining in the GCL are displaced amacrines. Dopaminergic amacrines are relatively rare, arborize extensively, and appear to have a neuromodulatory role (Masland, 1988). This amacrine subtype, identified by tyrosine hydroxylase immunostaining (Ballesta et al., 1984), appears to be missing in Math5−/− mice (data not shown). However, because the overall number of amacrines is largely unaffected in Math5−/− eyes, it is plausible that one or more untested subclass will exhibit an increase in cell number.
Loss of Math5 had no apparent effect on the number or arrangement of rods, assessed by rhodopsin immunostaining (data not shown), or horizontal cells, assessed by calbindin (data not shown) and 160 kDa neurofilament immunostaining (Fig. 4AB). However, we did observe a decrease in rod bipolar (Fig. 5G,H) and Müller glial (Fig. 5I,J) cells. The rod bipolars also did not form a defined sublayer in the IPL. A similar decrease in cone bipolars was noted using the marker recoverin (data not shown). This reduction in bipolars and Müller glia may result directly from loss of Math5 activity or indirectly from a depletion of progenitors earlier in development.
RGC development is abnormal in Math5−/− mice
Our initial assessment of the Math5−/− retinal phenotype focused on adult eyes so that all neuron and glial cell classes could be examined simultaneously. However, at birth, mutant retinae appeared thinner and less well laminated (data not shown). This suggested that Math5 is required at earlier, prenatal stages of retinal development. We therefore examined mutant retinae at E15.5, when Math5 mRNA is maximally expressed and at E13.5, which is 1–2 days after the onset of RGC differentiation. At both ages, Math5−/− retinae are noticeably thinner than those of Math5+/+ or Math5+/− littermates (compare Fig. 6A with B, 6E with F, and 6G with H). Hematoxylin and eosin staining at E15.5 showed that RGC axon bundles, normally present at the vitreal surface, are missing or greatly diminished in Math5−/− retinae (arrows in Fig. 6A,B). The reduction in RGC axons was also revealed by comparing the patterns of β-gal expression in Math5+/− and Math5−/− eyes at E15.5 (arrows in Fig. 6C,D). Intense β-gal staining of RGC axons was observed at the vitreal margin in Math5+/− but not in Math5−/− eyes. In addition, β-gal-positive cells, which are confined to two layers in Math5+/− ptic cups at E15.5, were distributed across the thickness of the retina (compare Fig. 6C with D). To determine if any differentiating RGCs are present transiently, we examined the E15.5 mutant retinae with the marker TUJ1, which specifically identifies RGCs at this age (Brittis and Silver, 1994; Watanabe et al., 1991). In contrast to the adult eye (Fig. 4C,D), we found that loss of Math5 caused a great reduction, but not a complete absence, of RGCs at E15.5 (Fig. 6E,F).
Fig. 6.
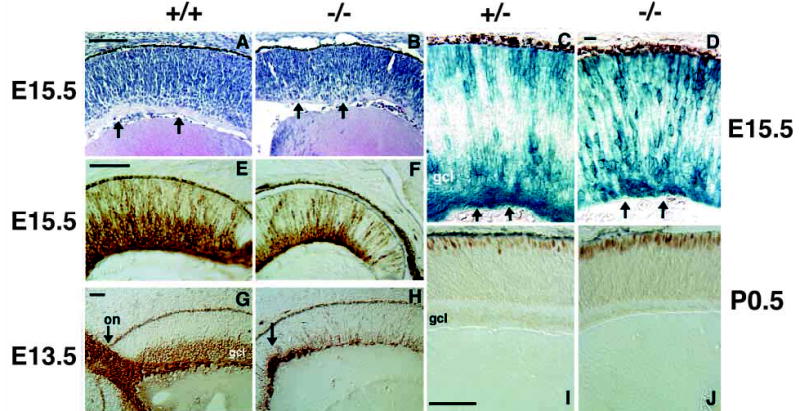
Early stages of RGC and cone formation are abnormal in Math5−/− retinae. Micrographs of retinal sections from E15.5 (A–F) and E13.5 (G,H) embryos and P0.5 newborn mice (I,J). (A,B) Hematoxylin and Eosin-stained sections of wild-type (A) and Math5−/− (B) retinae. Mutant retinae are thinner in the radial dimension. Arrows in both panels indicate the position of eosinophilic RGC axon fibers, which are apparent in A but largely absent in B. (C,D) β-gal expression in Math5+/− and Math5−/− retinal sections. Arrows indicate the RGC axon fibers in heterozygotes that are greatly reduced in the mutant. Math5−/− β-gal-expressing cells are distributed across the retinal thickness (top to bottom of D). (E,F) Anti-β-tubulin (TUJ1) immunostaining of E15.5 retinae. A well formed GCL is apparent in the wild-type (E), but is greatly diminished in the Math5−/− retina (F). (G,H) Neurofilament (160 kDa) immunostaining highlights the failure of Math5−/− retinae to form a significant number of RGCs at an early stage. Arrows indicate a normal optic nerve in wild-type (G) and abnormal axons in the Math5 mutant that do not exit the eye (H). (I,J) Recoverin immunostaining demonstrates an early increase in the density of cone photoreceptors in Math5−/− mice (J). on, optic nerve; gcl, ganglion cell layer. Bars are 100 μm in A,E,I; 20 μm in D; and 50 μm in G.
We also compared Math5−/−, Math5+/− and Math5+/+ eyes at E15.5 for expression of Brn3b by semi-quantitative RT-PCR (Fig. 1D). This POU domain transcription factor is expressed by nascent RGCs as they leave the cell cycle, migrate to the vitreal surface of the retina, and differentiate (Erkman et al., 1996; Xiang et al., 1993). Targeted deletion of Brn3b causes a 70% reduction in the number of RGCs in the adult eye (Erkman et al., 1996; Gan et al., 1996). This gene may thus be a downstream target of Math5 regulation. Our results show a severe reduction of Brn3b expression in Math5−/− eyes at E15.5 and an intermediate level in heterozygotes (Fig. 1D).
Finally, we assessed RGC formation in Math5−/− retinae at E13.5 using the 160 kDa neurofilament marker (Fig. 6G,H). Neurofilament 160 kDa expression in RGCs generally precedes TUJ1 reactivity (Macabe et al., 1999) and can be detected in migrating RGC precursors as well as post-migratory RGCs residing in the GCL. The results were similar to those obtained at E15.5 with TUJ1. In Math5−/− retinae at E13.5, there is a conspicuous deficiency of neurofilament-positive RGCs (Fig. 6G,H). A small number of nascent RGCs can be observed, but these are poorly organized. There is no well formed GCL and the few axons present are not bundled into nerve fibers (Fig. 6H). Normally the optic nerve is easily recognized at E13.5 (arrow in Fig. 6G) and RGC axons have already extended toward the optic chiasm. Serial sectioning of E13.5 and E15.5 mutant eyes (n=8) failed to detect even a rudimentary optic nerve (data not shown). Instead, we consistently saw a concentration of neurofilament staining in the central retina where the optic nerve normally exits (arrow in Fig. 6H). Because some RGCs appear to form in Math5−/− embryos, but do not remain in postnatal eyes, we surveyed mutant retinae for apoptotic cells by activated caspase-3 immunostaining. These experiments provided no evidence of increased programmed cell death in mutant retinae at E13.5, P0.5, P7, P14 and P21 (data not shown). The small number of RGCs that form independently of Math5 function must be eliminated from the eye between the ages of E15.5 and P7, but this may occur gradually.
The increase in cone cell differentiation in Math5−/− mice occurs early
In Math5−/− mice, an increase in cone photoreceptors is correlated with the deficiency of RGCs, suggesting that a shift in progenitor cell fate occurs during retinal histogenesis. To explore this hypothesis further, we sought to test Math5 mutant mice for an increase in cones at the earliest possible age that committed cone photoreceptors can be identified. This is difficult because there is a long delay between cone cell birthdates and overt differentiation. For example, the majority of cones in mice are born between E12 and E15 (Carter-Dawson and LaVail, 1979b), yet cone opsin expression does not begin until P5 (Cepko, 1996; Szel et al., 1993) and focal PNA reactivity is not observed in the outer retina until after P6 (Blanks and Johnson, 1983). To determine whether Math5 deficiency causes an early increase in cones, we therefore tested the developing retinae of newborn mutant and wild-type littermates with antibodies to recoverin, a calcium-binding protein that is expressed by rods, cones and cone bipolar cells (Dizhoor et al., 1991; Milam et al., 1993). Recoverin expression normally begins in the outer margin of the mouse retina between E17.5 and P0.5, and thus significantly precedes the appearance of cone opsin and PNA reactivity. Moreover, at P0.5 recoverin expression is almost exclusively restricted to cone photoreceptors, since few rods and no bipolar cells have differentiated at this stage (Belliveau and Cepko, 1999; Morrow et al., 1998). Our results demonstrate a substantial increase in the density of differentiating cones in Math5−/− mice at P0.5 compared to wild type (Fig. 6I,J). The shift in retinal cell populations is thus likely to be determined early during histogenesis.
DISCUSSION
We propose that Math5 acts positively to determine ganglion cell fate, for four reasons. First, Math5 expression in retinal progenitors closely matches RGC birthdates (Brown et al., 1998). Second, ectopic expression of the Xenopus homologue Xath5 or the chicken homologue Cath5 biases retinal progenitors toward an RGC fate (Kanekar et al., 1997; Liu et al., 2001). Third, this is consistent with the established proneural role of other bHLH genes such as atonal (Cepko, 1999; Jarman et al., 1994; Kageyama et al., 1995). Fourth, we now report that targeted disruption of Math5 in mice causes a specific loss of RGCs and optic nerves, and a concomitant increase in cone photoreceptors. While Math5 may also contribute to early RGC survival, it is not significantly expressed in adult eyes and thus cannot be continuously required for ganglion cell maintenance. Moreover, because mutant eyes are normal in size, Math5 also does not appear to significantly control growth of the optic cup, in contrast to Pax6 and Chx10 (Burmeister et al., 1996; Hogan et al., 1986).
Although other mutations are known that affect the quantity of RGCs in adult mice (Bonfanti et al., 1996; Burne et al., 1996; Erkman et al., 1996; Gan et al., 1996; Martinou et al., 1994; Rice et al., 1997; Williams et al., 1998), none completely deletes this neuronal cell class. In particular, the less severe phenotype of Brn3b−/− mice (Erkman et al., 1996; Gan et al., 1996), and the dosage-dependent decrease in Brn3b expression in Math5 mutants at E15.5 (Fig. 1D), strongly suggest that Brn3b is a downstream target of Math5 regulation. A comparable relationship has been demonstrated in Caenorhabditis elegans between the Math5 homologue lin32 (Portman and Emmons, 2000) and the POU domain transcription factor unc86 (Baumeister et al., 1996).
Loss of Math5 causes a change in cell fate
In Math5 mutant mice, RGCs are absent and cone photoreceptors increased. This relatively narrow phenotype contrasts sharply with the absence of all photoreceptors in atonal mutant flies (Jarman et al., 1994), which occurs because Drosophila eye development relies upon sequential inductive mechanisms that begin with R8 cells. We believe the cell type shift in Math5−/− mice reflects the continuously available pool of progenitors that underlies vertebrate retinogenesis (Cepko et al., 1996; Turner et al., 1990). A similar binary switch has been observed in developing cochlea of Math1−/− mice, between inner hair cell and supporting cell fates (Bermingham et al., 1999).
We propose that the majority of progenitors that would normally become RGCs switch their primary fate to cones when Math5 function is removed. Indeed, the changes we observed in ganglion and cone cell populations are similar in magnitude, and were detected as early as it is possible to reliably identify these two cell types (Fig. 6). The numerical surplus of cones, compared to the loss of RGCs, could arise because precursor cells undergo one to two additional mitoses before differentiating as cones, or because RGCs are normally overproduced and then selectively culled. A developmental connection between RGCs and cones is supported by independent gain- and loss-of-function experiments in which progenitor cell fate was altered by ectopic expression of Delta1. Retroviral transduction of chick retinae (Henrique et al., 1997) and mRNA injection of frog retinal precursors (Dorsky et al., 1997) with wild-type Delta1 biased progenitor cells to become cones. Conversely, infection of chick retinae with a dominant-negative Delta1 retrovirus produced an excess of RGCs at the expense of photoreceptor cells (Henrique et al., 1997).
Three models can explain the binary cell fate switch in Math5−/− mice (Fig. 7). Each is consistent with the overlapping early birthdates of RGCs and cones (Altshuler et al., 1991), their shared expression of β-gal in Math5+/− heterozygotes, and retroviral lineage studies (Turner et al., 1990). These models are conceptually distinct, but not mutually exclusive. First, although vertebrate retinogenesis does not utilize a strict lineage mechanism (Cepko et al., 1996), a subpopulation of precursors may emerge with relatively restricted developmental potential, through which particular cell fates are coupled. In the absence of Math5, for example, a bipotential RGC-cone precursor would be expected to differentiate exclusively into cone photoreceptors. Cones would thus represent a specific alternative fate for early progenitors whose RGC development is blocked (Waid and McLoon, 1995). According to this orthodox model, the persistance of β-gal activity in cones (Fig. 2G) may reflect the legacy of Math5 expression in this bipotential precursor cell. Second, RGCs may act extrinsically to inhibit cone cell determination (Belliveau and Cepko, 1999; Waid and McLoon, 1998), so that loss of Math5 (and RGCs) may indirectly increase the abundance of cones. Third, cones may represent a relatively generic default fate for early progenitors whose exit from the cell cycle is delayed (Henrique et al., 1997). According to this model, the essential and primordial role of Math5 is not to specify ganglion cells per se, but to direct commitment of early retinal neurons to the first appropriate fate, which in vertebrates is an RGC. In the absence of Math5, progenitors are forced to commit to the next available fate, which in mice is a cone photoreceptor. The last model is similar to the first but involves a more fluid and time-dependent view of developmental competence (Cepko et al., 1996). In this context, it is compelling that the lakritz mutation in the zebrafish Ath5 gene causes an equivalent deletion of all RGCs and a concomitant increase in amacrine, bipolar and Müller cells, but no change in cone photoreceptors (Kay et al., 2001). These observations correlate well with the relative birth order of retinal cell types in these two species (Altshuler et al., 1991), since cones and rods are the last-born cell types in the central zebrafish retina (Hu and Easter, 1999).
Fig. 7.
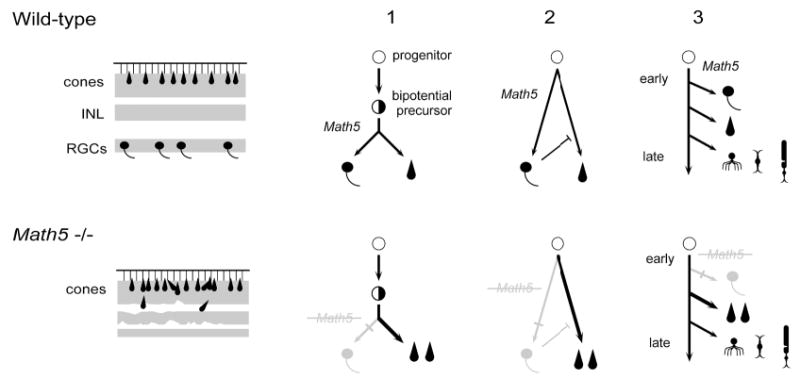
Binary cell fate switch between RGCs and cones in Math5−/− mice. On the left, diagrams summarize wild-type (upper) and mutant (lower) retinal phenotypes. RGCs are missing and cones are increased in the mutant. The nuclear layers are indicated by shading. The inner nuclear (INL) and plexiform layers are significantly thinner in the mutant and lamination is disrupted in places. On the right, diagrams depict three models that explain the shift from RGCs to cones, which are outlined in the text. In addition to the multipotent progenitor and hypothetical bipotential precursor cells, five differentiated retinal cell types are represented (RGCs, cones, amacrines, bipolars, and rods).
In addition to major changes in RGC and cone populations, we also observed less dramatic reductions in bipolar and Müller glial cell populations in Math5−/− mutants. There are two possible explanations for this phenotype. First, bipolar and Müller cells may require Math5 function directly. However, we have been unable to detect either endogenous Math5 mRNA or β-gal activity in these cell types. Instead, it is more likely that the deficiency of bipolar and Müller glial cells occurs because of an overall defect in retinal development, secondary to the loss of RGCs and depletion of progenitors, since these are the last two cell types to form in the mammalian retina. The reduced thickness of Math5 mutant retinae at E15.5 supports this idea. We also observed patches of laminar disorganization in Math5−/− mutants. This may reflect the deficiency of Müller glia, whose processes extend radially across the retina and which are required for laminar integrity (Dubois-Dauphin et al., 2000; Rich et al., 1995; Willbold et al., 2000), or the absence of an inherent patterning function provided by the RGCs during retinal development (Hinds and Hinds, 1974; Watanabe et al., 1991). Finally, although we observed no global change in amacrine cells, we believe it will be important in the future to investigate whether the relative abundance of specifc subtypes is significantly affected.
RGC development does not depend strictly upon Math5
Because adult Math5−/− retinae are missing virtually all RGCs, our finding that a subset of embryonic RGCs can differentiate was unexpected and suggests that the initial specification and development of some RGCs is Math5-independent. This situation is not unprecedented since mutations in other vertebrate bHLH genes, such as Mash1 and Ngn2, have been shown to cause a marked reduction, but not the complete deletion of particular neuron classes in the central nervous system (Fode et al., 2000). It will be interesting to study this small, transient class of RGCs further, which can initiate development in the absence of Math5. For example, it will be important to determine what becomes of these neurons between ages E15.5 and P21. Since they are so few in number, and unable to organize into an optic nerve or innervate their targets, it is likely that they are cleared from the retina through apoptosis. This occurs during normal postnatal development to a substantial fraction of RGCs whose axons fail to make appropriate synaptic connections in the brain (Bonfanti et al., 1996; Burne et al., 1996; Cowan et al., 1984; Martinou et al., 1994). In Math5 mutant mice, this apoptosis probably occurs in the interval between P0.5 and P7, since we observed no significant increase in programmed cell death outside this window. It will also be important to determine if these short-lived RGCs correspond to a particular functional or anatomical subclass of RGCs. Finally, it is also possible in theory that these cells are not true RGCs, but are a distinct immature cell type that transiently expresses part of the RGC differentiation program, including Brn3b, neural-specific β-tubulin (TUJ1), and neurofilament 160 kDa. We think this latter possibility is unlikely.
While this paper was under review, a similar Math5 knockout allele was reported, with several notable differences (Wang et al., 2001). First, the β-gal cassette was targeted to the nucleus, although the insertion site was not detailed. As a result, the descendants of Math5-expressing progenitors and their cellular processes were not histochemically detectable after differentiation. In contrast, the cytoplasmic β-gal cassette we introduced contains eighteen N-terminal amino acids from the Math5 polypeptide and was readily detected in the optic nerve at E17.5 and in differentiated cones at P21 (Fig. 2F,G). Since the mRNAs transcribed from these two alleles are likely to have similar half-lives, this limited comparison suggests that the β-galactosidase protein may be stabilized by the Math5 amino terminus or may have significantly greater stability in the cytoplasm of differentiated neurons (Callahan and Thomas, 1994; Schilling et al., 1991). Second, Wang et al. did not evaluate cone photoreceptors in the Math5−/− mice and thus did not observe a major cell fate shift. Third, these authors reported a small number of RGCs in adult Math5−/− mice but none in E13.5 embryos. This discrepancy can be explained if the limited survival of RGCs in Math5−/− mice is somewhat variable and is influenced by genetic background (Williams et al., 1998). This hypothesis is difficult to evaluate at present since detailed information regarding the mouse strains was not provided, and no systematic breeding scheme was described.
Together Math1 and Math5 reconstitute atonal function
Our results significantly advance the concept of functional conservation within the atonal gene family. In Drosophila, atonal controls development of retinal neurons and the chordotonal organs, which are internal mechanosensory structures that act as proprioceptors. In vertebrates, visual system and proprioceptive functions are separated, with Math5 controlling the former and Math1 (Atoh1) regulating the latter. Math1 is expressed in cochlear and vestibular hair cells, vibrissae, the dorsal spinal column, joint capsules, Merkel touch receptors, and the cerebellum (Ben-Arie et al., 1997; Ben-Arie et al., 2000; Bermingham et al., 1999). Each of these structures is associated with mechanosensory perception, locomotory coordination or the integration and processing of three-dimensional spatial information. Math5 and Math1 are the vertebrate bHLH genes most closely related in structure to atonal. This relationship, the absence of significant Math1 expression in the eye, and the findings we present here, strongly suggest that Math5 is the atonal orthologue, and thus the major proneural gene, for the mammalian eye. There are few other examples where two functions identified in the Metazoa are so cleanly partitioned by evolution in the vertebrate lineage. In other gene families, the expression of paralogous genes is largely overlapping so that phenotypes can arise only at the edges where incomplete redundancy is exposed. In contrast, Math1 and Math5 act in independent domains. Taken together they are functionally equivalent to atonal.
Math5 acts during retinal histogenesis, after primary pattern formation in the anterior neural tube, specification of the optic primordia, and the major period of the optic cup growth has occurred. Math5 expression is dependent upon Pax6 (Brown et al., 1998), which has been positioned near the top of a hierarchy for metazoan eye development (Gehring and Ikeo, 1999). Consequently, our results, like those of Neumann and Nüsslein-Volhard (Neumann and Nüsslein-Volhard, 2000) for the hedgehog genes, show that functional homology in visual system development extends more deeply than Pax6. Finally, our results further establish an evolutionary parallel between vertebrate RGCs and fly R8 photoreceptors (Austin et al., 1995; Macabe et al., 1999), the earliest-born neurons of bilaterian visual systems.
Acknowledgments
We thank Thomas Saunders and Patrick Gillespie of the University of Michigan Transgenic Animal Model Core for assistance in producing mutant mice, with support from the Center for Organogenesis; Grant Mastick, Meredithe Applebury, Jenny Robbins, and Alexander Dizhoor for antibody reagents; Ursula Dräger, Janet Blanks, Pete Hitchcock, Pamela Raymond, Greg Dressler, Sally Camper, Jim Lauderdale, Gracie Andrews, Grant Mastick, Connie Cepko and Bill Goossens for scientific and technical advice; and David Ginsburg, Monica Vetter, David Turner, Greg Dressler, Derek van der Kooy and Steve Easter for critically reading the manuscript. J. B. was supported by NIH genetics training grant GM07544. This work was supported by grants from the NIH (EY14259 and EY11729) and the Howard Hughes Medical Institute to T. G.
References
- Altshuler D, Cepko C. A temporally regulated, diffusible activity is required for rod photoreceptor development in vitro. Development. 1992;114:947–957. doi: 10.1242/dev.114.4.947. [DOI] [PubMed] [Google Scholar]
- Altshuler, D. M., Turner, D. L. and Cepko, C. L. (1991). Specification of cell type in the vertebrate retina. In Cell Lineage and Cell fate in Visual System Development (ed. D. M.-K. Lam and C. J. Shatz), pp. 37–58. Cambridge, MA: MIT Press.
- Applebury ML, Antoch MP, Baxter LC, Chun LL, Falk JD, Farhangfar F, Kage K, Krzystolik MG, Lyass LA, Robbins JT. The murine cone photoreceptor: a single cone type expresses both S and M opsins with retinal spatial patterning. Neuron. 2000;27:513–523. doi: 10.1016/s0896-6273(00)00062-3. [DOI] [PubMed] [Google Scholar]
- Austin CP, Feldman DE, Ida JA, Cepko CL. Vertebrate retinal ganglion cells are selected from competent progenitors by the action of Notch. Development. 1995;121:3637–3650. doi: 10.1242/dev.121.11.3637. [DOI] [PubMed] [Google Scholar]
- Ballesta J, Terenghi G, Thibault J, Polak JM. Putative dopamine-containing cells in the retina of seven species demonstrated by tyrosine hydroxylase immunocytochemistry. Neuroscience. 1984;12:1147–1156. doi: 10.1016/0306-4522(84)90009-5. [DOI] [PubMed] [Google Scholar]
- Barnstable CJ, Hofstein R, Akagawa K. A marker of early amacrine cell development in rat retina. Brain Res. 1985;352:286–290. doi: 10.1016/0165-3806(85)90116-6. [DOI] [PubMed] [Google Scholar]
- Baumeister R, Liu Y, Ruvkun G. Lineage-specific regulators couple cell lineage asymmetry to the transcription of the Caenorhabditis elegans POU gene unc-86 during neurogenesis. Genes Dev. 1996;10:1395–1410. doi: 10.1101/gad.10.11.1395. [DOI] [PubMed] [Google Scholar]
- Belecky-Adams T, Tomarev S, Li HS, Ploder L, McInnes RR, Sundin O, Adler R. Pax-6, Prox 1, and Chx10 homeobox gene expression correlates with phenotypic fate of retinal precursor cells. Invest Ophthalmol Vis Sci. 1997;38:1293–1303. [PubMed] [Google Scholar]
- Belliveau MJ, Cepko CL. Extrinsic and intrinsic factors control the genesis of amacrine and cone cells in the rat retina. Development. 1999;126:555–566. doi: 10.1242/dev.126.3.555. [DOI] [PubMed] [Google Scholar]
- Ben-Arie N, Bellen HJ, Armstrong DL, McCall AE, Gordadze PR, Guo Q, Matzuk MM, Zoghbi HY. Math1 is essential for genesis of cerebellar granule neurons. Nature. 1997;390:169–172. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- Ben-Arie N, Hassan BA, Bermingham NA, Malicki DM, Armstrong D, Matzuk M, Bellen HJ, Zoghbi HY. Functional conservation of atonal and Math1 in the CNS and PNS. Development. 2000;127:1039–1048. doi: 10.1242/dev.127.5.1039. [DOI] [PubMed] [Google Scholar]
- Bermingham NA, Hassan BA, Price SD, Vollrath MA, Ben-Arie N, Eatock RA, Bellen HJ, Lysakowski A, Zoghbi HY. Math1: an essential gene for the generation of inner ear hair cells. Science. 1999;284:1837–1841. doi: 10.1126/science.284.5421.1837. [DOI] [PubMed] [Google Scholar]
- Birgbauer E, Cowan CA, Sretavan DW, Henkemeyer M. Kinase independent function of EphB receptors in retinal axon pathfinding to the optic disc from dorsal but not ventral retina. Development. 2000;127:1231–1241. doi: 10.1242/dev.127.6.1231. [DOI] [PubMed] [Google Scholar]
- Blanks JC, Johnson LV. Selective lectin binding of the developing mouse retina. J Comp Neurol. 1983;221:31–41. doi: 10.1002/cne.902210103. [DOI] [PubMed] [Google Scholar]
- Bonfanti L, Stretto E, Chierzi S, Cenni MC, Liu XH, Martinou JC, Maffei L, Rabacci SA. Protection of retinal ganglion cells from natural and axotomy-induced cell death in neonatal transgenic mice overexpressing bcl-2. J Neurosci. 1996;16:4186–4194. doi: 10.1523/JNEUROSCI.16-13-04186.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan CA, Moses K. Determination of Drosophila photoreceptors: timing is everything. Cell Mol Life Sci. 2000;57:195–214. doi: 10.1007/PL00000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brittis PA, Silver J. Exogenous glycosaminoglycans induce complete inversion of retinal ganglion cell bodies and their axons within the retinal neuroepithelium. Proc Natl Acad Sci USA. 1994;91:7539–7542. doi: 10.1073/pnas.91.16.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NL, Kanekar S, Vetter ML, Tucker PK, Gemza DL, Glaser TM. Math5 encodes a murine basic helix-loop-helix transcription factor expressed during early stages of retinal neurogenesis. Development. 1998;125:4821–4833. doi: 10.1242/dev.125.23.4821. [DOI] [PubMed] [Google Scholar]
- Burmeister M, Novak J, Liang MY, Basu S, Ploder L, Hawes NL, Vidgen D, Hoover F, Goldman D, Kalnins VI, Roderick TH, Taylor BA, Hankin MH, McInnes RR. Ocular retardation mouse caused by Chx10 homeobox null allele: impaired retinal progenitor proliferation and bipolar cell differentiation. Nat Genet. 1996;12:376–384. doi: 10.1038/ng0496-376. [DOI] [PubMed] [Google Scholar]
- Burne JF, Staple JK, Raff MC. Glial cells are increased proportionally in transgenic optic nerves with increased numbers of axons. J Neurosci. 1996;16:2064–2073. doi: 10.1523/JNEUROSCI.16-06-02064.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan CA, Thomas JB. Tau-β-galactosidase, an axon-targeted fusion protein. Proc Natl Acad Sci USA. 1994;91:5972–5976. doi: 10.1073/pnas.91.13.5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter-Dawson LD, LaVail MM. Rods and cones in the mouse retina. I Structural analysis using light and electron microscopy. J Comp Neurol. 1979a;188:245–262. doi: 10.1002/cne.901880204. [DOI] [PubMed] [Google Scholar]
- Carter-Dawson LD, LaVail MM. Rods and cones in the mouse retina. II Autoradiographic analysis of cell generation using tritiated thymidine. J Comp Neurol. 1979b;188:263–272. doi: 10.1002/cne.901880205. [DOI] [PubMed] [Google Scholar]
- Cepko CL. The patterning and onset of opsin expression in vertebrate retinae. Curr Opin Neurobiol. 1996;6:542–546. doi: 10.1016/s0959-4388(96)80062-6. [DOI] [PubMed] [Google Scholar]
- Cepko CL. The roles of intrinsic and extrinsic cues and bHLH genes in the determination of cell fates. Curr Opin Neurobiol. 1999;9:37–46. doi: 10.1016/s0959-4388(99)80005-1. [DOI] [PubMed] [Google Scholar]
- Cepko CL, Austin CP, Yang X, Alexiades M, Ezzeddine D. Cell fate determination in the vertebrate retina. Proc Natl Acad Sci USA. 1996;93:589–595. doi: 10.1073/pnas.93.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien CT, Hsiao CD, Jan LY, Jan YN. Neuronal type information encoded in the basic helix-loop-helix domain of proneural genes. Proc Natl Acad Sci USA. 1996;93:13239–13244. doi: 10.1073/pnas.93.23.13239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan WM, Fawcett JW, O’Leary DD, Stanfield BB. Regressive events in neurogenesis. Science. 1984;225:1258–1265. doi: 10.1126/science.6474175. [DOI] [PubMed] [Google Scholar]
- Davis JA, Reed RR. Role of Olf-1 and Pax-6 transcription factors in neurodevelopment. J Neurosci. 1996;16:5082–5094. doi: 10.1523/JNEUROSCI.16-16-05082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deiner MS, Kennedy TE, Fazeli A, Serafini T, Tessier-Lavigne M, Sretavan DW. Netrin-1 and DCC mediate axon guidance locally at the optic disc: loss of function leads to optic nerve hypoplasia. Neuron. 1997;19:575–589. doi: 10.1016/s0896-6273(00)80373-6. [DOI] [PubMed] [Google Scholar]
- Dizhoor AM, Ray S, Kumar S, Niemi G, Spencer M, Brolley D, Walsh KA, Philipov PP, Hurley JB, Stryer L. Recoverin: a calcium sensitive activator of retinal rod guanylate cyclase. Science. 1991;251:915–918. doi: 10.1126/science.1672047. [DOI] [PubMed] [Google Scholar]
- Dokucu ME, Zipursky SL, Cagan RL. Atonal, rough and the resolution of proneural clusters in the developing Drosophila retina. Development. 1996;122:4139–4147. doi: 10.1242/dev.122.12.4139. [DOI] [PubMed] [Google Scholar]
- Dorsky RI, Chang WS, Rapaport DH, Harris WA. Regulation of neuronal diversity in the Xenopus retina by Delta signalling. Nature. 1997;385:67–70. doi: 10.1038/385067a0. [DOI] [PubMed] [Google Scholar]
- Drager UC, Olsen JF. Ganglion cell distribution in the retina of the mouse. Invest Ophthalmol Vis Sci. 1981;20:285–293. [PubMed] [Google Scholar]
- Dubois-Dauphin M, Poitry-Yamate C, de Bilbao F, Julliard AK, Jourdan F, Donati G. Early postnatal Muller cell death leads to retinal but not optic nerve degeneration in NSE-Hu-Bcl-2 transgenic mice. Neuroscience. 2000;95:9–21. doi: 10.1016/s0306-4522(99)00313-9. [DOI] [PubMed] [Google Scholar]
- Erkman L, McEvilly RJ, Luo L, Ryan AK, Hooshmand F, O’Connell SM, Keithley EM, Rapaport DH, Ryan AF, Rosenfeld MG. Role of transcription factors Brn-3.1 and Brn-3.2 in auditory and visual system development. Nature. 1996;381:603–606. doi: 10.1038/381603a0. [DOI] [PubMed] [Google Scholar]
- Evers P, Uylings HB. An optimal antigen retrieval method suitable for different antibodies on human brain tissue stored for several years in formaldehyde fixative. J Neurosci Meth. 1997;72:197–207. doi: 10.1016/s0165-0270(96)02204-2. [DOI] [PubMed] [Google Scholar]
- Fode C, Ma Q, Casarosa S, Ang SL, Anderson DJ, Guillemot F. A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev. 2000;14:67–80. [PMC free article] [PubMed] [Google Scholar]
- Gan L, Xiang M, Zhou L, Wagner DS, Klein WH, Nathans J. Pou domain factor Brn-3b is required for the development of a large set of retinal ganglion cells. Proc Natl Acad Sci USA. 1996;93:3920–3925. doi: 10.1073/pnas.93.9.3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring WJ, Ikeo K. Pax6: mastering eye morphogenesis and eye evolution. Trends Genet. 1999;15:371–7. doi: 10.1016/s0168-9525(99)01776-x. [DOI] [PubMed] [Google Scholar]
- Hall CV, Jacob PE, Ringold GM, Lee F. Expression and regulation of Escherichia coli lacZ gene fusions in mammalian cells. J Mol Appl Genet. 1983;2:101–109. [PubMed] [Google Scholar]
- Hassan BA, Bellen HJ. Doing the MATH: is the mouse a good model for fly development? Genes Dev. 2000;14:1852–1865. [PubMed] [Google Scholar]
- Hassan BA, Bermingham NA, He Y, Sun Y, Jan YN, Zoghbi HY, Bellen HJ. atonal regulates neurite arborization but does not act as a proneural gene in the Drosophila brain. Neuron. 2000;25:549–561. doi: 10.1016/s0896-6273(00)81059-4. [DOI] [PubMed] [Google Scholar]
- Henrique D, Hirsinger E, Adam J, Le Roux I, Pourquie O, Ish-Horowicz D, Lewis J. Maintenance of neuroepithelial progenitor cells by Delta-Notch signalling in the embryonic chick retina. Curr Biol. 1997;7:661–670. doi: 10.1016/s0960-9822(06)00293-4. [DOI] [PubMed] [Google Scholar]
- Hinds JW, Hinds PL. Early ganglion cell differentiation in the mouse retina: An electron microscopic analysis utilizing serial sections. Dev Biol. 1974;37:381–416. doi: 10.1016/0012-1606(74)90156-0. [DOI] [PubMed] [Google Scholar]
- Hitchcock PF, Macdonald RE, Van de Ryt JT, Wilson SW. Antibodies against Pax6 immunostain amacrine and ganglion cells and neuronal progenitors, but not rod precursors, in the normal and regenerating retina of the goldfish. J Neurobiol. 1996;29:399–413. doi: 10.1002/(SICI)1097-4695(199603)29:3<399::AID-NEU10>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Hogan BLM, Horsburgh G, Cohen J, Hetherington CM, Fisher G, Lyon MF. Small eyes (Sey): a homozygous lethal mutation on chormosome 2 which affects the differentiation of both lens and nasal placodes in the mouse. J Embryol Exp Morphol. 1986;97:95–110. [PubMed] [Google Scholar]
- Hu M, Easter SS. Retinal neurogenesis: the formation of the initial central patch of postmitotic cells. Dev Biol. 1999;207:309–321. doi: 10.1006/dbio.1998.9031. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Grell EH, Ackerman L, Jan LY, Jan YN. atonal is the proneural gene for Drosophila photoreceptors. Nature. 1994;369:398–400. doi: 10.1038/369398a0. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Sun Y, Jan LY, Jan YN. Role of the proneural gene, atonal, in formation of Drosophila chordotonal organs and photoreceptors. Development. 1995;121:2019–2030. doi: 10.1242/dev.121.7.2019. [DOI] [PubMed] [Google Scholar]
- Jeon CJ, Strettoi E, Masland RH. The major populations of the mouse retina. J Neurosci. 1998;18:8936–8946. doi: 10.1523/JNEUROSCI.18-21-08936.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama R, Sasai Y, Akazawa C, Makoto I, Takebayashi K, Shimizu C, Tomita K, Nakanishi S. Regulation of mammalian neural development by helix-loop-helix transcription factors. Crit Rev Neurobiol. 1995;9:177–188. [PubMed] [Google Scholar]
- Kanekar S, Perron M, Dorsky R, Harris WA, Jan LY, Jan YN, Vetter ML. Xath5 participates in a network of bHLH genes in the developing Xenopus retina. Neuron. 1997;19:981–994. doi: 10.1016/s0896-6273(00)80391-8. [DOI] [PubMed] [Google Scholar]
- Kay, J. N., Finger, K. C., Roeser, T., Staub, W. and Baier, H. (2001). Retinal ganglion cell determination requires lakritz, a zebrafish homolog of Drosophila atonal. Neuron (in press). [DOI] [PubMed]
- Koroma BM, Yang JM, Sundin OH. The Pax-6 homeobox gene is expressed throughout the corneal and conjunctival epithelia. Invest Ophthalmol Vis Sci. 1997;38:108–120. [PubMed] [Google Scholar]
- Lawson KA, Dunn NR, Roelen BA, Zeinstra LM, Davis AM, Wright CV, Korving JP, Hogan BL. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev. 1999;13:424–436. doi: 10.1101/gad.13.4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Mo Z, Xiang M. The Ath5 proneural genes function upstream of Brn3 POU domain transcription factor genes to promote retinal ganglion cell development. Proc Natl Acad Sci USA. 2001;98:1649–1654. doi: 10.1073/pnas.98.4.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macabe KL, Gunther EC, Reh TA. The development of the pattern of retinal ganglion cells in the chick retina: mechanisms that control differentiation. Development. 1999;126:5713–5724. doi: 10.1242/dev.126.24.5713. [DOI] [PubMed] [Google Scholar]
- MacNeil MA, Masland RH. Extreme diversity among amacrine cells: implications for function. Neuron. 1998;20:971–982. doi: 10.1016/s0896-6273(00)80478-x. [DOI] [PubMed] [Google Scholar]
- Martinou JC, Dubois-Dauphin M, Staple JK, Rodriguez I, Frankowski H, Missotten M, Albertini P, Talabot D, Catsicas S, Pietra C, Huarte J. Overexpression of Bcl-2 in transgenic mice protects neurons from naturally occurring cell death and experimental ischemia. Neuron. 1994;13:1017–1030. doi: 10.1016/0896-6273(94)90266-6. [DOI] [PubMed] [Google Scholar]
- Masai I, Stemple DL, Okamoto H, Wilson SW. Midline signals regulate retinal neurogenesis in zebrafish. Neuron. 2000;27:251–263. doi: 10.1016/s0896-6273(00)00034-9. [DOI] [PubMed] [Google Scholar]
- Masland RH. Amacrine cells. Trends Neurosci. 1988;11:405–410. doi: 10.1016/0166-2236(88)90078-1. [DOI] [PubMed] [Google Scholar]
- Massey SC, Mills SL. Antibody to calretinin stains AII amacrine cells in the rabbit retina: double-label and confocal analyses. J Comp Neurol. 1999;411:3–18. [PubMed] [Google Scholar]
- Mastick GS, Andrews GL. Pax6 Regulates the Identity of Embryonic Diencephalic Neurons. Mol Cell Neurosci. 2001;17:190–207. doi: 10.1006/mcne.2000.0924. [DOI] [PubMed] [Google Scholar]
- Matter-Sadzinski L, Matter JM, Ong MT, Hernandez J, Ballivet M. Specification of neurotransmitter receptor identity in developing retina: the chick ATH5 promoter integrates the positive and negative effects of several bHLH proteins. Development. 2001;128:217–231. doi: 10.1242/dev.128.2.217. [DOI] [PubMed] [Google Scholar]
- Milam AH, Dacey DM, Dizhoor AM. Recoverin immunoreactivity in mammalian cone bipolar cells. Vis Neurosci. 1993;10:1–12. doi: 10.1017/s0952523800003175. [DOI] [PubMed] [Google Scholar]
- Morrow EM, Belliveau MJ, Cepko CL. Two phases of rod photoreceptor differentiation during rat retinal development. J Neurosci. 1998;18:3738–3748. doi: 10.1523/JNEUROSCI.18-10-03738.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann CJ, Nusslein-Volhard C. Patterning of the zebrafish retina by a wave of sonic hedgehog activity. Science. 2000;289:2137–2139. doi: 10.1126/science.289.5487.2137. [DOI] [PubMed] [Google Scholar]
- Ng L, Hurley JB, Dierks B, Srinivas M, Salto C, Vennstrom B, Reh TA, Forrest D. A thyroid hormone receptor that is required for the development of green cone photoreceptors. Nat Genet. 2001;27:94–98. doi: 10.1038/83829. [DOI] [PubMed] [Google Scholar]
- Nixon RA, Lewis SE, Dahl D, Marotta CA, Drager UC. Early posttranslational modifications of the three neurofilament subunits in mouse retinal ganglion cells: neuronal sites and time course in relation to subunit polymerization and axonal transport. Brain Res Mol Brain Res. 1989;5:93–108. doi: 10.1016/0169-328x(89)90001-6. [DOI] [PubMed] [Google Scholar]
- Perry VH. Evidence for an amacrine cell system in the ganglion cell layer of the rat retina. Neuroscience. 1981;6:931–944. doi: 10.1016/0306-4522(81)90174-3. [DOI] [PubMed] [Google Scholar]
- Portman DS, Emmons SW. The basic helix-loop-helix transcription factors LIN-32 and HLH-2 function together in multiple steps of a C. elegans neuronal sublineage. Development. 2000;127:5415–5426. doi: 10.1242/dev.127.24.5415. [DOI] [PubMed] [Google Scholar]
- Ramón y Cajal, S. (1892). The Structure of the Retina. S. A. Thorpe and M. Glickstein, translators. 1972 edition Springfield, Illinois: Charles C. Thomas.
- Rice DS, Tang Q, Williams RW, Harris BS, Davisson MT, Goldowitz D. Decreased retinal ganglion cell number and misdirected axon growth associated with fissure defects in Bst/+ mutant mice. Invest Ophthalmol Vis Sci. 1997;38:2112–2124. [PubMed] [Google Scholar]
- Rich KA, Figueroa SL, Zhan Y, Blanks JC. Effects of Muller cell disruption on mouse photoreceptor cell development. Exp Eye Res. 1995;61:235–248. doi: 10.1016/s0014-4835(05)80043-0. [DOI] [PubMed] [Google Scholar]
- Rich KA, Zhan Y, Blanks JC. Migration and synaptogenesis of cone photoreceptors in the developing mouse retina. J Comp Neurol. 1997;388:47–63. [PubMed] [Google Scholar]
- Rockhill RL, Euler T, Masland RH. Spatial order within but not between types of retinal neurons. Proc Natl Acad Sci USA. 2000;97:2303–2307. doi: 10.1073/pnas.030413497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodieck, R. W. (1998). The First Steps in Seeing. Sunderland, MA: Sinauer.
- Rubin GM. Development of the Drosophila retina: Inductive events studied at single cell resolution. Cell. 1989;57:519–520. doi: 10.1016/0092-8674(89)90120-7. [DOI] [PubMed] [Google Scholar]
- Sanes JR, Rubenstein JL, Nicolas JF. Use of a recombinant retrovirus to study post-implantation cell lineage in mouse embryos. EMBO J. 1986;5:3133–3142. doi: 10.1002/j.1460-2075.1986.tb04620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling K, Luk D, Morgan JI, Curran T. Regulation of a fos-lacZ fusion gene: a paradigm for quantitative analysis of stimulus-transcription coupling. Proc Natl Acad Sci USA. 1991;88:5665–5669. doi: 10.1073/pnas.88.13.5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidman, R. L. (1961). Histogenesis of mouse retina studied with thymidine-H3. In Structure of the Eye (ed. G. K. Smelser), pp. 487–506. New York: Academic Press.
- Silver J, Sidman RL. A mechanism for the guidance and topographic patterning of retinal ganglion cell axons. J Comp Neurol. 1980;189:101–111. doi: 10.1002/cne.901890106. [DOI] [PubMed] [Google Scholar]
- Snow RL, Robson JA. Ganglion cell neurogenesis, migration and early differentiation in the chick retina. Neuroscience. 1994;58:399–409. doi: 10.1016/0306-4522(94)90046-9. [DOI] [PubMed] [Google Scholar]
- Sokal, R. R. and Rohlf, F. J. (1969). Biometry. The principles and practice of statistics in biological research. pp. 392–4. San Francisco: W. H. Freeman.
- Strettoi E, Masland RH. The number of unidentified amacrine cells in the mammalian retina. Proc Natl Acad Sci USA. 1996;93:14906–14911. doi: 10.1073/pnas.93.25.14906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Jan LY, Jan YN. Ectopic scute induces Drosophila ommatidia development without R8 founder photoreceptors. Proc Natl Acad Sci USA. 2000;97:6815–6189. doi: 10.1073/pnas.110154497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundin OH, Eichele G. A homeodomain protein reveals the metameric nature of the developing chick hindbrain. Genes Dev. 1990;4:1267–1276. doi: 10.1101/gad.4.8.1267. [DOI] [PubMed] [Google Scholar]
- Szel A, Rohlich P, Mieziewska K, Aguirre G, van Veen T. Spatial and temporal differences between the expression of short- and middle-wave sensitive cone pigments in the mouse retina: a developmental study. J Comp Neurol. 1993;331:564–577. doi: 10.1002/cne.903310411. [DOI] [PubMed] [Google Scholar]
- Tomlinson A. Cellular interactions in the developing Drosophila eye. Development. 1988;104:183–193. doi: 10.1242/dev.104.2.183. [DOI] [PubMed] [Google Scholar]
- Turner DL, Snyder EY, Cepko CL. Lineage-independent determination of cell type in the mouse retina. Neuron. 1990;4:833–345. doi: 10.1016/0896-6273(90)90136-4. [DOI] [PubMed] [Google Scholar]
- Waid DK, McLoon SC. Ganglion cells influence the fate of dividing retinal cells in culture. Development. 1998;125:1059–1066. doi: 10.1242/dev.125.6.1059. [DOI] [PubMed] [Google Scholar]
- Waid DK, McLoon SC. Immediate differentiation of ganglion cells following mitosis in the developing retina. Neuron. 1995;14:117–124. doi: 10.1016/0896-6273(95)90245-7. [DOI] [PubMed] [Google Scholar]
- Wang SW, Kim BS, Ding K, Wang H, Sun D, Johnson RL, Klein WH, Gan L. Requirement for math5 in the development of retinal ganglion cells. Genes Dev. 2001;15:24–29. doi: 10.1101/gad.855301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Rutishauser U, Silver J. Formation of retinal ganglion cell and optic fiber layers. J Neurobiol. 1991;22:85–96. doi: 10.1002/neu.480220109. [DOI] [PubMed] [Google Scholar]
- Willbold E, Rothermel A, Tomlinson S, Layer PG. Muller glia cells reorganize reaggregating chicken retinal cells into correctly laminated in vitro retinae. Glia. 2000;29:45–57. [PubMed] [Google Scholar]
- Williams RW, Strom RC, Goldwitz D. Natural variation in neuron number in mice is linked to a major quantitative trait locus on chromosome 11. J Neurosci. 1998;18:138–146. doi: 10.1523/JNEUROSCI.18-01-00138.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang M, Zhou H, Nathans J. Molecular biology of retinal ganglion cells. Proc Natl Acad Sci USA. 1996;93:596–601. doi: 10.1073/pnas.93.2.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang M, Zhou L, Peng YW, Eddy RL, Shows TB, Nathans J. Brn-3b: a POU domain gene expressed in a subset of retinal ganglion cells. Neuron. 1993;11:689–701. doi: 10.1016/0896-6273(93)90079-7. [DOI] [PubMed] [Google Scholar]
- Young RW. Cell differentiation in the retina of the mouse. Anat Rec. 1985;212:199–205. doi: 10.1002/ar.1092120215. [DOI] [PubMed] [Google Scholar]


