Abstract
In many organisms, mitochondria import nuclear DNA-encoded small RNAs. In yeast Saccharomyces cerevisiae, one out of two cytoplasmic isoacceptor tRNAsLys is partially addressed into the organelle. Mitochondrial targeting of this tRNA was shown to depend on interaction with the precursor of mitochondrial lysyl–tRNA synthetase, preMsk1p. However, preMsk1p alone was unable to direct tRNA targeting, suggesting the existence of additional protein factor(s). Here, we identify the glycolytic enzyme, enolase, as such a factor. We demonstrate that recombinant enolase and preMSK1p are sufficient to direct tRNA import in vitro and that depletion of enolase inhibits tRNA import in vivo. Enzymatic and tRNA targeting functions of enolase appear to be independent. Three newly characterized properties of the enolase can be related to its novel function: (1) specific affinity to the imported tRNA, (2) the ability to facilitate formation of the complex between preMsk1p and the imported tRNA, and (3) partial targeting toward the mitochondrial outer membrane. We propose a model suggesting that the cell exploits mitochondrial targeting of the enolase in order to address the tRNA toward peri-mitochondrially synthesized preMsk1p. Our results indicate an alternative molecular chaperone function of glycolytic enzyme enolase in tRNA mitochondrial targeting.
Keywords: Mitochondria, targeting, transfer RNA, enolase, glycolysis, yeast
A double lipid-bilayer membrane protects the mitochondrial matrix, the inner compartment of the mitochondria, where mtDNA replication, transcription, and mRNA translation occur. During a permanent cross-talk between nucleo-cytoplasmic and mitochondrial compartments of the cell, this barrier is crossed by a variety of molecules using controlled and, for the majority of cases, active mechanisms involving multiprotein complexes located at the mitochondrial outer membrane, inner membrane, or at both (Scheffler 2001). Mitochondrial DNA codes for only a small subset of organellar macromolecules, while the majority of proteins and some small RNAs are expressed from nuclear genes and targeted into mitochondria (Neupert 1997; Entelis et al. 2001b).
RNA import, far less understood than the protein mitochondrial import, may now be considered quasi-universal. Import of nuclear-encoded RNA into mitochondria has been identified in protozoans, fungi, animals, and plants; however, the type and number of imported RNAs vary from species to species (Schneider and Marechal-Drouard 2000; Entelis et al. 2001b). Among extensively studied cases, it appears that RNA import mechanisms are similar in human (5S rRNA) and yeast (tRNA) but differ from those operating in trypanosomes and plants (Mahapatra and Adhya 1996; Entelis et al. 2001a; Delage et al. 2003). One notable difference concerns the need of soluble (cytosolic) proteins to achieve the import. In fact, in our experimental systems with yeast and human isolated mitochondria, the presence of such factors was essential, while for plant or trypanosomal mitochondria the presence of soluble factors was dispensable in the in vitro assay. On the other hand, numerous indirect data favor the idea that such factors may be recruited in vivo to provide the selectivity of RNA targeting in plant or protozoan mitochondria (Dietrich et al. 1996; Rusconi and Cech 1996a, b; Schneider and Marechal-Drouard 2000). Although these pathways are diverse, their physiological significance and molecular mechanisms remain largely unknown.
In the yeast Saccharomyces cerevisiae, two nuclear DNA-encoded tRNAs were reported as mitochondrially targeted, tRNAGln (Rinehart et al. 2005) and tRNALysCUU (further referred to as tRK1) (Martin et al. 1979), whose import appears to proceed by different mechanisms. Mitochondrial functions of tRK1 are not completely clear, although our previous results demonstrated that its mutant versions are functional in mitochondrial translation, since they are able to suppress a nonsense mutation in mitochondrial DNA-localized gene (Kolesnikova et al. 2000). tRK1 import is highly specific, since the second cytosolic tRNALysUUU (tRK2) is not imported (Entelis et al. 1998). The mitochondrial targeting process requires interaction of the aminoacylated form of tRK1 with the cytosolic precursor of mitochondrial lysyl–tRNA synthetase (preMsk1p) (Tarassov et al. 1995a). Our hypothesis suggested that preMsk1p, which is able to recognize aminoacylated tRK1, targets this RNA to the outer mitochondrial membrane and can function as a tRNA mitochondrial carrier. Nevertheless, preMsk1p is not the only protein needed, because alone it was not able to direct the tRK1 import in vitro.
Here we present results of a search for other proteins implicated in the tRK1 mitochondrial import process. Unexpectedly, we identified a glycolytic enzyme, enolase, as a factor of tRK1 import. We show that this protein is able to recognize the imported tRNA and favors its binding to preMsk1p prior to the import. We also found that this enzyme, responsible for the last but one step of glycolysis, is partially targeted to the outer membrane surface. This mitochondrial targeting, probably aimed to compartmentalize glycolytic reactions, could also be exploited for tRK1 delivery toward the mitochondria. These findings give new insights into the mechanisms of RNA mitochondrial import and establish a link between two pathways previously thought to be independent.
Results
Identification of enolase as a factor of tRNA mitochondrial import
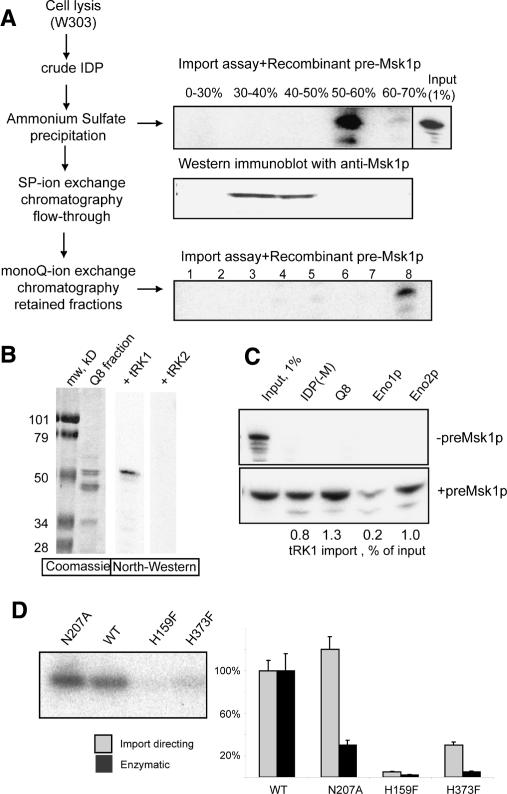
To search for proteins implicated in tRNA mitochondrial import, a crude protein extract from wild-type yeast strain W303 was separated by differential ammonium sulphate (SA) precipitation. Fractions were tested for their ability to direct the import of tRK1 into isolated mitochondria using 5′-[32P]-labeled and aminoacylated tRK1 (Fig. 1A). The proteins precipitated by 30%–40% and 40%–50% SA, contained a known import factor, preMsk1p (identified by Western analysis) (Fig. 1A), and were unable to direct tRK1 mitochondrial import. In contrast, proteins precipitated by 50%–60% SA did not contain any detectable preMsk1p though were active in the tRK1 mitochondrial import assay in the presence of recombinant preMsk1p. This protein fraction was subjected to further fractionation by chromatography on Sepharose SP and Mono-Q. One of the Mono-Q-retained fractions, Q8, which demonstrated a high yield of import-directing activity, still contained several proteins. North–Western analysis revealed a single protein of apparent molecular weight of 49 kDa interacting with [32P]-labeled aminoacylated tRK1 (Fig. 1B). This interaction was specific, since neither tRK2 nor deacylated tRK1 interacted with this protein. The 49-kDa protein was identified by N-terminal sequencing as the glycolytic enzyme, enolase (2-phospho-D-glycerate hydrolyase, EC 4.2.1.11).
Figure 1.
Identification of import factors. (A) Strategy of proteins fractionation. (IDP) Protein extract. (Top and bottom right panels) In vitro import assays and autoradiographs of protected tRK1 after the incubation with isolated yeast mitochondria. (Input) An aliquot (1%) of labeled tRK1 used in each import assay. Ammonium sulphate saturation percentage and the numbers of mono-Q fractions are indicated. The middle panel represents results of Western immunodetection of preMsk1p in the same ammonium sulphate fractions as above. (B) North–Western analysis of the proteins from the mono-Q fraction 8 (Q8). The left panel is the Coomassie-stained gel, and the two panels on the right show autoradiographs after incubation of proteins with 5′-[32P]-labeled tRK1 or tRK2. (C) In vitro tRK1 import assay with fraction Q8 proteins and recombinant Eno1p or Eno2p in the presence or absence of preMsk1p. [IDP(−M)] IDPs lacking preMsk1p. tRK1 import efficiencies are indicated below the autoradiograph. (D) Comparison of enzymatic and tRK1 import-directing activities of a normal (wild-type [WT]) and mutant recombinant versions of Eno2, containing the following substitutions: N207A, H159F, or H373F. On the left is the in vitro import assay in the presence of recombinant preMsk1p, with Eno2p versions indicated above; on the right is the quantification of import-directing efficiency and enolase enzymatic activity of the recombinant proteins.
Yeast cells contain two enolase isoforms: Eno1p encoded by the gene YGR254w, and Eno2p by YHR174w. They differ by 20 out of 437 amino acids and are characterized by different isoelectric points (pI) (6.6 and 5.9, respectively) and different modes of transcriptional regulation (McAlister and Holland 1982; Pancholi 2001). To demonstrate the participation of enolase in tRK1 import and to compare the import-directing capacities of the two isozymes, we overexpressed Eno1p and Eno2p in Escherichia coli. Tag-purified recombinant Eno2p was able to direct tRK1 import in vitro in the presence of preMsk1p (Fig. 1C), whereas Eno1p showed a significantly lower import-directing ability (10%–20% with respect to Eno2p). No tRK1 import was detected in the absence of preMsk1p. “Mock-import” tests without mitochondria allowed excluding artifactual protection of the tRNA by the recombinant proteins. These experiments demonstrate that Eno1/2p and preMsk1p are sufficient to direct tRK1 import in vitro and constitute a minimal tRNA import vehicle.
Determination of recombinant Eno1p and Eno2p enzymatic activities showed very similar specific activities for the two isozymes (53 U/mg for Eno1p and 58 U/mg for Eno2p, comparable to 65 U/mg for commercial bakers’ yeast enolase). In contrast, their abilities to direct tRK1 import were different, Eno1p being significantly less efficient than Eno2p. This suggests that their participation in glycolysis and in tRK1 import is distinct and unrelated functions. The absence of correlation between two enolase functions was further supported using several mutant versions of Eno2p. It has been previously shown that mutations N207A, H159F, and H373F in Eno1p affected its catalytic activity in a differential manner (Brewer et al. 2003). We introduced the same mutations in the Eno2p and found that the recombinant mutant proteins indeed demonstrated reduced enzymatic activity: 13 U/mg (22% of the wild-type protein activity) for the N207A version, 0.6 U/mg (1%) for H373F, and 0.2 U/mg (0.3%) for the H159F version. In contrast, the Eno2(N207A)p was as efficient as the wild-type Eno2p in tRK1 import (Fig. 1D). Eno2(H159F)p had completely lost the import-directing capacity while Eno2(H373F)p had intermediate import activity (30% vs. the wild-type Eno2p). It appears therefore that there was no direct relation between the enzymatic enolase activity and its tRK1 import-directing capacity.
To further investigate the mechanism of enolase participation in the tRNA import process, we have measured the affinity of enolase to imported and nonimported tRNAs and studied its intracellular localization.
Eno2p binds to tRK1 and enhances formation of tRK1–preMsk1p complex
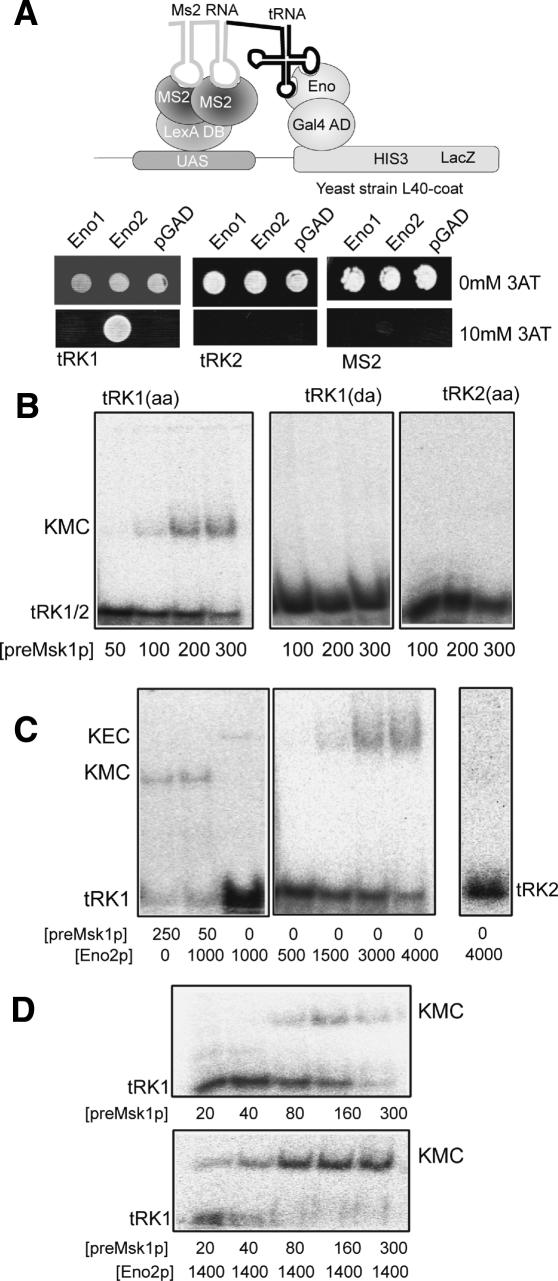
The first evidence of RNA-binding properties of enolase was obtained by using the North–Western analysis (Fig. 1B). To verify if tRK1–Eno2p interaction can also occur in vivo, we used the three-hybrid system (SenGupta et al. 1996). To this end, two hybrid proteins were expressed in yeast: The first consisted of the DNA-binding domain of LexA fused to the MS2 coat protein, the second contained the Gal4 transcription activation domain fused to either Eno1p or Eno2p (Fig. 2A). The two fusion proteins can be bridged by a bifunctional hybrid RNA molecule containing a fragment of bacteriophage MS2 RNA and tRK1 (or tRK2), providing that the tRNA moiety can interact with enolase. Only tRK1 revealed a positive reaction with Eno2p in the presence of 10 mM 3-amino-triazole (3-AT) in the medium (Fig. 2A), suggesting that enolase has RNA-binding capacity in vivo. This interaction proved to be specific, since neither tRK2 (which is not imported) nor MS2 RNA alone interacted with Eno2p. Furthermore, no interaction of tRK1 with Eno1p was detected, even at a lower concentration of 3-AT (2 mM). This was in agreement with our in vitro results and suggests that the capacity of Eno1p to direct tRK1 import is significantly lower than that of Eno2p.
Figure 2.
Interaction of Eno2p with tRK1. (A) Detection of the interaction between Eno1/2p and tRK1 in vivo by the three-hybrid approach. The strategy is presented in the upper panel. MS2 RNA-fused tRK1 or tRK2 (nonimported RNA) were assayed for interaction with either Eno1p or Eno2p. Empty MS2 RNA gene-containing vector (MS2) and empty pGAD-GH plasmid (pGAD) were used as controls. To detect interaction, yeast L40-coat strain transfected cells were grown in the absence or presence of 10 mM 3AT, as indicated. (B–D) Analysis of RNA–protein interactions by gel-shift. Autoradiographs of native PAGE-separations of labeled tRK1 in the presence of preMsk1p (B) and/or Eno2p (C). Deacylated tRK1 [tRK1(da)] and aminoacylated tRK2 [tRK2(aa)] were used as controls of the specificity of interaction. Concentrations of the recombinant proteins in nM are indicated below the panels. (KMC) tRK1–preMsk1 complex; (KEC) tRK1–Eno2p complex; (tRK1) unbound labeled tRNA. (D) The effect of Eno2p presence on tRK1–preMsk1p complex formation.
Using gel-retardation assay with labeled tRK1 and increasing amounts of recombinant proteins, we observed that aminoacylated tRK1 could form a stable complex with both preMsk1p (Fig. 2B) and Eno2p (Fig. 2C). In both cases, the interaction was tRK1-specific, since neither tRK2 nor deacylated tRK1 formed any complex. Similarly, no complexes were detected between tRK1 and Eno1p (data not shown). tRK1 interacted with preMsk1p with an apparent dissociation constant Kd = 180 ± 20 nM (dimer concentration), comparable with Kd reported for complexes of lysyl–tRNA synthetases (LysRS) with noncognate tRNAs (Francin et al. 2002). The affinity of tRK1 to Eno2p (apparent Kd = 2.5 ± 0.2 μM) was one order lower than preMsk1p. When both preMsk1p and Eno2p were present in the mixture, no ternary complex tRK1/preMsk1p–Eno2p was detected (Fig. 2C, left panel). Instead, in the presence of very low preMsk1p concentration, the tRK1/Eno2p complex was rapidly shifted to the tRK1/preMsk1p one. The presence of Eno2p significantly increased the efficiency of tRK1/preMsk1p complex formation (apparent Kd = 40 ± 10 nM opposed to Kd = 180 nM in the absence of enolase) (Fig. 2D).
One can therefore conclude that Eno2p could specifically bind the imported tRNA and enhance formation of the complex between this tRNA and its import carrier, preMsk1p.
Eno2p is partially localized on the surface of mitochondria
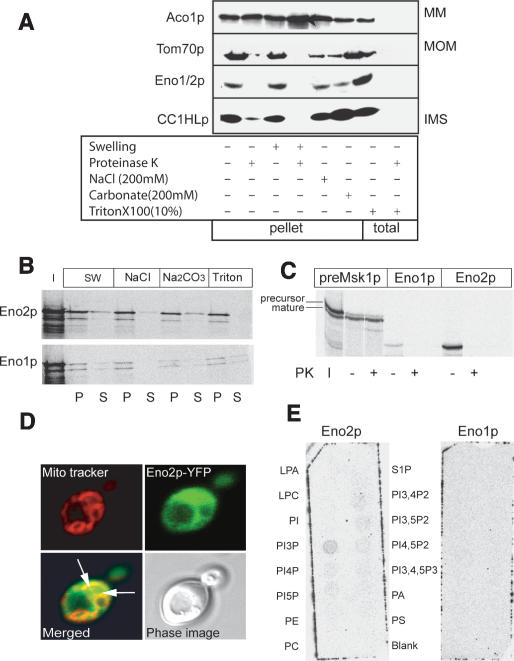
We have observed that 2%–7% of the total enolase glycolytic activity was copurified with mitochondria (data not shown). To verify if this copurification was not artifactual, we analyzed the cellular localization of Eno2p in details (Fig. 3). The submitochondrial localization of enolase was determined using antibodies against proteins of different mitochondrial compartments (Diekert et al. 2001). Proteins of mitochondrial pellets, after different treatments of mitochondria, were gel-separated and immunodetected (Fig. 3A). The copurification of enolase with mitochondria was resistant to swelling (hypotonic shock) and to treatment with NaCl or Na2CO3, but enolase was completely released from mitochondria by proteinase K treatment. In this respect, enolase behaves similarly to the outer membrane receptor Tom70p.
Figure 3.
A part of Eno2p colocalizes with mitochondria. (A) Western analyses of mitochondrially associated proteins. After the treatment indicated below the picture, the mitochondrial pellets or the total material were analyzed, as indicated. The antibodies used were Aco1p, aconite hydratase, 85 kDa, matrix (MM) localization; Tom70p, 70 kDa, outer membrane (MOM) associated; and CCH1HLp, Cytochrome c heme lyase, 30 kDa, inner membrane space (IMS) protein. Aco1p and CCH1HLp are known to be resistant to carbonate treatment (Diekert et al. 2001). (B,C) In vitro assay of mitochondrial localization using reticulocyte lysate-synthesized [35S]-labeled proteins incubated with isolated organelles. (B) Autoradiographs of gel-fractionated proteins after treatment of mitochondria as indicated. (I) Input; (SW) swelling; (NaCl) treatment with 200 mM of salt; (Na2CO3) washing with 200 mM carbonate; (Triton X-100) treatment with 10% detergent; (P) pellet; (S) supernatant. (C) Autoradiograph of the gel-fractionated proteins after treatment of the mitochondria with 50 μg/mL of proteinase K (PK). Mitochondrially imported preMsk1p was used as a reference. (D) Analysis of Eno2p localization in yeast cells by confocal microscopy. Eno2p was YFP-labeled, mitochondria were localized by mito-tracker red. Zones of colocalization are indicated by white arrows on the merge image. (E) Assay for Eno1/2p interaction with immobilized phospholipids. (LPA) Lisophosphatidic acid; (LPC) lisophosphocholine; (PI) phosphatidyl-inositol; (PI3/4/5) phosphatidyl-inositol-3/4/5-phosphate; (PE) phosphatidylethanolamine; (PC) phosphatidylcholine; (S1P) sphingosine-1-phosphate; (PI3, 4/3,5/4,5P2) phosphatidyl-inositol-3,4/3,5/4,5-diphosphate; (PI3, 4,5P3) phosphatidyl-inositol-3,4,5-triphosphate; (PA) phosphatidic acid; (PS) phosphatidylserine.
To further investigate the submitochondrial localization of the Eno1p and Eno2p isozymes, we used an assay of protein import into isolated mitochondria (Diekert et al. 2001; Ryan et al. 2001). For this purpose, [35S]-labeled enolases were incubated with isolated yeast mitochondria in conditions permitting protein import (preMsk1p, an imported mitochondrial protein, was used as a control) (Fig. 3B,C). As expected, preMsk1p was protected from proteinase K degradation and processed. Eno2p was not protected from proteinase K but was partially (10%–15% of input) associated with the mitochondrial pellet. Eno1p was also partially copurified with mitochondria, but to a lower extent (1%–2%). The association of enolase with mitochondria was resistant to NaCl treatment and was only partially disrupted by Na2CO3.
To investigate if the in vitro and mitochondrial fractionation data correlate with the in vivo situation, we analyzed subcellular localization of Eno2p after expression of the fusion of the ENO2 gene with yellow fluorescent protein (YFP) by confocal microscopy. As stressed above, only a small portion (<7%) of cellular enolase activity was associated with mitochondria, so it was expected that most of the protein would be distributed in the cytoplasm. It was indeed the case: We reproducibly observed nonhomogenous cytosolic distribution of YFP-Eno2p, while a fraction of enolase colocalized with the mitochondria, which became detectable on the merge image (Fig. 3D). Therefore, in vivo and in vitro data are not contradictory and support the hypothesis that a portion of the enolase is associated with mitochondria.
Since soluble or peripheral membrane proteins can usually be released from mitochondria by treatment with sodium carbonate in alkaline conditions (Diekert et al. 2001; Ryan et al. 2001), our results indicate that enolase could be anchored in the outer mitochondrial membrane. This anchoring may proceed either by interaction with other proteins or by direct interaction with the lipids of the mitochondrial membrane. Inspection of Eno1/2p amino acid sequences revealed the presence of potential transmembrane domains at positions 33–44 (Pancholi 2001) and 101–130 (Tmap program EMBOSS). To study if enolases possess any affinity to the lipids present in the mitochondrial membranes, we assayed the interaction of [35S]-labeled proteins with a set of membrane-immobilized lipids (Fig. 3E). Indeed, we found that Eno2p (but not Eno1p) has a specific affinity to phosphatidyl-inositol-3-phosphate (PI3P). No interaction with major mitochondrial phospholipids (phosphatidylinositol, phosphatidylcholine, phosphatydylserine, and phosphatidylethanolamine) (Fig. 3E) or with cholesterol or sphingomyelin (data not shown) was detected.
These results do not provide definitive proof by which mechanism enolase is anchored in the outer membrane; however, they strongly indicate that this protein might be tightly bound to mitochondrial surface. For Eno2p, the affinity to the mitochondrial surface was shown to be at least 10-fold higher than for Eno1p.
Inactivation of the two S. cerevisiae ENO genes impairs tRK1 import in vivo
To investigate the involvement of the two enolase isoforms in tRK1 import in vivo, we analyzed a yeast strain with deletions of the two ENO genes. A double-deletion strain was obtained by mating eno1 and eno2 haploids and subsequent tetrad analysis and was tested for mitochondrial phenotype (growth on a nonfermentable carbon source), respiration (measurement of oxygen consumption), and for the presence of tRK1 in the mitochondria (Northern hybridization).
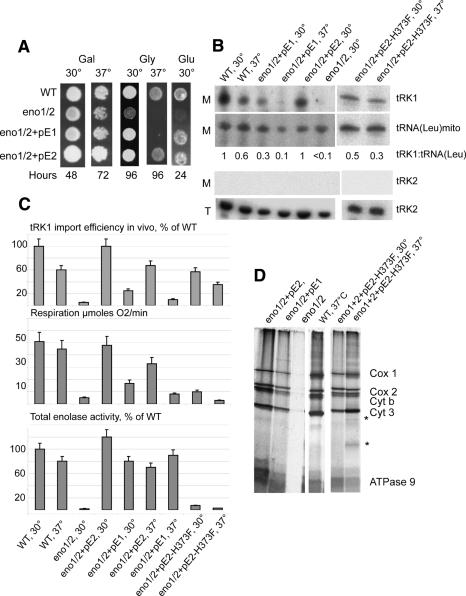
The double deletant strain eno1 eno2 (eno1/2) was characterized by the absence of growth on glucose-containing media and by a poor growth on glycerol at 30°C while, at 37°C, it failed to grow (Fig. 4A). Concomitantly, tRK1 was undetectable in the mitochondria of this strain (Fig. 4B). Glucose-positive growth could be restored by expression of either Eno1p or Eno2p from centromeric plasmids (pE1 or pE2), which correlated with restoration of the enolase enzymatic activity to a nearly wild-type level (Fig. 4A,C).
Figure 4.
Effect of ENO1/2 genes inactivation on tRK1 import and mitochondrial functions. (A) The effect of ENO1 and ENO2 genes deletion on yeast cells growth on nonfermentable carbon source (Glycerol [Gly] YPEG medium), YPGal medium (Gal), or YPD (Glu). Culture growing temperature is indicated above the panel, the duration of growing, at the bottom (hours). (WT) Wild-type strain W303; (+pE1 and +pE2) strains expressing Eno1p or Eno2p from centromeric plasmids pE1 or pE2. (B) The effect of ENO2 gene deletion or mutation on tRK1 import in vivo. Northern hybridization detection of tRK1 and control tRNAs [mitochondrial tRNA(Leu) and cytoplasmic nonimported tRNA(Lys), tRK2] are presented. On the left, M indicates RNA isolated from purified mitochondria, and T indicates total RNA. Strains and temperature of cultivation are indicated on top. (eno1/2) eno1 eno2 deletant strain; (pE2-H373F) plasmid expressing Eno2p with substitution H373F. Equal amounts of RNAs were analyzed for each strain. The ratio between the signals for tRK1 and mt tRNA (Leu) used as a criterion of import efficiency is presented in the middle. (C) Quantification of tRK1 import efficiency, respiration, and enolase enzymatic activity in the crude cell lysates of different strains (listed on the left side of the panel). (D) Effect of ENO2 gene deletion or mutation on mitochondrial translation. The autoradiograph of the PAGE-separated [35S]-labeled mitochondrially synthesized polypeptides is presented. Strains and the temperature of incubation during the translation reaction are indicated above. Polypeptides were identified by comparison with standard mitochondrial translation pattern (McKee et al. 1984). (Cox 1, 2, and 3) Subunits 1–3 of cytochrome c oxidase; (Cyt b) cytochrome c; (ATPase 9) subunit 9 of ATP synthase. Additional bands appearing at 37°C with the H373F Eno2p version are indicated with the asterisks.
In contrast, the thermosensitive growth on glycerol of the double deletant was complemented only by plasmid-driven expression of ENO2 but not of ENO1. Likewise, the low level of oxygen consumption of this strain was cured by ENO2 expression but only partially by ENO1 expression. Although wild-type levels of enolase enzymatic activity were reached in eno1/2 + pE1 cells, the absence of Eno2p was manifested by a slow growth on glycerol and by a decreased level of oxygen consumption, and this correlated with a low level of imported tRK1 at 30°C. At elevated temperature, strain eno1/2 + pE1 did not grow on glycerol media and was characterized by an extremely low level of respiration and by the absence of tRK1 in the mitochondria (Fig. 4A–C). We can conclude that Eno2p is the enolase isoform primarily involved in tRK1 import in vivo. In the absence of Eno2p, Eno1p can only partially substitute for the Eno2p function in tRK1 import, and this conclusion stands in line with our in vitro data.
To investigate if the reduced tRK1 import was related to the organellar translation, we compared patterns of mitochondrially synthesized polypeptides in the deletant strains. Nondetectable or a very low level of translation was found in organelles from the double deletant eno1 eno2 cells, while in Eno1p bearing cells mitochondrial translation was reduced with respect to the wild-type cells (Fig. 4D). These data suggest that the imported tRNA has a role in organellar protein synthesis. However, it could also mean that defects in mitochondrial import and translation in eno1/2 and eno1/2 + pE1 strains were due to secondary defects in respiration. To rule out this possibility, we tested Eno2p with H373F mutation, characterized in vitro by a very low enzymatic activity, but relatively good import-directing capacity (see above, Fig. 1D). Expression of the Eno2p–H373F in eno1/2 strain resulted, as expected, in a dramatically decreased enolase enzymatic activity in the cells (9 ± 3% of the wild-type activity at 30°C and <5% at 37°C) and in a low level of respiration (20 ± 4% and 9 ± 2%, respectively) (Fig. 4C). In contrast, tRK1 mitochondrial import was partially restored (58 ± 5% and 37 ± 4%, respectively) as well as the overall mitochondrial translation (Fig. 4B,D). Indeed, at 30°C mitochondrial translation pattern appeared as normal, while at 37°C we observed the appearance of minor additional bands, which may be explained by abortive termination of translation due to reduced tRK1 import at elevated temperature.
We observe therefore that mitochondrial translation is inhibited or perturbed when ENO2 gene is deleted and that this effect coincides with the inhibition of tRK1 import and not with the respiration defects. Furthermore, the in vivo data confirms the results of in vitro experiments and suggest that the two enolase functions (enzymatic and tRNA import) are distinct.
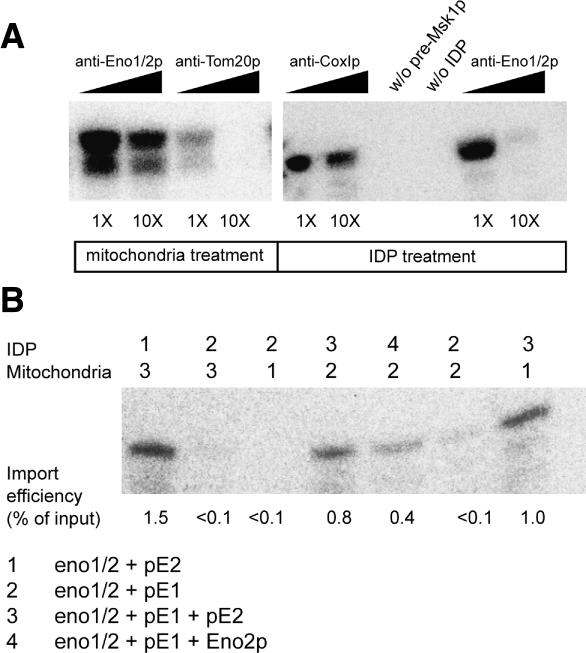
The soluble pool of Eno2p is required for tRK1 import
The fact that a fraction of cellular enolase is bound to the mitochondrial surface raises a question: Does the organelle-bound form of enolase participate in tRNA mitochondrial import? To this end, we tested the capacity of isolated mitochondria to import tRK1 after pretreatment of the organelles with anti-enolase antibodies. Such a treatment had no effect on tRK1 import, contrary to the anti-Tom20p antibodies, which, by obstructing the general insertion pore, were shown to inhibit the import (Tarassov et al. 1995b). In contrast, anti-enolase pretreatment of the soluble import-directing proteins (IDP) inhibited their tRNA import-directing capacity in a dramatic way (Fig. 5A). These results were confirmed by in vitro import assays with IDP or mitochondria isolated from eno1/2 + pE1 and eno1/2 + pE2 yeast strains (Fig. 5B). IDP and mitochondria from eno1/2 + pE2 strain were both active in tRK1 in vitro import. Mitochondria isolated from eno1/2 + pE1 cells had an import capacity comparable to the wild-type ones. In contrast, the IDP from this strain were unable to direct tRK1 import into mitochondria of all strains tested. Addition of purified recombinant Eno2p to IDP from eno1/2 + pE2 cells restored their import capacities. We therefore concluded that the soluble form of Eno2p is needed for tRK1 import.
Figure 5.
Soluble pool of Eno2p is required for tRK1 import. (A) Effect of antibodies on tRK1 mitochondrial import. The antibodies used are indicated at the top. As 1× concentration, we used the concentration of antibodies with the same immunological capacity (determined by Western analysis). The left panel corresponds to tRK1 import into antibody pretreated mitochondria; the right panel corresponds to tRK1 import directed by immunodepleted IDP. (B) tRK1 import assays directed by IDP isolated from yeast strains indicated at the first line on the top (see explanations below the panel) into mitochondria isolated from different strains (second line). (Bottom) Efficiency of tRK1 import. (+Eno2p) Addition of 0.1 μg of recombinant Eno2p to the IDP of the strain indicated.
What is the role of soluble enolase in tRK1 import? It has been shown that the mRNAs of numerous mitochondrially addressed proteins are translated in the vicinity of the mitochondria to facilitate their compartmentalization (Marc et al. 2002). This may also be the case for preMsk1p, which is essential for mitochondrial translocation of tRK1. If so, this precursor should not be available in the cytosol for tRK1 binding. Thus, soluble enolase, which was shown to have tRK1-binding capacity and ability to enhance tRK1–preMsk1p complex formation, may serve as a vehicle to address the tRNA toward the mitochondria where the main protagonists of tRK1 import form the “import-directing” complex.
To test this hypothesis, we have quantified the mRNAs for preMsk1p and Eno1/2p in mitochondria-associated and cytosolic polysomes. For this purpose, RNA was isolated from whole cells, cytosolic polysomes, and mitochondria after treatment of the cells with cycloheximide, which permits the fixing of ribosomes in their respective subcellular compartments (Corral-Debrinski et al. 2000). Real-time RT–PCR was next used to measure the amounts of mRNAs coding for Eno1/2p, preMsk1p, and two control proteins, Krs1p (cytosolic lysyl–tRNA synthetase) and Atp1p (previously reported to be peri-mitochondrially translated (Fig. 6; Marc et al. 2002). The percentage of mitochondria-associated mRNAs, compared with either cytosolic polysomes or total cell mRNA, was calculated. As expected, the Atp1 p.m.RNA was found to be associated with mitochondria to an extent of up to 45%. This value is in agreement with the previously reported one (Marc et al. 2002) and reflects the fact that a portion of mitochondria-bound polysomes were released during mitochondria isolation. Up to 35% of the preMsk1 p.m.RNA was found to be mitochondria-associated, which is comparable to the Atp1 p.m.RNA. In contrast, for Eno1/2 p.m.RNAs and Krs1p the percentage of mitochondrial association was <2%, which suggests cytosolic translation (Fig. 6). Withdrawing of cycloheximide during the isolation of mitochondria (expected to lead to a dissociation of the cytosolic ribosomes from mitochondria) results in a release of the most of mitochondrially associated preMsk1 p.m.RNA. This strongly suggests that the mitochondrially associated portion of preMsk1 p.m.RNA is a part of translationally active polysomes.
Figure 6.
Translation of MSK1 mRNA is compartmentalized. Quantification results obtained after RT–qPCR measurement of mRNAs coding for Atp1 (first subunit of the mitochondrial ATP synthase), Krs1 (cytoplasmic lysyl–tRNA synthetase), Msk1 (preMsk1p), and Eno1/2 (enolase 1 and 2; oligonucleotide primers used do not distinguish between the two mRNAs). To obtain the indicated values, the absolute amounts of mRNAs were first calculated using purified T7-transcripts corresponding to each analyzed mRNA, then the ratio between each mRNA amount in the mitochondrially associated RNA pool and in the total RNA was calculated. In the control experiment, the isolation of mitochondria for RNA purification was performed in the absence of cycloheximide (−CYH).
The mRNA quantification experiments led us to conclude that preMsk1 p.m.RNA localized mainly to mitochondria-associated polysomes, while Eno1/2 p.m.RNAs were translated by cytoplasmic ones. This result can explain the fact that the soluble form of enolase is needed for tRK1 import. The subpopulation of soluble enolase that is addressed toward the mitochondrial surface may be exploited to address the tRNA toward the organelle.
Discussion
Enolase, another protein with multiple functions
The total number of protein-coding genes does not increase from organism to organism commensurately with the increase in complexity of those organisms. One possibility to explain the relative dearth of protein-coding genes is that many proteins may have previously unsuspected or unappreciated “second jobs,” a situation commonly termed as “protein moonlighting.” An increasing number of proteins are found to have two or more different functions, which can be switched as a consequence of changing their cellular localization, oligomeric state, or ligand concentration (Jeffery 1999). Several glycolytic enzymes were also found to have additional functions. For example, glyceraldehyde-3-phosphate dehydrogenase acts as an uracil–DNA glycosylase, an activator of transcription, and is involved in tubulin assembly and in the regulation of apoptosis; lactate dehydrogenase has a DNA-unwinding activity and is implicated in the regulation of mRNA turnover (Pioli et al. 2002); phosphoglucose isomerase can be secreted by mammalian cells and plays a role of neuroleukin (Jeffery 1999).
Enolase is also supposed to have additional nonglycolytic functions. In yeast, it was identified as Hsp48p and was also found in association with the cell wall (Edwards et al. 1999). Enolase was identified as tau-crystallin of turtle eye lens and as a strong plasminogen-binding receptor expressed on the surface of a variety of eukaryotic cells (for review, see Pancholi 2001). Finally, in E.coli, enolase is an integral component of the RNA degradosome (Kuhnel and Luisi 2001; Carpousis 2002). Its role was hypothesized to direct subcellular compartmentalization of degradosome to the plasma membrane (Liou et al. 2001; Morita et al. 2004).
We show here that, in addition to its principal role in glycolysis, enolase allows addressing of a cytosolic tRNA toward the mitochondrial surface, possibly acting as an RNA chaperone. Although the two yeast enolase isoforms lack any known RNA-binding motif (analyzed by PROSITE), Eno2p can specifically bind the imported tRK1. Interaction of yeast Eno1/2p with single-stranded DNA was reported previously (al-Giery and Brewer 1992). Numerous other proteins, including glycolytic enzymes, were already found to have RNA- or tRNA-binding capacities (Singh and Green 1993; Hentze 1994; Bringaud et al. 1997; Pioli et al. 2002). In the majority of cases, there was no direct evidence of the functional importance of these RNA-binding capacities of well known enzymes, but additional roles in RNA turnover and compartmentalization were suggested.
Besides the specific affinity to the imported tRNA, Eno2p was found here to have the capacity to reduce the Kd of tRK1–preMsk1p complex formation. At the same time, in the presence of both proteins, no ternary complex was observed, and only tRK1–preMsk1p was detectable (Fig. 4C,D). As we suggested previously (Entelis et al. 1998; Kolesnikova et al. 2002), formation of this latter complex requires some conformational changes in the tRNA molecule. Now we can hypothesize that Eno2p induces these conformational changes in the tRNA or stabilizes the conformation of the tRNA that is better recognized or is more tightly bound by preMsk1p. If this hypothesis reveals true, Eno2p function in tRNA mitochondrial import would be not only as targeting factor, but also as an RNA-chaperone.
We have also found that a portion of the cellular enolase pool is tightly bound to the outer mitochondrial membrane. For the moment, we cannot be sure about the mechanism by which the enolase molecule is bound to the mitochondria. We show that it can specifically recognize PI3P. However, PI3P was not found in association with mitochondrial membranes in yeast (Gillooly et al. 2000). On the other hand, no interaction was detected with phospholipids and sphingolipids that are abundant in the outer mitochondrial membrane (Ardail et al. 1990). These results likely indicate an indirect interaction of the enolase with mitochondrial surface, which may be mediated by other proteins. What might be the role of the partial localization of enolase on the mitochondrial surface? In fact, enolase as well as other glycolytic enzymes have already been detected in association with mitochondria (Giege et al. 2003; Taylor et al. 2003). In Arabidopsis cells, the entire glycolytic pathway (3%–12% of enzyme activities) was localized on the cytosolic face of the outer membrane (Giege et al. 2003). We have also shown the presence of all the glycolytic enzymes activities on the surface of mitochondria in yeast and found that enolase takes part in a mitochondrial outer membrane-associated macromolecular complex (I. Brandina, J. Graham, C. Lemaitre-Guillier, N. Entelis, I. Krasheninnikov, L. Sweetlove, I. Tarassov, and R.P. Martin, in prep.). Glycolysis resumes in a pyruvate molecule, which is imported into mitochondria to serve as a substrate for respiration. The compartmentalization of this pathway may allow direct delivery of pyruvate to the site of its consumption. We hypothesize that yeast cells exploited the RNA-binding properties and the mitochondrial localization of enolase for the targeting of tRK1 toward the mitochondrial surface.
The mechanism of enolase action in tRNA import
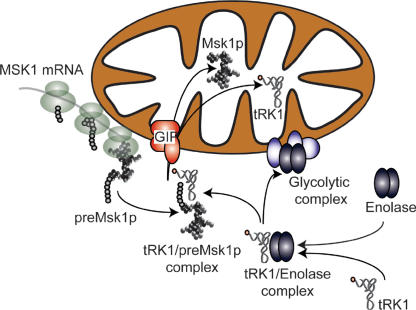
tRK1 mitochondrial import in yeast requires stable and specific interaction of the aminoacylated tRNA with the cytoplasmic precursor of mitochondrial lysyl–tRNA synthetase (preMsk1p), which is likely to act as a carrier for its mitochondrial transport (Tarassov et al. 1995a). Experiments of quantification of mRNA subcellular pools presented above permit us to hypothesize that the mRNA of preMsk1 is mainly translated peri-mitochondrially, which would facilitate mitochondrial import of this preprotein. This implies that there must be an extremely low amount of this precursor available in the cytoplasm. On the other hand, tRK1 takes part in the cytoplasmic translation and thus is expected to localize mainly in the vicinity of cytoplasmic polysomes. Therefore, to meet one another, tRK1 and preMsk1p need an additional targeting factor, enolase, to serve as a vehicle to deliver the tRNA to newly synthesized preMsk1p molecules at the mitochondrial surface (see Fig. 7).
Figure 7.
Proposed mechanism of enolase involvement in tRK1 mitochondrial targeting. GIP indicates the general insertion pore, the outer membrane preprotein import complex composed of TOM proteins. Aminoacylated tRK1 is recognized by enolase (most probably Eno2p) in the cytoplasm and addressed toward the mitochondrial surface, where the tRNA is transferred to the peri-mitochondrially synthesized preMsk1p, while enolase inserts in the hypothetical glycolytic multiprotein complex, whose role may be the channelling of the pyruvate to the organelle. Further targeting of tRK1 to the mitochondrial matrix involves the functional GIP (Tarassov et al. 1995b).
We found that masking of membrane-bound enolase with antibodies did not affect tRK1 mitochondrial import, while withdrawing the soluble portion of enolase from IDPs abolished it. We suggest that enolase can bind tRK1 in the cytosol and deliver it toward the mitochondrial surface. Enolase would then favor tRK1/preMsk1p complex formation for translocation into the mitochondrial matrix (Fig. 7). This model is in agreement with the fact that tRK1 has higher affinity to preMsk1p than to enolase (taking into account the 14-fold decrease in apparent Kd, the difference in free energy of binding is 1.6 kcal/mol), and in the presence of both proteins only the preMsk1p/tRK1 complex is formed. Besides preMsk1p and Eno2p, additional proteins may participate in tRK1 mitochondrial targeting in vivo, for example, to regulate tRK1–enolase interaction and to target the complex toward the mitochondria.
Our experiments show that Eno2p is much more efficient than Eno1p for directing tRK1 mitochondrial import in vitro, and its affinity to tRK1 is much higher. Furthermore, the mitochondrial localization of Eno2p is more obvious than the one of Eno1p. However, in the absence of Eno2p (strain eno1/2 + pE1), Eno1p can perform tRK1 targeting, but less efficiently. tRK1 mitochondrial import in vivo was completely abolished only in the strain with deletions of both enolase genes.
The model we propose for tRK1 import therefore involves a noncognate aminoacyl–tRNA synthetase (mitochondrial enzyme for a cytosolic-type tRNA) and a glycolytic enzyme. Why would the yeast cell use so strange a pair of actors to address a single tRNA species into mitochondria? When one compares the yeast system with tRNA import events in other organisms, it appears that only two imported tRNA species were detected in yeast and from several to all tRNA species in some other cases. It would be logical that, in such organisms as trypanosomatids that import all tRNA species into the organelle (Simpson et al. 1989), a mitochondrially bound tRNA-specific receptor was postulated (Mahapatra and Adhya 1996), while in yeast, two bifunctional proteins, preMsk1p and Eno2p—which possess the needed RNA-binding and compartmentalization properties—were adapted to perform the targeting. The second point is that this targeting was evolutionarily retained in S. cerevisiae, if even mtDNA encodes another tRNALys. In this context, the fact that the absence of tRK1 in the mitochondria of enolase-depleted strain correlates with inhibition of mitochondrial translation indicates participation of the imported tRNALys in the organellar protein synthesis. This effect proves not to depend on the cellular enolase enzymatic activity and to be more pronounced at 37°C. One could suggest therefore that inhibited or perturbed translation (appearance of additional translation products) is due to a decrease of imported tRK1. This is in agreement with our previous observation of suppression of a mtDNA mutation by imported tRNAs (Kolesnikova et al. 2000) and with a more recent hypothesis suggesting the need of several cytoplasmic tRNAs to provide a complete decoding in yeast mitochondria (Rinehart et al. 2005). It may occur that we obtained here the first evidence that, at least at higher temperature, tRK1 is recruited by mitochondrial translation machinery.
All the data presented confirm the new role of glycolytic enzyme enolase in the mitochondrial targeting of tRNA in yeast. We suppose that this pathway should be precisely regulated in yeast cell, and its regulation includes tRNA–enolase recognition, interaction, and transport of this complex to the vicinity of the mitochondria. On the mitochondrial surface, enolase might facilitate interaction between the imported tRNA and the carrier protein, the precursor of the mitochondrial aminoacyl–tRNA synthetase. We can hypothesize that enolase–tRNA interaction induces required conformational changes in the tRNA structure, suggesting an RNA-chaperone function as a “second job” for this glycolytic enzyme.
Material and methods
Strains, antibodies, and plasmids
E. coli strain BL21 RIL3 Codon Plus (Stratagene) was used for expression of recombinant proteins. Yeast strain W303 was used to isolate mitochondria and protein extracts. ENO1 gene was deleted in W303 yeast strain by KanMX4 cassette replacement (Wach et al. 1994). Deletion was PCR-verified at 3′ and 5′ ends. Yeast strain MJ196 with deletion of ENO2 gene (YM4587 YHR174W/yhr174w∷gfp-HIS3) was kindly provided by Dr. J. Hegemann (Heinrich-Heine-Universitat Dusseldorf, Dusseldorf, Germany) (Niedenthal et al. 1999). Strain with a double deletion of both enolase-coding genes was obtained in this work by mating eno1 and eno2 haploid strains and subsequent tetrad analysis. To allow the unequivocal comparison of isogenic strains, only the final eno1 eno2 strain (also referred to as eno1/2) was used for all genetic and phenotypic analyses. This strain, when transfected with plasmids expressing both enolases, does not demonstrate any phenotype on all media tested, as did the wild-type strain W303 used as a control. One can affirm, therefore, that the genetic background of the initial strains does not influence the observed phenotypes.
Antibodies against human α-enolase were from Molecular Probes (C-19: sc-7455, Santa Cruz Biotechnology); antibodies against yeast cytochrome oxidase I were from Calbiochem; antibodies against Msk1p were kindly provided by A. Tzagoloff (Columbia University, New York); antibodies against α-Aco1p and CC1HLp were kindly provided by R. Lill (Philipps-Universitat Marburg, Marburg, Germany); and antibodies against Tom20p, Tom70p, and ATP-ADP carrier were kindly provided by N. Pfanner (Universitat Freiburg, Freiburg, Germany).
For overexpression of yeast proteins in E. coli, MSK1, ENO1, and ENO2 genes containing C-terminal His6-tags were cloned in pET3a vector (Novogen).
For expression of proteins in yeast cells, ENO1 and ENO2 genes with 300-base-pair (bp) flanking regions were PCR-cloned into the centromeric vector pRS416 with URA3 marker, to give pE1 and pE2 plasmids.
For yeast three-hybrid assay, ENO1 and ENO2 genes were cloned into a 2-μm/LEU2 vector pGAD-GH, which contains the GAL4 (768–881) activation domain under ADH1 promoter. The shuttle plasmid pIIIA/MS2-1 with a URA3 marker was used to express hybrid RNA molecules in yeast (Kraemer et al. 2000). To create plasmid-coding hybrid RNAs, genes of imported tRNALys (tRK1) and cytoplasmic tRNALys (tRK2) were cloned into the plasmid pIIIA/MS2-1 in the 3′ position of phage MS2 RNA.
For cellular localization experiments, we used the plasmid pDH5 containing the ORF for a pH-tolerant YFP. Gene-coding YFP was inserted in the yeast genome by homologous recombination with 3′ flank of the ENO2 gene. Localization experiments were performed by visualizing of mitochondria using Mitotracker Red (Molecular Probes) and confocal microscope Zeiss LSM 510.
Isolation of mitochondria, import assays, translation, and oxymetry
Mitochondria were isolated and verified for intactness and functionality as described previously (Entelis et al. 2002). The tRNA import assay was done as described (Entelis et al. 2002): Each assay contained 50 μg of mitochondria (mitochondrial protein), 3 pmol of gel-purified 5′-[32P]-labeled tRNA (aminoacylated and refolded), and either 5 μg of crude IDPs or 0.2 μg of recombinant proteins in 100 μL, then mitochondria were RNase-treated and repurified by differential centrifugation. Import efficiency was quantified, after separation of mitochondrial RNA on denaturing polyacrylamide gels, by scanning autoradiography using PhosphorImager (Fuji, MacBAS2000 program).
For tRNA import immunoinhibition, we used a modified protocol developed for protein import (Ryan et al. 2001): Isolated mitochondria were suspended in binding buffer (0.6 M sorbitol, 10 mM HEPES-KOH at pH 6.7, 1 mM EDTA) without labeled RNA and IDP in the presence of the appropriate amount of antibodies for 30 min at 4°C, then purified by percoll gradient centrifugation. For IDPs, antibodies were added in the binding buffer without mitochondria and the mixture was incubated for 30 min at 4°C and either used directly for import or depleted by protein A Sepharose beads and then used for import. Both approaches gave similar results.
Protein import assays were done with [35S]-labeled proteins synthesized on PCR-amplified DNA fragments in a coupled transcription-translation reticulocyte lysate-dependent system TNT Quick for PCR DNA (Promega). The in vitro import assay was performed as described elsewhere (Ryan et al. 2001).
Mitochondrial translation was analyzed essentially as described (Fox et al. 1991) with minor modifications: Cells were grown in YPD to A600 = 1.0, transferred to synthetic complete medium lacking Met (0.67% yeast nitrogen base, 0.08% CSM-Met [BIO101], 2% raffinose) with addition of 200 μg/mL of cycloheximide and incubated for 10 min at 30°C with shaking. Cells were then labeled with 10 μCi of [35S]-methionine (1.4 Ci/mmol, Amersham) for 20 min and chased with unlabeled 2 mM methionine for 10 min before breaking cells with glass beads. The proteins were TCA-precipitated and analyzed on 15% SDS-PAGE.
The rate of oxygen consumption by yeast cells (A600 = 0.3) cultured in 1 mL of YPD medium was measured in thermostated chamber for 10 min at 30°C or 37°C using a Clark-type electrode and the SI oxygen meter followed by addition of myxothiasol (750 nM) to measure nonmitochondrial oxygen uptake, which was then subtracted from all values.
Isolation and analysis of proteins
Crude IDP fraction was isolated as described previously (Entelis et al. 2002). Proteins were then subjected to differential ammonium sulfate precipitation. Fractions were dialyzed against Pipes-NaOH buffer (pH 6.5) 20 mM, NaCl 20 mM and passed through the SP Sepharose column (Pharmacia). The flow-through fraction was dialyzed against 20 mM Tris-HCl (pH 7.5), 20 mM NaCl, 5 mM DTT, and 10% glycerol and fractionated on Mono-Q column with a linear NaCl gradient up to 1 M. Fractions were dialyzed against 20 mM buffer HEPES-KOH (pH 6.8), 20 mM KCl, 50% glycerol, and 2 mM DTT. N-terminal protein sequencing was performed by Neosystems.
Recombinant enolases 1 and 2 were expressed in E. coli BL21 strain and purified on Ni-NTA agarose (Qiagen) under native conditions. Enolase enzymatic activity was assayed as described (Sangadala et al. 1995): Protein solutions were added to assay medium containing 50 mM Tris-HCl (pH 7.5), 1 mM MgCl2, and 2 mM 2-phosphoglyceric acid at room temperature. Absorbance at 230 nm was monitored as a function of time and divided by enzyme concentration. One unit of activity measured as (ΔA230/ΔA280)/min corresponds to 0.32 U/mg (Westhead and McLain 1964).
For North–Western analysis, proteins from Laemmli SDS-PAGE were electro-blotted onto nitrocellulose membrane Protran (0.45 μm), renaturated in 100 mM Tris-HCl (pH 7.5) and 0.1% Nonidet P-40 (v/v) for 6 h at 4°C. The filter was blocked in 10 mM Tris-HCl (pH 7.5), 5 mM Mg acetate, 2 mM DTT, and 1% BSA for 30 min prior to incubation with the binding buffer (20 mM HEPES-KOH at pH 6.8, 20 mM KCl, 0.5 mM MgCl2, 1 mM ATP), containing 1000 Cpm/cm2 of labeled tRNA and 1 μg/cm2 of recombinant preMSK1p overnight at 4°C. Filters were washed with the binding buffer without ATP and exposed to Fuji Imaging Plate.
For Western blot analysis, the ECL-Plus Western Detection system (Amerscham Biosciences, Inc.) was used. Tests for interaction enolases with lipids was done by incubation of 50 kCpm of [35S]-labeled Eno1p or Eno2p with either PIP strips (P-6001) or SphingoStrips (S-6000) (Echelon Biosciences, Inc.) in PBS for 1 h at 30°C with shaking, followed by triple washing with PBS and autoradiography.
Isolation and analysis of RNA
Total and mitochondrial RNAs were isolated by use of Trizol-reagent (Invitrogen). For import, North–Western, and gel-shift analysis we used purified natural tRK1 and tRK2. tRNA was dephosphorylated, [32P]-labeled by T4 polynucleotide kinase, gel-purified, refolded, and aminoacylated with recombinant cytosolic lysyl–tRNA synthetase as described previously (Entelis et al. 1998).
For gel-shifts, appropriate amounts of proteins and labeled RNAs were mixed in the volume of 20 μL of buffer containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 10 mM MgCl2, 5 mM DTT, 10% Glycerol, and 0.1 mg/mL BSA and incubated for 10 min at 30°C. Separation was done in native 6% PAGE, 20 mM Tris-borate buffer (pH 8.3), and 5% Glycerol as described (Kaminska et al. 2000). Free and bound tRNA was quantified by Fuji PhosphorImager, and apparent dissociation constants for tRNA–protein complexes were estimated as in Francin et al. (2002).
The following oligonucleotide probes were used for Northern analyses: anti-tRK1, CCTAACCTTATGATTAAGAGT; anti-mt tRNA Leu, TGGTTGCTATTTAAAGGACTTG; anti-tRK2, GCCGAACGCTCTACCAACTCAGC.
To isolate cytoplasmic polysomes, we used the method proposed in Marc et al. (2002) with minor modifications. Yeast YPH500 cells were grown to A600 = 5–10 on YPGal medium. Spheroplasts were generated by zymolyase and treated with 300 μg/mL of cycloheximide (20 min at 30°C). After two washings with 1 M sorbitol and 200 μg/mL of cycloheximide, the spheroplasts were broken in a vibro-homogenizer with glass beads (0.5 mm) in the polysome buffer (PA): 0.6 M mannitol, 30 mM Tris-HCl (pH 7.5), 5 mM MgAcetate, 100 mM KCl, 5 mM DTT, 200 μg/mL of cycloheximide, 500 μg/mL of heparine, protease inhibitor cocktail without EDTA (Roche). Mitochondria were pelleted and purified by differential centrifugation as described previously (Entelis et al. 2002). Cytoplasmic polysomes were isolated by loading the post-mitochondrial lysate on a 2–0.5 M sucrose gradient in the PA buffer and centrifuged at 190,000g for 16 h at 6°C.
To quantify mRNAs, real time RT–qPCR was performed (MyQ cycler, Bio-Rad) using the one-step RT–PCR with SYBR-green system (Bio-Rad). All samples were analyzed in triplicate and in three serial dilutions. Before quantification, all the RNA isolates were tested by PCR for the absence of contamination with DNA. To calculate absolute amounts of mRNAs in the isolates, we used as calibration controls known amounts of transcripts corresponding to the tested mRNAs. For this, the fragments of corresponding ORFs were PCR-amplified under control of the T7 promoter, transcribed by T7 RNA-polymerase, the transcripts were gel-purified and their amount determined by spectroscopy.
Acknowledgments
We thank N. Pfanner, R. Lill, and A. Tzagoloff for providing antibodies against mitochondrial proteins; J. Hegemann and J. Brewer for providing yeast strains; and L. Sweetlove for helpful discussion and communication of results before publication. We are grateful to J. Mutterer (IBMP, Strasbourg) for confocal microscopy experiments assistance. This work was partially supported by the CNRS, ULP, AFM, ANR, RFFI (grant no. 00-04-48488) and GIS “Institut de Maladies Rares.” I.B. was supported by a Thesis Fellowship form French Government (BGF), P.K. was supported by an Eiffel Ph.D. fellowship.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.385706
References
- al-Giery A.G., Brewer J.M. Characterization of the interaction of yeast enolase with polynucleotides. Biochim. Biophys. Acta. 1992;1159:134–140. doi: 10.1016/0167-4838(92)90017-8. [DOI] [PubMed] [Google Scholar]
- Ardail D., Privat J.P., Egret-Charlier M., Levrat C., Lerme F., Louisot P.1990Mitochondrial contact sites. Lipid composition and dynamics. J. Biol. Chem. 26518797 – 18802 [PubMed] [Google Scholar]
- Brewer J.M., Glover C.V., Holland M.J., Lebioda L. Enzymatic function of loop movement in enolase: Preparation and some properties of H159N, H159A, H159F, and N207A enolases. J. Protein Chem. 2003;22:353–361. doi: 10.1023/a:1025390123761. [DOI] [PubMed] [Google Scholar]
- Bringaud F., Stripecke R., Frech G.C., Freedland S., Truck C., Byrne E.M., Simpson L. Mitochondrialglutamate dehydrogenase from Leishmania tarentolae is a guide RNA-binding protein. Mol. Cell. Biol. 1997;17:3915–3923. doi: 10.1128/mcb.17.7.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpousis A.J. The Escherichia coli RNA degradosome: Structure, function and relationship in other ribonucleolytic multienzyme complexes. Biochem. Soc. Trans. 2002;30:150–155. [PubMed] [Google Scholar]
- Corral-Debrinski M., Blugeon C., Jacq C. In yeast, the 3′ untranslated region or the presequence of ATM1 is required for the exclusive localization of its mRNA to the vicinity of mitochondria. Mol. Cell. Biol. 2000;20:7881–7892. doi: 10.1128/mcb.20.21.7881-7892.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delage L., Dietrich A., Cosset A., Marechal-Drouard L. In vitro import of a nuclearly encoded tRNA into mitochondria of Solanum tuberosum. Mol. Cell. Biol. 2003;23:4000–4012. doi: 10.1128/MCB.23.11.4000-4012.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekert K., de Kroon A.I.P.M., Kispal G., Lill R. Isolation and subfractionation of mitochondria from the yeast Saccharomyces cerevisiae. In: Pon L.A., Schon E.A., editors. Mitochondria. Academic Press; San Diego: 2001. pp. 37–51. [DOI] [PubMed] [Google Scholar]
- Dietrich A., Marechal-Drouard L., Carneiro V., Cosset A., Small I. A single base change prevents import of cytosolic tRNA(Ala) into mitochondria in transgenic plants. Plant J. 1996;10:913–918. doi: 10.1046/j.1365-313x.1996.10050913.x. [DOI] [PubMed] [Google Scholar]
- Edwards S.R., Braley R., Chaffin W.L. Enolase is present in the cell wall of Saccharomyces cerevisiae. FEMS Microbiol. Lett. 1999;177:211–216. doi: 10.1111/j.1574-6968.1999.tb13734.x. [DOI] [PubMed] [Google Scholar]
- Entelis N.S., Kieffer S., Kolesnikova O.A., Martin R.P., Tarassov I.A. Structural requirements of tRNALys for its import into yeast mitochondria. Proc. Natl. Acad. Sci. 1998;95:2838–2843. doi: 10.1073/pnas.95.6.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entelis N.S., Kolesnikova O.A., Dogan S., Martin R.P., Tarassov I.A. 5 S rRNA and tRNA import into human mitochondria. Comparison of in vitro requirements. J. Biol. Chem. 2001a;276:45642–45653. doi: 10.1074/jbc.M103906200. [DOI] [PubMed] [Google Scholar]
- Entelis N.S., Kolesnikova O.A., Martin R.P., Tarassov I.A. RNA delivery into mitochondria. Adv. Drug Deliv. Rev. 2001b;49:199–215. doi: 10.1016/s0169-409x(01)00135-1. [DOI] [PubMed] [Google Scholar]
- Entelis N., Kolesnikova O., Kazakova H., Brandina I., Kamenski P., Martin R.P., Tarassov I. Import of nuclear encoded RNAs into yeast and human mitochondria: Experimental approaches and possible biomedical applications. Genet. Eng. (N.Y.) 2002;24:191–213. doi: 10.1007/978-1-4615-0721-5_9. [DOI] [PubMed] [Google Scholar]
- Fox T.D., Folley L.S., Mulero J.J., McMullin T.W., Thorsness P.E., Hedin L.O., Costanzo M.C. Analysis and manipulation of yeast mitochondrial genes. Methods Enzymol. 1991;194:149–165. doi: 10.1016/0076-6879(91)94013-3. [DOI] [PubMed] [Google Scholar]
- Francin M., Kaminska M., Kerjan P., Mirande M. The N-terminal domain of mammalian Lysyl–tRNA synthetase is a functional tRNA-binding domain. J. Biol. Chem. 2002;277:1762–1769. doi: 10.1074/jbc.M109759200. [DOI] [PubMed] [Google Scholar]
- Giege P., Heazlewood J.L., Roessner-Tunali U., Millar A.H., Fernie A.R., Leaver C.J., Sweetlove L.J. Enzymes of glycolysis are functionally associated with the mitochondrion in Arabidopsis cells. Plant Cell. 2003;15:2140–2151. doi: 10.1105/tpc.012500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly D.J., Morrow I.C., Lindsay M., Gould R., Bryant N.J., Gaullier J.M., Parton R.G., Stenmark H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000;19:4577–4588. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze M.W. Enzymes as RNA-binding proteins: A role for (di)nucleotide-binding domains? Trends Biochem. Sci. 1994;19:101–103. doi: 10.1016/0968-0004(94)90198-8. [DOI] [PubMed] [Google Scholar]
- Jeffery C.J. Moonlighting proteins. Trends Biochem. Sci. 1999;24:8–11. doi: 10.1016/s0968-0004(98)01335-8. [DOI] [PubMed] [Google Scholar]
- Kaminska M., Deniziak M., Kerjan P., Barciszewski J., Mirande M. A recurrent general RNA binding domain appended to plant methionyl–tRNA synthetase acts as a cis-acting cofactor for aminoacylation. EMBO J. 2000;19:6908–6917. doi: 10.1093/emboj/19.24.6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnikova O.A., Entelis N.S., Mireau H., Fox T.D., Martin R.P., Tarassov I.A. Suppression of mutations in mitochondrial DNA by tRNAs imported from the cytoplasm. Science. 2000;289:1931–1933. doi: 10.1126/science.289.5486.1931. [DOI] [PubMed] [Google Scholar]
- Kolesnikova O., Entelis N., Kazakova H., Brandina I., Martin R.P., Tarassov I. Targeting of tRNA into yeast and human mitochondria: The role of anticodon nucleotides. Mitochondrion. 2002;2:95–107. doi: 10.1016/s1567-7249(02)00013-2. [DOI] [PubMed] [Google Scholar]
- Kraemer B., Zhang B., SenGupta D., Fields S., Wickens M. Using the yeast three-hybrid system to detect and analyze RNA–protein interactions. Methods Enzymol. 2000;328:297–321. doi: 10.1016/s0076-6879(00)28404-8. [DOI] [PubMed] [Google Scholar]
- Kuhnel K., Luisi B.F. Crystal structure of the Escherichia coli RNA degradosome component enolase. J. Mol. Biol. 2001;313:583–592. doi: 10.1006/jmbi.2001.5065. [DOI] [PubMed] [Google Scholar]
- Liou G.G., Jane W.N., Cohen S.N., Lin N.S., Lin-Chao S. RNA degradosomes exist in vivo in Escherichia coli as multicomponent complexes associated with the cytoplasmic membrane via the N-terminal region of ribonuclease E. Proc. Natl. Acad. Sci. 2001;98:63–68. doi: 10.1073/pnas.011535498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahapatra S., Adhya S.1996Import of RNA into Leishmania mitochondria occurs through direct interaction with membrane-bound receptors. J. Biol. Chem. 27120432 – 20437 [DOI] [PubMed] [Google Scholar]
- Marc P., Margeot A., Devaux F., Blugeon C., Corral-Debrinski M., Jacq C. Genome-wide analysis of mRNAs targeted to yeast mitochondria. EMBO Rep. 2002;3:159–164. doi: 10.1093/embo-reports/kvf025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R.P., Schneller J.M., Stahl A.J., Dirheimer G. Import of nuclear deoxyribonucleic acid coded lysine-accepting transfer ribonucleic acid (anticodon C-U-U) into yeast mitochondria. Biochemistry. 1979;18:4600–4605. doi: 10.1021/bi00588a021. [DOI] [PubMed] [Google Scholar]
- McAlister L., Holland M.J. Targeted deletion of a yeast enolase structural gene. Identification and isolation of yeast enolase isozymes. J. Biol. Chem. 1982;257:7181–7188. [PubMed] [Google Scholar]
- McKee E.E., McEwen J.E., Poyton R.O. Mitochondrial gene expression in Saccharomyces cerevisiae. II. Fidelity of translation in isolated mitochondria from wild type and respiratory-deficient mutant cells. J. Biol. Chem. 1984;259:9332–9338. [PubMed] [Google Scholar]
- Morita T., Kawamoto H., Mizota T., Inada T., Aiba H. Enolase in the RNA degradosome plays a crucial role in the rapid decay of glucose transporter mRNA in the response to phosphosugar stress in Escherichia coli. Mol. Microbiol. 2004;54:1063–1075. doi: 10.1111/j.1365-2958.2004.04329.x. [DOI] [PubMed] [Google Scholar]
- Neupert W. Protein import into mitochondria. Annu. Rev. Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- Niedenthal R., Riles L., Guldener U., Klein S., Johnston M., Hegemann J.H. Systematic analysis of S. cerevisiae chromosome VIII genes. Yeast. 1999;15:1775–1796. doi: 10.1002/(SICI)1097-0061(199912)15:16<1775::AID-YEA496>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Pancholi V. Multifunctional α-enolase: Its role in diseases. Cell. Mol. Life Sci. 2001;58:902–920. doi: 10.1007/PL00000910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pioli P.A., Hamilton B.J., Connolly J.E., Brewer G., Rigby W.F. Lactate dehydrogenase is an AU-rich element-binding protein that directly interacts with AUF1. J. Biol. Chem. 2002;277:35738–35745. doi: 10.1074/jbc.M204002200. [DOI] [PubMed] [Google Scholar]
- Rinehart J., Krett B., Rubio M.A., Alfonzo J.D., Soll D. Saccharomyces cerevisiae imports the cytosolic pathway for Gln–tRNA synthesis into the mitochondrion. Genes & Dev. 2005;19:583–592. doi: 10.1101/gad.1269305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusconi C.P., Cech T.R. The anticodon is the signal sequence for mitochondrial import of glutamine tRNA in Tetrahymena. Genes & Dev. 1996a;10:2870–2880. doi: 10.1101/gad.10.22.2870. [DOI] [PubMed] [Google Scholar]
- Rusconi C.P., Cech T.R. Mitochondrial import of only one of three nuclear-encoded glutamine tRNAs in Tetrahymena thermophila. EMBO J. 1996b;15:3286–3295. [PMC free article] [PubMed] [Google Scholar]
- Ryan M.T., Voos W., Pfanner N. Assaying protein import into mitochondria. Methods Cell Biol. 2001;65:189–215. doi: 10.1016/s0091-679x(01)65012-x. [DOI] [PubMed] [Google Scholar]
- Sangadala V.S., Glover C.V., Robson R.L., Holland M.J., Lebioda L., Brewer J.M. Preparation by site-directed mutagenesis and characterization of the E211Q mutant of yeast enolase 1. Biochim. Biophys. Acta. 1995;1251:23–31. doi: 10.1016/0167-4838(95)00049-z. [DOI] [PubMed] [Google Scholar]
- Scheffler I.E. Mitochondria make a come back. Adv. Drug Deliv. Rev. 2001;49:3–26. doi: 10.1016/s0169-409x(01)00123-5. [DOI] [PubMed] [Google Scholar]
- Schneider A., Marechal-Drouard L. Mitochondrial tRNA import: Are there distinct mechanisms? Trends Cell Biol. 2000;10:509–513. doi: 10.1016/s0962-8924(00)01854-7. [DOI] [PubMed] [Google Scholar]
- SenGupta D.J., Zhang B., Kraemer B., Pochart P., Fields S., Wickens M. A three-hybrid system to detect RNA–protein interactions in vivo. Proc. Natl. Acad. Sci. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson A.M., Suyama Y., Dewes H., Campbell D.A., Simpson L. Kinetoplastid mitochondria contain functional tRNAs which are encoded in nuclear DNA and also contain small minicircle and maxicircle transcripts of unknown function. Nucleic Acids Res. 1989;17:5427–5445. doi: 10.1093/nar/17.14.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Green M.R. Sequence-specific binding of transfer RNA by glyceraldehyde-3-phosphate dehydrogenase. Science. 1993;259:365–368. doi: 10.1126/science.8420004. [DOI] [PubMed] [Google Scholar]
- Tarassov I., Entelis N., Martin R. Mitochondrial import of a cytoplasmic lysine-tRNA in yeast is mediated by cooperation of cytoplasmic and mitochondrial lysyl–tRNA synthetases. EMBO J. 1995a;14:3461–3471. doi: 10.1002/j.1460-2075.1995.tb07352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarassov I., Entelis N., Martin R.P. An intact protein translocating machinery is required for mitochondrial import of a yeast cytoplasmic tRNA. J. Mol. Biol. 1995b;245:315–323. doi: 10.1006/jmbi.1994.0026. [DOI] [PubMed] [Google Scholar]
- Taylor S.W., Fahy E., Zhang B., Glenn G.M., Warnock D.E., Wiley S., Murphy A.N., Gaucher S.P., Capaldi R.A., Gibson B.W., et al. Characterization of the human heart mitochondrial proteome. Nat. Biotechnol. 2003;21:281–296. doi: 10.1038/nbt793. [DOI] [PubMed] [Google Scholar]
- Wach A., Brachat A., Pohlmann R., Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Westhead E.W., McLain G. A Purification of brewers’ and bakers’ yeast enolase yielding a single active component. J. Biol. Chem. 1964;239:2464–2468. [PubMed] [Google Scholar]