Abstract
In many human infections, hosts and pathogens coexist for years or decades. Important examples include HIV, herpes viruses, tuberculosis, leprosy, and malaria. With the exception of intensively studied viral infections such as HIV/AIDs, little is known about the extent to which the clonal expansion that occurs during long-term infection by pathogens involves important genetic adaptations. We report here a detailed, whole-genome analysis of one such infection, that of a cystic fibrosis (CF) patient by the opportunistic bacterial pathogen Pseudomonas aeruginosa. The bacteria underwent numerous genetic adaptations during 8 years of infection, as evidenced by a positive-selection signal across the genome and an overwhelming signal in specific genes, several of which are mutated during the course of most CF infections. Of particular interest is our finding that virulence factors that are required for the initiation of acute infections are often selected against during chronic infections. It is apparent that the genotypes of the P. aeruginosa strains present in advanced CF infections differ systematically from those of “wild-type” P. aeruginosa and that these differences may offer new opportunities for treatment of this chronic disease.
Keywords: chronic infection, positive selection, virulence, antibiotic resistance
Most cystic fibrosis (CF) patients acquire chronic Pseudomonas aeruginosa infections by their teenage years, if not earlier, and these respiratory infections are responsible for much of the morbidity and mortality caused by CF (1, 2). It has been established that most of these infections are clonal (3), and even among groups of CF patients treated in specific clinics the infections are acquired independently, presumably from diverse environmental reservoirs (4). Previous studies, particularly of the O-antigen biosynthetic locus and the transcriptional regulator mucA, indicate that some P. aeruginosa genes commonly incur loss-of-function mutations as the infections progress (5–7). Mutator phenotypes also arise frequently (8).
The overall picture is reminiscent of typical cancers: a clone of cells, albeit in this instance one of exogenous origin, experiences selection for an accumulation of genetic variants that promote long-term survival and clonal expansion. Our data validate this model for P. aeruginosa infections in CF and provide strong evidence for the role of selection in shaping the genotypes of the bacteria that are present during the late, life-threatening phase of the infections. Our data also focus attention on particular aspects of P. aeruginosa metabolism that are premier targets of selection, both in the patient we studied in most detail and in other, independently evolving, P. aeruginosa infections in additional CF patients.
Results and Discussion
Our basic approach was to construct a composite genome by using whole-genome shotgun sampling of two isolates from one CF patient (patient 1): (i) a single, clonally purified 6-month isolate and (ii) a comparable 96-month isolate. The depth of whole-genome sampling provided independent coverage of ≈97% of the composite genome in high-quality sequence data derived from each isolate. Hence, we expected to detect nearly all mutational differences between the two isolates. As is summarized in Table 1, we detected 68 mutations.
Table 1.
Whole-genome comparison between two P. aeruginosa isolates from patient 1
| Description | Value |
|---|---|
| Early-isolate collection age, mo | 6 |
| Late-isolate collection age, mo | 96 |
| Early-isolate genome size, bp | 6,492,423 |
| No. of mutations between 6- and 96-mo isolates | |
| Nonsynonymous (nonsense) | 33 (3) |
| Synonymous | 8 |
| Insertions/deletions (frameshifts and transposon insertions) | 19 (15) |
| Intergenic | 8 |
| Total | 68 |
Two Related Isolates from Patient 1 Collected 7.5 Years Apart Show a Genomewide Signal of Positive Selection.
The pattern of mutation between the genomes shows a signal of positive selection. This signal is most clearly seen in the ratio of nonsynonymous to synonymous changes per site, which is 1.4. As a control, we compared the two publicly available P. aeruginosa genome sequences, PAO1 and PA14; single-nucleotide differences within coding regions between these two strains display a nonsynonymous-to-synonymous ratio of 0.1. In nearly all cases of mutations in genes that have clear orthologs in the PAO1 reference strain (i.e., our best representative of the wild-type state), the sequence of the 6-month isolate matches the reference sequence. Thus most, if not all, mutations accumulated in patient 1 along the cellular lineage that led from the 6-month to the 96-month isolate. Most mutations are single base-pair changes or one to three base-pair insertions/deletions; an exception is one large deletion of 188 kb that removes ≈139 genes. Many of the mutations are likely to cause partial or complete loss of function in the altered genes because ≈25% of the mutations cause a premature translation stop or a shift in the reading frame, or involve either a transposon insertion or outright deletion of a gene. A standard computational method for predicting the effect of each nonsynonymous mutation on protein function suggests that approximately half (33 of 68) of all mutations in the 96-month isolate are likely to affect protein function (see Tables 4 and 5, which are published as supporting information on the PNAS web site).
Many Genes Functioning in Virulence and Multidrug Efflux Were Mutated During the Patient 1 Infection.
The functions of mutated genes cluster in specific categories (see Tables 2, 4, and 5). Particularly striking is the observation that a whole array of “virulence” factors, and their regulators, are mutated in the 96-month isolate. Virulence is classically defined in terms of establishment-of-infection assays, in which the loss of a virulence factor results in a decrease in the ability to cause disease. The virulence factors and regulators mutated in the 96-month isolate include genes functioning in O-antigen biosynthesis, type III secretion, twitching motility, exotoxin A regulation, multidrug efflux, osmotic balance, phenazine biosynthesis, quorum sensing, and iron acquisition. Phenotypes of the 96-month isolate, when compared with those of the 6-month isolate, are consistent with the expected effects of these mutations, including loss of serotype-specific antigenicity, loss of twitching motility, loss of pyoverdine production, loss of secreted protease activities (including elastase), and reduced biofilm formation (see Fig. 2, which is published as supporting information on the PNAS web site). Each of these phenotypes is associated with decreased virulence in models of acute infection (9–11).
Table 2.
Selected functional categories of mutated genes in the 96-month isolate from patient 1
| Functional category | No. of genes mutated |
|---|---|
| Virulence factors, virulence regulators | 13 |
| Small-molecule transport, antibiotic resistance | 12 |
| Iron acquisition | 5 |
| DNA replication, cell division, transcription, translation | 5 |
| Quorum sensing | 3 |
| Fatty-acid metabolism | 3 |
| DNA mismatch repair | 1 |
| Anaerobic metabolism | 1 |
Numerous multidrug-efflux-pump genes are mutated in the 96-month isolate. These pumps normally export small molecules from the cytoplasm and provide P. aeruginosa with natural resistance to some antibiotics. We tested both isolates for sensitivity to a panel of antibiotics and found that the 96-month isolate had increased resistance, relative to wild-type cells, to the aminoglycosides amikacin, gentamicin, and tobramycin. The 6-month isolate had increased resistance to the β-lactam antibiotic ticarcillan, perhaps as a result of mutation that occurred before our collection of this isolate. The resistance in these isolates may be the result of antibiotic treatments that patient 1 received, which included both aminoglycoside and β-lactam antibiotics.
The 96-month isolate has a nonsynonymous mutation in mutS, a DNA mismatch repair gene that, when disrupted, results in an increased rate of mutation. Mutators have been reported in isolates from numerous infectious diseases, including P. aeruginosa isolates from CF airways (8), and in cells isolated from many cancers. An increased mutation rate seems beneficial when there is a need for rapid accumulation of genetic adaptations. For example, when a bacterial culture is exposed to two antibiotics successively, the proportion of mutators in the surviving population is significantly higher than in the original culture (12). Analogously, the presence of a mutator mutation in the 96-month isolate makes our observation of a strong positive-selection signal particularly significant. We were unable to determine whether the 96-month isolate had an increased rate of mutation compared with the 6-month isolate by using the standard assay for a mutator phenotype. This assay measures the frequency of mutation to resistance to the antibiotic rifampicin, to which the 96-month isolate is already resistant. Because the mutS mutation occurred relatively late in the infection (see Fig. 1 and Table 6, which is published as supporting information on the PNAS web site), it is uncertain to what extent a mutator phenotype might have influenced the existing mutations in the 96-month isolate. Despite the expectation that a mutator may produce a large number of background mutations, we did not observe this effect. As discussed above, a strikingly high proportion of the mutations we did observe appear to affect gene function.
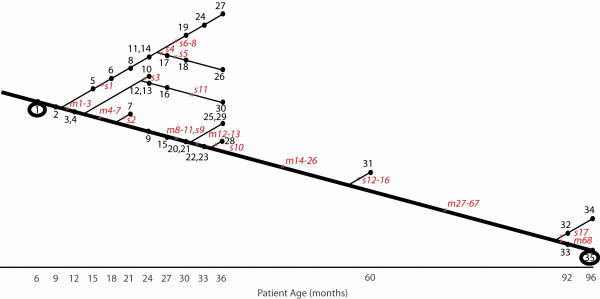
Fig. 1.
Tree showing mutations that occurred in patient 1 isolates during chronic airways infection. Isolates are numbered 1–35 in black type. Isolates 1 and 35, both circled, are the respective 6- and 96-month isolates whose genomes were sequenced. Mutations are shown in red italic type; mutations present in the 96-month isolate are numbered m1–m68, and mutations present only in intermediate isolates are numbered s1–s17. Mutations s9 and m52 are both in the same highly mutable repeat and appear to have occurred consecutively during the infection. Refer to Tables 4 and 5 for more information on individual mutations.
Multiple Related Lineages Coexisted During the Patient 1 Infection.
A full view of the sequence of events that led from the 6-month isolate to the 96-month isolate would require sequencing of the entire genomes of many intermediate isolates. Because this undertaking is impractical with current methods, we constructed a partial picture by tracing the mutations detected in the 96-month isolate back through a collection of intermediate isolates by means of PCR-based genotyping. Approximately one-third of the mutations (26 of 68) are present in the 60-month isolate, and half of these (13 of 26) are present in isolates collected after 30 months. Mutations accumulate in a parsimonious fashion, such that a self-consistent phylogenetic tree can be constructed showing the sequence of mutational events (see Fig. 1 and Table 6).
This retrospective tracing of mutations present in the 96-month isolate provides no information about the heterogeneity of the infection at any given time point. However, we were able to gain some insight into the degree of heterogeneity between isolates by finding mutations in intermediate isolates that were not present in the 96-month isolate. During the PCR-based genotyping of intermediate isolates, we discovered nine additional mutations that are present only in intermediate isolates. We also found three mutations specific to the 60-month isolate by comparing its partial genome sequence, published previously (13), to the composite genome. Multiple, independently acquired mutations in mucA have also been reported (13). Viewed together, the known mutations in patient 1 isolates indicate that parallel lineages existed during the early years of the infection and coexisted, in some cases, for several years (see Fig. 1 and Table 6). For example, mutations in mucA commonly occur during CF infections, and mucA mutations arose independently in three different lineages during the patient 1 infection. Isolates from a single time point are also heterogeneous, e.g., five of the six isolates collected at 36 months had detectably different genotypes.
Signals of Positive Selection and Loss of Function Are Typical in Isolates from Chronic CF Airway Infections.
A major issue is whether the mutations observed in the patient 1 infection are, to some extent, representative of the mutational change associated with P. aeruginosa infections in other CF patients with independently acquired infections. To address this issue, we tested other patients’ isolates for similar mutations. We assembled a collection of longitudinally collected isolates from 29 different CF patients by using the following criteria: (i) at least two clonally related isolates were available from each patient, (ii) the earliest collected isolate from each patient was collected before 11 years of age, and (iii) the latest collected isolate from each patient was collected >5 years (and in some cases 20 years) after the earliest collected isolate. In the resulting collection of 91 isolates, we could sequence a gene in each isolate and, by comparing isolates that were collected from the same patient, find all mutational differences in that gene that had occurred during 29 different CF infections. In each isolate of the collection, we chose to sequence the entire gene and regulatory region of 24 genes that had been mutated early in the patient 1 infection (genes from mutations m1, m3–m15, m17–m22, m24–m26, and s15, as shown in Fig. 1). We also sequenced the entire gene and regulatory region of 10 genes that were candidates for mutation in many CF infections, as determined from other studies (33–39). Table 3 shows the results of this sequencing, reporting a comprehensive view of all mutational differences in these genes that exist between isolates collected from the same patient.
Table 3.
Mutations that occurred in 34 genes during airway infections in 29 CF patients
| Gene name† | Gene annotation no. | Gene function | No. of nonsynonymous mutations (nonsense) | No. of synonymous mutations | No. of insertions/deletions (frameshifts and transposon insertions) | No. of intergenic mutations | Total no. of independent mutations‡ | No. of patients with at least one mutated isolate |
|---|---|---|---|---|---|---|---|---|
| mexZ | PA2020 | Transcriptional regulator of multidrug efflux | 13 (5) | 0 | 11 (9) | 0 | 24 | 18 |
| lasR | PA1430 | Transcriptional regulator of quorum sensing | 18 (3) | 0 | 4 (4) | 0 | 22 | 18 |
| PA0313 | Probable permease of ABC transporter | 4 (1) | 0 | 10 (2) | 1 | 15 | 13 | |
| mexA | PA0425 | RND multidrug efflux membrane fusion protein MexA precursor | 13 (4) | 0 | 1 (1) | 0 | 14 | 11 |
| accC | PA4848 | Biotin carboxylase | 6 | 0 | 3 (2) | 0 | 9 | 9 |
| *vfr | PA0652 | Transcriptional regulator | 4 | 0 | 3 (2) | 1 | 8 | 7 |
| mexS | PA2491 | Probable oxidoreductase | 7 | 0 | 1 | 0 | 8 | 6 |
| exsA | PA1713 | Transcriptional regulator of type III secretion | 6 | 0 | 2 (1) | 0 | 8 | 6 |
| PA0506 | Probable acyl-CoA dehydrogenase | 5 | 0 | 1 (1) | 1 | 7 | 7 | |
| *wspF | PA3703 | Probable methylesterase involved in chemosensory signal transduction | 4 (2) | 0 | 2 (1) | 0 | 6 | 6 |
| *rpoN | PA4462 | RNA polymerase σ-54 factor | 2 (1) | 0 | 3 (2) | 0 | 5 | 5 |
| *fleQ | PA1097 | Transcriptional regulator of flagella synthesis | 5 (1) | 0 | 0 | 0 | 5 | 5 |
| mexT | PA2492 | Transcriptional regulator of multidrug efflux | 4 | 0 | 0 | 1 | 5 | 5 |
| *nalD | PA3574 | Probable transcriptional regulator | 1 | 1 | 1 (1) | 1 | 4 | 4 |
| *ampD | PA4522 | β-Lactamase expression regulator | 3 | 0 | 1 | 0 | 4 | 4 |
| PA0366 | Probable aldehyde dehydrogenase | 1 | 0 | 2 (2) | 1 | 4 | 4 | |
| *cyaB | PA3217 | Adenylate cyclase | 2 | 1 | 0 | 0 | 3 | 3 |
| P1-001 | In 45-kb genomic island§ | Hypothetical protein | 1 | 0 | 2 (2) | 0 | 3 | 2 |
| PA3817 | Probable methyltransferase | 2 | 1 | 0 | 0 | 3 | 3 | |
| pilB | PA4526‡ | Type 4 fimbrial biogenesis protein | 1 | 1 | 0 | 0 | 2 | 2 |
| PA1333 | Hypothetical protein | 0 | 0 | 0 | 1 | 1 | 1 | |
| PA3565 | Probable transcriptional regulator | 1 | 0 | 0 | 0 | 1 | 1 | |
| PA2312 | Probable transcriptional regulator | 0 | 1 | 0 | 0 | 1 | 1 | |
| *anr | PA1544 | Transcriptional regulator of anaerobic metabolism | 0 | 0 | 0 | 1 | 1 | 1 |
| *rhlR | PA3477 | Transcriptional regulator of quorum sensing | 0 | 0 | 0 | 1 | 1 | 1 |
| *phoP | PA1179 | Two-component response regulator | 0 | 0 | 0 | 0 | 0 | 0 |
| rhlI | PA3476 | Autoinducer synthesis protein | 0 | 0 | 0 | 0 | 0 | 0 |
| PA4796 | Hypothetical protein | 0 | 0 | 0 | 0 | 0 | 0 | |
| pqsB | PA0997 | Homologous to β-keto-acyl-acyl-carrier protein synthase | 0 | 0 | 0 | 0 | 0 | 0 |
| toxR | PA0707 | Transcriptional regulator | 0 | 0 | 0 | 0 | 0 | 0 |
| PA2435 | Probable cation-transporting P-type ATPase | 0 | 0 | 0 | 0 | 0 | 0 | |
| PA4420 | Hypothetical protein | 0 | 0 | 0 | 0 | 0 | 0 | |
| None§ | Hypothetical protein ORF C/SG114 | 0 | 0 | 0 | 0 | 0 | 0 | |
| PA2121 | Hypothetical protein | 0 | 0 | 0 | 0 | 0 | 0 | |
| Total | 103 (17) | 5 | 47 (30) | 9 | 164 | 143 |
†The 10 gene names that are preceded by an asterisk were candidate genes based on other studies. The remaining 24 genes were mutated in the 96-month isolate of patient 1.
‡Each gene was amplified with PCR and completely sequenced in 91 isolates. Most patients had three representative isolates that had been collected longitudinally. A mutation was defined as any difference in sequence between clonally related isolates from the same patient. When the same mutation was present in multiple isolates from the same patient, we assumed that the mutation occurred once during the infection and was then propagated clonally. Some patients had two isolates with different mutations in the same gene; we considered these as two independent mutations.
§Two genes are not present in all strains: the gene P1-001 is present in isolates from five patients, and gene C/SG114 is present in isolates from 21 patients. pilA and pilB were only partially sequenced because the locus exhibits high strain-to-strain divergence.
The majority of genes identified as mutational targets in patient 1 or from our candidate-gene list accumulate few or no mutations during the infections in other patients. However, a few genes mutate during most CF infections (see Table 3 and Tables 7 and 8, which are published as supporting information on the PNAS web site). The effect of positive selection is overwhelming, especially in the commonly mutated genes: only five synonymous mutations accompany 103 nonsynonymous mutations. At least 25% of mutations are conspicuous loss-of-function mutations (nonsense mutations, shifts in the reading frame, or transposon insertions); if we also computationally predict the effect of each nonsynonymous mutation on protein function, then approximately two-thirds of all mutations are predicted to affect protein function (see Tables 7 and 8). Most mutations appear to occur relatively late in the infections, because the genotype of the earliest isolate usually matches the PAO1 reference genome, even though some of the “early” isolates were sampled at ages as late as 10 years.
The Multidrug Efflux Gene mexZ and the Quorum-Sensing Regulator lasR Are Common Mutational Targets During Chronic CF Airway Infections.
The most frequently mutated genes are multidrug-efflux pumps (see Tables 3, 7, and 8). Of the genes we sequenced, the most frequently mutated gene is mexZ, a negative regulator of mexX and mexY, which are components of the MexXY-OprM multidrug-efflux pump (14, 15). Isolates with resistance to aminoglycosides often have increased expression of mexX and mexY (16), and over half of the mutations we identified in mexZ are conspicuous loss-of-function mutations that lead to increased expression of mexX and mexY. CF patients are routinely treated with antibiotics such as the aminoglycoside tobramycin, and these treatments may select for mutations in multidrug-efflux pumps and their regulators.
Virulence-related genes are also mutated during most infections, most notably the primary regulator of quorum sensing, lasR (see Tables 3, 7, and 8). Bacteria use quorum sensing to communicate, by using the concentration of a secreted small molecule to measure the prevailing cell density. On reaching high cell density, the cells dramatically alter their pattern of gene expression. At high cell density, wild-type P. aeruginosa produces virulence factors, forms biofilms, and becomes more antibiotic resistant (17, 18). All of these phenotypes are disrupted in lasR mutants. P. aeruginosa is thought to exist in a biofilm state during CF airway infections (19); if so, our results are paradoxical because they indicate that there is intense selection to mutate lasR and thus impair biofilm formation during chronic airways infections. Other commonly mutated genes that contribute to virulence are vfr, a virulence regulator that also regulates lasR; exsA, the regulator of type III secretion, a system that injects virulence factors into eukaryotic cells (20); and mexA, a component of a multidrug-efflux pump (21).
Virulence Factors Are Selected Against During Chronic CF Airway Infections.
Virulence factors are, by definition, required for acute infections, but our results show that they are selected against during chronic infection of CF airways. The presumed explanation is immune evasion, because the immune system recognizes numerous virulence factors and attempts to kill those cells that express them (22), thereby selecting for a population of cells with mutations in virulence factors and virulence regulators. Other bacteria appear to reduce virulence-factor expression during chronic infections by regulation rather than by mutational mechanisms. An example is the regulated virulence system of some Bordetella species that appear to shut off virulence-factor expression and reduce biofilm development after sensing appropriate environmental conditions (23).
By sequencing 10 candidate genes in our panel of CF isolates from 29 patients, we could test whether we had enriched for commonly mutated genes by focusing on 24 genes that were mutated in the patient 1 infection. Genes from both of these groups show a pattern of mutation consistent with positive selection acting at many different genes, but only in the genes that were mutated during the patient 1 infection did we find genes such as mexA and lasR that are common mutational targets (see Table 3). Overall, we conclude that a surprisingly large number of genes in the genome are targets for mutation during adaptation to CF airways, although most of these genes are mutated in only a fraction of infections. The effect of the specific pattern of mutation in particular patients on clinical outcomes requires further study.
One concern with this study is whether the infection in patient 1 is typical of other CF infections. At the outset of this study, we did not have access to clinical information about patient 1, such as the severity of the patient’s disease, antibiotics used during treatment, and the CF transmembrane conductance regulator (CFTR) genotype of the patient. However, late in the study we were granted Institutional Review Board approval to learn this information. Patient 1 is homozygous for the most common CF disease allele, CFTR ΔF508, and was hospitalized for acute respiratory illness on multiple occasions in each of the first 7 years of life. Despite these infections, patient 1 had normal lung function at age 5 (measured by forced expiratory volume for 1 sec) but had decreased lung function by age 7, suggestive of the typical decline in lung function from chronic airways infection. Patient 1 has been treated with inhaled corticosteroids and numerous different i.v. and oral antibiotics, including β-lactam and aminoglycoside antibiotics, trimethoprim/sulfamethoxazole, and azithromycin.
Concluding Remarks.
Using a detailed, whole-genome analysis, we have shown that P. aeruginosa adapts genetically to CF airways. As the cost of large-scale DNA sequencing continues to drop, this analytical approach will become increasingly practical and, therefore, applicable to many types of chronic infection. Common loss-of-function mutations in genes such as lasR and mexZ may offer new therapeutic opportunities. Whereas mutations in these genes appear to favor the clonal expansion of P. aeruginosa in the airways of CF patients, the mutations may also create new vulnerabilities that can be used for treatment. This hope arises by analogy with the situation in typical cancers, in which mutational loss of DNA-repair checkpoints favors clonal expansion but also confers sensitivity to DNA-damaging agents. What is already clear is that the genetic properties of the bacterial cells present late in P. aeruginosa infections of CF airways differ greatly from the properties of the cells that initiated the infections many years before decline in lung function became life-threatening.
Materials and Methods
Isolates.
All isolates were grown on LB medium and incubated overnight at 37°C. All patient 1 isolates were collected by oropharyngeal swab, except for isolates 4, 12–14, and 28–30, which were collected by bronchoalveolar lavage. When multiple colony morphotypes were present at a time point, each was saved and numbered in order of prevalence on the isolation plate (e.g., at 21 months, isolate 7 was more numerous than isolate 8). Patient 1 isolates are the same as “patient-nine” isolates in ref. 24. The collection method is unknown for isolates from patients other than patient 1. All isolates were clonally purified from clinical specimens. We used multilocus sequence typing to determine clonal relatedness between isolates from the same patient. All isolates from the same patient are clonally related with three exceptions: patient 22 has a pair of clonally related isolates from 9.9 and 20.0 years that is not related to the 6.8-year isolate; patient 20 has a pair of clonally related isolates from 9.9 and 17.3 years that is not related to the 17.5-year isolate; and patient 16 has a pair of clonally related isolates from 7.0 and 23.4 years that is not related to the clonally related pair of isolates from 7.1 and 17.6 years.
Isolate Phenotyping.
To assay for mutation rate, overnight cultures were serially diluted and plated on solid agar media containing LB or LB + 300 μg/ml rifampicin; each assay was performed in triplicate. To assay for twitching motility, single colonies were stabbed into thin (3-mm) LB agar plates and incubated at room temperature for 72 hr (25). To assay for pyoverdine production, strains were streaked onto Casamino acid-containing medium (low-iron medium), incubated at room temperature for 48 hr, and visually scored under UV light for fluorescent pyoverdine production (26). Serotyping with O1-specific antibodies was performed as described in ref. 27. Antibiotic sensitivities of isolates were determined at the time of initial isolation by broth microdilution (28). The panel of antibiotics tested included amikacin, gentamicin, tobramycin, ceftazidime, ticarcillan, aztreonam, ciprofloxacin, meropenem, and piperacillin.
Secreted protease production was assayed as described in ref. 29. Briefly, single colonies of each strain were patched onto brain–heart infusion/milk agar [1.5% agar, 3.7% brain–heart infusion (Difco), and 1.5% nonfat powdered milk] and incubated at 37°C for 28 hr or 45 hr. Zones of clearing surrounding bacterial growth reflected degradation of casein by secreted proteases, including the lasB protease.
Biofilm formation was assayed as described in ref. 30. Briefly, overnight cultures were diluted 1:100 in Mueller–Hinton broth (Difco) and adjusted for equivalent cell densities. From each culture, 100 μl was inoculated into 6 different wells of a sterile polystyrene 96-well culture plate (Nunc), covered, and incubated at 37°C for 24 hr without shaking. The culture medium was then removed, the wells were washed with sterile water three times, and the wells were stained for 15 min with 125 μl of 0.1% crystal violet (Fisher). The wells were again washed with water three times and air-dried at room temperature. The crystal violet was dissolved from the wells with 125 μl of ethanol for 15 min at room temperature, and the solution was then removed to a fresh plate for spectrophotometry. Results are representative of three separate experiments with six replicates each.
Sequencing.
The genomes of isolates 1 and 35 from patient 1 were sequenced by the whole-genome shotgun method, by using standard protocols. Each genome was sequenced to 5.7× coverage, and a combined assembly was built by using reads from both isolates. Mutations were found by identifying high-quality discrepancies between the isolates. Each mutation was confirmed by whole-cell PCR and sequencing of the mutated site.
We used whole-cell PCR and sequencing to sequence the entire gene and regulatory region of 24 genes mutated early in the patient 1 infection, as well as 10 candidate genes in a panel of 91 isolates from 29 CF patients. We generated high-quality sequence data for ≈96% of isolates in each of these regions. In a few cases, one isolate produced no sequence for any PCR products in a given gene, suggesting that the gene was deleted; this result occurred for multiple isolates in mexZ, one isolate in lasR, and several isolates in both mexT and PA2312. When a mutation was found between clonally related isolates from the same patient, we again performed PCR and sequencing to confirm the mutation. wbpA (mutation m2) was not included in this survey because the O-antigen locus is highly divergent between strains, and dnaX (mutation m23) was not included because we identified the mutation late in the study.
Computational Tools.
genemark was used to make gene predictions (31). P1-001 is the temporary name for the gene in a 45-kb genomic island that is integrated at tRNA-Pro (PA2736.1) in patient 1 isolates. Gene annotations were obtained from www.pseudomonas.com.
The program sift was used to predict the effect of nonsynonymous mutations on protein function (32). As a control for neutral nonsynonymous mutations, we compared the single-nucleotide polymorphisms in 100 genes between the PAO1 and PA14 genome sequences. Approximately 10–15% of nonsynonymous mutations in these genes were predicted to lose function; this percentage does not differ from the estimated false-positive rate in the sift analysis. We used the default parameters of sift (median conservation of sequences = 3.00 and removal of sequences >90% identical to the query) and used the National Center for Biotechnology Information nonredundant database (www.ncbi.nlm.nih.gov) (October 2004) to find homologs. A mutation was scored as “not tolerated” when sift predicted that the probability of observing this mutation in other functional homologs was 0.06 or less.
Supplementary Material
Acknowledgments
We thank the staff of the University of Washington Genome Center. We also thank Dr. Peter Greenberg, Charla Lambert, David Spencer, and Marcella Harris for help in preparing the manuscript. This work was supported by the Canadian Cystic Fibrosis Foundation, University of Washington Center for Excellence in Genomic Science Grant 5P50HG002351 (to M.V.O.), Cystic Fibrosis Foundation Grant MILLEROOVO (to S.I.M. and M.V.O.), and National Institutes of Health Grant 1 R01 DK64954-01A1 (to S.I.M. and M.V.O.).
Abbreviation
- CF
cystic fibrosis.
Conflict of interest statement: No conflicts declared.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. DQ470842).
See Commentary on page 8305.
References
- 1.Cystic Fibrosis Foundation. Annual Report 2002. Bethesda: Cystic Fibrosis Foundation; 2002. [Google Scholar]
- 2.Govan J. R., Deretic V. Microbiol. Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Struelens M. J., Schwam V., Deplano A., Baran D. J. Clin. Microbiol. 1993;31:2320–2326. doi: 10.1128/jcm.31.9.2320-2326.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Romling U., Wingender J., Muller H., Tummler B. Appl. Environ. Microbiol. 1994;60:1734–1738. doi: 10.1128/aem.60.6.1734-1738.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin D. W., Schurr M. J., Mudd M. H., Govan J. R., Holloway B. W., Deretic V. Proc. Natl. Acad. Sci. USA. 1993;90:8377–8381. doi: 10.1073/pnas.90.18.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hancock R. E., Mutharia L. M., Chan L., Darveau R. P., Speert D. P., Pier G. B. Infect. Immun. 1983;42:170–177. doi: 10.1128/iai.42.1.170-177.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luzar M. A., Montie T. C. Infect. Immun. 1985;50:572–576. doi: 10.1128/iai.50.2.572-576.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oliver A., Canton R., Campo P., Baquero F., Blazquez J. Science. 2000;288:1251–1254. doi: 10.1126/science.288.5469.1251. [DOI] [PubMed] [Google Scholar]
- 9.Tang H. B., DiMango E., Bryan R., Gambello M., Iglewski B. H., Goldberg J. B., Prince A. Infect. Immun. 1996;64:37–43. doi: 10.1128/iai.64.1.37-43.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sato H., Okinaga K., Saito H. Microbiol. Immunol. 1988;32:131–139. doi: 10.1111/j.1348-0421.1988.tb01372.x. [DOI] [PubMed] [Google Scholar]
- 11.Meyer J. M., Neely A., Stintzi A., Georges C., Holder I. A. Infect. Immun. 1996;64:518–523. doi: 10.1128/iai.64.2.518-523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mao E. F., Lane L., Lee J., Miller J. H. J. Bacteriol. 1997;179:417–422. doi: 10.1128/jb.179.2.417-422.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spencer D. H., Kas A., Smith E. E., Raymond C. K., Sims E. H., Hastings M., Burns J. L., Kaul R., Olson M. V. J. Bacteriol. 2003;185:1316–1325. doi: 10.1128/JB.185.4.1316-1325.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aires J. R., Kohler T., Nikaido H., Plesiat P. Antimicrob. Agents Chemother. 1999;43:2624–2628. doi: 10.1128/aac.43.11.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Westbrock-Wadman S., Sherman D. R., Hickey M. J., Coulter S. N., Zhu Y. Q., Warrener P., Nguyen L. Y., Shawar R. M., Folger K. R., Stover C. K. Antimicrob. Agents Chemother. 1999;43:2975–2983. doi: 10.1128/aac.43.12.2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sobel M. L., McKay G. A., Poole K. Antimicrob. Agents Chemother. 2003;47:3202–3207. doi: 10.1128/AAC.47.10.3202-3207.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pearson J. P., Gray K. M., Passador L., Tucker K. D., Eberhard A., Iglewski B. H., Greenberg E. P. Proc. Natl. Acad. Sci. USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schuster M., Lostroh C. P., Ogi T., Greenberg E. P. J. Bacteriol. 2003;185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh P. K., Schaefer A. L., Parsek M. R., Moninger T. O., Welsh M. J., Greenberg E. P. Nature. 2000;407:762–764. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- 20.Galan J. E., Collmer A. Science. 1999;284:1322–1328. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- 21.Hirakata Y., Srikumar R., Poole K., Gotoh N., Suematsu T., Kohno S., Kamihira S., Hancock R. E., Speert D. P. J. Exp. Med. 2002;196:109–118. doi: 10.1084/jem.20020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hollsing A. E., Granstrom M., Vasil M. L., Wretlind B., Strandvik B. J. Clin. Microbiol. 1987;25:1868–1874. doi: 10.1128/jcm.25.10.1868-1874.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irie Y., Mattoo S., Yuk M. H. J. Bacteriol. 2004;186:5692–5698. doi: 10.1128/JB.186.17.5692-5698.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burns J. L., Gibson R. L., McNamara S., Yim D., Emerson J., Rosenfeld M., Hiatt P., McCoy K., Castile R., Smith A. L., Ramsey B. W. J. Infect. Dis. 2001;183:444–452. doi: 10.1086/318075. [DOI] [PubMed] [Google Scholar]
- 25.Semmler A. B., Whitchurch C. B., Mattick J. S. Microbiology. 1999;145:2863–2873. doi: 10.1099/00221287-145-10-2863. [DOI] [PubMed] [Google Scholar]
- 26.Meyer J. M., Stintzi A., De Vos D., Cornelis P., Tappe R., Taraz K., Budzikiewicz H. Microbiology. 1997;143:35–43. doi: 10.1099/00221287-143-1-35. [DOI] [PubMed] [Google Scholar]
- 27.Raymond C. K., Sims E. H., Kas A., Spencer D. H., Kutyavin T. V., Ivey R. G., Zhou Y., Kaul R., Clendenning J. B., Olson M. V. J. Bacteriol. 2002;184:3614–3622. doi: 10.1128/JB.184.13.3614-3622.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saiman L., Burns J. L., Whittier S., Krzewinski J., Marshall S. A., Jones R. N. J. Clin. Microbiol. 1999;37:2987–2991. doi: 10.1128/jcm.37.9.2987-2991.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sokol P. A., Ohman D. E., Iglewski B. H. J. Clin. Microbiol. 1979;9:538–540. doi: 10.1128/jcm.9.4.538-540.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoffman L. R., D’Argenio D. A., MacCoss M. J., Zhang Z., Jones R. A., Miller S. I. Nature. 2005;436:1171–1175. doi: 10.1038/nature03912. [DOI] [PubMed] [Google Scholar]
- 31.Lukashin A. V., Borodovsky M. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ng P. C., Henikoff S. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith R. S., Wolfgang M. C., Lory S. Infect. Immun. 2004;72:1677–1684. doi: 10.1128/IAI.72.3.1677-1684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Macfarlane E. L., Kwasnicka A., Hancock R. E. Microbiology. 2000;146:2543–2554. doi: 10.1099/00221287-146-10-2543. [DOI] [PubMed] [Google Scholar]
- 35.Mahenthiralingam E., Campbell M. E., Speert D. P. Infect. Immun. 1994;62:596–605. doi: 10.1128/iai.62.2.596-605.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hickman J. W., Tifrea D. F., Harwood C. S. Proc. Natl. Acad. Sci. USA. 2005;102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoon S. S., Hennigan R. F., Hilliard G. M., Ochsner U. A., Parvatiyar K., Kamani M. C., Allen H. L., DeKievit T. R., Gardner P. R., Schwab U., et al. Dev. Cell. 2002;3:593–603. doi: 10.1016/s1534-5807(02)00295-2. [DOI] [PubMed] [Google Scholar]
- 38.Bagge N., Ciofu O., Hentzer M., Campbell J. I., Givskov M., Hoiby N. Antimicrob. Agents Chemother. 2002;46:3406–3411. doi: 10.1128/AAC.46.11.3406-3411.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sobel M. L., Hocquet D., Cao L., Plesiat P., Poole K. Antimicrob. Agents Chemother. 2005;49:1782–1786. doi: 10.1128/AAC.49.5.1782-1786.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.