Abstract
Nonsegmented negative-sense (nsNS) RNA viruses typically replicate within the host cell cytoplasm and do not have access to the host mRNA capping machinery. These viruses have evolved a unique mechanism for mRNA cap formation in that the guanylyltransferase transfers GDP rather than GMP onto the 5′ end of the RNA. Working with vesicular stomatitis virus (VSV), a prototype nsNS RNA virus, we now provide genetic and biochemical evidence that its mRNA cap methylase activities are also unique. Using recombinant VSV, we determined the function in mRNA cap methylation of a predicted binding site in the polymerase for the methyl donor, S-adenosyl-l-methionine. We found that amino acid substitutions to this site disrupted methylation at the guanine-N-7 (G-N-7) position or at both the G-N-7 and ribose-2′-O (2′-O) positions of the mRNA cap. These studies provide genetic evidence that the two methylase activities share an S-adenosyl-l-methionine-binding site and show that, in contrast to other cap methylation reactions, methylation of the G-N-7 position is not required for 2′-O methylation. These findings suggest that VSV evolved an unusual strategy of mRNA cap methylation that may be shared by other nsNS RNA viruses.
Keywords: capping, evolution, methyltransferase, S-adenosyl-l-methionine
Eukaryotic mRNAs possess at their 5′ terminus a 7mGpppN cap structure that is essential for mRNA stability and efficient translation (1, 2). Formation of this structure requires a series of enzymatic reactions. First, the 5′ triphosphate end of the nascent mRNA chain is acted on by a RNA triphosphatase to yield the diphosphate 5′ ppN, which is capped by RNA guanylyltransferase (GTase). GTase transfers GMP through a 5′–5′ linkage to yield GpppN. The capping guanylate is then methylated by a guanine-N-7 (G-N-7) MTase to yield 7mGpppN. The cap structure can then be further methylated by a ribose-2′-O (2′-O) MTase to yield 7mGpppNmpNpN (3). The mRNA capping reactions are conserved among all eukaryotes (4). The structures of the enzymes that catalyze these reactions have been determined, and their mechanism of action has been examined (5–7).
Viruses of eukaryotes use the host translational machinery, and with the exception of viruses that use internal ribosome entry sites (8), they do so by mRNA cap-dependent mechanisms. Many viruses have evolved their own mRNA capping machinery, among the best-studied example of which is the poxvirus vaccinia virus (VV). For VV, the RNA triphosphatase, GTase, and G-N-7 MTase activities are provided by a complex of two viral proteins, D1R and D12L (9, 10). A separate viral enzyme, VP39, provides 2′-O MTase activity (11). Other viruses have evolved distinctive approaches to cap formation. For example, the orthomyxoviruses, such as influenza A, encode a cap-dependent endonuclease that steals host cell mRNA caps to prime viral mRNA synthesis (12–14). The alphaviruses, such as Sindbis, have evolved an S-adenosyl-l-methionine (AdoMet)-dependent GTase activity that results in the transfer of methylated GMP to form the 7mGpppN cap (15). The nonsegmented negative-sense (nsNS) RNA viruses use a unique method of cap formation. For vesicular stomatitis virus (VSV) (16), spring viremia of carp virus (17), and human respiratory syncytial virus (18), the GTase transfers GDP rather than GMP. The cap structure of these viruses is methylated, usually at both the G-N-7 and 2′-O positions (19–26), but the details of these reactions are poorly understood.
Capping reactions of nsNS RNA viruses have been difficult to dissect, in part because the machinery does not respond to exogenous transcripts (27). For VSV, complementation studies mapped the methylase activities to the L gene (19), which encodes the 241-kDa large subunit (L) of the viral RNA-dependent RNA polymerase. Sequence comparisons among L proteins of nsNS RNA viruses identified six conserved regions, I–VI (28). Signature polymerase motifs are present in region III of L protein, suggesting that it contains the active site for ribonucleotide polymerization, and this assignment was supported by L gene mutations (29). Sequence alignments between 2′-O MTases and region VI of L protein identified a binding site for the methyl donor AdoMet and suggested that the active site comprised a conserved K-D-K-E catalytic tetrad (30, 31). Consistent with a role in methylation, a fragment of Sendai virus (SeV) L protein that includes region VI was shown to exclusively G-N-7-methylate short, SeV-specific mRNAs (32). For VSV, amino acid substitutions in region VI of L were shown to disrupt both G-N-7 and 2′-O methylation (33, 34). Although these studies supported a role for region VI of L in cap methylation, they did not determine whether predicted catalytic or AdoMet-binding residues were required for 2′-O and/or G-N-7 methylation. The triphosphatase and GTase activities are less well understood, although recent studies with human respiratory syncytial virus showed that resistance mutations to a chemical inhibitor that affected formation of the GpppN cap mapped to region V of L (35).
In the present study, we evaluated the role of the predicted AdoMet-binding site in region VI of VSV L protein on mRNA cap methylation. We generated eight viruses with amino acid substitutions throughout this region. These viruses exhibit defects in cap methylation in vitro. Some substitutions resulted in defects only in G-N-7 methylation, whereas others prevented all cap methylation. These data support a unique strategy of cap methylation in which both methylases use a single AdoMet-binding site and in which G-N-7 methylation is not required for 2′-O methylation. These studies show that the cap methylase activities of VSV, like those of its GTase, are distinct to those of the host and provide evidence that the entire capping apparatus evolved a separate mechanism for mRNA cap formation.
Results
Amino Acid Changes to a Predicted AdoMet-Binding Site in VSV L Protein.
The AdoMet-dependent MTase superfamily contains a series of conserved motifs including a G-rich motif and an acidic residue (D/E) that is involved in binding AdoMet (36). Sequence alignments between nsNS RNA virus L proteins and MTases (30, 31) suggest that the AdoMet-binding residues of VSV L include G1670, G1672, G1674, G1675, and D1735 (Fig. 1). To test the role of these residues in mRNA cap methylation, we engineered the L gene of an infectious cDNA clone of VSV to introduce substitutions in the predicted AdoMet-binding site. Each residue was individually substituted for alanine; or, for G4A, all four G residues were replaced with A; for G4AD, residue D1735 was also replaced with A. We also chose to modify flanking amino acid residues D1671 and S1673, of which D1671V was shown to prevent cap methylation in vitro (34).
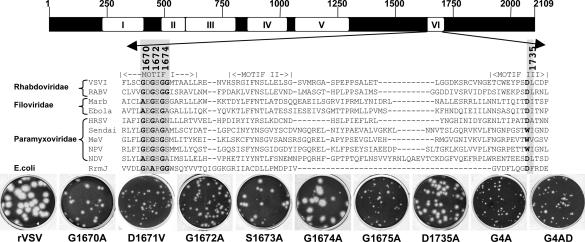
Fig. 1.
AdoMet-binding site alterations. (Upper) Amino acid sequence alignments of a predicted AdoMet-binding region of domain VI of nsNS RNA virus L proteins and known RNA methylases. The conserved motifs of nsNS RNA virus polymerases (I–VI) are shown (28). The AdoMet-binding residues modified in this study are shaded. VSVI, VSV Indiana; RABV, rabies virus; Marb, Marburg; HRSV, human respiratory syncytial virus; MeV, measles virus; NPV, Nipah virus; NDV, Newcastle disease virus; RrmJ, Escherichia coli 2′-O MTase. (Lower) Plaque morphology of recombinant viruses on Vero cells. Plaques of rVSV and G1674A were developed after 24 h; those of G1670A, G1672A, G1675A, D1735A, D1671V, and S1673A were developed after 48 h; those of G4A and G4AD were developed after 96 h.
Recovery of Recombinant VSV (rVSV) with L Gene Mutations.
Recombinant viruses were recovered from each of the L gene mutations. The viruses showed defects in viral growth as judged by their plaque morphology (Fig. 1 and Table 1). The entire L gene of each virus was amplified by RT-PCR, and sequence analysis confirmed the presence of the mutation in seven of eight viruses. Recombinant G4AD encoded wild-type glycine at amino acids 1670 and 1672. Multiple attempts to isolate a virus containing all five substitutions were unsuccessful. Recombinant D1671V showed a noncoding change, A5776C. No other substitutions were detected within the L gene of these viruses.
Table 1.
Summary of phenotypic properties of VSV L gene mutants
| Mutant | Plaque size, mm | Titer, 24 h, log10 pfu/ml | RNA synthesis |
Protein | 7mG% [AdoMet] |
GpppAm, % | ||
|---|---|---|---|---|---|---|---|---|
| Cells | In vitro | 1 mM | 0.2 mM | |||||
| rVSV | 4.1 ± 0.5* | 9.7 ± 0.2 | 100 | 100 | 100 | 97 | 94 | 2 |
| G1670A | 2.8 ± 0.4† | 8.8 ± 0.1 | 108 | 90 | 80 | 20 | <1 | 75 |
| G1672A | 2.2 ± 0.3† | 8.7 ± 0.2 | 110 | 100 | 90 | 10 | <1 | 80 |
| G1674A | 4.2 ± 0.6* | 9.8 ± 0.1 | 110 | 110 | 105 | 92 | 36 | 5 |
| G1675A | 1.5 ± 0.3† | 7.5 ± 0.2 | 50–250§ | 50 | 75 | 15 | <1 | <1 |
| G4A | 1.4 ± 0.3‡ | 6.1 ± 0.4 | 55–350§ | 40 | 55–110¶ | <1 | <1 | <1 |
| D1735A | 3.0 ± 0.4† | 8.6 ± 0.2 | 70 | 55 | 40–90¶ | 30 | 10 | 15 |
| D1671V | 1.8 ± 0.3† | 6.8 ± 0.1 | 70 | 75 | 85 | <1 | <1 | <1 |
| S1673A | 3.1 ± 0.4† | 8.4 ± 0.1 | 70 | 60 | 35–90¶ | 20 | 10 | 75 |
| G4AD | 1.6 ± 0.3‡ | 6.4 ± 0.1 | 75–370§ | 35 | 80–110¶ | <1 | <1 | <1 |
pfu, plaque-forming units.
*Plaque diameter was measured at 24 h after inoculation.
†Plaque diameter was measured at 48 h after inoculation.
‡Plaque diameter was measured at 96 h after inoculation.
§Percentage of RNA varied as follows: P/M (50%)-V (250%); P/M (55%)-V (350%); P/M (75%)-V (370%).
¶Percentage of protein varied as follows: L (55%)-N (110%); L (40%)-N (90%); L (35%)-N (90%); L (80%)-N (110%).
To examine the effect of these mutations on viral growth, we determined the yield of virus from infected cells. Briefly, BHK-21 cells were infected at a multiplicity of infection (moi) of 3, and the viral titer was determined at 24 h after inoculation. The average titers from three experiments are shown (Table 1). Virus yield correlated with plaque morphology in that recombinants G1670A, G1672A, D1735A, and S1673A had a 1–1.5 logarithmic growth defect compared with rVSV. Recombinants G1675A and D1671V had a 2–2.5 logarithmic growth defect, and G4A and G4AD had a >3 logarithmic growth defect.
L Gene Mutations Disrupt G-N-7 Methylation.
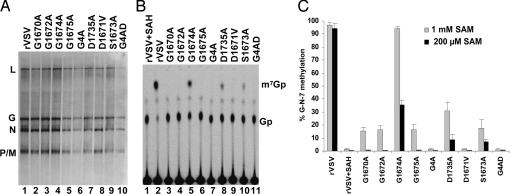
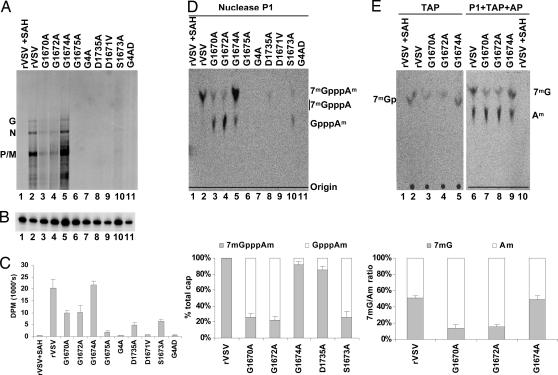
To determine whether the L gene mutations affect methylation, transcription reactions were performed in vitro in the presence of [α-32P]GTP or UTP. RNA was extracted and analyzed by electrophoresis on acid-agarose gels. Each virus synthesized the five viral mRNAs, although the yield from G1675A, G4A, D1735A, D1671A, S1673A, and G4AD was diminished compared with rVSV (Fig. 2A). To determine the effect of these mutations on G-N-7 methylation, the products were digested with tobacco acid pyrophosphatase (TAP) (33). TAP cleaves the pyrophosphate bond of the GpppN cap but does not cleave the mRNA, liberating Gp, or 7mGp if the cap was methylated (37). These products are resolved by TLC on polyethyleneimine (PEI) cellulose F sheets. For rVSV, when reactions were performed in the presence of 200 μM AdoMet, a single product of TAP cleavage was observed that comigrated with 7mGp (Fig. 2B, lane 2). Reactions performed in the presence of S-adenosyl-homocysteine (SAH), the byproduct formed upon methyl group transfer from AdoMet during methylation, yield Gp, indicating that the cap structure was not methylated (Fig. 2B, lane 1). Each of the mutants was defective in G-N-7 methylation (Fig. 2B, lanes 3–11). Quantitative analysis (Fig. 2C) showed that 7mGp accounted for 40% of the released cap structure for G1674A (Fig. 2B, lane 5) and 10% of the released cap structure for D1735A and S1673A (Fig. 2B, lanes 8 and 10). Recombinants G1670A, G1672A, G1675A, G4A, D1671A, and G4AD showed no detectable G-N-7 methylation, although each generated capped mRNA (Fig. 2B, lanes 3, 4, 6, 7, 9, and 11).
Fig. 2.
Effect of L gene mutations on G-N-7 methylation. (A) Transcription reactions were performed in the presence of [α-32P]GTP, RNA was analyzed by electrophoresis on acid-agarose gels, and products were detected by using a phosphoimager. The virus and the migration of the RNA are shown. (B) RNA was synthesized in the presence of 200 μM AdoMet or SAH and 15 μCi of [α-32P]GTP and digested with 2 units of TAP, and the products were analyzed by TLC on PEI cellulose F sheets. Plates were dried, and the spots were visualized with a phosphoimager. The migration of the markers 7mGp and Gp are shown. (C) Quantitative analysis of three independent experiments. For each virus, the released 7mGp (mean ± SD) was expressed as a percentage of the total released cap structure.
Alterations to the AdoMet-binding site might alter the binding affinity of L protein for AdoMet. To test this, reactions were also performed in the presence of 1 mM AdoMet. Under these conditions, 7mGp accounted for 96% of the cap structure for G1674A (Fig. 2C). For D1735A and S1673A, the extent of G-N-7 methylation was increased to 40% and 20%, respectively (Fig. 2C). In addition, G1670A, G1672A, and G1675A all showed a low level of G-N-7 methylation ranging from 10% to 22% of the total cap structure (Fig. 2C). Even at 1 mM AdoMet, G4A, D1671V, and G4AD failed to produce detectable levels of 7mGp (Fig. 2C). These data show that substitutions to the predicted AdoMet-binding site diminish G-N-7 methylation (Fig. 5, which is published as supporting information on the PNAS web site).
Effect of L Gene Mutations on G-N-7 and 2′-O Methylation.
To examine the effect of these mutations on both methylations, transcription reactions were performed in the presence of [3H]AdoMet, and RNA was analyzed by electrophoresis (Fig. 3A). The N, P, M, and G mRNAs were visualized for rVSV, but not when reactions were supplemented with SAH (Fig. 3A, lanes 1 and 2). Similar amounts of labeled RNA were observed for G1674A (Fig. 3A, lane 5), consistent with efficient methylation. Recombinants G1670A, G1672A, and S1673A generated detectable levels of RNA (Fig. 3A, lanes 3, 4, and10). Methylated RNA was not detected for G1675A, G4A, D1735A, D1671V, or G4AD (Fig. 3A, lanes 6–9 and 11).
Fig. 3.
Effect of L gene mutations on 2′-O and G-N-7 methylation. (A) RNA was synthesized in the presence of [3H]AdoMet and analyzed by electrophoresis on acid-agarose gels. The virus and the identity of the mRNAs are shown. (B) RNA from A was examined by primer extension assay by using a primer designed to anneal to the N mRNA. (C) [3H]AdoMet incorporation monitored by scintillation counting. Three independent experiments were used to generate the graph shown. (D and E) (Upper) RNA was digested with P1, TAP, and AP, and the products were analyzed by TLC on PEI cellulose F sheets. Plates were dried, and the spots were visualized with a phosphoimager. The identity of the virus and the migration of the markers 7mGpppA, GpppA, 7mG, and 2′-OmA are shown. (Lower) Quantitative analysis of three independent experiments is shown. For each virus, the fraction of the mRNA cap that was 7mGpppAm and GpppAm or 7mG and 2′-OmA is shown (mean ± SD).
To confirm that mRNA was present, the samples shown in Fig. 3A were examined by primer extension assay. For each virus, an 80-nt product was detected, which corresponded to the 5′ end of the N mRNA (Fig. 3B). Quantitative analysis (data not shown) showed that the levels of N mRNA detected were consistent with the [32P]GTP incorporation data (Fig. 2A). The amount of [3H] incorporated into the RNA was determined by scintillation counting and the dpm normalized to the amount of RNA synthesized (Fig. 3C). The level of incorporation of [3H]AdoMet into RNA by recombinants G1670A and G1672A was 50% that of rVSV. Based on the reduced G-N-7 methylation (Fig. 2B), these data suggested that the mRNAs synthesized by G1670A and G1672A might be fully 2′-O-methylated.
To examine this, the [3H]AdoMet-labeled products of transcription were subjected to nuclease P1 digestion followed by TLC on PEI cellulose F sheets. P1 cleaves the bond between the 3′-hydroxyl and 5′-phosphoryl group of adjacent nucleosides. Cleavage of VSV mRNAs by P1 should yield 7mGpppAm, GpppAm, 7mGpppA, or GpppA, depending on the extent of cap methylation. For rVSV, a single [3H] product of P1 cleavage was observed (Fig. 3D, lane 2), consistent with the fully methylated cap structure 7mGpppAm. No detectable products were seen when SAH was included in the reaction (Fig. 3D, lane 1). The extent of cap methylation varied for each mutant. For G1670A and G1672A, two products of P1 cleavage were visible. Approximately 20% of the released cap comigrated with the product obtained from rVSV, suggesting that it represented 7mGpppAm (Fig. 3D, lanes 3 and 4). The remaining 80% did not comigrate with a 7mGpppA marker, suggesting that it was GpppAm. For G1674A, 95% of the cap structure migrated with 7mGpppAm (Fig. 3D, lane 5). The remaining mutations affected all cap methylation (Fig. 3D, lanes 6–11). Low levels of 7mGpppAm were detected for D1735A (Fig. 3D, lane 8), and some 7mGpppAm and the potential GpppAm product was detected for S1673A (Fig. 3D, lane 10). Taken together, these experiments suggested that the mRNA caps of G1670A and G1672A are 2′-O-methylated but not efficiently G-N-7-methylated, that G1674A has a slight defect in G-N-7 methylation, and that all other substitutions affected both methylase activities. The in vitro synthesis reactions were performed in the presence of a cell lysate to increase RNA yields and facilitate detection of the 3H-labeled cap structures. These conditions did not significantly affect G-N-7 methylation (Fig. 6, which is published as supporting information on the PNAS web site).
To confirm that G1670A and G1672A were defective in G-N-7 methylation but not 2′-O methylation, we performed additional cap hydrolysis experiments. The [3H]AdoMet-labeled RNA of rVSV, G1670A, G1672A, and G1674A was digested with combinations of P1, TAP, and alkaline phosphatase (AP) (Fig. 3E). TAP digestion of rVSV and G1674A mRNA released 7mGp, 15% of which was observed on cleavage of G1670A and G1672A mRNA (Fig. 3E, lanes 1–5). Digestion of rVSV and G1674A RNA with P1, TAP, and AP yielded two spots of equal intensity that comigrated with unlabeled 7mG and 2′-OmA markers (Fig. 3E, lanes 6 and 9). By contrast, digestion of G1670A and G1672A mRNAs showed Am levels higher than 7mG (Fig. 3E, lanes 7 and 8). These data confirm that G1670A and G1672A are defective in G-N-7 methylation but not 2′-O methylation. Additional cap hydrolysis experiments provide further support for this finding (Fig. 7, which is published as supporting information on the PNAS web site).
Effect of L Gene Mutations on Viral Gene Expression.
The above experiments showed that alterations to a predicted AdoMet-binding motif in L protein diminished viral replication in cell culture and cap methylation in vitro. We expected that this reduction would be accompanied by a decrease in viral gene expression in infected cells.
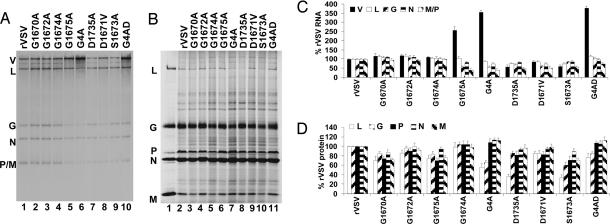
To examine viral RNA synthesis, BHK-21 cells were infected at an moi of 3, and RNA was labeled by incorporation of [3H]uridine in the presence of actinomycin D from 3 to 6 h after inoculation. Total cytoplasmic RNA was extracted, purified, and analyzed by electrophoresis on acid-agarose gels (Fig. 4A). Quantitative analysis showed that levels of replication were enhanced 2.5-fold for G1675A, yet transcription was reduced 2-fold (Fig. 4A, lane 5). This effect was more pronounced when G1675A was combined with substitutions G1674A and D1735A (Fig. 4A, lane 6) or G1670A, 1672A, and 1674A (Fig. 4A, lane 10) such that replication was enhanced 4-fold over rVSV levels. When mRNA levels were normalized to the replication products (V), recombinants G1675A, G4A, and G4AD synthesized 15–25% of the mRNA per genome compared with rVSV (Fig. 4C).
Fig. 4.
Effect of L gene mutations on viral gene expression in BHK-21 cells. (A) Cells were infected at an moi of 3, and RNAs were labeled with [3H]uridine, resolved by electrophoresis on acid-agarose gels, and visualized by fluorography. RNA extracted from an equivalent number of cells was loaded in each lane. The virus and the identity of the RNAs are shown. V, replication products; L, G, N, and P/M, mRNA. (B) Proteins were labeled by incorporation of [35S]Express, and cytoplasmic extracts were analyzed by SDS/PAGE and detected by using a phosphoimager. Extract from equivalent numbers of cells was loaded in each lane. The virus and the identity of the proteins are shown. (C and D) Quantitative analysis of RNA (C) and protein abundance (D). The mean ± SD was expressed as a percentage of that observed for rVSV from three independent experiments.
To examine viral protein synthesis, BHK-21 cells were infected at an moi of 3, and proteins were labeled by the incorporation of [35S]Met-Cys from 3 to 6 h after inoculation. Cytoplasmic extracts were prepared and analyzed by SDS/PAGE (Fig. 4B). Quantitative analysis showed 2-fold less L protein in cells infected with recombinants G4A, D1735A, and S1673A (Fig. 4D). Differences in the abundance of other viral proteins were modest. These data demonstrate that the levels of protein synthesized did not correlate well with the levels of mRNA synthesized.
Discussion
Using genetic and biochemical approaches, we determined the role of a predicted AdoMet-binding motif in region VI of VSV L protein in mRNA cap methylation. We generated eight recombinant viruses with amino acid changes throughout the predicted AdoMet-binding site and examined the effect of these alterations on cap methylation and gene expression. The data show that a single predicted AdoMet-binding site is required for both mRNA cap methylase activities and that G-N-7 methylation is not required for 2′-O methylation. These experiments provide evidence that the nsNS RNA viruses have evolved a unique strategy of cap methylation. The RNA GTase activities of these viruses are also unique, demonstrating that the entire capping apparatus of these viruses evolved a separate mechanism to that of their hosts.
A Single AdoMet-Binding Site for both mRNA Cap Methylases.
Sequence alignments suggested the presence of an AdoMet-binding site within region VI of the L protein of nsNS RNA viruses (30, 31). Alterations to this site reduced either G-N-7 or both G-N-7 and 2′-O methylation. However, none of the substitutions resulted in defects only in 2′-O methylation. The observation that G1670A and G1672A diminished G-N-7 methylation but not 2′-O methylation demonstrates that, in contrast to other mRNA cap methylation reactions (3), G-N-7 is not required for 2′-O methylation. Recombinant S1673A showed significantly reduced cap methylation, but the defect in G-N-7 was more pronounced than that in 2′-O. All other substitutions affected both methylations equally, suggesting that the two activities use the same AdoMet-binding site.
In prior work, the Km of the two methylase activities for AdoMet was shown to be 0.5 μM for 2′-O and 10 μM for G-N-7 (26). We show that substitutions at G1670 and G1672 inhibit G-N-7 methylation but not 2′-O methylation. A plausible explanation is that these changes reduce the efficiency of AdoMet binding such that only G-N-7 activity is affected, consistent with its higher Km. This finding could also explain the observation that G1675A had partial G-N-7 activity at 1 mM AdoMet but not at 200 μM AdoMet (Fig. 2C). The remaining substitutions might increase the Km for AdoMet to a level at which L protein no longer binds AdoMet even at 1 mM.
Biochemical and structural studies of other cap methylases suggest that the mechanism by which 2′-O and G-N-7 methylation are catalyzed are distinct (4, 7, 38–40), which seems incompatible with the use of a single AdoMet-binding site for both activities. However, in most systems, the two activities are catalyzed by separate proteins, each of which has its own AdoMet-binding site, or, in the case of reovirus, the two activities are catalyzed by separate domains of the same protein (41, 42). Here, we show that amino acid substitutions to a single predicted AdoMet-binding site affect either G-N-7 methylation or both 2′-O and G-N-7 methylation. These data are consistent with both methylases using the same AdoMet-binding site and suggest that, if there is an obligatory order of methylation for VSV, 2′-O occurs first. Perhaps AdoMet and RNA bind the methylase domain, and the mRNA is first methylated at the 2′-O position, which induces a conformational change in L such that a subsequent molecule of AdoMet binds and favors the G-N-7 activity.
A Different Order of mRNA Cap Methylation?
Conventional cap methylation occurs through a series of reactions where two separate enzymes sequentially methylate the RNA, with G-N-7 occurring first (3). Structural and biochemical analysis of the vaccinia virus 2′-O MTase, VP39, shows that the 7mG stabilizes RNA binding to VP39 (38–40, 43). A host range mutant of VSV, hr8, was shown to synthesize mRNA cap structures that lacked G-N-7 but were partially 2′-O-methylated (44). Here, we found that substitutions to a predicted AdoMet-binding site in VSV L protein affected either G-N-7 alone or both G-N-7 and 2′-O methylases. Two possible explanations are consistent with these observations: either there is no mandatory order to the cap methylation reactions of VSV, or 2′-O methylation is required for G-N-7 methylation. In prior work, 2′-O-methylated RNAs were chased into fully methylated mRNAs in vitro (26), consistent with a distinctive order of methylation. However, other studies with VSV also support the conventional order (45, 46). For other nsNS RNA viruses, a fragment of Sendai virus L protein that includes region VI was shown to exclusively G-N-7-methylate (32), and Newcastle disease virus produces mRNAs that are not 2′-O-methylated (47). These studies show that 2′-O is not required for G-N-7 methylation in these two paramyxoviruses, indicating that the order of cap methylation in nsNS RNA viruses is not mandatory. It will be of interest to determine whether the predicted catalytic residues, previously shown to be essential for all cap methylation in VSV (33), directly participate in both methylation reactions.
Model for 5′ End mRNA Modifications.
The details of the mechanism of VSV mRNA synthesis are beginning to emerge. We propose that our findings fit with the emerging model in the following way. The polymerase initiates mRNA synthesis in response to a specific gene-start sequence, and at some point shortly after initiation, the nascent RNA transcript gains access to the mRNA capping machinery, where a series of sequential reactions occurs. A yet to be identified phosphatase trims two phosphates from the 5′ end of the initiated RNA, and an unidentified GTase activity transfers GDP onto the nascent RNA chain to form a 5′–5′ GpppA cap structure. These two activities presumably reside within L protein. The nascent RNA chain is then methylated, likely first at the 2′-O position and second at the G-N-7 position. These two methylase activities have a unique property in that they appear to share a single binding site for the methyl donor, AdoMet.
A Role for AdoMet Binding in Regulating Polymerase.
Remarkably, viruses G1675A, G4A, and G4AD showed a significant alteration in the products of transcription and replication (Fig. 4A). Replication was enhanced 2.5- to 4-fold, and transcription decreased up to 8-fold compared with rVSV. Although we do not know the basis for this perturbation in polymerase activity, it is tempting to speculate that AdoMet binding directly influences the template activity of the polymerase. Perhaps binding of AdoMet to L protein favors a conformation that is adopted by the transcriptase, whereas L protein that lacks AdoMet adopts a conformation that favors formation of the replicase. Additional experiments are needed to clarify this intriguing phenotype.
Rational Attenuation of nsNS RNA Viruses Through Ablation of Their Methylase Activities.
These studies, combined with our earlier work, suggest that ablating nsNS RNA virus cap methylation might represent a useful way to rationally attenuate these viruses for development of live attenuated vaccines and their exploitation as viral vectors for vaccines (48), oncolytic therapy (49), and gene delivery (50). We showed previously that substitutions to the conserved MTase catalytic residues KDKE diminished virus yield 1–3 logs in cell culture (33), and in this study, we demonstrate that substitutions within the AdoMet-binding site similarly diminish virus replication. In both cases, these substitutions ablate the same function: mRNA cap methylation. By combining multiple substitutions within this region, it should be possible to generate an attenuated virus that is genetically stable, because reversion to wild type at any single amino acid should not provide a fitness gain.
In summary, we show that the two mRNA cap methylase activities of VSV use a single predicted AdoMet-binding site to methylate the viral mRNA and that 2′-O methylation can occur without G-N-7 methylation. These data add a significant dimension to the concept that the mRNA capping reactions of the nsNS RNA viruses are unique and represent attractive targets for antiviral intervention by providing genetic and biochemical evidence that demonstrates that the cap methylase activities of these viruses are also unusual.
Materials and Methods
Plasmid Construction and Recovery of VSV.
Plasmids encoding the VSV N, P, and L proteins and an infectious cDNA clone of VSV, pVSV1(+), were as described in ref. 51. Mutagenesis and sequence analysis of the L gene was performed as described in ref. 33. rVSV was recovered from cDNA by transfection of BSR-T7 cells (52) infected with a recombinant vaccinia virus expressing T7 RNA polymerase (53) as described in ref. 54. Cell culture fluids were harvested at 48–96 h after transfection, and the virus was isolated, purified, and sequenced as described in ref. 33.
Transcription of Viral RNA in Vitro.
Viral RNA was synthesized in vitro by using 10 μg of purified virus as described in refs. 55 and 56. Reactions were performed in the presence of 1 mM ATP; 0.5 mM CTP, GTP, and UTP; 0–1 mM AdoMet or SAH; and 15 μCi (1 Ci = 37 GBq) of [α-32P]GTP or UTP (3,000 Ci/mmol) or [3H]AdoMet (85 Ci/mmol, PerkinElmer) as described in ref. 33. A rabbit reticulocyte lysate was used to supplement reactions [30% (vol/vol)] to increase RNA yield for experiments performed with [3H]AdoMet, which also supplies additional AdoMet to the reaction.
Cap Methylase Assays.
Purified RNA was digested with TAP (Epicentre Technologies, Madison, WI), P1 (Sigma), AP (New England Biolabs), and RNase T2 (Invitrogen), and the products were analyzed by TLC on PEI cellulose F sheets (EM Science) as described in ref. 33. To examine G-N-7 or 2′-O methylation, reactions were performed in the presence of [α-32P]GTP and 0–1 mM AdoMet/SAH or 0–20 μM [3H]AdoMet, respectively. Cap markers 7mGpppA and GpppA (NEB, Beverly, MA) and 7mG and mA (Sigma) were visualized by UV shadowing.
Primer Extension Assays.
A minus sense oligonucleotide, nucleotides 130–115 of the VSV genome, was end-labeled by using [γ-32P]ATP and T4 polynucleotide kinase (Invitrogen). The labeled primer was purified, and 7.5 pmol was annealed with 1/25 of the total RNA from an in vitro transcription reaction and extended by Superscript III reverse transcriptase (Invitrogen) at 50°C (56). Products were analyzed by electrophoresis on denaturing 6% polyacrylamide gels and detected by phosphoimage analysis.
Scintillation Counting.
Aliquots of purified RNA were mixed with 4 ml of ReadySafe scintillation mixture (Beckman Coulter), and dpm was measured by using a 1414 series counter (PerkinElmer).
Gene Expression.
Viral RNA and protein synthesis were examined in BHK-21 cells. Cells were infected at an moi of 3 and exposed to [3H]uridine or [35S]Express from 3 to 6 h after inoculation; cytoplasmic extracts were prepared, and proteins and RNA were analyzed as described in ref. 33.
Quantitative Analysis.
Densitometric scanning of autoradiographs and phosphoimage analysis were as described in ref. 33. Statistical analysis was performed on three to five independent experiments, and the mean ± SD was expressed. The significance of the values was determined by a paired Student t test.
Supplementary Material
Acknowledgments
We thank D. Knipe, M. Nibert, and D. Cureton for critical reviews of the manuscript. This work was supported by National Institutes of Health Grant AI059371 (to S.P.J.W.). S.P.J.W. is the recipient of a Burroughs Wellcome Investigators in Pathogenesis of Infectious Disease Award.
Abbreviations
- AdoMet
S-adenosyl-l-methionine
- nsNS
nonsegmented negative sense
- VSV
vesicular stomatitis virus
- rVSV
recombinant VSV
- GTase
guanylyltransferase
- SAH
S-adenosyl-homocysteine
- 2′-O
ribose-2′-O
- moi
multiplicity of infection
- TAP
tobacco acid pyrophosphatase
- PEI
polyethyleneimine
- AP
alkaline phosphatase.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Muthukrishnan S., Morgan M., Banerjee A. K., Shatkin A. J. Biochemistry. 1976;15:5761–5768. doi: 10.1021/bi00671a012. [DOI] [PubMed] [Google Scholar]
- 2.Furuichi Y., LaFiandra A., Shatkin A. J. Nature. 1977;266:235–239. doi: 10.1038/266235a0. [DOI] [PubMed] [Google Scholar]
- 3.Furuichi Y., Shatkin A. J. Adv. Virus Res. 2000;55:135–184. doi: 10.1016/S0065-3527(00)55003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shuman S. Prog. Nucleic Acid Res. Mol. Biol. 2001;66:1–40. doi: 10.1016/s0079-6603(00)66025-7. [DOI] [PubMed] [Google Scholar]
- 5.Lima C. D., Wang L. K., Shuman S. Cell. 1999;99:533–543. doi: 10.1016/s0092-8674(00)81541-x. [DOI] [PubMed] [Google Scholar]
- 6.Hakansson K., Wigley D. B. Proc. Natl. Acad. Sci. USA. 1998;95:1505–1510. doi: 10.1073/pnas.95.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fabrega C., Hausmann S., Shen V., Shuman S., Lima C. D. Mol. Cell. 2004;13:77–89. doi: 10.1016/s1097-2765(03)00522-7. [DOI] [PubMed] [Google Scholar]
- 8.Pelletier J., Sonenberg N. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 9.Harris N., Rosales R., Moss B. Proc. Natl. Acad. Sci. USA. 1993;90:2860–2864. doi: 10.1073/pnas.90.7.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vos J. C., Sasker M., Stunnenberg H. G. EMBO J. 1991;10:2553–2558. doi: 10.1002/j.1460-2075.1991.tb07795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barbosa E., Moss B. J. Biol. Chem. 1978;253:7698–7702. [PubMed] [Google Scholar]
- 12.Bouloy M., Plotch S. J., Krug R. M. Proc. Natl. Acad. Sci. USA. 1978;75:4886–4890. doi: 10.1073/pnas.75.10.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plotch S. J., Bouloy M., Ulmanen I., Krug R. M. Cell. 1981;23:847–858. doi: 10.1016/0092-8674(81)90449-9. [DOI] [PubMed] [Google Scholar]
- 14.Li M. L., Rao P., Krug R. M. EMBO J. 2001;20:2078–2086. doi: 10.1093/emboj/20.8.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahola T., Kaariainen L. Proc. Natl. Acad. Sci. USA. 1995;92:507–511. doi: 10.1073/pnas.92.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abraham G., Rhodes D. P., Banerjee A. K. Cell. 1975;5:51–58. doi: 10.1016/0092-8674(75)90091-4. [DOI] [PubMed] [Google Scholar]
- 17.Gupta K. C., Roy P. J. Virol. 1980;33:292–303. doi: 10.1128/jvi.33.1.292-303.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barik S. J. Gen. Virol. 1993;74:485–490. doi: 10.1099/0022-1317-74-3-485. Part 3. [DOI] [PubMed] [Google Scholar]
- 19.Hercyk N., Horikami S. M., Moyer S. Virology. 1988;163:222–225. doi: 10.1016/0042-6822(88)90253-x. [DOI] [PubMed] [Google Scholar]
- 20.Keene J. D., Lazzarini R. Virology. 1976;69:364–367. doi: 10.1016/0042-6822(76)90229-4. [DOI] [PubMed] [Google Scholar]
- 21.Moyer S. A., Abraham G., Adler R., Banerjee A. K. Cell. 1975;5:59–67. doi: 10.1016/0092-8674(75)90092-6. [DOI] [PubMed] [Google Scholar]
- 22.Moyer S. A., Banerjee A. K. Virology. 1976;70:339–351. doi: 10.1016/0042-6822(76)90276-2. [DOI] [PubMed] [Google Scholar]
- 23.Rhodes D. P., Moyer S. A., Banerjee A. K. Cell. 1974;3:327–333. doi: 10.1016/0092-8674(74)90046-4. [DOI] [PubMed] [Google Scholar]
- 24.Rhodes D. P., Banerjee A. K. J. Virol. 1975;17:33–42. doi: 10.1128/jvi.17.1.33-42.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rose J. K. J. Biol. Chem. 1975;250:8098–8104. [PubMed] [Google Scholar]
- 26.Testa D., Banerjee A. K. J. Virol. 1977;24:786–793. doi: 10.1128/jvi.24.3.786-793.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shuman S. Virology. 1997;227:1–6. doi: 10.1006/viro.1996.8305. [DOI] [PubMed] [Google Scholar]
- 28.Poch O., Blumberg B. M., Bougueleret L., Tordo N. J. Gen. Virol. 1990;71:1153–1162. doi: 10.1099/0022-1317-71-5-1153. [DOI] [PubMed] [Google Scholar]
- 29.Sleat D. E., Banerjee A. K. J. Virol. 1993;67:1334–1339. doi: 10.1128/jvi.67.3.1334-1339.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bujnicki J. M., Rychlewski L. Protein Eng. 2002;15:101–108. doi: 10.1093/protein/15.2.101. [DOI] [PubMed] [Google Scholar]
- 31.Ferron F., Longhi S., Henrissat B., Canard B. Trends Biochem. Sci. 2002;27:222–224. doi: 10.1016/s0968-0004(02)02091-1. [DOI] [PubMed] [Google Scholar]
- 32.Ogino T., Kobayashi M., Iwama M., Mizumoto K. J. Biol. Chem. 2005;280:4429–4435. doi: 10.1074/jbc.M411167200. [DOI] [PubMed] [Google Scholar]
- 33.Li J., Fontaine-Rodriguez E. C., Whelan S. P. J. Virol. 2005;79:13373–13384. doi: 10.1128/JVI.79.21.13373-13384.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grdzelishvili V. Z., Smallwood S., Tower D., Hall R. L., Hunt D. M., Moyer S. J. Virol. 2005;79:7327–7337. doi: 10.1128/JVI.79.12.7327-7337.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liuzzi M., Mason S. W., Cartier M., Lawetz C., McCollum R. S., Dansereau N., Bolger G., Lapeyre N., Gaudette Y., Lagace L., et al. J. Virol. 2005;79:13105–13115. doi: 10.1128/JVI.79.20.13105-13115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schluckebier G., O’Gara M., Saenger W., Cheng X. J. Mol. Biol. 1995;247:16–20. doi: 10.1006/jmbi.1994.0117. [DOI] [PubMed] [Google Scholar]
- 37.Shinshi H., Miwa M., Sugimura T. FEBS Lett. 1976;65:254–257. doi: 10.1016/0014-5793(76)80492-9. [DOI] [PubMed] [Google Scholar]
- 38.Gershon P. D., Shi X., Hodel A. E. Virology. 1998;246:253–265. doi: 10.1006/viro.1998.9209. [DOI] [PubMed] [Google Scholar]
- 39.Hodel A. E., Gershon P. D., Quiocho F. Mol. Cell. 1998;1:443–447. doi: 10.1016/s1097-2765(00)80044-1. [DOI] [PubMed] [Google Scholar]
- 40.Lockless S. W., Cheng H. T., Hodel A. E., Quiocho F. A., Gershon P. D. Biochemistry. 1998;37:8564–8574. doi: 10.1021/bi980178m. [DOI] [PubMed] [Google Scholar]
- 41.Luongo C. L., Contreras C. M., Farsetta D. L., Nibert M. L. J. Biol. Chem. 1998;273:23773–23780. doi: 10.1074/jbc.273.37.23773. [DOI] [PubMed] [Google Scholar]
- 42.Reinisch K. M., Nibert M. L., Harrison S. C. Nature. 2000;404:960–967. doi: 10.1038/35010041. [DOI] [PubMed] [Google Scholar]
- 43.Hodel A. E., Gershon P. D., Shi X., Quiocho F. Cell. 1996;85:247–256. doi: 10.1016/s0092-8674(00)81101-0. [DOI] [PubMed] [Google Scholar]
- 44.Horikami S. M., Moyer S. Proc. Natl. Acad. Sci. USA. 1982;79:7694–7698. doi: 10.1073/pnas.79.24.7694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moyer S. Virology. 1981;112:157–168. doi: 10.1016/0042-6822(81)90621-8. [DOI] [PubMed] [Google Scholar]
- 46.Hammond D. C., Lesnaw J. Virology. 1987;159:229–236. doi: 10.1016/0042-6822(87)90459-4. [DOI] [PubMed] [Google Scholar]
- 47.Colonno R. J., Stone H. O. Nature. 1976;261:611–614. doi: 10.1038/261611a0. [DOI] [PubMed] [Google Scholar]
- 48.Roberts A., Buonocore L., Price R., Forman J., Rose J. K. J. Virol. 1999;73:3723–3732. doi: 10.1128/jvi.73.5.3723-3732.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Giedlin M. A., Cook D. N., Dubensky T. W., Jr Cancer Cell. 2003;4:241–243. doi: 10.1016/s1535-6108(03)00251-4. [DOI] [PubMed] [Google Scholar]
- 50.Neumann G., Whitt M. A., Kawaoka Y. J. Gen. Virol. 2002;83:2635–2662. doi: 10.1099/0022-1317-83-11-2635. [DOI] [PubMed] [Google Scholar]
- 51.Schubert M., Harmison G. G., Richardson C. D., Meier E. Proc. Natl. Acad. Sci. USA. 1985;82:7984–7988. doi: 10.1073/pnas.82.23.7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Buchholz U. J., Finke S., Conzelmann K. K. J. Virol. 1999;73:251–259. doi: 10.1128/jvi.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fuerst T. R., Niles E. G., Studier F. W., Moss B. Proc. Natl. Acad. Sci. USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whelan S. P., Ball L. A., Barr J. N., Wertz G. T. Proc. Natl. Acad. Sci. USA. 1995;92:8388–8392. doi: 10.1073/pnas.92.18.8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baltimore D., Huang A. S., Stampfer M. Proc. Natl. Acad. Sci. USA. 1970;66:572–576. doi: 10.1073/pnas.66.2.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whelan S. P., Wertz G. W. Proc. Natl. Acad. Sci. USA. 2002;99:9178–9183. doi: 10.1073/pnas.152155599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.