Abstract
Agents targeting topoisomerases are active against a wide range of human tumors. Stabilization of covalent complexes, converting topoisomerases into DNA-damaging agents, is an essential aspect of cell killing by these drugs. A unique aspect of the repair of topoisomerase-mediated DNA damage is the requirement for pathways that can remove protein covalently bound to DNA. Tyrosyl-DNA phosphodiesterase (Tdp1) is an enzyme that removes phosphotyrosyl moieties bound to the 3′ end of DNA. Cells lacking Tdp1 are hypersensitive to camptothecin, consistent with a role for Tdp1 in processing 3′ phosphotyrosyl protein–DNA covalent complexes. Because Top2p forms a 5′ phosphotyrosyl linkage with DNA, previous work predicted that Tdp1p would not be active against lesions involving Top2p. We found that deletion of the TDP1 gene in yeast confers hypersensitivity to Top2 targeting agents. Combining tdp1 mutations with deletions of genes involved in nonhomologous end joining, excision repair, or postreplication repair enhanced sensitivity to Top2 targeting drugs over the level seen with single mutants, suggesting that Tdp1 may function in collaboration with multiple pathways involved in strand break repair. tdp1 mutations can sensitize yeast cells to drugs targeting Top2 even when TOP1 is deleted. Finally, bacterially expressed yeast Tdp1p is able to remove a peptide derived from yTop2 that is covalently bound to DNA by a 5′ phosphotyrosyl linkage. Our results show that Tdp1 plays more general roles in DNA repair than repair of Top1 mediated DNA damage, and may participate in repairing many types of base damage to DNA.
Keywords: topoisomerase II, etoposide, RAD2, repair nuclease, RAD52
DNA topoisomerases (topos) carry out alterations in DNA structure by transiently introducing breaks in DNA. Topos use tyrosine residues to form a transient covalent intermediate between the enzyme and DNA, resulting in single or double strand DNA breaks (1). This mechanism of DNA breakage can create difficulties for a cell under conditions that interfere with the normal religation reaction of the enzyme, such as the presence of specific inhibitors. Then, the enzyme becomes trapped on DNA as a stable covalent adduct (2). Topos can become trapped by small molecule inhibitors such as camptothecin or etoposide, which are clinically used anticancer agents targeting topo I and topo II, respectively. The complete repair of this type of lesion requires DNA strand break repair pathways, and also requires activities that can remove covalent protein DNA adducts (3).
An enzyme described by Nash and colleagues (4), tyrosyl-DNA phosphodiesterase (Tdp1), can remove peptides covalently bound to DNA by a 3′ phosphotyrosine linkage. This enzyme participates in the repair of Top1 covalently trapped on DNA in the presence of camptothecin in both yeast (5, 6) and mammalian cells (7). Tdp1 is a phospholipase D superfamily enzyme (8), and the three-dimensional structure of the protein (9) as well as transition state intermediates have been reported (10). Tdp1 does not define the only pathway for removing Top1 that is trapped by camptothecin, because mutations in the genes encoding several different endonucleases have been shown in yeast to confer additive sensitivity to camptothecin when combined with tdp1 mutations (6, 11, 12). In addition to the removal of peptides bound via a 3′ phosphotyrosyl linkage, the Tdp1 enzyme can cleave other chemical bonds such as a phosphohistidine bond (13). The enzyme possesses DNA and RNA exonuclease activity, can act at abasic sites, and remove a phosphoglycolate or biotin linked substrate. All of these substrates were 3′ linked and left a 3′ phosphate on the DNA (13).
Here, we report that yeast cells lacking Tdp1 are hypersensitive to Top2 poisons. Although cells lacking TDP1 show minor etoposide sensitivity, overexpression of yeast Top2 resulted in greater etoposide sensitivity in tdp1 mutants than in isogenic wild-type cells. These results support recent data showing that overexpression of Tdp1 in cultured cells alters the processing of both Top1- and Top2-mediated DNA damage (14). We also show by a direct enzymatic assay that yeast Tdp1p can remove peptides covalently bound to DNA by a 5′ phosphotyrosyl linkage. Our results suggest that Tdp1 plays a more general role in repair than previously suggested, including the repair of DNA damage mediated by Top2.
Results
Saccharomyces cerevisiae Δtdp1 Cells Are Hypersensitive to Top2 Targeting Drugs.
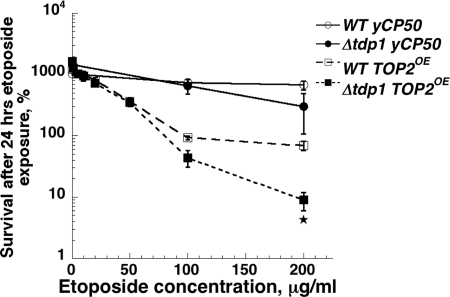
We previously showed that overexpression of Top2 in yeast confers hypersensitivity to Top2 targeting agents such as etoposide (15), and found that overexpression of Top2 provided a sensitive test for examining the importance of DNA repair genes in sensitivity to Top2-mediated DNA damage (16). We tested whether Tdp1 plays a role in survival after exposure to etoposide. Wild-type or tdp1 mutant yeast cells carrying an empty vector or the Top2 overexpression vector pDED1TOP2 were exposed to etoposide in liquid culture for 24 h, and then plated to determine cell viability. Wild-type cells carrying an empty vector were insensitive to etoposide, and the presence of a deletion of the TDP1 gene did not substantially increase sensitivity (Fig. 1). Wild-type cells overexpressing Top2 were much more sensitive to etoposide than cells with the empty vector. Cells deleted for tdp1 carrying pDED1TOP2 showed a further increase in sensitivity compared to wild-type cells carrying pDED1TOP2. At 200 μg/ml etoposide, survival of wild-type cells with pDED1TOP2 was ≈90% compared to the viable titer before drug addition. By contrast, in tdp1 mutants exposed to 200 μg/ml etoposide, cell survival was ≈10%, a difference that was statistically significant. These surprising results suggested that yeast Tdp1p may participate in the repair of Top2 as well as Top1 mediated DNA damage.
Fig. 1.
Δtdp1 cells overexpressing TOP2 enzyme are hypersensitive to etoposide. Etoposide sensitivity of Δtdp1 and wild-type strains carrying yCP50 (vector control) or a plasmid overexpressing (OE) wild-type yeast TOP2 enzyme was determined as described in Materials and Methods. The asterisk indicates a significant difference between wild type and Δtdp1 at the indicated drug concentration.
Δtdp1 Cell Hypersensitivity Is Enhanced by Defects in Double Strand Break and Postreplication Repair.
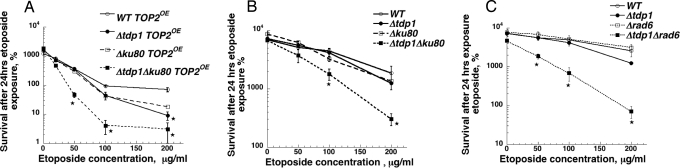
Mutations that confer sensitivity to DNA damaging agents have been used to test roles of Tdp1 in repairing Top1 mediated DNA damage (5, 11). We took a similar approach, and examined the sensitivity of tdp1 deletions combined with genes important for double strand break repair. When wild-type Top2p was overexpressed in Δyku80 cells, which are deficient in nonhomologous end joining, there was a significant increase in etoposide sensitivity compared with wild type cells (Fig. 2A). Deletion of tdp1 greatly increased the sensitivity of Δyku80 mutants. We also examined the sensitivity of Δyku80Δtdp1 double mutants with normal levels of Top2 expression (Fig. 2B). Neither Δyku80 nor Δtdp1 single mutants show appreciable etoposide sensitivity after a 24-h drug exposure without Top2 overexpression. However, the Δtdp1Δyku80 double mutant showed significant growth inhibition in the presence of etoposide. We also examined the effect of combining deletion of tdp1 with deletions in genes required for postreplication repair as shown in Fig. 2C. When rad6 deletions were combined with deletion of tdp1, there was substantial sensitivity to etoposide, whereas no sensitivity was seen with either single mutant. The data from Fig. 2 B and C indicate that Top2 overexpression is not needed to observe enhanced sensitivity to etoposide in Δtdp1 mutants. The effects of Δrad6 and Δyku80 mutants were confirmed by assessing sensitivity to a different Top2 targeting drug mAMSA, with similar results (Fig. 7, which is published as supporting information on the PNAS web site).
Fig. 2.
Combination of Δtdp1 with either Δku80 or Δrad6 increases sensitivity to etoposide. Etoposide sensitivity of wild-type, Δtdp1, Δku80, and Δtdp1Δku80 mutant cells overexpressing TOP2 (A) or in the absence of TOP2 overexpression (B) was determined. (C) Survival of Δtdp1, Δrad6, or Δtdp1Δrad6 strains after exposure to etoposide. An asterisk indicates a significant difference between the single mutant and the double mutant at the indicated drug concentration.
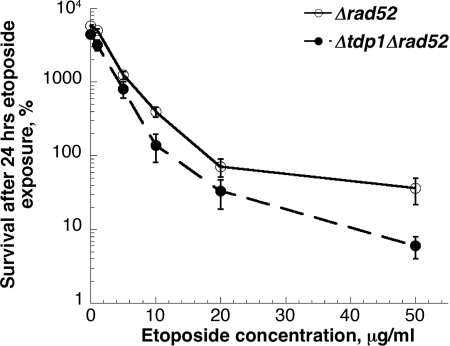
When Δtdp1 mutations were combined with mutations in genes such as rad52 that confer defects in homologous recombination, no increase in sensitivity to camptothecin was observed above the high levels of sensitivity seen with Δrad52 single mutants (17). Yeast cells defective in homologous recombination are also hypersensitive to Top2-targeting drugs (18, 19). We constructed strains carrying deletions in rad52 and/or tdp1 and examined etoposide sensitivity. Fig. 3 shows the sensitivity to etoposide of a Δrad52 strain compared to a Δrad52Δtdp1 double mutant strain, assessed by clonogenic survival after 24-h drug exposure. A small reproducible increase in sensitivity to etoposide was seen in Δrad52Δtdp1 strains compared to Δrad52 single mutants, although the difference was not significant. These results suggest that deletion of tdp1 does not substantially alter sensitivity to mutants with defects in homologous recombination, suggesting that lesions involving Top2 that are repaired by a Tdp1-dependent pathway are primarily substrates for homologous recombination.
Fig. 3.
Combining mutations conferring defects in homologous recombination with Δtdp1 results in minor increases in etoposide sensitivity. Strains with deletions in Δrad52 or Δrad52Δtdp1 were examined for sensitivity to etoposide. Differences in survival between the two strains at each etoposide concentration were not significant.
Sensitivity to Top2 Targeting Drugs in Δtdp1 Strains Does Not Require Active Top1.
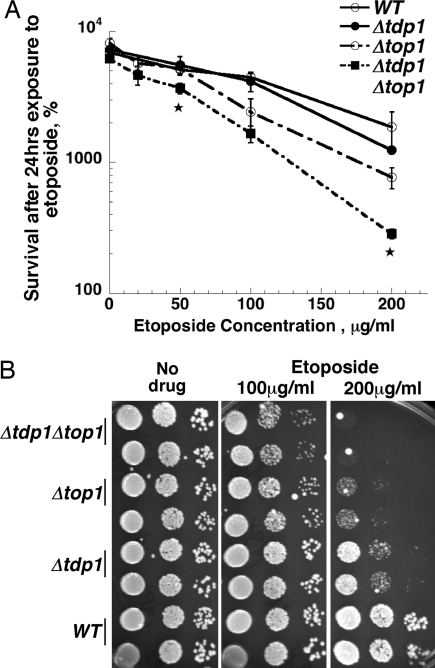
Because Tdp1p has not been shown to have any activity against peptides covalently bound to the 5′ end of DNA, a possible explanation of our results was that trapping of Top2 by etoposide results in DNA alterations that can trap Top1. We considered this unlikely because we had previously shown that yeast top1− mutants are hypersensitive to etoposide and other drugs that target Top2 (20). If Top1p were trapped directly or indirectly by the action of etoposide, we would have expected that deletion of TOP1 would reduce etoposide sensitivity. Nonetheless, we tested whether active Top1 was required for etoposide hypersensitivity in Δtdp1 strains. We compared the etoposide sensitivity of Δtdp1 TOP1+ versus Δtdp1 Δtop1 strains (Fig. 4A). When cells were exposed to etoposide in liquid culture, we observed a significant increase in sensitivity to etoposide for the Δtdp1 Δtop1 strain compared to the Δtop1 single mutant. We confirmed the enhanced sensitivity of Δtdp1 Δtop1 strains to etoposide by spotting dilutions of cells to plates containing etoposide. The double mutant was also more sensitive to etoposide in this assay (Fig. 4B) when compared to the wild-type, Δtdp1, or Δtop1 strains. These results show that the sensitivity to etoposide in Δtdp1 mutants does not arise from effects of Top1.
Fig. 4.
Increased sensitivity of Δtdp1 to etoposide does not require TOP1. (A) Sensitivity of wild-type, Δtdp1, Δtop1, or Δtdp1Δtop1 strains to etoposide was assessed by using the same approach as in Fig. 1. An asterisk indicates a significant difference between Δtop1 and Δtop1 Δtdp1 at the indicated drug concentration. (B) Sensitivity of wild-type, Δtdp1, Δtop1 or Δtdp1Δtop1 strains to etoposide was also determined by spotting diluted cell cultures onto etoposide containing YPDA agar plates. Relevant genotypes are indicated.
Mutations in Genes Encoding Nucleases Implicated in DNA Repair Enhance the Sensitivity of Δtdp1 Mutations to Etoposide.
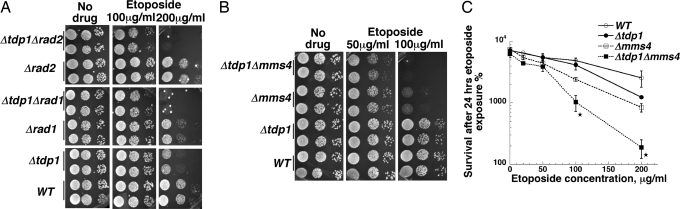
The results above clearly demonstrate that yeast cells lacking tdp1 have sensitivity to Top2-targeting agents either when Top2 is overexpressed (thus leading to elevated levels of DNA damage), or when cells lack one or more repair pathways. Yeast cells carrying deletions of TDP1 are also only weakly sensitive to camptothecin, a Top1-targeting drug (5). The lack of sensitivity of Δtdp1 single mutants was ascribed to the presence of alternate pathways for removal of Top1 covalently bound to DNA. Vance and Wilson showed that the Rad1–Rad10 endonuclease represented an alternate pathway for removing Top1 covalently bound to DNA (11). Similar results were obtained by Nash and colleagues (6), who also showed that that Mms4/Mus81 pathway defined a third pathway based on additive sensitivity of tdp1 mutants with both rad1 and mus81 mutants. We carried out a similar analysis to assess different pathways that may be involved in removing Top2 covalently bound to DNA. We constructed strains that combined tdp1 deletions with deletions of either rad1 or rad2. Deletion of rad2 does not confer additive sensitivity to camptothecin when combined with tdp1 deletions, presumably because the Rad2 endonuclease, cutting 3′ of DNA damage (21) does not possess the correct polarity to remove a 3′ phosphotyrosyl adduct. Fig. 5A shows the sensitivity obtained by spotting cultures of these mutant combinations onto plates containing different etoposide concentrations. Using this assay, reduced growth of Δtdp1 strains can be seen on plates containing 200 μg/ml etoposide. Neither Δrad1 nor Δrad2 strains show significant growth inhibition; however, both the Δrad1Δtdp1 and Δrad2Δtdp1 exhibit substantially less growth on etoposide containing plates than either single mutant. This effect is not general for all genes required for excision repair, because there is no enhanced sensitivity of a Δrad4Δtdp1 double mutant compared to the Δtdp1 single mutant (data not shown).
Fig. 5.
Combination of Δtdp1 with deletions of nuclease genes relevant to DNA repair leads to additive sensitivity to etoposide. (A) Survival of WT, Δtdp1, Δrad1, Δrad2, Δrad1Δtdp1, and Δrad2Δtdp1 strains was determined by spotting diluted cell cultures onto etoposide containing YPDA agar plates. (B) Survival of WT, Δtdp1, Δmms4, and Δmms4Δtdp1 strains to etoposide was assessed by spotting diluted cell cultures onto etoposide containing YPDA agar plates. (C) Survival of the same strains as shown in B was assessed after 24-h etoposide exposure in liquid culture assay followed by plating to determine survival. An asterisk indicates a significant difference between Δmms4 and Δmms4Δtdp1 at the indicated drug concentration.
A similar effect was seen when combining Δtdp1 mutations with deletions of the gene encoding a subunit of the endonuclease Mms4/Mus81 (22). Δmms4 strains have appreciable etoposide sensitivity (Fig. 5B), and the Δmms4Δtdp1 strain has greater etoposide sensitivity than either single mutant strain. This result was confirmed by examining clonogenic survival (Fig. 5C). A similar pattern of sensitivity was seen when the mutants were analyzed on plates containing mAMSA, a different Top2 targeting drug (Fig. 7). Taken together, these results show that combining repair nucleases with tdp1 mutations results in additive sensitivity to Top2-targeting drugs. Importantly, combination of Δtdp1 mutations with Δrad2 mutations also results in additive sensitivity, a result expected if the enhanced sensitivity reflects defects in processing Top2 covalent complexes.
Yeast Tdp1p Can Remove Peptides Bound to DNA by a 5′ Phosphotyrosyl Linkage.
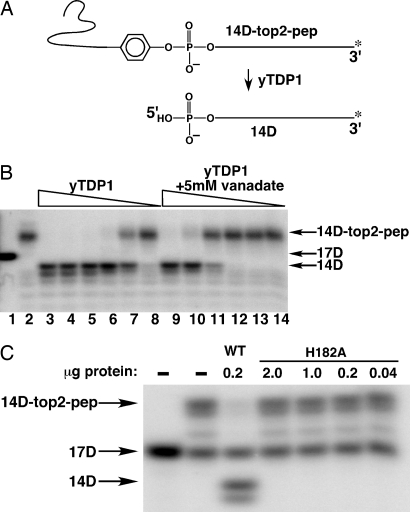
One model that can explain the results described above is that Tdp1p is also capable of processing phosphotyrosyl peptides linked to the 5′ end of DNA. Despite the failure to observe processing of a 5′ phosphotyrosine linked to an oligonucleotide (oligo) by Nash and colleagues (4), we tested the ability of yeast Tdp1p to remove peptides linked to DNA by a 5′ phosphotyrosyl linkage. A substrate including a 5′ phosphotyrosyl containing peptide linked to DNA was generated by using a partially double stranded “suicide” oligo that traps Top2 as a covalent complex (23, 24). The Top2 cleavage substrate consisted of a 16-nt top strand and a 28-nt bottom strand using the DNA sequence chosen by Anderson et al. (25). The top strand was labeled by the addition of an α-32P-labeled nucleotide to the 3′ end of the top strand (after annealing to the bottom strand), resulting in a 17-nt top strand. Treatment of this partially double stranded oligo with purified Top2 leads to trapping of the enzyme because the 3-nt fragment 5′ of the cleavage site diffuses from the covalent complex, and deprives the enzyme of an acceptor for religation (25); this results in a 14-mer oligo covalently bound to Top2 that is detectable in the experiment. After purification of the Top2–oligo covalent complex away from unreacted oligo, the protein–oligo complex was digested with trypsin, yielding the substrate shown in Fig. 6A labeled 14D-top2-pep. Trypsin digestion should yield an 8-aa peptide ending in a tyrosine residue linked to a single stranded oligo. As shown diagrammatically in Fig. 6A, reaction with Tdp1p is predicted to generate a free peptide and 5′ phosphate terminated oligo. The 14D-top2-pep substrate was reacted with yTdp1, and the products of the reaction were analyzed by PAGE. The autoradiogram of the reaction products is shown in Fig. 6B. Lane 1 shows the mobility of the labeled 17D oligo that was not reacted with Top2. The mobility of the trapped peptide (after trypsin digestion) bound to the oligo is shown in lane 2. In the absence of trypsin digestion, the substrate is trapped at the top of the gel (not shown). Lanes 3–7 show the formation of the 14D oligo following reaction with decreasing quantities of yTdp1. It should be noted that two product bands are formed after the reaction with Tdp1, one that migrates slightly faster than the 14D product. This second band may reflect some heterogeneity in the cleavage site by yeast Top2 with the suicide substrate oligo. Davies et al. (26) had observed that vanadate was a potent inhibitor of the Tdp1 reaction. In lanes 9–14, the same reaction conditions as in lanes 3–8 with the addition of 5 mM vanadate were examined. Whereas 8 ng of Tdp1p led to complete disjoining of the tyrosyl peptide from the oligo (lane 6 in Fig. 6B), 1 μg was required in the presence of 5 mM vanadate (lane 9 in Fig. 6B, lanes 10–14 show incomplete reaction in the presence of 200 ng or less of added Tdp1p). This result shows that the removal of the 5′ phosphotyrosyl peptide, like 3′ substrates, is strongly inhibited by vanadate, suggesting that the reaction mechanism is likely similar. To further demonstrate that a similar reaction mechanism is involved in removing 5′ linked peptides, a critical His residue (8), His-182 (His-263 in human Tdp1) was mutated to Ala, the mutant protein was purified from Escherichia coli, and tested for activity against the substrate described above. As shown in Fig. 6C, mutating His-182 completely eliminated activity against the 5′ linked substrate. (A PAGE analysis of the purified mutant protein is shown in Fig. 8, which is published as supporting information on the PNAS web site) This experiment also demonstrates that the activity against 5′ phosphotyrosyl substrates was due to the yeast Tdp1p, and not a contaminating activity. Our results demonstrate that yeast Tdp1 protein is able to remove 5′ phosphotyrosyl substrates as well as 3′ phosphotyrosyl substrates, and with the in vivo results presented above, suggest that Tdp1p is part of a repair pathway for repairing Top2-mediated DNA damage.
Fig. 6.
Removal of 5′ linked peptide bound to DNA by yeast TDP1 protein. (A) Schematic representation of Tdp1 enzymatic processing of a 5′-linked oligopeptide substrate. Top2 is covalently trapped on a 3′ end-labeled oligonucleotide duplex suicide substrate (17D) via the active site tyrosine. Digestion with trypsin leaves an 8-aa peptide bound to the labeled substrate (14D-top2-pep). If Tdp1 cleaves the peptide from DNA, the resulting product is a 14-mer oligonucleotide (14D). The peptide linked substrate used in B and C was boiled before use, and is therefore single stranded. (B) Serial 5-fold dilutions of yeast Tdp1p, starting with 1 μg protein were incubated with 0.5 fmol of substrate for 15 min (lanes 3–8) as well as in the presence of the inhibitor sodium orthovanadate (lanes 9–13). 17D is the original 3′ end-labeled suicide substrate (lane 1), and mock-treated substrate is in lane 2 (14D-top2-pep). (C) The 5′-linked oligopeptide (0.5 fmol of 14D-top2-pep) was incubated with the indicated amounts of wild-type or H182A (active site mutant) Tdp1p. The cleavage product (14D) was only observed after incubation with wild-type Tdp1p (lane 3).
Discussion
We present two lines of evidence that indicate that Tdp1 participates in the repair of Top2-mediated damage in yeast. We found that yeast cells lacking TDP1 are hypersensitive to Top2-targeting drugs under a variety of contexts. We also developed a peptide-linked oligo substrate derived from Top2 and showed that yeast Tdp1p can efficiently disjoin the peptide from the nucleic acid. These experiments greatly expand the types of DNA damage that can be processed by Tdp1.
Previous experiments assessing the enzymatic activities of Tdp1 have concentrated on substrates with peptides bound to the 3′ end of DNA (26–28) and on other 3′ adducts (13, 29, 30). Nash and colleagues (4), in their initial characterization of yeast Tdp1p failed to observe removal of 5′ phosphotyrosine, whereas the 3′ phosphotyrosine was efficiently removed. The apparent lack of reactivity against 5′ modified oligos originally observed, as compared to our results presented here, may be due to the advantages of using a recombinant, highly active enzyme preparation. Alternately, the presence of additional amino acids flanking the tyrosine may confer a significant binding advantage compared to a single tyrosine. When the yeast deletion of the TDP1 gene was constructed, no enhanced sensitivity to mitoxantrone, an intercalating Top2 poison, was seen in tdp1 deletions (17). Exposing yeast cells to mitoxantrone results in large decreases in cell viability over very small changes in drug concentration, making detection of drug hypersensitivity difficult (31). Our use of Top2 overexpression as a means of enhancing drug sensitivity in repair proficient strains was particularly valuable, because our experiments in repair proficient cells showed only a modest increase in sensitivity to etoposide in Δtdp1 cells when Top2 was expressed at normal levels.
Our results are in agreement with and extend recent results that showed that overexpression of Tdp1 in human cells reduced the DNA damage generated by both etoposide and camptothecin (14). Marko and colleagues (14) suggested hypotheses predicated on the lack of activity of purified Tdp1p against 5′ adducts, and suggested as one possibility that additional proteins acting in concert with Tdp1p resulted in a complex active against 5′ adducts in DNA. Our results indicate that no other protein is uniquely required for processing 5′ adducts, and that their observed reduction in DNA damage from etoposide and camptothecin arose directly from the enzymatic activity of Tdp1.
We observed additive sensitivity to etoposide when tdp1 deletions were combined with deletions of other DNA repair functions. It has been suggested that there are several repair pathways that can remove Top1 covalently trapped on DNA. Our results suggest that there are also multiple pathways that can remove Top2 from DNA. However, several of the DNA repair functions we examined do not have activities suggestive of removal of peptides bound to DNA. For example, whereas the Rad2 nuclease has an activity that could remove Top2 from DNA, the Rad1–Rad10 nuclease (32) and the Mms4–Mus81 (33) nuclease do not have the correct polarity for removing a peptide bound to 5′ end of DNA. Although it is possible that both Rad1-Rad10 and Rad2 nucleases must be present for processing Top2 bound to DNA, as occurs with incision of UV damaged DNA (32), such a scenario is less likely in the case of the Mms4–Mus81 nuclease. The additive sensitivity may instead arise from the specific recombination defects conferred by rad1 or mms4 deletions. Nonetheless, it is interesting that there is an additive sensitivity with tdp1 and rad2 deletions, because deletion of rad2 does not confer a recombination defect. We hypothesize that RAD2 plays a significant although not indispensable role in removing Top2 bound to DNA.
One reason that Tdp1 has provoked substantial interest recently has been the finding that Tdp1 mutations have been found in association with a hereditary neuropathy termed SCAN1 (34). Recent work has centered on the hypothesis that a loss of Tdp1 function confers a defect in single-strand break repair (35). It has also been suggested that the accumulation of Tdp1–DNA covalent intermediates, which occurs due to the H493R mutant found in SCAN1 cells, significantly contributes to cell killing (7). Our results greatly extend the lesions that may be unrepaired or misrepaired in human tdp1-deficient cells. Because DNA damage arising from Top2 can kill cells independent of DNA replication, and Top2 can be trapped by endogenous DNA damage (36), Top2-mediated cell killing could contribute to the SCAN1 phenotype. This hypothesis will require confirmation that Tdp1 plays a significant role in the repair of Top2-mediated damage in higher eukaryotic cells. It should be noted that Champoux and colleagues (7) reported that they did not observe etoposide sensitivity in SCAN1-derived cells, perhaps indicating that repair of Top2-mediated DNA damage in mammalian cells relies on multiple redundant pathways.
In conclusion, we have shown that deletion of the Tdp1 gene affects the survival of yeast cells exposed to drugs targeting Top2. The biochemical activities of Tdp1 include the ability to process damage at the 5′ as well as the 3′ end of DNA. These results indicate that Tdp1 may play diverse roles in repairing base damage to DNA that occurs at the sites of DNA breaks. Future studies should be directed toward characterizing the range of 5′ modified substrates that can be recognized by Tdp1 and determining the steps required for processing Top2 damage before Tdp1 action.
Materials and Methods
Strains.
All strains used in this work are derived from the S. cerevisiae strain JN362a (15). Individual yeast ORF deletion mutants marked with KANMX4 (tdp1, mms4, and rad52) were purchased from Open Biosystems (Invitrogen). The deletion cassettes were isolated from strains by PCR, using PCR primers chosen between 300 and 700 bp upstream and downstream of the KANMX4 deleted ORF. (Primer sequences used to isolate the deletion cassettes are available on request.) The PCR products (1–5 μg) were introduced in JN362a by lithium acetate transformation of JN362a cells (37), and after transformation, cells were plated to YPDA agar. Two days later, cells were scraped off and replated to YPDA agar plates containing 70 μg/ml G418 (GIBCO/BRL). The presence of deletions were verified by PCR amplification using a combination of primers external to the primers used to isolate the deletion cassette and either the KANB or KANC primers within the ORF deletion. For generating disruptions of rad1, rad2, rad6, tdp1, top1, and yku80, plasmids containing the cloned sequences were digested with restriction enzymes to create an internal deletion of the gene followed by replacement with either a LEU2 or URA3 cassette, as indicated in Table 1, which is published as supporting information on the PNAS web site. Disrupted alleles were PCR amplified, and the PCR product was used in transformation of JN362a.
Plasmids.
Strains were transformed with yCP50 as an empty vector, or a yTOP2 overexpression plasmid pDED1yTOP2 (15) in which yTOP2 expression is driven from the DED1 promoter to assess the effects of Top2 overexpression on etoposide sensitivity.
Drug Sensitivity.
For yeast survival studies, overnight cultures were diluted to 2 × 106 cells per ml in synthetic medium lacking uracil or yeast extract/peptone/glucose/adenine (YPDA), and the cells were exposed to etoposide at the indicated concentrations (μg/ml) for 24 h. Aliquots were removed, serial dilutions were performed, and cells were plated on appropriate media. Survival was determined after incubation for 3–4 days at 30°C. The survival rate is expressed as the percentage of cells able to form colonies after drug exposure relative to the number of viable colonies at the time of drug addition. The average (±SEM) of three to four experiments is shown. For growth on media containing drug, 3 μl of a serial dilution of growing cells was applied to YPDA plates containing the indicated concentrations of etoposide. Plates were incubated at 30°C for 2 days and photographed.
TDP1 Gene Cloning, Protein Expression, and Purification.
The cloning of the yeast TDP1 gene, construction of an E. coli expression vector, construction of the mutation to Ala at His-182, and the purification of the Tdp1 proteins are described in Supporting Text, which is published as supporting information on the PNAS web site.
Preparation of 5′ Phosphotyrosyl Peptide Substrate.
Purified yTop2 was prepared as described (38). The oligos for the suicide reaction were synthesized on an ABI 3900 synthesizer. The bottom strand has a three-carbon spacer modification to the 3′ end. Sequences, annealing, labeling, and reactions conditions were exactly as described by Wang et al. (23) with the following modification. DMSO, added to the reaction mixture to a final concentration of 6.6% (vol/vol), was found to enhance the reaction significantly. Reactions (150 μl) contained 1 pmol of labeled suicide substrate and 25 μg of yTOP2. After 4 h at 30°C, reactions were stopped by the addition of Laemmli buffer and boiled. The sample was run on a 7.5% acrylamide 2D/prep (Bio-Rad) gel for 1 h at 120 V, and the band migrating at ≈165 kDa was excised. The band was minced and shaken overnight in a microcentrifuge tube containing 0.25 M Tris·Cl (pH 6.8) and 0.1% SDS. The tubes were then centrifuged for 10 min at 20,000 × g, and the recovered supernatant was spun for another 10 min. The solution was treated with final concentration of 1 mg/ml trypsin for 3 h. Trypsin was inactivated by adding 4-(2-aminoethyl)-benzenesulfonyl fluoride to 0.7 μg/μl. Samples were TCA precipitated along with 20 μg of tRNA as carrier, acetone washed, air dried, and resuspended in yTDP1 reaction buffer (80 mM KCl/50 mM Tris·Cl, pH 8.0/2 mM EDTA/2 mM DTT).
yTDP1 Reactions.
Purified yTDP1 (1 μg) was 5-fold diluted in reaction buffer and incubated with 0.5 fmol of labeled substrate for 15 min at 30°C. Reactions were stopped by the addition of an equal volume of 2× TBE-Urea Sample buffer (Novex), boiled for 5 min, and run on a 15% urea-acrylamide gel. The gels were fixed and dried before exposure and analysis on a PhosphorImager (Molecular Dynamics).
Supplementary Material
Acknowledgments
We thank Drs. Shengli Huang (St. Jude Children's Research Hospital) and Anna Rogojina (St. Jude Children's Research Hospital) for providing purified yeast Top2 protein and the Hartwell Center for Bioinformatics and Biotechnology for synthesis of oligonucleotides. This work was supported by National Cancer Institute Grants CA52814 and CA82313, Core Grant CA21765, and the American Lebanese Syrian Associated Charities (ALSAC).
Abbreviations
- topo
DNA topoisomerase
- Tdp1
tyrosyl-DNA phosphodiesterase
- oligo
oligonucleotide.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Wang J. C. Nat. Rev. Mol. Cell. Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 2.Byl J. A., Cline S. D., Utsugi T., Kobunai T., Yamada Y., Osheroff N. Biochemistry. 2001;40:712–718. doi: 10.1021/bi0021838. [DOI] [PubMed] [Google Scholar]
- 3.Nitiss J. L. Curr. Opin. Invest. Drugs. 2002;3:1512–1516. [PubMed] [Google Scholar]
- 4.Yang S. W., Burgin A. B., Jr, Huizenga B. N., Robertson C. A., Yao K. C., Nash H. A. Proc. Natl. Acad. Sci. USA. 1996;93:11534–11539. doi: 10.1073/pnas.93.21.11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pouliot J. J., Yao K. C., Robertson C. A., Nash H. A. Science. 1999;286:552–555. doi: 10.1126/science.286.5439.552. [DOI] [PubMed] [Google Scholar]
- 6.Liu C., Pouliot J. J., Nash H. A. Proc. Natl. Acad. Sci. USA. 2002;99:14970–14975. doi: 10.1073/pnas.182557199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Interthal H., Chen H. J., Kehl-Fie T. E., Zotzmann J., Leppard J. B., Champoux J. J. EMBO J. 2005;24:2224–2233. doi: 10.1038/sj.emboj.7600694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Interthal H., Pouliot J. J., Champoux J. J. Proc. Natl. Acad. Sci. USA. 2001;98:12009–12014. doi: 10.1073/pnas.211429198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies D. R., Interthal H., Champoux J. J., Hol W. G. Structure (Cambridge, U.K.) 2002;10:237–248. doi: 10.1016/s0969-2126(02)00707-4. [DOI] [PubMed] [Google Scholar]
- 10.Davies D. R., Interthal H., Champoux J. J., Hol W. G. Chem. Biol. 2003;10:139–147. doi: 10.1016/s1074-5521(03)00021-8. [DOI] [PubMed] [Google Scholar]
- 11.Vance J. R., Wilson T. E. Proc. Natl. Acad. Sci. USA. 2002;99:13669–13674. doi: 10.1073/pnas.202242599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng C., Brown J. A., You D., Brown J. M. Genetics. 2005;170:591–600. doi: 10.1534/genetics.104.028795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Interthal H., Chen H. J., Champoux J. J. J. Biol. Chem. 2005;280:36518–36528. doi: 10.1074/jbc.M508898200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barthelmes H. U., Habermeyer M., Christensen M. O., Mielke C., Interthal H., Pouliot J. J., Boege F., Marko D. J. Biol. Chem. 2004;279:55618–55625. doi: 10.1074/jbc.M405042200. [DOI] [PubMed] [Google Scholar]
- 15.Nitiss J. L., Liu Y. X., Harbury P., Jannatipour M., Wasserman R., Wang J. C. Cancer Res. 1992;52:4467–4472. [PubMed] [Google Scholar]
- 16.Sabourin M., Nitiss J. L., Nitiss K. C., Tatebayashi K., Ikeda H., Osheroff N. Nucleic Acids Res. 2003;31:4373–4384. doi: 10.1093/nar/gkg497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pouliot J. J., Robertson C. A., Nash H. A. Genes Cells. 2001;6:677–687. doi: 10.1046/j.1365-2443.2001.00452.x. [DOI] [PubMed] [Google Scholar]
- 18.Nitiss J., Wang J. C. Proc. Natl. Acad. Sci. USA. 1988;85:7501–7505. doi: 10.1073/pnas.85.20.7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nitiss J. L., Rose A., Sykes K. Ann. N.Y. Acad. Sci. 1996;803:32–43. doi: 10.1111/j.1749-6632.1996.tb26374.x. [DOI] [PubMed] [Google Scholar]
- 20.Nitiss J. L., Liu Y. X., Hsiung Y. Cancer Res. 1993;53:89–93. [PubMed] [Google Scholar]
- 21.Habraken Y., Sung P., Prakash L., Prakash S. J. Biol. Chem. 1995;270:30194–30198. doi: 10.1074/jbc.270.50.30194. [DOI] [PubMed] [Google Scholar]
- 22.Ciccia A., Constantinou A., West S. C. J. Biol. Chem. 2003;278:25172–25178. doi: 10.1074/jbc.M302882200. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y., Knudsen B. R., Bjergbaek L., Westergaard O., Andersen A. H. J. Biol. Chem. 1999;274:22839–22846. doi: 10.1074/jbc.274.32.22839. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y., Thyssen A., Westergaard O., Andersen A. H. Nucleic Acids Res. 2000;28:4815–4821. doi: 10.1093/nar/28.24.4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anderson A. H., Sorensen B. S., Christiansen K., Svejstrup J. Q., Lund K., Westergaard O. J. Biol. Chem. 1991;266:9203–9210. [PubMed] [Google Scholar]
- 26.Davies D. R., Interthal H., Champoux J. J., Hol W. G. J. Mol. Biol. 2002;324:917–932. doi: 10.1016/s0022-2836(02)01154-3. [DOI] [PubMed] [Google Scholar]
- 27.Davies D. R., Interthal H., Champoux J. J., Hol W. G. J. Med. Chem. 2004;47:829–837. doi: 10.1021/jm030487x. [DOI] [PubMed] [Google Scholar]
- 28.Debethune L., Kohlhagen G., Grandas A., Pommier Y. Nucleic Acids Res. 2002;30:1198–1204. doi: 10.1093/nar/30.5.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inamdar K. V., Pouliot J. J., Zhou T., Lees-Miller S. P., Rasouli-Nia A., Povirk L. F. J. Biol. Chem. 2002;277:27162–27168. doi: 10.1074/jbc.M204688200. [DOI] [PubMed] [Google Scholar]
- 30.Zhou T., Lee J. W., Tatavarthi H., Lupski J. R., Valerie K., Povirk L. F. Nucleic Acids Res. 2005;33:289–297. doi: 10.1093/nar/gki170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nitiss J. L. Cancer Chemother. Pharmacol. 1994;34(Suppl):S6–S13. doi: 10.1007/BF00684857. [DOI] [PubMed] [Google Scholar]
- 32.Reardon J. T., Sancar A. Prog. Nucleic Acid Res. Mol. Biol. 2005;79:183–235. doi: 10.1016/S0079-6603(04)79004-2. [DOI] [PubMed] [Google Scholar]
- 33.Kaliraman V., Mullen J. R., Fricke W. M., Bastin-Shanower S. A., Brill S. J. Genes Dev. 2001;15:2730–2740. doi: 10.1101/gad.932201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takashima H., Boerkoel C. F., John J., Saifi G. M., Salih M. A., Armstrong D., Mao Y., Quiocho F. A., Roa B. B., Nakagawa M., et al. Nat. Genet. 2002;32:267–272. doi: 10.1038/ng987. [DOI] [PubMed] [Google Scholar]
- 35.El-Khamisy S. F., Saifi G. M., Weinfeld M., Johansson F., Helleday T., Lupski J. R., Caldecott K. W. Nature. 2005;434:108–113. doi: 10.1038/nature03314. [DOI] [PubMed] [Google Scholar]
- 36.Wilstermann A. M., Osheroff N. J. Biol. Chem. 2001;276:46290–46296. doi: 10.1074/jbc.M105733200. [DOI] [PubMed] [Google Scholar]
- 37.Gietz D., St. Jean A., Woods R. A., Schiestl R. H. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dong J., Walker J., Nitiss J. L. J. Biol. Chem. 2000;275:7980–7987. doi: 10.1074/jbc.275.11.7980. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.