Abstract
Persistent hepatitis B virus infection is a major risk factor for hepatocellular carcinoma, the most frequent cancer in some developing countries. Up to 95% of those infected at birth and 15% of those infected after the neonatal period fail to clear hepatitis B virus, together resulting in ≈350 million persistent carriers worldwide. Via a whole genome scan in Gambian families, we have identified a major susceptibility locus as a cluster of class II cytokine receptor genes on chromosome 21q22. Coding changes in two of these genes, the type I IFN receptor gene, IFN-AR2, and the IL-10RB gene that encodes a receptor chain for IL-10-related cytokines including the IFN-λs, are associated with viral clearance (haplotype P value = 0.0003), and in vitro assays support functional roles for these variants in receptor signaling.
Keywords: complex trait, IL-10, interferon, susceptibility, virus
There are ≈350 million people with persistent hepatitis B virus (HBV) infection worldwide. Of these infected people, up to one-quarter will die from complications of the infection including cirrhosis and primary liver cancer. However, infection with HBV does not invariably lead to chronic hepatitis: When infection is acquired during childhood, as is normally the case in subSaharan Africa, up to 90% will eliminate the virus spontaneously. When infection is acquired during the early neonatal period from an HBV-infected mother, only 10% of children will eliminate the virus, but this situation, known as vertical transmission, is rare in Africa.
Susceptibility to persistent infection is governed by a number of factors, in addition to the age at infection. Twin studies and limited genetic association study data indicate that host genetic background influences the outcome of infection (1). It is likely that genetic susceptibility to persistent HBV infection is polygenic, and that both MHC and non-MHC genes are involved (2).
Current treatment for persistent HBV infection is partially effective, expensive, and impractical in developing countries where persistent HBV infection rates are at their highest (3). It is therefore crucial to identify factors that determine whether HBV becomes a self-limiting or persistent infection because these factors may reveal new therapeutic opportunities for patients with chronic HBV infection. This study was conducted to identify genetic determinants of persistent HBV infection by using a genome-wide approach.
Results
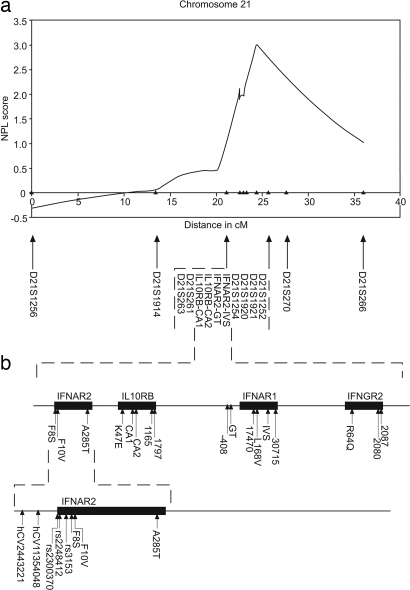
To search for major loci that might have some effect on viral persistence in a highly endemic area, 318 microsatellites were analyzed in 88 independent affected sibling pairs (ASPs) in 61 Gambian families. The most significant evidence of linkage was found on chromosome 21 [logarithm of odds (lod) = 1.66; P = 0.003] (Table 1, which is published as supporting information on the PNAS web site). This locus on chromosome 21 also showed evidence of linkage when a total of 31 markers (with SIBPAIR P value <0.1) were followed up in a further 106 independent ASPs in 74 families (D21S1252 screens 1 and 2 combined: lod = 2.46; P < 0.001). No other region retained evidence of linkage in the second screen (P > 0.1) (Table 1). These findings are intriguing because trisomy of chromosome 21 has been repeatedly associated with high rates of HBV viral persistence (4, 5). A further ten microsatellites were added to the region to fine map the locus. The resulting peak two-point lod score is located at D21S1254 (lod>3.16; P < 0.001), while the peak nonparametric linkage score, which was 3.00 (P = 0.001) (Fig. 1a), was found at marker D21S1921, located 1.3 Mb telomeric to D21S1254. This region was identified as chromosome 21q22 and found to contain a cluster of cytokine class II receptor (CRF2) genes (Fig. 1b).
Fig. 1.
Chromosome 21 analysis. (a) Nonparametric linkage (NPL) curve generated by genehunter (v2.5.1) for chromosome 21 (36, 37). Markers are indicated. Markers are shown from the p to q telomeres, and the CRF2 family is located at ≈22.5 cM. (b) Diagrammatic representation of the CRF2 family of genes found at Chr 21q22. Solid arrows represent polymorphisms typed in this study, and the dashed line represents the expanded region in which the genes are located. SNPs resulting in amino acid changes are shown in bold, and microsatellites are in italics.
Within this cluster lie genes encoding both subunits of the type I IFN receptor (IFN-AR1 and IFN-AR2) and the second subunits of both the IFN-γ receptor and IL-10 receptor (IFNGR2 and IL-10RB) (Fig. 1b). Polymorphisms in the members of the CRF2 family located on chromosome 21 were identified via literature searches and analysis of publicly available SNP databases such as the National Center for Biotechnology Information dbSNP (www.ncbi.nlm.nih.gov/SNP/). For our purposes, SNPs that were putatively functional, i.e., result in amino acid changes, were prioritized. A total of 18 SNPs in the four candidate genes were analyzed, and a further four microsatellites located either within introns or in the 5′ promoter region of IL-10RB and IFN-AR1 were also tested for association (Table 2, which is published as supporting information on the PNAS web site). All markers used were analyzed by using the pedigree disequilibrium test (PDT) (6, 7) and TRANSMIT (using only one sibling from each family) (8). SNPs in codon 8 of IFN-AR2 and codon 47 of IL-10RB, which both result in amino acid substitutions, were each significantly associated with outcome of persistent HBV infection (Table 3 a and b, which is published as supporting information on the PNAS web site). In the IFN-AR2 gene, the common variant phenylalanine (F), of the SNP IFN-AR2-F8S, was associated (PDT average χ2 = 6.63, P = 0.010; TRANSMIT P = 0.004) as was the common variant basic lysine (K) of the SNP IL-10RB-K47E (PDT average χ2 = 5.55; P = 0.019; TRANSMIT P = 0.085). None of the other SNPs or microsatellites showed any evidence of association with HBV persistence in the family study (P > 0.05) (Table 4, which is published as supporting information on the PNAS web site). Analysis of the haplotypes formed by the two SNPs revealed significant association (TRANSMIT P < 0.002) (Table 3c) with the common F (IFN-AR2-F8S)-K(IL-10RB-K47E) haplotype highly significantly associated with persistence of HBV infection (P = 0.0003) (Table 3c). Conditional transmission disequilibrium test analysis (9) indicates that the SNPs are significantly overtransmitted independently as well as the haplotype (data not shown). We found that the relative risks of persistent infection (i.e., the additional risk of viral persistence conferred by possession of the genotypes in question) are 2.8 [95% confidence interval (C.I.) 1.15–6.84] for individuals bearing the IFN-AR2-F8S-FF genotype, 2.1 (95% C.I. 0.77–5.65) for IL-10RB-K47E KK homozygotes, whereas the relative risk of persistent infection for individuals homozygous for both susceptibility alleles was 2.5 (95% C.I. 1.026–4.80).
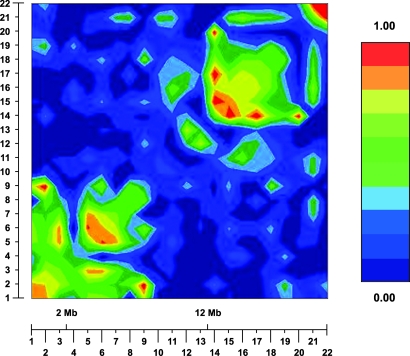
Extensive sequence analysis of IFN-AR2 (n = 20) and IL-10RB (n = 32) genes from Gambians and Europeans revealed no further polymorphisms in the coding region of these genes (data not shown). Analysis of the nonrandom distribution of alleles or linkage disequilibrium (LD) in this region in Gambians by using the graphical overview of linkage disequilibrium (GOLD) package (10) (Fig. 2) revealed a region of LD spanning most of the polymorphisms typed in the IFN-AR2 gene and a second spanning the 3′ terminal end of the IL-10RB gene and the IFN-AR1 gene. IFN-AR2-F8S lies within this first region of LD, while the 5′ end of IL-10RB lies between the two. LD between IFN-AR2-F8S and IL-10RB-K47E is moderately strong (D′ = 0.655), but this LD failed to reach statistical significance, probably because of the low minor allele frequencies of the two SNPs (Figure 5, which is published as supporting information on the PNAS web site). These relatively low-frequency protective alleles are more common in European Caucasians (IFN-AR2-S8 = 15%; IL-10RB-E47 = 35%, compared with 10% and 6% in The Gambia) where persistent HBV infection is relatively rare (Table 5, which is published as supporting information on the PNAS web site).
Fig. 2.
Linkage disequilibrium (LD) results from graphical overview of linkage disequilibrium (GOLD) (10). The key shows the color scale representing increasing D′ from dark blue (D′ < 0.1) through to red (D′ > 0.9). Markers shown from left to right areas follows: 1, IFN-α-R2-hCV2443221; 2, IFN-α-R2-hCV11354048; 3, IFN-α-R2-rs2300370; 4, IFN-α-R2-rs2448412; 5, IFN-α-R2-rs3153; 6, IFN-α-R2-F8S; 7, IFN-α-R2-F10V; 8, IFN-α-R2-A285T; 9, IL-10RB-K47E; 10, IL-10RB-CA1; 11, IL-10RB-CA2; 12, IL-10RB-1165; 13, IL-10RB-1797; 14, IFN-α-R1-(-408); 15, IFN-α-R1-GT; 16, IFN-α-R1–17470; 17, IFN-α-R1-L168V; 18, IFN-α-R1-IVS; 19, IFN-α-R1–30715; 20, IFN-γ-R2-R64Q; 21, IFN-γ-R2–2080; and 22, IFN-γ-R2–2087.
The amino acid substitution in the IL-10RB gene changes a K residue to an acidic glutamate (E), whereas, in the IFN-AR2 gene, the change is an F, which is hydrophobic, to a serine (S), which is hydrophilic. We therefore established functional assays to determine the impact of the IFAR2-F8S and IL-10RB-K47E genotypes on signal transduction through these receptors.
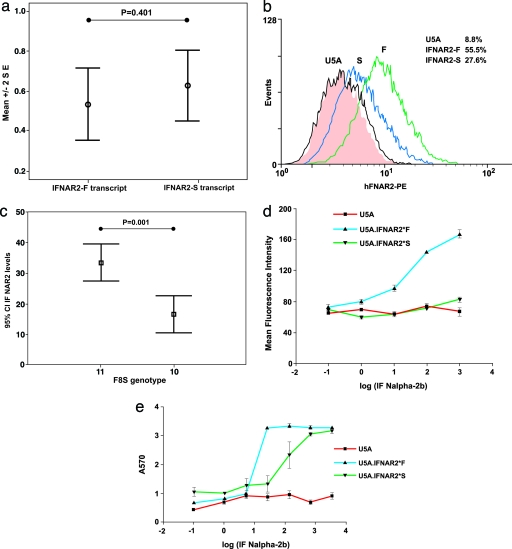
The effect of the IFN-AR2 polymorphism was explored in Epstein–Barr virus (EBV)-transformed B cell lines and in IFN-AR2-deficient U5A cells complemented with either of the two IFN-AR2 allelic variants. RNA from EBV-transformed B cells of normal heterozygous individuals was analyzed by a mass spectrometry-based method, to determine the relative levels of the two polymorphic transcripts, using genomic DNA to control for assay efficiency (11). There was no significant difference in RNA expression in the IFN-AR2-F8S variants (P = 0.401, Fig. 3a). However, analysis of the cell surface protein expression of IFN-AR2 by FACS revealed higher levels in lines homozygous for the variant encoding IFN-AR2-F (n = 22) compared with cell lines with both allelic variants (n = 10) (P = 0.001, Fig. 3c). Furthermore, IFN-AR2-deficient U5A cells transfected with each of the allelic variants restoring IFN-AR2 functionaltiy also revealed higher levels of the wild-type IFN-AR2-F receptor than the S receptor (Fig. 3b). To determine whether the increased receptor expression affected the level of signal transduction, we explored the induction of MHC class I and the antiviral response to IFN-α2. In the U5A cell lines, induction of MHC class I expression by IFN-α was greater in cells expressing the F than the S allele (P = 0.001; Fig. 3d). Furthermore, IFN-α-induced antiviral protection was significantly enhanced in U5A cells transfected with the IFN-AR2-F compared with the IFN-AR2-S allele (P < 0.001; Fig. 3e).
Fig. 3.
Evidence of functionality of IFN-AR2-F8S amino acid substitution. (a) Details of relative quantification of transcript levels in heterozygotes for the IFN-AR2 gene. Values used are the RNA expression level divided by the genomic DNA level to control for relative assay efficiency. The data are shown as plots describing the mean ± 2 SE. All data were analyzed by using a Student’s t test after testing for normality of data (version 12; SPSS, Chicago). (b) FACS analysis of cell surface receptor expression. The gating threshold is set at the level exceeded by <5% of untransfected cells, and the proportion of cells exceeding the threshold is given in each case. IFN-AR2 expression in U5a cells transfected with either IFN-AR2-F or IFN-AR2-S. (c) IFN-AR2 expression of EBV-transformed B cell lines of known genotypes. (d) MHC class I expression induced by IFN-α2b in U5a cells expressing either allele of the IFN-AR2 gene and controls. (e) Antiviral responses induced by IFN-α2b in U5a cells expressing either allele of the IFN-AR2 gene and controls.
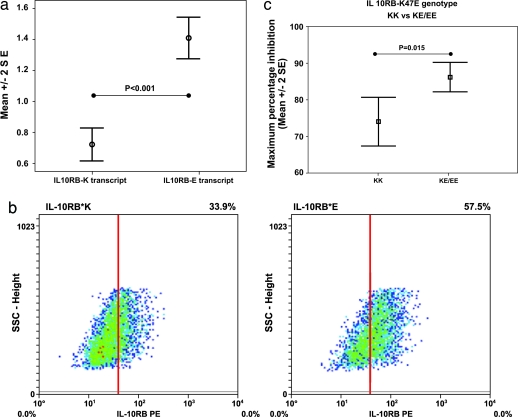
The effects of the polymorphism in IL-10RB were explored in the liver-derived cell line HUH7, which lacks expression of IL-10RB, and in monocytes obtained from healthy genotyped individuals. In heterozygous EBV-transformed B cell lines, the mRNA encoding the E allele of IL-10RB-K47E was expressed at significantly higher levels than the K allele (P < 0.001; Fig. 4a). In Huh7 cells transfected with either allele of IL-10RB, FACS analysis showed that the surface level of the variant IL-10RB-E receptor was higher than that of the K variant receptor (Fig. 4b). IL-10-mediated inhibition of LPS-induced TNF-α expression was measured in monocytes from individuals of differing genotypes. Monocytes homozygous for the glutamate-encoding allele, associated in the genetic study with viral clearance, showed significantly greater IL-10-mediated inhibition of TNF-α release (P = 0.015; Fig. 4c).
Fig. 4.
Evidence of functionality of IL-10RB-K47E amino acid substitution. (a) Details of relative quantification of transcript levels in heterozygotes for the IL-10RB gene. Values used are the RNA expression level divided by the genomic DNA level to control for relative assay efficiency. The data are shown as plots describing the mean ± 2 SE. All data were analyzed by using a Student’s t test after testing for normality of data (version 12; SPSS, Chicago). (b) Density plots showing FACS analysis of cell surface receptor expression. The gating threshold is set at the level exceeded by <5% of untransfected cells, and the proportion of cells exceeding the threshold is given in each case. IL-10RB expression on the cell surface of Huh7 cells transfected with either IL-10RB-K or IL-10RB-E. (c) IL-10-mediated inhibition of LPS-induced TNF-α expression by IL-10RB-K47E genotype. The concentration of IL-10 used was 5 ng/ml shown with the corresponding percentage inhibition (mean ± 2 SE). At maximum inhibition, KK vs. KE/EE shows a significant difference (P = 0.015). Mean percentage inhibition observed for each genotype were as follows (% ± 2 SE): KK, 74.0 ± 6.7; KE, 83.2 ± 4.7; and EE, 90.5 ± 5.8.
Discussion
The persistence or clearance of HBV represents a rare example in humans of an immunologically determined dichotomous outcome of infection that has a high prevalence of the less common outcome: 15% of Gambians develop persistent infection. The natural history of HBV infection in The Gambia is well documented with <10% of transmissions occurring perinatally and the remaining cases being acquired via family contact between the ages of 2 and 10. Therefore, there are unlikely to be many families in which vertical transmission had occurred, who might otherwise dilute the statistical power of the study. The identification of a major effect on this phenotype attributable to functional variation in the IFN-AR2 and IL-10RB genes, which are central to many immune responses, suggests that these loci may be relevant to other immunogically determined diseases. Conversely, the pleiotropic effects of these receptors and their ligands complicates analysis of underlying mechanisms. Although the linkage on chromosome 21 was observed in both populations in The Gambia, we were unable to demonstrate linkage of this region to persistent HBV infection in a population from Italy. This result may be explained by differences in the natural history of HBV infection between the two populations, but, more probably, there are other genes that exert influence on HBV persistence in the Italian population.
CRF2 receptors function as heterodimeric structures that usually consist of one long chain, with a second accessory chain (12). Signaling via the type I IFN receptor modulates the expression of >150 genes (13), influencing such processes as dendritic cell maturation, T cell survival, MHC class I antigen presentation, and stimulation of intracellular antiviral pathways such as those involving MxA and OAS (14–16). The F8S amino acid change in the IFN-AR2 gene is located in the signal peptide, suggesting that this alteration influences IFN-AR2 protein trafficking to the membrane or secretion of the alternatively spliced and truncated forms of the protein. This hypothesis is supported by the observed lack of difference in the RNA transcript levels and preliminary evidence that the two alleles lead to different levels of cell surface expression on B cells and to differential signaling effects. However, there is an apparent disparity between the enhanced cell surface expression and signal transduction of IFN-AR2-F and the susceptibility to persistent infection conferred by this allele. Possible explanations include premature down-regulation of the receptor in vivo or differential effects of the allele on tolerance to HBV antigens, both of which will require detailed evaluation (17–19).
The biological effects of IL-10 are mediated via the two subunits of the IL-10 receptor. IL-10RA is the long chain and major signaling component, whereas IL-10RB is the shorter component that spans the membrane (20, 21). IL-10RB is constitutively expressed in most cells and tissues, and several other members of the IL-10-related family of cytokines have recently been found to utilize IL-10RB in conjunction with specific second-receptor chains. IL-22 (22, 23) uses IL-22RA as the partner chain of IL-10RB. The newly identified IFN-λ (IFN-λ 1–3) family of proteins, also known as IL-28A, IL-28B, and IL-29, have recently been found to utilize IL-10RB in addition to a specific receptor chain IFN-λR1 (or IL-28RA) (12). The IFN-λs are distantly related to the type I IFNs and have antiviral activity. Although the IL-10RB-K47E change is located in the first extracellular FnIII domain repeat of the molecule, which is probably away from residues implicated in IL-10-ligand binding (24), we found clear evidence of a functional effect of this variant on mRNA levels, receptor surface expression and signaling. The association of the IL-10RB*E allele with viral clearance is consistent with amplified transduction of the IFN-λ signal, leading to more efficient viral clearance. The alternative explanation, that the IL-10 signal is amplified, is harder to reconcile with the immunoregulatory role of this cytokine and the requirement for a strong Th1 response to mediate viral elimination
Forty years ago, pedigree analysis of HBsAg positivity in the Philippines (25) and then elsewhere (26) supported an autosomal recessive basis for susceptibility to carriage, and we present the mapping and identification of a major locus for this phenotype. Studies of other infectious diseases suggest that susceptibility to HBV carriage is likely to be generally highly polygenic, consistent with our finding that the region of strongest linkage in a genome-wide scan includes two susceptibility genes with related functions. Genome-wide analysis of a cohort of Indian families with tuberculoid leprosy also identified a major susceptibility gene in which the minor allele showed a dominant protective effect, suggesting that these might be recent variants that are increasing in frequency through selection (K. Tosh, A.J.F., S. Meisner, R. M. Pitchappan, and A.V.S.H., unpublished data). We show that genome-wide analysis of a globally prevalent infectious disease can identify susceptibility loci with functionally relevant variation, providing avenues for identifying immunointervention strategies.
Methods
Ethical approval for this study was granted by the local scientific coordinating committee and the joint Gambian government/Medical Research Council ethical committee.
Patients-Family Study.
HBsAg positive index cases in households were identified from data collected during previous studies (27–29). Follow-up visits to the families of all of these chronic carriers were carried out, and all siblings of index cases (>10 years of age and not vaccinated against hepatitis B) and parents were asked to take part in the study. A questionnaire was completed to document the family status and vaccine history. Subsequent serology testing of these families was completed on all samples to ascertain the HBV status of all members. Reverse passive agglutination (Hepato-test, Wellcome Diagnostics, Dartford, U.K.) was used to detect HBsAg. All negative samples were later retested with an immunometric assay (Sorin Biomedica, Saluggia, Italy). An immunoradiometric assay kit (Dia, Sorin Biomedica) was used for the detection of IgM anticore, HBeAg, and anti-HBe. For the purpose of our statistical analysis, all individuals who tested positive for the HBsAg and negative for antiHBc-IgM were considered to be chronic HBV carriers (or affected), and those that tested positive for antiHBc-(total) and negative for HBsAg were considered to be recovered infections (unaffected) (recovered self limiting infection). Patients who tested positive for both HBsAg and antiHBc-IgM were classed as disease status unknown, being considered to be in the acute phase of infection before the outcome (self-limiting or persistent) is known. AntiHBc-(total) negative subjects were classified as showing no evidence of being infected. Routine HBV vaccination did not begin in The Gambia until 1996; however, any individuals who had been vaccinated were excluded on the basis of their serological profile [antiHBs positive, antiHBc-(total) negative]. A total of 135 families with 194 independent sibling pairs were recruited in this way, of which 94 have 2 affected siblings, 28 have 3 affected sibling pairs, and the remaining 13 have 4 or more affected siblings. Of these families, 55 (40.7%) families had both parents, 67 (49.6%) families were missing one parent, and a further 13 (9.6%) families were missing both parents. An additional 47 families with only one affected sibling were recruited for use in transmission disequilibrium test analysis. Of the total 182 families, in 74 families DNA was available from both parents, in 93 families DNA was available from only one parent (67 families with only maternal DNA available and 26 with paternal), and in the remaining 15 families parental DNA was not available. An additional 192 unaffected siblings from families with missing parents were genotyped to aid in the reconstruction of parental haplotypes. Transmission of the virus in these families is by an unknown route.
Genotyping.
Blood samples were taken, and genomic DNA was extracted by using the BACC2 kit from Nucleon Biosciences (Deeside, U.K.). For the genome-screen, genotypes of the samples were obtained by using fluorescence-based semiautomated method (30). PCR was carried out in 96-well microtiter plates in a final volume of 15 μl. This reaction mix contained 50 ng of genomic DNA, 10 mM Tris (pH 8.3), 50 mM KCl, 1–3 mM MgCl2, 200 μM dNTPs, 200 nM of each primer, and 0.5 units of Amplitaq gold polymerase. The PCR program involved an initial denaturation step of 14 min at 95°C, followed by 35 cycles (15 s at 95°C, 30 s at 50–61°C, and 30 s at 72°C), and a final extension step of 2 min at 72°C. The PCRs were performed in MJ Research (Hemel Hempstead, U.K.) thermocyclers, and the PCR products were then combined into pools and electrophoresed through 6% acrylamide gels on ABI373A-sequencing machines. DNA fragment sizing and genotyping of the microsatellite markers was performed by using the genescan and genotyper programs (Applied Biosystems). The LMSv2 marker set (MD10) from Applied Biosystems was used in this genome screen. A total of 318 markers were used in the genome scan, and the order and genetic distances used were taken from the Genethon map and additional web resources (31). The SNPs in this region were typed by using the ligase detection reaction (LDR) and MassExtend genotyping technology (Sequenom). The LDR method has been described in detail elsewhere (32, 33). Details of all PCR primers, LDR probe sequences, and extension primers are listed in Table 1.
Statistical Analysis.
Analysis for nonMendelian inheritance was performed by using the GAS package (version 2, A. Young, Oxford University) and checked by using pedcheck (34). Each microsatellite was analyzed by using the sibpair program (35), and maximum likelihood multipoint mapping was performed by using the nonparametric linkage function in genehunter v2.5.1 program (36, 37). Analysis of transmission disequilibrium was performed by using the PDT test (version 3.11) (6, 7) and the program transmit (8). transmit has the ability to construct missing parental genotypes, therefore increasing the number of completed families in the analysis. Information from multiple siblings is discarded in this test because only a single offspring is selected at random from the families to detect association independent of linkage. Unlike transmit, PDT can use data from multiple offspring whilst remaining a valid test for association in the presence of preexisting linkage, however, it does not try to infer missing parental genotypes. Haplotypes were generated and analyzed by using the program transmit (8). Bonferonni correction of markers for multiple testing was not performed, and it is widely recognized that this method is overconservative and is not applicable to nonindependent markers in a genomic region.
Comparison of TNF-α Suppression by IL-10 for Each Genotype.
IL-10-mediated inhibition of TNF-α expression was measured by using a standard assay as described in ref. 38. Peripheral blood mononuclear cells were separated from whole blood by using a density gradient centrifugation method as described in ref. 39. The macrophages were isolated from the white cell population by incubating overnight using the cytoadherent properties of this cell type. Levels of TNF-α secretion in the assay were measured by a “sandwich” type ELISA assay and carried out according the manufacturer’s instructions (Quantikine Immunoassay R&D Systems, Minneapolis). Increasing concentrations of Escherichia coli (serotype 0127B8) LPS were used to generate a dose-response curve for macrophage TNF-α secretion and a dose of 50 μg/ml at the midpoint of the curve selected for IL-10 inhibition studies. Macrophages were plated out to a concentration of 5 × 104 cells per well and treated with increasing concentrations of IL-10 followed 1 hour later by 50 μg/ml of LPS. Levels of TNF-α secretion were then measured after a 24-hour incubation. In total, the IL-10-mediated inhibition experiment was completed on 22 samples, of genotypes AA (7), AG (9), and GG (6).
Data produced from the experiment to compare the TNF-α suppression by IL-10 for each genotype was performed by using the Wilcoxon ranked-sum test (version 12; SPSS, Chicago).
Analysis of Relative Transcript Expression Levels.
DNA from the HRC1 panel of United Kingdom Caucasian blood donors [European Collection of Cell Cultures (ECACC), Porton, U.K.] was genotyped by a standard sequencing reaction for the F8S and K47E genotypes. EBV-transformed B cells (ECACC) were grown under standard conditions, and RNA was extracted from them by using RNeasy kits (Qiagen). The RNA was subsequently converted into cDNA by using the Omniscript kit (Qiagen). Levels of both IFN-AR2 and IL-10RB transcript levels within heterozygotes were analyzed by using the MassExtend genotyping system (Sequenom, San Diego), examining relative areas under the mass peak. Details of the PCR primers and extension primers can be found in Table 6, which is published as supporting information on the PNAS web site. Controlling for relative assay efficiency was performed by dividing all values by a corresponding genomic value for the same individual. Results were analyzed by using Student’s t test, after testing for the normality of the data (version 12; SPSS, Chicago).
Transfectants.
Huh7 cells were demonstrated to lack constitutive expression of IL-10-RB by using FACS analysis. Full-length cDNA for IL-10RB was extracted from peripheral blood mononuclear cells donated by a healthy heterozygous volunteer. cDNA was cloned into a pTarget expression system (Promega), which was transfected into Huh7 cells by electroporation.
U5a cells, a human fibroblast-derived cell line in which the IFN-AR2 gene has been disabled by chemical mutagenesis, were kindly provided by Ian Kerr (Cancer Research Campaign, London). The U5a cell line was previously complemented by stable transfection with pVADN1-IFN-AR2, which encodes the common IFN-AR2*F allele. Site-directed mutagenesis was used to produce pVADN1-IFN-AR2 encoding the S allele, and both plasmids were transfected into U5a cells.
Huh7 and U5a cells transfected with the corresponding empty vector were used as the baseline control in subsequent experiments. Stable transfectants for each allelic variant were obtained, and multiple clones from each line were subsequently tested in all subsequent experiments, confirming the original findings.
EBV cells derived from individuals of differing genotypes were also analyzed for cell-surface expression.
Cell Surface Cytokine Receptor Expression.
Huh7 cells were resuspended in PBS with 0.53 mM EDTA. After washing, cells were incubated with biotinylated goat anti-human IL-10Rβ (R & D Systems) followed by incubation with Streptavidin phycoerythrin (BD Biosciences). U5A cells were incubated with phycoerythrin-conjugated mouse anti-human IFN-AR2 antibody (AMS Biotech, Abingdon, U.K.). Cells were washed, fixed in BD Cytofix solution (BD Biosciences), and analyzed for labeling on a FACS calibur (Becton Dickinson) cell-sorting instrument.
MHC Class I Expression.
U5a cells transfected with either the IFN-AR2-F or IFN-AR2-S allele and control cells were incubated with IFN-α-2b at concentrations of 1, 10, 100, and 1000 units/ml for 48 h. The cells were stained with R-phycoerythrin (R-PE)-conjugated mouse anti-human HLA-A,B,C monoclonal antibody (BD Biosciences Pharmingen) at a concentration of 10 μl of antibody per 105 cells for 30 min at 4°C and fixed for 20 min in 100 μl Cytofix buffer (Becton Dickinson). Analysis of cell surface expression of MHC class I molecules was performed by FACS.
Antiviral Assay.
U5a cells transfected with either the IFN-AR2-F or IFN-AR2-S allele and control cells were incubated with IFN-α-2b for 24 h before challenge with encephalomyocarditis virus at a concentration of 0.5 plaque-forming unit (pfu)/cell. After 24 h, viable cells were detected by staining with 0.25% crystal violet in 4% paraformaldehyde and measured by absorbance at 570 nM.
Supplementary Material
Acknowledgments
We thank the patients and their families for their cooperation and participation in this study. We are grateful to Andy Hall for his contribution to the epidemiology of the study in The Gambia, Kevin Bottomley and Jonathan Sheldon for their support throughout, and Christophe Aucan for details of PCR and SNP assays. This work was funded by Roche Discovery Welwyn (Herts, U.K.) and the Wellcome Trust (London, U.K.). A.J.F. was supported by a Wellcome Trust Research Training Fellowship, and A.V.S.H. was supported by a Wellcome Trust Principal Research Fellowship.
Abbreviations
- E
acidic glutamate
- EBV
Epstein–Barr virus
- F
phenylalanine
- HBV
hepatitis B virus
- K
basic lysine
- PDT
pedigree disequilibrium test
- S
serine.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Thursz M. Antiviral Res. 2001;52:113–116. doi: 10.1016/s0166-3542(01)00175-9. [DOI] [PubMed] [Google Scholar]
- 2.Hohler T., Reuss E., Evers N., Dietrich E., Rittner C., Freitag C. M., Vollmar J., Schneider P. M., Fimmers R. Lancet. 2002;360:991–995. doi: 10.1016/S0140-6736(02)11083-X. [DOI] [PubMed] [Google Scholar]
- 3.Lee W. M. N. Engl. J. Med. 1997;337:1733–1745. doi: 10.1056/NEJM199712113372406. [DOI] [PubMed] [Google Scholar]
- 4.Rundle A. T., Atkin J., Sudell B. Clin. Genet. 1975;8:1–4. doi: 10.1111/j.1399-0004.1975.tb01946.x. [DOI] [PubMed] [Google Scholar]
- 5.Bayer M. E., Blumberg B. S., Werner B. Nature. 1968;218:1057–1059. doi: 10.1038/2181057a0. [DOI] [PubMed] [Google Scholar]
- 6.Martin E. R., Bass M. P., Kaplan N. L. Am. J. Hum. Genet. 2001;68:1065–1067. doi: 10.1086/319525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin E. R., Monks S. A., Warren L. L., Kaplan N. L. Am. J. Hum. Genet. 2000;67:146–154. doi: 10.1086/302957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clayton D. Am. J. Hum. Genet. 1999;65:1170–1177. doi: 10.1086/302577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koeleman B. P., Dudbridge F., Cordell H. J., Todd J. A. Ann. Hum. Genet. 2000;64:207–213. doi: 10.1017/S0003480000008095. [DOI] [PubMed] [Google Scholar]
- 10.Abecasis G. R., Cookson W. O. Bioinformatics. 2000;16:182–183. doi: 10.1093/bioinformatics/16.2.182. [DOI] [PubMed] [Google Scholar]
- 11.Knight J. C., Keating B. J., Rockett K. A., Kwiatkowski D. P. Nat. Genet. 2003;33:469–475. doi: 10.1038/ng1124. [DOI] [PubMed] [Google Scholar]
- 12.Pestka S., Krause C. D., Sarkar D., Walter M. R., Shi Y., Fisher P. B. Annu. Rev. Immunol. 2004;22:929–979. doi: 10.1146/annurev.immunol.22.012703.104622. [DOI] [PubMed] [Google Scholar]
- 13.Der S. D., Zhou A., Williams B. R., Silverman R. H. Proc. Natl. Acad. Sci. USA. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buelens C., Bartholome E. J., Amraoui Z., Boutriaux M., Salmon I., Thielemans K., Willems F., Goldman M. Blood. 2002;99:993–998. doi: 10.1182/blood.v99.3.993. [DOI] [PubMed] [Google Scholar]
- 15.Fellous M., Nir U., Wallach D., Merlin G., Rubinstein M., Revel M. Proc. Natl. Acad. Sci. USA. 1982;79:3082–3086. doi: 10.1073/pnas.79.10.3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marrack P., Kappler J., Mitchell T. J. Exp. Med. 1999;189:521–530. doi: 10.1084/jem.189.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manavalan J. S., Rossi P. C., Vlad G., Piazza F., Yarilina A., Cortesini R., Mancini D., Suciu-Foca N. Transplant Immunol. 2003;11:245–258. doi: 10.1016/S0966-3274(03)00058-3. [DOI] [PubMed] [Google Scholar]
- 18.Suciu-Foca N., Manavalan J. S., Cortesini R. Transplant Immunol. 2003;11:235–244. doi: 10.1016/S0966-3274(03)00052-2. [DOI] [PubMed] [Google Scholar]
- 19.Constantinescu S. N., Croze E., Murti A., Wang C., Basu L., Hollander D., Russell-Harde D., Betts M., Garcia-Martinez V., Mullersman J. E., et al. Proc. Natl. Acad. Sci. USA. 1995;92:10487–10491. doi: 10.1073/pnas.92.23.10487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fickenscher H., Hor S., Kupers H., Knappe A., Wittmann S., Sticht H. Trends Immunol. 2002;23:89–96. doi: 10.1016/s1471-4906(01)02149-4. [DOI] [PubMed] [Google Scholar]
- 21.Kotenko S. V., Krause C. D., Izotova L. S., Pollack B. P., Wu W., Pestka S. EMBO J. 1997;16:5894–5903. doi: 10.1093/emboj/16.19.5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie M. H., Aggarwal S., Ho W. H., Foster J., Zhang Z., Stinson J., Wood W. I., Goddard A. D., Gurney A. L. J. Biol. Chem. 2000;275:31335–31339. doi: 10.1074/jbc.M005304200. [DOI] [PubMed] [Google Scholar]
- 23.Kotenko S. V., Izotova L. S., Mirochnitchenko O. V., Esterova E., Dickensheets H., Donnelly R. P., Pestka S. J. Biol. Chem. 2001;276:2725–2732. doi: 10.1074/jbc.M007837200. [DOI] [PubMed] [Google Scholar]
- 24.Josephson K., Logsdon N. J., Walter M. R. Immunity. 2001;15:35–46. doi: 10.1016/s1074-7613(01)00169-8. [DOI] [PubMed] [Google Scholar]
- 25.Blumberg B. S., Melartin L., Guint R. A., Werner B. Am. J. Hum. Genet. 1966;18:594–608. [PMC free article] [PubMed] [Google Scholar]
- 26.Blumberg B. S., Friedlaender J. S., Woodside A., Sutnick A. I., London W. T. Proc. Natl. Acad. Sci. USA. 1969;62:1108–1115. doi: 10.1073/pnas.62.4.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayans M. V., Hall A. J., Inskip H. M., Lindsay S. W., Chotard J., Mendy M., Whittle H. C. Lancet. 1994;343:761–763. doi: 10.1016/s0140-6736(94)91838-4. [DOI] [PubMed] [Google Scholar]
- 28.Inskip H. M., Hall A. J., Chotard J., Loik F., Whittle H. Int. J. Epidemiol. 1991;20:764–769. doi: 10.1093/ije/20.3.764. [DOI] [PubMed] [Google Scholar]
- 29.Viviani S., Jack A., Hall A. J., Maine N., Mendy M., Montesano R., Whittle H. C. Vaccine. 1999;17:2946–2950. doi: 10.1016/s0264-410x(99)00178-4. [DOI] [PubMed] [Google Scholar]
- 30.Reed P. W., Davies J. L., Copeman J. B., Bennett S. T., Palmer S. M., Pritchard L. E., Gough S. C., Kawaguchi Y., Cordell H. J., Balfour K. M., et al. Nat. Genet. 1994;7:390–395. doi: 10.1038/ng0794-390. [DOI] [PubMed] [Google Scholar]
- 31.Dib C., Faure S., Fizames C., Samson D., Drouot N., Vignal A., Millasseau P., Marc S., Hazan J., Seboun E., et al. Nature. 1996;380:152–154. doi: 10.1038/380152a0. [DOI] [PubMed] [Google Scholar]
- 32.Day D. J., Speiser P. W., White P. C., Barany F. Genomics. 1995;29:152–162. doi: 10.1006/geno.1995.1226. [DOI] [PubMed] [Google Scholar]
- 33.Hennig B. J., Hellier S., Frodsham A. J., Zhang L., Klenerman P., Knapp S., Wright M., Thomas H. C., Thursz M., Hill A. V. Genes Immun. 2002;3:359–367. doi: 10.1038/sj.gene.6363883. [DOI] [PubMed] [Google Scholar]
- 34.O’Connell J. R., Weeks D. E. Am. J. Hum. Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Satsangi J., Parkes M., Louis E., Hashimoto L., Kato N., Welsh K., Terwilliger J. D., Lathrop G. M., Bell J. I., Jewell D. P. Nat. Genet. 1996;14:199–202. doi: 10.1038/ng1096-199. [DOI] [PubMed] [Google Scholar]
- 36.Kruglyak L., Daly M. J., Reeve-Daly M. P., Lander E. S. Am. J. Hum. Genet. 1996;58:1347–1363. [PMC free article] [PubMed] [Google Scholar]
- 37.Nyholt D. R. Am. J. Hum. Genet. 2000;67:282–288. doi: 10.1086/303029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marchant A., Deviere J., Byl B., De Groote D., Vincent J. L., Goldman M. Lancet. 1994;343:707–708. doi: 10.1016/s0140-6736(94)91584-9. [DOI] [PubMed] [Google Scholar]
- 39.Spencer S. D., Di Marco F., Hooley J., Pitts-Meek S., Bauer M., Ryan A. M., Sordat B., Gibbs V. C., Aguet M. J. Exp. Med. 1998;187:571–578. doi: 10.1084/jem.187.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.