Abstract
Recent studies by our group and others demonstrated a required and conserved role of Stim in store-operated Ca2+ influx and Ca2+ release-activated Ca2+ (CRAC) channel activity. By using an unbiased genome-wide RNA interference screen in Drosophila S2 cells, we now identify 75 hits that strongly inhibited Ca2+ influx upon store emptying by thapsigargin. Among these hits are 11 predicted transmembrane proteins, including Stim, and one, olf186-F, that upon RNA interference-mediated knockdown exhibited a profound reduction of thapsigargin-evoked Ca2+ entry and CRAC current, and upon overexpression a 3-fold augmentation of CRAC current. CRAC currents were further increased to 8-fold higher than control and developed more rapidly when olf186-F was cotransfected with Stim. olf186-F is a member of a highly conserved family of four-transmembrane spanning proteins with homologs from Caenorhabditis elegans to human. The endoplasmic reticulum (ER) Ca2+ pump sarco-/ER calcium ATPase (SERCA) and the single transmembrane-soluble N-ethylmaleimide-sensitive (NSF) attachment receptor (SNARE) protein Syntaxin5 also were required for CRAC channel activity, consistent with a signaling pathway in which Stim senses Ca2+ depletion within the ER, translocates to the plasma membrane, and interacts with olf186-F to trigger CRAC channel activity.
Keywords: capacitative calcium entry (CCE), genome-wide screen, CRAC channel, RNA interference, store-operated calcium (SOC) influx
Patch–clamp experiments have identified the biophysical characteristics of Ca2+ release-activated Ca2+ (CRAC) channels in lymphocytes and other human cell types (1, 2). Despite the acknowledged functional importance of store-operated Ca2+ (SOC) influx in cell biology (2) and of CRAC channels for immune cell activation (3), the intrinsic channel components and signaling pathways that lead to channel activation remain unidentified. In previous work (4), we demonstrated that SOC influx in S2 cells occurs through a channel that shares biophysical properties with CRAC channels in human T lymphocytes. In a medium-throughput RNA interference (RNAi) screen targeting 170 candidate genes in S2 cells, we discovered an essential conserved role of Stim and the mammalian homolog STIM1 in SOC influx and CRAC channel activity (5). STIM1 and STIM2 also were identified in an independently performed screen of HeLa cells by using the Drosophila enzyme Dicer to generate small interfering RNA species from dsRNA (6). Drosophila Stim and the mammalian homolog STIM1 appear to play dual roles in the CRAC channel activation sequence, sensing the luminal Ca2+ store content through an EF hand motif and trafficking from an endoplasmic reticulum (ER)-like localization to the plasma membrane to trigger CRAC channel activity (6–8). However, as single-pass transmembrane proteins, Stim and its mammalian homolog STIM1 are unlikely to form the CRAC channel itself. To search systematically for additional components of the CRAC channel, and to analyze the signaling network and other required factors that lead to SOC channel activity, we devised and performed a genome-wide screen on S2 cells based on a fluorescence assay of Ca2+ influx. The library at Harvard’s Drosophila RNAi Screening Center (DRSC) of 23,845 dsRNA amplicons has been used in several functional screens (9–14).
A very recent report identified a genetic defect in patients with severe combined immune deficiency (SCID) (15). The screen in this study made use of the ability of thapsigargin (TG) to send GFP-tagged nuclear factor of activated T cells (NFAT) to the nucleus in S2 cells, providing an assay for disruption of signaling anywhere in the cascade from elevated [Ca2+]i to calcineurin activation and nuclear relocalization of NFAT. The fly gene olf186-F (named Orai) was identified in the screen, and a human homolog on chromosome 12 was shown to be mutated in SCID patients, resulting in the loss of CRAC channel activity. Heterologous expression of the wild-type human homolog, which was named Orai1, restored CRAC channel activity in SCID T cell lines.
Here, based on direct Ca2+ influx measurements in a genome-wide screen, we identify several genes that are required for CRAC channel function in S2 cells. Our results confirm the functional requirement of olf186-F (Orai) for Ca2+ signaling and extend these results to investigate effects of knockdown and overexpression on CRAC channel activity. We also show that the sarco-/ER calcium ATPase (SERCA) pump and the trafficking protein Syntaxin 5 are required for CRAC channel activity.
Results
Genome-Wide Screen for SOC Influx.
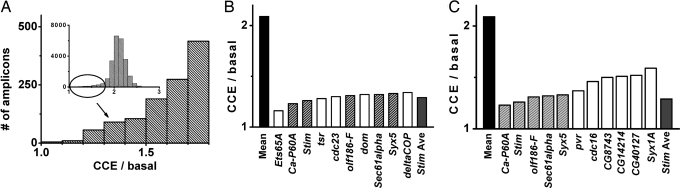
Each well of 63 separate 384-well plates contained an individual dsRNA amplicon. Ca2+-indicator fluorescence measurements were made in each well to monitor cytosolic Ca2+ ([Ca2+]i) before (basal) and after [capacitive calcium entry (CCE)] addition of TG. TG inhibits SERCA pump-mediated reuptake of Ca2+ into cellular stores, depleting them and triggering CCE in S2 cells (4, 16), as well as in mammalian cells (2). Hits in the screen were defined by significantly reduced CCE/basal values, as described in Methods and illustrated by a tail in the histogram shown in Fig. 1A. The “top 10 hits,” with strong suppressive effects comparable with the average value of the Stim positive control (CCE/basal ≈ 1.3), were selected for further evaluation (Fig. 1B; see also Table 1, which is published as supporting information on the PNAS web site). Among the 75 filtered hits with z-scores of CCE/basal < −3 (see Table 2, which is published as supporting information on the PNAS web site), only 11 contained transmembrane segments, as shown in Fig. 1C. Among these hits, the five strongest are annotated in Flybase (www.flybase.org) as Ca-P60A, Stim, olf186-F, sec61alpha, and Syx5.
Fig. 1.
Identification of genes involved in store-operated calcium entry. (A) The effect of individual gene silencing on TG-evoked Ca2+ entry (CCE) relative to basal Ca2+, displayed as a histogram. (Inset) The distribution of averaged CCE/basal values for each well. Low values of CCE/basal are enlarged to show the tail of the distribution, representing amplicons that dramatically suppressed TG-evoked calcium entry. (B) The top 10 hits with strongest effect on TG-evoked Ca2+ influx. Averaged values of CCE/basal are shown for all 48,384 wells tested in the assay (“mean”), for the top 10 hits from the screen, and for the positive control well that contained Stim dsRNA in each assay plate (“Stim Ave”). Striped bars represent hits with transmembrane regions. (C) Transmembrane (TM) protein hits.
The consistent suppressive effect of Stim dsRNA validates the present screen. However, Stim is unlikely to constitute the CRAC channel, because multiple transmembrane segments are found in all known ion-channel pore-forming subunits. The protein product of sec61alpha is a subunit of the translocon complex, which recognizes and delivers newly synthesized membrane proteins into ER, and may be a hit in this screen by altering synthesis or localization of other essential components. Ca-P60A is the SERCA pump gene in fly, whose products are located in the ER for filling/refilling the Ca2+ store. Syx5 generates a single transmembrane-soluble N-ethylmaleimide-sensitive (NSF) attachment receptor (SNARE) protein (Syntaxin 5), which is essential for vesicle fusion and may modulate CCE by altered protein trafficking rather than serving as the channel pore. Thus, among the top 10 hits, olf186-F is the only gene of unknown structure and function that is predicted to contain multiple transmembrane segments.
Effects of olf186-F Knockdown and Overexpression on Ca2+ Influx and CRAC Currents in Single Cells.
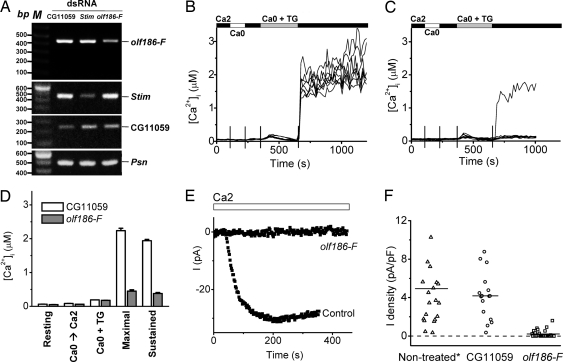
To clarify effects of suppressing olf186-F at the level of single cells, we examined Ca2+ signaling and CRAC currents in cells treated with dsRNA for olf186-F, in comparison with untreated cells or with cells treated with dsRNA for CG11059, an irrelevant cell adhesion molecule (5), as controls. RT-PCR showed >50% decrease of olf186-F mRNA expression, compared with controls (Fig. 2A). Fig. 2B illustrates ratiometric fura-2 [Ca2+]i measurements before and after TG-evoked store depletion in eight individual control cells. Addition of TG in zero-Ca2+ solution to deplete the store elicited a Ca2+ release transient caused by net leak of Ca2+ from the store when the reuptake pump is blocked. Upon readdition of external Ca2+, a robust Ca2+ signal was observed in every cell. In cells pretreated with olf186-F dsRNA, neither the resting [Ca2+]i level nor the release transient were significantly altered, but the rise in [Ca2+]i upon readdition of external Ca2+ was strongly suppressed in the vast majority of the individual cells (Fig. 2C). Fig. 2D clearly demonstrates that suppression of olf186-F effectively inhibits both the early and sustained components of Ca2+ entry evoked by TG at the single-cell level. Comparable inhibition was obtained in cells pretreated with Stim dsRNA as a positive control (data not shown), consistent with our previous report (5).
Fig. 2.
Suppression of TG-dependent Ca2+ influx and CRAC current by olf186-F dsRNA. (A) Reduction of olf186-F mRNA expression in olf186-F dsRNA-treated cells. RT-PCR analysis on olf186-F, Stim, CG11059, and a control gene, Presenilin (Psn). (B) [Ca2+]i in eight representative S2 cells treated with CG11059 dsRNA. Solution exchanges are indicated. (C) [Ca2+]i in eight cells treated with olf186-F dsRNA. (D) Averaged [Ca2+]i values ± SEM for control cells (n = 195 cells in three experiments; white bars) and olf186-F dsRNA-treated cells (n = 189 in four experiments; gray bars): resting [Ca2+]i, peak value upon readdition of 2 mM external Ca2+ before TG treatment (Ca0 → Ca2), peak [Ca2+]i during TG-evoked release transient (Ca0 + TG), and maximal and sustained (3 min) [Ca2+]i after readdition of 2 mM external Ca2+. (E) Representative time course of whole-cell currents recorded in control cells treated with CG11059 dsRNA and in cells treated with olf186-F dsRNA. (F) Suppression of CRAC current by olf186-F dsRNA pretreatment. Each point represents the maximal inward CRAC current density (pA/pF) in a single cell, plotted as absolute values in consecutive order from left to right within three groups of cells: untreated, cells treated with dsRNA to suppress CG11059, or olf186-F (P < 5 × 10−6 compared with either control group). The untreated cell group includes two cells each with current density >12 pA/pF. Horizontal lines indicate the mean value of current density in each group.
Patch–clamp experiments confirmed a dramatic suppression of CRAC currents after knockdown of olf186-F (Fig. 2 E and F). CRAC current normally develops after establishing the whole-cell recording configuration as the cytoplasm is dialyzed by a pipette solution containing a strong Ca2+ chelator to reduce cytosolic [Ca2+]i and deplete internal stores. With this method of “passive stores depletion,” current increases after an initial delay to a maximum value before declining slowly. However, in the majority of cells pretreated with olf186-F dsRNA, CRAC current was completely suppressed, as illustrated by the representative traces in Fig. 2E and by a chart of CRAC current densities (Fig. 2F). As we showed previously for Stim (5), olf186-F expression is required for normal CRAC channel activity.
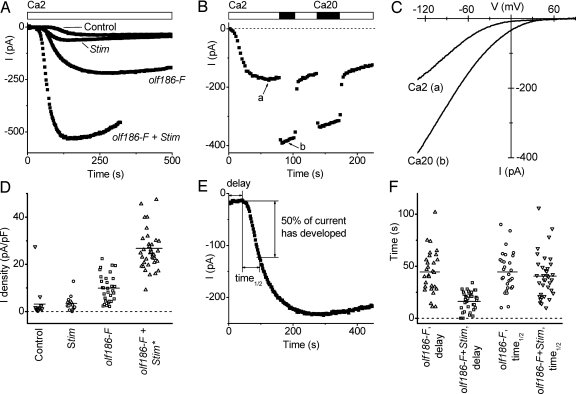
To examine further the function of olf186-F, we cloned its full-length cDNA from S2 cells and inserted it into a Drosophila expression vector. The olf186-F clone was overexpressed with or without a cotransfected Stim clone in S2 cells, by using a cotransfected GFP construct for identification of transfected cells. Increased expression levels of olf186-F and Stim after separate transfections or cotransfection were verified by RT-PCR (see Fig. 6A, which is published as supporting information on the PNAS web site). Fig. 3A illustrates the time course of current development after break-in to achieve whole-cell recording in four representative cells. Expression of Stim by itself had no significant effect on current amplitude compared with control, untransfected cells. However, when olf186-F was overexpressed, CRAC current increased significantly, and when olf186-F was coexpressed with Stim, CRAC current was further enhanced. The induced current after cotransfection of olf186-F with Stim exhibited Ca2+ selectivity and current–voltage shapes indistinguishable from native CRAC current (Fig. 3 B and C). When external Ca2+ was elevated 10-fold, the current magnitudes approximately doubled, as is the case for native CRAC current in S2 cells (4), and current–voltage curves had the same inwardly rectifying characteristic. Fig. 3D illustrates CRAC current densities for individual cells in each group of transfected cells. Overexpression of olf186-F increased the average current density 3-fold, and although Stim by itself did not alter current density, cotransfection with olf186-F produced a remarkable 8-fold enhancement. Interestingly, cotransfection with Stim also decreased the initial delay to the onset of current development (Fig. 3 A, E, and F). Together, these results show that overexpression of olf186-F is sufficient to increase CRAC current density, that coexpression with Stim produces a further enhancement, and that interaction with Stim may be the rate-limiting step for channel activation.
Fig. 3.
Overexpression of olf186-F leads to increased CRAC currents in S2 cells. (A) Representative CRAC currents in S2 cells transfected with GFP only (control), Stim, olf186-F, and olf186-F plus Stim. (B) Ca2+ current in olf186-F + Stim cotransfected cell. Arrows a and b indicate the time corresponding to current–voltage curves in C. (C) Current–voltage relationship of CRAC current in the same cell. (D) CRAC current density in transfected S2 cells, plotted as in Fig. 2F, within four groups of cells: GFP-transfected control; Stim and GFP cotransfected (not significantly different from controls); olf186-F and GFP cotransfected (P < 10−3); and olf186-F, Stim, and GFP cotransfected (P < 5 × 10−6). The group of cells cotransfected by olf186-F, Stim, and GFP includes one cell with current density >50 pA/pF. (E) Method to analyze kinetics of CRAC current development. (F) Effect of cotransfected Stim on delay kinetics. Delay times are significantly reduced (P < 5 × 10−6), but time1/2 values are not altered when Stim is expressed together with olf186-F, compared with olf186-F alone.
Apart from much larger current amplitudes, the Ca2+-selective current in cells cotransfected with olf186-F and Stim exhibited biophysical properties that were indistinguishable from native CRAC currents. Monovalent ion selectivity upon removal of external Ca2+ (divalent-free), Na+ current inactivation, and potentiation of Ca2+ current upon readdition of external Ca2+ were similar to that described for native CRAC current in lymphocytes and S2 cells (see Fig. 7A, which is published as supporting information on the PNAS web site) (4, 17–19). Current–voltage relations for the monovalent Na+ current also showed inward rectification and a reversal potential of +45 mV (Fig. 7B), the same as native monovalent CRAC current and consistent with low permeability to Cs+ (4). The response to voltage steps was also the same, with currents that increase slightly at very negative potentials (Fig. 7 C and D), as seen previously in S2 cells (4). Furthermore, the Ca2+ current in olf186-F + Stim transfectants was sensitive to pharmacological agents that act on native CRAC currents (Fig. 7 E and F). Gd3+ (50 nM) and 2-aminoethyldiphenyl borate (2-APB; 20 μM) blocked the enhanced Ca2+ currents, and at lower concentration (5 μM) 2-APB exhibited a characteristic potentiation of current before blocking. In summary, the ion selectivity, development and inactivation kinetics, and pharmacological profile of the large induced Ca2+ current after overexpression of olf186-F plus Stim match native CRAC currents. Because the current is not enhanced by overexpression of Stim alone, these findings support the possibility that olf186-F itself is part of the channel.
Effects of Ca-P60A, Syx5, and tsr dsRNA Treatment on Ca2+ Dynamics and CRAC Current.
The SERCA pump also emerged from the RNAi screen as a putative regulator of SOC influx. However, because the screen was based on Ca2+ influx induced by TG (which blocks the SERCA pump), we were concerned about the potential for a false-positive hit. We therefore performed single-cell Ca2+ imaging and patch–clamp experiments using alternative stimuli (ionomycin, passive stores depletion) to deplete the Ca2+ store. Selective lowering of Ca-P60A mRNA was first verified by RT-PCR (Fig. 6B). Knockdown of Ca-P60A significantly increased resting [Ca2+]i, reduced the store release transient upon addition of TG and strongly suppressed Ca2+ influx upon readdition of external Ca2+ (Fig. 4A and B). In addition, ionomycin in zero-Ca2+ solution applied to control cells evoked a sharp Ca2+ release transient with a peak that averaged ≈200 nM, but a greatly reduced release transient in Ca-P60A dsRNA-treated cells (Fig. 4 C and D), indicating reduced Ca2+ store content as a consequence of reduced SERCA pump activity. As shown by the summary of Ca2+ imaging experiments (Fig. 4E), knockdown of SERCA has a strong Ca2+ phenotype, raising resting [Ca2+]i, reducing release transients, and suppressing influx evoked by TG. Furthermore, patch–clamp experiments demonstrated that CRAC currents also were suppressed when stores were depleted passively by dialysis of a Ca2+ chelator (Fig. 4F), confirming a requirement of Ca-P60A for activation of functional CRAC channels.
Fig. 4.
Effects of Ca-P60A dsRNA on Ca2+ dynamics in individual S2 cells. (A) Averaged [Ca2+]i in cells treated with control CG11059 dsRNA. (B) Averaged [Ca2+]i in cells treated with Ca-P60A dsRNA. (C and D) Ca2+ release evoked by 1 μM ionomycin in control cells and in cells treated with Ca-P60A dsRNA to knock down SERCA expression. (E) Averaged [Ca2+]i values ± SEM for control cells (white bars) and Ca-P60A dsRNA-treated cells (gray bars) labeled as in Fig. 2D and including peak [Ca2+]i during ionomycin-evoked release transient (Ca0 + Iono). (F) Summary of inward CRAC current densities in control CG11059- and Ca-P60A dsRNA-treated cells (P = 0.002), using the same plotting format as in Fig. 2F.
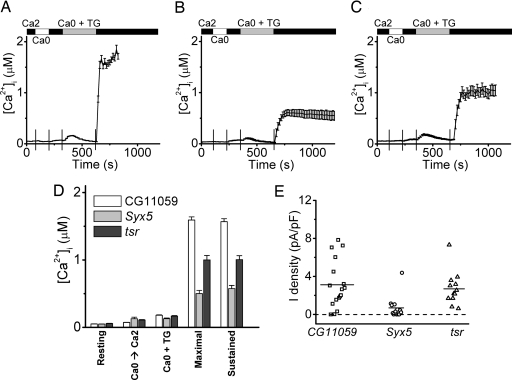
Several trafficking proteins also were identified as putative regulators of SOC activity (Table 2). Syx5 is a syntaxin, several of which have been implicated in SNARE complexes that regulate vesicle trafficking; and tsr is referred to as an actin-binding protein that regulates cytoskeleton remodeling. A putative role of its human homolog, cofilin, has been reported in activation of store-operated calcium entry in platelets (20). Both Syx5 and tsr dsRNA preincubation caused significant and selective lowering of mRNA levels (Fig. 6 C and D) and a corresponding inhibition of TG-dependent Ca2+ influx in S2 cells, without altering the resting [Ca2+]i or store release (compare Fig. 5A–C). Fig. 5D summarizes the inhibition of TG-evoked [Ca2+]i influx when Syx5 or tsr expression was knocked down. Patch–clamp experiments confirmed that CRAC currents were indeed suppressed during passive stores depletion when Syx5 was knocked down, but effects of tsr knockdown on CRAC currents did not achieve statistical significance (Fig. 5E).
Fig. 5.
Suppression of Ca2+ influx and CRAC current by Syx5 and tsr dsRNA. (A–C) Averaged [Ca2+]i in cells treated with control CG11059 dsRNA (A), Syx5 dsRNA (B), or tsr dsRNA (C). (D) Averaged [Ca2+]i values ± SEM for control cells (white bars), Syx5 dsRNA-treated cells (gray bars), and tsr dsRNA-treated cells (black bars) labeled as in Fig. 2D. (E) Summary of inward CRAC current densities in Syx5 and tsr dsRNA-treated cells, using the same plotting format as in Fig. 2F. Mean values for CG11059 and Syx5 are significantly different (P = 0.004). The mean values for CG11059 and tsr are not significantly different (P = 0.65).
Discussion
Our genome-wide screen, based on direct Ca2+ influx measurements, validated Stim and identified several additional genes that are required for CRAC channel activity. We independently identified olf186-F (Orai) as essential for Ca2+ signaling and activation of CRAC current in S2 cells, confirming two recent reports (15, 21). In addition, we provide evidence based on overexpression that it may form an essential part of the CRAC channel. In mammalian cells overexpression of STIM1 increases Ca2+ influx rates and CRAC currents by ≈2-fold (7, 8), but in S2 cells we show that overexpression of Stim alone does not increase CRAC current, consistent with Stim serving as a channel activator rather than the channel itself. In contrast, transfection of olf186-F by itself increased CRAC current densities 3-fold, and cotransfection of olf186-F with Stim resulted in an 8-fold enhancement and the largest CRAC currents ever recorded. These results support the hypothesis that olf186-F constitutes part of the CRAC channel and that Stim serves as the messenger for its activation. Consistent with this hypothesis, the CRAC channel activation kinetics during passive Ca2+ store depletion were significantly faster with cotransfected Stim. Many fundamental aspects of the mechanism of CRAC channel activation remain to be clarified, including the protein–protein interactions that underlie trafficking and channel activation. Site-directed mutagenesis in a heterologous expression system may help to define the putative pore-forming region.
Similar to Stim, knockdown of olf186-F did not produce a severe cell growth phenotype (data not shown). It was neither a hit in a previous screen of cell survival (9) nor in any other published Drosophila whole-genome RNAi screen (10–14). The olf186-F gene is a member of a highly conserved gene family that contains three homologs in mammals, two in chicken, three in zebrafish, and one member only in fly and worm (see Fig. 8A, which is published as supporting information on the PNAS web site). C09F5.2, the only homolog in Caenorhabditis elegans, is expressed in intestine, hypodermis, and reproductive system as well as some neuron-like cells in the head and tail regions (www.wormbase.org). Worms under RNAi treatment against C09F5.2 are sterile (22). Analysis of hydrophobic regions of the predicted protein from the fly gene and the three mammalian homologs (Fig. 8B) suggested the presence of four conserved transmembrane segments. Cytoplasmic C termini are suggested by the presence of coiled-coil motifs in each sequence. A predicted transmembrane topology and the sequence for the fly gene are shown in Fig. 8C. Sequence alignment between members from human, chicken, and fly revealed strong sequence conservation in putative transmembrane regions and conserved negatively charged residues in loops between transmembrane segments. All three human members are expressed in the immune system (GNF Symatlas; http://symatlas.gnf.org/SymAtlas). Mutation of a human homolog of Drosophila olf186-F, ORAI1 on chromosome 12, appears to be the cause of defective CRAC channel activity in severe combined immune deficiency patient T cells (15), consistent with a requirement for functional CRAC channels in the immune response. Interestingly, microarray data from public databases (GEO profiles; www.ncbi.nlm.nih.gov) combined with tissue-specific EST counts show that all three human members are expressed in a variety of nonexcitable tissues including thymus, lymph node, intestine, dermis, and many other tissues including the brain, although expression patterns and levels are different among the three members.
Ca-P60A has been proposed to be the only Drosophila SERCA gene (23). We validated its ER pump function by showing that ionomycin did not induce significant store release from S2 cells pretreated with dsRNA against Ca-P60A, consistent with a previous report (23). The elevation in resting [Ca2+]i and rapidly changing Ca2+ transients during changes in external Ca2+ before addition of TG may indicate a low level of constitutive CRAC channel activity induced by store depletion. In addition, SERCA knockdown inhibited CRAC channel activity after passive store depletion in whole-cell patch recordings. These results are consistent with the SERCA pump being required for normal activity of CRAC channels but do not rule out indirect inhibition of CRAC current as a consequence of residual high resting [Ca2+]i or store depletion. The role of SERCA in CRAC channel function merits further study.
Among the hits, several are known to be involved in protein trafficking. The gene products of both Syx5 and Syx1A are t-SNARE proteins involved in vesicle fusion in many cell types. We verified the RNAi effects of Syx5 at the single-cell level and demonstrated strong suppression of CRAC channel activity as well as the SOC influx. tsr may regulate SOC influx indirectly by controlling cell metabolism because RNAi of tsr did not significantly influence CRAC current density in whole-cell patch–clamp experiments. Membrane trafficking previously was suggested to be important for SOC channel activity in Xenopus oocytes, based on inhibition by botulinum toxin or by a dominant-negative SNAP-25 construct (24), and our results further suggest a requirement for syntaxins and SNARE-complex formation, possibly to mediate translocation of Stim to the plasma membrane (6, 7). The screen also revealed three other groups of hits that may influence calcium dynamics. These results set the stage for experiments targeting specific genes to understand the fine tuning of Ca2+ homeostasis and signaling.
Methods
Drosophila S2 cells were cultured in 384-well plates containing ≈0.25 μg of dsRNA (≈104 cells per well). Each plate included a well with dsRNA targeting Stim as a positive control. After 5 days, cells were loaded with a [Ca2+]i indicator fluo-4/AM (10 μM; Molecular Probes); free dye then was washed by Ringer solution containing 2 mM Ca2+ (see Table 3, which is published as supporting information on the PNAS web site, for all solution recipes). Three fluorescence measurements were systematically performed: basal (resting intracellular free Ca2+), CCE (TG-dependent Ca2+ influx assessed 4 min after addition of TG), and Fmax (maximal fluorescence 15 min after addition of Triton X-100 to a final concentration of ≈2% to detect changes in cell number). A schematic diagram is shown in Fig. 9A, which is published as supporting information on the PNAS web site. Values of “basal/Fmax” were calculated for each well to indicate the normalized resting [Ca2+]i level, and values of “CCE/basal” were computed to represent the relative CCE levels. The screen was carried out in duplicate. To correct for variation in dye loading or cell number, we computed ratios of fluorescence values (CCE/basal) as an index for Ca2+ influx evoked by TG. A scatter plot showed reasonable agreement for the replicate assays for most amplicons (Fig. 9B), particularly for hits with reduced Ca2+ influx reflected in lower CCE/basal values. Because most amplicons did not influence the dynamics of Ca2+ signaling, the average for a given plate was very close to that of nontreated wells. Therefore, z-scores of basal/Fmax and CCE/basal equal to the value of the well minus the average of the plate divided by the standard deviation for the plate were calculated for each well. The averaged z-scores (Fig. 9C) represent variations in the distribution of CCE/basal measurements for each amplicon. Hits in the screen, defined by values of >3 standard deviations from the mean (z-score < −3 or >3) fell into four categories: (i) decreased resting [Ca2+]i; (ii) increased resting [Ca2+]i; (iii) decreased CCE (Table 2); and (iv) increased CCE. To eliminate false-positive outcomes, putative hits with a z-score of Fmax < −2, or with more than five off-targets, were generally filtered out from the lists. Overlapping hits between groups i and iv and groups ii and iii were removed from group iv and iii, respectively.
The remaining methods can be found in Supporting Materials and Methods and Table 4, which are published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
We thank Sindy Wei for help with [Ca2+]i imaging; J. Ashot Kozak for helpful discussion; Karinne Cahalan for assistance with illustrations; Dr. Weihua Jiang for data processing; Dr. Luette Forrest for help with cell culture; and B. Mathey-Prevot, N. Perrimon, and staff at the Drosophila RNAi Screening Center at Harvard. This work was supported by National Institutes of Health Grant NS14609 (to M.D.C.), a George E. Hewitt Foundation fellowship (to S.L.Z.), and American Heart Association Scientist Development Grant 0630117N (to Y.Y.).
Abbreviations
- CCE
capacitive calcium entry
- CRAC
Ca2+ release-activated Ca2+
- ER
endoplasmic reticulum
- RNAi
RNA interference
- SERCA
sarco-/ER calcium ATPase
- SNARE
single transmembrane-soluble N-ethylmaleimide-sensitive attachment receptor
- SOC
store-operated Ca2+
- TG
thapsigargin.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Prakriya M., Lewis R. S. Cell Calcium. 2003;33:311–321. doi: 10.1016/s0143-4160(03)00045-9. [DOI] [PubMed] [Google Scholar]
- 2.Parekh A. B., Putney J. W., Jr Physiol. Rev. 2005;85:757–810. doi: 10.1152/physrev.00057.2003. [DOI] [PubMed] [Google Scholar]
- 3.Gallo E. M., Cante-Barrett K., Crabtree G. R. Nat. Immunol. 2006;7:25–32. doi: 10.1038/ni1295. [DOI] [PubMed] [Google Scholar]
- 4.Yeromin A. V., Roos J., Stauderman K. A., Cahalan M. D. J. Gen. Physiol. 2004;123:167–182. doi: 10.1085/jgp.200308982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roos J., DiGregorio P. J., Yeromin A. V., Ohlsen K., Lioudyno M., Zhang S., Safrina O., Kozak J. A., Wagner S. L., Cahalan M. D., et al. J. Cell Biol. 2005;169:435–445. doi: 10.1083/jcb.200502019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liou J., Kim M. L., Heo W. D., Jones J. T., Myers J. W., Ferrell J. E., Jr, Meyer T. Curr. Biol. 2005;15:1235–1241. doi: 10.1016/j.cub.2005.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang S. L., Yu Y., Roos J., Kozak J. A., Deerinck T. J., Ellisman M. H., Stauderman K. A., Cahalan M. D. Nature. 2005;437:902–905. doi: 10.1038/nature04147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spassova M. A., Soboloff J., He L. P., Xu W., Dziadek M. A., Gill D. L. Proc. Natl. Acad. Sci. USA. 2006;103:4040–4045. doi: 10.1073/pnas.0510050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boutros M., Kiger A. A., Armknecht S., Kerr K., Hild M., Koch B., Haas S. A., Consortium H. F., Paro R., Perrimon N. Science. 2004;303:832–835. doi: 10.1126/science.1091266. [DOI] [PubMed] [Google Scholar]
- 10.DasGupta R., Kaykas A., Moon R. T., Perrimon N. Science. 2005;308:826–833. doi: 10.1126/science.1109374. [DOI] [PubMed] [Google Scholar]
- 11.Agaisse H., Burrack L. S., Philips J. A., Rubin E. J., Perrimon N., Higgins D. E. Science. 2005;309:1248–1251. doi: 10.1126/science.1116008. [DOI] [PubMed] [Google Scholar]
- 12.Baeg G. H., Zhou R., Perrimon N. Genes Dev. 2005;19:1861–1870. doi: 10.1101/gad.1320705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bard F., Casano L., Mallabiabarrena A., Wallace E., Saito K., Kitayama H., Guizzunti G., Hu Y., Wendler F., Dasgupta R., et al. Nature. 2006;439:604–607. doi: 10.1038/nature04377. [DOI] [PubMed] [Google Scholar]
- 14.Nybakken K., Vokes S. A., Lin T. Y., McMahon A. P., Perrimon N. Nat. Genet. 2005;37:1323–1332. doi: 10.1038/ng1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feske S., Gwack Y., Prakriya M., Srikanth S., Puppel S.-V., Tanasa B., Hogan P. G., Lewis R. S., Daly M., Rao A. Nature. 2006;441:179–185. doi: 10.1038/nature04702. [DOI] [PubMed] [Google Scholar]
- 16.Yagodin S., Hardie R. C., Lansdell S. J., Millar N. S., Mason W. T., Sattelle D. B. Cell Calcium. 1998;23:219–228. doi: 10.1016/s0143-4160(98)90120-8. [DOI] [PubMed] [Google Scholar]
- 17.Lepple-Wienhues A., Cahalan M. D. Biophys. J. 1996;71:787–794. doi: 10.1016/S0006-3495(96)79278-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zweifach A., Lewis R. S. J. Gen. Physiol. 1996;107:597–610. doi: 10.1085/jgp.107.5.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christian E. P., Spence K. T., Togo J. A., Dargis P. G., Patel J. J. Membr. Biol. 1996;150:63–71. doi: 10.1007/s002329900030. [DOI] [PubMed] [Google Scholar]
- 20.Redondo P. C., Harper M. T., Rosado J. A., Sage S. O. Blood. 2006;107:973–979. doi: 10.1182/blood-2005-05-2015. [DOI] [PubMed] [Google Scholar]
- 21.Vig M., Peinelt C., Beck A., Koomoa D. L., Rabah D., Koblan-Huberson M., Kraft S., Turner H., Fleig A., Penner R., Kinet J. P. Science. 2006;312:1220–1223. doi: 10.1126/science.1127883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maeda I., Kohara Y., Yamamoto M., Sugimoto A. Curr. Biol. 2001;11:171–176. doi: 10.1016/s0960-9822(01)00052-5. [DOI] [PubMed] [Google Scholar]
- 23.Raymond-Delpech V., Towers P. R., Sattelle D. B. Cell Calcium. 2004;35:131–139. doi: 10.1016/j.ceca.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 24.Yao Y., Ferrer-Montiel A. V., Montal M., Tsien R. Y. Cell. 1999;98:475–485. doi: 10.1016/s0092-8674(00)81976-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.