Abstract
Since their discovery as key regulators of early animal development, microRNAs now are recognized as widespread regulators of gene expression. Despite their abundance, little is known regarding the regulation of microRNA biogenesis. We show that three highly conserved muscle-specific microRNAs, miR-1, miR-133 and miR-206, are robustly induced during the myoblast-myotube transition, both in primary human myoblasts and in the mouse mesenchymal C2C12 stem cell line. These microRNAs were not induced during osteogenic conversion of C2C12 cells. Moreover, both loci encoding miR-1, miR-1-1, and miR-1-2, and two of the three encoding miR-133, miR-133a-1 and miR-133a-2, are strongly induced during myogenesis. Some of the induced microRNAs are in intergenic regions, whereas two are transcribed in the opposite direction to the nonmuscle-specific gene in which they are embedded. By using CHIP analysis, we demonstrate that the myogenic factors Myogenin and MyoD bind to regions upstream of these microRNAs and, therefore, are likely to regulate their expression. Because miR-1 and miR-206 are predicted to repress similar mRNA targets, our work suggests that induction of these microRNAs is important in regulating the expression of muscle-specific proteins.
Keywords: development, myogenesis, transcription
Small ribonucleotide-based regulators of gene expression known as microRNAs play important roles in regulatory gene expression (see refs. 1 and 2 for reviews). Although initially identified through their role in the development of Caenorhabditis elegans larvae (3, 4), microRNA abundance (5–7) and conservation is suggestive of a much broader role within both the animal and plant kingdoms. Studies in vertebrates and invertebrates have confirmed this notion: microRNAs play a fundamental role in diverse biological and pathological processes including apoptosis (8), cell fate determination (9–11), metabolism (12, 13) and tumorigenesis (14–16). microRNAs are synthesized as longer primary microRNAs (17, 18) that are sequentially processed in the nucleus and cytoplasm by distinct multiprotein complexes containing Drosha/DGCR8 (19–23) and Dicer/TRBP (24–26), respectively. The resulting duplex is unwound by a helicase activity, and the mature single-stranded RNA is loaded onto the RNA-induced silencing complex, wherein interactions between microRNAs and mRNAs occur. microRNAs repress gene expression by cleaving target mRNAs or by inhibiting the ability of the target mRNA to be translated into protein. Computational approaches have yielded hundreds of putative mRNA targets for individual microRNAs (27–31), but the number of verified targets with biological relevance is still very small.
We are interested in studying the ability of microRNAs to regulate cell differentiation. Our work focuses on three microRNAs specifically and abundantly expressed in muscle tissue, miR-1, miR-133, and miR-206 (32–34). The overrepresentation of these and other microRNAs in various differentiated tissues implicates microRNAs in the determination or maintenance of the differentiated state (35). Studies aimed at the perturbation of miR-1 expression in mice and flies have suggested both types of function for this microRNA. Using a gain-of-function approach, a role for miR-1 in mice has been postulated in the regulation of cardiomyocyte proliferation, thereby implicating miR-1 in the determination of the differentiated state (36). Using the complementary loss-of-function approach in flies, Sokol and Ambros (37) have determined that miR-1 is dispensable for formation of the musculature and suggest that miR-1 may play a role in the maintenance of the differentiated state. Authors of a more recent study (38) have proposed a model in which miR-1 and miR-133 regulate myogenesis by controlling distinct aspects of the differentiation process.
Here we examine the myogenic regulation of microRNAs that belong to distinct families, the miR-1 family (miR-1-1, miR-1-2, and miR-206) and the miR-133 family (miR-133a-1, miR-133a-2, and miR-133b). We show that five of six microRNAs that belong to these families are specifically induced during myogenesis, suggesting these microRNAs play crucial roles during the process of skeletal muscle formation. The myogenic transcription factors myogenin and myogenic differentiation 1 (MyoD) bind to regions upstream of the miR-1 and miR-133 stem loop, thereby providing a molecular explanation for the observed induction during myogenesis. These results, along with similar studies by Chen et al. (38), lay the groundwork for analyzing putative contributions of microRNAs to the process of myogenesis.
Results
microRNA Expression During Myogenesis in C2C12 Cells and Primary Human Myoblasts.
Previous studies (32–35) as well as our own preliminary microRNA-array analysis (S.B. and P.K.R., unpublished data) had suggested that mirR-1, miR-133, and miR-206 are highly expressed in heart and skeletal muscle tissue. To gain insights into microRNA function in skeletal muscle, we used C2C12 mesenchymal stem cells and examined expression patterns of these microRNAs as they mature into myotubes. C2C12 cells were differentiated along the myogenic pathway by placing them in 2% horse serum media for 4 days. Differentiation under these conditions is very efficient, and the steps preceding myotube formation, including Myogenin expression, cell cycle arrest, and cell cycle withdrawal, are well characterized (39). As shown in Fig. 1A Upper, miR-1 and miR-206 are induced during the C2C12 myogenesis. miR-1 is readily detectable by day 2, by which time the majority of the cells are expressing the cdk inhibitor p21 (39) and are mitotically inactive. By day 4, levels of sarcomeric proteins such as myosin are abundant; miR-1 levels are also high. The ≈60-base precursor to miR-1 also is induced in a manner similar to mature miR-1, implying that the regulation of miR-1 occurs primarily at the transcriptional level. miR-206 expression is detectable when C2C12 cells are grown in proliferation media (d0/GM lane in Fig. 1A Middle; GM, growth medium), but its steady-state level increases manyfold as C2C12 cells differentiate. miR-133 (Fig. 1B) is undetectable at day 0 and abundant by day 4. Three distinct mature microRNA bands are seen in miR-133 blots and the exact identity of each band is not known, but many mature microRNAs are heterogeneous in size and vary from 17 to 24 nucleotides. Moreover, miR-133 is encoded by multiple loci (see below), and could also account for the size heterogeneity. Stripping and reprobing the blot with a U6 small nuclear RNA probe indicates equal loading of the lanes (Fig. 1A Lower and B Lower).
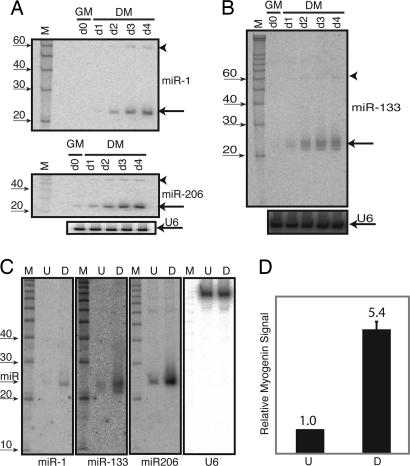
Fig. 1.
Myogenic miRs. (A) Time course of miR-1 and miR-206 induction during C2C12 myogenesis. Total RNA from C2C12 cells in GM or DM for zero (d0), one (d1), two (d2), three (d3), or four (d4) days was subject to Northern blot analysis with a 32P end-labeled miR-1, miR-206, or U6 probe. 33P-labeled 10-bp RNA ladder (Ambion) is shown on the left. The miR-1 blot (Top) was stripped and reprobed sequentially for miR-206 (Middle) and U6 (Bottom). Mature and precursor microRNAs are labeled with an arrow and an arrowhead, respectively. (B) Same as in A except that a new set of samples was probed with a 32P end-labeled miR-133 probe, stripped, and reprobed with a 32P end-labeled U6 probe. (C) RNA was isolated from growing, undifferentiated primary human myoblasts in SkGM2 proliferation media (U) or from primary human myoblasts differentiated for 2 weeks in 2% horse serum-containing media (D). The position of the microRNA is indicated on the left. The membrane first was probed with miR-1, stripped, and subsequently probed for miR-133, miR-206 and U6 as indicated below the blots. 33P-labeled 10-bp RNA ladder (Ambion) is shown on the left for the miR-1 blot. (D) Myogenin induction (by using quantitative RT-PCR) in the differentiated sample indicates efficient differentiation of the primary human myoblasts along the skeletal myogenic lineage. The same RNA samples was used for C and D.
We repeated these analyses by using primary human myoblasts. These cells required a longer time to differentiate; after two weeks in culture, these myoblasts demonstrate a myotube-like morphology. Differentiation along the skeletal myogenic lineage was confirmed by detecting increased expression of myogenin (Fig. 1D). miR-1 expression is barely detectable in undifferentiated primary myoblastic cells but is induced during myotube differentiation. miR-133 also is induced upon differentiation and displays the same heterogeneity in size as seen in C2C12 myotubes. Lastly, miR-206 is detectable in the myoblasts and is further up-regulated upon differentiation (Fig. 1C). The observation that miR-1, miR-133, and miR-206 are regulated similarly in primary human myoblasts and mouse mesenchymal stem cells implies that mechanisms underlying their expression have been conserved during mammalian evolution.
Myogenic Specificity in microRNA Expression Patterns.
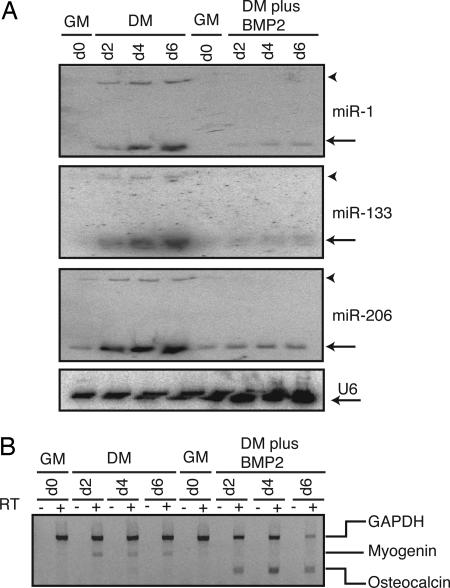
We next investigated whether miR-1, miR-133, and miR-206 are specifically induced during myogenesis or whether their expression was associated with differentiation in general. Accordingly, we induced osteogenic or myogenic differentiation of C2C12 cells by using low serum in the presence or absence of BMP2 (Bone Morphogenetic Protein 2), respectively. In this protocol, BMP2 overrides the myogenic signal induced by low serum and induces the cells along an osteoblastic lineage (40). As seen in Fig. 2A, neither miR-1, miR-133, nor miR-206 expression was up-regulated during C2C12 osteogenesis (“DM plus BMP2” lanes; DM, differentiation media). However, an increase in the levels of miR-1, miR-133, and miR-206 is seen during myogenesis (“DM” lanes). Parallel RT-PCR analyses (Fig. 2B) reveal appropriate induction of protein-encoding mRNA markers associated with myogenesis (myogenin) and osteogenesis (osteocalcin).
Fig. 2.
miR-1, miR-133, and miR-206 induction is specific to myogenesis. (A) C2C12 cells were grown in GM or 2% horse serum (DM) in the presence or absence of 300 ng/ml BMP2. RNA from treated cells were harvested at the indicated times and probed for miR-1, miR-133, miR-206, and U6 small RNAs. Mature and precursor microRNAs are labeled with an arrow and arrowhead, respectively. (B) The same RNA samples used in A was DNase I treated and subject to RT-PCR for the detection of a myogenic marker (myogenin) or a osteoblastic marker (osteocalcin) and a housekeeping gene (GAPDH). Separate RT-PCR reactions were performed but were all loaded in the same well for simultaneous detection by using ethidium bromide staining. RT reactions performed without reverse transcriptase yielded no signal after PCR, and these samples are loaded in the “−” lanes.
Identification of the Active Genomic Loci for Induced microRNAs.
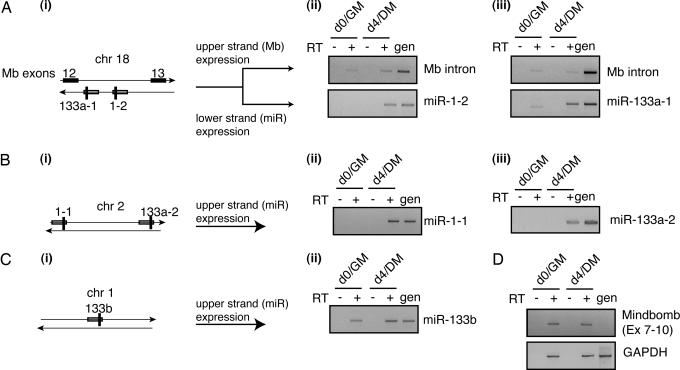
Of the three muscle-specific microRNAs, miR-1 and miR-133 are encoded by two and three loci, respectively. A schematic for these genomic loci is depicted in Fig. 3Ai, Bi, and Ci. In order to shed light on aspects of microRNA regulation that are still poorly understood, we set out to identify the specific miR-1 and miR-133 loci that are induced upon differentiation. To this end, we used RT-PCR transcribed sequences flanking the mature microRNAs, because these to amplify sequences differ in the multiple genes encoding the same mature microRNA.
Fig. 3.
Determination of the loci contributing to mature microRNAs. DNase I-treated RNA from either undifferentiated (day 0/GM) or differentiated (day 4/DM) C2C12 cells was used for RT-PCR. Genomic DNA (gen) was used as a positive control; lack of a signal from reactions without reverse transcriptase (− lanes) shows that there was no genomic DNA contamination. The RNA species being detected is indicated. (A) Intronic miRs. miR-1-2 and miR-133a-1 are located on chromosome 18 and are intronic (i); they are robustly induced during myogenesis (ii Lower and iii Lower). Mindbomb is not induced during myogenesis (ii Upper and iii Upper). (B and C) Intergenic microRNAs. (B) miR-1-1 and miR-133a-2 are located on chromosome 2 and are intergenic (i); they are also robustly induced during myogenesis (ii and iii). (C) miR-133b is located on chromosome 1 and is intergenic (i); it is not up-regulated as dramatically as the other miR-133 isoforms (ii). (D) Amplification of Mindbomb mRNA (Upper) confirms the lack of inducibility during myogenesis; GAPDH amplification (Lower) reveals equal loading of input RNA. Similar results were obtained in a duplicate experiment; results from one set are shown for consistency.
Using this assay for miR-1-1, miR-133a-2, and miR-133b is straightforward because these microRNAs are located in intergenic regions of the chromosome. However, both miR-1-2 and miR-133a-1 are located in an intron between exons 12 and 13 of Mindbomb (Mb), a protein-encoding gene. Moreover, miR-1-2 and miR-133a-1 are expressed from the same DNA strand, whereas Mb is expressed from the opposite strand (Fig. 3Ai). To distinguish expression from the Mb strand versus the miR-1-2/miR-133a-1 strand, we used strand-specific reverse transcription, followed by PCR to detect expression from either strand. Fig. 3 reveals the expression patterns of the miR-1 and miR-133 isoforms before (lanes labeled d0/GM) and after (lanes labeled d4/DM) differentiation of C2C12 cells along the myogenic lineage. miR-1-2 and miR-133a-1 are robustly induced during myogenesis (Fig. 3A ii Lower and iii Lower). Parallel RT-PCR reactions designed to detect expression of the Mb intron reveals constitutive expression (at low levels) from the Mb strand (Fig. 3A ii Upper and iii Upper). Expression from the other miR-1 and miR-133 loci is shown in Fig. 3 B and C. miR-1-1 and miR-133a-2 are robustly induced. In contrast, miR-133b is not; we detect some miR-133b expression in myoblasts, but the level is not significantly enhanced after myogenesis. We confirmed that Mb expression was indeed constitutive with another set of primers that amplified a fragment of mRNA encompassing exons 7–10 of Mb(Fig. 3D Upper). GAPDH mRNA was also amplified to demonstrate equal input of RNA in the various samples (Fig. 3D Lower). The relative physical orientation of Mb and miR-1-2/miR-133a-1, coupled with our expression data, collectively implies that distinct regulatory elements control the expression of miR-1-2/miR-133a-1 and Mb.
Binding of Myogenic Regulators to Induced microRNA Loci.
The observation that miR-1, miR-133, and miR-206 are all specifically up-regulated during myogenesis suggests that regulators of muscle differentiation might be responsible for their activation. MyoD and myogenin both are myogenic transcriptional regulatory factors that activate a number of muscle-specific structural genes and transcription factors to drive myogenesis (41). MyoD is expressed in proliferating C2C12 myoblasts, and upon induction of differentiation, it activates expression of myogenin and other muscle-specific genes. MyoD and myogenin share a number of common targets and participate in a feed-forward circuit that helps to temporally pattern gene expression during myogenesis (42, 43). We used chromatin immunoprecipitation (ChIP) coupled with tiled DNA arrays or promoter-specific PCR to examine the occupancy of these myogenic factors at genomic regions upstream of the induced microRNAs in differentiated C2C12 cells.
The ≈10 kb upstream of the induced microRNAs were tiled on DNA arrays, and immunoprecipitated DNA was labeled and hybridized to these arrays against a reference unenriched sample. Binding data from the arrays was confirmed by gene-specific PCR, or in cases where suitable probes were not identified for microRNA loci, PCR against conserved genomic regions was used as a proxy to assess binding of MyoD and myogenin at these sequences. Fig. 4A–D depicts unprocessed enrichment ratios from the tiled arrays at the chromosomal regions surrounding miR-133a-2, miR-206/miR-133b, miR-133a-1/miR-1-2, and miR-122. Peaks indicating MyoD and myogenin binding are observed in the upstream regions of all of the myogenic microRNAs; in contrast, no peaks are observed upstream of miR-133b and miR-122a, a liver-specific microRNA. For the miR-133a-2 locus, a single peak (region A) was detected for myogenin and MyoD binding at ≈−5 kb relative to the start of the miR-133a-2 stem loop. For miR-133a-1, a peak (region C) is observed at ≈−1.3 kb upstream of the stem loop. This peak lies in between miR133a-1 and miR-1-2, suggesting that miR-133a-1 and miR-1-2 may be independently regulated. Upstream of miR-1-2, we detect two peaks for myogenic factor occupancy that are ≈−2.1 kb (region D) and −3.9 kb (region E) from the start of the stem loop. Lastly, our ChIP analysis reveals that a region (region B) ≈0.8 kb away from the start of the miR-206 stem loop is occupied by MyoD and myogenin. We did not detect any MyoD or myogenin occupancy in the ≈3.7-kb region between the miR-133b and miR-206 stem loops. Sequence information for these genomic regions and the embedded E boxes (with conservation information) are indicated in Data Set 1, which is published as supporting information on the PNAS web site. Because our tiled array did not have probes upstream of miR-1-1, we performed PCR on MyoD and myogenin immunoprecipitates with primers flanking conserved E boxes found in the upstream region of miR-1-1. As shown in Fig. 4E (blots labeled miR-1-1 S1 and S2), an enrichment indicative of MyoD and myogenin binding is detectable at or near two of these conserved E boxes. Confirmatory PCRs for the peaks shown in Figs. 4 A–D are shown in Fig. 4E. Based on these results, we conclude that MyoD and myogenin are likely mediating the up-regulation of miR-1-1, miR-1-2, miR-133a-1, miR-133a-2, and miR-206 in differentiated myotubes.
Fig. 4.
Myogenin and MyoD binding to upstream regions of microRNAs. ChIP-on-chip analyses detects specific binding of MyoD (black lines) and myogenin (gray lines) in the upstream regions of myogenic microRNAs: miR133a-2 (A), miR-206 (B), miR-133a-1 and miR-1-2 (C), and lack thereof in the upstream region of a hepatic microRNA, miR-122a (D). Arrows below the microRNA indicate the direction of transcription. (E) The results of a PCR-based assay to confirm enrichment of sequences bound by myogenin and MyoD in the upstream regions of all of the microRNAs listed in A–D plus two conserved E boxes upstream of miR-1-1 (bottom set). Myogenin (Myog) and pancreatic amylase (Amy2) promoters were used as positive and negative controls, respectively, for myogenic factor binding. In each of the sets of four lanes, the first three lanes correspond to genomic DNA inputs of 90, 30, and 10 ng of DNA from whole-cell lysates. The last lane corresponds to an input of 10 ng of immunoenriched DNA from MyoD or myogenin immunoprecipitations. Shaded boxes under peak regions indicate likely binding sites for the myogenic factors and are listed in Data Set 1. Because the average size of the labeled fragments hybridized to the DNA arrays is ≈300 bp, we cannot resolve sites below this distance.
Discussion
Here we describe the regulated expression patterns of five microRNAs (miR-1-1, miR-1-2, miR-133a-1, miR-133a-2, and miR-206) during the myogenic conversion of C2C12 cells. We also provide evidence suggestive of similar regulation in primary human myoblasts. To understand the molecular mechanism underlying such regulation, we performed ChIP analyses and demonstrated that MyoD and myogenin binded to regions upstream of the induced microRNAs. Taken together, these data strongly implicate MyoD and myogenin as transcription factors that are crucial for the biogenesis of these myogenic microRNAs. A concurrent study (43) also demonstrated the presence of MyoD/myogenin binding to upstream regions of miR-100, miR-191, miR-138-2, and miR-22. Hence, the total number of microRNAs putatively regulated by MyoD/myogenin is nine.
Earlier studies have shed some light on the transcriptional regulation of miR-1 expression in muscle tissue. The sole miR-1 gene in Drosophila is transcriptionally up-regulated by Twist and Mef2 during myogenesis (37). Transgenic mice expressing reporter genes fused to sequences upstream of miR-1-1 and miR-1-2 have revealed the importance of SRF, Mef2, and MyoD for miR-1-1 and miR-1-2 expression (36). By demonstrating that MyoD and myogenin bind to regions upstream of miR-1-1, miR-1-2, miR-133a-1, miR-133a-2, and miR-206, we conclude that they are directly involved in the regulated expression of these microRNAs. A polyadenylated transcript encompassing miR-206/133b was previously identified (in rats) as 7H4, a synapse-associated noncoding RNA (44). The expression patterns of 7H4 closely follows that of myogenin and MyoD, both during embryonic/postnatal development and during reinnervation (45). Hence, these earlier studies provide strong corroborative in vivo evidence of the direct regulation of miR-206 by myogenin and MyoD. Moreover, amongst these two myogenic factors, myogenin is likely to be a more crucial determinant in miR-206 induction because 7H4 is also induced in cells that are unable to activate MyoD (46).
Understanding microRNA function is critically dependent on the identification of its targets. Nucleotides 2–8 (referred to as a “seed”) of the microRNA are crucial in determining target specificity (27, 28). The fact that three microRNAs with identical seeds (miR-1-1, miR-1-2, and miR-206) are all expressed during myogenic differentiation suggests that the down-regulation of the putative targets of miR-1/206 is critical for myotube function. Similarly, induction of two distinct miR-133 (miR-133a-1 and miR-133a-2) genes points to the importance of inhibiting miR-133 targets. Examining the list of miR-1 and miR-133 targets reveals potential targets whose activation results in inhibition of myogenesis. These potential targets include components of the Notch pathway (CSL, mastermind, and Notch3) and BMP2 pathways (BMPR2) (27, 28). miR-1 has been shown to enhance C2C12 differentiation through the down-regulation of a HDAC4, a known negative regulator of myogenesis (38). Other examples of verified miR-1 targets include Hand2 (in mammalian cardiac tissue; ref. 36) and components of the Notch pathway (in flies; ref. 47). A microRNA-centric analysis of microarray expression data derived from differentiating C2C12 cells has implicated miR-1 and miR-133 in the repression of the myoblast transcriptome to allow for the activation of the myotube gene expression program (48). microRNAs also target genes within the context of feedback loops as has been demonstrated for miR-223 and miR-17–5p/miR-20a (49, 50). miR-133 also may act in a regulatory loop because some of the predicted targets for miR-133 include promyogenic transcription factors such as MyoD and Mef2C (27, 28). Hence, myogenic microRNAs also may be involved in keeping the expression levels of promyogenic proteins within a confined window.
Clues pertaining to miR-206 function also can be derived from examining its expression pattern. As Vellaca et al. (44) note, miR-206/133b (7H4) expression during development suggests its role in synapse elimination, the process by which multiple axonal inputs, are withdrawn from a muscle fiber in early postnatal life. Postsynaptic, muscle-derived factors have been implicated in this process (51), and ectopic expression of glial cell-derived neurotrophic factor (GDNF), in particular, causes polyinnervation (52). Intriguingly, GDNF [and brain-derived neurotrophic factor (BDNF)] possess miR-1/206-binding sites in their 3′ UTRs (27, 28). Hence, an attractive model would invoke miR-1/206-mediated down-regulation of either BDNF or GDNF in the process of synapse elimination. Further studies will be needed to address these complex regulatory roles of microRNAs, and the identification of bona fide, biologically relevant targets for microRNAs will be an important goal for microRNA researchers in the coming future.
Materials and Methods
Cell Culture.
C2C12 cells were obtained from ATCC (Manassas, VA) and grown in growth media (GM/10% FCS in DMEM supplemented with glutamine and antibiotics). Before the day of the experiment, five plates of cells were seeded so as to reach ≈75% confluence the next day. RNA was harvested from one of these plates the next day and designated day 0/GM. Concurrently, the four additional plates were treated with differentiation media (DM-2% horse serum in DMEM supplemented with glutamine and antibiotics). RNA was harvested from one plate every day and, respectively, designated day 1/DM, day 2/DM, day 3/DM and day 4/DM. New DM was added to those plates that were not used for RNA extraction. Under these conditions, syncytial myotube formation was evident by the beginning of the second day, and numerous myotubes could be detected on the fourth day. Desmin-positive primary human myoblasts and appropriate media for their culture was obtained from Cambrex (Walkersville, MD). The serum component was changed to 2% horse serum for differentiating primary human myoblasts. Media was changed every other day, and RNA was extracted after 2 weeks of differentiation. For the osteogenesis experiments, C2C12 cells were seeded and placed in fresh GM the next day. RNA was harvested after an additional day of culture (for the d0/GM samples); parallel cultures were treated with 300 ng/ml BMP2 (R & D Systems) diluted into differentiation media for 6 days. Fresh media with BMP2 was added every other day. Differentiation along the myogenic lineage was carried out in an identical manner except that no BMP2 was added to the differentiation media.
RNA Analysis.
Total RNA was extracted by using TRIzol (Invitrogen), of which 10 μg was DNase I treated by using DNA-free (Ambion, Austin, TX), and subject to reverse transcription by using SuperScript II (Invitrogen). For data presented in Fig. 3, the RT reaction was performed by using a single primer to generate strand-specific cDNA. RNase H then was used to degrade RNA, and PCR was performed on the cDNA by using dual primers (one of these primers was the same as the one used for the RT reaction). For data shown in Fig. 2B, random primers were used to generate cDNA and an aliquot of the RT reaction was used directly for PCR by using gene-specific primers to detect myogenin, osteocalcin and GAPDH. For quantitative RT-PCR, DNase I treated total RNA was subject to reverse transcription and real-time PCR by using Myogenin and GAPDH primers in conjunction with SYBR Green Detection real-time PCR kit (Applied Biosystems). The protocol established by Lau et al. (5) was used for Northern blotting of microRNAs. Oligonucleotide sequences, as well as annealing temperatures for PCR and hybridization temperatures for Northern blotting, can be found in Table 1, which is published as supporting information on the PNAS web site. All PCR products were sequenced to verify the amplification product.
Tiling of microRNA Regions on DNA Arrays, ChIP, and Promoter-Specific PCR.
Sequences upstream of miR-1, miR-133, miR-206, and miR-122 loci were analyzed, and probes were designed for the ≈10kb upstream of each microRNA by the methods described in Boyer et al., (53). Probes were spaced ≈250 bp apart. Suitable probes were not identified upstream of the miR-1-1 locus by this method, so promoter-specific PCR was used to study this site. Arrays were manufactured by Agilent Technologies (Palo Alto, CA). ChIP was performed as described in Boyer et al. (53). Chromatin fragments from 48-h differentiated C2C12 cells were immunoprecipitated overnight at 4°C by using either anti-MyoD sc-760 or anti-myogenin sc-576 (Santa Cruz Biotechnology). After reversal of crosslinks, enriched fragments were amplified by using a two-step ligation-mediated PCR protocol and fluorescently labeled with Cy5-dUTP (Amersham Pharmacia). Labeled fragments were hybridized to DNA arrays in Agilent Technologies hybridization chambers at 40°C for ≈40 h against a Cy3-dUTP labeled reference sample of unenriched DNA. Arrays were washed and scanned by using an Agilent Technologies DNA microarray scanner BA as described in ref. 53. Unprocessed enrichment ratios were examined to look for evidence of MyoD or myogenin binding, and putative bound sequences were subjected to promoter-specific PCR to confirm enrichment. PCR reactions were performed as described in ref. 54. PCRs were performed on biological duplicate immunoprecipitations to confirm results. Primers used for miR-1-1 and miR-122a ChIP-PCR were designed so as to flank conserved (between human and mouse) E boxes found in a 5,000-bp region upstream of the microRNA stem loop. The alignment and E box identification was performed by using rvista (http://rvista.dcode.org). All primer sequences used in Fig. 4E are listed in Table 2, which is published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
We thank Drs. Dave Bartel, Richard Young, and I-Hung Shih for their intellectual support. This work was supported by Muscular Dystrophy Association Grant MDA-3882 (to P.K.R.), an American Cancer Society grant (to R.M.K.), National Institutes of Health Grant R01 DK068348 (to H.F.L.), and Richard A. Young Grants GM069400 and HG002668.
Abbreviations
- ChIP
chromatin immunoprecipitation
- DM
differentiation media
- GM
growth media
- MyoD
myogenic differentiation 1.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Bartel D. P. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Ambros V. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 3.Lee R. C., Feinbaum R. L., Ambros V. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 4.Wightman B., Ha I., Ruvkun G. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 5.Lau N. C., Lim L. P., Weinstein E. G., Bartel D. P. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 6.Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 7.Lee R. C., Ambros V. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 8.Xu P., Vernooy S. Y., Guo M., Hay B. A. Curr. Biol. 2003;13:790–795. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- 9.Chang S., Johnston R. J., Jr, Frokjaer-Jensen C., Lockery S., Hobert O. Nature. 2004;430:785–789. doi: 10.1038/nature02752. [DOI] [PubMed] [Google Scholar]
- 10.Chen C. Z., Li L., Lodish H. F., Bartel D. P. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 11.Giraldez A. J., Cinalli R. M., Glasner M. E., Enright A. J., Thomson J. M., Baskerville S., Hammond S. M., Bartel D. P., Schier A. F. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 12.Poy M. N., Eliasson L., Krutzfeldt J., Kuwajima S., Ma X., Macdonald P. E., Pfeffer S., Tuschl T., Rajewsky N., Rorsman P., Stoffel M. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 13.Esau C., Kang X., Peralta E., Hanson E., Marcusson E. G., Ravichandran L. V., Sun Y., Koo S., Perera R. J., Jain R., et al. J. Biol. Chem. 2004;279:52361–52365. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 14.He L., Thomson J. M., Hemann M. T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S. W., Hannon G. J., Hammond S. M. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu J., Getz G., Miska E. A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B. L., Mak R. H., Ferrando A. A., et al. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 16.Johnson S. M., Grosshans H., Shingara J., Byrom M., Jarvis R., Cheng A., Labourier E., Reinert K. L., Brown D., Slack F. J. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 17.Lee Y., Kim M., Han J., Yeom K. H., Lee S., Baek S. H., Kim V. N. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai X., Hagedorn C. H., Cullen B. R. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Radmark O., Kim S., Kim V. N. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 20.Gregory R. I., Yan K. P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., Shiekhattar R. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 21.Landthaler M., Yalcin A., Tuschl T. Curr. Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 22.Han J., Lee Y., Yeom K. H., Kim Y. K., Jin H., Kim V. N. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Denli A. M., Tops B. B., Plasterk R. H., Ketting R. F., Hannon G. J. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 24.Saito K., Ishizuka A., Siomi H., Siomi M. C. PLoS Biol. 2005;3:e235. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forstemann K., Tomari Y., Du T., Vagin V. V., Denli A. M., Bratu D. P., Klattenhoff C., Theurkauf W. E., Zamore P. D. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chendrimada T. P., Gregory R. I., Kumaraswamy E., Norman J., Cooch N., Nishikura K., Shiekhattar R. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis B. P., Burge C. B., Bartel D. P. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 28.Lewis B. P., Shih I. H., Jones-Rhoades M. W., Bartel D. P., Burge C. B. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 29.Enright A. J., John B., Gaul U., Tuschl T., Sander C., Marks D. S. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krek A., Grun D., Poy M. N., Wolf R., Rosenberg L., Epstein E. J., MacMenamin P., da Piedade I., Gunsalus K. C., Stoffel M., Rajewsky N. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 31.Kiriakidou M., Nelson P. T., Kouranov A., Fitziev P., Bouyioukos C., Mourelatos Z., Hatzigeorgiou A. Genes Dev. 2004;18:1165–1178. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sempere L. F., Freemantle S., Pitha-Rowe I., Moss E., Dmitrovsky E., Ambros V. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baskerville S., Bartel D. P. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lagos-Quintana M., Rauhut R., Yalcin A., Meyer J., Lendeckel W., Tuschl T. Curr. Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 35.Wienholds E., Kloosterman W. P., Miska E., Alvarez-Saavedra E., Berezikov E., de Bruijn E., Horvitz H. R., Kauppinen S., Plasterk R. H. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 36.Zhao Y., Samal E., Srivastava D. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 37.Sokol N. S., Ambros V. Genes Dev. 2005;19:2343–2354. doi: 10.1101/gad.1356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen J. F., Mandel E. M., Thomson J. M., Wu Q., Callis T. E., Hammond S. M., Conlon F. L., Wang D. Z. Nat. Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andres V., Walsh K. J. Cell Biol. 1996;132:657–666. doi: 10.1083/jcb.132.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Katagiri T., Yamaguchi A., Komaki M., Abe E., Takahashi N., Ikeda T., Rosen V., Wozney J. M., Fujisawa-Sehara A., Suda T. J. Cell Biol. 1994;127:1755–1766. doi: 10.1083/jcb.127.6.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tapscott S. J. Development (Cambridge, U.K.) 2005;132:2685–2695. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- 42.Blais A., Tsikitis M., Acosta-Alvear D., Sharan R., Kluger Y., Dynlacht B. D. Genes Dev. 2005;19:553–569. doi: 10.1101/gad.1281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cao Y., Kumar R. M., Penn B. H., Berkes C. A., Kooperberg C., Boyer L. A., Young R. A., Tapscott S. J. EMBO J. 2006;25:502–511. doi: 10.1038/sj.emboj.7600958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Velleca M. A., Wallace M. C., Merlie J. P. Mol. Cell. Biol. 1994;14:7095–7104. doi: 10.1128/mcb.14.11.7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eftimie R., Brenner H. R., Buonanno A. Proc. Natl. Acad. Sci. USA. 1991;88:1349–1353. doi: 10.1073/pnas.88.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brennan T. J., Edmondson D. G., Olson E. N. J. Cell Biol. 1990;110:929–937. doi: 10.1083/jcb.110.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kwon C., Han Z., Olson E. N., Srivastava D. Proc. Natl. Acad. Sci. USA. 2005;102:18986–18991. doi: 10.1073/pnas.0509535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Farh K. K., Grimson A., Jan C., Lewis B. P., Johnston W. K., Lim L. P., Burge C. B., Bartel D. P. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 49.Fazi F., Rosa A., Fatica A., Gelmetti V., De Marchis M. L., Nervi C., Bozzoni I. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 50.O’Donnell K. A., Wentzel E. A., Zeller K. I., Dang C. V., Mendell J. T. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 51.Sanes J. R., Lichtman J. W. Annu. Rev. Neurosci. 1999;22:389–442. doi: 10.1146/annurev.neuro.22.1.389. [DOI] [PubMed] [Google Scholar]
- 52.Nguyen Q. T., Parsadanian A. S., Snider W. D., Lichtman J. W. Science. 1998;279:1725–1729. doi: 10.1126/science.279.5357.1725. [DOI] [PubMed] [Google Scholar]
- 53.Boyer L. A., Lee T. I., Cole M. F., Johnstone S. E., Levine S. S., Zucker J. P., Guenther M. G., Kumar R. M., Murray H. L., Jenner R. G., et al. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee T. I., Jenner R. G., Boyer L. A., Guenther M. G., Levine S. S., Kumar R. M., Chevalier B., Johnstone S. E., Cole M. F., Isono K., et al. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.