Abstract
The craniate head is innervated by cranial sensory and motor neurons. Cranial sensory neurons stem from the neurogenic placodes and neural crest and are seen as evolutionary innovations crucial in fulfilling the feeding and respiratory needs of the craniate “new head.” In contrast, cranial motoneurons that are located in the hindbrain and motorize the head have an unclear phylogenetic status. Here we show that these motoneurons are in fact homologous to the motoneurons of the sessile postmetamorphic form of ascidians. The motoneurons of adult Ciona intestinalis, located in the cerebral ganglion and innervating muscles associated with the huge “branchial basket,” express the transcription factors CiPhox2 and CiTbx20, whose vertebrate orthologues collectively define cranial motoneurons of the branchiovisceral class. Moreover, Ciona's postmetamorphic motoneurons arise from a hindbrain set aside during larval life and defined as such by its position (caudal to the prosensephalic sensory vesicle) and coexpression of CiPhox2 and CiHox1, whose orthologues collectively mark the vertebrate hindbrain. These data unveil that the postmetamorphic ascidian brain, assumed to be a derived feature, in fact corresponds to the vertebrate hindbrain and push back the evolutionary origin of cranial nerves to before the origin of craniates.
Keywords: Ciona intestinalis, development, evolution, hindbrain
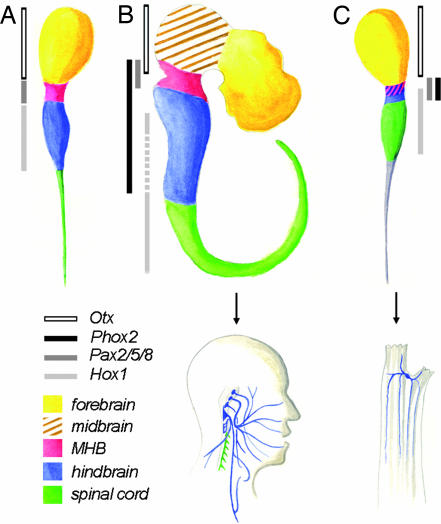
More than a century after Kowalevski (1) noted the chordate affinities of ascidians, homologies between the rostrocaudal divisions of their CNS and that of vertebrates have been explicitly formulated (see Fig. 5A) (2–7). These efforts, mostly based on gene expression, have focused on the embryonic and tadpole larval stages during which the chordate body plan is evident. The rostral sensory vesicle expressing CiOtx has been equated with a forebrain, the “visceral ganglion” expressing CiHox genes has been equated with a hindbrain, and the intervening narrow region expressing CiPax2/5/8A has been equated with a midhindbrain boundary (MHB) variously assigned anatomically to the “neck” (a constriction detectable in Ciona larvae between vesicle and ganglion) (2, 7) or to the caudal sensory vesicle (6). Orthologues of other MHB markers, such as En and Fgf10, also are expressed close to the putative ascidian MHB but in ways difficult to reconcile with the vertebrate pattern (8). Neither midbrain nor metencephalon are recognized around the MHB because of the lack of CiDmbx expression rostral to it (4) and the lack of an ascidian Gbx2 orthologue (9), respectively. Caudal to the ganglion, CiHox5 expression, presumably in ependymal cells (10), has led to liken the “tail nerve cord” (however devoid of neuronal cell bodies) to the vertebrate spinal cord (e.g., refs. 3 and 4).
Fig. 5.
Homologies between rostrocaudal regions of the ascidian and vertebrate CNS and between their motoneuronal derivatives. (A) Previously proposed version of the ascidian tripartite brain model (4), based on gene expression in Halocynthia rorertzii. (B Upper) Rostrocaudal partition of the vertebrate CNS. (C Upper) Revised version of the ascidian larval brain model based on the present study. Regions of the CNS are color-coded as indicated in the key, according to the vertebrate nomenclature. Boxes on the left side of each model, color-coded as indicated in the key, demarcate neuroepithelial gene expression patterns used to define these regions. In mouse, Hox1 expression initially extends from rhombomere 4 to the caudal end of the spinal cord (41, 42) and is secondarily extinguished in the caudal myelencephalon (stippled light-gray box). In the previous version of the tripartite model (A), Phox2 was not examined and no overlap was detected between Pax2/5/8 and Hox1 (2). In the new model (C), both the pattern of Phox2 expression and its overlap with Hox1 lead to redefine the middle part of the larval brain as a hindbrain and to subdivide it into a posterior myelencephalon (blue) and an anterior metencephalon and/or MHB (pink and blue hatching). The trunk ganglion is homologous to the spinal cord, whereas the correspondence of the larval caudal cord (shaded in gray) with parts of the vertebrate CNS, if any, is uncertain. (B Lower and C Lower) Schematic of vertebrate (B Lower) and adult ascidian (C Lower) motoneurons color-coded according to, simultaneously, their origin and nature. Blue indicates branchiovisceral motoneurons born in the hindbrain; green indicates somatic motoneurons born in the spinal cord. No somatic motoneuron is detected in the ganglion of adult Ciona. In vertebrates, all branchiovisceral motor nuclei are born in the hindbrain, and somatic ones are born in the spinal cord except for the abducens (VI) and hypoglossal (XII), which were omitted for clarity.
Most of the larval CNS degenerates during metamorphosis, and the origin of the adult CNS (the cerebral ganglion), positioned between oral and atrial siphon (11), is controversial. On the basis of morphological data in Clavelina, Ciona, and Ecteinascidia, the origin of the cerebral ganglion has been variably traced to the dorsolateral wall of the sensory vesicle (12–14), the caudal sensory vesicle (15), or a “placode” rostral to the sensory vesicle and contiguous with the stomodeum (16). A. S. Romer (17) saw this “small ganglion from which radiate a few nerves” as the “somatic nervous system” of an animal that he otherwise deemed “almost purely visceral.” Aside from this cursory and paradoxical mention, and Berrill's (18) thinly argued parallel with the vertebrate hypothalamus and thalamus, no homology has been proposed so far to our knowledge.
Here, we reexamine comparisons between the ascidian and vertebrate CNS by using CiPhox2, the orthologue of the vertebrate Phox2a and Phox2b paired-like homeobox genes. Phox2b expression and function is highly specific for neurons of the visceral nervous system (19). In the CNS, Phox2b is required for the differentiation of visceral motoneurons (20), both “special” (also called branchial motoneurons and innervating branchial-arch-derived muscles of the face, jaw, neck, and pharynx) and “general” (presynaptic to parasympathetic and enteric neurons), as well as relay visceral sensory neurons of the nucleus of the solitary tract (21), all born and located in the hindbrain, more specifically the myelencephalon. Phox2a, coexpressed with Phox2b throughout the branchiovisceral nervous system (22), specifies three nuclei at the MHB: the locus coeruleus and the oculomotor and trochlear nuclei (19). Thus, within the CNS, Phox2 genes are restricted to the hindbrain and MHB. The characterization of CiPhox2 expression at both larval and adult stages, and throughout metamorphosis leads us to (i) revise the tripartite model of the ascidian larval brain by assigning a hindbrain status to the neck rather than to the visceral ganglion; (ii) show that the larval neck gives rise to motoneurons in the adult; and (iii) establish the homology of the latter with cranial motoneurons of vertebrates.
Results
We identified exons 1–3 of CiPhox2 by searching the Ciona intestinalis genome (JGI, Department of Energy Joint Genome Institute database, available at http://genome.jgi-psf.org/Cioin2/Cioin2.home.html) for similarity with the vertebrate Phox2a and Phox2b homeobox sequence and the Ciona savignyi genome (Ciona savignyi Database, available at www.broad.mit.edu/annotation/ciona) for phylogenetic footprints with C. intestinalis. Orthology was tested by alignment of homeodomain sequences (Fig. 6, which is published as supporting information on the PNAS web site) and gene tree analysis (data not shown). There is no sequence identity outside the homeodomain between CiPhox2 and its vertebrate orthologues, except for a stretch of six amino acids N-terminal to the homedomain that are equally conserved with both Phox2a and Phox2b.
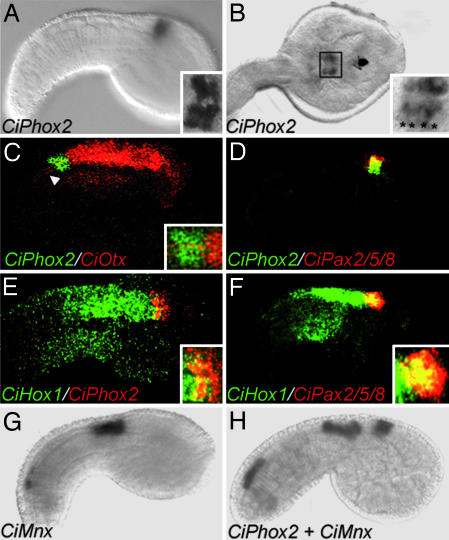
In situ hybridization showed CiPhox2 to be expressed in the embryonic CNS starting at the mid-tailbud stage, in a small patch of cells (Fig. 1A and B) that caudally abuts the CiOtx domain (Fig. 1C) and coincides with the CiPax2/5/8A-positive domain (Fig. 1D). Although CiPax2/5/8 expression was previously described as rostral to and mutually exclusive with that of Hox1 in Halocynthia roretzi (2), we found that the caudal half of the CiPhox2;CiPax2/5/8A-positive region coexpresses CiHox1 (Fig. 1 E and F). Whether this discrepancy reflects differences in species, embryo stage, or detection threshold is unknown. Caudally to the CiPhox2/CiPax2/5/8 domain a patch of cells expressed CiMnx (the orthologue of the vertebrate somatic motoneurons markers Mnr2 and Hb9) and CiChAT (encoding choline acetyltransferase), prefiguring the motoneuronal contingent of the so-called visceral ganglion of the larva (Fig. 1 G and H) (see Discussion for further details about this terminology).
Fig. 1.
Expression of CiPhox2 in embryos of C. intestinalis. Shown are lateral views (anterior to the right) of mid-tailbud-stage embryos (A and C–H) and a dorsal view of a late-tailbud-stage embryo (B) hybridized with the indicated probes. In the insets are shown higher magnifications of the CiPhox2-positive region. (A–F) CiPhox2 is coexpressed in a few cells behind the anlage of the sensory vesicle (A–C), together with CiPax2/5/8 (D), and partially overlapping caudally with CiHox1 (E and F). A Inset and B Inset feature magnifications illustrating the presence of four and eight CiPhox2-positive cells, respectively (asterisks). (C) Note that CiOtx is detected in axons projecting caudally from the sensory vesicle (arrowhead). (G and H) CiMnx is expressed in two domains, one caudal in unidentified cells and one rostral, marking part of the future visceral ganglion, where it is coexpressed with CiChAT (data not shown) in, presumably, the tail motoneurons. (H) Note the gap between CiMnx and CiPhox2, which may correspond to presumptive larval interneurons.
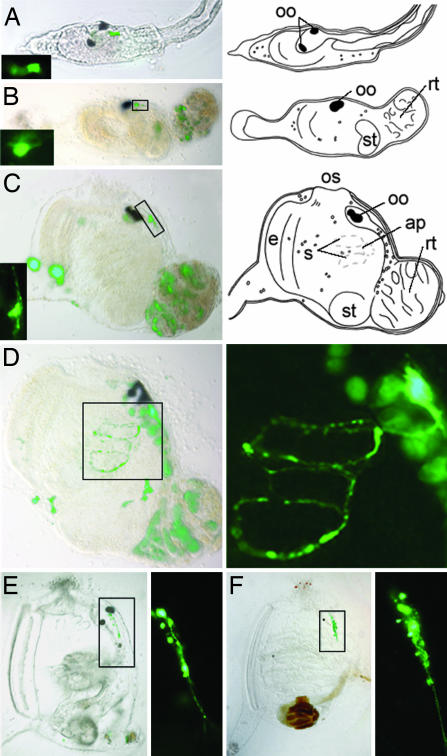
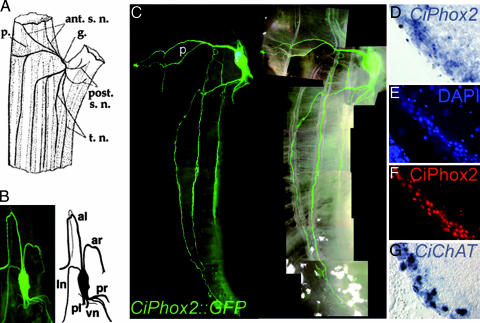
To explore the fate of the CiPhox2-positive cells after hatching and during metamorphosis, we followed the expression of a yellow fluorescent protein (YFP) transgene driven by 3 kb of CiPhox2 promoter sequences that cover three phylogenetic footprints with C. savignyi (Fig. 6). We found transgene expression spatially and temporally continuous between the neck of the larva and the cerebral ganglion of the postmetamorphic animal. CiPhox2::YPF-positive cells were found in the neck of the swimming larva, which did not appear to be neurons based on the absence of neurites (Fig. 2A). YFP expression persisted at the same position (easily spotted caudal to the melanized otolith and ocellus) after settlement of the larva and during its rotation (Fig. 2 B–D). Soon after the beginning of metamorphosis, the YFP-expressing cells started growing neurites (Fig. 2 B and C), and, somewhat later, they had elaborated axons that circumnavigated the first pharyngeal slits (Fig. 2D). At the transition between the first and second ascidian stages, i.e., just before and after the fusion of the atriopores (23) and onset of feeding, the neurite-bearing, YFP-positive cells increased in number (Fig. 2 E and F). In transgenic animals grown to adulthood, the cerebral ganglion was brightly stained together with all major nerve roots (Fig. 3A–C). YFP-positive axons were found in the paired anterior and posterior nerves (Fig. 3B) as well as in the trunk and siphonal nerves, which leave them to run along longitudinal “body wall” muscle bands (Fig. 3C) or project toward the siphons (data not shown) (11). With the caveat that mosaicism of the transgene may have obscured some projections, no neurite could be seen on the surface of the heart or postbranchial digestive tract, whose innervation is controversial and at best sparse (data not shown) (11, 24). Widespread CiPhox2 expression in the cerebral ganglionic cortex, where most neuronal cell bodies are found (11) was confirmed by in situ hybridization (Fig. 3D) and immunohistochemistry (Fig. 3 E and F). Counts on sections double-stained with DAPI and anti-CiPhox2 showed that >75% of the nuclei in the cortex, presumably neuronal, were CiPhox2-positive. Finally, YFP expression did not reveal CiPhox2 expression outside the cerebral ganglion, whether in neurons or any other cell type (data not shown). Notably, we could not confirm by this method the diffuse atrial expression of CiPhox2 recently reported in addition to an earlier CNS expression (25).
Fig. 2.
Expression of a CiPhox2::YFP transgene in the larva and during metamorphosis. (A Left–F Left) YFP-positive cells are detected in the larval neck (A Left) and at an equivalent position in neuronal precursors throughout metamorphosis, from early rotational (23) (B Left) to late rotational (23) (C Left and D), and juvenile (E and F) stages. (A Right–C Right) Schematic of larva and rotational stages of metamorphosis. (A Inset–C Inset and D Right–F Right) Magnifications of CiPhox2-positive cells. The type of long axon navigating around gill slits seen in D is no longer seen at juvenile stages, likely obscured by the thickness of the animal body wall. Fluorescence other than in neuronal precursors was either due to autofluorescence of apoptotic cells [e.g., in the degenerating tail (B–D) and CNS (D)] or spurious YFP expression (such as in the tunic in C and D). Neuronal expression was found in 80–90% of electroporated animals at all stages examined (51 larvae, 29 rotational stage animals, 8 juveniles, and 5 adults). ap, atriopore; e, endostyle; oo, otolith and ocellus; os, oral siphon; rt, tail in the process of resorption; s, stigmata; st, stomach.
Fig. 3.
Expression of the CiPhox2::YFP transgene in the cerebral ganglion of adult Ciona. (A) Diagram of the upper half of the adult Ciona showing the body wall muscle bands, the cerebral ganglion and major nerves (adapted from ref. 11). Note that some variability has been described in the arrangement of both nerves and muscle bands (11). ant.s.n., anterior siphonal nerves; g., cerebral ganglion; p., pericoronal nerve; post.s.n., posterior siphonal nerves; t.n., trunk nerves. (B) (Left) Dorsal view of the cerebral ganglion and its five canonical roots, plus an additional root rostrally on the left, all labeled with YFP. (Right) Schematic of the ganglion and roots. al, anterior left nerve ar, anterior right nerve; ln, lateral nerve (inconstant); pl, posterior left nerve; pr, posterior right nerve; vn, visceral nerve. (C) Lateral view of a transgenic adult animal showing the YFP-labeled cerebral ganglion, longitudinal nerves coursing along body wall muscle bands (superimposed on the right), and the pericoronal nerve (p) running across the oral siphon. (D–G) Magnifications of sections through the cortex of the ganglion hybridized with CiPhox2 (D), stained with DAPI (E), immunostained with an anti-CiPhox2 antibody (F), and hybridized with CiChAT (G).
The axonal projections and cholinergic phenotype (Fig. 3G) of YFP-positive neurons argue that these neurons are motoneurons, and their expression of CiPhox2 argues that they are homologous with vertebrate branchiovisceral motoneurons (see the Introduction). We further tested this homology by assessing cerebral ganglionic neurons for the expression of the Ciona orthologue (26) of the murine T-box transcription factor Tbx20, which was previously described as a vertebrate motoneuron marker (27) and which we found to be entirely specific for branchial and visceral motoneurons of the hindbrain (Fig. 4A–E). The ganglionic cortex indeed widely expressed CiTbx20 (Fig. 4F), and, in contrast with the larval visceral ganglion, did not express CiMnx (data not shown). CiTbx20 was not expressed in the larval neck. This situation is reminiscent of that in the vertebrate hindbrain, where Phox2b expression preceeds that of Tbx20, which is mostly confined to differentiated neurons (27).
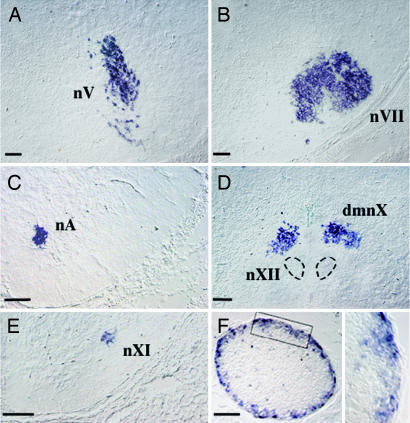
Fig. 4.
Expression of Tbx20 in the CNS of mouse and Ciona. (A–D) Sections through the hindbrain of an embryonic-day-16.5 mouse embryo hybridized with a Tbx20 probe showing expression restricted to branchial and visceral motor nuclei: the trigeminal nucleus (nV) (A), the facial nucleus (nVII) (B), the nucleus ambiguus (nA) (C), the dorsal motor nucleus of the vagus nerve (dmnX) (D), and the accessory nucleus (nXI) (E), to the exclusion of somatic ones such as the hypoglossal (nXII in D) and abducens (data not shown). (F) (Left) Section through Ciona's cerebral ganglion hybridized with CiTbx20. (Right) Magnification of boxed area. The sense probe gave no signal (data not shown). (Scale bar, 100 μm.)
Discussion
The ascidian larva has long been recognized as having a chordate body plan (1). The ascidian larva shares extensive morphological and molecular homologies with vertebrates, and correspondences have been proposed between the rostrocaudal divisions of its brain and those of vertebrates (reviewed in ref. 28). Expression of CiPhox2 in the neck of the ascidian larva leads us to revise the current model of the ascidian “tripartite brain” (2–7) so that its middle part, previously homologized with the MHB (Fig. 5A and B), is now homologous with the hindbrain (possibly including a MHB), to which Phox2b and Phox2a expression is confined in vertebrates (Fig. 5 B and C). The caudal half of this region coexpresses CiHox1, whose vertebrate orthologues of the paralogue 1 group have anterior expression limits in the middle of the embryonic hindbrain (29), and would thus be homologous with the caudal hindbrain or myelencephalon, whereas the Hox-free rostral half would be homologous with the metencephalon and/or MHB. The caudal coexpression of CiPax2/5/8A with CiHox1 has no equivalent in vertebrates, whereas its rostral coexpression with CiPhox2 could be interpreted as a MHB feature, as in the previous model and despite the lack of overlap with CiOtx. However, the absence of Pax gene expression at an equivalent position in either Amphioxus (30) or Oikopleura (31) casts doubt on the chordate origin of a bona fide MHB.
This reassignment of a hindbrain status to the central narrow domain abutting the presumptive sensory vesicle leads, in turn, to homologize the caudally situated visceral ganglion with the spinal cord of vertebrates rather than their hindbrain, as previously proposed. This view is compatible with the expression of Hox genes [CiHox1 and CiHox3 (ref. 32 and this paper)] and of CiMnx, the orthologue of Hb9 and Mnr2, two markers of somatic motoneurons, the vast majority of which are in the spinal cord. Indeed, the ill-named visceral ganglion of the larva (which has no viscera) innervates the tail muscles (7) and, thus, has locomotor function like the spinal cord. Accordingly, we propose to change its name back to that initially given by Kowalevski (1): rumpfganglion, i.e., trunk ganglion. Finally, it is unclear how legitimate it is to homologize the tail nerve cord (which is devoid of neurons) with any part of the vertebrate CNS.
By monitoring the expression of a CiPhox2::YFP transgene from the larval stage on, we show a spatiotemporal continuity of expression between the larval neck region and the motoneurons in the cerebral ganglion of the filter-feeding adult. Although at this stage we cannot formally exclude that the larval CiPhox2-expressing cells turn off CiPhox2 or die, whereas other cells, either born or having migrated in their close vicinity, acquire it, the fine-grain time course of our monitoring (approximately every half day) pleads in favor of a lineage relationship between the larval neck and the postmetamorphic cerebral ganglion. Thus, the seemingly radical remodeling of the ascidian CNS during metamorphosis boils down to the resorption of its anterior (prosencephalic) and posterior (spinal) parts (the sensory vesicle and trunk ganglion, respectively) and the expansion and differentiation of its middle part, which can be seen as a set-aside or prospective hindbrain. These changes logically parallel the shedding of the “somatic animal” in Romer's (17) terms (i.e., the sensory and motor structures required for navigation and locomotion of the larva) and the differentiation and expansion of the “visceral animal” (i.e., the pharynx and postpharyngeal digestive tract required for feeding and breathing in the sessile adult).
Not only do our data reduce the gap between the ascidian and vertebrate adult neuroanatomy but they also shed light on the origin of cranial nerves. Anatomical considerations had previously suggested a homology of vertebrate branchiovisceral motoneurons with the so-called “visceromotor neurons” of cephalochordates (33). We now show that the transcriptional code Phox2/Tbx20, which uniquely defines branchiovisceral motoneurons in the vertebrate CNS, also characterizes ascidians adult motoneurons. This finding strongly argues that cranial nerves V, VII, IX, X, and XI that serve the feeding and breathing purposes of the vertebrate “new head” (34) are elaborations on the nerves of a chordate, filter-feeding “old throat.” With hindsight, this homology could have been envisaged based on the anatomy of the target muscles. The muscle bands innervated by CiPhox2/CiTbx20-positive neurons, although separated from the pharynx and its few intrinsic muscle fibers by the ectodermal invagination of the atrial cavity, are coextensive with them and indeed continuous on their edges and through multiple bridges (trabeculae) across the cavity (11). The function of these muscles is to close the pharynx (by contracting the oral siphon) or to contract it together with the surrounding atrium. Despite their name, body wall muscles are, thus, branchial on the basis of both connectivity and function. These considerations raise the intriguing possibility that the mesoderm, which gives rise to the ascidian body wall muscles, is homologous to the paraxial mesoderm of the vertebrate head, which gives rise to branchial-arch-associated muscles and whose evolutionary status is, so far, totally unresolved.
Materials and Methods
Animals, Embryos, and Transgenesis.
Adult animals were purchased from the Station de Biologie Marine (Roscoff, France), and embryos were obtained as described in ref. 35. Dechorionated eggs were electroporated according to ref. 36. At stage 6 (23), animals were transferred to an aquarium filled with artificial sea water and fed for up to 3 months the unicellular alga Isochrysis galbana and the diatom Chaetoceros gracilis.
In Situ Hybridization on Embryos.
Antisense digoxigenin (DIG) and fluorescein-labeled riboprobes were synthesized with a transcription kit (Roche Diagnostics) by following the manufacturer's instructions. For in situ hybridization, embryos were incubated overnight at 4°C in fixative (4% paraformaldehyde/0.1 M Mops buffer, pH 7.5/2 mM Mg2SO4/1 mM EGTA/0.5 M NaCl) and washed in PBS/0.1% Tween 20 (PBT); permeabilized for 30 min with 2 μg/ml proteinase K in PBT at 37°C; treated with 2 mg/ml glycine in PBT and washed in PBT; and treated 15 min with 0.1 M triethanolamine/0.15% acetic anhydride and washed in PBT. Prehybridization lasted 2 h, and hybridization was conducted overnight at 55°C in 50% formamide/5× SSC (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7)/5× Denhardt's solution (0.02% polyvinylpyrrolidone/0.02% Ficoll/0.02% BSA)/500 mg/ml herring sperm DNA/250 mg/ml yeast RNA, followed by washes at 55°C with 50% formamide/2× SSC, washes at room temperature with 0.2× SSC/0.1% Tween 20, and, finally, washes at room temperature with TNT (0.1 M Tris·HCl, pH 7.5/0.15M NaCl/0.1% Tween 20). The hybridized embryos were blocked in TNB (0.1 M Tris·HCl, pH 7.5/0.15 M NaCl/0.5% blocking reagent; PerkinElmer) for 1 h, then incubated for 6 h in alkaline phosphatase-coupled anti-DIG Fab fragments (1/2,000) (Roche Diagnostics) in TNB, washed in TNT, then treated for standard detection as described by Roche. For simultaneous detection of two probes, the glycine treatment was followed by an incubation with 2% H2O2 in ethanol, and hybridization was performed in the presence of DIG and FITC probes, followed by washing steps and incubation with horseradish peroxidase-coupled anti-FITC Fab fragments (Roche Diagnostics). The embryos were then washed with TNT and incubated for 5 min in the CY3-tyramide working solution (PerkinElmer). After washing steps in TNT and incubation for 10 min with 50% formamide/2× SSC/0.1% Tween 20 at 55°C, they were blocked once more in TNB and incubated overnight at 4°C with horseradish-coupled anti-DIG Fab fragments (Roche Diagnostics), washed in TNT, and incubated for 5 min in the FITC-tyramide working solution (PerkinElmer). Specimens were washed in TNT, mounted in Fluorsave (Calbiochem), and analyzed with a LEICA TCS SP2 confocal microscope.
Immunohistochemistry and in Situ Hybridization on Sections.
Mouse embryos were treated as described in ref. 21. Neural complexes (i.e., cerebral ganglion and neural gland) of adult ciona were fixed for 1 h (for immunohistochemistry) or overnight (for in situ hybridization) at 4°C, then cryoprotected in 20% sucrose in PBS and embedded in Tissue-Tek (Sakura, Zoeterwoude, The Netherlands). For immunohistochemistry, 14-μm sections were blocked in PBT/10% FCS for 30 min and then incubated overnight at 4°C with the rabbit anti-Phox2 antiserum (1/500 in PBT/10% FCS). The sections were then incubated with a Cy3-conjugated donkey anti-IgG antiserum (Jackson ImmunoResearch) for 2 h (1/200 in PBT/10% FCS). Nonimmune rabbit serum was used as a control. The rabbit antiserum was produced (Neosystem, Strasbourg, France) by using a BSA-coupled 19-mer corresponding to the N terminus (MPTAAAYGLNSLRDQSPYC) of the CiPhox2 protein. In situ hybridization was done as described in ref. 21, except that hybridization was carried out at 60°C instead of 70°C. Sense riboprobes were used as controls.
Cloning and Constructs.
We identified exons 1–3 of CiPhox2 by searching the C. intestinalis genome (37) for similarity with the vertebrate Phox2 homeobox sequence and the C. savignyi genome for phylogenetic footprints with C. intestinalis. The complete coding sequence was PCR-amplified from total RNA of adult animals with the following primers: CGGAAACTGCCAGCCACCGATG (forward); CTCGTCGGAATCGTCAGATAACC (reverse). The 589-bp fragment was cloned in pGEMT, then shortened to 486 bp to eliminate a repetitive sequence in the 3′ UTR (GenBank database accession no. DQ530507).
Probes used for in situ hybridization were CiPhox2 (see above), CiChAT (38), CiOtx (39), and CiPax2/5/8-A (gene collection assembled at Kyoto University by N. Satoh and collaborators). Others were amplified by RT-PCR from total larval RNA: CiHox1 (715 bp; forward primer, TCACGTGACTATATTCATGTCCGCCTC; reverse primer, CAATGAATCGTACCCAACCTCCAATCC), CiMnx (660 bp; forward primer, CAGACACGACGCCCCATCACTTGG; reverse primer, AAGACAAGTTCGTGTGTACACTGAACACAGTG), CiTbx20 (641 bp; forward primer, GTATTCTGGAAACAAAAGATTTGTGGGG; reverse primer, TTATAAAAACATAACCAACCTTTCAAATTCGTTC). The mouse Tbx20 probe was isolated from an E10.5 rhombomere 4-specific cDNA library (N. Grillet, C.G., and J.-F.B., unpublished data).
To construct the CiPhox2::YFP transgene, C. intestinalis and C. savignyi genomic sequences were compared with the vista algorithm (40) with 80-bp windows and a 65% identity threshold. A 3,015-bp genomic DNA region (ending in 3′ at the CiPhox2 ATG codon) was PCR-amplified with the primers CATCAGAAGCTTTTCGTGAAGCGGACGTTTTCT (forward) and CGGGATCCTGTAGGCATCGGGGGTTG (reverse) and cloned in pSD-YFP (35), producing an in-frame fusion of the ATG with YFP.
Supplementary Material
Acknowledgments
We thank N. Satoh and the Japanese In Situ Consortium for sharing the gene collection plates with the ascidian community; P. Lemaire and U. Rothbächer (both from Institut de Biologie du Développement, Marseille, France) for advice and materials at early phases of the project; L. Christiaen for advice on electroporation; C. Hudson and H. Yasuo (both from Observatoire Océanologique, Villefranche-sur-Mer, France) for probes; L. Legendre for animal husbandry; A. Pattyn for initial data on mouse Tbx20 expression; and T. Lacalli and G. O. Mackie for discussions. This work was supported by grants from Association Française contre les Myopathies (to C.G.) and Agence Nationale de la Recherche (to J.-F.B.) and by French Ministry of Research Grant ACI2002 (to J-S.J.) and European Community Grant QLK3-CT-2001-01890 (to C.D.).
Abbreviations
- YFP
yellow fluorescent protein
- MHB
midhindbrain boundary.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. DQ530507).
References
- 1.Kowalevsky A. Mem. Acad. Imp. Sci. St. Petersbourg. 1866;7:1–19. [Google Scholar]
- 2.Wada H., Saiga H., Satoh N., Holland P. W. Development (Cambridge, U.K.) 1998;125:1113–1122. doi: 10.1242/dev.125.6.1113. [DOI] [PubMed] [Google Scholar]
- 3.Wada H., Satoh N. Curr. Opin. Neurobiol. 2001;11:16–21. doi: 10.1016/s0959-4388(00)00168-9. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi T., Holland P. W. Development (Cambridge, U.K.) 2004;131:3285–3294. doi: 10.1242/dev.01201. [DOI] [PubMed] [Google Scholar]
- 5.Satoh N. Nat. Rev. Genet. 2003;4:285–295. doi: 10.1038/nrg1042. [DOI] [PubMed] [Google Scholar]
- 6.Meinertzhagen I. A., Okamura Y. Trends Neurosci. 2001;24:401–410. doi: 10.1016/s0166-2236(00)01851-8. [DOI] [PubMed] [Google Scholar]
- 7.Meinertzhagen I. A., Lemaire P., Okamura Y. Annu. Rev. Neurosci. 2004;27:453–485. doi: 10.1146/annurev.neuro.27.070203.144255. [DOI] [PubMed] [Google Scholar]
- 8.Imai K. S., Satoh N., Satou Y. Gene Expression Patterns. 2002;2:319–321. doi: 10.1016/s0925-4773(02)00383-0. [DOI] [PubMed] [Google Scholar]
- 9.Wada S., Tokuoka M., Shoguchi E., Kobayashi K., Di Gregorio A., Spagnuolo A., Branno M., Kohara Y., Rokhsar D., Levine M., et al. Dev. Genes Evol. 2003;213:222–234. doi: 10.1007/s00427-003-0321-0. [DOI] [PubMed] [Google Scholar]
- 10.Gionti M., Ristoratore F., Di Gregorio A., Aniello F., Branno M., Di Lauro R. Dev. Genes Evol. 1998;207:515–523. doi: 10.1007/s004270050142. [DOI] [PubMed] [Google Scholar]
- 11.Millar R. H. LMBC Mem. Typ. Br. Mar. Plants Anim. 1953;35:1–123. [Google Scholar]
- 12.van Beneden E., Julin C. Arch. Biol. 1884;V:317–367. [Google Scholar]
- 13.Willey A. Q. J. Microsc. Sci. 1894;35:295–316. [Google Scholar]
- 14.Elwyn A. Bull. Neurol. Inst. N.Y. 1937;6:163–177. [Google Scholar]
- 15.Takamura K. Rep. Res. Inst. Mar. Bioresour. Fukuyama Univ. 2002;12:27–35. [Google Scholar]
- 16.Manni L., Lane N. J., Joly J. S., Gasparini F., Tiozzo S., Caicci F., Zaniolo G., Burighel P. J. Exp. Zool. B. 2004;302:483–504. doi: 10.1002/jez.b.21013. [DOI] [PubMed] [Google Scholar]
- 17.Romer A. S. Evol. Biol. 1972;6:121–156. [Google Scholar]
- 18.Berrill N. J. The Origin of Vertebrates. Oxford: Clarendon; 1955. [Google Scholar]
- 19.Brunet J.-F., Pattyn A. Curr. Opin. Genet. Dev. 2002;12:435–440. doi: 10.1016/s0959-437x(02)00322-2. [DOI] [PubMed] [Google Scholar]
- 20.Pattyn A., Hirsch M.-R., Goridis C., Brunet J.-F. Development (Cambridge, U.K.) 2000;127:1349–1358. doi: 10.1242/dev.127.7.1349. [DOI] [PubMed] [Google Scholar]
- 21.Dauger S., Pattyn A., Lofaso F., Gaultier C., Goridis C., Gallego J., Brunet J.-F. Development (Cambridge, U.K.) 2003;130:6635–6642. doi: 10.1242/dev.00866. [DOI] [PubMed] [Google Scholar]
- 22.Tiveron M.-C., Hirsch M.-R., Brunet J.-F. J. Neurosci. 1996;16:7649–7660. doi: 10.1523/JNEUROSCI.16-23-07649.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiba S., Sasaki A., Nakayam A., Takamura K., Satoh N. Zool. Sci. 2004;21:285–298. doi: 10.2108/zsj.21.285. [DOI] [PubMed] [Google Scholar]
- 24.Bone Q., Whitear M. Publ. Stn. Zool. Napoli. 1958;30:337–341. [Google Scholar]
- 25.Mazet F., Hutt J. A., Millard J., Graham A., Shimeld S. M. Dev. Biol. 2005;282:494–508. doi: 10.1016/j.ydbio.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 26.Takatori N., Hotta K., Mochizuki Y., Satoh G., Mitani Y., Satoh N., Satou Y., Takahashi H. Dev. Dyn. 2004;230:743–753. doi: 10.1002/dvdy.20082. [DOI] [PubMed] [Google Scholar]
- 27.Kraus F., Haenig B., Kispert A. Mech. Dev. 2001;100:87–91. doi: 10.1016/s0925-4773(00)00499-8. [DOI] [PubMed] [Google Scholar]
- 28.Corbo J. C., Di Gregorio A., Levine M. Cell. 2001;106:535–538. doi: 10.1016/s0092-8674(01)00481-0. [DOI] [PubMed] [Google Scholar]
- 29.Keynes R., Krumlauf R. Annu. Rev. Neurosci. 1994;17:109–132. doi: 10.1146/annurev.ne.17.030194.000545. [DOI] [PubMed] [Google Scholar]
- 30.Kozmik Z., Holland N. D., Kalousova A., Paces J., Schubert M., Holland L. Z. Development (Cambridge, U.K.) 1999;126:1295–1304. doi: 10.1242/dev.126.6.1295. [DOI] [PubMed] [Google Scholar]
- 31.Canestro C., Bassham S., Postlethwait J. Dev. Biol. 2005;285:298–315. doi: 10.1016/j.ydbio.2005.06.039. [DOI] [PubMed] [Google Scholar]
- 32.Ikuta T., Yoshida N., Satoh N., Saiga H. Proc. Natl. Acad. Sci. USA. 2004;101:15118–15123. doi: 10.1073/pnas.0401389101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fritzsch B., Northcutt R. G. Acta Anat. 1993;148:96–109. doi: 10.1159/000147529. [DOI] [PubMed] [Google Scholar]
- 34.Northcutt R. G. J. Exp. Zool. B. 2005;304:274–297. [Google Scholar]
- 35.Moret F., Christiaen L., Deyts C., Blin M., Joly J. S., Vernier P. Eur. J. Neurosci. 2005;21:3043–3055. doi: 10.1111/j.1460-9568.2005.04147.x. [DOI] [PubMed] [Google Scholar]
- 36.Corbo J. C., Levine M., Zeller R. W. Development (Cambridge, U.K.) 1997;124:589–602. doi: 10.1242/dev.124.3.589. [DOI] [PubMed] [Google Scholar]
- 37.Dehal P., Satou Y., Campbell R. K., Chapman J., Degnan B., De Tomaso A., Davidson B., Di Gregorio A. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- 38.Takamura K., Egawa T., Ohnishi S., Okada T., Fukuoka T. Dev. Genes Evol. 2002;212:50–53. doi: 10.1007/s00427-001-0205-0. [DOI] [PubMed] [Google Scholar]
- 39.Hudson C., Lemaire L. Mech. Dev. 2001;100:189–203. doi: 10.1016/s0925-4773(00)00528-1. [DOI] [PubMed] [Google Scholar]
- 40.Mayor C., Brudno M., Schwartz J. R., Poliakov A., Rubin E. M., Frazer K. A., Pachter L. S., Dubchak I. Bioinformatics. 2000;16:1046–1047. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- 41.Marshall H., Nonchev S., Sham M. H., Muchamore I., Lumsden A., Krumlauf R. Nature. 1992;360:737–741. doi: 10.1038/360737a0. [DOI] [PubMed] [Google Scholar]
- 42.Murphy P., Davidson D. R., Hill R. E. Nature. 1989;341:156–159. doi: 10.1038/341156a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.