Abstract
The enormous complexity of biological networks has led to the suggestion that networks are built of modules that perform particular functions and are “reused” in evolution in a manner similar to reusable domains in protein structures or modules of electronic circuits. Analysis of known biological networks has revealed several modules, many of which have transparent biological functions. However, it remains to be shown that identified structural modules constitute evolutionary building blocks, independent and easily interchangeable units. An alternative possibility is that evolutionary modules do not match structural modules. To investigate the structure of evolutionary modules and their relationship to functional ones, we integrated a metabolic network with evolutionary associations between genes inferred from comparative genomics. The resulting metabolic–genomic network places metabolic pathways into evolutionary and genomic context, thereby revealing previously unknown components and modules. We analyzed the integrated metabolic–genomic network on three levels: macro-, meso-, and microscale. The macroscale level demonstrates strong associations between neighboring enzymes and between enzymes that are distant on the network but belong to the same linear pathway. At the mesoscale level, we identified evolutionary metabolic modules and compared them with traditional metabolic pathways. Although, in some cases, there is almost exact correspondence, some pathways are split into independent modules. On the microscale level, we observed high association of enzyme subunits and weak association of isoenzymes independently catalyzing the same reaction. This study shows that evolutionary modules, rather than pathways, may be thought of as regulatory and functional units in bacterial genomes.
Keywords: clustering, evolution, modules
Recent studies of biological networks have revealed structural modules and ubiquitous motifs, many of which have transparent biological functions. However, it remains to be shown that identified structural modules constitute evolutionary building blocks, independent and easily interchangeable units. An alternative possibility is that evolutionary modules do not match structural modules. Comparative genomics and analysis of biological networks provide tools to address this question. Here, we study one of the most accurately assembled networks, the metabolic network of Escherichia coli. To reveal evolutionary modules, we integrate metabolic network with evolutionary associations between genes inferred by comparative genomics of multiple bacterial species. Two genes are associated if (i) they have conserved proximity in distantly related genomes; and/or (ii) demonstrate co-occurrence (i.e., both present or both absent) in most genomes; and/or (iii) have been found fused together. The frequency of these events provides a measure of evolutionary association between the genes. We combine this measure with the structure of the metabolic network to identify evolutionary modules as regions of the network that are highly linked by metabolic reactions and highly associated in related organisms.
Several studies have explored the link between metabolic pathways and conservation of genomic context. Ogata et al. (1), von Mering et al. (2), and Glazko and Mushegian (3) have demonstrated that clusters of chromosomal proximity, co-occurrence, or genomic association are enriched in functionally related enzymes. Several studies reported chromosomal proximity (4–7), grouping into operons and coexpression (8–11) of enzymes of the same metabolic pathway. Kharchenko et al. (8, 11) and Green and Karp (12) used this observation to identify missing enzymes. Li et al. (13) used functional associations to identify parallel modules (sets of proteins in an organism that catalyze the same or similar biochemical reactions but act on different substrates or use different cofactors). Zheng et al. (14) used proximity on the genome and on the metabolic reaction network to predict operons and map them onto metabolic pathways. The same group (15) used phylogenetic profiles and proximity to detect conserved gene clusters and predict protein function. Snel and Huynen (16) examined evolution of protein complexes and metabolic pathways, suggesting, consistent with our results, that traditional pathways lack modularity from the evolutionary point of view. See recent reviews (17–19) for more references.
Although several studies have explored the evolution and organization of the metabolic network, most of them have predominantly studied either the large-scale structure (20) of the network, e.g., degree and flux distribution (21–24) or mean clustering coefficient (21) or small motifs of 2–5 genes (8, 9, 11). Our focus, in contrast, is on the multiscale nature of the relationships between the metabolic network and genomic associations and, particularly, on the modules of 5–30 enzymes, similar to our study of the network of protein–protein interactions (25).
Here, we systematically analyze the metabolic network on three scales. Macroscale analysis explores the patterns of evolutionary association between metabolic enzymes, studied by introducing a graph–theoretical measure of cross-correlation coefficient. Mesoscale analysis focuses on identification of evolutionary modules and their relationships to traditional biochemical pathways. Microscale analysis studies patterns of associations of isoenzymes and subunits of enzymes. Consistent with previous studies (16), we find that traditional metabolic pathways do not match discovered functional modules. Uniquely, we identify such modules and show that they can be parts of pathways or span across pathways.
Results and Discussion
We start by mapping the metabolic network and genomic associations (2, 26) on a graph with vertices representing reactions and two types of edges: metabolic ones that connect reactions sharing a metabolite and edges representing genomic associations (see above) weighted according to the association score S. The two reactions are connected by a genomic edge with weight S if at least one pair of enzymes catalyzing these reactions (or their subunits) is associated with score S. For comparison, we generate control networks by randomly shuffling gene-to-reaction assignments. Such control preserves topologies of both metabolic and association networks individually and randomly assigns one to the other (see Methods).
Cross-Clustering Coefficient.
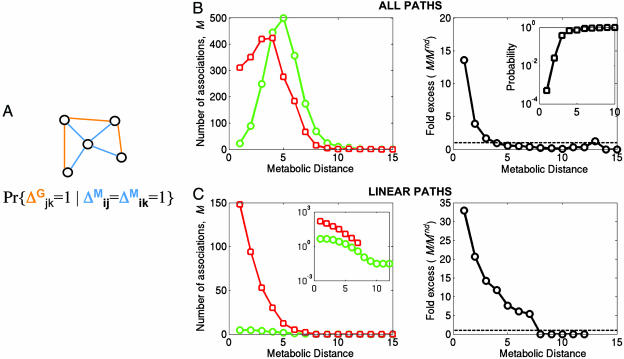
An important question about the macroscale level is whether genomic association brings some clustering to the metabolic network. For a network with one type of edges, the degree of clustering can be estimated by the local clustering coefficient (27) as the probability of a link among neighbors of node i : ci = Pr{Δjk = 1|Δij = Δik = 1}, where Δij is the adjacency matrix of the network. Here, we generalize the clustering coefficient for networks with two types of edges (e.g., edges of type M and G: ΔijM and ΔijG) by introducing cross-clustering coefficient (see Fig. 1A). Consider all M neighbors of node i, i.e., nodes connected to i by edges of type M. We define a cross-clustering coefficient of node i as the probability of a G-edge between M-neighbors of i.
Fig. 1.
The excess of genomic associations at various metabolic distances. (A) The cross-clustering coefficient as defined in the text with metabolic (blue) and genomic (orange) links. (B Left) The number of observed (red) and expected (green) associations M between reactions vs. metabolic distance D between reactions. (B Right) The ratio of observed and expected number of associations (M/Mrnd) between reactions at distance D. Notice several-fold excess of observed over expected associations at distance D ≤ 3. (Inset) Probability of observing M > Mrnd in random controls. (C) Same as above but for reactions in linear paths (Inset, log-scale). A pathway is said to be linear if it contains no major “intersections,” i.e., all of its metabolites participate in four or fewer reactions. Notice significant excess of associations between reactions as far as D = 7 metabolic steps apart on linear pathways. Compare B (all pathways) and C (linear pathways) to see that linear pathways demonstrate long-range associations.
In the case of the genomic–metabolic network, the cross-clustering coefficient ciG|M is calculated as frequency of genomic association ΔjkG between nodes j and k, which are metabolic neighbors of node i (see Fig. 1A). By averaging over all nodes i, we obtain an average cross-clustering coefficient of the integrated network.
The average cross-clustering coefficient of the integrated network is 16 times higher than in the control network, demonstrating that neighboring reactions are 16 times more likely to be genomically associated than expected at random (P < 0.001), suggesting a great deal of “cliquishness” introduced by genomic associations into a mostly branched metabolic network.
Proximal Reactions Are Genomically Associated.
Does abundance of genomic associations decrease with the distance between reactions in the metabolic network?
We find that reactions closer than three intermediate reactions on the metabolic network are much more likely to be catalyzed by genomically associated enzymes than are random controls (Fig. 1B). The tendency to be associated (and hence coregulated and/or coinherited) decays as the number of intermediate reactions increases, with no significant abundance of associated reactions over what would be randomly expected when separated by three or more intermediate reactions (see Fig. 1B Inset). Hence, on average, genomic associations are short-range. Our results are consistent with earlier studies (6, 15) that demonstrated that enzymes close on a metabolic network tend to be close in the genome, and vice versa.
Linear Pathways Demonstrate Long Reach of Association.
The metabolic network contains several linear or weakly branched pathways that contain metabolites with small degree. High-degree metabolites can be considered as “major intersections” of several pathways. Fig. 1C shows that linear pathways contain many more associations than expected. Strikingly, such excessive associations span metabolic distances of up to D = 7. Such long-range associations in linear pathways contrast with fairly short ranges (up to D = 3) if all pairs of reactions are considered indiscriminate of the branching degree of their metabolites.
In summary, our macroscale analysis of the integrated genomic–metabolic network suggests several design principles: (i) genomic associations tend to link nearby reactions (D = 1–3) and (ii) reactions along linear pathways tend to be linked even if they are far apart on the metabolic networks (up to seven intermediates).
Despite significant local clustering, the vast majority (≈70%) of functional associations are among reactions separated by three or more intermediate metabolites (Fig. 1B), suggesting that associations bring together distant reactions of a metabolic pathway or pathways to each other. Such long-range associations give rise to large modules of metabolically and genomically associated reactions.
The Network Contains Several Evolutionary and Regulatory Metabolic Modules.
On the mesoscale level, we identify evolutionary/regulatory modules of highly associated and metabolically proximal enzymes.
The modules are identified as clusters of enzymes that operate on common substrates (i.e., reactions that are a small metabolic distance apart) and have strong genomic associations (i.e., likely to be colocalized, coinherited, or fused). We developed algorithms and statistical techniques to find subgraphs that contain significantly more edges of both types than expected in randomized controls (see Methods). Each of these modules, possibly consisting of parts of different linear biochemical pathways, tends to be regulated and inherited together and, thus, can be treated as the basic building blocks of the cell’s metabolic network.
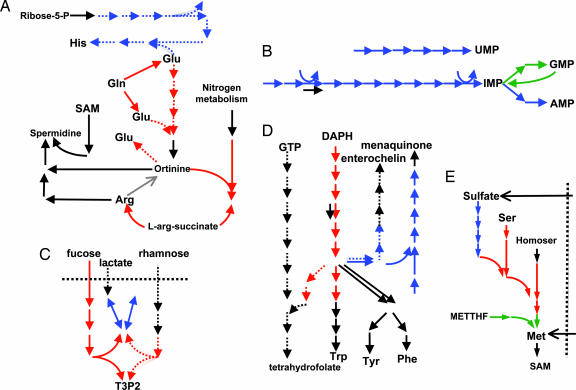
We discovered >20 nonredundant modules. Fig. 2 presents examples of these modules mapped on metabolic pathways. Whereas some pathways contain several dense modules (e.g., biosynthesis of amino acids, purines and pyrimidines, cell-wall components, and certain cofactors), others contain only a few (e.g., central metabolism, salvage, and catabolism). We observe that (i) a module can map on the whole pathway, (ii) a pathway can break into nonoverlapping modules, and (iii) a hybrid module can bring together pieces of two or more pathways. Such diversity indicates a different mechanism of regulation and the extent of structural and evolutionary constraints that a pathway exhibits.
Fig. 2.
Examples of inter- and intrapathway modules of genomically associated reactions. (A) Arginine and histidine pathways. Red, arginine biosynthesis module; blue, histidine biosynthesis module; dotted line, arginine plus histidine biosynthesis module; black (spe genes), spermidine/putrescine biosynthesis, not in any cluster. (B) Purine (Lower) and pyrimidine (Upper) pathways. Blue, hybrid purine–pyrimidine module; green, GMP module; black, nonassociated isoenzyme. (C) Fucose and rhamnose pathways and clusters. Fucose pathway, solid lines; rhamnose pathway, dotted lines; colored (red and blue), two clusters. (D) Aromatic amino acids and folate pathways. Folate pathway: dotted (Left); aromatic amino acids, solid (Left); enterochelin, dotted (Right); menaquinone, solid (Right). Interpathway clusters: aromatic/folate, red; enterochelin/menaquinone, blue. (E) Cystein and methionine biosynthesis. (Left and Center) Cysteine. (Right) Methionine. Horizontal, one-carbon metabolism (partial). Clusters are colored in red, blue, and green.
Modules Do Not Necessarily Coincide with Metabolic Pathways.
For example, modules contained within amino acid biosynthetic pathways rarely coincide with traditional pathways (see Fig. 2). Fig. 2A presents modular structures of arginine and histidine pathways. One module contains the arginine biosynthesis part, another the histidine pathway, and the third hybrid module links the two pathways together by the genomic associations. The third module links the initial part of the arginine pathway (glutamate to ornithine) with the histidine pathway, leaving the rest of the arginine pathway to a separately regulated module. The main metabolite keeping together these pathways is glutamate (source compound for argA, argD, and hisC and product for ygjGH and hisFH), a likely reason for coclustering of the glutamate-pathway gene gltBD with the arginine-biosynthesis genes (see Supporting Appendix, which is published as supporting information on the PNAS web site, for more examples).
Similarly, the cysteine pathway breaks into two modules (cysDN, cysC, cysH, and cysIJ) and (cysE, cysK, cysM, metA, and metB), the latter containing two genes of the methionine pathway. This way, the cysteine and methionine pathways are redistributed between the modules that look reasonable from the biochemical point of view (Fig. 2B). Another unexpected mode of genomic association is observed in the pathways of purine and pyrimidine biosynthesis. These pathways are linked together by a single module (Fig. 2C). Such fusion of purine and pyrimidine pathways can be due to coregulation of their genes by PurR transcription factor. Purine biosynthesis is also split at the IMP junction, revealing the IMP-to-GMP production line as a single module (guaA, guaB, and guaC). This separation is surprising, because guaA and guaB are also regulated by PurR. However, weak genomic associations with other genes in the pathway bring guaA–guaB–guaC into a separate module.
Most of the pathways have not been detected as modules. To make sure that this result is not because of deficiency of our algorithm to detect pathways as modules, we computed the statistical significance of all pathways. We found that 75% of traditional pathways of three or more reactions do not form statistically significant modules (as judged by Eevd > 1; see Methods). The remaining 25% (13 pathways) have been identified as parts or whole modules (e.g., histidine and murein biosynthesis, see Supporting Appendix for details). In summary, the observed discrepancy between modules and pathways in not because of limitations of the algorithm but rather reflect the complex modular structure and evolution of the metabolic network.
Diversity of Central Metabolism.
Few modules are present in the large pathways of the central metabolism [glycolysis, pentosephosphate pathway, the Krebs (TCA) cycle, and respiration]. Although strict thresholds yield only small clusters of associated reactions (e.g., a module of the nonoxidative branch of the pentose phosphate pathway), large superpathway modules containing representatives from several pathways are obtained at low thresholds. For example, part of the EMP pathway, degradation of several carbon sources and the nonoxidative branch of the pentose phosphate pathway form a single superpathway module (Fig. 2D). The lack of modules mapping to traditional pathways in the central metabolism suggests high diversity in its structure and evolution in different bacteria as well as the complexity of its regulation [e.g., a cascade of 11 transcription factors regulating three genes, aslL, zwf, and gnd, in the pentose phosphate pathway (28)]. This finding agrees with observations of Glazko and Mushegian (3) and earlier analyses of Dandekar et al.(40) and Huynen et al.(29) who demonstrated the high diversity of the Krebs cycle and the glycolysis pathway.
A Module May Include Several Pathways.
Examples of superpathway modules (obtained mostly by Monte Carlo search) include cell wall and membrane biosynthesis, biosynthesis of certain amino acid whose genes demonstrate strong linkage, central metabolism (see above), enterochilin, and tetrapyrrole pathways, thus corresponding to large functional systems.
Although observed differences between pathways and modules could not be systematically explained by gene regulation, pathways coregulated in E. coli tend to cluster into modules more than do pathways without a common regulator. We observe this tendency in biosynthetic pathways (e.g., modular arginine, branched chain and aromatic amino acids, histidine, threonine and lysine, and methionine pathways vs. nonmodular glutamine/glutamate, asparagine/aspartate, serine and glycine, and proline pathways) and vitamin biosynthetic pathways [modular biotin pathway regulated by BirA vs. other vitamin pathways, such as riboflavin and thiamin (30–34)]. In the same vein, we notice that purB, the gene breaking the purine biosynthesis pathway, is regulated in a unique way, by a transcription roadblock mechanism with the binding site for the PurR repressor deep within the coding region (35).
In summary, discovered modules demonstrate that regulation and evolutionary mechanisms operate on metabolic pathways by rules that are far more complicated than “one pathway–one regulator.” In other words, the “cell’s definition” of a pathway as a regulatory and evolutionary unit can be dramatically different from those commonly accepted in metabolic biochemistry. See Supporting Appendix for systematic comparison of pathways and modules.
Isoenzymes and Enzymatic Subunits Demonstrate Distinct Patterns of Regulation and Evolution.
On a microscale, we analyze patterns of association between isoenzymes and subunits. A reaction can be catalyzed by one enzyme that consists of several polypeptide chains (subunits) or by several enzymes, each capable of catalyzing this reaction (isoenzymes).
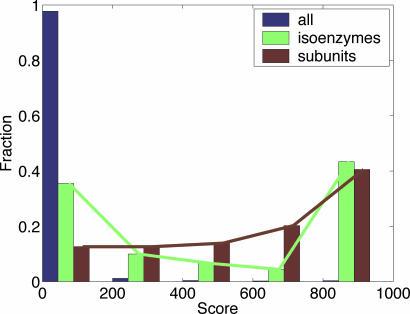
Fig. 3 shows distribution of association scores between subunits, isoenzymes, and, for control, enzymes catalyzing different reactions. Subunits are highly associated, with 72% of them having association score of S > 300. For comparison, only 1.5% of enzymes catalyzing different reactions have S > 300. The biological importance of strong association between subunits is apparent. If all subunits are required for normal operation of an enzyme, then the subunits (i) have to be coregulated/coexpressed, and (ii) loss of one subunit is likely to affect the enzyme’s function and reduce the fitness of the organism. Requirements (i) and (ii) lead to strong genomic association; chromosomal proximity and gene fusion provide coexpression and genetic linkage. Coinheritance shows that the requirements are satisfied in several genomes. This result supports the “balance hypothesis,” which suggests that imbalance in the concentration of proteins that constitute a single complex is deleterious (36). Weak association between some subunits indicates structural flexibility of a multisubunit enzyme (see Supporting Appendix).
Fig. 3.
Distribution of association scores in isoenzymes (green), subunits (brown), and, for control, all enzymes (blue). Notice that isoenzymes exhibit bimodal distribution, with most isoenzymes being either strongly associated (S > 800) or not associated (S < 200). In contrast, subunits have one peak at S > 800, tending to be much more associated than any other enzymes.
Isoenzymes show a pattern of association different from that of subunits. Only 52% of isoenzymes are associated with a score S > 300, whereas the remaining 48% are weakly associated. Isoenzymes demonstrate bimodal distribution, with most of them having S > 800 or S < 100 (Fig. 3). This pattern of association reflects two modes of isoenzyme operation. Associated isoenzymes provide increased flux through the catalyzed reaction and have somewhat different specificities (see example in Supporting Appendix). Weakly associated isoenzymes can be differently regulated in response to different stimuli or conditions (9) and/or participate in different pathways (e.g., speA and adiA). Such isoenzymes have no tendency to be close on the genome, coinherited, or fused.
Summarizing results obtained at different scales for the integrated metabolic-genomic network, we can suggest design principles behind the complex organization, regulation, and evolution of the metabolic network. Our analysis suggests that (i) modules of high genomic association and metabolic proximity do not necessarily match traditional metabolic pathways, and, thus, such modules, rather than the traditional pathways, can be thought of as evolutionary and regulatory units. We also see that genomic associations favor linear metabolic pathways, breaking at branching points. This observation suggests that (ii) linear pathways are regulated and inherited as a single “building block” of the metabolic network. Finally, we see that (iii) although enzymatic subunits are strongly associated, suggesting a persistent coregulation and coevolution, regulation and evolution of isoenzymes depends on their role in providing alternative specificity or differential expression.
Individual Contributions of Chromosomal Proximity and Co-Occurrence.
Several studies used these characteristics and their combinations to predict protein function (see refs. 17–19 for reviews). A recent study has also demonstrated that coregulation rather than horizontal gene transfer drives chromosomal proximity (37).
It is important to understand the individual contributions of gene fusion, proximity, and co-occurrence to functional metabolic modules and linear pathways. In fact, when considered separately, these characteristics exhibit similar patterns on the metabolic network (see Supporting Appendix) For example, there are similar relationships between metabolic distance and proximity and metabolic distance and co-occurrence. In addition, metabolic modules found by using only proximity or only co-occurrence are very similar to those obtained by using a combined score as described above (see http://insilico.mit.edu/METABOLIC, Full Set of Clusters). These results suggest that proximity on the chromosome and co-occurrence are reflections of some general functional association (e.g., participation in the same metabolic module), thus allowing us to look at organization of the metabolic network from the cell’s “point of view.”
Contribution of Operons and Divergent Gene Pairs.
Do functional associations contain more information about modularity of the metabolic network than simply E. coli operons? To what extent can operon structure explain observed modularity and long-range associations in linear pathways?
To investigate the effect of operon organization, we excluded all functional edges between genes that belong to the same operon (38) and repeated our analysis on the modified network.
Macroscale results remain the same within a statistical error (Supporting Appendix). This result comes as no surprise, because the original graph contained ≈2,000 functional links, whereas 356 operons of two or more genes provide only as many as ≈200 links between metabolic enzymes.
Mesoscale analysis shows a more complicated picture of relationships between modules and operons. We found 23 modules (of 182 nonidentical modules) that are built primarily of genes coming from a single operon: histidine, murein, and thiamin biosynthesis and smaller modules. Most (87%) of identified modules, however, contain genes from several operons. In summary, although operon organization is known to be correlated with metabolic pathways and proximity on the metabolic network (3, 6, 15, 18), genomic associations between genes go far beyond operons in revealing functional modules.
Recently, Korbel et al. (39) argued that adjacent bidirectionally transcribed genes with conserved gene orientation are strongly coregulated. They reported 391 divergent gene pairs. Because of a much smaller number of these pairs compared with the total number of functional edges on the metabolic–genomic graph, we expect the effect of these divergent genes to be limited as well.
Biological Implications.
This analysis has a number of implications. First, we expect that the genes forming a module would be strongly coregulated (even when they are not part of the same operon). The analysis of expression data for bacteria, by using as a control a random group of metabolically proximal enzymes, can test such a hypothesis. It would be interesting to see whether such coexpression of genes within a module exceeds coexpression of metabolic pathways.
Furthermore, because identified modules are detected using evolutionary information obtained across several bacterial genomes, we would expect to have modules coregulated (and hence coexpressed) in different close species. In other words, we expect modules to show conservation of coexpression. This conservation, again, can be tested by using bacterial expression data.
As we pointed out above, modules do not necessarily correspond to operons nor are they known to be regulated by the same transcription factor. However, the hypothesis of coexpression suggests searching for a common regulatory site, motif, or combination of sites in promoters of a single module.
Methods
Construction of an Integrated Metabolic–Genomic Network.
We first map the network on a graph with vertices representing reactions and two types of edges. Edges of the first type connect reactions sharing a metabolite. Edges of the second type connect reactions that are catalyzed by genomically associated enzymes (2, 26). Such edges carry a weight that equals the association score (0 ≤ S ≤ 1,000; see below). The weight of an edge between reactions is taken as the maximal of the scores between their enzymes (or subunits). Because genomic association indicates coregulation and/or evolutionary coinheritance, such representation allows one to identify metabolic modules and reveals principles of regulation and evolution of the metabolic networks.
Two reactions are connected by an edge of the first type if they have at least one common metabolite as a substrate or product. Common (nonspecific) metabolites, such as water, CO2, phosphate, etc. have been excluded (see Supporting Appendix for a complete list). The same reactions catalyzed by isoenzymes are considered as different reactions. Two reactions are connected by an edge of the second type with weight S if at least one pair of enzymes catalyzing these reactions (or their subunits) are associated with score S. The weight of an edge between reactions is taken as the maximal of the scores between their enzymes (or subunits).
Macroscale.
For every pair of reactions, we computed the shortest distance along the metabolic edges (metabolic distance D). We grouped association links into strong (S > 700), moderate (400 < S < 700), and weak (100 < S < 400). For each category k = 1, 2, 3, we calculated the number of association links Mk(D) between reactions at metabolic distance D in the metabolic network and average Mkrnd(D) in control networks.
We define a degree of each metabolite as the number of reactions in which this metabolite participates. Two reactions are said to be connected by a linear path if all metabolites along the shortest metabolic path between them have a degree of four or less. We compute MLINEAR(D) as the number of strong and moderate associations (S > 400) between enzymes connected by a linear path and average the same quantity in random controls MLINEARrnd(D).
Search Algorithms.
We searched for clusters that contain large numbers of metabolic and association links. We developed a Monte Carlo algorithm that searches for a set of nodes to maximize the number of edges of both types between them. The score to be maximized is s = mm+ a·ma, where mm is the number of metabolic edges, ma is the number of association edges (edges with S > Scutoff), and a is a relative weight of association edges. The algorithm is similar to the one we developed to search for protein complexes in the network of protein–protein interactions (25). By varying the relative contribution of the metabolic vs. genomic edges, we can steer our search toward modules that are richer in a particular type of edge (see Supporting Appendix).
We also exactly enumerated clusters within which every pair of nodes is connected with a path through both metabolic and association edges. There are two types of such clusters. In the first type, every edge is a product of metabolic and association links, and the metabolic and association paths between each pair of nodes are exactly the same. In the second type, although every pair of nodes is connected through both metabolic and association paths, these paths may be different. The cluster enumeration is a search for connected components on the networks with appropriately constructed edges (see Supporting Appendix for details).
Statistical Significance.
The statistical significance of each Monte Carlo cluster is evaluated by using extreme value statistics with parameters obtained by running the same search algorithm on the random control networks (see Supporting Appendix). Control networks have been obtained by randomly assigning gene names to the enzymes on the metabolic network. Such random controls preserve the structure of both metabolic and association networks, randomly assigning one to the other. Monte Carlo clusters with an E value <0.1 (see Supporting Appendix for details) have been retained for further analysis.
For clusters found by exact enumeration, we estimated the statistical significance by using random control networks (see above) 10,000 times and enumerating all clusters in each reshuffled graph. A cluster from the original network was statistically significant if we found, at most, 100 clusters with a higher density of metabolic and association links in the 10,000 control networks, corresponding to an E value of 0.01 (see Supporting Appendix for details).
Web Access.
Additional information is available from http://insilico.mit.edu/METABOLIC.
Supplementary Material
Acknowledgments
This work was supported, in part, by Howard Hughes Medical Institute Grant 55000309 (to M.S.G.) and Russian Academy of Sciences Programs “Molecular and Cellular Biology” and “Origins and Evolution of Biosphere.” L.A.M. is an Alfred P. Sloan Research Fellow.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Ogata H., Fujibuchi W., Goto S., Kanehisa M. Nucleic Acids Res. 2000;28:4021–4028. doi: 10.1093/nar/28.20.4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.von Mering C., Zdobnov E. M., Tsoka S., Ciccarelli F. D., Pereira-Leal J. B., Ouzounis C. A., Bork P. Proc. Natl. Acad. Sci. USA. 2003;100:15428–15433. doi: 10.1073/pnas.2136809100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glazko G. V., Mushegian A. R. Genome Biol. 2004;5:R32. doi: 10.1186/gb-2004-5-5-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Snel B., Bork P., Huynen M. A. Proc. Natl. Acad. Sci. USA. 2002;99:5890–5895. doi: 10.1073/pnas.092632599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pal C., Hurst L. D. Trends Genet. 2004;20:232–234. doi: 10.1016/j.tig.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 6.Rison S. C., Teichmann S. A., Thornton J. M. J. Mol. Biol. 2002;318:911–932. doi: 10.1016/S0022-2836(02)00140-7. [DOI] [PubMed] [Google Scholar]
- 7.Light S., Kraulis P. BMC Bioinformatics. 2004;5:15. doi: 10.1186/1471-2105-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kharchenko P., Vitkup D., Church G. M. Bioinformatics. 2004;20:I178–I185. doi: 10.1093/bioinformatics/bth930. [DOI] [PubMed] [Google Scholar]
- 9.Ihmels J., Levy R., Barkai N. Nat. Biotechnol. 2004;22:86–92. doi: 10.1038/nbt918. [DOI] [PubMed] [Google Scholar]
- 10.Snel B., van Noort V., Huynen M. A. Nucleic Acids Res. 2004;32:4725–4731. doi: 10.1093/nar/gkh815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen L., Vitkup D. Genome Biol. 2006;7:R17. doi: 10.1186/gb-2006-7-2-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green M. L., Karp P. D. BMC Bioinformatics. 2004;5:76. doi: 10.1186/1471-2105-5-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H., Pellegrini M., Eisenberg D. Nat. Biotechnol. 2005;23:253–260. doi: 10.1038/nbt1065. [DOI] [PubMed] [Google Scholar]
- 14.Zheng Y., Anton B. P., Roberts R. J., Kasif S. BMC Bioinformatics. 2005;6:243. doi: 10.1186/1471-2105-6-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng Y., Szustakowski J. D., Fortnow L., Roberts R. J., Kasif S. Genome Res. 2002;12:1221–1230. doi: 10.1101/gr.200602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Snel B., Huynen M. A. Genome Res. 2004;14:391–397. doi: 10.1101/gr.1969504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gelfand M. S. Curr. Opin. Struct. Biol. 2006;16:1, 10. doi: 10.1016/j.sbi.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 18.Rogozin I. B., Makarova K. S., Wolf Y. I., Koonin E. V. Brief. Bioinform. 2004;5:131–149. doi: 10.1093/bib/5.2.131. [DOI] [PubMed] [Google Scholar]
- 19.Bowers P. M., O’Connor B. D., Cokus S. J., Sprinzak E., Yeates T. O., Eisenberg D. FEBS Lett. 2005;272:5110–5118. doi: 10.1111/j.1742-4658.2005.04946.x. [DOI] [PubMed] [Google Scholar]
- 20.Ouzounis C. A., Karp P. D. Genome Res. 2000;10:568–576. doi: 10.1101/gr.10.4.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagner A., Fell D. A. Proc. Biol. Sci.; 2001. pp. 1803–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Almaas E., Kovacs B., Vicsek T., Oltvai Z. N., Barabasi A. L. Nature. 2004;427:839–843. doi: 10.1038/nature02289. [DOI] [PubMed] [Google Scholar]
- 23.Edwards J. S., Covert M., Palsson B. Environ. Microbiol. 2002;4:133–140. doi: 10.1046/j.1462-2920.2002.00282.x. [DOI] [PubMed] [Google Scholar]
- 24.Segre D., Vitkup D., Church G. M. Proc. Natl. Acad. Sci. USA. 2002;99:15112–15117. doi: 10.1073/pnas.232349399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spirin V., Mirny L. A. Proc. Natl. Acad. Sci. USA. 2003;100:12123–12128. doi: 10.1073/pnas.2032324100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.von Mering C., Huynen M., Jaeggi D., Schmidt S., Bork P., Snel B. Nucleic Acids Res. 2003;31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dorogovtsev S. N., Mendes J. F. F. 2004. arXiv: cond-mat/0404593.
- 28.Keseler I. M., Collado-Vides J., Gama-Castro S., Ingraham J., Paley S., Paulsen I. T., Peralta-Gil M., Karp P. D. Nucleic Acids Res. 2005;33:D334–D337. doi: 10.1093/nar/gki108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huynen M. A., Dandekar T., Bork P. Trends Microbiol. 1999;7:281–291. doi: 10.1016/s0966-842x(99)01539-5. [DOI] [PubMed] [Google Scholar]
- 30.Robison K., McGuire A. M., Church G. M. J. Mol. Biol. 1998;284:241–254. doi: 10.1006/jmbi.1998.2160. [DOI] [PubMed] [Google Scholar]
- 31.Rodionov D. A., Vitreschak A. G., Mironov A. A., Gelfand M. S. J. Biol. Chem. 2002;277:48949–48959. doi: 10.1074/jbc.M208965200. [DOI] [PubMed] [Google Scholar]
- 32.Rodionov D. A., Mironov A. A., Gelfand M. S. Genome Res. 2002;12:1507–1516. doi: 10.1101/gr.314502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodionov D. A., Vitreschak A. G., Mironov A. A., Gelfand M. S. Nucleic Acids Res. 2004;32:3340–3353. doi: 10.1093/nar/gkh659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vitreschak A. G., Rodionov D. A., Mironov A. A., Gelfand M. S. Nucleic Acids Res. 2002;30:3141–3151. doi: 10.1093/nar/gkf433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He B., Zalkin H. J. Bacteriol. 1992;174:7121–7127. doi: 10.1128/jb.174.22.7121-7127.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Papp B., Pal C., Hurst L. D. Nature. 2003;424:194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- 37.Price M. N., Huang K. H., Arkin A. P., Alm E. J. Genome Res. 2005;15:809–819. doi: 10.1101/gr.3368805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salgado H., Gama-Castro S., Peralta-Gil M., Diaz-Peredo E., Sanchez-Solano F., Santos-Zavaleta A., Martinez-Flores I., Jimenez-Jacinto V., Bonavides-Martinez C., Segura-Salazar J., et al. Nucleic Acids Res. 2006;34:D394–D397. doi: 10.1093/nar/gkj156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Korbel J. O., Jensen L. J., von Mering C., Bork P. Nat. Biotechnol. 2004;22:911–917. doi: 10.1038/nbt988. [DOI] [PubMed] [Google Scholar]
- 40.Dandekar T., Schuster S., Snel B., Huynen M., Bork P. Biochem. J. 1999;343:115–124. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.