Abstract
In the present study, we show that E2Fs (E2 promoter-binding factors) regulate the expression of ASK-1 (apoptosis signal-regulating kinase 1), which encodes a mitogen-activated protein kinase kinase kinase, also known as MAP3K5. Its mRNA expression is cell-cycle-regulated in human T98G cells released from serum starvation. Moreover, overexpression and RNA interference experiments support the requirement of endogenous E2F/DP (E2F dimerization partner) activity for ASK-1 expression. Characterization of the human ASK-1 promoter demonstrates that the −95/+11 region is critical for E2F-mediated up-regulation. Chromatin immunoprecipitation assays show that E2F1–E2F4 are bound in vivo to the ASK-1 promoter in cycling cells, probably through a non-consensus E2F-binding site located 12 bp upstream of the transcription start site. Mutation of this site completely abolishes the ASK-1 promoter response to E2Fs as well as the E2F1 binding in electrophoretic mobility-shift experiments. Our results indicate that E2Fs modulate the expression of ASK-1 and suggest that some of the cellular functions of ASK-1 may be under the control of E2F transcription factors. Moreover, the up-regulation of ASK-1 may also favour the p53-independent E2F1 apoptotic activity.
Keywords: apoptosis, apoptosis signal-regulating kinase 1 (ASK-1), cell cycle, E2 promoter-binding factor (E2F), mitogen-activated protein kinase (MAPK), promoter analysis
Abbreviations: Ad, adenovirus; ARPP-P0, acidic ribosomal phosphoprotein P0; ASK-1, apoptosis signal-regulating kinase 1; BS, binding site; CCNA2, cyclin A2 gene; ChIP, chromatin immunoprecipitation; CREB, cAMP-response-element-binding protein; E2F, E2 promoter-binding factor; DP, E2F dimerization partner; EMSA, electrophoretic mobility-shift assay; ER, endoplasmic reticulum; GFP, green fluorescent protein; HA, haemagglutinin; ICAM1, intracellular adhesion molecule 1; MAPK, mitogen-activated protein kinase; MAPKKK, MAPK kinase kinase; MBP, myelin basic protein; MKK, MAPK kinase; MOI, multiplicity of infection; poly(A)+, polyadenylated; Rb, retinoblastoma protein; RDA, representational difference analysis; RNAi, RNA interference; RT, reverse transcription; shRNA, short hairpin RNA
INTRODUCTION
E2F1 (E2 promoter-binding factor 1) was initially identified as a cellular DNA-binding factor required for the E1A (adenovirus 5 early region 1A product)-mediated activation of the adenoviral E2 gene [1]. It belongs to the E2F family, which to date comprises ten genes that encode the E2F1–E2F8 and DP1/2 (E2F dimerization partner 1/2 proteins (for review see [2,3]). Except for E2F7 and E2F8 proteins, the heterodimerization of E2F and DP subunits is essential to generate functional E2F complexes [4,5]. All E2F/DP family members recognize variants of the nucleotide sequence 5′-TTTSSCGC-3′ found in the E2 enhancer element [6]. E2F1 is known primarily for its role in the co-ordination of cell-cycle progression. Particularly, E2F1 regulates transition of the cell cycle from G1- to S-phase by transactivating genes required for DNA synthesis and cell-cycle control. Thus E2F1 overexpression is sufficient to drive quiescent cells into the S-phase of the cell cycle [7]. Moreover, by co-operation with the Ras oncogene, E2F1 can transform rat embryo fibroblasts and generate tumours in nude mice, providing evidence that E2F1 has oncogenic capacity [7,8]. However, E2F1 can also act as a tumour suppressor as shown by E2F1-knockout mice which developed numerous tumours [9,10]. Consistent with these observations, E2F1 overexpression has been shown to induce apoptosis in a p53-dependent or -independent manner [11]. However, the mechanisms of E2F1-mediated apoptosis have not been completely elucidated [12].
In order to identify new E2F1 target genes, we performed an RDA (representational difference analysis) of cDNA, comparing p53-deficient Saos-2 cells overexpressing either E2F1 or GFP (green fluorescent protein). One of the E2F1-induced genes identified in this screen is ASK-1 (apoptosis signal-regulating kinase 1), also designated MAP3K5. This kinase is an ubiquitously expressed MAPKKK [MAPK (mitogen-activated protein kinase) kinase kinase], initially identified as an apoptosis-inducing kinase [13,14]. It activates both the MKK4 (MAPK kinase 4)/MKK7–JNK (c-Jun N-terminal kinase) and MKK3/MKK6–p38 MAPK pathways, and is implicated in various cellular processes such as proliferation, differentiation and apoptosis in response to pro-inflammatory and stress signals [15,16]. Moreover, ASK-1-knockout mice revealed that ASK-1 plays essential roles in oxidative-stress- and ER (endoplasmic reticulum)-stress-induced apoptosis [17].
In the present study, we show that overexpression of E2F1 induces the expression of ASK-1 mRNA and protein, thus leading to increased kinase activity levels in the cells. Conversely, reduction of endogenous E2F/DP activity via DP1 RNAi (RNA interference) considerably decreases ASK-1 expression. To understand the mechanism by which ASK-1 expression is regulated by E2F, we characterized the ASK-1 promoter. Transfection and EMSAs (electrophoretic mobility-shift assays) demonstrated the crucial role of a non-consensus E2F-binding site located 12 bp upstream of the putative start site. E2F2, E2F3 and, to a lesser extent, E2F4 also enhanced ASK-1 promoter activity. Mutation within this site induced a critical decrease in the E2F-responsiveness of the ASK-1 promoter. Moreover, we demonstrated by ChIP (chromatin immunoprecipitation) assays that E2F1–E2F4 were associated in vivo with the regulatory region of the ASK-1 gene in cycling cells. Our results demonstrate that E2Fs modulate the expression of the apoptotic molecule ASK-1 and suggest that some of the cellular functions of ASK-1 may be under the control of E2F transcription factors. Moreover, up-regulation of ASK-1 may also favour E2F1-induced apoptosis.
EXPERIMENTAL
Cell culture and RNA analysis
Human U2OS (A.T.C.C. number HTB-96), Saos-2 (A.T.C.C. number HTB-85), T98G (A.T.C.C. number CRL-1690), MDA-MB-468 (A.T.C.C. number HTB-132), HeLa (0) (A.T.C.C. number CCL-2) and WI-38 (A.T.C.C. number CCL-75) cell lines were routinely maintained in Dulbecco's medium with 10% foetal calf serum at 37 °C in a water-saturated 5% CO2 atmosphere. The MCF10A (A.T.C.C. number CRL-10317) cell line was maintained in a 1:1 (v/v) mixture of Ham's F12 medium and Dulbecco's medium supplemented with 2 mM L-glutamine, 20 ng/ml EGF (epidermal growth factor), 100 ng/ml cholera toxin, 0.01 mg/ml insulin, 500 ng/ml hydrocortisone and 5% horse serum. Synchronized T98G cells were obtained after serum starvation for 72 h. Total RNA and poly(A)+ (polyadenylated) RNA were purified as described previously [18] using RNA PLUS reagent (Qbiogene) and oligo(dT)–cellulose columns (APBiotech). Cell-cycle analyses were performed by flow cytometry after DNA staining with propidium iodide (50 μg/ml). Poly(A)+ RNA from synchronized T98G cells were subjected to Northern blot analysis using 32P-labelled probes corresponding to the human full-length E2F1 cDNA, the human full-length CCNA2 (cyclin A2) cDNA, a 900 bp EcoRI fragment of the pcDNA3-ASK-1 construct [19] corresponding to the human coding region +360 to +1258. As a loading control, the blots were also hybridized with a human ARPP-P0 (acidic ribosomal phosphoprotein P0) probe amplified by PCR using the ARPP-P0-5′ and ARPP-P0-3′ primers (shown in Table 1).
Table 1. Primers used in semi-quantitative PCRs.
| Name | Sequence |
|---|---|
| ARPP-P0-5′ | 5′-GGCATCACCACGAAAATCTCCA-3′ |
| ARPP-P0-3′ | 5′-GGTTGCTTTGGCCGGGATTAGTC-3′ |
| ASK1-5′ | 5′-TGAATCTGAGCCAACACTACAG-3′ |
| ASK1-3′ | 5′-CATCAGGAAGCACGTGCCAAA-3′ |
| DP1-5′ | 5′-ATGGCAAAAGATGCCGGTC-3′ |
| DP1-3′ | 5′-CAGACCAAGGTGAGGAGT-3′ |
| DP2-5′ | 5′-GCTGATTCGCAGGCTTAT-3′ |
| DP2-3′ | 5′-AAGTAGTCCCTGATTTAACC-3′ |
| CyclinA2-5′ | 5′-TTGGGCAACTCTGCGCCG-3′ |
| CyclinA2-3′ | 5′-CAGATTTAGTGTCTCTGGTGG-3′ |
RDA is a PCR-based differential cloning method that allows selective amplification of differentially expressed genes upon hybridization of two populations of cDNA. RDA was performed according to the procedure described precisely in [20] on cDNA obtained after retrotranscription of poly(A)+ RNA isolated from human Saos-2 cells infected for 18 h with recombinant adenoviruses expressing GFP (Ad-GFP) or FLAG-tagged E2F1 transcription factor (Ad-E2F1) at an MOI (multiplicity of infection) of 100, as described previously [21]. Briefly, double-stranded cDNAs were digested with DpnII, adaptor-ligated and amplified by PCR using adaptor primers to generate ‘representative amplicons’. Among the two populations of cDNA, cDNA from Ad-E2F1-infected Saos-2 cells was used as a ‘tester’, containing potential target genes, and cDNA from Ad-GFP-infected Saos-2 cells was used as a ‘driver’, driving the process of subtraction. After three rounds of subtraction, the amplified cDNAs were subcloned and DNA sequencing was performed.
Retroviral infections and semi-quantitative RT (reverse transcription)–PCR analyses
Retrovirus productions and infections were performed as described previously [22]. HeLa (0) and MCF10A cells were infected and selected with 1 μg/ml puromycin for 96 h. Total RNA (2 μg) was treated for 30 min with RQ1 RNase-free DNase (Promega) and was reverse-transcribed for 50 min at 37 °C with MMLV (mouse Moloney leukaemia virus) (Invitrogen). PCRs were performed using Taq Gold Star (Eurogentec) with various amounts of the RT reaction mixture to determine the linear range of amplification. ARPP-P0 was used as a control. The sequences of primers used in the PCRs are listed in Table 1.
Western blot analyses
Cells were lysed in RIPA buffer (50 mM Tris/HCl, pH 7.4, 150 mM NaCl, 1% Nonidet P40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM EDTA, 50 mM NaF, 4 mM Na3VO4, 1 mM PMSF and 1 μM each of aprotinin and leupeptin) 18 h after adenoviral infections. Equal amounts of protein (25 μg) from each of the lysates were resolved by standard SDS/PAGE on 8% and 12% polyacrylamide gels and then transferred on to nitrocellulose. The membranes were incubated overnight with antibodies directed against ASK-1 (sc-7931), p21 (sc-397), E2F1 (sc-193), p53 (sc-126) or actin (sc-1616) (all from Santa Cruz Biotechnology) or directed against the HA (haemagglutinin) tag (BAbCO).
ASK-1 kinase assays
ASK-1 kinase assays were performed using the MBP (myelin basic protein) as a substrate according to the previously described protocol [23]. Briefly, to eliminate non-specific kinase activity immunoprecipitation, total protein lysates (400 μg) were first pre-cleared with an irrelevant antibody directed against ICAM1 (intracellular adhesion molecule 1) (sc-7891; Santa Cruz Biotechnology). ASK-1 was then immunoprecipitated with 2 μg of anti-ASK-1 antibody in RIPA buffer. The immunoprecipitates were then mixed with 10 μg of MBP-1 (Sigma) in kinase buffer (20 mM Hepes, pH 7.6, 25 mM 2-glycerophosphate, 100 μM Na3VO4, 2 mM dithiothreitol and 20 μM ATP) containing 10 μCi of [γ-32P]ATP. The kinase assay was performed at 25 °C for 30 min. The reactions were terminated by the addition of Laemmli sample buffer, and samples were separated on 10% polyacrylamide gels.
Isolation of the 5′-flanking region of the human ASK-1 gene and deletion promoter constructs
A human female genomic λDash Sau3A partial DNA library was screened with the 900 bp EcoRI fragment of the pcDNA3-ASK-1 construct radiolabelled with [α-32P]CTP using the Megaprime DNA labelling system (APBiotech). Nine positive clones were purified and two clones containing ASK-1 sequences upstream from the ATG were subcloned into pZero vector (Invitrogen). A 3257 bp EcoRI/NaeI fragment and a 1685 bp SmaI/BsiHKaI fragment from the ASK-1 genomic clones were first cloned into the pGL3-basic vector (Promega). These clones were entirely sequenced using a BigDye Terminator Cycle Sequencing Ready Reaction kit (Amersham Biosciences). The presence of putative transcription-factor-binding sites was obtained using MatInspector software (http://www.genomatix.de) and Transcription Element Search Software (http://www.cbil.upenn.edu/tess). Deletion constructs were produced by a series of digestions. Mutated constructs were derived from the corresponding deletion one by introducing mutated nucleotides with the QuikChange® site-directed mutagenesis kit (Stratagene) and the oligonucleotides D-M1 and D-M2 (see Table 2).
Table 2. Oligonucleotides used in binding assays.
Mutation sites are underlined.
| Name | Sequence |
|---|---|
| E2F-WT (wild-type) | 5′-ATTTAAGTTTCGCGCCCTTTCTCAA-3′ |
| E2F (mutated) | 5′-ATTTAAGTTTCGATCCCTTTCTCAA-3′ |
| A(−95/−72) | 5′-CGCCCTTCCTGGCCCCTCCCAGAG-3′ |
| B(−74/−54) | 5′-GAGCGGCCGGGACGGAAGGAG-3′ |
| C(−56/−20) | 5′-GAGCTGCGCGGCGCGGCGCGGCGCGGCGCGGCGGGTG-3′ |
| D(−22/+11) | 5′-GTGGCGAGGGCGGGCTGCACCCCGAGCGCGGCG-3′ |
| D-M1 | 5′-GTGGCGAGGGTAGGCTGCACCCCGAGCGCGGCG-3′ |
| D-M2 | 5′-GTGGCGAGGGCGGGCTGCACCCCGAGCAAGGCG-3′ |
| D-M1+2 | 5′-GTGGCGAGGGTAGGCTGCACCCCGAGCAAGGCG-3′ |
Transfections and expression vector constructs
Transfections were performed by following the Ex Gen 500 (Euromedex) procedure in 12-well plates. All assays were normalized to the β-galactosidase activity of the pCMV-βGal vector in order to take into account transfection efficiency variations. All the experiments were performed at least twice using two different plasmid preparations. For co-transfection experiments, the pcDNA3-HA-E2F1, pcDNA3-HA-E2F2, pcDNA3-HA-E2F3, pcDNA3-HA-E2F4 and pcDNA3-HA-DP1 plasmids were constructed by cloning the coding sequence of the human E2F and DP1 genes generated by PCR into the pcDNA3-HA expression vector (Invitrogen), in-frame with the sequence encoding the C-terminal HA tag. E2F1(1–374)ΔTA is a C-terminal 63-amino-acid-truncated transactivation-defective E2F1 mutant generated by PCR. This mutant is competent for DNA binding but defective for Rb (retinoblastoma protein) binding and transcription. E2F1(Δ120–190)ΔDBD is an internal DNA-binding-domain deletion mutant of residues 120–190.
EMSAs
EMSAs were performed using nuclear extracts from U2OS Ad-E2F1-infected cells as described previously [24]. Briefly, extracts were incubated for 20 min in a final volume of 20 μl containing 20 mM Hepes, pH 7.9, 20% (v/v) glycerol, 0.1 mM EDTA, 1 mM dithiothreitol, 1 μg of sheared salmon sperm DNA, 50 mM NaCl and 50000 c.p.m. of probe. The reaction mixtures were then loaded on a 4.5% polyacrylamide gel in 0.5× TBE (Tris/borate/EDTA) and run for 4 h at 170 V (4 °C). For supershift experiments, 1 μl of anti-FLAG M2 monoclonal antibody (Sigma), 1 μl of anti-E2F1 antibody or 1 μl of anti-CREB2 (cAMP-response-element-binding protein; sc-200; Santa Cruz Biotechnology), as a non-relevant antibody, were added to binding reactions. When indicated, a 200-fold molar excess of unlabelled double-stranded oligonucleotides was added. Oligonucleotides employed in binding assays after hybridization to obtain the corresponding DNA duplex are shown in Table 2.
ChIP assays
In vivo detection of ASK-1 promoter-associated E2F was performed on HeLa (0) cells by ChIP as described previously [21]. The E2F-containing complexes were immunoprecipitated with polyclonal antibodies obtained from Santa Cruz Biotechnology, anti-E2F1 (sc-193), anti-E2F2 (sc-633), anti-E2F3 (sc-878) and anti-E2F4 (sc-1082). The anti-ICAM1 (sc-7891) antibody was used to control the immunoprecipitation specificity. The following oligonucleotides were designed to amplify by PCR a 297 bp fragment of the human ASK-1 gene: 5′ASK-1, 5′-GT-GCTGGACCGCTTTTACAATGC-3′, and 3′ASK-1, 5′-CAG-TTCAAGTCGATCGCATGGAC-3′. Specific primers were also designed to amplify a 296 bp fragment of the fifth intron of the ASK-1 gene as a negative control: 5′ASK-1Int5, 5′-GTTCCCTTCCAGAATGCGTTAAC-3′, and 3′ASK-1Int5, 5′-GATA-ACATCATATTCATGTAAC-3′. Finally, as positive controls, specific primers were designed to amplify a 124 bp fragment of the human E2F1 promoter and a 169 pb fragment of the human CCNA2 promoter: 5′E2F1p, 5′-CGCCGTTGTTCCCGTCA-CGGC-3′, and 3′E2F1p, 5′-CGGCCGCCGCTGCCTGCAAA-GTCC-3′; 5′CyA2, 5′-CTGCTCAGTTTCCTTTGGTTTACC-3′, and 3′CyA2, 5′-CAAAGACGCCCAGAGATGCAG-3′.
RESULTS
Isolation of ASK-1 in an RDA screen
An RDA strategy was used to identify new E2F1-regulated genes in p53-null Saos-2 cells. After three rounds of subtraction, potential E2F1 target genes were specifically isolated. Among them, approx. 5% of the identified cDNA corresponded to the well-characterized MAP3K5 gene, also known as ASK-1 [13]. Among the 12 other genes isolated by RDA, we found previously described E2F target genes (encoding cyclin E and enolase 2 [25]). Analysis of the other genes that were isolated is currently underway.
ASK-1 expression is cell-cycle-regulated
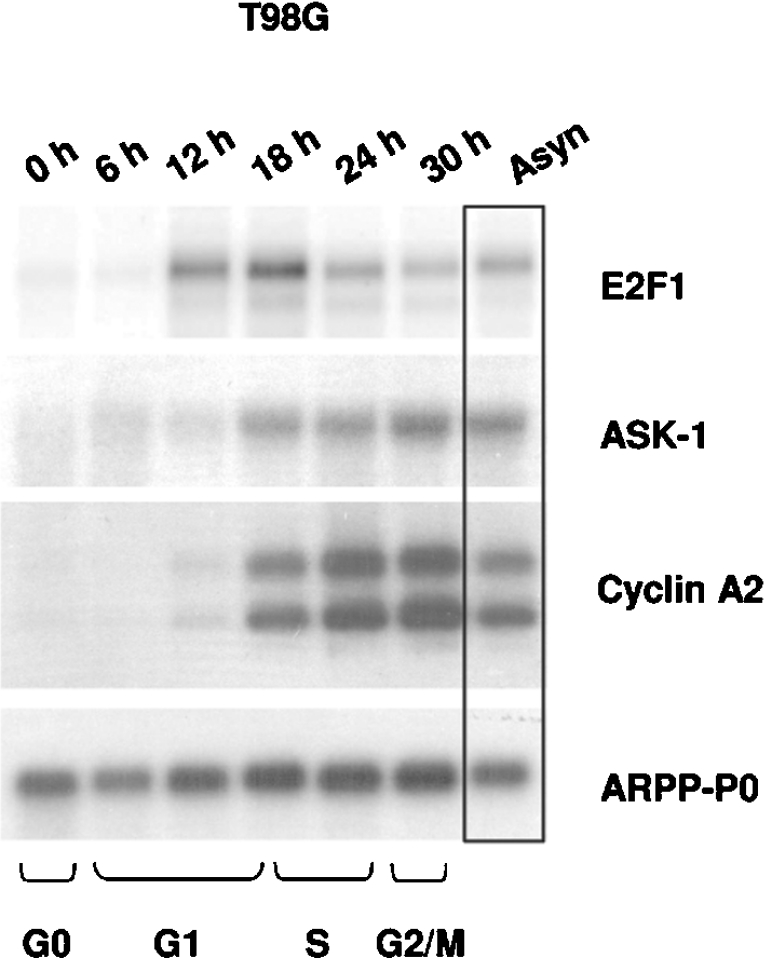
Since the activity of E2F factors plays a major role in inducing cell-cycle progression through the transcriptional control of their target genes, we analysed the endogenous ASK-1 expression during cell-cycle progression of T98G neuroblastoma cells as follows. The cells were made quiescent by serum starvation and were then stimulated with serum and harvested at different time points from 0 to 30 h. Cell-cycle distribution was monitored by flow cytometry after propidium iodide staining. As expected, we observed a drastic increase in endogenous E2F1 levels around the G1-to-S-phase transition (Figure 1). The expression level of ASK-1 began to increase when the E2F1 expression level was maximal, temporally correlated with cell entry into S-phase. It is important to note that the cell-cycle-expression profile of ASK-1 perfectly matches that of CCNA2, a well-known E2F target gene ([26], but see [26a]). Therefore our results indicate that ASK-1 expression is cell-cycle-regulated in T98G cells leading us to hypothesize that its expression is, in part, under the control of E2F transcription factors.
Figure 1. Cell-cycle expression of E2F1, ASK-1, CCNA2 and ARPP-P0 mRNAs in synchronized human T98G neuroblastoma cells.
T98G cells, serum-starved for 3 days, were harvested at the indicated times after 20% serum stimulation. Northern blot analysis was performed on poly(A)+ RNA and probed sequentially with 32P-labelled cDNAs encoding E2F1, ASK-1 and cyclin A2, and ARPP-P0 as a loading control. Cell positions in the cell cycle were determined by flow-cytometric analyses. Asyn, asynchronized cells.
E2F1 up-regulates ASK-1 mRNA and protein expression and increases ASK-1 enzymatic activity
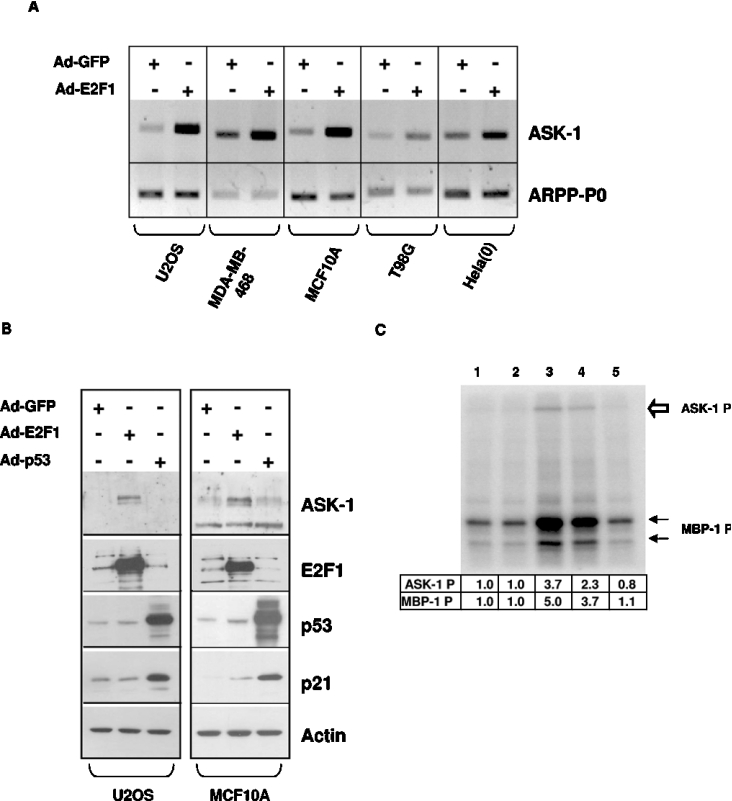
As shown in Figure 2(A), RT–PCR detection of ASK-1 mRNA indicates that ASK-1 expression in various cell lines is increased dramatically upon expression of E2F1 in comparison with cells expressing GFP. Moreover, the ASK-1 mRNA increase upon Ad-E2F1 infection leads to an increase in ASK-1 protein levels as observed by Western blot analyses of protein extracts from U2OS and MCF10A cells (Figure 2B). Comparable results were also obtained in WI38 and MDA-MB-468 cells (results not shown). Infection of U2OS or MCF10A cells by recombinant adenoviruses expressing p53 (Ad-p53) does not affect the ASK-1 protein level (Figure 2B) or the ASK-1 kinase activity present in the cells (Figure 2C, compare lane 5 with lane 1). In contrast, p53 dramatically increases the expression level of its well-known target gene p21, indicating that p53 is fully functional in our experiments (Figure 2B). The level of exogenous E2F1 and p53 proteins were also detected by Western blot to compare their levels with those of the endogenous proteins. Detection of actin was performed as a loading control.
Figure 2. Endogenous ASK-1 mRNA, protein levels and kinase activity are increased upon E2F1 overexpression.
(A) ASK-1 mRNA expression was assessed by semi-quantitative RT-PCR using ASK-1-specific primers in human cells infected with Ad-GFP or Ad-E2F1. The amount of endogenous ARPP-P0 transcripts was determined as a control. (B) Western blot analysis of ASK-1, E2F1, p53, p21 and actin proteins in human cells expressing wild-type p53 (U2OS and MCF10A) infected with Ad-GFP, Ad-E2F1 or Ad-p53 (MOI 100). (C) For ASK-1 kinase activity determination, the endogenous ASK-1 protein was immunoprecipitated from U2OS cells not infected (lane 1), or infected with Ad-GFP MOI 100 (lane 2), Ad-E2F1 MOI 500 (lane 3) or MOI 100 (lane 4) or Ad-p53 MOI 100 (lane 5) after pre-clearing with an irrelevant isotype control antibody. MBP-1 phosphorylation is indicated by black arrows, while ASK-1 autophosphorylation is indicated by a white arrow. Phosphorylation was quantified using a Molecular Dynamics PhosphorImager.
To gauge whether the effect of E2F1 on ASK-1 expression results in functional ASK-1 synthesis, ASK-1 kinase activity was measured in vitro using MBP as a substrate [23]. After adenoviral infections, endogenous ASK-1 was immunoprecipitated from U2OS cell lysates and its kinase activity was determined. As shown in Figure 2(C), a 3–5-fold increase of ASK-1 kinase activity was observed upon infection by Ad-E2F1 at an MOI of 500 and 100 (lanes 3 and 4 respectively). This increased activity also led to the previously described ASK-1 autophosphorylation process [27]. In contrast, the ASK-1 kinase activity observed upon infections by Ad-GFP (Figure 2C, lane 2) or Ad-p53 (Figure 2C, lane 5) at an MOI of 100 is the same as that observed in non-infected cells (Figure 2C, lane 1). Therefore the ASK-1 protein expressed upon E2F1 activation is apparently functional, since more ASK-1 expression leads to more apparent kinase activity in the cells.
Reduction of E2F/DP activity causes decreased ASK-1 expression
We next examined whether a reduction of endogenous E2F/DP activity through RNAi-mediated depletion of DP1 affects ASK-1 expression. HeLa (0) and MCF10A cells were infected with retrovirus encoding an shRNA (short hairpin RNA) directed against DP1 (DP1-shRNA) or a scramble control sequence (control shRNA), RNA was prepared 4 days after infection and was subjected to RT–PCR. As reported previously [28], DP1-specific shRNA significantly diminished the endogenous DP1 mRNA level (Figure 3). There was no significant changes in the levels of DP2, thus demonstrating the specificity of the DP1-shRNA. In contrast, similarly to the well known E2F-target gene CCNA2, expression of ASK-1 transcripts was reduced in DP1-shRNA-infected samples, whereas no changes were observed in ARPP-P0 expression levels. These results indicate that E2F/DP1 activity is partially responsible of the endogenous ASK-1 gene expression.
Figure 3. Reduction of E2F/DP activity causes decreased ASK-1 expression.

HeLa (0) and MCF10A cells were infected with retrovirus encoding DP1-shRNA or control shRNA. The amounts of endogenous DP1, DP2, ASK-1 and CCNA2 transcripts and ARPP-P0 transcripts as a control were determined by semi-quantitative RT–PCR analysis using RNA prepared 4 days after infection. The results shown are representative of three independent experiments.
Functional analysis of the ASK-1 promoter
As a first step to isolate the ASK-1 promoter sequence, we screened a human genomic DNA library by colony hybridization using a radiolabelled fragment of the ASK-1 cDNA (+360/+1258). Among the nine positive clones isolated and size-characterized, the two 5′-longest clones were digested by EcoRI, and the resulting 4.2 kb fragments were subcloned into the pZero vector and sequenced. Computer analysis revealed that this putative promoter sequence has neither TATA nor CAAT boxes. No perfect consensus E2F-binding site was identified in the ASK-1 5′-flanking region. Despite numerous attempts using both RNase protection assays and primer extension experiments, we were unable to identify precisely the transcription start site, probably due to the high GC content of the region surrounding the putative transcription start site. We thus consider the first nucleotide of the longest mRNA sequence found in the GenBank® database (accession number NM_005923) as the potential transcription start site located 362 bp upstream of the ATG. A 3257 bp EcoRI/NaeI fragment and a 1685 bp SmaI/BsiHKaI fragment of the 5′ flanking sequence of the ASK-1 gene [designated A(−3200/+57) and B(−1319/+366) respectively] were cloned into the promoterless reporter plasmid pGL3-basic and tested for their ability to act as functional promoters in transient transfection assays.
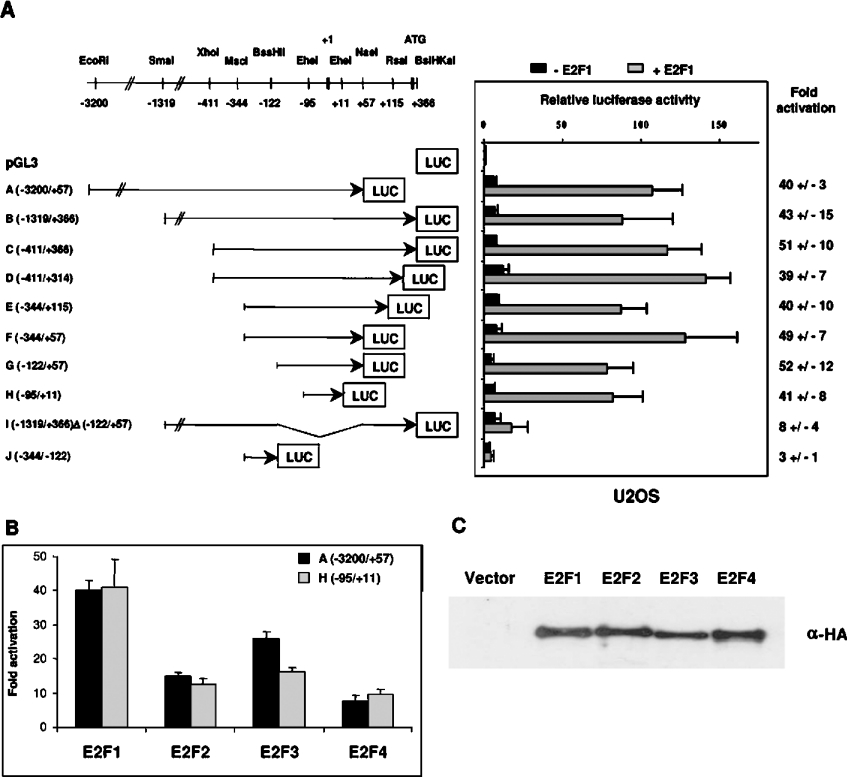
As shown in Figure 4(A), upon transfection into U2OS cells, these constructs exhibited a low level of activity with approx. 6-fold increase in luciferase expression above the promoterless vector. These constructs were also tested in other cell lines [T98G and HeLa (0)] and exhibited comparable basal transcriptional activity (results not shown). To determine whether these sequences can provide E2F-responsiveness to the ASK-1 promoter, the reporter constructs were transfected into U2OS cells together with vectors expressing E2F1 and DP1 proteins. As expected, E2F1 caused approx. 40-fold increase in ASK-1 luciferase activity. Deletion constructs derived from A and B were generated to define clearly the minimal ASK-1 promoter region necessary for E2F1 activation. Other 5′ and 3′ deletion plasmids C, D, E, F and G, as outlined in Figure 4(A), presented almost the same basal transactivation level as the A and B promoter constructs and their transcriptional activity was actively stimulated upon co-transfection with pcDNA3-HA-E2F1 and pcDNA3-HA-DP1 in the U2OS cell line. Progressive deletion of sequences 5′ to position −95 (construct H) resulted in transcriptional activity similar to that of the −3200 bp fragment (A), implying that the ASK-1 promoter region spanning nucleotides −3200 to −95 is dispensable for induction by E2F1. In addition, deletion of the 3′ region up to position +11 had no significant effect on the E2F1-dependent activation, thus suggesting that the minimal sequence necessary for E2F1-dependent ASK-1 gene transactivation in U2OS cells approximately maps to −95/+11. Accordingly, the E2F-responsiveness of the promoter was dramatically reduced (by approx. 80%) in construct I, corresponding to the B construct deleted of fragment (−122/+57). Likewise, no activation was detected upon co-transfection of the J(−344/−122) construct and the E2F1 expression vector.
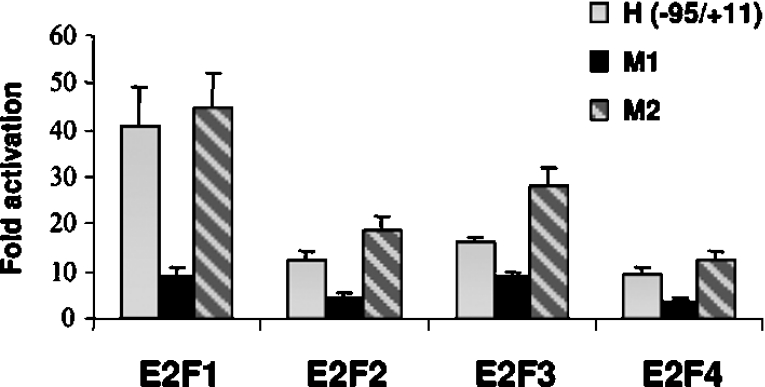
Figure 4. Activation of the ASK-1 promoter by different E2Fs.
(A) Constructs harbouring progressive 5′ and 3′ deletions of the ASK-1 promoter were cloned into the pGL3-basic vector. Transfections were carried out in U2OS cells with empty pcDNA3-HA vector or pcDNA3-HA-E2F1 and pcDNA3-HA-DP1 co-transfection. After β-galactosidase normalization, the basal activity of each construct was compared with that of the pGL3-basic set to 1. The responsiveness of each construct to E2F1 is indicated as a fold increase (right-hand panel). Results are the means±S.D. for three independent experiments. (B) Comparison of the responsiveness of the A(−3200/+57) and H(−95/+11) ASK-1 promoter constructs to E2F1, E2F2, E2F3 and E2F4. Results are expressed as a fold increase with respect to the pcDNA3-HA control plasmid alone. (C) E2F expression levels determined by Western blot analyses of cell lysates with an anti-HA (α-HA) antibody.
We next found that E2F1–E2F4 were able to activate the promoter (Figure 4B). Although they were expressed at similar protein levels (Figure 4C), E2F1 and E2F3 were stronger activators of the ASK-1 promoter compared with E2F2 and E2F4 (Figure 4B).These results indicate that the −95 to +11 ASK-1 promoter sequence contains a strong E2F positive regulatory element.
E2F1 activation of human ASK-1 promoter requires the DNA-binding and transactivation domains of E2F1
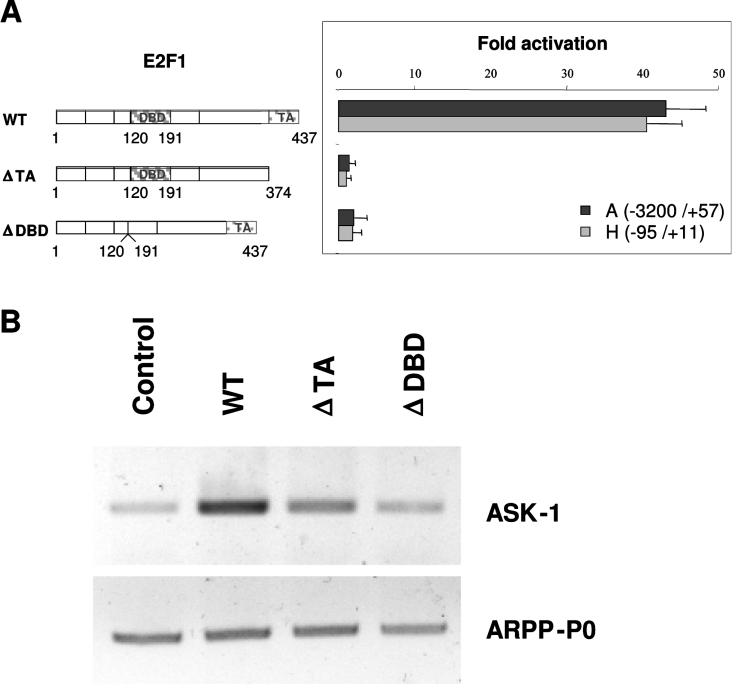
To determine the E2F1 functional domains that are required for the ASK-1 promoter activation, we performed transfections using various E2F1 mutants. As illustrated in Figure 5(A), when compared with cells transfected with the pcDNA3-HA plasmid (control), the relative luciferase activity from the ASK-1 promoter constructs A(−3200/+57) and H(−95/+11) were increased 43-fold and 40-fold respectively in U2OS cells transfected with the plasmid encoding the full-length E2F1. E2F1 constructs lacking the DNA-binding or transactivation domains, i.e. the E2F1(Δ120–190)ΔDBD and E2F1(1–374)ΔTA proteins, were unable to significantly stimulate transactivation over the basal value. We also determined the endogenous ASK-1 promoter activity by RT–PCR analysis of the ASK-1 mRNA level upon transfection of the same E2F1 constructs. As shown in Figure 5(B), the results obtained on the endogenous ASK-1 gene matched perfectly those obtained with the reporter constructs. These data therefore indicate that the DNA-binding domain and the transactivation domain of E2F1 are necessary for efficient E2F1 transactivation of the ASK-1 promoter, thus suggesting a direct up-regulation of ASK-1 by E2F1.
Figure 5. E2F1 DNA-binding and transactivation domains are necessary for ASK-1 transcriptional activation.
(A) Expression vectors for full-length (WT, wild-type) or two truncated forms of the E2F1 protein (ΔTA, transactivation-domain-deleted, and ΔDBD, DNA-binding-domain-deleted) were co-transfected with the A(−3200/+57) and H(−95/+11) ASK-1 reporter constructs in U2OS cells. As in Figure 3, the responsiveness of each construct to E2F1 vectors is indicated as a fold increase with respect to the pcDNA3-HA control plasmid alone (right-hand panel). (B) The amounts of endogenous ASK-1 transcripts, and ARPP-P0 transcripts as a control, were determined after transfection of U2OS cells with the same E2F1 constructs.
Binding of E2F1 to the human ASK-1 promoter
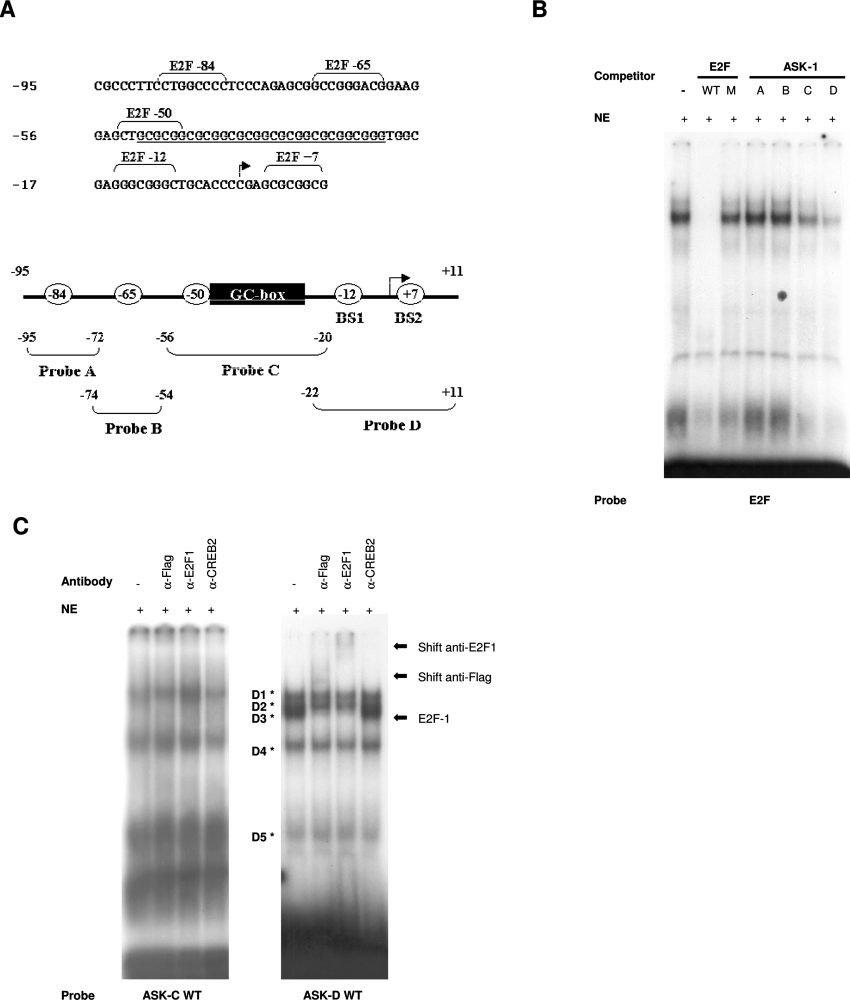
Analysis of the minimal ASK-1 promoter region responsible for E2F1 transactivation, i.e. −95 to +11 bp upstream from the translation site, with MatInspector and Transcription Element Search System software revealed no perfect consensus E2F-binding site. Nevertheless, as illustrated in Figure 6(A), five non-consensus E2F binding sites named E2F −84 (nucleotides −88 to −80), E2F−65 (nucleotides −69 to −61) E2F −50 (nucleotides −54 to −46), E2F −12 (nucleotides −15 to −8) and E2F+7 (nucleotides +4 to +11) and a GC-box (nucleotides −50 to −22) were identified. To determine whether E2F1 can bind these sequences, we synthesized four double-stranded oligonucleotides covering this region and performed EMSAs with Ad-E2F1-infected U2OS nuclear extracts. Incubation of the E2F consensus probe with these extracts resulted in the formation of distinct protein–DNA complexes (Figure 6B). These complexes were eliminated by the addition of a 200-fold excess of unlabelled oligonucleotide containing a functional E2F consensus site, but not by an oligonucleotide containing a mutated E2F-binding site in which the core CG was replaced by AT (see Table 2). On the other hand, the migrating complexes were unaffected by competition with the ASK-A and ASK-B unlabelled oligonucleotides, but were strongly competed by either the ASK-C or ASK-D unlabelled oligonucleotides. To determine whether E2F1 binds sequences ASK-C and ASK-D, EMSAs were performed using these oligonucleotides as probes. Four bands were observed when the ASK-C probe was incubated with U2OS nuclear extracts overexpressing E2F1 proteins (Figure 6C, left-hand panel). Supershift assays revealed that the anti-E2F1 and the anti-FLAG antibody did not generate any supershifted band, neither did the non-relevant anti-CREB2 control antibody. Conversely, when the ASK-D probe was incubated with the same extracts, five bands (D1–D5) were observed (Figure 6C, right-hand panel). Interestingly, among them, the D3 complex was supershifted by either the anti-E2F1 or the anti-FLAG antibody, but not by the anti-CREB2 control antibody, which was used to assess the shift specificity. Taken together, these results confirm that E2F1 protein associates with the ASK-D sequence of the minimal E2F1-responding ASK-1 promoter fragment. Since the −22 to +11 sequence of the ASK-D oligonucleotide contains two putative non-consensus E2F-BS (binding sites), we mutated these sites by replacing the CG with TA at positions −12 and −11 (BS1), thus leading to the ASK-D M1 oligonucleotide, and by replacing the GC with AA at positions +6 and +7 (BS2), leading to the ASK-D M2 oligonucleotide, or mutated both sites, leading to the ASK-D M1+2 oligonucleotides. Competition experiments were performed against the wild-type ASK-D probe with an excess of unlabelled oligonucleotides that carry respectively the wild-type putative E2F-binding sites or the mutated versions of these sites (Figure 7A). These results clearly show that the five protein–DNA complexes were competed by a 200-fold molar excess of the wild-type ASK-D unlabelled competitor, as well as by an excess of ASK-D M2 unlabelled competitor. In contrast, competition with the ASK-D M1 or M1+2 unlabelled oligonucleotides resulted in a major loss of complexes D1, D2, D4 and D5, indicating that they originate from protein binding outside of the putative E2F-binding sites. Conversely, complex D3 was unaffected by excess unlabelled oligonucleotides M1 or M1+2. Moreover, the presence of E2F1 in the D3 complex was confirmed in the presence of unlabelled M1 competitor by the shift observed upon addition of anti-E2F1 antibody (Figure 7B). As a control, no significant effect was observed by the addition of anti-CREB2 antibody. As expected, incubation of U2OS nuclear extracts with labelled mutated M1 or M1+2 ASK-D oligonucleotides resulted in the absence of the D3 complex (Figure 7C). Similar results were obtained when EMSAs were performed using purified recombinant bacterially expressed GST–E2F1 in combination with GST–DP1 proteins (results not shown). These results demonstrated that the non-consensus E2F-binding site at position −15 to −8 is responsible for E2F1 binding on the ASK-1 minimal E2F1-responsive promoter region.
Figure 6. DNA-binding activity of E2F1 on ASK-1 promoter.
(A) The ASK-1 106 bp promoter region activated by E2F1 expression. The arrow indicates the position of the putative transcription initiation site corresponding to the longest published cDNA sequence (GenBank® accession number NM_005923). The GC box is underlined and the putative non-consensus E2F transcription-factor-binding sites are indicated. The four oligonucleotide probes used in EMSAs, corresponding to regions −95 to −72, −74 to −54, −56 to −20 and −22 to +11, are depicted in the schematic representation of the ASK-1 promoter. (B) DNA-binding activity of Ad-E2F1-infected U2OS nuclear extracts on the E2F-binding site. Gel-shift analyses were performed with a wild-type (WT) E2F consensus site as a probe. Competition experiments with wild-type or mutated (M) form of consensus E2F-binding sequences or with the ASK-1 sequences are indicated. (C) Binding on ASK-C (left-hand panel) or ASK-D (right-hand panel) probes. Supershift experiments were performed with the anti-FLAG antibody (α-Flag), anti-E2F1 antibody (α-E2F1) and a non-relevant isotype antibody (α-CREB2). The complexes formed with the ASK-D probe are indicated by asterisks. NE, nuclear extract.
Figure 7. Identification of the E2F1-binding element on the ASK-D probe by EMSA.
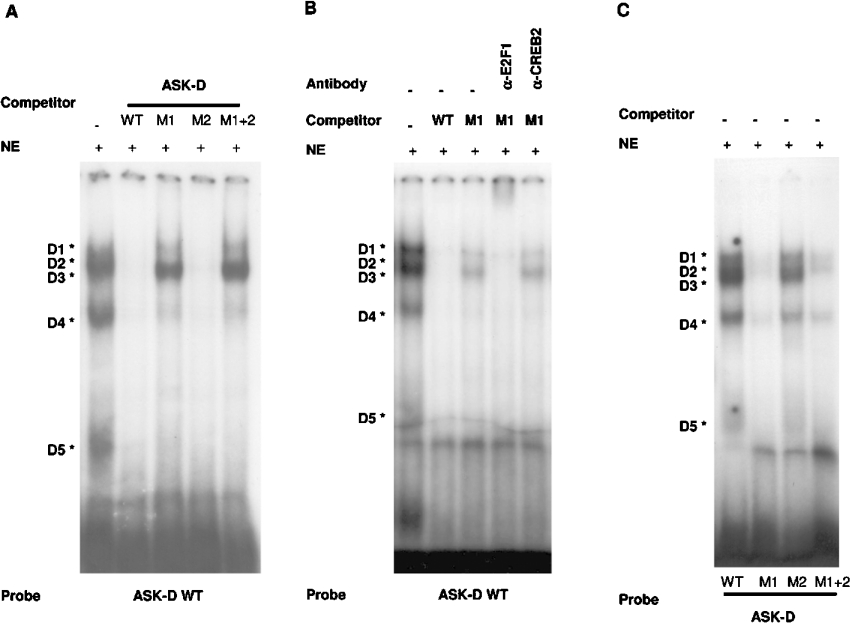
(A) Gel-shift analysis with the ASK-D oligonucleotide (−22 to +11) as a probe. Competition experiments were performed with the ASK-D sequence wild-type (WT) or mutated in the putative E2F binding site BS1 (M1), BS2 (M2) or both (M1+2). (B) Supershift experiments of the D3 complex were performed with the anti-E2F1 antibody (α-E2F1) and the anti-CREB2 antibody (α-CREB2). (C) DNA-binding activity of Ad-E2F1-infected U2OS nuclear extracts on wild-type ASK-D probe or mutated versions of ASK-D oligonucleotide. NE, nuclear extract.
Mutation of the ASK-1 promoter at position −12 abolishes transactivation by E2F
In order to test the effect of the mutation of the identified E2F-binding sites BS1 and BS2 on the ASK-1 promoter activity, the above-mentioned mutations were introduced in the ASK-1 reporter construct H(−95/+11). The activities of the resulting constructs (M1-H and M2-H) were then compared with the activity of the wild-type H(−95/+11) reporter construct upon transient transfection in U2OS cells (Figure 8). The results, expressed as fold activity relative to that obtained in absence of E2F expression vectors, indicate that only the M1 mutation dramatically decreased the E2F effect on the ASK-1 reporter construct. These results indicate that E2F-BS1 is the sole functional binding site present in the −95/+11 region of the ASK-1 gene.
Figure 8. Mutation of the ASK-1 promoter at positions −11 and −12 abolishes the E2F transactivation.
U2OS cells were transfected with 0.15 μg of expression vector (pcDNA3-HA control or the pcDNA3-HA-E2F and pcDNA3-HA-DP1 vectors) and 0.3 μg of the ASK-1 reporter construct H(−95/+11) wild-type or mutated in the putative E2F-binding site BS1 (M1) or BS2 (M2). The responsiveness of the various forms of ASK-1 promoter to E2F1 induction is presented relative to that of the empty pGL3-basic, arbitrarily set to 1.
E2Fs associate in vivo with the ASK-1 promoter
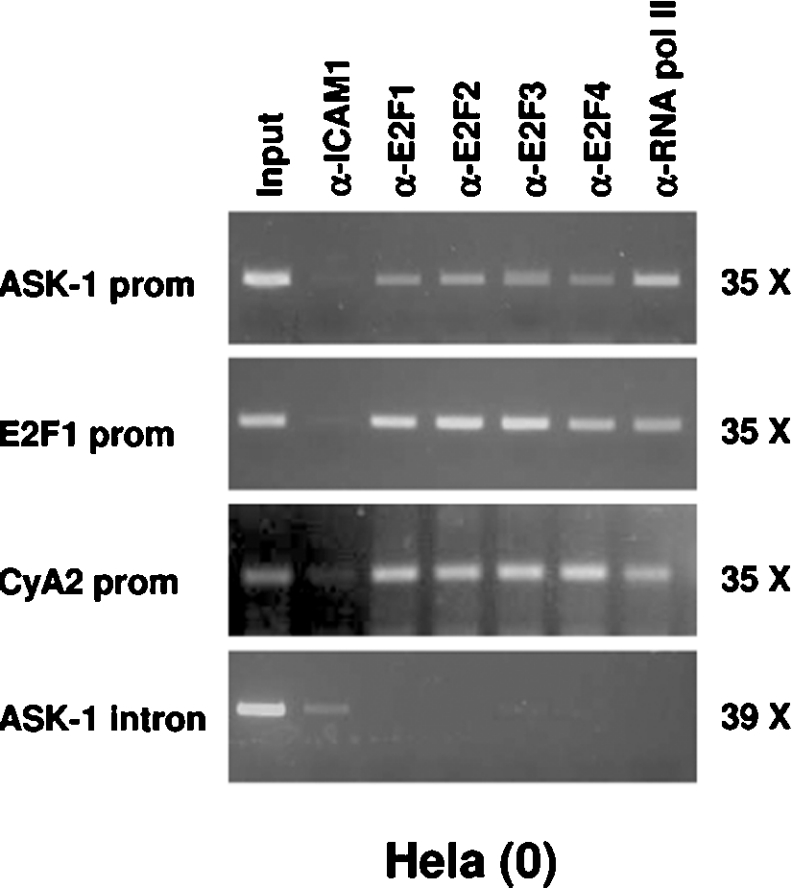
To confirm the implication of the E2Fs in the ASK-1 promoter control, we performed ChIP experiments using cycling HeLa (0) cells. As shown in Figure 9, the cross-linked ASK-1 promoter–E2Fs complexes immunoprecipitated with polyclonal antibodies directed against E2F1, E2F2, E2F3 and E2F4 were detected by PCR amplification with oligonucleotides spanning the first exon of the human ASK-1 gene. As positive controls, in vivo association of the E2F factors with E2F1 and CCNA2 promoters were detected by PCR with specific primers on the same samples, as described previously [29].
Figure 9. E2F1–E2F4 bind to the ASK-1 promoter in vivo.
Formaldehyde cross-linked chromatin from He La (0) cells were subjected to ChIP experiments. Immunoprecipitations of E2F containing complexes were performed using anti-E2F1 (α-E2F1), anti-E2F2 (α-E2F2), anti-E2F3 (α-E2F3) and anti-E2F4 (α-E2F4) antibodies. Anti-ICAM1 polyclonal antibody (α-ICAM1) was used as negative control and anti-(RNA pol II) (α-RNA pol II) as a positive control. After isolation of bound DNA, PCR was performed for a 297 bp region of the endogenous ASK-1 promoter. As positive controls, PCRs were also performed using E2F1-promoter-specific primers and CCNA2-promoter-specific primers, two well-documented E2F target genes, and ASK-1-intron 5-specific primers as a negative control. Input indicates PCR performed on DNA (diluted 1/300) without any immunoprecipitation. Values on the right indicate the number of PCR cycles performed. prom, promoter.
The absence of PCR amplification using primers spanning the fifth intron of the ASK-1 gene rules out spurious interaction between E2F factors and the ASK-1 promoter region. As a negative control, ChIPs with an irrelevant antibody directed against ICAM1 failed to detect genomic DNA fragments for the ASK-1 promoter as well as for the E2F1 and CCNA2 promoters. Inversely, RNA pol II association with both promoters provided an additional positive control. These results demonstrate that E2F1, E2F2, E2F3 and E2F4 bind the ASK-1 promoter in vivo and provide further evi-dence for E2F transcriptional control of ASK-1 promoter activity.
DISCUSSION
In our experiments, ASK-1 was identified as a potential E2F1 target gene by RDA comparison of cDNA from Saos-2 cells infected by E2F1-expressing recombinant adenovirus compared with cells infected with a GFP-expressing one. Despite the fact that ASK-1 was also identified as a potential E2F target gene by others in U2OS cells [30] and in melanoma cells [31], the mechanism for such a regulation has remained uninvestigated, particularly in a normal cellular context. In the present study, we demonstrated the isolation and characterization of the promoter region of the human ASK-1 gene and analysed the mechanisms involved in its regulation by the E2Fs.
ASK-1 is a MAPKKK that is involved in the cellular apoptotic response resulting from oxidative stress, ER stress as well as TNF (tumour necrosis factor) exposure [16]. However, the functions of ASK-1 are not limited to the apoptotic pathway, since it is also involved in the control of cellular proliferation, differentiation and survival [16,32,33].
Our results indicate that E2Fs increase ASK-1 expression at the mRNA and protein levels in various mammalian cell lines. Besides, inhibition of DP1 expression by RNAi confirmed the role of E2F/DP1 activity in maintaining ASK-1 gene expression in these cell lines. The fact that ASK-1 mRNA is only partially diminished upon DP1-shRNA expression is probably due to the residual DP1 expression level as well as the presence of endogenous DP2 in the cells. Moreover, additional pathways may be involved in the regulation of ASK-1.
According to the cell-cycle regulation of bona fide E2F target genes, the expression pattern of ASK-1 during T98G cell-cycle progression is similar to that of CCNA2, an extensively characterized E2F target gene. Involvement of p53 in E2F1-induced up-regulation of some genes has been described previously [34]. Concerning ASK-1, such a mechanism was excluded since ectopic expression of E2F1 in p53-deficient human cells (Saos-2) or p53-mutated human cells (MDA-MB-468) led to an increase of ASK-1 levels, whereas overexpression of p53 in wild-type p53-expressing cells (U2OS and MCF10A) never affected the ASK-1 levels or kinase activity. Moreover, our results also rule out the possibility that ASK-1 is a p53 target gene in these cells. Activation of the promoter of ASK-1 requires the DNA-binding and transactivation domains of E2F1 and is direct, since ER–E2F1 fusion protein overexpression can induce ASK-1 mRNA expression even in the presence of protein synthesis inhibitors (results not shown).
To understand the mechanism by which ASK-1 expression is regulated by E2F, we characterized the ASK-1 promoter. The 5′-flanking region of ASK-1 gene is highly GC-rich and lacks a consensus TATA box or CAAT box upstream of the putative transcription initiation site. As revealed by a computer study of E2F targets, only a few E2F-regulated genes contain a TATA box, and a common feature of most of the E2F target genes is the presence of E2F sites located within the first 150 bp upstream of the transcription start site [35]. Interestingly, the human ASK-1 minimal promoter fragment H (−95/+11) does not contain any consensus E2F-binding sites. Nevertheless, several E2F-regulated genes (e.g. CCNE1, CCNA2 and MYBL2) do not contain perfect consensus E2F DNA-binding sites [36]. As estimated by Weinmann et al. [37], 25% of E2F-target promoters recruit E2F via a mechanism distinct from the defined consensus site [37]. Using deletion and site-directed mutants, we found that a non-consensus E2F site BS1 (GGGCGGGC) is strictly necessary for the up-regulation of the ASK-1 promoter. Gel-mobility shift and supershift experiments involving BS1 located at position −15 to −8 of the ASK-1 promoter indicate that E2F1 binds ASK-1 in vitro. The ASK-1 promoter is activated by the growth-promoting E2F transcription factors (E2F1–E2F3) and, to a lesser extent, by E2F4, indicating that ASK-1 is the target of various E2F family members. Moreover, ChIP assays performed on cycling cells revealed in vivo binding of E2F1–E2F4 to the ASK-1 promoter, assessing the physiological relevance of the effect that E2F family members have on the ASK-1 promoter activity.
Since E2F1 and ASK-1 apoptotic functions are well documented, it is temping to implicate ASK-1 as a component of the E2F1-induced apoptotic response. Nevertheless, in our hands, neither overexpression of a kinase death dominant-negative ASK-1 form (ASK-1-K709R) nor knockdown of ASK-1 expression by RNAi were able to significantly reduce E2F1-induced apoptosis (results not shown). Our results may be accounted for by various, non-mutually exclusive, possibilities. First, the resulting levels of ASK-1 activity, although reduced significantly, could remain sufficient to allow apoptosis. Secondly, ASK-1 involvement in E2F1-induced apoptosis might be cell-type-specific. Thirdly, because ASK-1 represents only one of the several known E2F target genes involved in parallel redundant pathways of cell death, inhibition of just one of these may have little effect. Nevertheless, in addition to its stress-induced apoptotic functions, ASK-1 is also implicated in other cellular processes (proliferation, differentiation and survival). Therefore it is possible that the E2F transcription factors that are highly implicated in such cellular processes control some of the ASK-1 functions through its transcriptional regulation. In this context, Yamamoto et al. [38] demonstrated that ASK-1 kinase activity is highly enhanced during the G2/M-phase. So, as suggested by the ASK-1 cell-cycle expression profile observed, E2F might be responsible for the cyclic accumulation of ASK-1 proteins, allowing their activation during the G2/M-phase. Moreover, Dasgupta et al. [39] demonstrated a physical and functional interaction between ASK-1 and Rb proteins in response to specific apoptotic stimuli. This interaction inactivates Rb and enhances the transcriptional activity of E2F. Therefore, under stress conditions, the up-regulation of ASK-1 may contribute to a positive-feedback loop by which E2Fs inhibit their own repression by Rb, allowing the activation of their target genes.
Acknowledgments
We thank Dr J. R. Woodgett for providing the ASK-1 and ASK-1-K709R expression vectors, and Dr E. Hara for providing the DP1-shRNA and scrambled shRNA. We thank J. L. Baert for his critical reading of the manuscript, and A. Bègue for valuable technical discussions. This work has been carried out on the basis of grants awarded in part by the Centre National de la Recherche Scientifique, the Institut Pasteur de Lille, the Université des Sciences et Technologies de Lille 1, the Conseil Régional Nord/Pas-de-Calais, the Association pour la Recherche sur le Cancer, the Ligue contre le Cancer (France) and the European Regional Developmental Fund. A.B. is supported by a postdoctoral fellowship from the Canadian Institutes of Health Research.
References
- 1.Kovesdi I., Reichel R., Nevins J. R. Role of an adenovirus E2 promoter binding factor in E1A-mediated coordinate gene control. Proc. Natl. Acad. Sci. U.S.A. 1987;84:2180–2184. doi: 10.1073/pnas.84.8.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blais A., Dynlacht B. D. Hitting their targets: an emerging picture of E2F and cell cycle control. Curr. Opin. Genet. Dev. 2004;14:527–532. doi: 10.1016/j.gde.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 3.Dimova D. K., Dyson N. J. The E2F transcriptional network: old acquaintances with new faces. Oncogene. 2005;24:2810–2826. doi: 10.1038/sj.onc.1208612. [DOI] [PubMed] [Google Scholar]
- 4.Helin K., Harlow E. Heterodimerization of the transcription factors E2F-1 and DP-1 is required for binding to the adenovirus E4 (ORF6/7) protein. J. Virol. 1994;68:5027–5035. doi: 10.1128/jvi.68.8.5027-5035.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu C. L., Zukerberg L. R., Ngwu C., Harlow E., Lees J. A. In vivo association of E2F and DP family proteins. Mol. Cell. Biol. 1995;15:2536–2546. doi: 10.1128/mcb.15.5.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yee A. S., Reichel R., Kovesdi I., Nevins J. R. Promoter interaction of the E1A-inducible factor E2F and its potential role in the formation of a multi-component complex. EMBO J. 1987;6:2061–2068. doi: 10.1002/j.1460-2075.1987.tb02471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson D. G., Schwarz J. K., Cress W. D., Nevins J. R. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature (London) 1993;365:349–352. doi: 10.1038/365349a0. [DOI] [PubMed] [Google Scholar]
- 8.Singh P., Wong S. H., Hong W. Overexpression of E2F-1 in rat embryo fibroblasts leads to neoplastic transformation. EMBO J. 1994;13:3329–3338. doi: 10.1002/j.1460-2075.1994.tb06635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Field S. J., Tsai F. Y., Kuo F., Zubiaga A. M., Kaelin W. G., Jr, Livingston D. M., Orkin S. H., Greenberg M. E. E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell. 1996;85:549–561. doi: 10.1016/s0092-8674(00)81255-6. [DOI] [PubMed] [Google Scholar]
- 10.Yamasaki L., Jacks T., Bronson R., Goillot E., Harlow E., Dyson N. J. Tumor induction and tissue atrophy in mice lacking E2F-1. Cell. 1996;85:537–548. doi: 10.1016/s0092-8674(00)81254-4. [DOI] [PubMed] [Google Scholar]
- 11.Ginsberg D. E2F1 pathways to apoptosis. FEBS Lett. 2002;529:122–125. doi: 10.1016/s0014-5793(02)03270-2. [DOI] [PubMed] [Google Scholar]
- 12.Denchi E. L., Helin K. E2F1 is crucial for E2F-dependent apoptosis. EMBO Rep. 2005;6:661–668. doi: 10.1038/sj.embor.7400452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ichijo H., Nishida E., Irie K., ten Dijke P., Saitoh M., Moriguchi T., Takagi M., Matsumoto K., Miyazono K., Gotoh Y. Induction of apoptosis by ASK1, a mammalian MAPKKK that activates SAPK/JNK and p38 signaling pathways. Science. 1997;275:90–94. doi: 10.1126/science.275.5296.90. [DOI] [PubMed] [Google Scholar]
- 14.Chang H. Y., Nishitoh H., Yang X., Ichijo H., Baltimore D. Activation of apoptosis signal-regulating kinase 1 (ASK1) by the adapter protein Daxx. Science. 1998;281:1860–1863. doi: 10.1126/science.281.5384.1860. [DOI] [PubMed] [Google Scholar]
- 15.Matsuzawa A., Nishitoh H., Tobiume K., Takeda K., Ichijo H. Physiological roles of ASK1-mediated signal transduction in oxidative stress- and endoplasmic reticulum stress-induced apoptosis: advanced findings from ASK1 knockout mice. Antioxid. Redox Signaling. 2002;4:415–425. doi: 10.1089/15230860260196218. [DOI] [PubMed] [Google Scholar]
- 16.Takeda K., Matsuzawa A., Nishitoh H., Ichijo H. Roles of MAPKKK ASK1 in stress-induced cell death. Cell Struct. Funct. 2003;28:23–29. doi: 10.1247/csf.28.23. [DOI] [PubMed] [Google Scholar]
- 17.Tobiume K., Matsuzawa A., Takahashi T., Nishitoh H., Morita K., Takeda K., Minowa O., Miyazono K., Noda T., Ichijo H. ASK1 is required for sustained activations of JNK/p38 MAP kinases and apoptosis. EMBO Rep. 2001;2:222–228. doi: 10.1093/embo-reports/kve046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kherrouche Z., De Launoit Y., Monte D. Human E2F6 is alternatively spliced to generate multiple protein isoforms. Biochem. Biophys. Res. Commun. 2004;317:749–760. doi: 10.1016/j.bbrc.2004.03.099. [DOI] [PubMed] [Google Scholar]
- 19.Hoeflich K. P., Yeh W. C., Yao Z., Mak T. W., Woodgett J. R. Mediation of TNF receptor-associated factor effector functions by apoptosis signal-regulating kinase-1 (ASK1) Oncogene. 1999;18:5814–5820. doi: 10.1038/sj.onc.1202975. [DOI] [PubMed] [Google Scholar]
- 20.Hubank M., Schatz D. G. Identifying differences in mRNA expression by representational difference analysis of cDNA. Nucleic Acids Res. 1994;22:5640–5648. doi: 10.1093/nar/22.25.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blais A., Monte D., Pouliot F., Labrie C. Regulation of the human cyclin-dependent kinase inhibitor p18INK4c by the transcription factors E2F1 and Sp1. J. Biol. Chem. 2002;277:31679–31693. doi: 10.1074/jbc.M204554200. [DOI] [PubMed] [Google Scholar]
- 22.Brummelkamp T. R., Bernards R., Agami R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. 2002;2:243–247. doi: 10.1016/s1535-6108(02)00122-8. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Q., Zhang G. Activation and autophosphorylation of apoptosis signal-regulating kinase 1 (ASK1) following cerebral ischemia in rat hippocampus. Neurosci. Lett. 2002;329:232–236. doi: 10.1016/s0304-3940(02)00650-x. [DOI] [PubMed] [Google Scholar]
- 24.Kherrouche Z., De Launoit Y., Monte D. The NRF-1/α-PAL transcription factor regulates the human E2F6 promoter activity. Biochem. J. 2004;383:529–536. doi: 10.1042/BJ20040935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stanelle J., Stiewe T., Theseling C. C., Peter M., Putzer B. M. Gene expression changes in response to E2F1 activation. Nucleic Acids Res. 2002;30:1859–1867. doi: 10.1093/nar/30.8.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeGregori J., Kowalik T., Nevins J. R. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol. Cell. Biol. 1995;15:4215–4224. doi: 10.1128/mcb.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26a.Erratum. Mol. Cell. Biol. 1995. pp. 5846–5847.
- 27.Tobiume K., Saitoh M., Ichijo H. Activation of apoptosis signal-regulating kinase 1 by the stress-induced activating phosphorylation of pre-formed oligomer. J. Cell. Physiol. 2002;191:95–104. doi: 10.1002/jcp.10080. [DOI] [PubMed] [Google Scholar]
- 28.Maehara K., Yamakoshi K., Ohtani N., Kubo Y., Takahashi A., Arase S., Jones N., Hara E. Reduction of total E2F/DP activity induces senescence-like cell cycle arrest in cancer cells lacking functional pRB and p53. J. Cell Biol. 2005;168:553–560. doi: 10.1083/jcb.200411093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takahashi Y., Rayman J. B., Dynlacht B. D. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 2000;14:804–816. [PMC free article] [PubMed] [Google Scholar]
- 30.Muller H., Bracken A. P., Vernell R., Moroni M. C., Christians F., Grassilli E., Prosperini E., Vigo E., Oliner J. D., Helin K. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 2001;15:267–285. doi: 10.1101/gad.864201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jamshidi-Parsian A., Dong Y., Zheng X., Zhou H. S., Zacharias W., McMasters K. M. Gene expression profiling of E2F-1-induced apoptosis. Gene. 2005;344:67–77. doi: 10.1016/j.gene.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 32.Chibon F., Mariani O., Derre J., Mairal A., Coindre J. M., Guillou L., Sastre X., Pedeutour F., Aurias A. ASK1 (MAP3K5) as a potential therapeutic target in malignant fibrous histiocytomas with 12q14-q15 and 6q23 amplifications. Genes Chromosomes Cancer. 2004;40:32–37. doi: 10.1002/gcc.20012. [DOI] [PubMed] [Google Scholar]
- 33.Faigle R., Brederlau A., Elmi M., Arvidsson Y., Hamazaki T. S., Uramoto H., Funa K. ASK1 inhibits astroglial development via p38 mitogen-activated protein kinase and promotes neuronal differentiation in adult hippocampus-derived progenitor cells. Mol. Cell. Biol. 2004;24:280–293. doi: 10.1128/MCB.24.1.280-293.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kowalik T. F., DeGregori J., Leone G., Jakoi L., Nevins J. R. E2F1-specific induction of apoptosis and p53 accumulation, which is blocked by Mdm2. Cell Growth Differ. 1998;9:113–118. [PubMed] [Google Scholar]
- 35.Kel A. E., Kel-Margoulis O. V., Farnham P. J., Bartley S. M., Wingender E., Zhang M. Q. Computer-assisted identification of cell cycle-related genes: new targets for E2F transcription factors. J. Mol. Biol. 2001;309:99–120. doi: 10.1006/jmbi.2001.4650. [DOI] [PubMed] [Google Scholar]
- 36.Schulze A., Zerfass K., Spitkovsky D., Middendorp S., Berges J., Helin K., Jansen-Durr P., Henglein B. Cell cycle regulation of the cyclin A gene promoter is mediated by a variant E2F site. Proc. Natl. Acad. Sci. U.S.A. 1995;92:11264–11268. doi: 10.1073/pnas.92.24.11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weinmann A. S., Yan P. S., Oberley M. J., Huang T. H., Farnham P. J. Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes Dev. 2002;16:235–244. doi: 10.1101/gad.943102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamamoto K., Ichijo H., Korsmeyer S. J. BCL-2 is phosphorylated and inactivated by an ASK1/Jun N-terminal protein kinase pathway normally activated at G2/M. Mol. Cell. Biol. 1999;19:8469–8478. doi: 10.1128/mcb.19.12.8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dasgupta P., Betts V., Rastogi S., Joshi B., Morris M., Brennan B., Ordonez-Ercan D., Chellappan S. Direct binding of ASK1 to Rb: novel links between apoptotic signaling and cell cycle machinery. J. Biol. Chem. 2004;279:38762–38769. doi: 10.1074/jbc.M312273200. [DOI] [PubMed] [Google Scholar]