Abstract
Yersinia enterocolitica, an important cause of human gastroenteritis generally caused by the consumption of livestock, has traditionally been categorized into three groups with respect to pathogenicity, i.e., nonpathogenic (biotype 1A), low pathogenicity (biotypes 2 to 5), and highly pathogenic (biotype 1B). However, genetic differences that explain variation in pathogenesis and whether different biotypes are associated with specific nonhuman hosts are largely unknown. In this study, we applied comparative phylogenomics (whole-genome comparisons of microbes with DNA microarrays combined with Bayesian phylogenies) to investigate a diverse collection of 94 strains of Y. enterocolitica consisting of 35 human, 35 pig, 15 sheep, and 9 cattle isolates from nonpathogenic, low-pathogenicity, and highly pathogenic biotypes. Analysis confirmed three distinct statistically supported clusters composed of a nonpathogenic clade, a low-pathogenicity clade, and a highly pathogenic clade. Genetic differences revealed 125 predicted coding sequences (CDSs) present in all highly pathogenic strains but absent from the other clades. These included several previously uncharacterized CDSs that may encode novel virulence determinants including a hemolysin, a metalloprotease, and a type III secretion effector protein. Additionally, 27 CDSs were identified which were present in all 47 low-pathogenicity strains and Y. enterocolitica 8081 but absent from all nonpathogenic 1A isolates. Analysis of the core gene set for Y. enterocolitica revealed that 20.8% of the genes were shared by all of the strains, confirming this species as highly heterogeneous, adding to the case for the existence of three subspecies of Y. enterocolitica. Further analysis revealed that Y. enterocolitica does not cluster according to source (host).
Yersinia enterocolitica is an important gastrointestinal pathogen that can cause a range of human diseases from mild diarrhea to mesenteric lymphadenitis (8). The clinical manifestations of the gastrointestinal disease can vary according to the age and health of the host. Children often present with acute enteritis, fever, and watery diarrhea which can be occasionally bloody, and the illness can last between a couple of days and 4 weeks (7, 52). Adults typically present with an illness lasting between 7 and 14 days of fever, diarrhea, acute terminal ileitis, and mesenteric lymphadenitis.
The incidence of Y. enterocolitica is apparently increasing worldwide (7). In parts of Europe, Y. enterocolitica rivals salmonellae as a cause of gastrointestinal disease (7, 8, 21). Y. enterocolitica can be isolated from a wide variety of sources, i.e., surface water; many food products, such as seafood and dairy products; and livestock, particularly pigs (7, 27, 28, 52, 58). This may be due to the consumption of uncooked and undercooked meat of porcine origin, such as chitterlings or tongue (7, 30, 38, 52). The transmission of Y. enterocolitica from pigs to humans is thought to occur through the contamination of pork products; however, the relationship between isolates from livestock and human disease is poorly understood.
A recent abattoir survey in the United Kingdom (1999 and 2000) revealed the fecal carriage rates of Y. enterocolitica in cattle, sheep, and pigs sent for slaughter as 6.3, 10.7, and 26.1%, respectively (43). The surprisingly high carriage rate of Y. enterocolitica in livestock was unexpected and suggests that this livestock may contribute to human yersiniosis.
Y. enterocolitica is classified into six biotypes (1A, 1B, 2, 3, 4, and 5) based upon varied biochemical properties. The six biotypes can be separated into three groups according to their lethality in a murine infection model which closely resembles the naturally acquired human infection: biotype 1A, which is considered to be mostly nonpathogenic (8); biotypes 2 to 5, which are low in pathogenicity; and biotype 1B, which is highly pathogenic (4, 14). Biotypes with low pathogenicity are generally isolated in Europe and Japan and are termed Old World strains. By contrast, highly pathogenic strains are most commonly isolated in North America and are termed New World strains.
Both the low- and high-pathogenicity groups carry a 70-kb plasmid that has been termed virulence plasmid pYV (29, 51, 61). This plasmid is usually absent from the nonpathogenic 1A biogroup. In addition, highly pathogenic group 1B also possess a high-pathogenicity island (HPI) (13). The importance of this gene cluster was demonstrated by transferring the HPI core genes of WA-C serotype O:8 biotype 1B to strain MRS40 O:9 biotype 2 of Y. enterocolitica (low pathogenicity), resulting in increased virulence (49). More recently, a novel virulence-associated type II secretion system unique to high-pathogenicity Y. enterocolitica strains has been described (36). However, it is unlikely that these genetic differences alone can explain the relative pathogenicity of the three groups.
Comparative genomic DNA (gDNA) microarray analysis has been used to investigate several pathogenic bacterial species in relation to pathogenesis and host specificity (17, 19, 20, 23, 32, 39). Microarray technology, allied to complex mathematical analysis to determine phylogeny, has provided a sensitive and robust method to examine the genetic relatedness of bacterial populations. The genetic relationships described by Bayesian phylogeny of a DNA-DNA microarray data set can then be correlated against the known phenotypes and ecological behavior of each bacterial strain in the analysis; this is particularly useful when studying the epidemiology and host association of pathogens (17). Comparison of strains isolated from different hosts, as well as pathogenic and nonpathogenic strains, can reveal predicted coding sequences (CDSs) that may be important for virulence, pathogen-host interactions, and transmission. For example, within the two other human pathogenic Yersinia species, microarray studies of Yersinia pseudotuberculosis revealed 11 DNA loci that were absent or highly divergent compared to the Yersinia pestis C092 genome (32). Acquisition of these loci may help to explain why Y. pestos can cause such a vastly different disease compared to its close enteropathogenic relative Y. pseudotuberculosis, from which it evolved an estimated 1,500 to 20,000 years ago (1).
In this study, we have carried out a whole-genome analysis of 94 isolates of Y. enterocolitica from human and livestock sources with a whole-genome microarray based on the recently sequenced genome of Y. enterocolitica 8081, biotype 1B serotype O8. The DNA microarray data have been combined with sensitive Bayesian method-based algorithms to gain new insight into the population structure of Y. enterocolitica, which revealed several new potential virulence factors, suggests livestock as an important source of human yersiniosis and provided basic information on the evolution of the species Y. enterocolitica.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The 94 strains used in this study are summarized elsewhere (43). Y. enterocolitica 8081 1B was sequenced and annotated at the Pathogen Sequencing Unit, The Wellcome Trust Sanger Institute, Cambridge, United Kingdom. Y. enterocolitica 8081 1B was originally isolated from a patient suffering from septicemia (51), is highly pathogenic in the murine yersiniosis infection model, and has been widely studied.
Y. enterocolitica strains were cultured either in Luria-Bertani (LB) broth with constant shaking at 200 rpm or on solid medium prepared with LB agar. Incubation of both liquid cultures and agar plates was carried out at 28°C. All bacterial strains used in this study were stored in LB broth containing glycerol (15%) at −80°C. Bacterial chromosomal DNA was prepared with the Wizard gDNA purification kit (Promega). DNA samples were then analyzed by agarose gel electrophoresis and quantified with a GeneQuant spectrophotometer (Amersham). All gDNA was stored at −20°C in distilled H2O.
Microarray construction and hybridization.
The Y. enterocolitica-specific microarray was designed to include 4,208 predicted CDSs from the Y. enterocolitica 8081 chromosome and 83 from plasmid pYV. PCR products were designed by the approach described by Hinds et al. (33). Ten PCR products for each CDS were designed with Primer3 (56), and then, based on BLAST analysis, the optimum PCR product to represent each CDS was selected. PCR primers were synthesized by MWG Biotech (Ebersberg, Germany), and high-throughput PCR amplification was carried out with a liquid-handling and PCR amplification robot (RoboAmp 9600; MWG Biotech). PCR products were analyzed by agarose gel electrophoresis to ensure a unique band of the correct size and 5% of the products were confirmed by DNA sequencing. Microarrays were constructed by robotic spotting of the PCR products in duplicate on UltraGaps amino-silane-coated glass slides (Corning) with a MicroGrid II (BioRobotics). Further details of microarray construction methodology can be found in reference 34.
All of the strains in the collection were competitively hybridized with the Y. enterocolitica 8081 1B microarray, which had duplicate spots of the 4,291 CDSs. Hybridizations were performed by modifying a protocol previously described by Dorrell et al. (20). Test gDNA was labeled with Cy5-dCTP, and Cy3-dCTP-labeled Y. enterocolitica 8081 gDNA was used as a common reference for all hybridizations. UltraGap slides were prehybridized in 3.5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate (SDS)-10 mg/ml bovine serum albumin for at least 20 min at 65°C. After prehybridization, the slides were rinsed in distilled water for 1 min and then in isopropanol for 1 min. The slides were dried by centrifugation at 1,200 rpm for 5 min and stored in the dark until ready for use. Control Cy3-labeled and test Cy5-labeled gDNA samples were mixed together and purified with QIAGEN PCR purification columns. The mixed control and test sample was eluted from the column with 71.5 μl of distilled H2O. The labeled DNA was mixed with hybridization solution at a final concentration of 4× SSC-0.3% SDS. The sample was then denatured and added to a prehybridized microarray slide. Two Lifter Slips (22 by 22 mm; Eyrie Scientific) were placed onto each array before sealing in a humidified hybridization chamber (Telechem International). The chambers were immersed in a water bath at 65°C for 16 to 20 h. The slides were washed in 400 ml of 1× SSC-0.06% SDS at 65°C for 2 min, followed by two separate washes in 400 ml of 0.06% SSC at room temperature. The microarray slides were then scanned with a GMS 418 Scanner (Genetic Microsystems). Spot fluorescence intensities were acquired with ImaGene 5.5 (BioDiscovery Inc.).
Microarray data analysis and comparative phylogenomics.
Whole-genome comparisons were carried out with GeneSpring v6.1 (Silicon Genetics). Samples were normalized in GeneSpring by the following criteria. Intensity values below 0.01 were set to 0.01, and a ratio was calculated by dividing the signal channel intensity by the control channel intensity for each gene in each sample; if the control channel was below 0.01, then 0.01 was used instead, and if the control channel and the signal channel were both below 0.01, then no data were reported. Each sample was normalized by dividing each measurement by the 50th percentile of all measurements in that sample with only genes flagged present or marginal by ImaGene to calculate the percentile. Normalized raw and control data for each array were exported from GeneSpring, transformed into log ratio data, and analyzed with GACK software to determine whether genes were (i) present or (ii) absent or highly divergent (40). This software determines dynamic cutoffs for each array individually. The default GACK settings for trinary analysis were used (data histogram bin size, 0.10; data smoothing, none; peak modeling, normal curve; trinary output, trinary %EPP cutoff 1 = 0; trinary %EPP cutoff 2 = 100). The output of GACK was transformed into NEXUS format, and the relationship of the strains was determined with Bayesian method-based algorithms implemented through Mr Bayes v3.0 software (53) as described in reference 17. Mr Bayes requires data to be in a binary format, so any genes designated as marginal were attributed to the present category for further analysis.
All analytical procedures were identical to those described by Champion et al. (17). In summary, microarray data analysis was undertaken by Bayesian methods performed with a Metropolis-coupled Markov chain Monte Carlo (Mr Bayes (35) with a 16-category gamma distribution to model the presence or absence rate heterogeneity per gene throughout the genome of the strain. A four-chain Metropolis-coupled Markov chain Monte Carlo was performed for 1.1 × 106 generations, and a burn in of 0.1 × 106 generations was used.
The resulting tree was viewed by TREEVIEW (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html). In addition, for phylogenetic analysis, if 5% of the CDSs were flagged (poor-quality spot) in a strain, then the strain was removed. If 10% of the strains had a CDS flagged, the CDS was removed. By the 5% criterion, strains Y14/02 (cattle, biotype 1A serotype O6,30) and Y13/02 (cattle, biotype 1A serotype O6,30) were not analyzed; therefore, 92 strains were included in the phylogenomic analysis.
Identification of genes contributing to clusters.
To identify genes which contributed to the generation of the pathogenic and nonpathogenic clusters, MacClade 4 analysis of phylogeny and character evolution software was used (42). The NEXUS file for all strains and the one-millionth tree from Mr Bayes were loaded into MacClade. The tree was then rooted with all of the 1B strains. Characters were then traced with the “trace all changes calculations” set to unambiguous changes only (minimum number changes). If a gene was absent or highly divergent in a particular strain, then the branch representing that strain was colored yellow. If the gene was present, then the branch was colored blue.
Nucleotide sequence accession numbers.
Fully annotated microarray data have been deposited in BμG@Sbase (accession number E-BUGS-36; http://bugs.sgul.ac.uk/E-BUGS-36) and also ArrayExpress (accession number E-BUGS-36).
RESULTS AND DISCUSSION
Strain selection.
Strains selected for comparative phylogenomic analysis included 35 human (12 nonpathogenic, 15 low-pathogenicity, and 8 highly pathogenic), 35 pig (10 nonpathogenic and 25 low-pathogenicity), 15 sheep (8 nonpathogenic and 7 low-pathogenicity), and 9 bovine (7 nonpathogenic and 2 low-pathogenicity) isolates (43). Eighty-six of these isolates are well characterized and were selected to represent a cross-section of the serotypes and biotypes prevalent between 1999 and 2000 in the United Kingdom. These isolates came from patients presenting to their general practitioners (GPs) with diarrhea or were collected during the Infectious Intestinal Diseases (IID) study (60). Strains were biotyped and serotyped at the Health Protection Agency according to the modified scheme of Wauters (7). Further details of the methodology used in this survey and livestock sampling are described in reference 43. No biotype 5 strains were tested, as they were absent from the study samples, and environmental biotype 1A isolates were not included in this study. A further eight human biotype 1B isolates of different serotypes originating from the United States were also included for analysis (5).
Core set of genes present in all test strains identified.
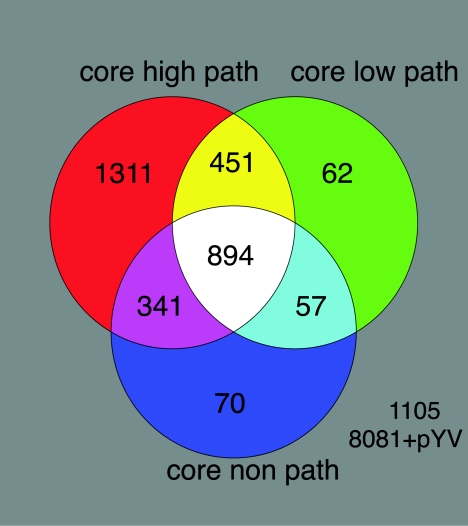
Genomic comparisons of 94 isolates were used to calculate the minimal core gene set. This was achieved by calculating the total number of genes that had a GACK score of “present” in every strain and control strain Y. enterocolitica 8081 1B. The minimal core gene set for Y. enterocolitica was 894 CDSs; this low core gene value validates that we have sampled a diverse collection of strains. The core gene set represents a surprisingly low value of 20.8% of the total genome (Fig. 1), suggesting that the pangenome of Y. enterocolitica is vast and that the size of the variable component dwarfs that of the core set of genes. By contrast, in similar sample size studies of C. jejuni and S. aureus the reported core set values were 59.2% and 78%, respectively (17, 25). As expected, many of the functional categories that are involved in essential housekeeping functions, such as DNA and RNA metabolism, protein processing and secretion, cell structure, cellular processes, and energetic and intermediary metabolism, were represented in the core gene set. Many of the accessory genes composed of CDSs likely to have been acquired by lateral gene transfer, such as transposons, insertion sequences, phages, and pathogenicity islands.
FIG. 1.
Venn diagrams showing core gene sets calculated with CDSs that were classified as present by GACK analysis. A core gene set of 894 CDSs for Y. enterocolitica is shown in white. Core gene sets for high-pathogenicity (path), low-pathogenicity, and nonpathogenic Y. enterocolitica strains are shown in the red (2,997 CDSs), green (1,464 CDSs), and dark blue (1,362 CDSs) circles with overlapping regions, respectively. This excludes 1,105 chromosomal and pYV CDSs that are not core to any of the biogroups.
Strains were designated high pathogenicity, low pathogenicity, or nonpathogenic depending on the clade they fell into by phylogenomic analysis. Individual core gene sets for high-pathogenicity, low-pathogenicity, and nonpathogenic Y. enterocolitica strains were calculated as 71.2% (2,997 CDSs), 34.8% (1,464 CDSs), and 32.4% (1,362 CDSs), respectively (Fig. 1).
Comparative phylogenomic analysis.
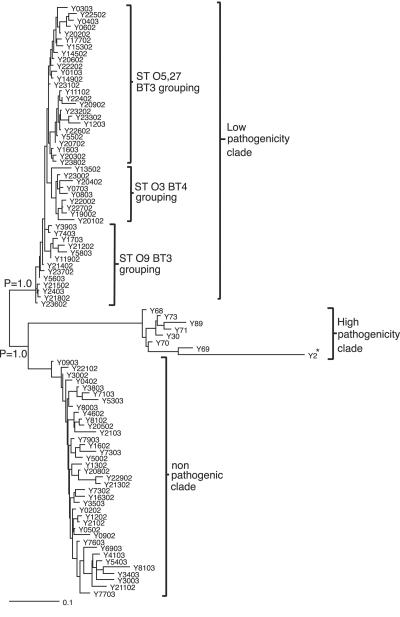
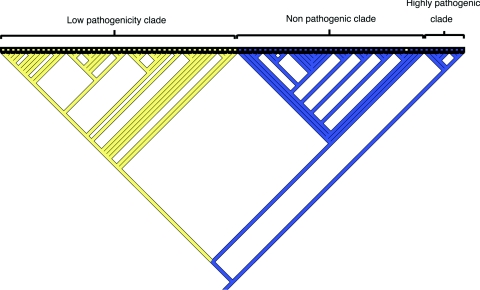
Phylogenomic analysis was carried out with microarray data generated from a strain collection consisting of 35 human isolates, 35 pig isolates, 15 sheep isolates, and 7 bovine isolates. Phylogenomic analysis was initially carried out with all 4,208 chromosomal CDSs of Y. enterocolitica 8081 and 83 CDSs from virulence plasmid pYV. This resulted in division of the low-pathogenicity clade into further clades containing either strains that possessed pYV or those which had lost pYV. Because pYV can be readily lost during laboratory passage, the CDSs present in pYV were excluded from the subsequent analysis (8). These data showed that by Bayesian method-based phylogeny the Y. enterocolitica isolates fell into three distinct clades, high pathogenicity, low pathogenicity, and nonpathogenic, one lineage supported by Bayesian probabilities (P = 1.0) (shown in Fig. 2). These distinct clades formed irrespective of pYV, thus confirming that the presence of this plasmid is not the sole discriminatory factor for virulence in this species. These results also confirmed that the traditional biotyping is a useful method for distinguishing strains of Y. enterocolitica.
FIG. 2.
Bayesian phylogenomic relationships of all 92 strains following DNA-DNA microarray hybridization against the genome strain (*). Numerical values represent the probability (P) of support for each internal branch, where only values above 0.95 are considered robust. Serotypes and biotypes are designated ST and BT, respectively. A more detailed analysis will be presented elsewhere.
The highly pathogenic clade contained 8.7% of the strains analyzed (8/92) and was composed of eight biotype 1B strains isolated from patients in the United States. In the analysis, strain Y2 represents sequenced strain Y. enterocolitica 8081 1B. However, 1B strains of the same serotype did not cluster together.
The strains in the low-pathogenicity clade contained 51% of the strains analyzed (47/92) and were all of pathogenic biotype 2, 3, or 4. These strains had been isolated from either livestock (cattle, pigs, and sheep) sent for slaughter for human consumption or were human isolates from patients presenting to their GPs with diarrhea or collected during the IID study. Within the low-pathogenicity clade, the isolates have partially separated according to biotype and serotype. Twenty-five isolates comprising 12 biotype 3 serotype O:5,27 strains, 4 biotype 3 unserotypeable strains, 3 biotype 3 serotype O:5 strains, 2 biotype 3 serotype O-rough strains, 2 biotype 4 serotype O:5,27 strains, and 2 biotype 4 serotype O:3 strains clustered together. Nine isolates comprising one biotype 3 serotype O:5 strain and eight biotype 4 serotype O:3 strains clustered together. Thirteen isolates which were slightly more heterogeneous, comprising 10 biotype 3 serotype O:9 strains, 2 biotype 2 serotype O:9 strains, and 1 biotype 3 unserotypeable strain clustered together.
The third major clade that formed was the nonpathogenic clade. This clade contained 40.3% of the strains tested (35/92); 94.6% of these strains were biotype 1A strains. These strains were isolated from either livestock (cattle, pigs, and sheep) sent for slaughter for human consumption or were human clinical isolates from patients presenting to their GPs with diarrhea or collected during the IID study. Two strains of pathogenic biotypes Y213/02 (biotype 4 serotype O:3) and Y21/03 (biotype 1B serotype O:19) fell within this clade. These two isolates have been retyped and their biotypes confirmed; in addition, it was also noted that strain 21/03 was completely noninvasive in tissue culture (A. McNally, personal communication). Thus, this analysis has highlighted strains of particular interest that warrant further investigation. The distribution of isolates within the nonpathogenic clade was more heterogeneous than in the low-pathogenicity clade. As with the low-pathogenicity strains, human and livestock isolates were distributed throughout the clade and did not cluster according to source (host).
Although Y. enterocolitica 1A strains are traditionally considered to be nonpathogenic, three of the human isolates were recovered from patients presenting to their GPs with diarrhea or vomiting. Biotype 1A strains are generally thought to be avirulent because of the lack of virulence plasmid pYV and classical virulence determinants such as attachment-and-invasion locus (ail), myf, and ystA genes and a functional inv gene, which is typical of invasive isolates (45, 50). However, there is growing epidemiological evidence suggesting that 1A strains can cause disease. Y. enterocolitica biotype 1A strains have been isolated from patients presenting with gastrointestinal illness in Australia, New Zealand, South Africa, Chile, Switzerland, Canada, and the United States (6, 11, 47). Direct comparisons of a clinical 1A isolate against an environmental 1A isolate by subtractive hybridization identified 54 sequences that were present in the clinical isolate but absent from the environmental isolate (59). Genetic differences between asymptomatic and case isolates in this study could not be found. It may be that host immune factors play a role in the clinical outcome of infection rather than genetic differences between isolates. Together, this evidence suggests that additional analysis of biotype 1A isolates from the environment, livestock, and clinical settings is required before further assumptions about the pathogenicity of this biotype can be made. The Y. enterocolitica 8081 microarray represents more than 4,200 CDSs; however, one disadvantage of using single-genome microarrays is that it is only possible to detect CDSs that are present in the strain used to make the array.
Genes present exclusively in American highly pathogenic (biotype 1B) strains.
CDSs that were present in all eight highly pathogenic biotype 1B strains were identified with CDSs that had been designated as present by GACK analysis. GACK analysis designates CDSs present, marginal, or absent without the use of defined arbitrary cutoffs. These CDSs were present in all eight highly pathogenic U.S. strains and absent from or highly divergent in all other isolates. One hundred twenty-five chromosomal CDSs were identified by this method as detailed elsewhere (http://bugs.sgul.ac.uk/E-BUGS-36). Biotype 1B strains are highly pathogenic in mice, and genetic regions have previously been identified which have been shown to be unique to this biogroup. The list of 125 CDSs included regions of the previously characterized HPI (YE2611 to YE2622) (12, 13), chromosomal type III secretion system Ysa (YE3536 to YE3561) (26, 31), and a recently identified type II secretion system, Yts1 (YE3562 to YE3579) (36), all of which are unique to highly pathogenic biotype 1B strains. These results validate our microarray methodology and analysis. In addition, several of the 125 CDSs identified were insertion elements, phage-related proteins, and 32 hypothetical proteins (representing 40% of the total number of CDSs). Of the remaining high-pathogenicity-specific CDSs, seven were highlighted as particularly noteworthy. BLASTX (3) analysis of the seven previously uncharacterized proteins revealed that these CDSs had highly significant similarities to other bacterial virulence determinants and thus may contribute to the virulence of biotype 1B strains (Table 1). YE0126 showed amino acid similarity to hemophores present in Y. pestos, Y. pseudotuberculosis, and Erwinia carotovora (16); YE0344 was similar to the MceH protein in Klebsiella pneumoniae (16, 41); YE2408 was similar to hemolysin activator proteins from many other bacteria (16); YE4052 was similar to metalloproteases found in several other bacteria; and YE4088, which is encoded by a pseudogene in Y. pestos, was similar to sensor kinase proteins found in many other bacterial species (16). YE2447 showed similarity to OspG, a protein which is secreted by the Mxi-Spa type III secretion machinery in Shigella flexneri (10, 37), and YE3614 was similar to a probable SPI2 translocated effector protein found in Chromobacterium violaceum and phospholipases found in many bacteria (9). Effector proteins have been shown in many bacteria, including yersiniae, to be important in bacterial virulence. YopE and YopH are intracellular effectors encoded by virulence plasmid pYV of Y. enterocolitica. YopH is a phosphotyrosine phosphatase which is thought to protect Y. enterocolitica from phagocytosis, contribute to inhibition of cytokines produced by T cells, and prevent the ability of B cells to upregulate surface expression of the costimulatory molecule B7.2. YopE disrupts actin filaments (54) by depolymerization of actin stress fibers via activation of Rho GTPase (55). We believe that these additional CDSs could help to explain why biotype 1B strains exhibit high virulence in mice and warrant further investigation.
TABLE 1.
Selected CDSs that are present in all 1B biotype strains and absent from all other isolates
| Gene | Sequencing annotation | BLASTX annotation | Score (no. of bits) | E value |
|---|---|---|---|---|
| YE0126 | Probable hemophore | Extracellular heme acquisition hemophore | 95.5 | 9.0E − 19 |
| YE0344 | Putative HlyD family secretion protein | MceH protein (Klebsiella pneumoniae) | 427 | 1.0E − 188 |
| YE2408 | Putative hemolysin activator protein | Hemolysin activator protein | 711 | 0E + 00 |
| YE2447 | Putative type III secreted effector protein | OspG, secreted by the Mxi-Spa secretion machinery, function unknown | 367 | 5.0E − 101 |
| YE3614 | Putative lipase-acylhydrolase | Probable SPI2 translocated effector protein | 207 | 3.0E − 52 |
| YE4052 | Metalloprotease | Metalloprotease | 365 | 2.0E − 99 |
| YE4088 | Two-component system, sensor kinase | Sensor protein | 342 | 2.0E − 92 |
Genes present in highly pathogenic Y. enterocolitica 8081 1B and low-pathogenicity biotypes.
To identify further genes which may be important virulence factors, sequences that were present in all low-pathogenicity strains and the highly pathogenic Y. enterocolitica 8081 1B strain but absent from all nonpathogenic strains were identified. Only CDSs designated present by GACK analysis were included. Twenty-seven CDSs were identified as shown in Table 2. There were many hypothetical proteins and three transposases for the insertion element IS1667. Several CDSs which may contribute to the virulence of these low-pathogenicity strains were identified, such as YE2922, which shows amino acid similarity to a putative protease. Microbial proteases are widespread in many pathogenic bacteria, where they play a critical role in functions related to colonization and evasion of host immune defenses, acquisition of nutrients for growth and proliferation, facilitation of dissemination, and tissue damage during infection. Both Proteus mirabilis and Neisseria meningitidis have proteases which can cleave immunoglobulin A1 molecules, resulting in loss of mechanisms for the elimination of antigens (2). The YE3636-to-YE3643 region, which shares similarities with proteins encoding type IV secretion proteins and tight adherence protein A (TadA), which is present in many bacterial species, such as Vibrio vulnificus (18), was also absent from any nonpathogenic biotype. The YE3636-to-YE3643 region encodes proteins that are similar to both type II and type IV secretion system proteins and to tight adherence protein A from Haemophilus ducreyi. Type II secretion systems exclusively drive the translocation of exoproteins across the outer membrane. Many bacteria use this system to excrete various toxins and hydrolytic enzymes such as exotoxin A and elastase in Pseudomonas aeruginosa (24). Type IV secretion systems are known to be important for both transfer of DNA between species via conjugation and the translocation of effector proteins directly into target cells. CagA is an effector protein which is translocated by a type IV secretion system and has been shown to play a role in the virulence of Helicobacter pylori (15). Transfer of DNA between species can contribute to genome plasticity disseminating genes that can aid survival.
TABLE 2.
CDSs present in only all biotype 2 to 4 strains and control strain Y. enterocolitica 8081 1B
| Gene | Product |
|---|---|
| YE0321 | Conserved hypothetical protein |
| YE0583 | Conserved hypothetical protein |
| YE0733 | Nitrite extrusion protein |
| YE0799 | Putative aminotransferase |
| YE0965 | Putative oxidoreductase |
| YE1359 | Putative transposase for IS1667 |
| YE1360 | Hypothetical protein |
| YE1450 | Putative regulatory protein |
| YE1453 | Chaperone protein PsaB precursor |
| YE1870 | Putative transposase for IS1667 |
| YE1957 | Putative purine and pyrimidine permease |
| YE2423 | Putative exported protein |
| YE2531 | Hypothetical protein |
| YE2922 | Putative protease |
| YE3064 | Putative transposase for IS1667 |
| YE3328 | Hypothetical protein |
| YE3636 | Putative secretion system protein |
| YE3637 | Conserved hypothetical protein |
| YE3638 | Putative type II secretion protein |
| YE3639 | Putative membrane protein |
| YE3641 | Conserved hypothetical protein |
| YE3643 | Conserved hypothetical protein |
| YE3842 | Putative HipB transcriptional regulator |
| YE4001 | Putative MFS family transport protein |
| YE4002 | Hypothetical protein |
| YE4109 | Hypothetical protein |
| YE4110 | Hypothetical protein |
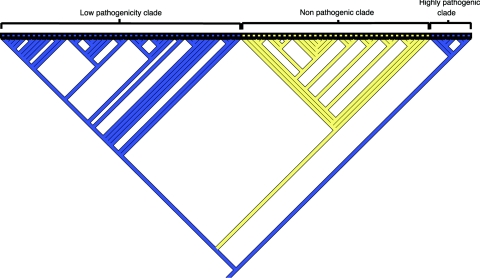
In addition, MacClade 4 software was used to identify further CDSs that were clade specific. This was achieved by tracing the character evolution within the hypothesized phylogenomic tree. Tracing displays on the branches the most parsimonious number of losses or gains of CDSs that have occurred in order for that particular clade to form. The tree was rooted against all U.S. 1B isolates. This method was particularly useful as it displayed genes which may not be present or absent from every single strain but still contributes to the genetic differences which cause clade formation. Because of the binary format of Mr Bayes in this analysis, both present and marginal GACK-designated CDSs were classified as present. Analysis revealed 82 CDSs (a full list is available at http://bugs.sgul.ac.uk/E-BUGS-36), of which 81 were absent or highly divergent from strains within the nonpathogenic clade. Thirty-three of these CDSs were hypothetical proteins, and 14 were associated with insertion elements or phage. Such sequences are often associated with virulence determinants. With MacClade, further CDSs were identified which could contribute to pathogenicity. YE1322, which has amino acid similarity to a putative repeats in toxin (RTX) family protein was shown to be absent from all nonpathogenic isolates. BLASTX analysis of this protein also showed similarities to putative virulence determinants found in Y. pestos and hemolysins. In addition, the YE1818-to-YE1820 region was absent from all 35 biotype 1A strains (Fig. 3 contains a MacClade analysis of YE1820), confirming previous studies showing that the attachment-and-invasion locus (Ail) protein (YE1820) is absent from nonpathogenic isolates (5, 45). The YE1818-to-YE1820 region encodes a transposase for the insertion sequence element IS1328, a hypothetical phage-related protein, and the well-characterized Ail protein (44).
FIG. 3.
Distribution of YE1820 among Y. enterocolitica strains. A parsimony-based gene analysis for determining the distribution of individual CDS YE1820 throughout the phylogenetic tree is shown. Strains from which YE1820 is absent are yellow, and strains in which YE1820 is present are blue. Strains in the low-pathogenicity clade and the high-pathogenicity clade all contain YE1820. YE1820 is absent from strains in the nonpathogenic clade.
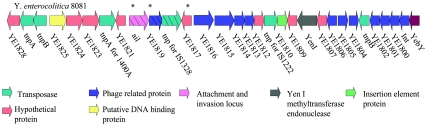
Closer examination of CDSs both up- and downstream of this region revealed a large number of transposases and phage-related proteins in sequenced strain Y. enterocolitica 8081 (Fig. 4). The majority of these regions were absent from biotypes 1A and biotypes 2 to 4.
FIG. 4.
Schematic of CDSs up- and downstream of ail in Y. enterocolitica 8081. The CDSs are written below with arrows denoting the direction of transcription. CDSs filled with hatched lines were absent from all 1A isolates. CDSs with asterisks were present in all biotype 2 to 4 isolates. The majority of CDSs from YE1799 to YE1827 were absent from both biotype 1A and 2 to 4 isolates.
Genes present in highly pathogenic Y. enterocolitica 8081 and nonpathogenic strains but absent from low-pathogenicity strains.
Genes that were present in nonpathogenic strains and highly pathogenic strain Y. enterocolitica 8081 1B and absent from low-pathogenicity isolates were identified. Twenty-four CDSs were present in all nonpathogenic strains and highly pathogenic strain Y. enterocolitica 8081 but absent from all low-pathogenicity strains (Table 3). By MacClade analysis, 102 CDSs were associated with the low-pathogenicity clade, of which 95 were absent, as detailed elsewhere (http://bugs.sgul.ac.uk/E-BUGS-36). A large proportion of the CDSs were involved in fatty acid or sugar metabolism and arsenic resistance. One notable cluster of genes is YE0896 to YE0910, which appears to encode several proteins involved in fatty acid biosynthesis. This gene cluster is conserved in a range of other bacteria, including E. carotovora and Escherichia coli. Within the YE0896-to-YE0910 region, YE0904, which is annotated as a putative thioesterase, was present in all biotype 1A and 1B strains but absent from biotype 2, 3, and 4 strains (Fig. 5 contains a MacClade analysis of YE0904). Several biochemical tests have been established to distinguish the various biotypes of Y. enterocolitica. One such test is the Tween esterase test, in which bacterial cells are spotted onto Tween 80 plates and incubated at 28°C for 24 h. A cloudy zone indicates the presence of this enzyme. The absence of this region from biotypes 2 to 4, which gives a negative result for this test, may explain this observation. CDSs which may be involved in the following biochemical tests were identified as follows: indole, YE1222; xylose, YE2511 and YE4122; trehalose, YE3690, YE3773, YE3774, and YE3775. However, these CDSs were present in all test isolates; thus, if they are involved in such biochemical tests it may be due to small deletions, point mutations, or frameshift mutations in the genes which result in loss of function. Such mutations would not be detected in our study because of the limitations of microarrays based on amplified products.
TABLE 3.
CDSs present only in all biotype 1A strains and control strain Y. enterocolitica 8081 1B
| Gene | Product |
|---|---|
| YE0306 | Putative hydrolase |
| YE0443 | Multidrug efflux protein |
| YE0467 | Hypothetical protein |
| YE0703 | AraC family transcriptional regulator |
| YE0752 | Superoxide dismutase (Cu-Zn) |
| YE0801 | ABC transporter, solute-binding component |
| YE0802 | ABC transporter, inner membrane component |
| YE0804 | ABC transporter, ATP-binding component |
| YE0896 | Conserved hypothetical protein |
| YE0897 | Putative acyltransferase |
| YE0900 | Putative inner membrane protein |
| YE0902 | Putative dehydratase |
| YE0903 | Putative glycosyltransferase |
| YE0904 | Putative thioesterase |
| YE0906 | Putative inner membrane protein |
| YE0908 | Putative beta-ketoacyl-(ACP)a synthase |
| YE0909 | Putative dehydratase |
| YE0910 | 3-Oxoacyl-(acyl carrier protein) reductase |
| YE1143 | Putative acetyltransferase |
| YE2109 | Hypothetical protein |
| YE2286 | Acyl-(acyl carrier protein)-UDP-N-acetylglucosamine O-acyltransferase |
| YE2812 | Putative formate transporter |
| YE2823 | Putative membrane protein |
| YE3528 | Putative membrane protein |
ACP, acyl carrier protein.
FIG. 5.
Distribution of YE0904 among Y. enterocolitica strains. A parsimony-based gene analysis for determining the distribution of individual CDS YE0904 throughout the phylogenetic tree is shown. Strains from which YE0904 is absent are yellow, and strains in which YE0904 is present are blue. Strains in the nonpathogenic clade and the high-pathogenicity clade all contain YE0904. YE0904 is absent from strains in the low-pathogenicity clade.
YE0752, which encodes a superoxide dismutase cofactored by copper and zinc, was also absent from all low-pathogenicity strains but present in highly pathogenic and nonpathogenic isolates. Analysis of the completed but unpublished genome sequence of Y. enterocolitica strain 8081 revealed that it is the only copper-zinc superoxide dismutase in Y. enterocolitica 8081. Copper-zinc superoxides are located in the periplasm or anchored by lipids in the outer envelope (57) and have been shown to be important in the protection of bacterial cells such as N. meningitidis from the action of phagocytic cells (22). pYV encodes proteins that aid in resistance to phagocytosis during infection; as nonpathogenic strains do not possess this plasmid, the presence of this superoxide dismutase may contribute to protecting the bacteria against the action of phagocytic cells. In addition, YE0444, encoding a multidrug efflux protein, and the YE0801-to-YE0804 region, which encodes several ATP-binding cassette proteins, were also absent from low-pathogenicity strains.
Evolution of Y. enterocolitica.
In evolutionary terms, Y. enterocolitica is considered to be distantly related to Y. pseudotuberculosis and Y. pestos. Indeed, it has been suggested that Y. enterocolitica is as closely related to the other pathogenic yersiniae as E. coli is to Salmonella species (48). The data obtained from microarray analysis were used to generate a phylogenetic tree based on a Bayesian method-based algorithm incorporating a gamma distribution to model rate heterogeneity across the genome. Each horizontal line on this tree represents the average number of gains or losses of a gene per gene of the genome strain; thus, the genetic distances among the strains in the three clades can be measured. Determining the exact evolutionary order of speciation or subspeciation for the three clades of Y. enterocolitica is not possible without an appropriate outgroup (a species which unequivocally evolved before the Y. enterocolitica complex). However, a brief analysis shows that low-pathogenicity and nonpathogenic strains were the most closely related genetically (approximately 0.19 gain or loss of a gene per gene) and the nonpathogenic clade was more closely related genetically to the highly pathogenic clade than the low-pathogenicity strains (approximately 0.33 compared to 0.37 gain or loss of a gene per gene). With the assumption that all genetic distances should be reasonably equidistant from the common ancestor (a molecular clock type of assumption), the highly pathogenic clade is a direct descendant of the most ancient common ancestor, and this could imply that the formation of mildly pathogenic and nonpathogenic strains resulted from a biogeographic movement between the New World and the Old World. The alternative rooting is presented herein and provides the most parsimonious evolutionary explanations, where the common ancestor is slightly more compatible with an Old World origin and is also the progenitor of the virulence plasmid, which would then be lost on a single occasion in the ancestor to the nonpathogenic and highly pathogenic clades.
In either scenario, a pathogenic phenotype could be present in the ancestral Y. enterocolitica strain where pathogenic determinants, for example, ail, are retained in both highly pathogenic and mildly pathogenic clades but lost in the formation of the nonpathogenic clade. Furthermore, the cumulative loss of both the virulence plasmid and the pathogenicity islands would then have resulted in the nonpathogenic strain phenotype. Alternatively, islands of pathogenicity could have been acquired independently through horizontal transmission and the large number of phage-related proteins surrounding ail for example, which were revealed during the sequencing of the Y. enterocolitica 8081 1B strain. The order of evolution of these three Y. enterocolitica clades is important because it provides a model for the evolution of pathogenicity and how over time an organism may become more or less virulent. Biotype 1B strains are termed New World strains as they are rarely reported outside of the United States. It is possible to speculate that, because of a shift in geographical location, the biotype 1A-1B intermediary was able to acquire virulence determinants from its new environment, resulting in the highly pathogenic 1B biotype, which is genetically much more diverse in comparison with the other biotypes. The split between 1A and 1B strains appears to be ancient, and is unlikely to be due to recent human migratory movements to North America and possibly is more likely due to ancient geographical land mass movements.
Our results provide the first detailed whole-genome comparison of Y. enterocolitica. A microarray was constructed with duplicate reporter elements representing all chromosomal and plasmid-predicted (4,291) CDSs of sequenced strain Y. enterocolitica 8081 1B. DNA microarray analysis of 94 biotype 1A, 1B, and 2 to 4 strains revealed extensive genome diversity within Y. enterocolitica species. The core minimal gene set in Y. enterocolitica was 894 CDSs. Bayesian method-based algorithms were then used alongside tracing of character evolution to reveal the distinct population structure of the 94 Y. enterocolitica strains. Based upon DNA-DNA hybridizations and 16S rRNA data, Neubauer et al. proposed the division of Y. enterocolitica into two subspecies, Y. enterocolitica subsp. enterocolitica for strains of American origin and Y. enterocolitica subsp. palearctica for strains of European origin (46). However, given the small core genome, unequivocal distinction between clades and the genetic distance between them, we believe that our data confirm that Y. enterocolitica is a highly heterogeneous species and adds weight to the case for the existence of three subspecies.
This method has also allowed a large proportion of CDSs to be identified that contribute to the formation of each clade and thereby identifying several new potential virulence determinants, which may help to explain the differences in pathogenicity observed in the various biotypes of this species. The approach described in this study provides a methodological prototype of robust phylogenomics that should be applicable to the study of other microbes.
Acknowledgments
We acknowledge BμG@S (the Bacterial Microarray Group at St George's University of London) for advice and supply of the microarray and The Wellcome Trust for funding the multicollaborative microbial pathogen microarray facility under its Functional Genomics Resources Initiative and The Pathogen Sequencing Unit at The Wellcome Trust Sanger Institute, Cambridge, for sequencing and annotation of Y. enterocolitica 8081. We acknowledge Ozan Gundogdu for excellent technical support and Mike Prentice, Alan McNally, Diane Newell, and Nicholas Thomson for stimulating discussions. Human and animal strains used in this study were donated by Virginia Miller (Washington University School of Medicine), Tom Cheasty (Health Protection Agency, London, United Kingdom), and Diane Newell (Veterinary Laboratory Agency, United Kingdom).
This work was funded by the Department for Environment, Food and Rural Affairs, United Kingdom.
REFERENCES
- 1.Achtman, M., K. Zurth, G. Morelli, G. Torrea, A. Guiyoule, and E. Carniel. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 96:14043-14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almogren, A., B. W. Senior, L. M. Loomes, and M. A. Kerr. 2003. Structural and functional consequences of cleavage of human secretory and human serum immunoglobulin A1 by proteinases from Proteus mirabilis and Neisseria meningitidis. Infect. Immun. 71:3349-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Autenrieth, I. B., R. Reissbrodt, E. Saken, R. Berner, U. Vogel, W. Rabsch, and J. Heesemann. 1994. Desferrioxamine-promoted virulence of Yersinia enterocolitica in mice depends on both desferrioxamine type and mouse strain. J. Infect. Dis. 169:562-567. [DOI] [PubMed] [Google Scholar]
- 5.Beer, K. B., and V. L. Miller. 1992. Amino acid substitutions in naturally occurring variants of ail result in altered invasion activity. J. Bacteriol. 174:1360-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bissett, M. L., C. Powers, S. L. Abbott, and J. M. Janda. 1990. Epidemiologic investigations of Yersinia enterocolitica and related species: sources, frequency, and serogroup distribution. J. Clin. Microbiol. 28:910-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bottone, E. J. 1999. Yersinia enterocolitica: overview and epidemiologic correlates. Microbes Infect. 1:323-333. [DOI] [PubMed] [Google Scholar]
- 8.Bottone, E. J. 1997. Yersinia enterocolitica: the charisma continues. Clin. Microbiol. Rev. 10:257-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brazilian National Genome Project Consortium. 2003. The complete genome sequence of Chromobacterium violaceum reveals remarkable and exploitable bacterial adaptability. Proc. Natl. Acad. Sci. USA 100:11660-11665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buchrieser, C., P. Glaser, C. Rusniok, H. Nedjari, H. D'Hauteville, F. Kunst, P. Sansonetti, and C. Parsot. 2000. The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol. Microbiol. 38:760-771. [DOI] [PubMed] [Google Scholar]
- 11.Burnens, A. P., A. Frey, and J. Nicolet. 1996. Association between clinical presentation, biogroups and virulence attributes of Yersinia enterocolitica strains in human diarrhoeal disease. Epidemiol. Infect. 116:27-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carniel, E. 1999. The Yersinia high-pathogenicity island. Int. Microbiol. 2:161-167. [PubMed] [Google Scholar]
- 13.Carniel, E., I. Guilvout, and M. Prentice. 1996. Characterization of a large chromosomal “high-pathogenicity island” in biotype 1B Yersinia enterocolitica. J. Bacteriol. 178:6743-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carter, P. B. 1975. Pathogenicity of Yersinia enterocolitica for mice. Infect. Immun. 11:164-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Censini, S., C. Lange, Z. Xiang, J. E. Crabtree, P. Ghiara, M. Borodovsky, R. Rappuoli, and A. Covacci. 1996. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. USA 93:14648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chain, P. S., E. Carniel, F. W. Larimer, J. Lamerdin, P. O. Stoutland, W. M. Regala, A. M. Georgescu, L. M. Vergez, M. L. Land, V. L. Motin, R. R. Brubaker, J. Fowler, J. Hinnebusch, M. Marceau, C. Medigue, M. Simonet, V. Chenal-Francisque, B. Souza, D. Dacheux, J. M. Elliott, A. Derbise, L. J. Hauser, and E. Garcia. 2004. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 101:13826-13831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Champion, O. L., M. W. Gaunt, O. Gundogdu, A. Elmi, A. A. Witney, J. Hinds, N. Dorrell, and B. W. Wren. 2005. Comparative phylogenomics of the food-borne pathogen Campylobacter jejuni reveals genetic markers predictive of infection source. Proc. Natl. Acad. Sci. USA 102:16043-16048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen, C.-Y., K.-M. Wu, Y.-C. Chang, C.-H. Chang, H.-C. Tsai, T.-L. Liao, Y.-M. Liu, H.-J. Chen, A. B.-T. Shen, J.-C. Li, T.-L. Su, C.-P. Shao, C.-T. Lee, L.-I. Hor, and S.-F. Tsai. 2003. Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res. 13:2577-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dorrell, N., S. J. Hinchliffe, and B. W. Wren. 2005. Comparative phylogenomics of pathogenic bacteria by microarray analysis. Curr. Opin. Microbiol. 8:620-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dorrell, N., J. A. Mangan, K. G. Laing, J. Hinds, D. Linton, H. Al-Ghusein, B. G. Barrell, J. Parkhill, N. G. Stoker, A. V. Karlyshev, P. D. Butcher, and B. W. Wren. 2001. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 11:1706-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doyle, M. P. 1985. Food-borne pathogens of recent concern. Annu. Rev. Nutr. 5:25-41. [DOI] [PubMed] [Google Scholar]
- 22.Dunn, K. L. R., J. L. Farrant, P. R. Langford, and J. S. Kroll. 2003. Bacterial [Cu,Zn]-cofactored superoxide dismutase protects opsonized, encapsulated Neisseria meningitidis from phagocytosis by human monocytes/macrophages. Infect. Immun. 71:1604-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dziejman, M., E. Balon, D. Boyd, C. M. Fraser, J. F. Heidelberg, and J. J. Mekalanos. 2002. Comparative genomic analysis of Vibrio cholerae: genes that correlate with cholera endemic and pandemic disease. Proc. Natl. Acad. Sci. USA 99:1556-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Filloux, A., G. Michel, and M. Bally. 1998. GSP-dependent protein secretion in gram-negative bacteria: the Xcp system of Pseudomonas aeruginosa. FEMS Microbiol. Rev. 22:177-198. [DOI] [PubMed] [Google Scholar]
- 25.Fitzgerald, J. R., D. E. Sturdevant, S. M. Mackie, S. R. Gill, and J. M. Musser. 2001. Evolutionary genomics of Staphylococcus aureus: insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. Proc. Natl. Acad. Sci. USA 98:8821-8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foultier, B., P. Troisfontaines, D. Vertommen, M. N. Marenne, M. Rider, C. Parsot, and G. R. Cornelis. 2003. Identification of substrates and chaperone from the Yersinia enterocolitica 1B Ysa type III secretion system. Infect. Immun. 71:242-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fredriksson-Ahomaa, M., S. Hallanvuo, T. Korte, A. Siitonen, and H. Korkeala. 2001. Correspondence of genotypes of sporadic Yersinia enterocolitica bioserotype 4/O:3 strains from human and porcine sources. Epidemiol. Infect. 127:37-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fredriksson-Ahomaa, M., and H. Korkeala. 2003. Low occurrence of pathogenic Yersinia enterocolitica in clinical, food, and environmental samples: a methodological problem. Clin. Microbiol. Rev. 16:220-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gemski, P. L. J., and T. Casey. 1980. Plasmid associated with pathogenicity and calcium dependency of Yersinia enterocolitica. Infect. Immun. 27:682-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gourdon, F., J. Beytout, A. Reynaud, J. P. Romaszko, D. Perre, P. Theodore, H. Soubelet, and J. Sirot. 1999. Human and animal epidemic of Yersinia enterocolitica O:9, 1989-1997, Auvergne, France. Emerg. Infect. Dis. 5:719-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haller, J. C., S. Carlson, K. J. Pederson, and D. E. Pierson. 2000. A chromosomally encoded type III secretion pathway in Yersinia enterocolitica is important in virulence. Mol. Microbiol. 36:1436-1446. [DOI] [PubMed] [Google Scholar]
- 32.Hinchliffe, S. J., K. E. Isherwood, R. A. Stabler, M. B. Prentice, A. Rakin, R. A. Nichols, P. C. F. Oyston, J. Hinds, R. W. Titball, and B. W. Wren. 2003. Application of DNA microarrays to study the evolutionary genomics of Yersinia pestis and Yersinia pseudotuberculosis. Genome Res. 13:2018-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hinds, J., A. A. Witney, and J. K. Vass. 2002. Microarray design for bacterial genomes. Academic Press, London, United Kingdom.
- 34.Hinds, J., K. G. Laing, J. A. Mangan, and P. D. Butcher. 2002. Glass slide microarrays for bacterial genomes, p. 83-99 In B. Wren and N. Dorrell (ed.), Methods in microbiology: functional microbial genomics. Academic Press, Inc., New York, N.Y.
- 35.Huelsenbeck, J. P., and F. Ronquist. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 36.Iwobi, A., J. Heesemann, E. Garcia, E. Igwe, C. Noelting, and A. Rakin. 2003. Novel virulence-associated type II secretion system unique to high-pathogenicity Yersinia enterocolitica. Infect. Immun. 71:1872-1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin, Q., Z. Yuan, J. Xu, Y. Wang, Y. Shen, W. Lu, J. Wang, H. Liu, J. Yang, F. Yang, X. Zhang, J. Zhang, G. Yang, H. Wu, D. Qu, J. Dong, L. Sun, Y. Xue, A. Zhao, Y. Gao, J. Zhu, B. Kan, K. Ding, S. Chen, H. Cheng, Z. Yao, B. He, R. Chen, D. Ma, B. Qiang, Y. Wen, Y. Hou, and J. Yu. 2002. Genome sequence of Shigella flexneri 2a: insights into pathogenicity through comparison with genomes of Escherichia coli K12 and O157. Nucleic Acids Res. 30:4432-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones, T. F. 2003. From pig to pacifier: chitterling-associated yersiniosis outbreak among black infants. Emerg. Infect. Dis. 9:1007-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kato-Maeda, M., J. T. Rhee, T. R. Gingeras, H. Salamon, J. Drenkow, N. Smittipat, and P. M. Small. 2001. Comparing genomes within the species Mycobacterium tuberculosis. Genome Res. 11:547-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim, C. C., E. A. Joyce, K. Chan, and S. Falkow. 2002. Improved analytical methods for microarray-based genome-composition analysis. Genome Biol. 3:1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lagos, R., J. E. Villanueva, and O. Monasterio. 1999. Identification and properties of the genes encoding microcin E492 and its immunity protein. J. Bacteriol. 181:212-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maddison, D. R., and W. P. Maddison. 2001. MacClade 4: analysis of phylogeny and character evolution. Version 4.03. Sinauer Associates, Sunderland, Mass.
- 43.McNally, A., T. Cheasty, C. Fearnley, R. W. Dalziel, G. A. Paiba, G. Manning, and D. G. Newell. 2004. Comparison of the biotypes of Yersinia enterocolitica isolated from pigs, cattle and sheep at slaughter and from humans with yersiniosis in Great Britain during 1999-2000. Lett. Appl. Microbiol. 39:103-108. [DOI] [PubMed] [Google Scholar]
- 44.Miller, V. L., and S. Falkow. 1988. Evidence for two genetic loci in Yersinia enterocolitica that can promote invasion of epithelial cells. Infect. Immun. 56:1242-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller, V. L., J. J. Farmer III, W. E. Hill, and S. Falkow. 1989. The ail locus is found uniquely in Yersinia enterocolitica serotypes commonly associated with disease. Infect. Immun. 57:121-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neubauer, H., S. Aleksic, A. Hensel, E. J. Finke, and H. Meyer. 2000. Yersinia enterocolitica 16S rRNA gene types belong to the same genospecies but form three homology groups. Int. J. Med. Microbiol. 290:61-64. [DOI] [PubMed] [Google Scholar]
- 47.Noble, M. A., R. L. Barteluk, H. J. Freeman, R. Subramaniam, and J. B. Hudson. 1987. Clinical significance of virulence-related assay of Yersinia species. J. Clin. Microbiol. 25:802-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parkhill, J. T., and N. R. Thompson. 2004. The Yersinia pestis chromosome, p. 1-11. In E. Carniel and B. J. Hinnebusch (ed.), Yersinia: molecular and cellular biology. Horizon Bioscience, Norfolk, United Kingdom.
- 49.Pelludat, C., M. Hogardt, and J. Heesemann. 2002. Transfer of the core region genes of the Yersinia enterocolitica WA-C serotype O:8 high-pathogenicity island to Y. enterocolitica MRS40, a strain with low levels of pathogenicity, confers a yersiniabactin biosynthesis phenotype and enhanced mouse virulence. Infect. Immun. 70:1832-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pierson, D. E., and S. Falkow. 1990. Nonpathogenic isolates of Yersinia enterocolitica do not contain functional inv-homologous sequences. Infect. Immun. 58:1059-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Portnoy, D. A., S. L. Moseley, and S. Falkow. 1981. Characterisation of plasmids and plasmid-associated determinants of Yersinia enterocolitica pathogenesis. Infect. Immun. 31:775-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prentice, M., D. Cope, and R. A. Swann. 1991. The epidemiology of Yersinia enterocolitica infection in the British Isles 1983-1988. Contrib. Microbiol. Immunol. 12:17-25. [PubMed] [Google Scholar]
- 53.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 54.Rosqvist, R., A. Forsberg, M. Rimpilainen, T. Bergman, and H. Wolf-Watz. 1990. The cytotoxic protein YopE of Yersinia obstructs the primary defence. Mol. Microbiol. 4:657-667. [DOI] [PubMed] [Google Scholar]
- 55.Rosqvist, R., A. Forsberg, and H. Wolf-Watz. 1991. Intracellular targeting of the Yersinia YopE cytotoxin in mammalian cells induces actin microfilament disruption. Infect. Immun. 59:4562-4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 57.St John, G., and H. M. Steinman. 1996. Periplasmic copper-zinc superoxide dismutase of Legionella pneumophila: role in stationary-phase survival. J. Bacteriol. 178:1578-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tauxe, R. V., J. Vandepitte, G. Wauters, S. M. Martin, V. Goossens, P. De Mol, R. Van Noyen, and G. Thiers. 1987. Yersinia enterocolitica infections and pork: the missing link. Lancet i:1129-1132. [DOI] [PubMed] [Google Scholar]
- 59.Tennant, S. M., T. H. Grant, and R. M. Robins-Browne. 2003. Pathogenicity of Yersinia enterocolitica biotype 1A. FEMS Immunol. Med. Microbiol. 38:127-137. [DOI] [PubMed] [Google Scholar]
- 60.Wheeler, J. G., D. Sethi, J. M. Cowden, P. G. Wall, L. C. Rodrigues, D. S. Tompkins, M. J. Hudson, and P. J. Roderick. 1999. Study of infectious intestinal disease in England: rates in the community, presenting to general practice, and reported to national surveillance. BMJ 318:1046-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zink, D. L., J. C. Feeley, J. G. Wells, C. Vanderzant, J. C. Vickery, W. D. Roof, and G. A. O'Donovan. 1980. Plasmid-mediated tissue invasiveness in Yersinia enterocolitica. Nature 283:224-226. [DOI] [PubMed] [Google Scholar]