Abstract
Tautomycin (TTM), a potent protein phosphatase inhibitor, consists of a polyketide chain containing a spiroketal moiety and an acyl chain bearing a dialkylmaleic anhydride structure. PCR using degenerate primers was used to clone genes from Streptomyces spiroverticillatus for formation of the methoxymalonyl-acyl carrier protein. This locus was found to contain five genes (ttmC, ttmA, ttmD, ttmB, and ttmE), one of which was used as a probe to clone the 110-kb TTM biosynthetic gene cluster. The involvement of the ttmA gene in TTM biosynthesis was confirmed by gene inactivation and mutation complementation experiments.
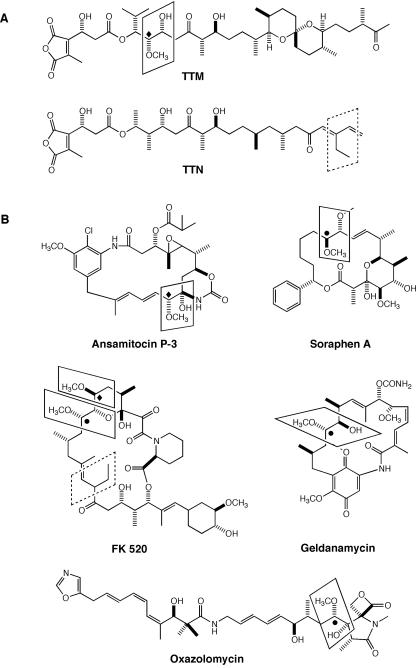
Tautomycin (TTM) was first isolated from Streptomyces spiroverticillatus in 1987 (3), and its structure and relative and absolute stereochemistry were established on the basis of chemical degradation and spectroscopic evidence (5, 23, 24). TTM consists of a polyketide chain containing a spiroketal moiety and an acyl chain bearing a dialkylmaleic anhydride unit (Fig. 1A). The latter moiety under neutral conditions exists as two interconverting anhydride and diacid forms in approximately a 5:4 ratio (3, 5), which is reflected in the name of this polyketide. TTM was originally found to exhibit potent antifungal activity and induce morphological change (bleb formation) in human leukemia cells (12). Bleb formation was correlated with the inhibition of protein phosphatases (PPs), leading to the eventual finding that TTM is a potent inhibitor of both PP1 and PP2A with a 50% inhibitory concentration (IC50) of 22 to 32 nM in weak preference to PP1 (11, 18, 22). The closely related tautomycetin (TTN) (Fig. 1A) (4, 6) exhibits nearly 40-times-higher specificity for PP1 (IC50 = 1.6 nM) than PP2A (IC50 = 62 nM) (14, 18). PP1 and PP2A are the catalytic subunits of two of the four major protein Ser/Thr phosphatase complexes that regulate diverse cellular events such as cell division, transcription, translation, muscle contraction, glycogen synthesis, and neuronal signaling in eukaryotic cells. Because of their ability to readily permeate cells, small-molecule inhibitors of PPs have become useful tools for studying such intracellular events in vivo and represent leads to discover drugs for the treatment of several human diseases (8, 18, 19).
FIG. 1.
(A) Structures of TTM and TTN and (B) selected examples of polyketides that incorporate methoxymalonate as an extender unit. The methoxymalonate or ethylmalonate-derived moieties are highlighted by solid- or dashed-line boxes, respectively. The methoxymalonate extender is predicted to be incorporated into the polyketide with either inversion (⧫) or retention (•) of configuration on the assumption that (R)-methoxymalonyl-ACP is the common substrate for their biosynthesis.
Assembly of the right half of TTM and TTN from short carboxylic acid precursors can be readily envisaged according to the current type I polyketide synthase (PKS) paradigm with the TTM PKS featuring a methoxymalonyl-acyl carrier protein (ACP) extender unit and the TTN PKS featuring an ethylmalonyl coenzyme A (CoA) extender unit (Fig. 1A) (25). Here we report the amplification of putative β-hydroxyacyl-CoA dehydrogenase (HADH) and acyl-CoA dehydrogenase (ADH) genes from the TTM-producing S. spiroverticillatus strain by PCR, utilization of the resulting ADH fragment as a probe to clone the methoxymalonyl-ACP biosynthesis locus, and confirmation of the necessity of this locus for TTM biosynthesis by gene replacement and complementation experiments.
Cloning of putative HADH and ADH genes for methoxymalonyl-ACP biosynthesis from S. spiroverticillatus by PCR.
Because it was inexpedient to attempt to identify the TTM gene cluster by correlation with the numerous ketosynthase loci cloned by PCR from S. spiroverticillatus using conventional primers (13, 15) (unpublished work), we turned to another cloning strategy. The biosynthetic gene clusters of PKSs that incorporate methoxymalonate as an extender unit have been cloned and characterized (10, 16, 26-30) (Fig. 1B). Two highly conserved regions, VLGAGVMG and GRKSGRGF in HADH and QGMAAWTV and DAKLMEII in ADH, both of which are products of the known methoxymalonyl-ACP-specific genes (10, 16, 26-30), were used to design primers FP1 (5′-GTC CTG GGC GCC GGS GTS ATG GG-3′) and RP1 (5′-GAA GCC GCG GCC SGA CTT SCG NCC-3′) for HADH and FP2 (5′-CAG GGC ATG GCC GCS TGG ACS GT-3′) and RP2 (5′-GAT GAT CTC CAT SAG CTT SGC RTC-3′) (N, G/A/T/C; R, A/G; S, C/G) for ADH by using the CODEHOP (consensus-degenerate hybrid primer) strategy (17). Distinct products with the expected size (0.8 kb) were amplified by PCR from S. spiroverticillatus genomic DNA using typical conditions for high-GC-content genes (9) with both pairs and cloned into pGEM-T Easy (Promega, Madison, WI) to yield pBS6001 (for HADH) and pBS6002 (for ADH), respectively. Both plasmids were sequenced. While the PCR-amplified HADH product exhibited lower homology to HADHs involved in methoxymalonyl-ACP biosynthesis, such as GdmK (28% identity) (16) and FkbK (27% identity) (27), than those for phenylacetate catabolism (PhaC [NP_628022, 41% identity] from Streptomyces coelicolor or PaaH [NP_825536, 41% identity] from Streptomyces avermitilis), the PCR-amplified ADH fragment (253 amino acids) was most similar to known ADHs for methoxymalonyl-ACP biosynthesis, such as GdmI (73% identity) (16) and FkbI (61% identity) (27).
Cloning and sequencing of a methoxymalonyl-ACP biosynthesis locus from S. spiroverticillatus.
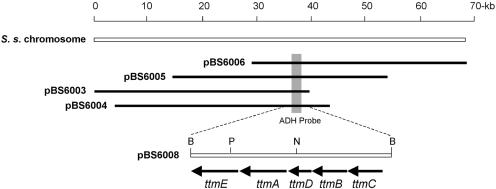
An internal 0.55-kb ADH fragment was amplified from pBS6002 by PCR and used directly as a probe to screen the S. spiroverticillatus cosmid library prepared in SuperKos1 (a modified SuperCos1 [Stratagene, La Jolla, CA] that lacks the neomycin resistance marker) by standard methods (9, 20). Of the 5,000 colonies screened by colony hybridization, 12 positive clones were identified and confirmed by PCR and Southern analysis. Restriction mapping and end sequencing revealed that these overlapping cosmids covered approximately 70 kb of DNA as exemplified by pBS6003, pBS6004, pBS6005, and pBS6006 (Fig. 2). Chromosome walking was later used to extend the size of the TTM cluster to 110 kb (not shown).
FIG. 2.
A 70-kb DNA region from S. spiroverticillatus (S. s.) that harbors a part of the tautomycin biosynthetic gene cluster as represented by four overlapping cosmids and the genetic organization of the methoxymalonyl-ACP biosynthetic locus. ADH probe, the PCR-amplified ADH fragment; B, BamHI; N, NcoI; P, PstI.
The 4.9-kb BamHI fragment that hybridized to the PCR-amplified ADH probe was subcloned from pBS6004 as a BamHI restriction fragment into the same site of pBluescript II SK(+) (Stratagene) to give pBS6008. DNA sequencing revealed five open reading frames, designated ttmA, ttmB, ttmC, ttmD, and ttmE (Fig. 2). The ttmA gene, which contains the PCR-amplified ADH fragment, encodes an ADH that is highly homologous to GdmI (74% identity) (16), FkbI (63% identity) (27), Asm15 (60% identity) (28), SorE (41% identity) (10, 30), and OzmE (62% identity) (29), respectively. The ttmB gene encodes an HADH, distinct from one that was amplified from the genomic DNA by PCR, that shows high sequence homology to HADHs from known methoxymalonyl-ACP loci, such as GdmK (75% identity) (16), FkbK (59% identity) (27), Asm13 (15% identity) (28), SorD (35% identity) (10, 30), and OzmG (62% identity) (29). The deduced gene products of ttmC, ttmD, and ttmE share high sequence homology to three other enzymes, an O-methyl transferase (TtmC), an ACP (TtmD), and a protein with no functional homologs that is proposed to play a role in glyceryl-ACP formation (TtmE), all of which are common in known methoxymalonyl-ACP biosynthesis loci (16, 26-30).
Inactivation of ttmA by gene replacement and complementation of the ttmA::aac(3)IV mutation by ttmA expression.
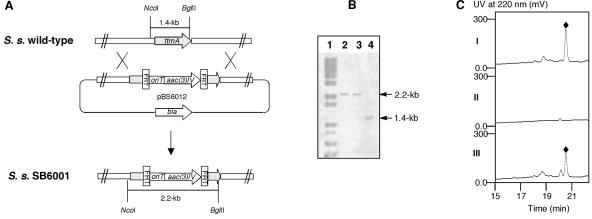
To confirm that the cloned methoxymalonyl-ACP locus is involved in TTM biosynthesis, ttmA was inactivated by gene replacement using REDIRECT Technology (7). The mutated cosmid, pBS6012, in which an internal 414-bp fragment of ttmA was replaced with the apramycin (Apr) resistance aac(3)IV/oriT gene cassette, was introduced into S. spiroverticillatus by conjugation using protocols for genetic manipulation of this organism (unpublished work). Exconjugants that were resistant to Apr (30 μg/ml, International Streptomyces Project 4 [ISP-4] agar plates freshly supplemented with 10 mM MgCl2, incubated several days at 28°C) were selected as the desired double-crossover gene replacement mutants and named S. spiroverticillatus SB6001. Genomic DNAs from both the S. spiroverticillatus wild-type and SB6001 mutant strains were digested with NcoI and BglII and probed with the 1.4-kb NcoI-BglII fragment of ttmA. While the wild-type strain yielded a distinct signal at 1.4 kb (Fig. 3A), this fragment was shifted to 2.2 kb in the SB6001 mutant strain as would be predicted for the replacement of the internal 414-bp fragment of ttmA by the aac(3)IV/oriT cassette (Fig. 3B).
FIG. 3.
ttmA inactivation by gene replacement and ΔttmA complementation. (A) Construction of the ttmA gene replacement mutant and restriction maps of S. spiroverticillatus wild-type and SB6001 mutant strains showing predicted fragment sizes upon BglII and NcoI digestion. (B) Southern analysis of the wild-type (lane 4) and SB6001 (lanes 2 and 3 are two individual isolates) genomic DNAs digested with BglII and NcoI using the 1.4-kb ttmA fragment as a probe. Lane 1, molecular weight standard. (C) HPLC analysis of tautomycin (TTM) production in wild-type (I) and recombinant strains SB6001 (II) and SB6002 (III). TTM, ⧫.
To complement the ttmA::aac(3)IV mutation in the SB6001 strain, the ttmA expression construct pBS6009 was generated in the integrative plasmid pSET152 (1) by placing the ErmE* promoter in front of the ttmA gene to ensure efficient expression as follows. A 1.6-kb NcoI-PstI DNA fragment from pBS6008 containing the entire ttmA gene was treated with the Klenow DNA polymerase large fragment and ligated into the HincII site of pWHM79 that harbors the ErmE* promoter upstream (21) to yield pBS6010. Upon digestion with EcoRI and HindIII, the 2.0-kb ErmE*::ttmA fragment was isolated from pBS6010, blunt ended with the Klenow DNA polymerase large fragment, and ligated into the EcoRV site of pSET152 to yield pBS6011. Since SB6001 carries aac(3)IV, the 1.5-kb neomycin resistance gene (neo) was amplified from SuperCos1 and ligated into the EcoRI site of pBS6011 to furnish pBS6009 with an alternative marker for selection in SB6001. The latter was introduced into SB6001 by conjugation to yield Apr-resistant strain SB6002, in which pBS6009 is integrated into the S. spiroverticillatus chromosome via the φC31 att site (1) and the expression of ttmA is under the control of the constitutive ErmE* promoter.
Characterization of TTM production in S. spiroverticillatus SB6001 and SB6002.
Fermentations of S. spiroverticillatus SB6001 and SB6002 were carried out under identical conditions with the wild-type strain as a positive control. Optimized TTM production in the wild-type strain ranged from 10 to 15 mg/liter. The identity of the isolated TTM was confirmed by mass spectrometry analysis, yielding the characteristic molecular ions (m/e for [M+H]+) of 767.1, consistent with the molecular formula of C41H66O13, and 1H and 13C nuclear magnetic resonance analyses, affording spectroscopic data that are identical to those reported in the literature (5). High-pressure liquid chromatography analysis of the fermentation cultures showed that inactivation of ttmA completely abolished TTM production in SB6001 and TTM production was partially restored (∼60% to that of the wild type) by expressing a functional copy of ttmA in trans (i.e., SB6002) (Fig. 3C).
The hypothesis that the biosynthesis of methoxymalonyl-ACP starts from a glycolytic pathway intermediate (2, 26, 27) raises an interesting stereochemistry question. Since glycolytic pathway intermediates are known to have a d (i.e., 2R) configuration, a corollary of this hypothesis would be the prediction of (R)-methoxymalonyl-ACP as the sole extender unit for the biosynthesis of all methoxymalonate-containing polyketides, regardless of the stereochemistry of the final methoxymalonate-derived unit in the natural products. The PKS modules that incorporate the (R)-methoxymalonyl-ACP unit must, therefore, control the stereochemistry of the methoxymalonate-derived units in the resultant natural products, as exemplified by the TTM PKS with the retention of configuration for TTM biosynthesis and the oxazolomycin PKS with the inversion of configuration for oxazolomycin biosynthesis (29), respectively (Fig. 1).
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper has been deposited in the GenBank database under accession number DQ088168.
Acknowledgments
We thank the Analytical Instrumentation Center of the School of Pharmacy, UW-Madison, for support in obtaining mass spectrometry and nuclear magnetic resonance data, John Innes Center, Norwich, United Kingdom, for providing the REDIRECT Technology kit, and C. Richard Hutchinson for critical reading of the manuscript.
This work is supported in part by the Graduate School and the School of Pharmacy, University of Wisconsin—Madison, a UWCCC Pilot Project program (CA014520), and an NCI National Cooperative Drug Discovery Group grant (CA113297). B.S. is the recipient of an NIH Independent Scientist Award (AI51689).
REFERENCES
- 1.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 2.Carroll, B. J., S. J. Moss, L. Bai, Y. Kato, S. Toelzer, T. W. Yu, and H. G. Floss. 2002. Identification of a set of genes involved in the formation of the substrate for the incorporation of the unusual “glycolate” chain extension unit in ansamitocin biosynthesis. J. Am. Chem. Soc. 124:4176-4177. [DOI] [PubMed] [Google Scholar]
- 3.Cheng, X. C., T. Kihara, H. Kusakabe, J. Magae, Y. Kobayashi, R. P. Fang, Z. F. Ni, Y. C. Shen, K. Ko, I. Yamaguchi, and K. Isono. 1987. A new antibiotic, tautomycin. J. Antibiot. 40:907-909. [DOI] [PubMed] [Google Scholar]
- 4.Cheng, X. C., T. Kihara, X. Ying, M. Uramoto, H. Osada, H. Kusakabe, B. N. Wang, Y. Kobayashi, K. Ko, I. Yamaguchi, Y. C. Shen, and K. Isono. 1989. A new antibiotic, tautomycetin. J. Antibiot. 42:141-144. [PubMed] [Google Scholar]
- 5.Cheng, X. C., M. Ubukata, and K. Isono. 1990. The structure of tautomycin, a dialkylmaleic anhydride antibiotic. J. Antibiot. 43:809-819. [DOI] [PubMed] [Google Scholar]
- 6.Cheng, X. C., M. Ubukata, and K. Isono. 1990. The structure of tautomycetin, a dialkylmaleic anhydride antibiotic. J. Antibiot. 43:890-896. [DOI] [PubMed] [Google Scholar]
- 7.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 100:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Honkanen, R. E., and T. Golden. 2002. Regulators of serine/threonine protein phosphatases at the dawn of a clinical era? Curr. Med. Chem. 9:2055-2075. [DOI] [PubMed] [Google Scholar]
- 9.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, United Kingdom.
- 10.Ligon, J. M., S. Hill, J. Beck, R. Zirkle, I. Molnar, J. Zawodny, S. Money, and T. Schupp. 2002. Characterization of the biosynthetic gene cluster for the antifungal polyketide soraphen A from Sorangium cellulosum So ce26. Gene 285:257-267. [DOI] [PubMed] [Google Scholar]
- 11.MacKintosh, C., and S. Klumpp. 1990. Tautomycin from the bacterium Streptomyces verticillatus: another potent and specific inhibitor of protein phosphatases 1 and 2A. FEBS Lett. 277:137-140. [DOI] [PubMed] [Google Scholar]
- 12.Magae, J., C. Watanabe, H. Osada, X. C. Cheng, and K. Isono. 1988. Induction of morphological change of human myeloid leukemia and activation of protein kinase C by a novel antibiotic, tautomycin. J. Antibiot. 41:932-937. [DOI] [PubMed] [Google Scholar]
- 13.Metsa-Ketela, M., V. Salo, L. Halo, A. Hautala, J. Hakala, P. Mantsala, and K. Ylihonko. 1999. An efficient approach for screening minimal PKS genes from Streptomyces. FEMS Microbiol. Lett. 180:1-6. [DOI] [PubMed] [Google Scholar]
- 14.Mitsuhashi, S., N. Matsuura, M. Ubukata, H. Oikawa, H. Shima, and K. Kikuchi. 2001. Tautomycetin is a novel and specific inhibitor of serine/threonine protein phosphatase type 1, PP1. Biochem. Biophys. Res. Commun. 287:328-331. [DOI] [PubMed] [Google Scholar]
- 15.Nicholson, T. P., B. A. Rudd, M. Dawson, C. M. Lazarus, T. J. Simpson, and R. J. Cox. 2001. Design and utility of oligonucleotide gene probes for fungal polyketide synthases. Chem. Biol. 8:157-178. [DOI] [PubMed] [Google Scholar]
- 16.Rascher, A., Z. Hu, N. Viswanathan, A. Schirmer, R. Reid, W. C. Nierman, M. Lewis, and C. R. Hutchinson. 2003. Cloning and characterization of a gene cluster for geldanamycin production in Streptomyces hygroscopicus NRRL3602. FEMS Microbiol. Lett. 218:223-230. [DOI] [PubMed] [Google Scholar]
- 17.Rose, T. M., E. R. Schultz, J. G. Henikoff, S. Pietrokovski, C. M. McCallum, and S. Henikoff. 1998. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res. 26:1628-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oikawa, H. 2002. Synthesis of specific protein phosphatase inhibitors, tautomycin and tautomycetin toward structure-activity relationship study. Curr. Med. Chem. 9:2033-2054. [DOI] [PubMed] [Google Scholar]
- 19.Sakoff, J. A., and A. McCluskey. 2004. Protein phosphatase inhibition: structure based design. Towards new therapeutic agents. Curr. Pharm. Des. 10:1139-1159. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 21.Shen, B., and C. R. Hutchinson. 1996. Deciphering the mechanism for the assembly of aromatic polyketides by a bacterial polyketide synthase. Proc. Natl. Acad. Sci. USA 93:6600-6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takai, A., K. Sasaki, H. Nagai, G. Mieskes, M. Isobe, K. Isono, and T. Yasumoto. 1995. Inhibition of specific binding of okadaic acid to protein phosphatase 2A by microcystin-LR, calyculin-A and tautomycin: method of analysis of interactions of tight-binding ligands with target protein. Biochem. J. 306:657-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ubukata, M., X. C. Cheng, and K. Isono. 1990. The structure of tautomycin, a regulator of eukaryotic cell growth. J. Chem. Soc. Chem. Commun. 1990:244-245. [Google Scholar]
- 24.Ubukata, M., X. C. Cheng, M. Isobe, and K. Isono. 1993. Absolute configuration of tautomycin, a protein phosphatase inhibitor from a streptomycete. J. Chem. Soc. Perkin Trans. I 1990:617-624. [Google Scholar]
- 25.Ubukata, M., X. C. Cheng, J. Uzawa, and K. Isono. 1995. Biosynthesis of the dialkylmaleic anhydride-containing antibiotics, tautomycin and tautomycetin. J. Chem. Soc. Perkin Trans. I 1995:2399-2404. [Google Scholar]
- 26.Watanabe, K., C. Khosla, R. M. Stroud, and S. C. Tsai. 2003. Crystal structure of an Acyl-ACP dehydrogenase from the FK520 polyketide biosynthetic pathway: insights into extender unit biosynthesis. J. Mol. Biol. 334:435-444. [DOI] [PubMed] [Google Scholar]
- 27.Wu, K., L. Chung, W. P. Revill, L. Katz, and C. D. Reeves. 2000. The FK520 gene cluster of Streptomyces hygroscopicus var. ascomyceticus (ATCC14891) contains genes for biosynthesis of unusual polyketide extender units. Gene 251:81-90. [DOI] [PubMed] [Google Scholar]
- 28.Yu, T. W., L. Bai, D. Clade, D. Hoffmann, S. Toelzer, K. Q. Trinh, J. Xu, S. J. Moss, E. Leistner, and H. G. Floss. 2002. The biosynthetic gene cluster of the maytansinoid antitumor agent ansamitocin from Actinosynnema pretiosum. Proc. Natl. Acad. Sci. USA 99:7968-7973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao, C., J. Ju, S. D. Christenson, W. C. Smith, D. Song, X. Zhou, B. Shen, and Z. Deng. 2006. Utilization of the methoxymalonyl-acyl carrier protein biosynthesis locus for cloning the oxazolomycin biosynthetic gene cluster from Streptomyces albus JA3453. J. Bacteriol. 188:4141-4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zirkle, R., J. M. Ligon, and I. Molnar. 2004. Heterologous production of the antifungal polyketide antibiotic soraphen A of Sorangium cellulosum So ce26 in Streptomyces lividans. Microbiology 150:2761-2774. [DOI] [PubMed] [Google Scholar]