Abstract
The C-terminal P5 domain of the histidine kinase CheA is essential for coupling CheA autophosphorylation activity to chemoreceptor control through a binding interaction with the CheW protein. To locate P5 determinants critical for CheW binding and chemoreceptor control, we surveyed cysteine replacements at 39 residues predicted to be at or near the P5 surface in Escherichia coli CheA. Two-thirds of the Cys replacement proteins exhibited in vitro defects in CheW binding, either before or after modification with a bulky fluorescein group. The binding-defective sites were widely distributed on the P5 surface and were often interspersed with sites that caused no functional defects, implying that relatively minor structural perturbations, often far from the actual binding site, can influence its conformation or accessibility. The most likely CheW docking area included loop 2 in P5 folding subdomain 1. All but four of the binding-defective P5-Cys proteins were defective in receptor-mediated activation, suggesting that CheW binding, as measured in vitro, is necessary for assembly of ternary signaling complexes and/or subsequent CheA activation. Other Cys sites specifically affected receptor-mediated activation or deactivation of CheA, demonstrating that CheW binding is not sufficient for assembly and/or operation of receptor signaling complexes. Because P5 is quite similar to CheW, whose structure is known to be dynamic, we suggest that conformational flexibility and dynamic motions govern the signaling activities of the P5 domain. In addition, relative movements of the CheA domains may be involved in CheW binding, in ternary complex assembly, and in subsequent stimulus-induced conformational changes in receptor signaling complexes.
The histidine autokinase CheA plays a central role in bacterial chemotaxis signaling pathways (see references 8, 33, and 35 for reviews of chemotactic signaling). The CheA protein of Escherichia coli, the best-studied example, forms a ternary signaling complex with transmembrane chemoreceptors, known as methyl-accepting chemotaxis proteins, and the small cytoplasmic protein CheW. In the absence of chemoeffector gradients, receptor complexes activate CheA autophosphorylation several hundred fold (20, 34). CheA in turn donates its phosphoryl groups to the CheB and CheY response regulators, which respectively control sensory adaptation and locomotor behavior. Phospho-CheB comprises a negative feedback loop that adjusts the methylation states of receptor molecules to modulate their signaling sensitivity. Phospho-CheY binds to the switching machinery at the base of the flagellar motors to promote clockwise rotation, which produces turning or tumbling episodes as the cell swims. Attractant increases and repellent decreases deactivate receptor-coupled CheA to slow the flux of phosphoryl groups to CheY and CheB, thereby promoting forward swimming and net methylation of the signaling receptor molecules.
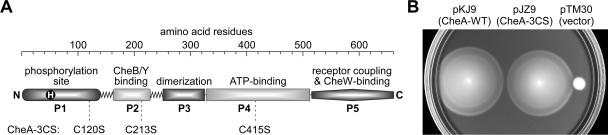
CheA functions as a homodimer with a modular subunit architecture (Fig. 1). The phosphorylation site, His-48, resides in the N-terminal P1 domain, whereas the ATP-binding site and important catalytic determinants reside in the P4 domain, near the C terminus of the molecule (3, 23, 32). The C-terminal P5 domain of CheA plays no role in the autophosphorylation reaction but is essential for CheW binding (6, 28) and for receptor-coupled control of phosphorylation activity (7). CheW also binds to the signaling domain of chemoreceptors (6, 11, 19, 30) and is required to couple CheA to receptor control. Because the CheA autophosphorylation reaction requires interaction between the P1 and P4 domains, CheW might serve to transmit stimulus-induced conformational changes between the receptor and CheA that allosterically regulate the P1-P4 interaction.
FIG. 1.
Structure and function of a cysteine-less CheA protein. (A) Domain organization of CheA. CheA functions as a homodimer; each subunit contains five domains (P1 to P5), each with a distinctive signaling role. Wild-type CheA contains cysteines at three locations that were converted to serine residues in the cysteine-less protein (CheA-3CS). (B) In vivo test of CheA-3CS function. Colonies of RP9535 (ΔcheA) carrying pKJ9 (CheA-WT), pJZ9 (CheA-3CS), or control vector pTM30 (ΔcheA) were tested for chemotactic ability on tryptone soft agar (0.2%) plates containing 50 μg/ml ampicillin. Incubation was at 32.5°C for 9 h.
A better understanding of the CheA-P5 domain, particularly its binding interaction with the CheW coupling factor, could shed considerable light on the mechanism of CheA control in receptor signaling complexes. Weak (Kd ≈ 15 μM) binding interactions between CheW and CheA have been demonstrated by several in vitro methods (6, 10, 28). This binding is dependent on the P5 domain (6, 28), and P5 missense mutations that reduce CheW binding also impair or abrogate receptor-coupling control (36). In this study, we adopted a cysteine-scanning strategy to locate critical CheW-binding and receptor-coupling determinants in the P5 domain. Our findings suggest that P5-CheW interactions are influenced by the dynamic motion of the P5 domain and its orientation relative to other domains in the CheA molecule. We propose that the CheA-P5 domain is not simply a tethering site for CheW but that it also manipulates other CheA domains to activate and deactivate the autophosphorylation reaction in response to conformational changes in the ternary signaling complex.
MATERIALS AND METHODS
Bacterial strains.
Strains used in this study were RP3098 [Δ(flhD-flhB)4] (31) and RP9535 [Δ(cheA)1643] (29). Both are derivatives of E. coli K-12 strain RP437, which is wild type for chemotaxis (27).
Plasmids.
Plasmid pPA113 is a pACYC184 derivative that confers resistance to chloramphenicol and expresses wild-type CheA under salicylate-inducible control (36). Optimal complementation of RP9535 by pPA113 occurred at 0.4 μM sodium salicylate. Plasmid pTM30 is a pBR322 derivative that confers ampicillin resistance and carries a ptac promoter inducible by isopropyl β-d-thiogalactopyranoside (IPTG) (23). Plasmid pKJ9 is a pTM30 derivative that expresses wild-type CheA under IPTG control (9, 17). Optimal complementation of RP9535 by pKJ9 occurred at 0 μM IPTG. Plasmid pJC3 (2) was used to prepare Tsr-containing membranes, pAR1.CheY (provided by Rick Stewart, University of Maryland) was used for preparing CheY, and pPA770 (36) was used to prepare CheW. Proteins for pull-down binding assays were prepared from pPA117, which encodes a glutathione S-transferase (GST)-CheW fusion protein (36) and pGEX-3X (Amersham Biosciences), which encodes a GST control protein. Plasmid pSN10 is a pPA113 derivative encoding a cysteine-less CheA (CheA-3CS) (S. Nishiyama and J. S. Parkinson, unpublished results). The coding region for CheA-3CS was amplified by PCR and substituted for the wild-type cheA gene of pKJ9 to create plasmid pJZ9.
Growth media.
Chemotactic ability was assessed on semisolid tryptone medium (T soft agar) consisting of tryptone broth (10 g tryptone, 5 g NaCl per liter) and 0.20 to 0.27% agar. L broth (T broth plus 0.5% yeast extract) was generally used for growth of bacterial strains. The growth medium for protein expression and stability tests was H1 minimal salts (25) containing 1% Casamino Acids, 0.4% glycerol, and required amino acids (1 mM each). IPTG and sodium salicylate were purchased from Promega Corp. Ampicillin and chloramphenicol were purchased from Sigma Chemical Co. and used at 100 μg/ml and 25 μg/ml, respectively, in solid and liquid media, except in soft agar chemotaxis assay plates, where their concentrations were halved.
Construction of CheA-P5 cysteine replacements.
The three wild-type cheA cysteine codons in pPA113 (Fig. 1) were sequentially changed to serine codons in producing plasmid pSN10 (Nishiyama and Parkinson, unpublished), whose cheA coding region was transferred to pKJ9, yielding pJZ9. Cysteine codons were introduced into the Cys-less cheA gene of pJZ9 by QuikChange site-directed mutagenesis (Stratagene) to create individual cysteine replacements at various CheA-P5 residue positions.
Protein purification.
CheA (14), CheW (1), and CheY (21) were expressed in RP3098 and purified following published protocols. Tsr-containing membranes were prepared as previously described (4).
Modification of cysteine-bearing CheA proteins.
Purified CheA-Cys proteins (50 μM) were mixed with 1 mM fluorescein-5-maleimide (5-FM; Amersham Biosciences) and incubated at room temperature in the dark for 10 min. Reactions were quenched with 200 mM dithiothreitol, and samples were subjected to denaturing gel electrophoresis (sodium dodecyl sulfate-polyacrylamide gel electrophoresis). The fluorescent protein label was quantified with a Typhoon variable mode imager (Molecular Dynamics/Amersham Biosciences).
In vitro assays of CheA-Cys proteins.
CheA autophosphorylation assays were performed as previously described (1). Receptor-coupling assays were done with Tsr-containing membranes, as previously described (22). The CheA-CheW binding interaction was measured with a GST-CheW pull-down assay, as previously described (36).
RESULTS
Construction of a cysteine-less CheA.
Wild-type CheA contains three cysteine residues, one each in the P1, P2, and P4 domains (Fig. 1A). To create unique reporter sites for a cysteine-scanning analysis of the P5 domain, the native cheA cysteine codons were converted to serine codons and the mutant cheA gene was transferred to an IPTG-inducible plasmid (pJZ9). The cysteine-less CheA protein (CheA-3CS) encoded by pJZ9 was tested for the ability to support chemotactic behavior in host strain RP9535 (ΔcheA) in soft agar assays (Fig. 1B). The CheA-3CS colonies expanded at more than 90% of the wild-type rate, and their morphology was indistinguishable from the wild type (Fig. 1B), indicating that the cysteine-less protein retains essentially wild-type signaling properties in vivo. For in vitro studies, CheA-3CS could be readily purified by following the scheme for wild-type CheA. We chose not to append an affinity tag to facilitate CheA-3CS purifications because preliminary experiments indicated that a six-His sequence at the N terminus compromised the in vivo function of wild-type CheA (data not shown). In vitro, CheA-3CS exhibited wild-type activities in autophosphorylation, CheW binding, and receptor-coupling assays (see below). In this report, we use the term “wild type” to refer to the CheA-3CS protein.
In vivo function of CheA-P5 cysteine replacement mutants.
Single cysteine replacements were created at 39 residues in the P5 domain of CheA-3CS by site-directed mutagenesis. The reporter sites were chosen on the basis of surface-exposed positions in the X-ray crystal structure of Thermotoga maritima CheA (3), whose P5 domain has over 30% sequence identity to its E. coli counterpart and presumably a similar tertiary structure, as well. All but six of the Cys-marked proteins supported at least 75% of the wild-type chemotactic ability in soft agar assays (Fig. 2A). Three mutant proteins (Q586C, G629C, and S630C) supported less than 25% of the wild-type chemotactic ability (Fig. 2A), indicating that their cysteine replacements severely impaired in vivo CheA function. Three other replacement mutants (R555C, I581C, and G588C) exhibited moderate reductions in signaling ability, with about 50% of wild-type function (Fig. 2A). These latter three residues were also identified in a study of random P5 missense mutants (36). Those mutants had different amino acid replacements at the critical positions (R555Q, I581T, I581V, and G588S) but in vivo functional defects similar to those of the Cys replacement proteins.
FIG. 2.
In vivo phenotypes and in vitro functions of CheA-P5-Cys proteins. (A) Ability of P5-Cys proteins to support chemotactic behavior. The chemotactic ability of RP9535/pJZ9 P5-Cys strains was assessed by colony size after 7 h of incubation at 32.5°C on tryptone soft agar (containing no IPTG). Columns indicate the averages and standard deviations for two to four measurements relative to pJZ9 (CheA-3CS) controls. The CheA residues scanned with cysteine replacements are listed below their respective data columns. Dark gray columns indicate P5-Cys proteins with less than 70% of wild-type function; black columns indicate proteins with less than 25% of wild-type function. The dashed line indicates the behavior of wild-type CheA-3CS. (B) CheW binding by P5-Cys proteins. Binding was assessed by pull-down assays (see Materials and Methods). Columns indicate the normalized averages and standard deviations of at least two measurements for each mutant protein. Dark gray columns indicate proteins with less than 40% of wild-type activity; black columns indicate proteins with less than 25% of wild-type binding ability. The dashed line indicates the behavior of wild-type CheA-3CS. (C) Ability of P5-Cys proteins to undergo receptor-mediated activation. Mutant CheA proteins were mixed with CheW and receptor-containing membranes and tested for autophosphorylation in the absence and presence of a saturating serine stimulus (see Materials and Methods). Columns show the normalized averages and standard deviations of at least two prestimulus measurements for each mutant protein. Dark gray columns indicate mutant proteins with less than 40% of wild-type activity; black columns indicate proteins with less than 20% of wild-type activity. Striped columns denote proteins with significant defects in CheA deactivation following presentation of the serine stimulus (see text). The dashed line indicates the behavior of wild-type CheA-3CS.
In vitro properties of cysteine-containing CheA proteins.
Each reporter protein was purified (always by the wild-type protocol) and tested in vitro for autophosphorylation, CheW binding, and receptor-coupling behavior. All proteins purified readily, implying that none had substantial defects in expression or stability. All mutant proteins, including those unable to support chemotaxis in vivo, also exhibited essentially wild-type autophosphorylation rates (data not shown), confirming that the proteins were not grossly misfolded. However, the set of Cys-containing proteins did exhibit a wide range of CheW-binding (Fig. 2B) and receptor-coupling (Fig. 2C) behaviors.
CheW binding was evaluated with pull-down assays, using CheW tagged at the N terminus with a glutathione S-transferase domain (36). The GST-CheW fusion protein supports chemotactic ability (36) and in BIAcore assays binds to CheA with a Kd of ∼15 μM (28), comparable to the CheW-CheA affinity measured by other methods (6, 10). In the pull-down assay, which is not an equilibrium method, CheA and GST-CheW were present at threefold and twofold, respectively, of the Kd value. Under these assay conditions most of the proteins (30/39) exhibited binding values between 0.4 and 1.4 times the wild type. Although the binding activities at both extremes of this range were statistically different from the wild type, the differences were not large and we consider this entire group essentially normal for CheW binding (Fig. 2B). In contrast, six proteins (D513C, G556C, A593C, R615C, G629C, and L633C) bound CheW very poorly, with binding signals less than 0.25 times the wild type (Fig. 2B). Three other proteins (D521C, R555C, and A622C) had intermediate binding defects, with binding signals of 0.25 to 0.4 times the wild type (Fig. 2B).
CheW-dependent receptor-coupling control of the Cys-marked CheA proteins was evaluated with membranes containing the serine chemoreceptor Tsr. A majority of the Cys proteins (23/39) were activated to nominally normal levels (0.4 to 1.6 times the wild type) in this assay (Fig. 2C). Eight proteins (D513C, D521C, G556C, G588C, V607C, N609C, A622C, and S630C) had reduced but nevertheless substantial levels of activation, 0.2 to 0.4 times the wild type (Fig. 2C). Another eight proteins (D541C, R555C, Y592C, R615C, G627C, D628C, G629C, and D636C) had activation levels below 0.2 times the wild type (Fig. 2C). Of the nine proteins deemed defective in CheW binding, seven also exhibited activation defects, consistent with the idea that CheW binding is essential for assembling active ternary receptor complexes. However, two of the binding-defective proteins (A593C and L633C) had activation levels within the normal range. Conceivably, their binding defects might have been alleviated by the presence of receptor molecules. Conversely, 9 of the 16 proteins with evident activation defects (D541C, G588C, Y592C, V607C, N609C, G627C, D628C, S630C, and D636C) had CheW-binding values within the normal range. We conclude that CheW binding by CheA is necessary, but not sufficient, for assembly of active receptor signaling complexes.
Cys-containing proteins with “normal” activation properties were also examined for the ability to undergo deactivation of autophosphorylation in response to a receptor-saturating level of serine (see Materials and Methods). Eight proteins exhibited substantial deactivation defects (Fig. 2C). Two (M532C and V562C) retained over 20% of their activated autophosphorylation rate in the presence of serine; six others (H543C, G547C, G548C, I581C, R590C, and A593C) retained more than 10% of their activated rate. In contrast, the other activatable CheA proteins, including the wild type, typically retained less than 5% (4.75% ± 3.3%) of their activated autophosphorylation ability after a serine stimulus.
Modification-dependent changes in in vitro behaviors of CheA-Cys proteins.
To identify P5 surface determinants involved in CheW binding and receptor-coupling control, we modified the purified CheA-Cys proteins with 5-FM, whose bulky fluorescein group ought to disrupt protein-protein interactions when attached to a cysteine reporter located in a critical contact region. To assess the completeness of the modification reactions, we compared the extent of 5-FM labeling of the native proteins with their sodium dodecyl sulfate-denatured counterparts. The difference in fluorescence intensity of the native and denatured sample was less than 10% for all reporter proteins (data not shown). Thus, the 5-FM modification reactions of the native proteins were effectively complete. Moreover, the autophosphorylation activities of all modified proteins were at least 50% of their unmodified rates, indicating that none of the proteins had been grossly damaged by the fluorescein modification (data not shown).
We evaluated the functional consequences of the 5-FM modifications by comparing the CheW-binding and receptor-coupling properties of the modified and unmodified CheA-Cys proteins. Modification with 5-FM produced less than a twofold change in CheW-binding behavior at most of the tested sites (Fig. 3A). The P5 sites at which a cysteine replacement itself impaired CheW binding (Fig. 3A) showed no additional decrement in binding upon 5-FM modification. In fact, 5-FM modification at one of those sites (A593) actually seemed to repair the binding defect caused by the cysteine replacement. Similarly, the partial CheW-binding defects caused by cysteine replacements at I600 and S630 (∼60% of wild type [Fig. 2B]) were alleviated by 5-FM modification (Fig. 4B). These findings suggest that A593, I600, and S630 are not critical determinants for CheW binding, but rather that cysteine replacements at those sites alter overall P5 conformation or dynamic behavior in a way that interferes with CheW binding. Evidently, attachment of a bulky fluorescein group largely reverses the detrimental effects of cysteines at these positions. In contrast, modification caused modest detrimental effects at nine positions (L512, V562, N569, G588, R591, V607, N609, I634, and V637) where CheW-binding values were over twofold less than those of the unmodified proteins (Fig. 3A). Some of these sites could be important determinants for CheW binding.
FIG. 3.
In vitro activities of 5-FM-modified CheA-P5-Cys proteins. (A) CheW binding by modified P5-Cys proteins. Binding values for the modified proteins were determined by pull-down assays and normalized to those of the corresponding unmodified protein. Columns indicate the averages and standard deviations of at least two measurements for each unmodified/modified protein pair. Dark gray columns indicate modified proteins that showed less than 45% of their unmodified binding activity. Squares above some of the columns flag proteins with low binding values before modification (Fig. 2B), dark gray squares denote unmodified proteins with less than 40% of wild-type binding ability, and black squares denote proteins with less than 25% of wild-type binding. The dashed line indicates the behavior of wild-type CheA-3CS. (B) Ability of modified P5-Cys proteins to undergo receptor-mediated activation. Activation values were determined as for Fig. 2C and normalized to those of the corresponding unmodified protein. Columns indicate the averages and standard deviations for at least two measurements for each unmodified/modified protein pair. Dark gray columns indicate modified proteins that had less than 45% of the unmodified activation level; black columns indicate proteins that had less than 20% of the unmodified activation level. None of the proteins capable of receptor-mediated activation showed significant changes in deactivation behavior upon modification (not shown). Triangular flags indicate proteins that had activation defects before modification (Fig. 2C). Dark gray triangles denote proteins that had less than 40% of wild-type activity before modification; black triangles indicate proteins that had less than 20% of wild-type activity before modification. The dashed line indicates the behavior of wild-type CheA-3CS.
FIG. 4.
Proposed location of CheW-binding determinants on the P5 surface. The P5 domain is shown in a space-filled representation. The upper left image shows the position of P5 relative to the P3 and P4 domains of the same subunit in the Thermotoga X-ray structure (3). Cysteine replacement sites that had no effect on CheW binding, either before or after modification with 5-FM, are shown in white; sites that caused significant reductions in CheW binding, either before or after modification with 5-FM, are shown in black. The boxed regions show three different views of residues that may define the binding target for CheW.
The effects of 5-FM modification on the receptor-coupling behavior of the reporter proteins were more diverse (Fig. 3B). About half of the proteins (21/39) showed less than twofold changes in activation ability after modification (Fig. 3B). This group included six proteins whose activation ability was substantially reduced by the cysteine replacement itself (Fig. 3B). However, 10 other reporter proteins with cysteine-caused activation defects experienced more than a twofold-further reduction in activation after modification (Fig. 3B). These sites may not be central determinants for the activation process, but rather P5 positions that modulate transitions between the activated and deactivated conformations of the receptor-signaling complex. In contrast, eight other sites (G548, V562, R591, A593, S612, L633, I634, and V637) showed substantial activation defects only after modification, ranging from 0.2 to 0.45 to less than 0.2 of the unmodified values. These sites may define P5 structural determinants that are more directly involved in the activation process.
Modification with 5-FM did not obviously affect the deactivation ability of any CheA-Cys proteins (data not shown). Those with deactivation defects before modification remained defective after modification. Those with no deactivation defect remained so after modification. Note that this test can only be conducted on proteins that undergo receptor-mediated activation, which excluded more than half (24/39) of the test set (Fig. 3B).
DISCUSSION
The premise underlying our cysteine-scanning analysis was that the binding interaction between CheW and the P5 domain of CheA would involve regions of mutual surface complementarity on both proteins. Accordingly, we expected that a bulky fluorescein modification on any P5 residue at or near a contact site for CheW would impair the binding interaction. Based on the X-ray structure of the closely related CheA protein of Thermotoga maritima (3), we surveyed cysteine replacements at 39 residues predicted to be at or near the P5 surface in E. coli CheA. Many of those Cys replacement proteins (26/39) exhibited defects in CheW binding, either before or after 5-FM modification. The functionally critical residues proved to be widely distributed on the P5 surface (Fig. 4) and usually interspersed with sites that caused no functional defects (Fig. 4). The probable CheW-docking area, based on a study of P5 missense mutants (36) and the recent X-ray structure of a P4-P5/CheW complex from Thermotoga (24), involves loop 2 in subdomain 1 (Fig. 4). In the crystal structures (3, 24), this region lies close to the P3 and P4 domains. In solution the P5 domain may adopt an orientation(s) that makes the CheW docking surface more accessible. The preponderance of surface sites throughout P5 that affect CheW binding suggests that relatively minor structural perturbations, often far from the actual binding site, can influence its conformation or accessibility. As discussed below, we propose that conformational flexibility and dynamic motions govern the signaling activities of the P5 domain and that most mutational lesions affect P5 function by altering its dynamic behavior.
CheW-related structure of the P5 domain.
The nuclear magnetic resonance structure of Thermotoga maritima CheW, which exhibits dynamic character in solution, closely resembles the X-ray structure of its P5 partner in CheA (3, 13). CheW and P5 consist of two strand-swapped SH3-like subdomains, each of which contains five beta strands, with extended loops joining some of the beta segments (Fig. 5). Both proteins begin with two beta strands in subdomain 1, then cross over to form the entirety of subdomain 2, and then back to complete subdomain 1. The C terminus of both proteins extends from subdomain 1 back to subdomain 2 and might serve to conformationally couple the two subdomains (13). Loops 1 and 2 in CheW are known to be highly dynamic (12); those in P5 may also be flexible but are well-resolved in the X-ray structure owing to extensive crystal contacts (3). The CheW and P5 structures also differ in several important respects. First, loop 2 in P5 contains a short alpha-helix that is absent in CheW loop 2 (Fig. 5) (3, 13). Second, CheW has a longer alpha-helical extension at its C terminus that makes more extensive contacts with subdomain 2 (13). The C terminus of P5 contains a shorter helix, followed, in the E. coli protein, by 17 unconserved residues that are not essential to CheA function (7) (Fig. 5).
FIG. 5.
Comparison of CheW and P5 structural organizations. The two proteins are similar in structure and are shown in the same relative orientations. Each contains two subdomains comprised of five beta strands (broad arrows) with large loops at both ends of the molecule. P5 has a short alpha-helix in the middle of loop 2; CheW does not. CheW has a prominent C-terminal helix; P5 has a shorter C-terminal helix that is not depicted (indicated by a bold dashed line).
Identification of CheW-binding determinants in the P5 domain.
The functional properties of the P5-Cys proteins and the distribution of scanned sites relative to P5 structural elements are summarized in Fig. 6. Nine of the Cys replacement proteins had substantial CheW-binding defects before modification; nine additional proteins exhibited binding defects after 5-FM modification. The sites that produced CheW-binding defects fell into both P5 subdomains (Fig. 6). Their positions in the P5 crystal structure suggest three structure-function classes (Fig. 7A).
FIG. 6.
Locations and functional properties of P5 Cys-scanning and missense mutation sites. (A) Summary of the Cys-scanning mutations. P5 secondary structure elements are shown schematically below the residue numbers for the E. coli protein. Dashed segments indicate turns and loops that are highly dynamic in the CheW solution structure (13). Symbols in the bottom half of the panel summarize the in vivo function and in vitro activities of CheA proteins containing a cysteine residue at the indicated positions. “Cys effects” rows indicate properties of the unmodified proteins; “5-FM effects” rows indicate properties of the proteins after modification. The shading conventions for severity of activation and CheW-binding defect match those used in Fig. 2 and 3. For serine-induced deactivation, black triangles denote more than 20% of the prestimulus activity; dark gray triangles denote more than 10% of the prestimulus activity. (B) Summary of P5 missense mutations characterized in a previous study (36). The shading conventions for severity of defect follow those used in panel A. Symbols with grayed borders denote in vitro tests that could not be done because the mutant proteins were unstable and recalcitrant to purification (36).
FIG. 7.
Structure-function relationships in P5-Cys mutants. All images are alpha-carbon backbone traces of P5, with key residues in space-filled representation. The same shading convention is used throughout: light gray residues indicate biochemical defects caused by the cysteine replacement itself, and black residues indicate that modification of a cysteine at that position caused the biochemical defect. The upper left and middle images correspond to the P5 orientation in the upper middle image of Fig. 4; the next three images correspond to the P5 orientation in the upper left image of Fig. 4. The image at bottom right corresponds to the P5 orientation in the bottom right image of Fig. 4. (A) P5-Cys sites that cause CheW-binding defects. Left: residues in loops and turns. The N and C termini and the subdomain interface are indicated. Both of the next two images show all other P5 residues implicated in CheW binding. Middle: same orientation as the left image. The four labeled sites may modulate conformational motions between subdomains. The boxed residues correspond to the putative CheW target site shown in the upper middle image of Fig. 4. Right: the boxed area corresponds to the putative binding target for CheW shown in the upper left image of Fig. 4. Some of the labeled sites may be direct binding targets; others may modulate the conformation of the binding surface. (B) P5-Cys sites that specifically affect receptor-mediated activation of CheA (see text for discussion). Underlined labels denote sites that cause a substantial activation defect before 5-FM modification and another substantial reduction in activity after modification. (C) P5-Cys sites that primarily affect receptor-mediated deactivation. The unmodified mutant proteins assemble CheA-activating ternary complexes but cannot deactivate in response to an attractant stimulus. All such sites are located in subdomain 2. Labeled residues in the first image reside in loop 1; labeled residues in the second image reside near the subdomain interface. In the right image, the loop 1 residues are not shown, to better illustrate the packing interactions and backbone connections of the other residues.
(i) Six sites occur at short loops between adjacent beta strands in the same subdomain (Fig. 7A, left). With the exception of large loops 1 and 2, every subdomain turn yielded sites with CheW-binding defects: D521 at β1-β2, R555 and G556 at β4-β5, N569 at β5-β6, G588 at β6-β7, and G629 at β9-β10 (Fig. 6). Cysteine replacements and bulky modifications at these residues might hinder loop flexibility or distort the packing interactions of the flanking beta strands. This implies that local conformational changes can produce global structural changes or dynamic motions that influence entry to the binding-competent state. The functional importance of several of these turn residues was also noted in our previous study of CheA-P5 missense mutants (36) (summarized in Fig. 6B): proteins with R555Q or -W, G588S, G629D, or V631M replacements could not support chemotactic behavior. We suggest that an altered range or rate of dynamic motion is responsible for the CheW-binding defects exhibited by sites of this class.
(ii) Four binding-defective sites may influence conformational interactions between the two P5 subdomains (Fig. 7A, center). Subdomain 2 residues V562 and A593 pack against one another at the interface with subdomain 1. In addition, A593 adjoins the interdomain connecting strand β7-β8; R591 is just a few residues away. The fourth member of this class of sites is V637, near the end of the C-terminal strand that returns from subdomain 1 and packs against β6 of subdomain 2 (Fig. 7A, center). The cysteine replacements at V562, R591, and V637 reduce side chain volume, whereas cysteine would cause a volume increase at A593. The former sites exhibit binding defects only upon modification, whereas A593C is binding defective without modification. This pattern suggests that the binding defects in this class of sites arise through distorted packing interactions, which could in turn lead to global conformational changes or altered dynamic behavior.
(iii) The third group of binding-defective sites is located in subdomain 1, which most likely contains the contact residues for CheW binding (Fig. 7A, right) (24, 36). However, some of the subdomain 1 sites may affect CheW binding indirectly. For example, V607 and A622 lie at each end of the large loop 2 and pack against each other. These residues also pack against L633 and I634. The interactions among these four sites probably control the conformation and dynamic behavior of loop 2, which contains residues that contact CheW directly. The N609 residue in loop 2, which disrupts CheW binding only after modification, may not be an actual contact site but at least must lie close to a critical residue. In contrast, R615 may be an actual contact residue because its cysteine replacement eliminates CheW binding. R615 lies near one end of the short alpha-helix embedded within loop 2. A charge reversal mutation at the adjacent helix residue (K616E) also disrupts CheW binding (36). Both of these basic residues may be important binding determinants. S612, another helix residue, does not seem important for CheW binding because it tolerates a cysteine replacement and modification. No other residues in the loop 2 helix were scanned in this study, but this segment clearly merits additional attention.
All but four of the P5-Cys proteins with CheW-binding defects were also defective in receptor-mediated activation (Fig. 6). We conclude that CheW binding, as measured in vitro, is necessary for assembly of ternary signaling complexes and/or subsequent CheA activation. The four apparent exceptions may be proteins whose binding defects are effectively suppressed in the presence of receptors. This compensatory effect need not be a direct one. For example, interaction of CheW with receptors might stabilize its structure and thereby enhance its affinity for CheA-P5 (11, 19, 30).
P5 determinants involved in receptor-mediated activation.
Proteins with CheW-binding lesions are most likely defective in receptor activation because they cannot form ternary signaling complexes. However, eight P5 sites exhibited activation-specific defects, indicating that although CheW binding is necessary, it is not sufficient for activation (Fig. 7B). Because we did not directly measure complex assembly in this study, these CheA mutants could either be defective in complex assembly or in the function of fully assembled complexes. In general, the activation-defective sites are close to binding-defective sites, suggesting that binding and activation are influenced by similar P5 conformational changes and dynamic behaviors. Two notable exceptions are D541 and G548, both in loop 1. Perhaps loop 1 plays a direct role in triggering CheA activation upon conformational changes transmitted to P5 through bound CheW and chemoreceptors. For example, activation might involve movement of the CheA ATP-binding domain (P4) relative to the phosphorylation site domain (P1). P5 is close to P4 in the Thermotoga crystal structure and could conceivably manipulate P4 through loop 1 contacts.
Other activation-specific sites indicate that global conformational changes or dynamic motions are probably instrumental in receptor control of CheA, as they are in CheW binding. Y592 and D636 are close together at the subdomain 1-2 interface (Fig. 7B) and probably affect interdomain communication. The cysteine replacements at these sites impair activation, but modification further impairs activation, consistent with the idea that these lesions act through changes in the dynamic movements of P5 rather than altogether blocking entry to the activated conformation. Another critical site for activation is the β9-β10 turn, which also influences CheW binding (see above). All turn residues scanned (G627, D628, G629, and S630) were critical for activation, whereas only G629 affected CheW binding (Fig. 6), which may mean that activation requires more frequent or more extravagant conformational changes than those that allow CheW binding.
P5 determinants involved in receptor-mediated deactivation.
Eight cysteine replacement proteins (M532C, H543C, G547C, G548C, V562C, I581C, R590C, and A593C) were able to activate CheA in receptor signaling complexes but had defects in down-regulating CheA in response to an attractant stimulus (Fig. 6). The structural lesions in these proteins presumably retard ternary complex conformational changes that accompany CheA deactivation. All eight sites fall in P5 subdomain 2, whose dynamic behavior also influences CheW binding and CheA activation (Fig. 7C). Before modification, these structural lesions probably cause relatively modest changes in conformation or dynamic behavior that somehow trap the protein in the activated state upon ternary complex formation. Upon modification with 5-FM, three of the deactivation-defective proteins acquired additional defects in receptor-mediated activation (G548C) and CheW binding (V562C and A593C), implying that the binding and activation processes are either less sensitive than is deactivation to changes in P5 dynamics or their mechanisms involve distinct but overlapping conformational changes. We conclude that conformational flexibility of the ternary complex may be more important for deactivation than for activation.
Role(s) of P5-CheW interaction(s) in ternary signaling complexes.
The P5-binding targets in CheW, defined by mutational and nuclear magnetic resonance studies, reside in its loop 1 region and C-terminal alpha helix (5, 13) (Fig. 8). This part of CheW most likely interacts with the loop 2 region of P5, including the short alpha-helix within the loop (Fig. 8) (24, 36). The slow-on, fast-off kinetics of the binding interaction (28) are consistent with the possibility that one or both targets may need to undergo local conformational changes that expose or properly align the critical binding determinants. Hence, dynamic motions of the P5 and CheW surfaces probably play an important role in modulating their relative affinity.
FIG. 8.
Model of P5-CheW interaction. The P5-CheW binding interaction needed to assemble functional receptor signaling complexes occurs between loop 2 of P5 and the loop 1 region of CheW (this study and references 24 and 36). Chemoreceptors, most likely arranged in trimers of dimers (26), may bind across the subdomain interface of CheW to manipulate P5 and thereby exert allosteric control over CheA activity. Whether P5 loop 1 and CheW loop 2 have important functional roles as well is not known.
Dynamic motions of CheW probably modulate its interaction with chemoreceptors as well. The probable receptor interaction surface bridges the subdomain interface on CheW (Fig. 8). Relative motions of the two domains could, for example, expand or constrict the binding pocket, modulating CheW-receptor affinity. Receptor binding would in turn be expected to stabilize CheW structure, including the conformation of its P5 target surface. Cooperative binding interactions between the ternary complex components probably account for P5 mutants with substantially reduced binding affinity for CheW that nevertheless form functional signaling complexes when chemoreceptors are present.
Similarly, binding to CheW should slow P5's dynamic motions. This might stabilize other interaction surfaces on P5 that participate in the activation-deactivation mechanisms. For example, activation might involve direct interaction between CheA and the chemoreceptors. Alternatively, P5 might promote activation by interacting with another part of the CheA molecule, for example, the adjacent ATP-binding (P4) domain. In both scenarios, activation might simply involve constraining the relative movements of the P1 and P4 domains to promote more frequent productive collisions between them. The best candidate for a P5 control surface that might interact with a regulatory target appears to be the domain interface between loops 1 and 2 (Fig. 8). Here, we found four sites at which 5-FM modification blocked activation but not CheW binding (Fig. 7B). In contrast, the P5 region analogous to the receptor-binding surface of CheW exhibited no sites critical to in vivo or in vitro function (Fig. 4 and 7).
The stimulus-elicited conformational change that causes CheA deactivation might involve breaking some of the binding interactions of the activated ternary complex. According to this scenario, P5 alterations that reduce interdomain flexibility might cause deactivation-specific defects by trapping ternary complexes in the activated conformation, as suggested for certain mutations at the interdomain interface (Fig. 7C). One consequence of this deactivation mechanism might be to destabilize the receptor cluster, an effect that has been reported by several groups following application of an attractant stimulus (15, 16, 18).
Disparities between in vivo and in vitro chemotaxis behaviors.
We found many P5 alterations that impaired CheW binding and/or receptor-mediated control in vitro but did not abrogate chemotactic behavior in vivo. Similarly, Boukhvalova et al. have described a number of mutant CheW proteins with altered binding affinities for CheA or receptors that still supported chemotactic behavior in vivo (6). At least two mechanisms might ameliorate the in vivo signaling consequences of P5 and CheW lesions: (i) molecular crowding probably lessens the effects of reduced binding affinities among ternary complex components, and (ii) the cell's sensory adaptation system can compensate for changes in receptor coupling efficiency. These factors probably explain why 26/39 CheA-P5 cysteine replacement proteins had demonstrable in vitro defects but still supported normal chemotactic behavior in vivo. Some of the six proteins that failed to support chemotaxis (e.g., G629C) could have functional lesions too drastic to overcome in vivo. However, others (e.g., Q586C) had no or only modest functional defects in vitro and seemingly should have supported chemotaxis. Boukhvalova et al. also found a mutant CheW protein with essentially normal in vitro activities that could not support chemotactic behavior (6). Perhaps these proteins are defective in signaling functions for which no in vitro assays exist. Clearly, we need to learn a great deal more about receptor signaling complexes and their underlying protein-protein interactions if we want to understand the sources of robustness and signal gain in bacterial chemotaxis.
Acknowledgments
We thank Peter Ames, Rick Stewart, and So-Ichiro Nishiyama for generously providing plasmids and antisera crucial to this study and Brian Crane for communication of unpublished results.
This work was supported by research grant GM19559 from the National Institute of General Medical Sciences. The Protein-DNA Core Facility at the University of Utah receives support from National Cancer Institute grant CA42014 to the Huntsman Cancer Institute.
REFERENCES
- 1.Ames, P., and J. S. Parkinson. 1994. Constitutively signaling fragments of Tsr, the Escherichia coli serine chemoreceptor. J. Bacteriol. 176:6340-6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ames, P., C. A. Studdert, R. H. Reiser, and J. S. Parkinson. 2002. Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc. Natl. Acad. Sci. USA 99:7060-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bilwes, A. M., L. A. Alex, B. R. Crane, and M. I. Simon. 1999. Structure of CheA, a signal-transducing histidine kinase. Cell 96:131-141. [DOI] [PubMed] [Google Scholar]
- 4.Borkovich, K. A., and M. I. Simon. 1991. Coupling of receptor function to phosphate-transfer reactions in bacterial chemotaxis. Methods Enzymol. 200:205-214. [DOI] [PubMed] [Google Scholar]
- 5.Boukhvalova, M., R. VanBruggen, and R. C. Stewart. 2002. CheA kinase and chemoreceptor interaction surfaces on CheW. J. Biol. Chem. 277:23596-23603. [DOI] [PubMed] [Google Scholar]
- 6.Boukhvalova, M. S., F. W. Dahlquist, and R. C. Stewart. 2002. CheW binding interactions with CheA and Tar: importance for chemotaxis signaling in Escherichia coli. J. Biol. Chem. 277:22251-22259. [DOI] [PubMed] [Google Scholar]
- 7.Bourret, R. B., J. Davagnino, and M. I. Simon. 1993. The carboxy-terminal portion of the CheA kinase mediates regulation of autophosphorylation by transducer and CheW. J. Bacteriol. 175:2097-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bourret, R. B., and A. M. Stock. 2002. Molecular information processing: lessons from bacterial chemotaxis. J. Biol. Chem. 277:9625-9628. [DOI] [PubMed] [Google Scholar]
- 9.Garzón, A., and J. S. Parkinson. 1996. Chemotactic signaling by the P1 phosphorylation domain liberated from the CheA histidine kinase of Escherichia coli. J. Bacteriol. 178:6752-6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gegner, J. A., and F. W. Dahlquist. 1991. Signal transduction in bacteria: CheW forms a reversible complex with the protein kinase CheA. Proc. Natl. Acad. Sci. USA 88:750-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gegner, J. A., D. R. Graham, A. F. Roth, and F. W. Dahlquist. 1992. Assembly of an MCP receptor, CheW, and kinase CheA complex in the bacterial chemotaxis signal transduction pathway. Cell 70:975-982. [DOI] [PubMed] [Google Scholar]
- 12.Griswold, I. J., and F. W. Dahlquist. 2002. The dynamic behavior of CheW from Thermotoga maritima in solution, as determined by nuclear magnetic resonance: implications for potential protein-protein interaction sites. Biophys. Chem. 101-102:359-373. [DOI] [PubMed] [Google Scholar]
- 13.Griswold, I. J., H. Zhou, M. Matison, R. V. Swanson, L. P. McIntosh, M. I. Simon, and F. W. Dahlquist. 2002. The solution structure and interactions of CheW from Thermotoga maritima. Nat. Struct. Biol. 9:121-125. [DOI] [PubMed] [Google Scholar]
- 14.Hess, J. F., K. Oosawa, N. Kaplan, and M. I. Simon. 1988. Phosphorylation of three proteins in the signaling pathway of bacterial chemotaxis. Cell 53:79-87. [DOI] [PubMed] [Google Scholar]
- 15.Homma, M., D. Shiomi, and I. Kawagishi. 2004. Attractant binding alters arrangement of chemoreceptor dimers within its cluster at a cell pole. Proc. Natl. Acad. Sci. USA 101:3462-3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inoue, I., D. Shiomi, I. Kawagishi, and K. Yasuda. 2004. Simultaneous measurement of sensor-protein dynamics and motility of a single cell by on-chip microcultivation system. J. Nanobiotechnol. 2:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jahreis, K., T. B. Morrison, A. Garzon, and J. S. Parkinson. 2004. Chemotactic signaling by an Escherichia coli CheA mutant that lacks the binding domain for phosphoacceptor partners. J. Bacteriol. 186:2664-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamanna, A. C., G. W. Ordal, and L. L. Kiessling. 2005. Large increases in attractant concentration disrupt the polar localization of bacterial chemoreceptors. Mol. Microbiol. 57:774-785. [DOI] [PubMed] [Google Scholar]
- 19.Levit, M. N., T. W. Grebe, and J. B. Stock. 2002. Organization of the receptor-kinase signaling array that regulates Escherichia coli chemotaxis. J. Biol. Chem. 277:36748-36754. [DOI] [PubMed] [Google Scholar]
- 20.Levit, M. N., Y. Liu, and J. B. Stock. 1999. Mechanism of CheA protein kinase activation in receptor signaling complexes. Biochemistry 38:6651-6658. [DOI] [PubMed] [Google Scholar]
- 21.Matsumura, P., J. J. Rydel, R. Linzmeier, and D. Vacante. 1984. Overexpression and sequence of the Escherichia coli cheY gene and biochemical activities of the CheY protein. J. Bacteriol. 160:36-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morrison, T. B., and J. S. Parkinson. 1997. A fragment liberated from the Escherichia coli CheA kinase that blocks stimulatory, but not inhibitory, chemoreceptor signaling. J. Bacteriol. 179:5543-5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morrison, T. B., and J. S. Parkinson. 1994. Liberation of an interaction domain from the phosphotransfer region of CheA, a signaling kinase of Escherichia coli. Proc. Natl. Acad. Sci. USA 91:5485-5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park, S., P. P. Borbat, G. Gonzalez-Bonet, J. Bhatagar, A. M. Pollard, J. H. Freed, A. M. Bilwes, and B. R. Crane. Reconstruction of the chemotaxis receptor:kinase assembly. Nat. Struct. Mol. Biol., in press. [DOI] [PubMed]
- 25.Parkinson, J. S. 1976. cheA, cheB, and cheC genes of Escherichia coli and their role in chemotaxis. J. Bacteriol. 126:758-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parkinson, J. S., P. Ames, and C. A. Studdert. 2005. Collaborative signaling by bacterial chemoreceptors. Curr. Opin. Microbiol. 8:116-121. [DOI] [PubMed] [Google Scholar]
- 27.Parkinson, J. S., and S. E. Houts. 1982. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J. Bacteriol. 151:106-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rasmussen, R. P. 1998. Biochemical phenotypes of mutations in the chemotactic signaling complex of E. coli. Ph.D. thesis. University of Utah, Salt Lake City.
- 29.Sanatinia, H., E. C. Kofoid, T. B. Morrison, and J. S. Parkinson. 1995. The smaller of two overlapping cheA gene products is not essential for chemotaxis in Escherichia coli. J. Bacteriol. 177:2713-2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shrout, A. L., D. J. Montefusco, and R. M. Weis. 2003. Template-directed assembly of receptor signaling complexes. Biochemistry 42:13379-13385. [DOI] [PubMed] [Google Scholar]
- 31.Smith, R. A., and J. S. Parkinson. 1980. Overlapping genes at the cheA locus of Escherichia coli. Proc. Natl. Acad. Sci. USA 77:5370-5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swanson, R. V., S. C. Schuster, and M. I. Simon. 1993. Expression of CheA fragments which define domains encoding kinase, phosphotransfer, and CheY binding activities. Biochemistry 32:7623-7629. [DOI] [PubMed] [Google Scholar]
- 33.Szurmant, H., and G. W. Ordal. 2004. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol. Mol. Biol. Rev. 68:301-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tawa, P., and R. C. Stewart. 1994. Kinetics of CheA autophosphorylation and dephosphorylation reactions. Biochemistry 33:7917-7924. [DOI] [PubMed] [Google Scholar]
- 35.Wadhams, G. H., and J. P. Armitage. 2004. Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 5:1024-1037. [DOI] [PubMed] [Google Scholar]
- 36.Zhao, J., and J. S. Parkinson. 2006. Mutational analysis of the chemoreceptor-coupling domain of the Escherichia coli chemotaxis signaling kinase. J. Bacteriol. 188:3299-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]