Abstract
Sinorhizobium meliloti exists either in a free-living state in the soil or in symbiosis within legume nodules, where the bacteria differentiate into nitrogen-fixing bacteroids. Expression of genes involved in nitrogen fixation and associated respiration is governed by two intermediate regulators, NifA and FixK, respectively, which are controlled by a two-component regulatory system FixLJ in response to low-oxygen conditions. In order to identify the FixLJ regulon, gene expression profiles were determined in microaerobic free-living cells as well as during the symbiotic life of the bacterium for the wild type and a fixJ null-mutant strain. We identified 122 genes activated by FixJ in either state, including 87 novel targets. FixJ controls 74% of the genes induced in microaerobiosis (2% oxygen) and the majority of genes expressed in mature bacteroids. Ninety-seven percent of FixJ-activated genes are located on the symbiotic plasmid pSymA. Transcriptome profiles of a nifA and a fixK mutant showed that NifA activates a limited number of genes, all specific to the symbiotic state, whereas FixK controls more than 90 genes, involved in free-living and/or symbiotic life. This study also revealed that FixJ has no other direct targets besides those already known. FixJ is involved in the regulation of functions such as denitrification or amino acid/polyamine metabolism and transport. Mutations in selected novel FixJ targets did not affect the ability of the bacteria to form nitrogen-fixing nodules on Medicago sativa roots. From these results, we propose an updated model of the FixJ regulon.
The aerobic soil bacterium Sinorhizobium meliloti may encounter oxygen limitation conditions in two different ecological niches: in a free-living state in the soil or in symbiotic association with certain legume species (e.g., Medicago sativa) inside nitrogen-fixing root nodules. Indeed, oxygen limitation inside legume nodules is necessary to induce expression and maintain activity of the nitrogenase enzyme. The expression of S. meliloti genes required for nitrogen fixation and for microaerobic respiration inside nodules is coordinated via a two-component regulatory system, FixL/FixJ. Under microaerobic conditions, FixL autophosphorylates and transmits phosphate to the FixJ response regulator. Once phosphorylated, FixJ activates transcription of the nifA and fixK genes, encoding two intermediate regulators which induce expression of nif and fix genes involved in respiration and nitrogen fixation (44). In total, about 35 FixJ targets (direct or indirect) have been described, including FixM, a flavoprotein whose physiological function is yet unknown (14), and two homologues of ORF277 (Bradyrhizobium japonicum and Rhodobacter capsulatus) presenting homology with the family of universal stress protein (48). FixJ also controls, via FixK, the expression of a gene, fixT, that negatively affects expression of FixLJ-dependent genes by inhibiting FixL autophosphorylation (29). More recently, an in vitro selection approach, genomic Selex, was used to systematically identify FixJ-binding sites in the S. meliloti genome (24). This study led to the identification of two novel FixJ-regulated genes, proB2 and SMc03253, both involved in proline metabolism but for which no role in symbiosis was found.
FixLJ, FixK, and NifA are conserved among rhizobia, but they differ in connectivity and targets (20, 25). In Azorhizobium caulinodans, nifA is under the direct control of FixK, whereas in B. japonicum, nifA expression is largely independent of FixJ. Morever, two copies of fixK are present in the latter species, with FixJ activating fixK2 which in turn activates fixK1. In this bacterium, microaerobic transcription of the nir, nor, and nos genes, involved in the denitrification pathway, depends on FixLJ and FixK2, indicating that FixJ participates also in functions important in free-living conditions. In Rhizobium etli, FixL (differing in structure from other known FixL proteins) is not involved in nitrogen fixation (18, 19). Caulobacter crescentus, a non-N2-fixing α-proteobacterium, presents homologues of fixLJ in its genome (13), which indicates that FixLJ is clearly not restricted to rhizobia. Very recently (15), a global transcriptional analysis of C. crescentus identified about 70 genes regulated by FixJ. In this study it was shown that the core signaling pathway of the Fix network was conserved between rhizobia and C. crescentus.
Transcriptome analysis is a method of choice to study bacterial gene expression on a genome-wide scale, and this approach has been successfully used to analyze two-component system targets in Escherichia coli (39) and Corynebacterium glutamicum (37) as examples. The availability of the entire annotated sequence of the S. meliloti genome has opened new perspectives in the study of its biology through the use of DNA microarrays. A set of publications has provided strong evidence for the usefulness of transcription profiling in S. meliloti to identify genes induced in a variety of situations, including microaerobic and symbiotic conditions (1, 5, 7, 10), oxidative stress (3), osmotic stress (45), nitrogen metabolism (16, 41), quorum sensing (31), phosphate starvation (33), and response to flavonoids (5, 11). Macroarrays for Mesorhizobium loti were also developed, and transcriptional dynamics of this bacterium were monitored in symbiosis, microaerobiosis, and carbon starvation (49). Very recently, a partial-genome microarray was designed to study transcriptional profiling of B. japonicum grown aerobically or microaerobically (30).
We have used oligonucleotide-based pan-genomic S. meliloti 1021 microarrays with the aim of deciphering the FixJ regulon. Toward this goal, we have studied the expression profiling of all S. meliloti genes in a wild-type strain and in a fixJ null mutant, in two relevant physiological situations, i.e., microaerobic free-living conditions and in M. sativa nodule bacteroids. Our approach identified numerous targets not described so far as being activated by FixJ. The analysis of NifA and FixK regulons permitted us to position these genes in the FixJ regulatory cascade and thus to provide a more complete vision of this important regulon.
MATERIALS AND METHODS
Bacterial strains and in vitro growth conditions.
The bacterial strains used in this study are listed in Table 1.
TABLE 1.
Strains used in this work
| S. meliloti strains | Relevant characteristics | Reference or source |
|---|---|---|
| Rm1021 | SU 47 (2011), str-21 | 35 |
| EK 101 | 1021 derivative, Nod+ Fix−, fixJ::Tn5 | Erno Kiss |
| GMI 292 | 1021 derivative, Nod+ Fix−, nifA::Tn5 | 51 |
| GMI 296 | 1021 derivative, Nod+ Fix−, nifH::Tn5 | 46 |
| GMI 942 | 2011 derivative, Nod+ Fix−, ΩfixK1 ΔJB8; ΔJB8 is an approx 5-kb-long deletion removing fixK2 and the adjacent duplicated sequence fixNOQP2 | 26 |
Bacteria were grown at 28°C. Erlenmeyer flasks (250 ml) containing 30 ml of Vincent minimal medium (7) were inoculated to give a starting optical density at 600 nm (OD600) of 0.05 and then shaken until an OD600 of 0.2 was reached. Then, half of the culture was filtered (0.2-μm-pore-size membranes), immediately frozen in liquid nitrogen, and stored at −80°C until RNA extraction. The other half of the cultures (15 ml) was transferred to a 50-ml Erlenmeyer flask and cultured in a 2% oxygen atmosphere. After 2 h, cultures were filtered, immediately frozen in liquid nitrogen, and stored at −80°C until RNA extraction. Two hours in microaerobic conditions is sufficient to induce the full expression of FixK while avoiding any growth limitation which could impair further transcriptome analysis.
Cultivation of plants for bacteroid isolation.
Seeds of M. sativa cv. Europe were surface sterilized, germinated, and grown in aeroponic tanks as previously described (28). Plants were first grown in a medium supplemented with 5 mM ammonium nitrate. After 15 days of growth in the presence of nitrogen, the medium was changed to nitrogen-free medium. After two more days, plants were inoculated with S. meliloti (wild type and fixJ, nifA, fixK, and nifH mutants) to promote nodulation. Fourteen days after inoculation, nodules were harvested, immediately frozen in liquid nitrogen, and stored at −80°C until RNA extraction.
RNA extraction.
Bacteria from cultures were resuspended and incubated for 20 min at 65°C in 3 ml of prewarmed lysis solution (1.4% sodium dodecyl sulfate, 4 mM EDTA, 15 mg of proteinase K). Proteins were precipitated by adding 1.5 ml of 5 M NaCl at 4°C. Nucleic acids were precipitated from the supernatant by the addition of 2 volumes of ethanol, and the pellet was resuspended in 100 μl of RNase-free water.
For preparation of bacteroid RNA, nodules were crushed in liquid nitrogen and resuspended in a prewarmed (60°C) mixture of 50% phenol, 25 mM sodium acetate, 5 mM EDTA, and 0.5% sodium dodecyl sulfate, pH 5.2, and incubated at 60°C for 5 min. RNA was then extracted with phenol-chloroform and ethanol precipitated. For both bacteria and bacteroid RNA, contaminating DNA was removed from RNA preparations by DNase I using QIAGEN columns (clean-up procedure in QIAGEN protocol).
RNA quality and proportions of bacterial and plant rRNAs extracted from nodules were estimated with Agilent RNA nanochips, by using an Agilent 2100 bioanalyzer (Agilent Technologies). Bacterial rRNAs represented about 25% of total rRNA.
Microarrays.
Microarrays used in this study were described by Ruberg et al. (45) and were purchased from A. Becker (Bielefeld University, Germany). Each microarray carries 70-mer oligonucleotides representative of the 6,207 predicted open reading frames (ORFs) of S. meliloti 1021, spotted in triplicate, as well as a number of control spikes.
Just before hybridization, microarrays were submerged successively as follows: 5 min in a Triton aqueous solution (0.1%), twice for 2 min each in a HCl aqueous solution (0.037 ‰), 10 min in a KCl aqueous solution (0.1 M), 1 min in water, 20 min (50°C) in an aqueous blocking solution (0.074 ‰ HCl, 1× QMT blocking solution [Quantifoil]), and 1 min in water. All the steps were performed at room temperature except where otherwise mentioned.
Synthesis of cDNAs, labeling, and hybridization.
Fluorescent-labeled cDNAs were prepared according to the method of De Risi et al. (17) from 10 μg of RNA. Hybridizations were performed at 42°C for 15 h in DIG Easy hybridization solution (Roche) on blocked microarrays, with a Lucidea module 1 automated slide processor (Amersham Pharmacia Biotech).
The amount of RNA extracted from nodules was adjusted in order to label the equivalent of 10 μg of bacterial RNAs. Unless otherwise indicated, two independent RNA extractions were performed from each of two independent biological repeats (aeroponic tank). For experiments with free-living culture, four RNA extractions were performed, each from an independent biological repeat.
Transcriptomic data analysis.
Signal intensities were detected with a GenePix 4000 scanner (Axon Instrument) and quantification of median signal and median local background intensities for each spot were performed using GenePix Pro version 3.0 (Axon). Data from the replicas were then processed using GeneSight version 3.5 (Biodiscovery Inc.) (normalization by mean of the signal; statistical test, t test). Genes were regarded as differentially expressed if M was ≥1.00 or M was ≤−1.00 [M = log2(experiment signal/control signal)] and significantly up- or downregulated if the P value from the t test was ≤0.05. Only genes with signals superior to three times local background signal in the control (wild type) were considered. Data are available in the Array Express database.
Quantitative PCR.
RNAs used for quantitative PCR (qPCR) were the same as those used for microarray hybridizations.
Prior to reverse transcription, 1 μg of RNA was treated with 2 μl of DNase (Amersham Biosciences) for 15 min at 37°C. DNase inactivation was done 10 min at 75°C. Reverse transcription was carried out using Superscript II reverse transcriptase and random hexamers as primers, during 1 h at 42°C. qPCR was carried out with 1 μl of a 1/10 dilution of the cDNA, by using a FastStart DNA Master plus SYBR Green I kit. Denaturation of cDNA (8 min at 95°C) and 45 cycles of denaturation annealing and elongation were run on a Lightcycler system (Roche).
Transcripts were quantified in two biological replicates for both wild type (wt1 and wt2) and fixJ (fixJ1 and fixJ2) mutant bacteroids. Relative transcript abundances were calculated for each possible pair (i.e., fixJ1/wt1, fixJ2/wt1, fixJ1/wt2, and fixJ2/wt2). Only when the four ratios were higher than two was the gene assessed as dependent upon FixJ. 16S RNA was chosen as a reference for ratio normalization.
In microaerobic conditions, transcripts were quantified from RNA extracted from a wild-type, a fixJ mutant, and/or a fixK mutant culture used for transcriptomic analysis. Relative transcript abundances were calculated for the wild type/mutant pairs. When the ratio was higher than 2, the gene was validated as dependent upon FixJ and/or FixK. The gene rplM (SMc01804) was chosen as a reference for ratio normalization, since its expression level remained almost constant in all conditions tested. Every reaction was made in at least three replicates. Standard deviation between replicates was generally around 10%. The primers used for qPCR are listed in the Table S1 (posted at http://bioinfo.genopole-toulouse.prd.fr/annotation/iANT/bacteria/rhime/DOC/Bobik2006/index.html).
Analysis of stress sensitivity. (i) Oxidative stress.
Bacteria were inoculated (OD600, 0.05) in Luria-Bertani (LB) medium and shaken at 28°C until an OD600 of 0.2 was reached. Then, 100 μl of cultures were diluted in 5 ml of LB medium-0.75% agar (wt/vol) and immediately spread on LB plates. Finally, Whatman sterile paper disks (6 mm in diameter) were placed onto the plates and spotted with 5 μl of 3% H2O2.
Zones of inhibition were measured after 2 days of incubation at 28°C in a 2% oxygen atmosphere. The experiment was repeated three times.
(ii) Osmotic stress.
Bacteria were inoculated in Vincent minimal medium (OD600, 0.05) and shaken until an OD600 of 0.1 was reached. Then, half of the culture was incubated in a 2% oxygen atmosphere while the other part of the culture was shaken in aerated conditions. After 2 h, cultures were diluted with a 5 M NaCl sterile solution in order to obtain a 2.5 M final concentration. Cells were plated (three independent series of dilutions 10−2 and 10−3) just before and 30 min after NaCl addition on tryptone-yeast extract supplemented with appropriate antibiotics. For each strain (wt and fixJ), cell mortality was estimated after 3 days (28°C), by calculating the number of colonies on plates spread after 30-min exposure to NaCl compared to the control without NaCl (zero time). The sensitivity of the fixJ mutant to salt stress was estimated by the ratio of cell mortality in the mutant compared to the wild type. The experiment was repeated three times.
(iii) Microaerobic stress.
Cultures were performed in the same conditions as described above (for the microarray experiments) in a 2% oxygen atmosphere or in aerobic conditions. At different times, OD600 was measured and three independent dilutions (10−5) were spread onto tryptone-yeast plates supplemented with appropriate antibiotics to determine the number of CFU after 3 days of incubation at 28°C.
In planta mutant tests.
Seeds of M. sativa cv. Europe were surface sterilized, germinated, and grown on nitrogen-free agar slants (22) in test tubes. After 3 days, plantlets were inoculated (100 μl/plant) with S. meliloti strains resuspended in sterile water in order to have 2.103 bacteria per tube. A series of 10 plants were inoculated either with the wild type or with the different mutants. The fixJ mutant was used as a negative control. Plants were grown in a culture room (22°C) with day and night cycles of 16 and 8 h, respectively. Dry weight of the plants was measured 32 days after inoculation. The experiment was repeated twice. The difference between the wild type and each mutant was regarded as significant when observed in both series of plants (t test; P value of ≤0.05).
Comparative genomics.
Presence of putative orthologues of members of the FixJ regulon of S. meliloti in other α-proteobacterial genomes was assessed using the Phylopro interface (http://bioinfo.genopole-toulouse.prd.fr/comparative/bcg/PhyloPro/index.html) searching for global hits (%Q and %S > 80%). A table reporting the results will be available upon request.
RESULTS
A genome-wide appraisal of the FixJ regulon in microaerobic free-living cultures.
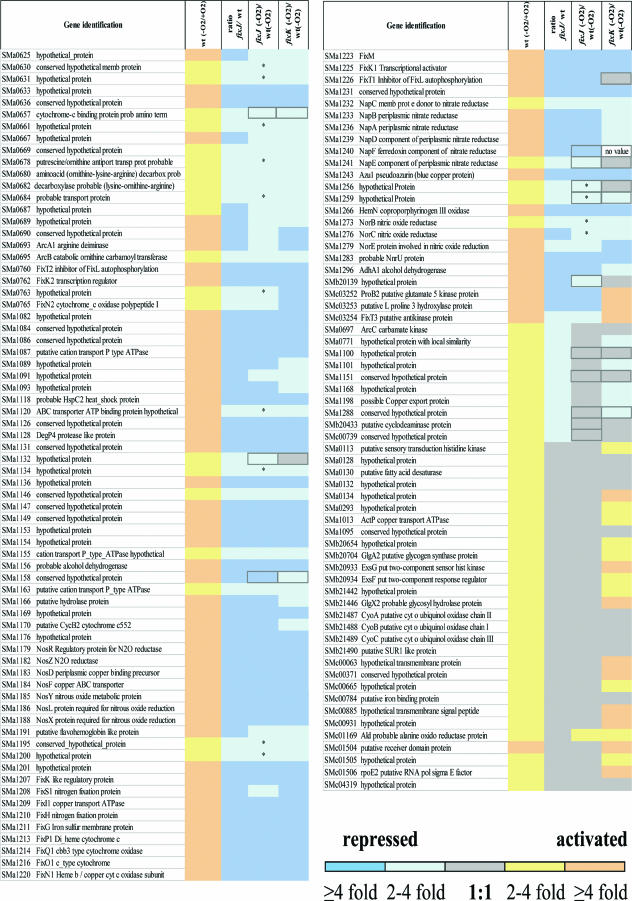
To gain insight into the FixJ regulon, we conducted genome-wide microarray analyses, comparing the transcriptome profiles of a fixJ mutant and the nearly isogenic wild-type strain Rm1021, exposed to normal and limited (2%) concentrations of oxygen. In the first series of experiments, we compared the transcript levels of the wild-type strain, grown in microaerobic and aerobic conditions. We considered as induced only genes showing an M value (log2 values of the ratio of intensities) of ≥1 and P values of ≤0.05 according to t statistics. One hundred thirty-two genes were upregulated two- or more than twofold (M ≥ 1) in microoxic conditions compared to aerobic conditions (Table S2, column 2; posted at http://bioinfo.genopole-toulouse.prd.fr/annotation/iANT/bacteria/rhime/DOC/Bobik2006/index.html). In order to determine which of these genes were dependent upon FixJ, we carried out a similar experiment using the fixJ mutant strain (Table S2, column 3). We considered FixJ-dependent genes those for which the induction level in microaerobiosis was reduced at least twofold (M ≤−1) in the mutant compared to the wild type (Table S2, column 4). The results showed that 103 genes out of the 132 genes overexpressed in microaerobic conditions were regulated by FixJ (Fig. 1, columns 2 and 3).
FIG. 1.
FixJ-upregulated genes in microaerobic free-living conditions. Genes with a twofold increase in their level of expression in microaerobic compared to aerobic conditions for the wild-type strain are presented in this figure (column 2). For each gene, the induction level in the wild type is compared to the induction level in the same condition (oxygen limitation) in the fixJ mutant strain; the ratio is represented in column 3. Columns 4 and 5 indicate the relative expression of the genes in the fixJ or fixK mutant, respectively, versus that of the wild-type strain, both under oxygen limitation. Yellow indicates genes that are upregulated in the assay compared to the control condition. Blue indicates genes that are downregulated in the mutant compared to the wild-type strain. A color scale is included at the bottom of the figure. An asterisk indicates a P value above 0.05. A dark frame indicates that the expression level of the gene has been verified by qPCR. The list of genes with M values obtained in each condition can be found in Table S2 (posted at http://bioinfo.genopole-toulouse.prd.fr/annotation/iANT/bacteria/rhime/DOC/Bobik2006/index.html).
In a second (direct) approach, we directly compared the transcript levels of the wild-type and the fixJ mutant strains, both grown in microaerobic conditions. The results indicated that 79 genes were dependent upon FixJ (Table S2, column 5; posted at http://bioinfo.genopole-toulouse.prd.fr/annotation/iANT/bacteria/rhime/DOC/Bobik2006/index.html). These genes were found in both approaches and with similar repression levels in the fixJ mutant strain. The first (indirect) approach allowed us to detect 24 additional potential FixJ targets. Most of these genes were not identified in the direct method due to a P value slightly above 0.05 (Fig. 1).
Both copies (fixK1 and fixK2) of the gene encoding transcription regulator FixK, known to be activated by FixJ, were recovered in this study as well as two other direct FixJ targets (SMc03252 and SMc03253) (24). The gene coding for the other intermediate regulator, NifA, was induced in microaerobic conditions with an M value of 0.8 and therefore was not included in our list of induced genes.
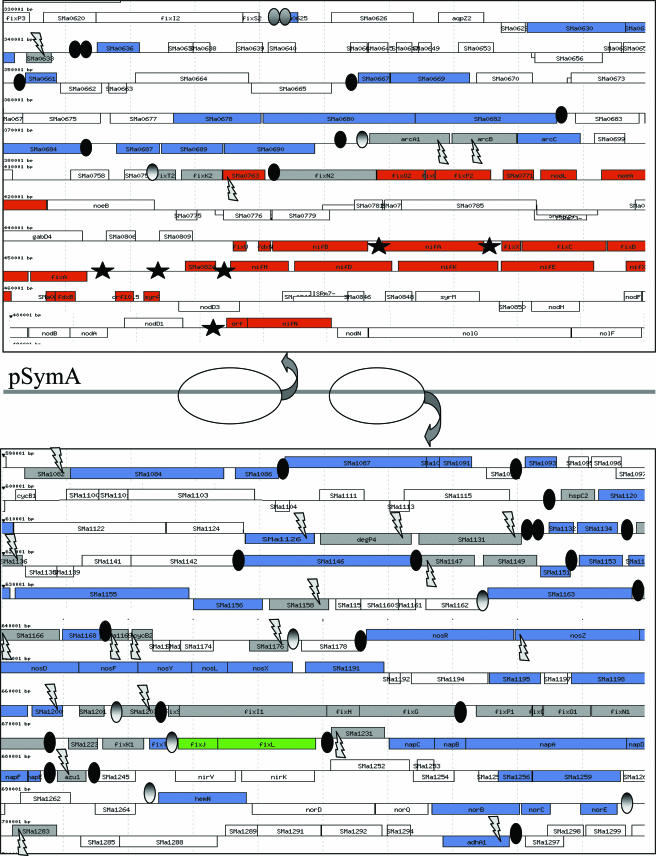
In order to determine which of the FixJ-dependent genes were activated by the intermediate transcriptional regulator FixK, we performed microarray experiments to compare the wild-type strain to a fixK1 fixK2 double mutant, both grown in microaerobic conditions. Among the FixJ-dependent genes, 87 were activated by FixK (Fig. 1) (Table S2, column 6; posted at http://bioinfo.genopole-toulouse.prd.fr/annotation/iANT/bacteria/rhime/DOC/Bobik2006/index.html). These genes included those already known to be regulated by FixK such as the genes encoding the respiratory oxidase complex fixNOQP1 (6), fixT2 (26), and fixM (14), thus validating our microarray approach. Two genes (SMa1147 and SMa1158), homologous to ORF277 from B. japonicum and R. capsulatus, which have previously been shown to be dependent upon FixJ (48) were also FixJ- as well as FixK-dependent in this study. Strikingly, all genes regulated by FixK were located on pSymA, in two main clusters (Fig. 2). Analysis of the pSymA intergenic sequences revealed the presence of 28 consensus FixK boxes (TTGA-N6-TCAA), 26 of which are located upstream of FixK-dependent genes, thus reinforcing the results obtained with the transcriptomic study. Additionally, 10 noncanonical FixK boxes (Table S2, column 7; posted at http://bioinfo.genopole-toulouse.prd.fr/annotation/iANT/bacteria/rhime/DOC/Bobik2006/index.html) were found in front of FixK-dependent genes. One of these modified boxes (TTGA-N6-GCAA), upstream of fixT, has indeed been shown to be functionally active (26). Altogether, FixK boxes could account for the activation of about 80 of the 87 genes regulated by FixK, if we consider putative operons.
FIG. 2.
FixJ targets on the symbiotic plasmid pSymA. Genes induced by FixJ in symbiosis, in microaerobiosis, or in both conditions are shown in red, blue, or gray, respectively. fixL and fixJ are in green. Putative NifA boxes (TGT-N10-ACA) are represented by a black star. Putative FixK boxes are represented by black ovals when they are consensus sequences (TTGA-N6-TCAA) and by gray ovals when they are noncanonical sequences (see Table S2, column 7 for the exact sequence; posted at http://bioinfo.genopole-toulouse.prd.fr/annotation/iANT/bacteria/rhime/DOC/Bobik2006/index.html). The gray flash signs indicate genes for which a mutant has been tested on M. sativa.
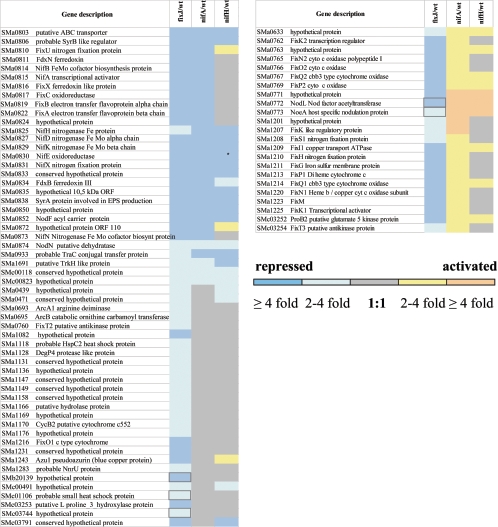
Genes for which regulation by FixJ and/or FixK was doubtful were analyzed by qPCR (Table S2; posted at http://bioinfo.genopole-toulouse.prd.fr/annotation/iANT/bacteria/rhime/DOC/Bobik2006/index.html) (Fig. 1).These genes could be classified in three groups: (i) genes regulated by FixJ but not by FixK although presenting a putative FixK box in their promoter (fixT1, SMa1100, SMa1132, and SMa1151), (ii) genes regulated by FixJ and FixK but for which we failed to find a putative FixK box (SMa0657, SMa1158, SMa1259, and SMa1288), and (iii) genes regulated by FixJ but not by FixK and which were not displaying a FixK box (SMa1256, SMb20139, SMb20433, and SMc00739). The results are presented in Tables 2 and 3. Six of these genes were validated as FixKtargets by qPCR: fixT1, SMa1132, SMa1151, SMa1158, SMa1256, and SMa1259. Six genes were not validated as FixJ dependent: SMa0657, SMa1100, SMa1288, SMb20139, SMb20433, and SMc00739. napABCD but not napEF (the two first genes of the presumed operon) were shown to be FixK dependent with the transcriptomic approach. The FixK dependency of napE and napF was validated by qPCR (Tables 2 and 3).
TABLE 2.
Relative transcript levels of selected genes under microaerobic conditionsa
| Gene | FC for wt/fixJ
|
FC for wt/fixK
|
||
|---|---|---|---|---|
| qPCR | Microarray | qPCR | Microarray | |
| SMa0657 | 1.43 | 2.93 | 1.03 | 2.00 |
| SMa1100 | 1.58 | 1.47 | 4.59 | 1.62 |
| SMa1132 | 21.5 | 2.09 | 23.34 | 1.62 |
| SMa1151 | 7.21 | 1.79 | 3.88 | 1.30 |
| SMa1158 | 12.75 | 7.43 | 21.07 | 3.48 |
| SMa1176 | 19.93 | 5.78 | 3.75 | 4.00 |
| fixT1 | 46.51 | 0.55 | ||
| napF | 6.95 | 4.71 | 5.78 | 1.00 |
| napE | 5.7 | 2.09 | 6.92 | 1.74 |
| SMa1256 | 3.19 | 2.50 | 6.37 | 1.46 |
| SMa1259 | 40.48 | 2.11 | 13.3 | 2.00 |
| SMa1288 | 1.29 | 1.86 | 3.26 | 3.36 |
| SMb20139 | 0.76 | 3.03 | ||
| SMb20433 | 1.31 | 1.80 | ||
| SMc00739 | 0.94 | 1.56 | ||
| SMb20026 | 0.44 | 0.31 | ||
| SMb20043 | 1.03 | 0.89 | ||
| SMb20518 | 1.22 | 1.39 | ||
| SMc03744 | 1.01 | 0.89 | ||
Transcript levels for the selected genes were determined for the wild type, the fixJ mutant, and/or the fixK mutant in microaerobic conditions using qPCR. Each reaction was made in triplicate. Relative transcript abundance is indicated as gene expression fold change (FC) between the wild type and the mutant strain. Values obtained with microarrays are also indicated.
TABLE 3.
Relative transcript levels of selected genes under bacteroid conditionsa
| Gene | FC in qPCR for:
|
|||||
|---|---|---|---|---|---|---|
| wt1/fixJ1 | wt1/fixJ2 | wt2/fixJ1 | wt2/fixJ2 | wt1/fixK1 | wt1/fixK2 | |
| nodL | 9.22 | 6.45 | 15.76 | 11.03 | 2.64 | 5.45 |
| noeA | 5.76 | 3.49 | 7.69 | 4.66 | ||
| SMb20139 | 0.47 | 0.31 | 0.47 | 0.31 | ||
| SMc01106 | 1.10 | 1.15 | 1.36 | 1.42 | ||
| SMc03744 | 2.59 | 3 | 3.34 | 3.86 | ||
Transcript levels for the selected genes were determined for the wild type, the fixJ mutant, and/or the fixK mutant in bacteroid conditions using qPCR. Each reaction was made in triplicate. Relative transcript abundance is indicated as gene expression fold change (FC) between the wild type and the mutant strain. wt1 and -2 and fixJ1 and -2 are biological replicates; fixK1 and -2 are independent RNA extractions from one aeroponic tank.
Interestingly, only three genes of pSymB (SMb20026, SMb20043, and SMb20518) displayed FixK boxes upstream of their coding sequence. These genes were not assigned as FixK induced by either the microarray approach or qPCR (Tables 2 and 3), hence indicating that all FixK targets are on pSymA.
Finally, 97 genes were dependent upon FixJ, in microaerobic conditions, of which 91 were dependent upon FixK. Seventy-six novel FixK targets were identified. fixT1 was the only gene already shown to be dependent upon FixK that we were not able to reveal with the transcriptomic approach.
The FixK dependency could be either direct or indirect, since a potential FixK-like regulator SMa1207 was itself regulated by FixK. In order to test this possibility, we conducted genome-wide microarray analyses, comparing the transcriptome profiles of a SMa1207 mutant and the wild-type strain Rm1021, both exposed to a limited (2%) concentration of oxygen. The results indicated that none of the genes regulated by FixK were dependent upon the FixK-like regulator SMa1207 (data not shown). Therefore, the whole set of genes is most likely activated directly by FixK. The results show that FixK affects not only respiration but also different functions such as denitrification (nap, nor, and nos genes), arginine and ornithine metabolism (arcABC, SMa0678, SMa0680, and SMa0682), stress response (hspC2, SMa1147, SMa1149, SMa1158, and SMa1231), transport (SMa1087, SMa1120, SMa1155, and SMa1163), and heme biosynthesis (hemN). About 40 novel FixK targets are of completely unknown function.
Twenty-nine genes were induced in microaerobic (2% oxygen) conditions in a FixJ-independent manner. Two-thirds of these genes showed a modest (less than three times) induction level in microoxic conditions. One gene displayed a sevenfold increase in its level of expression and encodes a putative two-component receiver domain (SMc01504). Eight genes had been shown previously to be induced by microoxic conditions independently of FixJ (9, 24, 48). Seven of these genes (SMa1447, SMb20238, SMb20481, SMb20482, SMb21441, SMc04299, and SMc03247) were not induced under microoxic conditions in our study. This result may be explained by a slightly different genetic background, shorter exposure to a low oxygen concentration (2 h instead of 6 or 8 h in the former studies [9, 48]), or a low level of induction of these genes (48) which did not allow us to detect their induction in microarray experiments. However, the gene cyoC, encoding a ubiquinol oxidase subunit protein, was shown to be induced independently of FixJ in our study as well as in a previous one (48). Additionally, both of the other subunits of the ubiquinol oxidase (cyoA and cyoB) displayed the same expression profile (i.e., induced in microaerobic conditions) and independence of FixJ.
Transcriptome analysis of the FixJ regulon in symbiosis.
To assess the extent of the FixJ regulon under symbiotic conditions, transcript profiles of 14-day-old nodules of M. sativa infected with either the wild-type strain 1021 or the fixJ mutant strain were compared. The transcriptomic approach revealed 79 genes which were dependent upon FixJ in symbiotic life (Fig. 3) (Table S3, column 2; posted athttp://bioinfo.genopole-toulouse.prd.fr/annotation/iANT/bacteria/rhime/DOC/Bobik2006/index.html). To identify genes whose expression could be affected by the Fix− phenotype of the mutant, we also determined the transcriptome of a null mutant for the nitrogenase structural gene, nifH. Among the potential targets of FixJ, 21 were also found using the nifH mutant (Fig. 3) (Table S3, column 4) and thus could have been eliminated from the list of FixJ-dependent genes. However, 8 out of these 21 genes, located downstream from nifH (nifDKE, nifX, SMa0833, fdxB, orf10.5, and syrA) are affected by the polar effect of nifH mutation itself (4), and they were considered bona fide FixJ-dependent genes. Additionally, the gene encoding the transcriptional regulator NifA was clearly FixJ dependent in symbiotic conditions in our analysis.
FIG. 3.
FixJ-dependent genes in symbiosis. Genes with a twofold decrease of their level of expression in the fixJ mutant compared to the wild-type bacteroids are presented in this figure (columns 1 and 2). Relative expressions of these genes in nifA and nifH mutant versus wild-type bacteroids are shown in columns 3 and 4, respectively. Yellow indicates genes that are upregulated in the mutant compared to the wild-type bacteria. Blue indicates genes that are downregulated in the mutant compared to the wild-type bacteria. A color scale is included at the bottom of the figure. An asterisk indicates a P value above 0.05. A dark frame indicates that the expression level of the gene has been verified by qPCR. The list of genes with M values obtained for each mutant can be found in Table S3 (posted at http://bioinfo.genopole-toulouse.prd.fr/annotation/iANT/bacteria/rhime/DOC/Bobik2006/index.html).
In order to identify the NifA regulon, we compared the transcript profiles of 14-day-old nodules elicited by a nifA mutant and the wild-type strain (Table S3, column 3; posted at http://bioinfo.genopole-toulouse.prd.fr/annotation/iANT/bacteria/rhime/DOC/Bobik2006/index.html) (Fig. 3). Among the genes regulated by FixJ, 19 appeared to be NifA targets. The analysis of the intergenic sequence upstream of these genes showed the presence of NifA boxes (TGT-N10-ACA) upstream of nifB, fixA, SMa0824, nifH, and SMa0872. An additional NifA box was also found upstream of nifA itself. The sequence analysis of the upstream regions of SMa0833, fdxB, orf10.5, and syrA did not reveal any consensus NifA box as would be expected if these genes were cotranscribed with the nifHDKE operon.
All genes known to be NifA targets were found in this study, and four novel ORFs (SMa0833, fdxB, orf10.5, and syrA) were confirmed to be NifA targets. All NifA targets are located on pSymA, close to the regulator itself (Fig. 2). Surprisingly, a set of genes including fixK1 and fixK2, fixNOQP1 and fixNOQP2, fixM, and fixGHIS were overexpressed in the nifA mutant strain (Fig. 3) together with fixLJ (data not shown), suggesting an interaction between the NifA and FixK regulons that we have not elucidated yet.
Five FixJ-dependent genes, nodL, noeA, SMb20139, SMc01106, and SMc03744 were not dependent upon NifA and were not FixK dependent in microaerobic conditions either. qPCR analyses were conducted to determine whether these genes were dependent upon FixJ. nodL and noeA, which form an operon (2), were validated as being part of the FixJ regulon (Table 3). To verify whether nodL could be FixK dependent in conditions where its expression was easily detected, qPCR analysis was performed on RNA extracted from nodules containing fixK bacteroids. The results indicated that nodL expression was diminished by a factor of 2.5 to 5.5 in a fixK mutant (Table 3). It remains to be demonstrated whether this is due to a cotranscription of nodL and fixNOQP2 or whether FixK activates nodL expression at its own promoter. SMc03744 was also validated as FixJ dependent in bacteroids, by using qPCR (Table 2). However, this gene was not upregulated in microaerobic conditions and its expression was FixJ independent (microarray approach [data not shown] and qPCR [Table 2]). Hence, it seems unlikely that SMc03744 is a genuine FixJ target. Altogether, these results indicate that FixJ did not possess any other direct target besides those already known (i.e., fixK, nifA, proB2, and SMc03253), ruling out the possibility of finding additional intermediate regulators activated by FixJ.
Biological significance of functions regulated by FixJ.
Among FixJ targets, several genes were related to stress response (hspC2, SMa1147, SMa1149, SMa1158, and SMa1231). This may imply that the fixJ mutant is more sensitive to stress than the wild-type strain. In order to test this hypothesis, the sensitivity to oxidative (H2O2) and osmotic (NaCl) stresses was assayed for the wild type and the fixJ mutant. The results show that, in the conditions used, both strains displayed the same sensitivity (data not shown).
Additionally, we tested the effect of microaerobic stress on the wild type and the fixJ mutant. Both strains presented the same behavior regarding cell growth (OD600) or cell capacity to form colonies (data not shown). Mutants for three of the genes mentioned above (SMa1147, SMa1158, and SMa1231) were inoculated onto M. sativa plantlets and examined for nodule formation and dry weight of the plants. All mutants displayed a wild-type phenotype, suggesting that these genes were not essential for symbiotic nitrogen fixation, in agreement with previous work (48). However, the lack of a symbiotic phenotype may be due to the presence of functional redundancy in the genome.
FixJ also affects functions such as denitrification and arginine metabolism. The symbiotic properties of mutants affected in genes involved in these functions (nosZ, arcA1, and arcB) or other functions (cycB2, nnrU, adhA1, azu1, and degP4) as well as by the FixK-like regulator (SMa1207) were also tested on M. sativa. Additionally, in order to test the biological relevance of some FixJ targets coding for hypothetical proteins, 10 mutants indicated in Fig. 2 were tested in planta. All the mutants tested were Nod+ Fix+ and showed a wild-type phenotype with regard to the dry weight of the plants measured 32 days after infection (data not shown).
Comparative genomics of the FixJ regulon.
Besides S. meliloti, the FixLJ two-component system exists in a number of other α-proteobacteria, including most α-rhizobia (B. japonicum, M. loti, and A. caulinodans but not Rhizobium leguminosarum bv. Viciae) and nonrhizobia such as C. crescentus, Rhodopseudomonas palustris, and Novosphingobium aromaticivorans (13).
B. japonicum and R. palustris carry the same proportion (ca. 50%) of the FixJ-dependent genes of S. meliloti, whereas M. loti has a lower proportion, for it lacks the whole denitrification pathway. Brucella, Agrobacterium, and R. leguminosarum bv. Viciae do not contain fixLJ orthologues although they carry the fixNOQP/ccoNOQP genes encoding the cbb3-type respiratory terminal oxidase. Bartonella carries neither fixLJ nor ccoNOQP genes and essentially none of the genes that constitute the FixJ regulon in S. meliloti.
Recently, the FixJ regulon of C. crescentus, a free-living aerobe that does not fix nitrogen, was determined experimentally from global transcriptional analysis (15). In C. crescentus FixJ upregulates 33 genes, of which only 9 are also FixJ activated in S. meliloti. C. crescentus only has ca. 20 likely orthologues of the genes belonging to the FixJ regulon of S. meliloti and, for example, lacks the whole denitrification pathway (nap, nor, and nos). The common FixJ-dependent genes in the two bacteria correspond to fixK, fixNOP, fixT, fixK-like (SMa1207), hemN, fixG, and SMa1168 (coding for an hypothetical protein).
Conversely, a number of genes that are fixJ-dependent in C. crescentus have not been detected as such in S. meliloti (e.g., the cytochrome bd terminal oxidase-encoding genes) or may not have orthologues in this bacterium (e.g., flmH).
The situation regarding FtrB, a FixJ-dependent regulator of carbon and amino acid metabolism in C. crescentus (15), is not clear. SMa1207 of S. meliloti displays only limited sequence similarity (29% identity) to FtrB of C. crescentus but is located in the vicinity of fixNOQP1 and fixGHIS operons as in Caulobacter. SMa1207 likely controls a subset of genes in yet-unidentified physiological conditions. However, a SMa1207 mutant displayed a wild-type phenotype in planta, indicating that this regulator may not have a major role in symbiosis.
Altogether, data in C. crescentus and S. meliloti point to a conserved FixJ-signaling pathway mainly involved in microaerobic respiratory functions and a species-specific part of the FixJ regulon with a variable size and functions that probably allow the bacteria to cope with additional constraints of the environment besides microoxic conditions.
DISCUSSION
The FixJ regulon is large and pSymA specific.
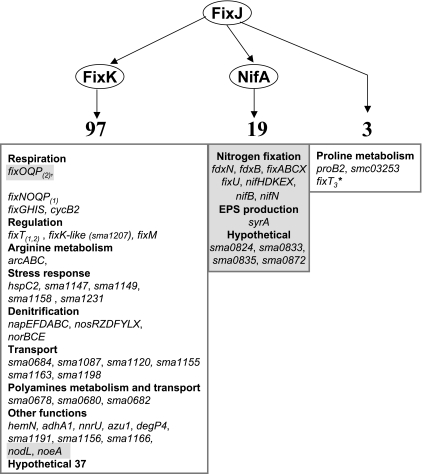
S. meliloti FixJ was known, from previous studies, to regulate the expression of ca. 35 genes in response to a limitation of oxygen. From the transcriptome and qPCR analyses described here, the size of the FixJ regulon was increased to, at least, 122 genes when considering genes activated by FixJ in either symbiotic or microaerobic life. As a point of comparison, most of the two-component regulatory systems of E. coli regulate less than 50 genes, and the ArcAB regulon is the only one that counts more than 100 members (39). The FixJ regulon of S. meliloti is thus among the largest known.
Behind the statistical evaluation of the transcriptome data, three circumstantial evidences indicate that the genes that we have identified are genuine FixJ targets. First, independent assessment of the nifA, fixK, and fixJ mutant transcriptomes led to an overall consistent picture of the FixJ regulon. Second, 97.5% of the genes activated by FixJ mapped in two clusters on pSymA, one of which contains fixLJ itself (Fig. 2). Only three genes were located on the chromosome—two of which were known already (24)—whereas pSymB had no FixJ target. Third, a majority of FixJ/FixK-dependent genes possessed a FixK box upstream of their coding sequence.
Conversely, although all the genes that were known to be FixJ targets were recovered in this study, it is however possible that a number of FixJ-dependent genes have been missed, due to their low level of expression, for example. This might be the case for 10 to 15 genes that were not detected as FixJ dependent in our assays, whereas neighbor genes in putatively the same operon had been detected. In a previous study, Barnett et al. found 750 genes downregulated in nodules of a fixJ mutant strain compared to wild-type S. meliloti (5). Most of these genes, all but 51, were not assigned as FixJ targets in our study. A number of reasons can account for this apparent discrepancy. First, it should be emphasized that the fixJ mutant was not used with the purpose of mapping the FixJ regulon in the Barnett et al. study. Hence, the two sets of data were analyzed differently. In our work, genes that were downregulated in both the fixJ and nifH mutants—both of which are Fix− in plants—were disregarded as bona fide FixJ targets. Second, nodules collected in the work by Barnett et al. were 35 days old instead of 14 as in our work, a difference which may be very significant in terms of bacterial gene expression, as nodules induced by the fixJ mutant are severely nitrogen starved and early senescing. Lastly, we noticed that the host plant differed in the two studies (Medicago truncatula versus M. sativa), although we do not know the impact of this.
We also identified in the course of this study genes whose expression was potentially negatively regulated by FixJ. These genes were very few in microaerobic conditions but were very numerous (ca. 280) in symbiosis (data not shown). However, most of them displayed a low expression level and/or a low expression ratio (less than twofold) between the wild type and the fixJ mutant, which made their genuine FixJ regulation doubtful. For this reason we have chosen to focus in this study on FixJ-activated genes, which could be identified without ambiguity, and left open the question of whether FixJ has a weak yet real negative impact on overall gene expression in bacteroids.
We did not find new FixJ direct targets besides the five that were already known (fixK1 and fixK2, nifA, proB2, and SMc03253). This confirms that the set of potential FixJ-binding sites that have been identified in S. meliloti by a Selex-based approach might not be functional in terms of gene activation (24). The FixJ-binding sites did not map upstream of or within genes that could be negatively regulated by FixJ from our analysis, thus making it unlikely that the cryptic FixJ-binding sites contribute to negative regulation either.
FixJ is a major regulator of microoxic and symbiotic life.
Among the 132 genes whose expression was upregulated by 2% oxygen limitation in free-living cultures of the wild-type strain in our assays, 74% of them were FixJ dependent. Hence, FixJ is clearly a major regulator of the microaerobic response, at least in this range of oxygen concentration. Nevertheless, these results also confirm the existence in S. meliloti of other regulatory systems that respond to low oxygen concentrations (24, 48). In previous work, 266 genes were shown to be upregulated by oxygen limitation (7). Yet oxygen limitation was stronger and longer in that previous work. In the present work, the transcript profiles were observed after only 2 h of microaerobiosis to avoid bacterial growth limitation and its side effect on overall gene expression.
Our data also indicate that FixJ is a major regulator of symbiotic gene expression, although the precise contribution of FixJ in this case is difficult to quantify precisely. In a separate study from our group aiming to describe the global transcription pattern of S. meliloti at different stages of the symbiosis, Capela et al. identified about 100 genes that were expressed more highly in mature than in young nodules (12). Sixty percent of these genes turned out to be FixJ dependent whereas, conversely, less than 2% of the genes expressed at an earlier symbiotic stage were FixJ dependent. Altogether, these results indicate that FixLJ is, at least, a major regulator of mature bacteroids. An even more surprising conclusion is raised by analyzing the FixJ transcriptome in the context of the cytology of fixJ-induced nodules. Earlier studies have shown that fixJ-dependent genes such as nifA and fixK express specifically in the so-called nitrogen fixation zone (ZIII) of mature nodules that contain type 3 and type 4 bacteroids (47, 50). Consistently, fixJ mutants infect nodules normally but do not differentiate into (or do not survive as) mature type 3-type 4 bacteroids (50). To our surprise, we found that a large majority of genes (ca. 80%) that appeared downregulated in nodules of the fixJ mutant were, in fine, genuine FixJ targets. This would imply that most of the genes activated in type 3-type 4 bacteroids are FixJ dependent, thus reinforcing the importance of FixJ regulation in symbiosis. However, we cannot rule out the possibility that the mutant strain used here presents a different cytology than the mutant described earlier (50). Actually, the strain backgrounds were not strictly identical and the nodules were grown for 4 weeks after inoculation in test tubes in the former study, whereas in our study, plants were grown in aeroponic tanks for only 2 weeks.
Another intriguing issue was the fact that only 38 FixJ-dependent genes were expressed (or at least detected) under both microaerobic and symbiotic conditions. Genes detected only in symbiotic conditions included the NifA regulon (19 genes) plus 5 additional genes, fixOQP2, nodL, and noeA. As for NifA-dependent genes, symbiotic specificity was expected since the NifA protein is extremely sensitive to O2 and requires the almost anoxic environment of the nodule for activity. Conversely, a large number of FixJ-dependent genes were only detected under microaerobic conditions, thus suggesting that a significant part of the FixJ regulation could be specific for the free-living lifetime of the bacterium. This would be consistent with the fact that many mutations in FixJ-dependent genes (Fig. 2) do not affect symbiosis. There is, however, an alternative explanation: most of these genes for which we did not detect expression in symbiosis were expressed at a low level under microaerobic conditions (data not shown). Hence we do not exclude that the lack of detection of these genes under symbiotic conditions might actually be due to a technical limitation, given that the overall sensitivity of the arrays is better with free-living than with symbiotic RNAs. Hence, whether FixJ contributes to biological/biochemical functions specific for free-living life in S. meliloti remains to be ascertained.
Biological functions controlled by FixJ.
Experimental assessment of the FixJ regulon has provided us with an almost exhaustive view of the biological functions that this two-component system controls in S. meliloti and that are described below. Yet our understanding is limited for a significant proportion (40%) of FixJ-dependent genes of unknown function. Seven transport genes have also been identified, the substrates of which are unknown.
(i) Microoxic respiration and oxidative phosphorylation.
No doubt the primary and ubiquitous function of the fixJ regulon in bacteria is to drive synthesis of the ccoNOQP-encoded respiratory oxidase, thus allowing efficient respiration and ATP generation in low oxygen environments. Yet it was noted before that ccoNO(Q)P genes could either be fnrN or fixLJ/fixK regulated in bacteria (13).
S. meliloti has three copies of the fixNOQP operon. Copy 1 was regulated by FixJ under both microoxic and symbiotic functions, whereas copy 2, which is located next to the NifA regulon (Fig. 2), was only detectable in bacteroids. In Sinorhizobium medicae, a second two-component system, ActR/ActS, modulates the expression of fixK fixN2, in addition to FixLJ, in response to a low pH in the medium (23). A similar situation in S. meliloti could account for higher expression of copy 2 in bacteroids. fixNOQP3, which was not induced under either condition tested, is partially phoB regulated (33) and does not depend on fixJ for expression. Copy 3 might thus contribute to growth of the fixJ mutant under microoxic conditions. Hence, the three copies of fixNOQP undergo differential regulation in S. meliloti. It was already shown in M. loti that the two fixNOQP operons were not expressed in the same physiological context; the copy located out of the symbiotic island functions preferentially in microaerobic environments, whereas bacteroids likely use both copies for symbiotic respiration (49).
As a difference with C. crescentus, we did not detect regulation by FixJ of the second high-affinity oxidase (cydABCD) that is also present in S. meliloti and that is likely to contribute to microoxic respiration in addition to or in the absence of the cbb3 oxidase. cyo genes encoding cytochrome o ubiquinol oxidase were found to be induced under microaerobic conditions, but independently of FixJ. Hence, several differentially regulated oxidases contribute to microoxic respiration in S. meliloti.
(ii) N2 fixation and denitrification are both controlled by FixLJ.
FixJ controls two major and opposite aspects of N2 metabolism in S. meliloti.
On the one hand, FixJ controls all genes involved in nitrogenase synthesis and functioning via nifA. Expression of these genes, and thus N2 fixation, specifically takes place under symbiotic conditions in S. meliloti. On the other hand, FixJ in S. meliloti and in other bacteria controls a large set of genes involved in denitrification, a pathway that is opposite to N2 fixation, as its end product is N2. Denitrification is a widespread respiratory pathway in bacteria in which oxides of nitrogen are used as electron acceptors in the absence of oxygen. We detected here low expression of nap, nor, and nos genes in both free-living microoxic and symbiotic conditions, suggesting that the overall denitrifying activity is low in bacteroids. Yet, denitrification activity was detected in bacteroids in a former study (38). Expression of denitrification genes is dependent upon FixJ and FixK (Fig. 4) as in B. japonicum (8, 36), although we did not find that nnrR was induced by FixJ in S. meliloti or induced under low oxygen. There is indeed increasing evidence for a role of denitrification in the successful interaction of pathogenic or symbiotic bacteria with their hosts (40). Denitrification may therefore serve an energetic function and/or as a protection against NO generated during infection. A third role was suggested in rhizobia in which denitrification could protect nitrogenase activity from inhibition by nitrate (34). Inactivation of nosZ (data not shown) and large deletions of the denitrification cluster of S. meliloti (32) did not prevent successful infection or nitrogen fixation. It is however possible that denitrification contributes to symbiosis only in specific environments (e.g., nitrate-containing or oxygen-depleted soils) that, to our knowledge, were not tested so far.
FIG. 4.
Revised FixLJ regulatory cascade. Genes in a gray frame were detected only in bacteroids. *, fixT3 is most likely a pseudogene.
We have found that cycB2, encoding a putative cytochrome c552, was dependent on fixJ for expression. The biological function of c552 is so far unknown.
(iii) Arginine and proline metabolism.
We have found here that FixJ controls arginine degradation via at least two pathways. FixJ controls expression of arcA1BC which belong to the arginine deiminase pathway, the most widespread anaerobic route for arginine degradation in bacteria. Although it was suggested previously that ornithine itself was required for symbiosis (42), we have found that mutations in arcA1 and arcB did not affect the ability of bacteria to form nitrogen-fixing nodules on M. sativa. SMa0680 and SMa0682 likely encode arginine and ornithine decarboxylases, respectively, and thus may be part of the arginine decarboxylase (ADC) pathway. The primary role of the ADC pathway may actually be putrescine (polyamine) synthesis. SMa0678, which is probably in the same operon as SMa0680 and SMa0682, encodes a putative putrescine/ornithine antiport transport protein. Polyamines are ubiquitously found in living organisms. Interrelationships between polyamine metabolism and cell growth and/or pH or osmotic stress response have already been suggested for Rhizobium fredii P220 and B. japonicumA1017 (27).
We have also found that FixJ controls expression of two genes involved in proline metabolism (proB2 and SMc03253) in agreement with previous findings (24).
(iv) Stress response.
We have found that FixJ regulates several genes related to stress response. This is the case for four homologues of the ORF277 of B. japonicum and R. capsulatus and ORF280 of Azospirillum brasilense (SMa1147, SMa1149, SMa1158, and SMa1231) which belong to the family of universal stress protein (43). A fifth gene (hspC2) encoding a heat shock protein was also regulated by FixJ. The presence of two of these stress proteins (SMa1231 and HspC2) was confirmed in the S. meliloti bacteroid proteome (21). In an attempt to identify the type of stress that FixJ controls, we tested three different stresses, i.e., oxidative, osmotic, and microaerobic, for a difference between the wild type and the fixJ mutant. Both strains displayed the same general behavior toward these stresses. More work would be needed to identify the stress to which FixJ responds.
(v) Infection-related functions.
Among FixJ targets identified in bacteroids are syrA, nodL, and noeA. SyrA affects exopolysaccharide biosynthesis (4). However a syrA mutant forms normal N2-fixing nodules on alfalfa, and the functional consequence of coupling the expression of a gene involved in exopolysaccharide production to the expression of those involved in N2 fixation is unknown. nodL and noeA form a flavonoid-inducible operon (2). nodL is responsible for the O acetylation of Nod factors and noeA displays homology with a methyltransferase, both being host-specific nodulation genes. Our observations may suggest an implication of nodL and noeA in late symbiotic steps. Yet the significance of this is obscured by the fact that expression of nodABC structural genes is turned down at the early stages of infection.
Acknowledgments
We are grateful to Claude Bruand, Delphine Capela, Anne-Marie Garnerone, and Julie Cullimore for critical reading of the manuscript and to Sébastien Carrère and Jerôme Gouzy for bioinformatic support. We thank the members of the group for help in harvesting nodules and helpful discussions.
We thank Erno Kiss for constructing the Rm1021 fixJ mutant strain.
We thank Anke Becker (University of Bielefeld, Germany) for providing S. meliloti microarrays and Rm1021 mutants. These mutants were constructed by Manuela Meyer and Eva Schulte-Berndt; we would like to thank them here. We thank D. Capela for verifying, by Southern blotting, all the mutants used.
Eliane Meilhoc was supported by the National Institute for Applied Sciences (INSA-Toulouse). Christine Bobik was supported by a doctoral fellowship from the French Ministère de l'Enseignement Supérieur et de la Recherche.
This study was partly supported by the Genopole Toulouse Midi-Pyrénées and the Departments SPE of INRA and CNRS.
REFERENCES
- 1.Ampe, F., E. Kiss, F. Sabourdy, and J. Batut. 2003. Transcriptome analysis of Sinorhizobium meliloti during symbiosis. Genome Biol. 4:R15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ardourel, M., G. Lortet, F. Maillet, P. Roche, G. Truchet, J. C. Prome, and C. Rosenberg. 1995. In Rhizobium meliloti, the operon associated with the nod box n5 comprises nodL, noeA and noeB, three host-range genes specifically required for the nodulation of particular Medicago species. Mol. Microbiol. 17:687-699. [DOI] [PubMed] [Google Scholar]
- 3.Barloy-Hubler, F., A. Cheron, A. Hellegouarch, and F. Galibert. 2004. Smc01944, a secreted peroxidase induced by oxidative stresses in Sinorhizobium meliloti 1021. Microbiology 150:657-664. [DOI] [PubMed] [Google Scholar]
- 4.Barnett, M. J., J. A. Swanson, and S. R. Long. 1998. Multiple genetic controls on Rhizobium meliloti syrA, a regulator of exopolysaccharide abundance. Genetics 148:19-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnett, M. J., C. J. Toman, R. F. Fisher, and S. R. Long. 2004. A dual-genome symbiosis chip for coordinate study of signal exchange and development in a prokaryote-host interaction. Proc. Natl. Acad. Sci. USA 101:16636-16641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Batut, J., M. L. Daveran-Mingot, M. David, J. Jacobs, A. M. Garnerone, and D. Kahn. 1989. fixK, a gene homologous with fnr and crp from Escherichia coli, regulates nitrogen fixation genes both positively and negatively in Rhizobium meliloti. EMBO J. 8:1279-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becker, A., H. Berges, E. Krol, C. Bruand, S. Ruberg, D. Capela, E. Lauber, E. Meilhoc, F. Ampe, F. J. de Bruijn, J. Fourment, A. Francez-Charlot, D. Kahn, H. Kuster, C. Liebe, A. Puhler, S. Weidner, and J. Batut. 2004. Global changes in gene expression in Sinorhizobium meliloti 1021 under microoxic and symbiotic conditions. Mol. Plant-Microbe Interact. 17:292-303. [DOI] [PubMed] [Google Scholar]
- 8.Bedmar, E. J., E. F. Robles, and M. J. Delgado. 2005. The complete denitrification pathway of the symbiotic, nitrogen-fixing bacterium Bradyrhizobium japonicum. Biochem. Soc. Trans. 33:141-144. [DOI] [PubMed] [Google Scholar]
- 9.Berges, H., C. Checroun, S. Guiral, A. M. Garnerone, P. Boistard, and J. Batut. 2001. A glutamine-amidotransferase-like protein modulates FixT anti-kinase activity in Sinorhizobium meliloti. BMC Microbiol. 1:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berges, H., E. Lauber, C. Liebe, J. Batut, D. Kahn, F. J. de Bruijn, and F. Ampe. 2003. Development of Sinorhizobium meliloti pilot macroarrays for transcriptome analysis. Appl. Environ. Microbiol. 69:1214-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Capela, D., S. Carrere, and J. Batut. 2005. Transcriptome-based identification of the Sinorhizobium meliloti NodD1 regulon. Appl. Environ. Microbiol. 71:4910-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Capela, D., C. Filipe, C. Bobik, J. Batut, and C. Bruand. 2006. Sinorhizobium meliloti differentiation during symbiosis with alfalfa: a transcriptomic dissection. Mol. Plant-Microbe Interact. 19:363-372. [DOI] [PubMed] [Google Scholar]
- 13.Cosseau, C., and J. Batut. 2004. Genomics of the ccoNOQP-encoded cbb3 oxidase complex in bacteria. Arch. Microbiol. 181:89-96. [DOI] [PubMed] [Google Scholar]
- 14.Cosseau, C., A. M. Garnerone, and J. Batut. 2002. The fixM flavoprotein modulates inhibition by AICAR or 5′AMP of respiratory and nitrogen fixation gene expression in Sinorhizobium meliloti. Mol. Plant-Microbe Interact. 15:598-607. [DOI] [PubMed] [Google Scholar]
- 15.Crosson, S., P. T. McGrath, C. Stephens, H. H. McAdams, and L. Shapiro. 2005. Conserved modular design of an oxygen sensory/signaling network with species-specific output. Proc. Natl. Acad. Sci. USA 102:8018-8023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davalos, M., J. Fourment, A. Lucas, H. Berges, and D. Kahn. 2004. Nitrogen regulation in Sinorhizobium meliloti probed with whole genome arrays. FEMS Microbiol. Lett. 241:33-40. [DOI] [PubMed] [Google Scholar]
- 17.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. [DOI] [PubMed] [Google Scholar]
- 18.D'Hooghe, I., J. Michiels, and J. Vanderleyden. 1998. The Rhizobium etli FixL protein differs in structure from other known FixL proteins. Mol. Gen. Genet. 257:576-580. [DOI] [PubMed] [Google Scholar]
- 19.D'Hooghe, I., J. Michiels, K. Vlassak, C. Verreth, F. Waelkens, and J. Vanderleyden. 1995. Structural and functional analysis of the fixLJ genes of Rhizobium leguminosarum biovar phaseoli CNPAF512. Mol. Gen. Genet. 249:117-126. [DOI] [PubMed] [Google Scholar]
- 20.Dixon, R., and D. Kahn. 2004. Genetic regulation of biological nitrogen fixation. Nat. Rev. Microbiol. 2:621-631. [DOI] [PubMed] [Google Scholar]
- 21.Djordjevic, M. A. 2004. Sinorhizobium meliloti metabolism in the root nodule: a proteomic perspective. Proteomics 4:1859-1872. [DOI] [PubMed] [Google Scholar]
- 22.Fahraeus, G. 1957. The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J. Gen. Microbiol. 16:374-381. [DOI] [PubMed] [Google Scholar]
- 23.Fenner, B. J., R. P. Tiwari, W. G. Reeve, M. J. Dilworth, and A. R. Glenn. 2004. Sinorhizobium medicae genes whose regulation involves the ActS and/or ActR signal transduction proteins. FEMS Microbiol. Lett. 236:21-31. [DOI] [PubMed] [Google Scholar]
- 24.Ferrieres, L., A. Francez-Charlot, J. Gouzy, S. Rouille, and D. Kahn. 2004. FixJ-regulated genes evolved through promoter duplication in Sinorhizobium meliloti. Microbiology 150:2335-2345. [DOI] [PubMed] [Google Scholar]
- 25.Fischer, H. M. 1994. Genetic regulation of nitrogen fixation in rhizobia. Microbiol. Rev. 58:352-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foussard, M., A. M. Garnerone, F. Ni, E. Soupene, P. Boistard, and J. Batut. 1997. Negative autoregulation of the Rhizobium meliloti fixK gene is indirect and requires a newly identified regulator, FixT. Mol. Microbiol. 25:27-37. [DOI] [PubMed] [Google Scholar]
- 27.Fujihara, S., and T. Yoneyama. 1993. Effects of pH and osmotic stress on cellular polyamine contents in the soybean rhizobia Rhizobium fredii P220 and Bradyrhizobium japonicum A1017. Appl. Environ. Microbiol. 59:1104-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gallusci, P., A. Dedieu, E. P. Journet, T. Huguet, and D. G. Barker. 1991. Synchronous expression of leghaemoglobin genes in Medicago truncatula during nitrogen-fixing root nodule development and response to exogenously supplied nitrate. Plant Mol. Biol 17:335-349. [DOI] [PubMed] [Google Scholar]
- 29.Garnerone, A. M., D. Cabanes, M. Foussard, P. Boistard, and J. Batut. 1999. Inhibition of the FixL sensor kinase by the FixT protein in Sinorhizobium meliloti. J. Biol. Chem. 274:32500-32506. [DOI] [PubMed] [Google Scholar]
- 30.Hauser, F., A. Lindemann, S. Vuilleumier, A. Patrignani, R. Schlapbach, H. M. Fischer, and H. Hennecke. 2006. Design and validation of a partial-genome microarray for transcriptional profiling of the Bradyrhizobiumjaponicum symbiotic gene region. Mol. Genet. Genomics 275:55-67. [DOI] [PubMed] [Google Scholar]
- 31.Hoang, H. H., A. Becker, and J. E. Gonzalez. 2004. The LuxR homolog ExpR, in combination with the Sin quorum sensing system, plays a central role in Sinorhizobium meliloti gene expression. J. Bacteriol. 186:5460-5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.House, B. L., M. W. Mortimer, and M. L. Kahn. 2004. New recombination methods for Sinorhizobium meliloti genetics. Appl. Environ. Microbiol. 70:2806-2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krol, E., and A. Becker. 2004. Global transcriptional analysis of the phosphate starvation response in Sinorhizobium meliloti strains 1021 and 2011. Mol. Genet. Genomics 272:1-17. [DOI] [PubMed] [Google Scholar]
- 34.Lucinski, R., W. Polcyn, and L. Ratajczak. 2002. Nitrate reduction and nitrogen fixation in symbiotic association Rhizobium-legumes. Acta Biochim. Pol. 49:537-546. [PubMed] [Google Scholar]
- 35.Meade, H. M., S. R. Long, G. B. Ruvkun, S. E. Brown, and F. M. Ausubel. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mesa, S., E. J. Bedmar, A. Chanfon, H. Hennecke, and H. M. Fischer. 2003. Bradyrhizobium japonicum NnrR, a denitrification regulator, expands the FixLJ-FixK2 regulatory cascade. J. Bacteriol. 185:3978-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moker, N., M. Brocker, S. Schaffer, R. Kramer, S. Morbach, and M. Bott. 2004. Deletion of the genes encoding the MtrA-MtrB two-component system of Corynebacterium glutamicum has a strong influence on cell morphology, antibiotics susceptibility and expression of genes involved in osmoprotection. Mol. Microbiol. 54:420-438. [DOI] [PubMed] [Google Scholar]
- 38.O'Hara, G. W., R. M. Daniel, and K. W. Steele. 1983. effect of oxygen on the synthesis, activity and breakdown of the rhizobium denitrification system. J. Gen. Microbiol. 129:2405-2412. [Google Scholar]
- 39.Oshima, T., H. Aiba, Y. Masuda, S. Kanaya, M. Sugiura, B. L. Wanner, H. Mori, and T. Mizuno. 2002. Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol. Microbiol. 46:281-291. [DOI] [PubMed] [Google Scholar]
- 40.Philippot, L. 2005. Denitrification in pathogenic bacteria: for better or worst? Trends Microbiol. 13:191-192. [DOI] [PubMed] [Google Scholar]
- 41.Puskas, L. G., Z. B. Nagy, J. Z. Kelemen, S. Ruberg, M. Bodogai, A. Becker, and I. Dusha. 2004. Wide-range transcriptional modulating effect of ntrR under microaerobiosis in Sinorhizobium meliloti. Mol. Genet. Genomics 272:275-289. [DOI] [PubMed] [Google Scholar]
- 42.Randhawa, G. S., and R. Hassani. 2002. Role of rhizobial biosynthetic pathways of amino acids, nucleotide bases and vitamins in symbiosis. Indian J. Exp. Biol 40:755-764. [PubMed] [Google Scholar]
- 43.Revers, L. F., L. M. Passaglia, K. Marchal, J. Frazzon, C. G. Blaha, J. Vanderleyden, and I. S. Schrank. 2000. Characterization of an Azospirillum brasilense Tn5 mutant with enhanced N(2) fixation: the effect of ORF280 on nifH expression. FEMS Microbiol. Lett. 183:23-29. [DOI] [PubMed] [Google Scholar]
- 44.Reyrat, J. M., M. David, C. Blonski, P. Boistard, and J. Batut. 1993. Oxygen-regulated in vitro transcription of Rhizobium meliloti nifA and fixK genes. J. Bacteriol. 175:6867-6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruberg, S., Z. X. Tian, E. Krol, B. Linke, F. Meyer, Y. Wang, A. Puhler, S. Weidner, and A. Becker. 2003. Construction and validation of a Sinorhizobium meliloti whole genome DNA microarray: genome-wide profiling of osmoadaptive gene expression. J. Biotechnol. 106:255-268. [DOI] [PubMed] [Google Scholar]
- 46.Ruvkun, G. B., V. Sundaresan, and F. M. Ausubel. 1982. Directed transposon Tn5 mutagenesis and complementation analysis of Rhizobium meliloti symbiotic nitrogen fixation genes. Cell 29:551-559. [DOI] [PubMed] [Google Scholar]
- 47.Soupene, E., M. Foussard, P. Boistard, G. Truchet, and J. Batut. 1995. Oxygen as a key developmental regulator of Rhizobium meliloti N2-fixation gene expression within the alfalfa root nodule. Proc. Natl. Acad. Sci. USA 92:3759-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trzebiatowski, J. R., D. M. Ragatz, and F. J. de Bruijn. 2001. Isolation and regulation of Sinorhizobium meliloti 1021 loci induced by oxygen limitation. Appl. Environ. Microbiol. 67:3728-3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uchiumi, T., T. Ohwada, M. Itakura, H. Mitsui, N. Nukui, P. Dawadi, T. Kaneko, S. Tabata, T. Yokoyama, K. Tejima, K. Saeki, H. Omori, M. Hayashi, T. Maekawa, R. Sriprang, Y. Murooka, S. Tajima, K. Simomura, M. Nomura, A. Suzuki, Y. Shimoda, K. Sioya, M. Abe, and K. Minamisawa. 2004. Expression islands clustered on the symbiosis island of the Mesorhizobium loti genome. J. Bacteriol. 186:2439-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vasse, J., F. de Billy, S. Camut, and G. Truchet. 1990. Correlation between ultrastructural differentiation of bacteroids and nitrogen fixation in alfalfa nodules. J. Bacteriol. 172:4295-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zimmerman, J. L., W. W. Szeto, and F. M. Ausubel. 1983. Molecular characterization of Tn5-induced symbiotic (Fix−) mutants of Rhizobium meliloti. J. Bacteriol. 156:1025-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]