Abstract
Ralstonia solanacearum GMI1000 is a gram-negative plant pathogen which contains an hrp gene cluster which codes for a type III protein secretion system (TTSS). We identified two novel Hrp-secreted proteins, called PopF1 and PopF2, which display similarity to one another and to putative TTSS translocators, HrpF and NopX, from Xanthomonas spp. and rhizobia, respectively. They also show similarities with TTSS translocators of the YopB family from animal-pathogenic bacteria. Both popF1 and popF2 belong to the HrpB regulon and are required for the interaction with plants, but PopF1 seems to play a more important role in virulence and hypersensitive response (HR) elicitation than PopF2 under our experimental conditions. PopF1 and PopF2 are not necessary for the secretion of effector proteins, but they are required for the translocation of AvrA avirulence protein into tobacco cells. We conclude that PopF1 and PopF2 are type III translocators belonging to the HrpF/NopX family. The hrpF gene of Xanthomonas campestris pv. campestris partially restored HR-inducing ability to popF1 popF2 mutants of R. solanacearum, suggesting that translocators of R. solanacearum and Xanthomonas are functionally conserved. Finally, R. solanacearum strain UW551, which does not belong to the same phylotype as GMI1000, also possesses two putative translocator proteins. However, although one of these proteins is clearly related to PopF1 and PopF2, the other seems to be different and related to NopX proteins, thus showing that translocators might be variable in R. solanacearum.
Ralstonia solanacearum is a soilborne bacterium that causes bacterial wilt disease in more than 200 plant species, including several economically important crops (30). Strain GM1000, whose complete genome sequence is known (64), is pathogenic on several solanaceous plants, such as tomato, pepper, eggplant, and petunia. On tobacco, this strain induces a typical defense reaction, called the hypersensitive response (HR), which is associated with resistance and is characterized by a rapid, localized programmed cell death of plant cells (53). Strain GMI1000 possesses a type III protein secretion system (TTSS) which controls both the ability to induce disease on host plants and to elicit the HR on tobacco. Components of this TTSS are encoded by a gene cluster, named the hrp (HR and pathogenicity) gene cluster, which comprises more than 20 genes and which is located on a megaplasmid (26).
hrp gene clusters have also been identified in other gram-negative plant pathogenic bacteria, such as Xanthomonas sp., Erwinia sp., Pantoea stewartii, and Pseudomonas syringae (1). Eleven hrp genes, renamed hrc genes, are conserved in all of these clusters, suggesting that they form the core of the Hrp TTSS secretion machinery (7). Two lineages of hrp gene clusters have been identified on the basis of hrp gene similarities and cluster organization: group I includes Erwinia sp., Pantoea stewartii, and Pseudomonas syringae, while group II includes Xanthomonas sp. and R. solanacearum (1, 27, 31).
Besides their conservation in phytopathogenic bacteria, TTSSs have also been found in a wide variety of gram-negative bacteria interacting with animals or insects as pathogens or as symbionts (18, 31). The key denominator explaining the wide conservation of these TTSSs certainly resides in the fact that they control the development of machineries capable of delivering proteins into the cytosol of eukaryotic cells (18, 31). Two classes of proteins are believed to be secreted by TTSSs: effectors that are translocated inside host cells and helper or accessory proteins that support the translocation of effectors (17, 56). Altogether, over 100 effectors or candidate effector genes have been identified in plant-pathogenic bacteria (2, 15, 21, 58). These effectors have a dual role (2). They are believed to control disease progression by countering plant defenses or by inducing nutrient release from the host cell. However, in some cases effectors trigger HR defense responses upon recognition by specific plant surveillance systems involving resistance (R) genes. In these cases, effectors are named avirulence (Avr) proteins, since they prevent pathogenicity on host plants containing a corresponding R gene (39).
TTSSs have been extensively analyzed in animal pathogens (18, 31). Their structure resembles the flagellar basal body, and they are associated with an extracellular needle complex, which might function as a conduit for the transport of effector and accessory proteins. In plant pathogens, extracellular appendages associated with TTSSs have also been found (60). These so-called Hrp pili are much longer (several micrometers) than needle complexes, but they have the same diameter (∼8 nm) (31). They are thought to penetrate the plant cell wall, which represents a major obstacle for the translocation process. Hrp pili have been described for P. syringae, R. solanacearum, and Erwinia amylovora (37, 59, 74) and more recently for Sinorhizobium fredii, Rhizobium sp. strain NGR234, and Xanthomonas campestris pv. vesicatoria (43, 63, 76). Mutations in genes encoding Hrp pili prevent secretion of effectors and other accessory proteins in vitro (31). This result and elegant immunogold labeling observations suggest that the Hrp pili appendages form a channel allowing protein transport across the plant cell wall (36, 49). However, needle complexes and Hrp pili are not sufficient to deliver proteins inside animal or plant cells. Other accessory proteins that interact with host membranes are necessary. In Yersinia TTSS, three accessory proteins, LcrV, YopB, and YopD, called translocators, seem to associate to form a translocon required for the translocation of effector proteins across the plasma membrane into mammalian host cells (16, 29, 33, 52, 65). The existence of translocators has also been proposed in phytopathogenic bacteria. HrpF of Xanthomonas campestris pv. vesicatoria is required for translocation of AvrBs3 or AvrBS2 effector proteins into the plant cell but not for their secretion in the extracellular medium (14, 61, 69). Moreover, HrpF can form pores in a planar lipid bilayer system (12); this protein may thus act as a translocator. HrpF displays similarity to NolX (recently renamed NopX [63]), a TTSS-secreted protein of S. fredii and Rhizobium sp. strain NGR234 (34, 42, 51). However, the possible role of NopX as a translocator has not yet been studied. More recently the HrpK protein, secreted via the Pseudomonas syringae pv. tomato strain DC3000 TTSS and which displays a weak homology with HrpF, was shown to be involved in protein translocation and might therefore be a putative translocator (56).
Recently, using genomic and functional approaches, five candidate effectors of strain GMI1000 were shown to be translocated into host-plant cells via the R. solanacearum TTSS (21). Although an Hrp pilus has been characterized in R. solanacearum (74), translocators have not yet been identified. Here we report the identification of two proteins, PopF1 and PopF2, which are secreted via the GMI1000 TTSS. These proteins display homology to one another and to HrpF and NopX, and we present evidence that they are translocators for the R. solanacearum TTSS. We also show that double mutants carrying mutations in popF1 and popF2 can be partially complemented by HrpF from X. campestris pv. campestris. The contributions of PopF1 and PopF2 to virulence and translocation differs. PopF1 plays a more important role in virulence and HR elicitation than PopF2 under our experimental conditions. These differences might be due to differences in amino acid (aa) sequences and/or expression levels.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used for this study are described in Table 1. Escherichia coli cells were grown in Luria-Bertani medium at 37°C. R. solanacearum was grown at 28°C in complete medium B (8) or minimal medium supplemented with 20 mM glutamate (MMG) (5). When indicated, MMG was complemented with Congo red (100 μg ml−1). Xanthomonas campestris pv. campestris cells were grown at 28°C in Kado's medium (5). Antibiotics were used at the following concentrations (in milligrams liter−1): tetracyclin (Tc), 10 (E. coli and R. solanacearum) and 5 (X. campestris pv. campestris); ampicillin (Amp), 50; rifampin (Rif), 50; spectinomycin (Sp) 40; streptomycin (Sm) 200; kanamycin (Km) 25; gentamicin (Gm) 10 (E. coli) and 20 (R. solanacearum); and chloramphenicol (Cm), 12.5.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | F−recA lacZΔM15 | BRL |
| R. solanacearum | ||
| GMI1000 | Wild-type strain | 8 |
| GMI1402 | hrcS::Tn5-B20 mutant | 4 |
| GMI1410 | hrpY::Tn5-B20 mutant | 4 |
| GMI1475 | hrpB::Tn5-B20 mutant | 4 |
| GMI1551 | PopA::ΩSp/Sm | 5 |
| GMI1575 | prhA::Ω mutant | 50 |
| GMI1663 | popF1::ΩSp/Sm mutant | This study |
| GMI1664 | popF2::ΩCm mutant | This study |
| GMI1665 | popF2:: aprA (Gm) mutant | This study |
| GMI1666 | popF1::ΩSp/SmpopF2::ΩCm double mutant | This study |
| GMI1667 | popF1::ΩSp/SmpopF2::aprA (Gmr) double mutant | This study |
| GMI1668 | popF2-His6, Spr/Smr | This study |
| GMI1669 | popF2-His6hrcS::Tn5-B20, Spr/Smr Kmr | This study |
| GMI1670 | popF2-His6 (Sp/Sm) hrpY::Tn5-B20, Spr/Smr Kmr | This study |
| X. campestris pv. campestris 8004 | Wild type, Rifr | 73 |
| Plasmids | ||
| pBluescript II KS(−) | Cloning vector, Ampr | Stratagene |
| pGEM-T | Cloning vector, Ampr | Promega |
| pHP45Ω Sp/Sm | Spr, Smr, Ω cassette | 57 |
| pHP45ΩCm | Cmr, Ω cassette | 57 |
| pLAFR6 | Tcr | 35 |
| pMS107 | pIC20H containing the cyaA gene | 67 |
| pUC1318 Apra/Gm | Gmr, aprA cassette | 9 |
| pAM5 | pLAFR3 with a 2-kb insert containing hrpB, Tcr | 28 |
| pCL1 | pGEM T with a 2.5-kb insert containing popF2, Ampr | This study |
| pCL3 | pGEM-T with a 1.5-kb fragment containing popF1 N-terminal DNA region, Ampr | This study |
| pMG6 | pBluescript KS(−) with a 1-kb XbaI HindIII fragment containing popF2 C-terminal DNA region, Ampr | This study |
| pCL86 | pCZ205 with a 712-bp HindIII XbaI fragment carrying popF2 promoter, Ampr Gmr | This study |
| pCL94 | pLAFR6 carrying a 4.7-kb fragment carrying popF2 promoter and promoterless lacZ, Tcr Gmr | This study |
| pCZ205 | pUC19 carrying lacZ in KpnI-XbaI, Ampr Gmr | 46 |
| pCZ367 | pUC18-derived vector used for insertional mutagenesis, Ampr Gmr | 21 |
| pCZ406 | pCZ205 with a 600-bp HindIII XbaI fragment carrying popF1 promoter, Ampr Gmr | This study |
| pCZ408 | pLAFR6 carrying a 4.6-kb fragment carrying popF1 promoter and promoterless lacZ, Tcr Gmr | This study |
| pHrpF | pLAFR6 with a 3.5-kb HindIII-EcoRI fragment containing hrpFXcc, Tcr | This study |
| pMG7 | pMG6 with a 1-kb HindIII XhoI fragment containing popF2 C-terminal DNA region, Ampr | This study |
| pMDI | pGEM-T with a 3.5-kb HindIII-EcoRI fragment containing HrpFXcc, Ampr | This study |
| pPopF1 | pLAFR6 with a 2.5-kb HindIII-KpnI fragment carrying popF1 | This study |
| pPopF2 | pLAFR6 with a 2.5-kb HindIII-KpnI fragment carrying popF1 | This study |
| pSC154 | pET-26b(+)-derived vector with the cyaA′ gene from pMS107 used for translational fusion constructs; the T7 promoter is replaced by a ptac promoter, Kmr | 21 |
| pSC163 | PLAFR6 carrying a translational fusion between avrA1-99 and cyaA′ | 19 |
Bacterial conjugation, electroporation, and transformation.
Conjugations and triparental matings were performed as described previously by Arlat and colleagues to introduce plasmids into R. solanacearum (4) or X. campestris pv. campestris (3). Plasmids were also introduced into R. solanacearum strains by electroporation as previously described (46). Transformation of strains of R. solanacearum leading to homologous recombination of the incoming DNA was performed according to the method of Boucher et al. (8).
Molecular biology techniques, DNA sequencing, and sequence analysis.
Standard recombinant DNA techniques were performed as described previously (6). Chromosomal DNA was extracted from R. solanacearum or X. campestris pv. campestris as previously described (4). Restriction enzymes, DNA ligase, Taq polymerase (Invitrogen), and other DNA enzymes were used according to the manufacturer's recommendations. PCR amplifications were done in 50-μl volumes using genomic DNA of R. solanacearum strain GMI1000 or X. campestris pv. campestris strain 8004 (73). Sequences of oligonucleotide primers are listed in Table S1 in the supplemental material. To sequence the X. campestris pv. campestris 8004 hrpF gene and to verify junctions of all the constructs, including the His6 epitope fusion constructs, double-stranded plasmid templates were sequenced automatically with a Perkin Elmer/ABI3700 Sequencer.
DNA and protein sequences were analyzed using the GCG software package (Wisconsin Package version 9.0; Genetics Computer Group) or the Vector NTI package. Homology searches were performed using the NCBI BLAST server. The putative terminator sequences were found using the GCG program terminator.
Phylogenetic analyses.
Amino acid sequences were aligned and phylogenetic trees were reconstructed by the neighbor-joining method as implemented in ClustalX (71).
β-Galactosidase assays.
After collecting the bacteria by centrifugation and resuspending the pellet in water, the β-galactosidase activity was measured as described previously (4).
Cloning the hrpF gene of X. campestris pv. campestris strain 8004.
A 3.5-kb DNA fragment containing the hrpF gene sequences and promoter region (757 bp upstream of the ATG codon) was PCR amplified using primers PFXcc1 and FFXcc2 and cloned into the pGEM-T vector, giving plasmid pMD1. The cloned DNA fragment was sequenced using PFXcc1 and FFXcc2 primers and additional internal primers HS-10, HR-6, FS-2, FR-2 HR-4bis, and HS. The sequence of this DNA fragment matches, with 98% identity, the corresponding sequence of X. campestris pv. campestris strain ATCC33913 (22) (from nucleotides 1423578 to 1427109). The HrpF protein deduced from the 8004 strain is 99% identical to HrpF from strain ATCC33913. Plasmid pMD1 was double digested by HindIII and EcoRI to subclone the hrpF sequence into the pLAFR6 polylinker (35), generating plasmid pHrpF.
Cloning of the popF1 and popF2 genes.
A 2,506-bp DNA fragment containing the popF1 gene sequences and the 317-bp DNA region located immediately upstream of the popF1 start codon was PCR amplified using GMI1000 genomic DNA as a template and primers POPF1H and POPF1X. This fragment was cloned into the pCZ367 plasmid. The HindIII-KpnI fragment carried by this new construct was then subcloned into pLAFR6, generating plasmid pPopF1 (see Fig. 3B). A very similar strategy was used to clone the popF2 gene into pLAFR6. Primers POPF2H and POPF2X were used to amplify a 2,486-bp fragment using GMI1000 DNA as a template. This DNA fragment, which carries popF2 coding sequences and the upstream DNA region (265 bp), was cloned into pCZ367 and subcloned into pLAFR6 after HindIII/KpnI digestion, thus generating the pPopF2 plasmid (see Fig. 3B).
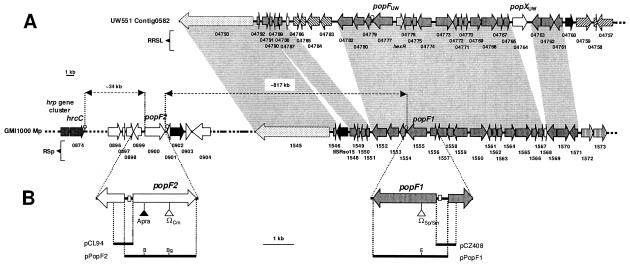
FIG. 3.
Genetic organization and relationships between genomic regions encompassing popF1 and popF2 in strain GMI1000 and popFUW551 and popXUW551 in strain UW551. (A) Representation of a GMI1000 megaplasmid (Mp) DNA region located at the righthand end of the hrp gene cluster and encompassing popF1 and popF2 (top) as well as genetic organization of part of contig0582 (accession no. NZ_AAKL01000001) of the strain UW551 draft genome sequence, which contains popFUW551 and popXUW551. Big arrows represent the predicted protein-coding genes. Arrows filled in dark gray correspond to orthologous genes having the same relative position in both strains. Hatched arrows represent genes having orthologs in the genome of the other strain but at different sites. A light gray arrow corresponds to hexR in UW551, which is present in strain GMI1000 (Rsp1556) and was not annotated in UW551. The dotted arrows represent Rsp1545 and RRSL04793 ORFs that display significant homologies but might not be true orthologs (data not shown). Putative integrase/recombinase or transposase genes are represented by black arrows. White arrows correspond to ORFs unique to each strain. Open circles show the position of putative Rho-independent transcription terminators. Colinear regions are shown by gray boxes. (B) Mutations and plasmids. The popF1 and popF2 DNA regions have been enlarged. Triangles show Ω or aprA cassette insertion sites. Horizontal black bars indicate sequences used in plasmid constructions. B, BamHI; Bg, BglII; E, EcoRI.
Construction of popF1 and popF2 promoter-reporter plasmids.
The popF1 promoter region (600 bp), corresponding to positions −601 to −1 relative to the A nucleotide of the popF1 start codon, was PCR amplified using primers HINDF1 and PF1X and cloned into the pGEM-T vector. The HindIII and XbaI restriction sites carried by HINDF1 and PF1X primers, respectively (boldface type in Table S1 in the supplemental material), were used to clone this fragment into the pCZ205 vector, which contains the lacZ gene sequences without a promoter. This construct, named pCZ406, was digested by HindIII and KpnI, generating a 4.6-kb DNA fragment carrying the popF1 promoter region, lacZ coding sequence, and a gene coding for gentamicin resistance. This fragment was cloned into pLAFR6 digested by HindIII and KpnI, thus generating the pCZ408 reporter plasmid (see Fig. 3B).
The promoter region of popF2 was cloned into a pGEM-T vector after PCR amplification of a 712-bp DNA fragment spanning positions −712 to −1 relative to the A nucleotide of the ATG start codon. GMI1000 genomic DNA was used as a template for primers PF2H and PF2X, which contain HindIII or XbaI restriction sites, respectively (Table S1 in the supplemental material). The XbaI/HindIII fragment was then cloned into the pCZ205 vector, thus generating plasmid pCL86. This plasmid was digested by HindIII and KpnI, generating a 4.7-kb DNA fragment carrying the popF2 promoter region, lacZ coding sequence, and a gene coding for gentamycin resistance, which was cloned into pLAFR6 digested by HindIII and KpnI, thus generating the pCL94 reporter plasmid (see Fig. 3B). pCL94 and pCZ408 were introduced in R. solanacearum strains by triparental matings.
Construction of popF1 or popF2 single mutants and popF1 popF2 double mutants.
The popF1 gene was disrupted by insertion of the ΩSp/Sm interposon. A 1,570-bp DNA fragment, corresponding to position −213 to +1345 relative to the ATG start codon of popF1, was PCR amplified using primers MFD and MFB2 and cloned into pGEM-T, thus generating pCL3. This plasmid was linearized by EcoRI and ligated with an EcoRI-digested ΩSp/Sm interposon, generating plasmid pCL3::ΩSp/Sm, which was then linearized by XbaI and used to transform various R. solanacearum strains. Transformants were selected on media containing spectinomycin and streptomycin.
The popF2 gene was disrupted by insertion of the ΩCm interposon (57) or by insertion of the aprA cassette, which confers resistance to gentamicin (Gmr) (9). A 2.4-kb DNA fragment, corresponding to positions −220 to +2211 relative to the ATG start codon of popF2, was PCR amplified using GMI1000 genomic DNA as template and primers MF2D and MF2F, which contain XbaI and HindIII restriction sites, respectively (underlined in the primer sequences). This DNA fragment was cloned into pGEM-T, generating plasmid pCL1. This plasmid was linearized by BglII and then ligated with the Cmr ΩCm interposon obtained by BamHI digestion of the plasmid pHP45ΩCm, thus generating plasmid pCL1::ΩCm. This plasmid was linearized by XbaI and used to transform various R. solanacearum strains. Transformants were selected on media containing chloramphenicol.
Alternatively, pCL1 was linearized by SmaI and ligated with the aprA cassette obtained by digestion of plasmid pUC1318 by SmaI. This new construct, named pCL1::aprA, was linearized by XbaI and used to transform various R. solanacearum strains. Transformants were selected on media containing gentamicin.
To obtain popF1 popF2 double mutants, the popF1::ΩSp/sm mutant GMI1663 was transformed with plasmid pCL1::ΩCm or pCL1::aprA linearized by XbaI. Transformants were either selected on media containing spectinomycin and chloramphenicol or spectinomycin and gentamicin, thus generating mutants GMI1666 and GMI1667, respectively.
Gene replacements were checked by PCR and by Southern hybridizations.
Modifications of popF2.
A DNA sequence encoding a His6 epitope (HHHHHH) was added at the C terminus of popF2 by PCR. For this purpose, GMI1000 genomic DNA was used to amplify the popF2-His6 fusion gene using the primers CF2D and CF2F. Primer CF2D contains an XbaI restriction site (underlined in the primer sequence) followed by the sequence of 5 amino acid codons of popF2 (codons 392 to 397); CF2F contains the sequence of the five carboxy-terminal amino acid codons of popF2, followed by the His6 coding sequence, a stop codon, and a HindIII restriction site (in boldface type in the primer sequence). The PCR product was digested by XbaI and HindIII, and the 1,062-bp fragment was cloned into pBluescript KS(−), generating plasmid pMG6.
A 970-bp HindIII/XhoI fragment corresponding to the DNA sequence located immediately downstream of the popF2 stop codon was generated by PCR amplification from GMI1000 genomic DNA using the primers PF2D and PF2F. After digestion by HindIII and XhoI (underlined in the PF2D and PF2F primer sequences, respectively), the PCR product was subcloned into pMG6 digested by HindIII and XhoI, producing plasmid pMG7. This plasmid was linearized by HindIII and then ligated with the Smr/Spr ΩSp/Sm interposon obtained by HindIII digestion of the plasmid pHP45ΩSp/Sm (57), giving pMG7::ΩSp/Sm. This plasmid was then linearized by XbaI and used to transform strain GMI1000. Transformants were selected on media containing streptomycin and spectinomycin. Gene replacements were checked by Southern hybridization.
Detection of Pop proteins in the supernatant and cell lysate.
Strain GMI1000 and derivatives were grown at 28°C for 16 h in MMG or MMG supplemented with Congo red (MMG-CR). Concentrated supernatant preparation (CSP or CSP-CR) and concentrated cell-free lysate (CLP) were prepared as described by Gueneron et al. (28) and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
SDS-PAGE and immunoblotting.
Electrophoresis of proteins in 12.5% polyacrylamide gels in the presence of sodium dodecyl sulfate (SDS-PAGE) was performed as described by Laemmli (45). After electrophoresis, proteins were either silver stained using the Silver Stain Plus kit (Bio-Rad, Calif.) or transferred onto a nitrocellulose membrane. Immunoblotting experiments were performed with rabbit polyclonal antibodies raised against PopA (data not shown) or with the mouse anti-His6 monoclonal antibody (Roche Molecular Biochemicals, Germany) which allows the detection of His6-tagged recombinant proteins. Peroxidase-conjugated sheep anti-mouse or goat anti-rabbit was used as secondary antibody and detected by using the ECL chemoluminescence kit (Amersham Pharmacia Biotech, Orsay, France).
Hrp pilus proteins.
Bacterial cell exostructures were isolated as previously described in Van Gijsegem et al. (74). Bacteria were grown in MMG solid medium and were resuspended in 10 mM Tris-HCl, pH 8, buffer, and their exostructures were shaved off by passing the bacterial suspension several times through a 28-gauge needle. Bacteria were then pelleted by centrifugation, and exostructures in the supernatant were partially purified by centrifugation in a Beckman SW40.1 swinging bucket rotor, first for 20 min at 6,000 × g. The supernatant was then collected and submitted to a subsequent 3-h centrifugation at 85,000 × g. The pellet containing the Hrp pili was resuspended in 10 mM Tris-HCl buffer and analyzed by SDS-PAGE using 10 to 20% Tris-Tricine Ready gels (Bio-Rad).
Protein sequencing.
After SDS-PAGE electrophoresis, proteins present in the CSP-CR of the wild-type strain GMI1000 were transferred onto polyvinylidene difluoride (PVDF) membranes (Problott Applied Biosystem). Protein bands on PVDF membranes were detected by Amido Black staining (N-3393; Naphtol Blue Black; Sigma). The 80-kDa protein band was cut out and microsequenced.
Automated Edman degradation of proteins was performed using an Applied Biosystems 475A sequencer and its online phenylthiohydantoin amino acid analyzer (model 120A); the reagents and methods were as specified by the manufacturer (Applied Biosystems, Roissy, France).
SDS-PAGE and immunodetection of AvrA1-99::CyaA fusion protein.
The construction of the avrA1-99::cyaA gene fusion has been described in detail by Cunnac (19). Briefly, a DNA fragment corresponding to codons 1 to 99 of avrA was generated by PCR amplification using GMI1000 DNA as a template and primers AvrA1 and AvrA2. This fragment was cloned into plasmid pSC154 to generate a translational fusion between AvrA1-99 and the CyaA′ coding sequences. The fusion construct was then subcloned into pLAFR6, generating plasmid pSC163. Immunodetection of CyaA fusion proteins was conducted as previously described by Cunnac et al. (21).
Adenylate cyclase (CyaA) translocation assay.
The procedures for adenylate cyclase assay were previously described in Schechter et al. (66) and in Cunnac et al. (21).
GMI1000 strain and derivatives carrying plasmid pSC163 were infiltrated into Nicotiana benthamania leaves at an optical density of 1.0 (21). Plants were sampled 7 h later with a cork borer. Leaf disks were transferred into Eppendorf tubes and immediately frozen in liquid nitrogen. For cyclic AMP (cAMP) extraction, samples in Eppendorf tubes were kept frozen while grinding by shaking with 2-mm tungsten beads shaken in a QIAGEN Mixer Mill MM300 for two runs of 2 min at 30 Hz. Samples were stored at −80°C before cAMP quantitation. Protein concentration was assessed with the Bio-Rad protein assay kit. Cyclic AMP (cAMP) levels were monitored with a cAMP enzyme immunoassay kit (Biotrak; Amersham Pharmacia Biotech) according to the manufacturer's instructions.
Plant tests.
HR tests on tobacco leaves and pathogenicity assays on tomato plants were performed as previously described (50).
RESULTS
Characterization of popF1 and popF2, two genes encoding novel putative Hrp-secreted proteins.
In a previous study we compared, by SDS-PAGE and silver staining analysis, concentrated supernatant preparations (CSPs) of strain GMI1000 and hrp mutants carrying pAM5, a plasmid harboring the hrpB regulatory gene which allows overexpression of hrp genes. After growth in minimal medium supplemented with Congo red (CSP-CR), this work allowed the detection of PopB and of four additional putative Hrp-secreted proteins (28). Among these proteins, we identified an ∼80-kDa protein which was also present in CSP-CRs of mutants altered in the popABC operon, suggesting that it might correspond to a novel Hrp-secreted protein. Interestingly, this band (apparent Mr, 80,000) was also detectable by SDS-PAGE and Coomassie blue staining analysis in CSPs or CSP-CRs of the wild-type strain carrying pAM5, but it was never visible in CSPs or CSP-CR of hrp mutants harboring pAM5 (Fig. 1). Amino-terminal sequencing of the putative 80-kDa Hrp-secreted protein gave a 24-amino-acid sequence with 4 ambiguous residues (Fig. 2). This N-terminal sequence was used as a query and compared with sequences in the protein database deduced from the annotation of the complete genome sequence of strain GMI1000 (http://bioinfo.genopole-toulouse.prd.fr/annotation/iANT/bacteria/ralsto/). This allowed the identification of two genes, RSp0900 and RSp1555, coding for proteins that match the 24-amino-acid (aa) query sequence. Whereas the amino acid sequence of residues 2 to 25 of the putative protein encoded by gene RSp1555 matched the N-terminal amino acid sequence of the 80-kDa protein exactly (Fig. 2), that of the protein deduced from the RSp0900 gene matched the 24-aa query sequence partially. The RSp1555 and RSp0900 genes were subsequently named popF1 and popF2, respectively. Both genes are located on the megaplasmid of strain GMI1000; popF2 maps approximately 34 kb to the right-hand side of the hrp gene cluster, whereas popF1 lies ∼817 kb to the right of popF2 (Fig. 3A). The popF2 and popF1 genes seem to be surrounded by genes that have no apparent direct relationship with the TTSS.
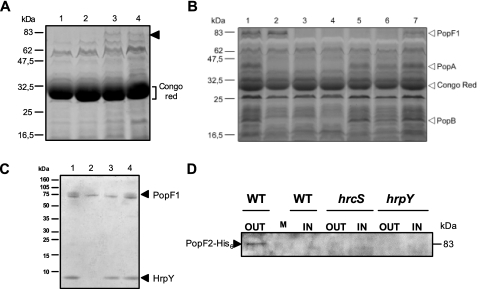
FIG. 1.
Secretion of PopF1 and PopF2 and detection of PopF1 in Hrp pili preparations. (A) Protein secretion after growth of various R. solanacearum strains carrying pAM5 in the presence of Congo red. Proteins present in CSP-CRs prepared from (lane 1) hrp mutant strain GMI1410(hrpY)/pAM5, (lane 2) hrp mutant strain GMI1402(hrcS)/pAM5, (lane 3) GMI1551(popA)/pAM5, and (lane 4) GMI1000/pAM5 grown in MMG supplemented with Congo red were separated by SDS-PAGE and visualized by Coomassie blue staining. The arrowhead shows the new putative 80-kDa Hrp-secreted protein. The bracket indicates the position of Congo red, which in SDS-PAGE exhibits the same electrophoretic mobility as a 28-kDa protein. (B) Proteins present in CSP-CRs prepared from GMI1000/pAM5 (lane 1), GMI1551(popA)/pAM5 (lane 2), hrp mutant strain GMI1402(hrcS)/pAM5 (lane 3), GMI1410 (hrpY)/pAM5 (lane 4), GMI1663 (popF1)/pAM5 (lane 5), GMI1666 (popF1 popF2)/pAM5 (lane 6), and GMI1664 (popF2)/pAM5 (lane 7), grown in MMG supplemented with Congo red, were separated by SDS-PAGE and visualized by silver staining. Positions of PopA, PopB, PopF1, and Congo red are shown by arrowheads. (C) Characterization of PopF1 in Hrp pili preparations. Shown is a Tris-Tricine SDS-PAGE electrophoresis gel (10 to 20% acrylamide) loaded with protein samples prepared from the surface of the following strains grown on MMG: (lane 1) GMI1000, (lane 2) GMI1584(hrcV) mutant, (lane 3) GMI1663(popF1) mutant, and (lane 4) GMI1664(popF2) mutant. Proteins were revealed by Coomassie blue staining. Arrowheads show PopF1 and HrpY. (D) Secretion of PopF2-His6 in wild-type (WT) or hrp background. Bacterial strains GMI1668(popF2-His6), GMI1669(hrcS popF2-His6), and GMI1670(hrpY popF2-His6) were cultivated in MMG supplemented with Congo red. Concentrated supernatant preparations (CSP-CR) (OUT) and concentrated lysate preparations (CLP-CR) (IN) were prepared from these cultures, and their protein contents were separated by SDS-PAGE and visualized by immunoblotting with an anti-His6 monoclonal antibody. M, size marker.
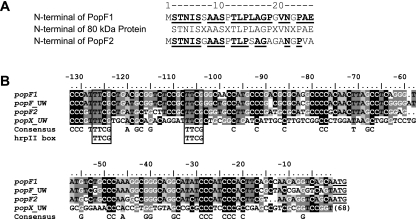
FIG. 2.
Sequence alignments of PopF1 and PopF2 N-terminal regions and of promoter DNA regions of popF1, popF2, popFUW551, and popXUW551. (A) Sequence alignment of the N-terminal sequences of the putative 80-kDa Hrp-secreted proteins PopF1 and PopF2. Amino acids from PopF1 or PopF2 present in the N-terminal sequence of the 80-kDa Hrp-secreted protein are shown in boldface and underlined. The ruler on the top of the sequences indicates amino acid position in PopF1 and PopF2. X, unknown amino acid. (B) Sequence alignment of popF1, popF2, popFUW551, and popXUW551 promoters. Nucleotides that are highlighted at a given position in the alignment are conserved in at least 50% of the sequences. The underlined three final nucleotides of popF1, popF2, and popFUW551 correspond to the initiation codon. The first adenine of this codon was used as the origin for the numbering of the sequence ruler. The number to the right of popXUW551 sequences represents the distance from the initiation codon. The open boxes indicate the hrpII box.
PopF1 and PopF2 belong to the HrpF/NopX family.
popF1 and popF2 code for putative proteins of 726 and 736 amino acids, with molecular masses of 77.28 and 78.08 kDa, respectively. These calculated molecular masses are in agreement with the apparent size of the sequenced protein. The theoretical pIs of PopF1 and PopF2 are significantly different, since that of PopF1 is 5.82 whereas that of PopF2 is 7.14.
The amino acid sequences of PopF1 and PopF2 share significant homology to one another, with 81% identity and 86% similarity (Table S2 in the supplemental material). Compared with sequences in the databases using BlastP, both PopF1 and PopF2 showed significant homology with the putative HrpF translocator from Xanthomonas campestris pv. vesicatoria and other Xanthomonas species or pathovars. Significant homologies were also observed with NopX from the symbiotic rhizobia S. fredii, Rhizobium sp. strain NGR234, and Mesorhizobium loti, which are related to HrpF (11). A summary of these results is presented in Fig. 4 and in Table S2 in the supplemental material.
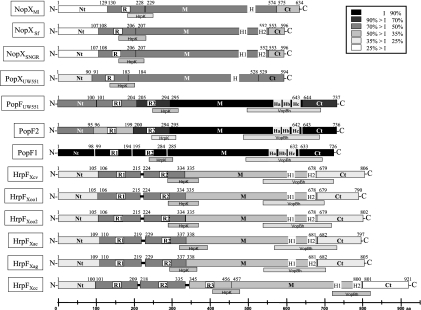
FIG. 4.
Schematic diagram of the structural organization and homologies between the HrpF, NopX, and PopF proteins. Proteins are shown by boxes and are subdivided into domains Nt, R, M, and Ct. Numbers located above each protein correspond to amino acid positions relative to their initiation codon and show domain borders. The level of identity (I) of each protein domain with the equivalent domain of PopF1 is presented with a black/gray color code. Putative transmembrane regions (H) are represented by gray boxes with white borders. Regions with homologies to HrpK or VopBh are indicated by adjacent gray bars (HrpK, Pseudomonas syringae pv. tomato DC3000, accession no. AAF771489; VopBh, Vibrio harveyi, accession no. AAS13307). Shown are Ralstonia solanacearum PopF1 (accession no. NP_523114); PopF2 (accession no. NP_522461); strain UW551 PopFUW551 (accession no. EAP74907) and PopXUW551 (accession no. EAP74895); Mesorhizobium loti NopX (NopXMl; accession no. NP_106860); Sinorhizobium fredii NopX (NopXSf; accession no. AAB19229); Rhizobium NGR234 NopX (NopXNGR; accession no. AAB91942); Xanthomonas campestris pv. vesicatoria HrpF (HrpFXcv; accession no. AAB86527); Xanthomonas oryzae pv. oryzae strain MA311018 HrpF (HrpFXoo1; accession no. BAB07869); X. oryzae pv. oryzae PXO99A HrpF (HrpFXoo2; accession no. AAS48653); Xanthomonas axonopodis pv. citri HrpF (HrpFXac; accession no. NP_640749); X. axonopodis pv. glycines HrpF (HrpFXag; accession no. AAP34358); X. campestris pv. campestris HrpF (HrpFXcc; accession no. NP_636591).
Characterization of PopF1 and PopF2 homologs in R. solanacearum strain UW551.
In fact, PopF1 and PopF2 showed the most significant similarities with RRSL_04777 protein from R. solanacearum strain UW551, for which a draft genome was recently obtained and annotated (25). Both proteins also displayed similarities with another protein of strain UW551, named RRSL_04764 (Table S2 in the supplemental material). RRSL_04777 displayed the best homologies with PopF2 (81% identity, 89% similarity) and PopF1 (78% identity, 85% similarity), whereas RRSL_04764 showed the best homologies with NopX from S. fredii and Rhizobium sp. strain NGR234 (63% identity, 77% similarity). The level of homology of RRSL_04764 with PopF1, PopF2, and RRSL_04777 was significantly lower (Table S2 in the supplemental material). RRSL_04777 and RRSL_04764 were subsequently named PopFUW551 and PopXUW551, respectively. It is worth noting that the theoretical pIs of PopFUW551 and PopXUW551 are both acidic, with values of 5.73 and 4.62, respectively. popFUW551 and popXUW551 genes are located on the same contig (Cont0582, accession no. NZ_AAKL01000001) (Fig. 3A). popFUW551 is surrounded by genes orthologous to those bordering popF1 in strain GMI1000, whereas popXUW551, which has no direct homolog in GMI1000, maps between two open reading frames (ORFs) which have orthologs in strain GMI1000. Interestingly these two GMI1000 orthologous ORFs border ORF RSp1568, which has no ortholog in the UW551 genome (Fig. 3A). The popF2 gene is located in a region which seems to be absent in strain UW551 (Fig. 3A). Therefore, it appears that, as observed for strain GMI1000, there are two genes coding for putative TTSS translocators in UW551. However, if one gene, popFUW551, seems to be well conserved with those of GMI1000, the other one, popXUW551, seems to be different (see below).
PopF and HrpF proteins display similarities to animal pathogen translocators of the YopB family and to HrpK translocator of P. syringae.
PopF1, PopF2, and PopFUW551 also display some degree of similarity with putative translocators from animal pathogens related to YopB of Yersinia spp. The highest score was obtained with VopB from Vibrio harveyi (VopBVh), which is part of the TTSS of this marine pathogen (32). The similarity was restricted to an ∼200-aa region located at the C terminus of PopF proteins (Fig. 4; Table S2 in the supplemental material). Other homologous proteins of this family include AopB from Aeromonas hydrophila (77), PopB from Pseudomonas aeruginosa (24), and YopB from Yersinia enterocolitica, Yersinia pseudotuberculosis, or Yersinia pestis (16), which gave less significant scores (Table S2 in the supplemental material). A multiple alignment between representatives of the YopB translocator family and the homologous sequence of PopF1 showed that the similarity occurs in one of the most conserved regions among YopB family members (Fig. 5). A BlastP search queried with the VopBVh region (aa 113 to 319) displaying homology to PopF1 and PopF2 showed that this region is also conserved in Xanthomonas HrpF proteins (E values ranging from 6e-4 to 7.6e-1) but not significantly in PopXUW551 or in Rhizobia NopX proteins (Fig. 4).
FIG. 5.
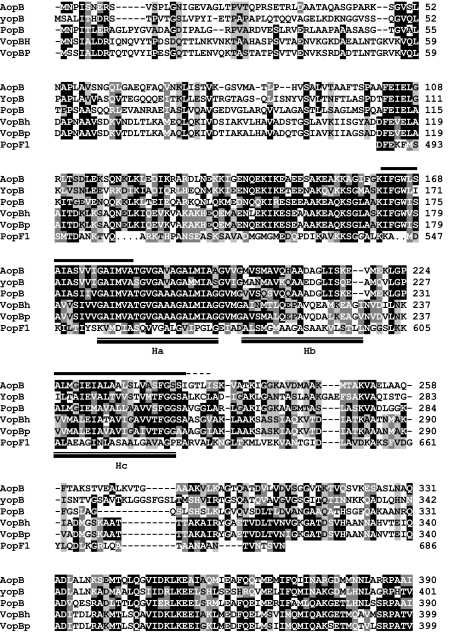
Amino acid sequence alignment of the region within PopF1 that is similar to YopB homologues. The PopF1 amino acid sequence from aa 487 to aa 686 was aligned with YopB from Yersinia enterolitica (accession no. AAD16813) and related TTSS putative translocators: AopB from Aeromonas hydrophila (accession no. AAS91821), PopB from Pseudomonas aeruginosa PAO1 (accession no. NP_250399), VopBh from Vibrio harveyi (accession no. AAS13307), and VopBp from Vibrio parahaemolyticus (accession no. NP_798036). Black boxes correspond to identical amino acids or conserved residues present in at least 50% of the sequences, dark gray indicates conserved substitutions, and light gray indicates semiconserved substitutions (following the ClustalW alignment sequence criteria [72]). Bars above the sequence of AopB represent the putative TM helices identified in YopB family proteins. The dashed line corresponds to a TM extension in YopB, PopB, VopBh, and VopBp. Double bars below the sequence of PopF1 show the position of Ha, Hb, and Hc putative TM of this protein. The ClustalW program was used for protein alignment.
Recently, using the PSI-BLAST program, Petnicki-Ocwieja et al. (56) showed that HrpK, a putative translocator of P. syringae pv. tomato TTSS, is similar to HrpF, NopX, PopF1, and PopF2 proteins over a 97-aa region located in the central part of these proteins (Fig. 4). This region is very well conserved among NopX, HrpF, PopF1, PopFUW551, and PopXUW551 proteins, but it is less conserved in PopF2.
Altogether, these similarities with the YopB family and with HrpK suggest that PopF1 and PopF2 are putative type III translocators of R. solanacearum strain GMI1000.
PopF1 and PopF2 contain putative transmembrane domains.
HrpF and HrpK proteins are known to share common structural features, such as transmembrane domains (TM), with the Yersinia YopB translocator and other translocators from animal pathogens (11, 56). Two putative TM domains, named H1 and H2, have been characterized in HrpF of X. campestris pv. vesicatoria (hereafter named HrpFXcv) and NopX of Rhizobium sp. NGR234 (hereafter named NGR234 and NopXNGR) (11, 34) (Fig. 4).
We used the TMpred program (http://www.ch.embnet.org/software/TMPRED_form.html) to identify putative TM segments in PopF1 and PopF2 and in related proteins. Three hydrophobic domains, called Ha, Hb, and Hc, located near the C terminus of PopF1, PopF2, and PopFUW551, were predicted to form putative membrane-spanning segments (Fig. 4). The TMpred scores for Ha (PopF1 score, 818; PopF2 score, 776; PopFUW551 score, 811), Hb (PopF1 score, 827; PopF2 score, 827; PopFUW551 score, 850), and Hc (PopF1 score, 641; PopF2 score, 641; PopFUW551 score, 639) domains in PopF1, PopF2, and PopFUW551 were lower than those obtained for HrpFXcv and NopXNGR H1 and H2 domains. Multiple alignments between PopF1, PopF2, PopFUW551, HrpFXcv, and NopXNGR using the Multalin program suggested that the Ha TM domain of PopF proteins might correspond to the H1 domain of HrpFXcv and NopXNGR, whereas the Hc domain might correspond to H2 domains (data not shown). Moreover, the Ha, Hb, and Hc putative TM domains are located in the region of PopF1 and PopF2 which displays similarity to the YopB family. Interestingly, the conserved region in YopB family members contains the two TM domains that have been characterized in these proteins (11) (Fig. 5). The Hc domain of PopF1/PopF2 overlaps the second TM of YopB family proteins, whereas the Ha domain only partially overlaps the first TM of YopB (Fig. 5).
PopF1 and PopF2 both control the interactions with plants.
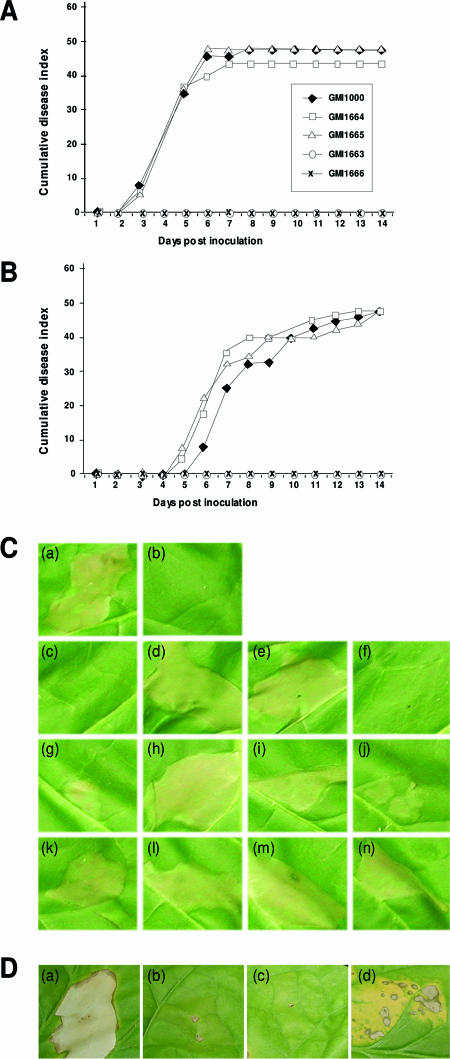
We constructed popF1 and popF2 single mutants and popF1 popF2 double mutants (Fig. 3; see Material and Methods). When inoculated on tomato plants, the popF1 mutant and the popF1 popF2 double mutants were unable to cause any wilting symptoms, thus behaving like hrp mutants (Fig. 6). In contrast, popF2 mutants were not affected in pathogenicity compared to the wild-type strain (Fig. 6). This was also true when a lower bacterial inoculum concentration was used (Fig. 6B).
FIG. 6.
Pathogenicity, HR, and complementation tests. (A and B) Average disease indices following root inoculation of 12 tomato plants per treatment by bacterial suspensions containing 3 × 108 CFU ml−1 (A) or 3 × 107 CFU ml−1 (B) of the wild-type strain GMI1000, popF1 mutant GMI1663, popF2 mutants GMI1664 and GMI1665, and popF1 popF2 double mutant GMI1666. These assays were repeated at least three times for each strain. Mutant GMI1667 (popF1 popF2) was also tested and gave results similar to those for mutant GMI1666, but it was not shown for clarity of the figure. (C) HR tests and complementation experiments with pPopF1 and pPopF2. Tobacco (cultivar Bottom Special) leaf shows responses 24 h after infiltration by bacterial suspensions (3 × 107 CFU ml−1) of R. solanacearum: (a) wild-type strain GMI1000, (b) hrcS mutant GMI1402, (c) popF1 popF2 mutant GMI1666, (d) GMI1666 carrying pPopF1 plasmid (GMI1666/pPopF1), (e) GMI1666/pPopF2, (f) GMI1666/pLAFR6, (g) popF1 mutant GMI1663, (h) GMI1663/pPopF1, (i) GMI1663/pPopF2, (j) GMI1663/pLAFR6, (k) popF2 mutant GMI1665, (l)GMI1665/pPopF1, (m) GMI1665/pPopF2, and (n) GMI1665/pLAFR6. (D) Complementation experiments with pHrpFXcc. Tobacco (cultivar Bottom Special) leaf shows responses 36 h after infiltration by suspensions (3 × 107 CFU ml−1) of R. solanacearum wild-type strain GMI1000 (a), hrpB mutant GMI1525 (b), popF1 popF2 double mutant GMI1666 (c), and mutant GMI1666 carrying pHrpFXcc plasmid (d).
Following infiltration into tobacco leaves, popF2 mutants did not show any alteration in the HR-inducing ability, but the popF1 mutant induced a partial and delayed HR. The popF1 popF2 double mutants did not induce any symptoms on tobacco leaves (Fig. 6C).
To confirm these results, we conducted complementation experiments. popF1 and popF2 were cloned into pLAFR6, generating pPopF1 and pPopF2 plasmids (Fig. 3B). These plasmids were introduced in trans into popF1 mutant (GMI1663), popF2 mutants (GMI1664 and GMI1665), and popF1 popF2 double mutants (GMI1666 and GMI1667). Interestingly, both pPopF1 and pPopF2 plasmids complemented the popF1 and popF1 popF2 mutants for HR induction (Fig. 6C). Disease complementation tests were not carried out, since previous experiments have shown that these kinds of tests only give partial phenotypes due to plasmid loss (50).
We also tested whether the HrpF protein of Xanthomonas campestris pv. campestris, hereafter called HrpFXcc, could complement the null HR phenotype of popF1 popF2 mutants. We chose HrpFXcc because previous experiments showed trans-complementation between X. campestris pv. campestris and GMI1000 hrp gene clusters (3). pHrpFXcc, a pLAFR6 derivative carrying hrpFXcc, was introduced into popF1 popF2 double mutants (strain GMI1666 or GMI1667). Interestingly, we observed a leaky but reproducible HR when strain GMI1666(pHrpFXcc) or GMI1667(pHrpFXcc) was infiltrated into tobacco leaves at 3 × 107 cells/ml, whereas strains GMI1666 and GMI1667 did not induce any HR at this inoculum concentration (Fig. 6D). No HR was ever observed when pHrpFXcc was introduced in trans into hrc mutant strains (data not shown).
Altogether, these results show that the popF1 popF2 double mutants behave like hrp mutants and that the functions of PopF1 and PopF2 are partially redundant and very similar to that of HrpF. However, the relative phenotypes of the popF1 and popF2 mutants on tomato and tobacco suggest that popF1 plays a more important role than popF2.
Do popF1 and popF2 belong to the hrpB regulon?
On the megaplasmid, both popF1 and popF2 ORFs are preceded by ORFs having a reverse orientation compared to their own orientation. Moreover, popF1 and popF2 coding sequences are followed by clear Rho-independent transcription termination sequences (Fig. 3). Altogether, these data and complementation results suggest that popF1 and popF2 are monocistronic operons transcribed from their own promoters.
The start codon of PopF1 is preceded by 313 nucleotides of noncoding DNA, whereas a 264-nucleotide noncoding DNA region precedes the PopF2 ORF. The sequence spanning −130 to 0 nucleotides upstream of both ORFs shares very significant homology (82% identity), whereas DNA regions further upstream are not similar. Interestingly, the promoter region of popFUW551 is highly similar to those of popF1 and popF2, whereas that of popXUW551 does not show significant similarities (Fig. 2B).
A conserved DNA motif, called the hrpII box, required for the regulation of hrp genes by the HrpB regulatory protein (20), was detected in the conserved upstream regions of popF1 and popF2 three nucleotides downstream of the beginning of the homologous region (Fig. 2B). The hrpII boxes of popF1 and popF2 are located 101 bp and 98 bp upstream of their respective initiation codons, suggesting that popF1 and popF2 belong to the hrpB regulon.
In order to study the regulation of these genes, DNA fragments located upstream of popF1 (600 bp) and popF2 (771 bp) start codons were cloned upstream of a promoterless lacZ gene in a reporter plasmid, giving pCZ408 and pCL94, respectively. These constructs were introduced into the wild-type strain or into the hrpB mutant GMI1525, and the production of β-galactosidase activity was measured after growth in rich medium (B) or minimal medium (MMG).
popF1 is not expressed in rich medium and is activated in minimal medium in an hrpB-dependent manner (Table 2). However, popF2 showed a similar and low level of transcription in both rich and minimal media, which was reduced by a twofold factor in the hrpB background in minimal medium but was unchanged in B medium. In addition, recently, a transcriptome analysis conducted in minimal medium clearly established that popF1 and popF2 are both positively regulated by hrpB (55). Clearly, popF1 belongs to the hrpB regulon, but the regulation of popF2 may be more complex, as our results obtained in minimal medium suggest that this gene is only partially regulated by hrpB (Table 2).
TABLE 2.
β-Galactosidase activity (Miller's units) of GMI1000 derivatives carrying pCZ408 or pCL94
| Gene and strain | Activity in medium:
|
|
|---|---|---|
| B | MMG | |
| popF1 | ||
| GMI1000(pCZ408) | 6 ± 1 | 54.6 ± 4.0 |
| GMI1525hrpB(pCZ408) | 3.3 ± 1.5 | 2.3 ± 0.6 |
| popF2 | ||
| GM1000(pCL94) | 18.3 ± 1.5 | 20.3 ± 1.2 |
| GMI1525hrpB(pCL94) | 19 ± 1.0 | 8.7 ± 0.6 |
Values are the averages of at least three independent experiments.
PopF1 and PopF2 are Hrp secreted but are not required for Pop secretion.
Comparison by SDS-PAGE analysis of CSP-CRs of the wild-type strain with popF1, popF2, or popF1 popF2 mutants carrying pAM5 indicated that the 80-kDa band might correspond to PopF1, since this band disappeared in CSP-CRs of the popF1 or popF1 popF2 mutants but not from those of the popF2 mutants (Fig. 1B), thus confirming our previous observations based on N-terminal sequence comparisons.
Furthermore, the analysis of CSP-CRs of these mutants showed that the secretion of PopA and PopB, two effector proteins of strain GMI1000, was not impaired, thus showing that popF1 and popF2 are not required for the secretory process in vitro (Fig. 1B). This observation was confirmed by Western analysis using an anti-PopA polyclonal antibody (data not shown).
In order to study PopF2 secretion, we introduced an immunological His6 tag at the C-terminal end of PopF2. The PopF2-His6 gene construct was marker exchanged into strain GMI1000 and in the two mutants GMI1402 (hrcS) and GMI1410 (hrpY), generating strains in which the popF2 gene was replaced by the His6-tagged derivative (see Materials and Methods). These strains were grown in MMG supplemented with Congo red, and the presence of PopF2-His6 was analyzed by Western blotting using an anti-His6 monoclonal antibody. A signal, corresponding in size (∼83 kDa) to popF2-His6, was detected in the CSP-CR from the wild-type derivative but not from the hrp mutant derivatives (Fig. 1D). This band was never detected in the CSP-CR of GMI1000, GMI1631 (popB-His6), or GMI1626 (popC-His6), showing that this signal seems specific for the PopF2-His6 fusion protein. We also noticed that this band was not detectable in the cell lysates prepared from the wild-type derivative or the hrp mutants carrying the popF2-His6 construct. This differs from previous observations of Pop proteins, which were always detected in the lysates of the wild type and of the hrp mutants (28, 46).
PopF1 and PopF2 are required for the translocation of AvrA.
Since PopF1 and PopF2 seem to belong to the HrpF family of translocators, we tested whether these proteins control the translocation of the avirulence protein AvrA. The avrA gene was characterized from R. solanacearum strain AW1 and was supposed to control HR development on tobacco plants, possibly at the species level (13). The strain GMI1000 possesses an avrA homologue named avrAGMI1000. This gene, previously referred to as RSc0608/brg46, is hrpB regulated, as shown in a study of strain GMI1000 TTSS candidate effectors (21). A GMI1000 avrA mutant is unable to induce the HR on Nicotiana benthamiana leaves (19). Transient expression of avrAGMI1000 in N. benthamiana cells results in HR induction, suggesting that this protein acts inside plant cells. Furthermore, experiments using the CyaA translocation reporter system in our laboratory have shown that this protein, and several other TTSS candidate effectors of strain GMI1000, are translocated into plant cells (19, 21). Translocation is tested by fusing the coding sequence of a candidate type III translocated protein to cyaA, the calmodulin-dependent adenylate cyclase domain of Bordetella pertussis cyclolysin (14, 66, 67). Since cAMP production by CyaA is dependent on calmodulin, which is only present in eukaryotic cells, the production of cAMP indicates that the CyaA domain was translocated into eukaryotic cells.
The plasmid pSC163 containing the translational fusion AvrA1-99::CyaA′ was introduced into strains GMI1000, GMI1402 (hrcS), GMI1663 (popF1), GMI1664 (popF2), and GMI1666 (popF1 popF2). The strains were grown in MMG supplemented with Congo red, and the CSP-CR and CLP-CR were analyzed by Western blotting with anti-CyaA antibodies. The fusion protein was found to be stable in the different strains (Fig. 7), and it was secreted in the extracellular medium in an Hrp-dependent manner (Fig. 7A). The AvrA1-99::CyaA′ construct was still secreted in popF1, popF2, and popF1 popF2 mutants.
FIG. 7.
Translocation of AvrA::CyaA′ fusion proteins into plant cells through the TTSS. (A) Immunoblot analysis of AvrA1-99::CyaA′ protein fusions in supernatants and cellular lysates. Bacterial strains were cultivated in MMG supplemented with Congo red. Concentrated supernatant preparations (CSP-CR) (OUT) and concentrated lysate preparations (CLP-CR) (IN) were prepared from these cultures, and their protein contents were separated by SDS-PAGE and visualized by immunoblotting with a monoclonal antiserum raised against adenylate cyclase for the following strains: (lane 1) GMI1666 popF1 popF2 carrying pSC163 (pLAFR6::AvrA1-99::CyaA′), (lane 2) GMI1663popF1/pSC163, (lane 3) GMI1000/pSC163, (lane 4) GMI1000, (lane 5) GMI1664popF2/pSC163, and (lane 6) GMI1402hrcS/pSC163. Control experiments with an anti-LacI antibody that detected the intracellular LacI protein demonstrated that cell lysis could not account for the signals detected with anti-CyaA′ antibody (data not shown). (B) Cya translocation assays showing adenylate cyclase activity of the AvrA1-99::CyaA′ fusion protein. Measurements of cAMP detected in bottom special tobacco leaf cells 7 h postinoculation with derivatives of the wild-type strain GMI1000 and hrp and popF mutants. For a particular strain, the means and standard deviations were calculated from measurements of three independent samples. Bacterial strains were the following: (lane 1) GMI1666 carrying pSC163 (pLAFR6::AvrA1-99::CyaA′), (lane 2) GMI1663popF1/pSC163, (lane 3) GMI1000/pSC163, (lane 4) GMI1000, (lane 5) GMI1664popF2/pSC163, and (lane 6) GMI1402hrcS/pSC163.
To study translocation, each strain was infiltrated into Nicotiana benthamiana leaves. Plant cell extracts were prepared 7 h later and assayed for cAMP. When expressed in the wild-type strain, the AvrA1-99::CyaA′ fusion protein resulted in higher cAMP levels than in the controls. The cAMP levels produced by the hrp mutant and the popF1 popF2 double mutant producing AvrA1-99::CyaA′ were very low and comparable to those of negative controls (Fig. 7). cAMP levels found using the popF1 mutant were low but significantly higher than those of hrp mutants and controls. popF2 mutants produced slightly lower levels than those obtained with the wild-type strain. The levels of cAMP produced by the mutants reflected the intensity of the HR they induced on tobacco leaves (Fig. 6B). We conclude that AvrA is translocated into plant cells and that full translocation activity requires both a functional type III secretion machinery and the presence of PopF1 and PopF2.
Is PopF1 associated with Hrp pili?
SDS-PAGE analysis of GMI1000 Hrp pili preparations showed that the hrpY gene encodes an abundantly present 7-kDa protein (75). A closer analysis revealed the presence of a double band in the upper part of the gel corresponding to proteins with an apparent mass of 80 kDa. Interestingly, the more diffuse upper band of this doublet seems to disappear in preparations of hrp mutants, whereas the lower band does not. In order to test whether this upper 80-kDa protein could correspond to PopF1 or PopF2, we compared pili preparations of the wild-type strain with those of popF1 or popF2 mutants by SDS-PAGE and Coomassie blue staining (Fig. 1C). The double band was clearly visible in preparations of strain GMI1000. The upper band disappeared in preparations of hrcV and popF1 mutants but was still present in preparations of popF2 mutants. The 7-kDa HrpY protein which is absent in the hrcV preparation was still visible in preparations of popF1, popF2, and popF1 popF2 mutants. Electron microscopic examinations of negatively stained popF1, popF2, and popF1 popF2 mutants showed that the production of Hrp pili was not impaired in these mutants (data not shown).
These results suggest that PopF1 protein might be associated with the Hrp pilus. They also indicate that PopF1 and PopF2 are not required for its assembly.
PopF, HrpF, and NopX proteins can be classified into three distinct groups.
The existence of four putative translocators in R. solanacearum prompted us to analyze the relationships between these proteins and HrpF or NopX protein. The comparison of whole-protein aa sequences, using alignments generated by ClustalW (72), showed that these proteins are well conserved. In addition, they can be subdivided into three major groups corresponding to their taxonomic origin: the NopX and HrpF proteins form two distinct groups, whereas PopF1, PopF2, and PopFUW551 are associated in a third group, indicating that these three latter proteins are more closely related to one another (Fig. S1 in the supplemental material). Interestingly, PopXUW551 is not in the PopF group but is positioned between PopF and NopX groups, apparently more closely related to the NopX group. Furthermore, this analysis places the PopF group closer to NopX proteins than to HrpF proteins.
Differential evolution of PopF, HrpF, and NopX domains.
A closer analysis reveals that the degree of similarity between PopF, PopX, HrpF, and NopX proteins varies greatly in different parts of these proteins. Multiple alignment and similarity comparisons provide strong evidence that PopF, PopX, NopX, and HrpF proteins can in fact be subdivided into the following four domains: an N-terminal domain (Nt), a repeat domain (R), a median domain (M), and a C-terminal domain (Ct) (Fig. 4). Interestingly, ClustalW analysis (Fig. S1 in the supplemental material) reveals that the grouping for each of these domains does not always match that for the entire protein, thus suggesting a certain independence in the evolution of the various domains and probably reflecting their different functions.
The R domain corresponds to repeated sequences, named R1 and R2, found in HrpFXcv (34) (Fig. 4). The analysis of PopF1, PopF2, and PopFUW551 revealed two 90- to 95-aa direct imperfect repeats in the N-terminal regions that partially match the R1 and R2 repeats of HrpFXcv. The three NopX proteins and PopXUW551 have only one copy of this region, whereas all HrpF proteins have two copies of the direct repeat, with the exception of HrpFXcc, which contains a third copy, named R3 (Fig. 4). The R domains of NopX, HrpF, and PopF proteins form three separate groups which are in line with the taxonomic classification (Fig. S1C in the supplemental material). The particularly large branch lengths of the R1 repeats of PopF2 and PopFUW551 suggest that the evolution of these repeats is relatively rapid (Fig. S1E in the supplemental material). Furthermore, the R1 and R2 repeats of PopF2 appear more divergent than those of PopF1.
The Nt domain is the most divergent in all groups (Fig. S1C in the supplemental material), suggesting that this domain is also subject to rapid evolution. The Nt domain of PopFUW551 appears more closely related to the PopF2 Nt domain than to the PopF1 Nt domain, in contrast to the M, Ct, and R domains of these proteins. Furthermore, it is worth noting a closer relationship between the Nt domain of PopXUW551 and the PopF Nt domains compared to the NopX Nt domain, whereas the other domains of this protein are more closely related to NopX proteins. Therefore, it is possible that the evolution of this domain is influenced by factors specific to each bacterial species. Taken together, these comparative analyses will certainly be very useful in future structure/function studies and in understanding the evolution of translocons in phytopathogenic bacteria.
DISCUSSION
In this study, we characterized two novel proteins, PopF1 and PopF2, secreted through the TTSS of R. solanacearum strain GMI1000. These two proteins are homologous to one another, and they share high similarities to NopX proteins from nitrogen-fixing symbiotic rhizobia and to HrpF proteins from Xanthomonas species. popF1 and popF2 do not seem to have the same importance in interactions with plants in our experimental system. popF1 plays a crucial role for disease development on tomato plants, whereas popF2 is not required for pathogenicity on this plant. The situation was slightly different for HR elicitation on tobacco leaves. The popF2 mutants gave HR very similar to those induced by the wild-type strain, whereas the popF1 mutant elicited a leaky and delayed HR. The popF1 popF2 double mutants did not induce any HR symptoms. This suggests that although popF1 seems to play a major part in HR induction, popF2 also seems to contribute to the development of this defense reaction. This hypothesis was confirmed by complementation experiments showing that when provided in trans, popF1 or popF2 can restore the ability to induce a normal HR to popF1 popF2 double mutants. The number of copies of pLAFR6 derivatives carrying popF1 or popF2 in these experiments might explain the appearance of normal rather than leaky HR. Are the differences observed between tomato and tobacco tests, concerning popF1 and popF2, due to plant or tissue specificities or to the type of symptoms that are studied (disease versus HR)? It will be interesting to extend our analysis to a broader range of plants to try to answer to these questions.
Our data strongly suggest that both PopF1 and PopF2 act as translocators for the R. solanacearum TTSS. These two proteins share characteristics specific to TTSS translocators: popF1 popF2 double mutants display an Hrp− phenotype on plants, but they retain the ability to secrete Pop and AvrA1-99::CyaA′ proteins in culture via the TTSS. However, both PopF1 and PopF2 are required for efficient translocation of AvrA1-99::CyaA′ protein into tobacco cells. This assay reflects the in planta response, since we observed a good correlation between the translocation efficiency of AvrA1-99::CyaA′ and the HR phenotype of popF1, popF2, and popF1 popF2 mutants. Finally, a popF1 popF2 mutant was partially complemented by the HrpF translocator of X. campestris pv. campestris, thus showing the functional similarity of R. solanacearum and Xanthomonas putative translocators.
In xanthomonads, the hrpF gene is unique and located very close to the hrp cluster in a region showing a high degree of variability, called the HrpF Peninsula (41, 47, 68). In S. fredii NGR234 and M. loti, nopX exists as a single copy and is located inside the TTSS gene cluster carried by symbiotic islands or plasmids (23, 38, 44). The situation seems slightly different in R. solanacearum, since we identified two genes coding for putative translocators. In strain GMI1000, these genes are not located in close vicinity of the hrp gene cluster. The popF2 gene is located 34 kb to the righthand end of the hrp gene cluster, and popF1 maps 817 kb further on. This region harbors at least 40 complete or truncated insertion elements, several transposase or integrase genes, and 12 DNA regions classified as ACURs (alternative codon usage regions), which may have been acquired through horizontal transfer (46, 64). In R. solanacearum, the hrp locus and its flanking regions cannot be considered a pathogenicity island, but it has most likely coevolved with the genome (64). Therefore, it is possible that the presence of popF1 and popF2 reflects the long evolution of this region, which has been submitted to several rearrangements. Is the presence of popF1 and popF2 the result of a duplication of an ancestral gene? Interestingly, strain UW551 also possesses two genes, popFUW551 and popXUW551, which code for putative related translocators. However, although PopFUW551 is closely related to PopF1 and PopF2, PopXUW551 is clearly different and seems to be more related to NopX proteins. Our phylogenetic study showed that NopX, HrpF, and PopF proteins form three distinct branches corresponding to their taxonomic origin, whereas PopXUW551 is positioned between PopF and NopX groups, thus forming a distinct and specific branch. No direct ortholog of PopXUW551 was found in strain GMI1000. Therefore, this gene seems unique to strain UW551. Interestingly, its location in the UW551 genome corresponds to a gene of GMI1000 which is absent in the UW551 genome (Fig. 3). This suggests that this locus is variable in R. solanacearum strains. It also raises the question of the origin of this gene. Concerning popFUW551, although it displays the best homologies with popF2, its location in the UW551 genome suggests that it corresponds to popF1 of GMI1000. This observation, and the fact that the DNA region carrying popF2 is not present in strain UW551, strongly favors the duplication hypothesis. Moreover, the importance of popF1 in the interaction with plants could suggest that this gene corresponds to the ancestral gene. However, we cannot rule out that the evolution of this gene after the putative duplication event has contributed to this gain of function. Alternatively, the fact that PopF2 and PopFUW551 seem to be more related to one another could suggest that they correspond to the ancestral gene. In GMI1000, it seems that the evolution of popF2 is accelerated compared to that of popF1. Thus, the presence of popXUW551 may also have influenced the evolution of popFUW551. This raises the following question: does the presence of two translocator genes in one genome influence their evolution? It will now be interesting to see whether popFUW551 and popXUW551 are both functional and serve as translocators for the TTSS and if they play similar roles in pathogenicity. It is worth noting that both genes possess an hrpII box in their promoter region (see reference 25 and Fig. 2), suggesting that they belong to the HrpB regulon and thus that they are related to the TTSS. Why do the two R. solanacearum strains studied so far carry two putative TTSS translocators, and why are these translocators different? Are there any relationships with their host range, their pathogenicity, or their phylogeny? R. solanacearum strains are classified into four phylotypes which seem to have evolved separately due to geographical isolation (25). Strain GMI1000 belongs to phylotype I, whereas UW551 is a member of phylotype II. In general, strains of phylotype I are thought to have a broader host range than strains of phylotype II, although this has recently been questioned (25). It will now be interesting to compare several R. solanacearum strains of different origins and phylotypes to see whether the presence of two sets of translocators is the rule or the exception in R. solanacerum strains and whether this is related to their phylogeny.
As mentioned above, the two translocators of strain GMI1000 do not seem to have the same importance for pathogenicity. Several explanations can be put forward to explain the differential behavior of these two genes. The first one is related to differences in expression of these genes. The expression of popF1 follows a typical hrp pattern; this gene is not expressed in rich medium and is induced in minimal medium in an hrpB-dependent manner. In contrast, by using transcriptional fusion with promoter of popF2, the level of transcription of this gene appeared similar in rich and minimal media and lower than that observed for popF1 in minimal medium. Our data and transcriptome analyses (55) also showed that popF2 is only partially regulated by HrpB in minimal medium. Did we introduce a bias when studying the expression of genes by cloning their promoter into reporter plasmids? Further experiments are needed to answer this question. The promoter regions of popF1 and popF2 share high similarity with one another, and they both contain an hrpII box, the presumed HrpB binding site. However, the hrpII boxes of these two genes differ by one nucleotide, and there are other differences in the promoter regions. A comparative analysis of these two promoters might help to characterize the molecular basis explaining their differential behavior in HrpB regulation.
A difference in expression is not the only possibility that can be put forward to explain the differences between popF1 and popF2 mutants. PopF1 and PopF2 proteins are very similar, but they display differences that might explain their differential behavior. Proteins in this translocator family can be subdivided into four domains, called Nt, R, M, and Ct, showing different levels of conservation. Each of these domains might correspond to different functions that are not subjected to the same selective pressure. The N termini of PopF1, PopF2, and PopFUW551, encompassing Nt and R1 domains, appears to be less conserved than the C termini (R2, M, and Ct domains), affecting the pIs of these proteins (pIs of PopF1, PopF2, and PopFUW551 are 5.82, 7.14, and 5.73, respectively). The pIs of PopF1 and PopFUW551 mostly correspond to the pI of HrpF/NopX family members, which ranges from 5.20 to 5.52, the only exception being HrpFXcc, with a pI of 6.01. Therefore, it seems that PopF2 occupies a particular place in this family. Do the differences observed between PopF1 and PopF2 N-terminal regions have an effect on the functionality or stability of PopF2, thus explaining its minor role in the interaction with plants? The Nt domain might be required for TTSS secretion by analogy to HrpFXcv, since a type III secretion signal was identified in the N terminus of this protein (12). The R domain is composed of two imperfect repeats (R1 and R2) in PopF1, PopF2, and PopFUW551 and in most HrpF proteins, whereas this region is not duplicated in NopX or PopXUW551 proteins. We observed that HrpFXcc carries three repeats, suggesting a high variability of this region. Is there any biological significance for the presence of these repeats and their number? It is worth noting that removal of R1 repeat in HrpFXcv does not affect its function (34).
The M domain is the most conserved domain among proteins of the HrpF/NopX/PopX/PopF family. The TM regions present in the C-terminal part of this domain might be related to the putative function of these proteins. The number of these domains varies from one in PopXUW551 and NopXMl of M. loti to two in other NopX proteins and in HrpF members and to three in PopF1, PopF2, and PopFUW551. In Xanthomonas campestris pv. vesicatoria, Büttner and colleagues (12) showed that the TM domains are essential for HrpF function. HrpFXcv has a lipid binding activity and induces pore formation in lipid bilayers (12). However, TM regions were not required for the lipid binding activity, suggesting that HrpF interacts with lipids via nonhydrophobic regions. TM domains have also been detected in translocators of animal pathogens such as IpaB, YopB, SipB, and EpsD of type III translocons of Shigella, Yersinia, Salmonella, and E. coli, respectively (see reference 11 for a review). Interestingly, the region of PopF and HrpF proteins carrying the putative TM domains shares a weak but significant similarity with a central region highly conserved in proteins belonging to the YopB translocator family. In YopB family members this region contains two TM domains (11). Multiple sequence alignments indicated that the TM domains of PopF and YopB proteins partially overlap. Although the occurrence of structural homologies between HrpF and YopB has already been mentioned (11, 31), this work is the first showing the existence of similarities between TTSS translocators of animal and plant pathogens. YopB can integrate into synthetic membranes, where it is associated with channel formation (70). However, this protein does not act alone to translocate effectors inside animal cells but does so in association with YopD and LcrV (33). Considering additionally the analogy to animal pathogen translocons, PopF1 and PopF2 are most likely type III translocators, and the possible involvement of accessory proteins should now be investigated. In P. syringae, proteins displaying high similarities with HrpF, NopX, or PopF1/PopF2 were not reported (10), but two candidate translocators, HrpK and HrpZ, have been identified. HrpK possesses all the characteristics of translocators: it has a TM domain and displays a very weak similarity to PopF, HrpF, and NopX. It is required for the translocation of Avr proteins (56). However, an hrpK mutation was not complemented by the hrpF gene of X. campestris pv. vesicatoria provided in trans. HrpZ, the other putative translocator of P. syringae, is a harpin which is capable of forming pores in lipid bilayers (48). Whether HrpK and HrpZ interact with one another to form a translocon has not yet been tested. Harpin-like proteins have been characterized in R. solanacearum and Xanthomonas sp. (5, 41, 78). In X. campestris pv. vesicatoria, XopA, which is secreted by the TTSS and which displays homologies with HpaG and Hpa1 harpin-like proteins of Xanthomonas axonopodis pv. glycines and Xanthomonas oryzae pv. oryzae (40, 68), has been suggested to be a component of the type III translocon (54). Indeed, a xopA mutant is reduced in pathogenicity without affecting type III secretion in vitro. It will be interesting to test whether the R. solanacearum PopA harpin-like elicitor is a putative translocator and interacts with PopF1 or PopF2.
Another question concerns whether the PopF1/PopF2 translocon forms a continuous conduit with the Hrp pilus or whether it functions at a distance to mediate effector delivery. In Y. pseudoturberculosis, YopB provided in trans cannot promote YopE translocation, thus suggesting that YopB must remain in the vicinity of the needle complex or even that there is continuous contact between YopB and the needle (62). In X. campestris pv. vesicatoria, immunoelectron microscopy experiments showed that HrpFXcv is in close contact with the Hrp pilus (76). However, it was not possible to conclude whether this localization was due to the detection of secreted HrpFXcv through the pilus conduit or whether it reflects an association between the translocon and the pilus. In S. fredii, Western blot analysis with antibodies raised against NopX revealed that this protein is associated with the TTSS pili (43). In NGR234, Saad and colleagues (63) showed that the TTSS pili are mainly composed of three proteins: NopA, the major component of the TTSS-pilus, NopB, a protein resembling FlgK, a protein involved in flagellum assembly, and NopX (63). In R. solanacearum, PopF1 is also associated with Hrp pili, as determined by SDS-PAGE analysis of purified pili preparations. Is the association of PopF, HrpF, or NopX with TTSS pili fortuitous, depending on specific properties of these proteins that allow nonspecific attachment to pili? Alternatively, does the presence of these proteins reflect continuity between the pilus and the translocon? Using the two-hybrid system, we were unable to detect an interaction between PopF1 and HrpY, the major component of R. solanacearum Hrp pilus. However, we cannot exclude that Hrp pili contain minor components that have not yet been detected and which connect the pilus and the translocon. Further work is needed to answer these questions.
Supplementary Material
Acknowledgments
We thank N. Grimsley for critical comments on the manuscript and J. Gouzy and S. Carrere for help in sequence analysis.
M. Guéneron, S. Cunnac, and D. Meyer were supported by grants from the Ministère de la Recherche et de l'Enseignement Supérieur. We gratefully acknowledge financial support from the Département Santé des Plantes et Environnement de L'Institut National de la Recherche Agronomique.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alfano, J. R., and A. Collmer. 1997. The type III (Hrp) secretion pathway of plant pathogenic bacteria: trafficking harpins, Avr proteins, and death. J. Bacteriol. 179:5655-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfano, J. R., and A. Collmer. 2004. Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42:385-414. [DOI] [PubMed] [Google Scholar]
- 3.Arlat, M., C. L. Gough, C. E. Barber, C. Boucher, and M. J. Daniels. 1991. Xanthomonas campestris contains a cluster of hrp genes related to the larger hrp cluster of Pseudomonas solanacearum. Mol. Plant-Microbe Interact. 4:593-601. [DOI] [PubMed] [Google Scholar]
- 4.Arlat, M., C. L. Gough, C. Zischek, P. A. Barberis, A. Trigalet, and C. A. Boucher. 1992. Transcriptional organization and expression of the large hrp gene cluster of Pseudomonas solanacearum. Mol. Plant-Microbe Interact. 5:187-193. [DOI] [PubMed] [Google Scholar]
- 5.Arlat, M., F. Van Gijsegem, J. C. Huet, J. C. Pernollet, and C. A. Boucher. 1994. PopA1, a protein which induces a hypersensitivity-like response on specific petunia genotypes, is secreted via the Hrp pathway of Pseudomonas solanacearum. EMBO J. 13:543-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1996. Current protocols in molecular biology, Wiley, New York, N.Y.
- 7.Bogdanove, A. J., S. V. Beer, U. Bonas, C. A. Boucher, A. Collmer, D. L. Coplin, G. R. Cornelis, H. C. Huang, S. W. Hutcheson, N. J. Panopoulos, and F. Van Gijsegem. 1996. Unified nomenclature for broadly conserved hrp genes of phytopathogenic bacteria. Mol. Microbiol. 20:681-683. [DOI] [PubMed] [Google Scholar]
- 8.Boucher, C. A., P. A. Barberis, A. P. Trigalet, and D. A. Demery. 1985. Transposon mutagenesis of Pseudomonas solanacearum: isolation of Tn5-induced avirulent mutants. J. Gen. Microbiol. 131:2449-2457. [Google Scholar]
- 9.Brau, B., U. Pilz, and W. Piepersberg. 1984. Genes for gentamicin-(3)-N-acetyltransferases III and IV: I. Nucleotide sequence of the AAC(3)-IV gene and possible involvement of an IS140 element in its expression. Mol. Gen. Genet. 193:179-187. [DOI] [PubMed] [Google Scholar]
- 10.Buell, C. R., V. Joardar, M. Lindeberg, J. Selengut, I. T. Paulsen, M. L. Gwinn, R. J. Dodson, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, S. Daugherty, L. Brinkac, M. J. Beanan, D. H. Haft, W. C. Nelson, T. Davidsen, N. Zafar, L. Zhou, J. Liu, Q. Yuan, H. Khouri, N. Fedorova, B. Tran, D. Russell, K. Berry, T. Utterback, S. E. Van Aken, T. V. Feldblyum, M. D'Ascenzo, W. L. Deng, A. R. Ramos, J. R. Alfano, S. Cartinhour, A. K. Chatterjee, T. P. Delaney, S. G. Lazarowitz, G. B. Martin, D. J. Schneider, X. Tang, C. L. Bender, O. White, C. M. Fraser, and A. Collmer. 2003. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 100:10181-10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buttner, D., and U. Bonas. 2002. Port of entry-the type III secretion translocon. Trends Microbiol. 10:186-192. [DOI] [PubMed] [Google Scholar]
- 12.Buttner, D., D. Nennstiel, B. Klusener, and U. Bonas. 2002. Functional analysis of HrpF, a putative type III translocon protein from Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 184:2389-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carney, B. F., and T. P. Denny. 1990. A cloned avirulence gene from Pseudomonas solanacearum determines incompatibility on Nicotiana tabacum at the host species level. J. Bacteriol. 172:4836-4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casper-Lindley, C., D. Dahlbeck, E. T. Clark, and B. J. Staskawicz. 2002. Direct biochemical evidence for type III secretion-dependent translocation of the AvrBs2 effector protein into plant cells. Proc. Natl. Acad. Sci. USA 99:8336-8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang, J. H., J. M. Urbach, T. F. Law, L. W. Arnold, A. Hu, S. Gombar, S. R. Grant, F. M. Ausubel, and J. L. Dangl. 2005. A high-throughput, near-saturating screen for type III effector genes from Pseudomonas syringae. Proc. Natl. Acad. Sci. USA 102:2549-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cornelis, G. R. 2002. Yersinia type III secretion: send in the effectors. J. Cell Biol. 158:401-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cornelis, G. R., A. Boland, A. P. Boyd, C. Geuijen, M. Iriarte, C. Neyt, M. P. Sory, and I. Stainier. 1998. The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 62:1315-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cornelis, G. R., and F. Van Gijsegem. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54:735-774. [DOI] [PubMed] [Google Scholar]
- 19.Cunnac, S. 2004. Genomewide identification of type III secretion system effectors from the bacterial plant pathogen Ralstonia solanacearum. Ph.D. thesis. Paul Sabatier University, Toulouse, France.
- 20.Cunnac, S., C. Boucher, and S. Genin. 2004. Characterization of the cis-acting regulatory element controlling HrpB-mediated activation of the type III secretion system and effector genes in Ralstonia solanacearum. J. Bacteriol. 186:2309-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cunnac, S., A. Occhialini, P. Barberis, C. Boucher, and S. Genin. 2004. Inventory and functional analysis of the large Hrp regulon in Ralstonia solanacearum: identification of novel effector proteins translocated to plant host cells through the type III secretion system. Mol. Microbiol. 53:115-128. [DOI] [PubMed] [Google Scholar]
- 22.Da Silva, A. C., J. A. Ferro, F. C. Reinach, C. S. Farah, L. R. Furlan, R. B. Quaggio, C. B. Monteiro-Vitorello, M. A. Van Sluys, N. F. Almeida, L. M. Alves, A. M. do Amaral, M. C. Bertolini, L. E. Camargo, G. Camarotte, F. Cannavan, J. Cardozo, F. Chambergo, L. P. Ciapina, R. M. Cicarelli, L. L. Coutinho, J. R. Cursino-Santos, H. El-Dorry, J. B. Faria, A. J. Ferreira, R. C. Ferreira, M. I. Ferro, E. F. Formighieri, M. C. Franco, C. C. Greggio, A. Gruber, A. M. Katsuyama, L. T. Kishi, R. P. Leite, E. G. Lemos, M. V. Lemos, E. C. Locali, M. A. Machado, A. M. Madeira, N. M. Martinez-Rossi, E. C. Martins, J. Meidanis, C. F. Menck, C. Y. Miyaki, D. H. Moon, L. M. Moreira, M. T. Novo, V. K. Okura, M. C. Oliveira, V. R. Oliveira, H. A. Pereira, A. Rossi, J. A. Sena, C. Silva, R. F. de Souza, L. A. Spinola, M. A. Takita, R. E. Tamura, E. C. Teixeira, R. I. Tezza, M. Trindade dos Santos, D. Truffi, S. M. Tsai, F. F. White, J. C. Setubal, and J. P. Kitajima. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459-463. [DOI] [PubMed] [Google Scholar]
- 23.Freiberg, C., R. Fellay, A. Bairoch, W. J. Broughton, A. Rosenthal, and X. Perret. 1997. Molecular basis of symbiosis between Rhizobium and legumes. Nature 387:394-401. [DOI] [PubMed] [Google Scholar]
- 24.Frithz-Lindsten, E., A. Holmstrom, L. Jacobsson, M. Soltani, J. Olsson, R. Rosqvist, and A. Forsberg. 1998. Functional conservation of the effector protein translocators PopB/YopB and PopD/YopD of Pseudomonas aeruginosa and Yersinia pseudotuberculosis. Mol. Microbiol. 29:1155-1165. [DOI] [PubMed] [Google Scholar]
- 25.Gabriel, D. W., C. Allen, M. Schell, T. P. Denny, J. T. Greenberg, Y. P. Duan, Z. Flores-Cruz, Q. Huang, J. M. Clifford, G. Presting, E. T. Gonzalez, J. Reddy, J. Elphinstone, J. Swanson, J. Yao, V. Mulholland, L. Liu, W. Farmerie, M. Patnaikuni, B. Balogh, D. Norman, A. Alvarez, J. A. Castillo, J. Jones, G. Saddler, T. Walunas, A. Zhukov, and N. Mikhailova. 2006. Identification of open reading frames unique to a select agent: Ralstonia solanacearum race 3 biovar 2. Mol. Plant-Microbe Interact. 19:69-79. [DOI] [PubMed] [Google Scholar]
- 26.Genin, S., and C. Boucher. 2004. Lessons learned from the genome analysis of Ralstonia solanacearum. Annu. Rev. Phytopathol. 42:107-134. [DOI] [PubMed] [Google Scholar]
- 27.Gophna, U., E. Z. Ron, and D. Graur. 2003. Bacterial type III secretion systems are ancient and evolved by multiple horizontal-transfer events. Gene 312:151-163. [DOI] [PubMed] [Google Scholar]
- 28.Gueneron, M., A. C. Timmers, C. Boucher, and M. Arlat. 2000. Two novel proteins, PopB, which has functional nuclear localization signals, and PopC, which has a large leucine-rich repeat domain, are secreted through the hrp-secretion apparatus of Ralstonia solanacearum. Mol. Microbiol. 36:261-277. [DOI] [PubMed] [Google Scholar]
- 29.Hakansson, S., K. Schesser, C. Persson, E. E. Galyov, R. Rosqvist, F. Homble, and H. Wolf-Watz. 1996. The YopB protein of Yersinia pseudotuberculosis is essential for the translocation of Yop effector proteins across the target cell plasma membrane and displays a contact-dependent membrane disrupting activity. EMBO J. 15:5812-5823. [PMC free article] [PubMed] [Google Scholar]
- 30.Hayward, A. C. 1991. Biology and epidemiology of bacterial wilt caused by Pseudomonas Solanacearum. Annu. Rev. Phytopathol. 29:65-87. [DOI] [PubMed] [Google Scholar]
- 31.He, S. Y., K. Nomura, and T. S. Whittam. 2004. Type III protein secretion mechanism in mammalian and plant pathogens. Biochim. Biophys. Acta 1694:181-206. [DOI] [PubMed] [Google Scholar]
- 32.Henke, J. M., and B. L. Bassler. 2004. Quorum sensing regulates type III secretion in Vibrio harveyi and Vibrio parahaemolyticus. J. Bacteriol. 186:3794-3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holmstrom, A., J. Olsson, P. Cherepanov, E. Maier, R. Nordfelth, J. Pettersson, R. Benz, H. Wolf-Watz, and A. Forsberg. 2001. LcrV is a channel size-determining component of the Yop effector translocon of Yersinia. Mol. Microbiol. 39:620-632. [DOI] [PubMed] [Google Scholar]
- 34.Huguet, E., and U. Bonas. 1997. hrpf of Xanthomonas campestris pv. vesicatoria encodes an 87-kDa protein with homology to NoIX of Rhizobium fredii. Mol. Plant-Microbe Interact. 10:488-498. [DOI] [PubMed] [Google Scholar]
- 35.Huynh, T. V., D. Dahlbeck, and B. J. Staskawicz. 1989. Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science 245:1374-1377. [DOI] [PubMed] [Google Scholar]
- 36.Jin, Q., and S. Y. He. 2001. Role of the Hrp pilus in type III protein secretion in Pseudomonas syringae. Science 294:2556-2558. [DOI] [PubMed] [Google Scholar]
- 37.Jin, Q., W. Hu, I. Brown, G. McGhee, P. Hart, A. L. Jones, and S. Y. He. 2001. Visualization of secreted Hrp and Avr proteins along the Hrp pilus during type III secretion in Erwinia amylovora and Pseudomonas syringae. Mol. Microbiol. 40:1129-1139. [DOI] [PubMed] [Google Scholar]
- 38.Kaneko, T., Y. Nakamura, S. Sato, E. Asamizu, T. Kato, S. Sasamoto, A. Watanabe, K. Idesawa, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, Y. Mochizuki, S. Nakayama, N. Nakazaki, S. Shimpo, M. Sugimoto, C. Takeuchi, M. Yamada, and S. Tabata. 2000. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 7:331-338. [DOI] [PubMed] [Google Scholar]
- 39.Keen, N. T. 1990. Gene-for-gene complementarity in plant-pathogen interactions. Annu. Rev. Genet. 24:447-463. [DOI] [PubMed] [Google Scholar]
- 40.Kim, J. G., E. Jeon, J. Oh, J. S. Moon, and I. Hwang. 2004. Mutational analysis of Xanthomonas harpin HpaG identifies a key functional region that elicits the hypersensitive response in nonhost plants. J. Bacteriol. 186:6239-6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim, J. G., B. K. Park, C. H. Yoo, E. Jeon, J. Oh, and I. Hwang. 2003. Characterization of the Xanthomonas axonopodis pv. glycines Hrp pathogenicity island. J. Bacteriol. 185:3155-3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krishnan, H. B. 2002. NopX of Sinorhizobium fredii USDA257, a type III-secreted protein involved in host range determination, is localized in the infection threads of cowpea (Vigna unguiculata [L.] Walp) and soybean (Glycine max [L.] Merr.) nodules. J. Bacteriol. 184:831-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krishnan, H. B., J. Lorio, W. S. Kim, G. Jiang, K. Y. Kim, M. DeBoer, and S. G. Pueppke. 2003. Extracellular proteins involved in soybean cultivar-specific nodulation are associated with pilus-like surface appendages and exported by a type III protein secretion system in Sinorhizobium USDA257. Mol. Plant-Microbe Interact. 16:617-625. [DOI] [PubMed] [Google Scholar]
- 44.Krishnan, H. B., and S. G. Pueppke. 1993. Flavonoid inducers of nodulation genes stimulate Rhizobium fredii USDA257 to export proteins into the environment. Mol. Plant-Microbe Interact. 6:107-113. [DOI] [PubMed] [Google Scholar]
- 45.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 46.Lavie, M., E. Shillington, C. Eguiluz, N. Grimsley, and C. Boucher. 2002. PopP1, a new member of the YopJ/AvrRxv family of type III effector proteins, acts as a host-specificity factor and modulates aggressiveness of Ralstonia solanacearum. Mol. Plant-Microbe Interact. 15:1058-1068. [DOI] [PubMed] [Google Scholar]
- 47.Lee, B. M., Y. J. Park, D. S. Park, H. W. Kang, J. G. Kim, E. S. Song, I. C. Park, U. H. Yoon, J. H. Hahn, B. S. Koo, G. B. Lee, H. Kim, H. S. Park, K. O. Yoon, J. H. Kim, C. H. Jung, N. H. Koh, J. S. Seo, and S. J. Go. 2005. The genome sequence of Xanthomonas oryzae pathovar oryzae KACC10331, the bacterial blight pathogen of rice. Nucleic Acids Res. 33:577-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee, J., B. Klusener, G. Tsiamis, C. Stevens, C. Neyt, A. P. Tampakaki, N. J. Panopoulos, J. Noller, E. W. Weiler, G. R. Cornelis, J. W. Mansfield, and T. Nurnberger. 2001. HrpZ(Psph) from the plant pathogen Pseudomonas syringae pv. phaseolicola binds to lipid bilayers and forms an ion-conducting pore in vitro. Proc. Natl. Acad. Sci. USA 98:289-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li, C. M., I. Brown, J. Mansfield, C. Stevens, T. Boureau, M. Romantschuk, and S. Taira. 2002. The Hrp pilus of Pseudomonas syringae elongates from its tip and acts as a conduit for translocation of the effector protein HrpZ. EMBO J. 21:1909-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marenda, M., B. Brito, D. Callard, S. Genin, P. Barberis, C. Boucher, and M. Arlat. 1998. PrhA controls a novel regulatory pathway required for the specific induction of Ralstonia solanacearum hrp genes in the presence of plant cells. Mol. Microbiol. 27:437-453. [DOI] [PubMed] [Google Scholar]
- 51.Marie, C., W. J. Deakin, V. Viprey, J. Kopcinska, W. Golinowski, H. B. Krishnan, X. Perret, and W. J. Broughton. 2003. Characterization of Nops, nodulation outer proteins, secreted via the type III secretion system of NGR234. Mol. Plant-Microbe Interact. 16:743-751. [DOI] [PubMed] [Google Scholar]
- 52.Neyt, C., and G. R. Cornelis. 1999. Insertion of a Yop translocation pore into the macrophage plasma membrane by Yersinia enterocolitica: requirement for translocators YopB and YopD, but not LcrG. Mol. Microbiol. 33:971-981. [DOI] [PubMed] [Google Scholar]
- 53.Nimchuk, Z., T. Eulgem, B. F. Holt III, and J. L. Dangl. 2003. Recognition and response in the plant immune system. Annu. Rev. Genet. 37:579-609. [DOI] [PubMed] [Google Scholar]
- 54.Noel, L., F. Thieme, D. Nennstiel, and U. Bonas. 2002. Two novel type III-secreted proteins of Xanthomonas campestris pv. vesicatoria are encoded within the hrp pathogenicity island. J. Bacteriol. 184:1340-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Occhialini, A., S. Cunnac, N. Reymond, S. Genin, and C. Boucher. 2005. Genome-wide analysis of gene expression in Ralstonia solanacearum reveals that the hrpB gene acts as a regulatory switch controlling multiple virulence pathways. Mol. Plant-Microbe Interact. 18:938-949. [DOI] [PubMed] [Google Scholar]
- 56.Petnicki-Ocwieja, T., K. van Dijk, and J. R. Alfano. 2005. The hrpK operon of Pseudomonas syringae pv. tomato DC3000 encodes two proteins secreted by the type III (Hrp) protein secretion system: HopB1 and HrpK, a putative type III translocator. J. Bacteriol. 187:649-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 58.Puhler, A., M. Arlat, A. Becker, M. Gottfert, J. P. Morrissey, and F. O'Gara. 2004. What can bacterial genome research teach us about bacteria-plant interactions? Curr. Opin. Plant Biol. 7:137-147. [DOI] [PubMed] [Google Scholar]
- 59.Roine, E., W. Wei, J. Yuan, E. L. Nurmiaho-Lassila, N. Kalkkinen, M. Romantschuk, and S. Y. He. 1997. Hrp pilus: an hrp-dependent bacterial surface appendage produced by Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 94:3459-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Romantschuk, M., E. Roine, and S. Taira. 2001. Hrp pilus - reaching through the plant cell wall. Eur. J. Plant Pathol. 107:153-160. [Google Scholar]
- 61.Rossier, O., G. Van den Ackerveken, and U. Bonas. 2000. HrpB2 and HrpF from Xanthomonas are type III-secreted proteins and essential for pathogenicity and recognition by the host plant. Mol. Microbiol. 38:828-838. [DOI] [PubMed] [Google Scholar]
- 62.Ryndak, M. B., H. Chung, E. London, and J. B. Bliska. 2005. Role of predicted transmembrane domains for type III translocation, pore formation, and signaling by the Yersinia pseudotuberculosis YopB protein. Infect. Immun. 73:2433-2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saad, M. M., H. Kobayashi, C. Marie, I. R. Brown, J. W. Mansfield, W. J. Broughton, and W. J. Deakin. 2005. NopB, a type III secreted protein of Rhizobium sp. strain NGR234, is associated with pilus-like surface appendages. J. Bacteriol. 187:1173-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Salanoubat, M., S. Genin, F. Artiguenave, J. Gouzy, S. Mangenot, M. Arlat, A. Billault, P. Brottier, J. C. Camus, L. Cattolico, M. Chandler, N. Choisne, C. Claudel-Renard, S. Cunnac, N. Demange, C. Gaspin, M. Lavie, A. Moisan, C. Robert, W. Saurin, T. Schiex, P. Siguier, P. Thebault, M. Whalen, P. Wincker, M. Levy, J. Weissenbach, and C. A. Boucher. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415:497-502. [DOI] [PubMed] [Google Scholar]
- 65.Sarker, M. R., C. Neyt, I. Stainier, and G. R. Cornelis. 1998. The Yersinia Yop virulon: LcrV is required for extrusion of the translocators YopB and YopD. J. Bacteriol. 180:1207-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schechter, L. M., K. A. Roberts, Y. Jamir, J. R. Alfano, and A. Collmer. 2004. Pseudomonas syringae type III secretion system targeting signals and novel effectors studied with a Cya translocation reporter. J. Bacteriol. 186:543-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sory, M. P., and G. R. Cornelis. 1994. Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol. Microbiol. 14:583-594. [DOI] [PubMed] [Google Scholar]
- 68.Sugio, A., B. Yang, and F. F. White. 2005. Characterization of the hrpF pathogenicity peninsula of Xanthomonas oryzae pv. oryzae. Mol. Plant-Microbe Interact. 18:546-554. [DOI] [PubMed] [Google Scholar]
- 69.Szurek, B., O. Rossier, G. Hause, and U. Bonas. 2002. Type III-dependent translocation of the Xanthomonas AvrBs3 protein into the plant cell. Mol. Microbiol. 46:13-23. [DOI] [PubMed] [Google Scholar]
- 70.Tardy, F., F. Homble, C. Neyt, R. Wattiez, G. R. Cornelis, J. M. Ruysschaert, and V. Cabiaux. 1999. Yersinia enterocolitica type III secretion-translocation system: channel formation by secreted Yops. EMBO J. 18:6793-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Turner, P., C. Barber, and M. Daniels. 1984. Behavior of the transposons Tn5 and Tn7 in Xanthomonas campestris pv. campestris. Mol. Gen. Genet. 195:101-107. [Google Scholar]
- 74.Van Gijsegem, F., J. Vasse, J. C. Camus, M. Marenda, and C. Boucher. 2000. Ralstonia solanacearum produces hrp-dependent pili that are required for PopA secretion but not for attachment of bacteria to plant cells. Mol. Microbiol. 36:249-260. [DOI] [PubMed] [Google Scholar]
- 75.Van Gijsegem, F., J. Vasse, R. De Rycke, P. Castello, and C. Boucher. 2002. Genetic dissection of Ralstonia solanacearum hrp gene cluster reveals that the HrpV and HrpX proteins are required for Hrp pilus assembly. Mol. Microbiol. 44:935-946. [DOI] [PubMed] [Google Scholar]
- 76.Weber, E., T. Ojanen-Reuhs, E. Huguet, G. Hause, M. Romantschuk, T. K. Korhonen, U. Bonas, and R. Koebnik. 2005. The type III-dependent Hrp pilus is required for productive interaction of Xanthomonas campestris pv. vesicatoria with pepper host plants. J. Bacteriol. 187:2458-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yu, H. B., P. S. Rao, H. C. Lee, S. Vilches, S. Merino, J. M. Tomas, and K. Y. Leung. 2004. A type III secretion system is required for Aeromonas hydrophila AH-1 pathogenesis. Infect. Immun. 72:1248-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhu, W., M. M. MaGbanua, and F. F. White. 2000. Identification of two novel hrp-associated genes in the hrp gene cluster of Xanthomonas oryzae pv. oryzae. J. Bacteriol. 182:1844-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.