Abstract
In Bacillus subtilis, Spx was recently characterized as a novel type of global regulator whose activity is regulated by the redox status of the cells. In the present study, we demonstrate that inactivation of Spx in the important pathogen Staphylococcus aureus renders the cells hypersensitive to a wide range of stress conditions including high and low temperature, high osmolarity, and hydrogen peroxide. Moreover, growth was restricted under nonstress conditions. Two-dimensional gel electrophoresis revealed that the proteome of the spx mutant differs substantially from the proteome of wild-type cells, supporting the finding that Spx is also a global regulator in S. aureus. More specifically, we demonstrated that Spx is required for transcription of trxB, encoding thioredoxin reductase, under all growth conditions examined. As trxB is essential in S. aureus, we speculate that the severely reduced trxB transcription could account for some of the growth defects of the spx mutant. Inactivation of spx also enhanced biofilm formation. S. aureus biofilm formation is associated with the production of the polysaccharide intercellular adhesin encoded by the ica operon. Interestingly, our data indicate that the augmented capacity of the spx mutant to form biofilms is due to Spx modulating the expression of icaR, encoding a repressor of the structural ica genes (icaABCD). In summary, we conclude that Spx fulfills an important role for growth, general stress protection, and biofilm formation in S. aureus.
Staphylococcus aureus is an opportunistic pathogen capable of forming a wide spectrum of infections ranging from folliculitis and food-borne intoxications to life-threatening endocarditis, osteomyelitis, and septicemia. The unique ability of S. aureus to persist in the environment, coupled with its ability to adhere to the surfaces of medical devices, has made this bacterium one of the major causes of hospital-acquired infections (49). On indwelling surfaces such as orthopedic devices or heart valves, the bacteria are associated in sessile, multicellular communities, also referred to as biofilms (4, 8). Bacteria living in biofilms are well protected from the host immune response as well as antibiotic agents and can therefore be very difficult to treat clinically.
To maintain cell viability under adverse environmental conditions, bacteria have evolved complex regulatory networks. An essential element of these networks is the controlled intracellular proteolysis performed by energy-dependent proteases. In gram-positive bacteria, the multisubunit ClpP proteolytic complexes play a central role in these adaptive processes (11, 12, 13, 35, 43). Functional ClpP proteolytic complexes have a bipartite molecular structure: a central proteolytic chamber is formed by two heptameric rings of the ClpP subunit (51). Access to the proteolytic chamber is controlled by two narrow, axial pores, and proteolytic processing of substrates can occur only after they have been unfolded and translocated into the central cavity by an associated Clp ATPase (22, 23). Clp ATPases form hexameric rings that either can dock to one or both sides of the ClpP double ring, forming a Clp proteolytic complex, or, independently of ClpP, can serve as molecular chaperones (52, 54). Substrate specificity of the ClpP proteolytic complexes is determined by the Clp ATPase component, and in the gram-positive model organism Bacillus subtilis, three Clp ATPases have the ability to associate with ClpP, namely, ClpE, ClpC, and ClpX (15). While no obvious phenotype was associated with a clpE mutation, inactivation of clpC, clpP, or clpX causes highly pleiotropic phenotypes in B. subtilis (7, 14, 34, 35). Increased stress sensitivity and defects in developmental pathways including sporulation and competence are common for all three mutants (14, 34, 35). In a series of very interesting studies, it was shown that many developmental phenotypes associated with mutations in clpP or clpX are due to the accumulation of Spx (suppressor of clpP and clpX), a transcriptional regulator that is normally degraded by the ClpXP protease (36, 37, 38). Detailed studies of Spx activity have revealed that it interacts directly with the α subunit of the RNA polymerase (RNAP) and thereby controls global transcription initiation, either negatively or positively, by an apparently unique mechanism (39, 40, 55). The negative regulation is mediated by the binding of Spx to Tyr263 in the C terminus of the RNAP α subunit, a residue that is highly conserved among gram-positive bacteria, whereby interactions between transcriptional activators and the RNA polymerase are blocked (39, 55). Examples of transcriptional activators, whose activities are reduced by Spx-RNAP interactions, are the response regulators ComA, activating transcription of genes required for developing genetic competence, and ResD, inducing genes involved in the adaptation to oxygen limitation (39, 55). In the absence of ClpXP, Spx accumulating in the cell will block complex formation between RNAP and ComA or ResD, thus resulting in the defects of the clpX and clpP mutants for developing competence and for growing under oxygen-restricted conditions.
DNA microarray analysis revealed that Spx also functions as a positive regulator of genes encoding proteins primarily involved in maintaining thiol homeostasis (40). Accordingly, a mutant lacking Spx is hypersensitive to diamide, a thiol-specific oxidant that promotes disulfide bond formation (40). The N-terminal end of Spx contains a Cys-X-X-Cys (CXXC) motif, and intriguingly, a thiol/disulfide switch seems to regulate the activity of Spx. During thiol-oxidizing conditions, Spx directly activates the transcription of trxA and trxB, encoding thioredoxin and thioredoxin reductase, respectively (41). This activation requires a direct interaction between the RNAP α subunit and the active form of Spx that has an intramolecular disulfide bond in the CXXC motif. This mechanism of transcriptional activation is unique in that it does not involve an initial Spx-DNA interaction (41).
Homologues of Spx are highly conserved among gram-positive bacteria with low G+C content (55). Interestingly, a gene homologous to spx, trmA, was previously identified as the site of mutations that could alleviate the heat-sensitive phenotype of a clpP mutant as well as of a recA mutant in Lactococcus lactis (9, 10). In S. aureus, cells lacking ClpP are hypersensitive to heat, cold, oxidative, and salt stress (11, 12). Moreover, the absence of ClpX or ClpC reduced biofilm formation, whereas it was enhanced in the absence of ClpP (12). Finally, the ClpXP proteolytic complex seems to play a direct role in controlling virulence regulation of S. aureus (11, 13). Thus, the ClpP protease controls several key processes important to the success of S. aureus as a pathogen. The present study was undertaken to examine the role of Spx in S. aureus and to assess whether any of the phenotypes associated with the clp mutations are Spx dependent.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. aureus 8325-4 (42) was used as the parental strain. The restriction-deficient strain S. aureus RN4220 (29, 42) was used as a primary recipient for plasmids. To study the expression of Spx in the absence of the ClpXP proteolytic complex, we used strain 8325-4ΔclpP or 8325-4ΔclpX (11). SH1000 was used as an rsbU-positive derivative of 8325-4 (21). S. aureus strains were grown in tryptic soya broth (TSB; Oxoid) medium under conditions of vigorous agitation (200 rpm). Usually, 20 ml of medium was inoculated into 200-ml flasks to allow efficient aeration of the medium. Erythromycin (5 μg ml−1) or tetracycline (4 μg ml−1) was added as required. For anaerobic conditions, screw-cap bottles were replenished to the maximum level with liquid culture (optical density at 600 nm [OD600] of 0.5) and incubated with shaking at 37°C. Escherichia coli strain DH5α was grown in Luria-Bertani medium (Oxoid) or on LB agar at 37°C. Erythromycin (150 μg ml−1) or ampicillin (50 μg ml−1) was added as required.
DNA manipulations.
Chromosomal DNA from S. aureus was isolated using the FAST DNA kit (Bio101). Cells were lysed by incubation for 1 h at 37°C in 20% sucrose-0.01 M NaPi (pH 7.0) containing 50 μg ml−1 lysostaphin (Sigma). Staphylococcal plasmid DNA was purified using the QiaMiniprep kit (QIAGEN). The plasmid DNA isolation procedure was modified by incubating the cell suspension in P1 buffer containing 50 μg ml−1 lysostaphin (Sigma) for 1 h at 37°C. Plasmid DNA from E. coli was purified by the boiling method (19). DNA restriction enzyme digestions and modifications were performed according to the manufacturer's instructions (New England BioLabs). Competent S. aureus cells were prepared as described previously by Frees et al. (11). Transduction was performed as described previously by McNamara and Iandolo (33).
Standard PCR conditions were as follows: 30 rounds for 30 s at 94°C, 30 s at 52°C, and 45 s at 72°C. PCR products were cloned using the TOPO TA cloning kit (Invitrogen).
Strain constructions. (i) 8325-4Δspx.
In order to create an spx deletion mutant, a 1,228-bp fragment containing the spx gene was amplified from chromosomal S. aureus 8325-4 DNA using the primers SayjbD196f (5′-GTAATTGCATGTTGATCTACG) and SayjbD1432r (5′-CAGTTGTATCATCTACTCG). The PCR product was cloned into pCR2.1-TOPO (Invitrogen), digested with AccI and HincII, and blunted before religation to make a 312-bp internal deletion in the spx PCR fragment. The remaining fragment was cloned into the temperature-sensitive shuttle vector pAUL-A (44) via HindIII and XbaI digestions. Subsequent transformation and double crossover were performed as described previously (11), except that primers SayjbD123f (5′-GTCTGGATCTATACCAGAAG) and SayjbD1432r were used to verify the chromosomal deletion in the spx gene. The wild-type spx gene comprises 396 bp; thus, the spx gene containing the 312-bp in-frame deletion is only 84 bases long. The resulting strain was named S. aureus 8325-4Δspx.
(ii) 8325-4Δspx-c.
8325-4Δspx was complemented with a chromosomally integrated copy of spx expressed from its own promoter using the single-copy integration vector system described previously by Lee et al. (31). spx was amplified from 8325-4 chromosomal DNA using the primers Saorf15test (5′-GTATAACTTACTACTAACAAAGGG, annealing 200 bp upstream of the spx start codon) and yjbD1249r (5′-CAGTTAAAGCATTAAAGCAC, annealing downstream of the predicted rho-independent transcriptional terminator). The PCR product was cloned into pCR2.1-TOPO (Invitrogen). The fragment was then sequenced to verify that it did not contain any mutations and was then cloned into PCL25 (kindly provided by Chia Y. Lee) via HindIII and XbaI digestions. The resulting plasmid was transformed into RN4220 containing the pYL112Δ19 plasmid encoding the integrase from the staphylococcal phage L54a that catalyzes site-specific integration of the plasmid (PCL25 + spx) into the 3′ end of the chromosomal geh gene (31). The integrated copy of PCL25 + spx was then transduced into 8325-4Δspx using the ϕ11 phage. PCR confirmed that the strain contained both the spx deletion and the intact copy of spx inserted into the geh gene. The strain was denoted 8325-4Δspx-c.
Western blot analysis.
S. aureus strains were grown in TSB as specified above. At an OD600 of 0.5 (exponential phase) and an OD600 of 2.5 (transition to stationary phase), cells were harvested for isolation of intracellular proteins. For heat-shocked cells, liquid cultures grown at 37°C were transferred to 46°C for 40 min prior to sampling. Cell pellets were resuspended in 50 mM Tris-HCl (pH 8.0) and lysed by the addition of 50 μg ml−1 lysostaphin (Sigma) and incubation at 37°C for 20 min. Cell debris was removed by centrifugation (15,000 × g for 15 min). The protein concentration was determined by using a Bio-Rad protein assay (Bio-Rad Laboratories), and 20 μg of each sample was separated on NuPAGE 4 to 12% Bis-Tris gels (Invitrogen) using MES buffer (Invitrogen). The gels were stained with Coomassie blue using Safestain (Invitrogen) or transferred onto a polyvinylidene difluoride membrane (Invitrogen) using an XCell SureLock Mini-Cell system (Invitrogen) as recommended by the supplier. Spx was probed with a 1:1,500 dilution of Spx antibody (a generous gift from P. Zuber) and the WesternBreeze Chemiluminescent Anti-Rabbit kit (Invitrogen).
Preparation of S. aureus cells for analysis by scanning electron microscopy (SEM).
Cells were grown in sterile filtered TSB medium at 37°C and harvested at an OD600 of 3.0. After the addition of natrium azide (final concentration, 25 mM), cells were bound to gold- and poly-l-lysine-coated glass for 10 min. Cells were subsequently rinsed in washing buffer (0.1 M cacodylate buffer, pH 7.4, 1 mM CaCl2, 25 mM NaN3) and prefixed in 2.5% glutaraldehyde-5 mM HEPES (pH 7.4)-25 mM NaN3 for 2 h at room temperature and overnight at 4°C. After rinsing cells in washing buffer (see above), further preparation steps occurred according to the microwave procedure described previously by Jongebloed and Kalicharan (26): cells were transferred on ice in 1 ml of a solution containing 2% arginine-HCl, glycine, Na-glutamate, and sucrose. Cells were then exposed to microwaves by the Microwave Processor H2500 (Energy Beam Sciences Inc., Agawam, MA) for 5 min; temperature upper limit was set at 40°C. After cells were rinsed in washing buffer (see above), cells were fixed with 2% guanidine-HCl and 2% tannic acid (Mallinckrodt Inc.) twice for 2 min by the microwave procedure. Cells were rinsed again in washing buffer (see above), fixed with osmium tetroxide (1% in washing buffer) using microwaves, and subsequently rinsed in water twice for 3 min. Dehydration of the cells was carried out by using ascending ethanol concentrations, successive transfer to acetic acid isoamyl ester, and critical point drying from liquid CO2 (Polaron CPD7501; VG Microtech, East Sussex, United Kingdom). Cells were sputtered with 10 nm gold (Polaron SC7640; VG Microtech, East Sussex, United Kingdom) and investigated with a DSM 940A scanning electron microscope (Zeiss, Oberkochen, Germany) using 7 kV and an operating distance of 7 mm.
Biofilm assay.
Bacteria were grown in TSB at 37°C with vigorous shaking up to an OD600 of 0.2 and were serially diluted from 10−1 to 10−4 in TSB medium. Subsequently, 100 μl of 10−2 to 10−4 dilutions were transferred into individual wells of a sterile, flat-bottomed, polystyrene 96-well microtiter plate (Sterilin) and incubated for 24 h at 37°C. Microtiter plates were washed twice with phosphate-buffered saline (pH 7.4), dried, and stained with 0.1% crystal violet to evaluate biofilm formation.
Preparation of intracellular proteins and two-dimensional (2D) polyacrylamide gel electrophoresis.
Bacteria were grown in TSB medium at 37°C with vigorous shaking. Cells were harvested at an OD600 of 0.5 (exponential phase), at an OD600 of 3.0 (transition to stationary phase), and 2 h after the onset of stationary phase (∼OD600 of 5.2) and washed twice in Tris-EDTA (pH 7.5). Cell disruption was achieved by using a Ribolyser (Hybaid) for 30 s at 6.5 m s−2. To remove cell debris and insoluble proteins, the lysate was centrifuged for 10 min at 9,000 × g and again for 30 min at 21,000 × g at 4°C. The supernatant was finally stored at −20°C.
Preparative 2D polyacrylamide gel electrophoresis was performed by using an immobilized pH gradient technique described previously (3). The protein samples were separated by using immobilized pH gradient strips (Immobiline IPG-Strips; Amersham Biosciences Europe GmbH, Freiburg, Germany) covering a pH range of 4 to 7 and stained using the colloidal Coomassie staining procedure. The 2D gel images were analyzed with Delta2D software (Decodon GmbH, Greifswald, Germany). Identification of proteins occurred by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry, and the resulting peptide mass fingerprints were analyzed by using GPMAW 4.10 (Lighthouse data) and the genome sequence of S. aureus N315 (http://www.tigr.org).
Northern blot analyses. (i) spx and icaA transcription analysis.
Total RNA of S. aureus strains was isolated according to the modified acid phenol procedure described previously by Gertz et al. (16). Northern blot analyses were performed as described previously by Wetzstein et al. (53). The specific digoxigenin-labeled probes were produced by using a DIG RNA labeling kit (Roche). To measure the transcription intensity of icaA, a PCR product was generated by using chromosomal DNA of S. aureus 8325-4 and the primer pair icaAf (5′-GTGGATAATTAGAAGGCAT)-icaAT7r (5′-CTAATACGACTCACTATAGGGAGATAGAAGCACTATATATACCC [the underlined sequence indicates the recognition sequence for T7 RNA polymerase]). To measure transcription of spx, we used primers yjbDf (5′-AGAGTGAGATGTATGGTAAC) and yjbDT7r (5′-CTAATACGACTCACTATAGGGAGACGTTGTGCTTCTTG [the underlined sequence indicates the recognition sequence for T7 RNA polymerase]).
Chemiluminescence signals were detected with a Lumi-Imager and were analyzed by using the appropriate software (LumiAnalyst; Boehringer Mannheim). The size of the detected transcripts was estimated by using the digoxigenin-labeled RNA molecular weight marker I (Roche).
(ii) Performance of trxB and icaR transcription analysis.
RNA extraction and Northern blot analysis were performed as described previously by Frees et al. (13). The specific radiolabeled probes were produced by using Ready-To-Go DNA labeling beads (-dCTP) (Amersham Biosciences). Internal fragments of the following genes (amplified with the primers given in parentheses) were used as templates in the labeling reactions: trxB (5′-CAGGTCCAGCTGGTATGACTG and 5′-ACCATCGCCAGTAGCAGTGAC) and icaR (5′-ACTTTCTTCCACTGCTCCAA and 5′-GGTATGACGGTACAACACTTG).
RESULTS
Spx accumulates in the absence of ClpX and ClpP.
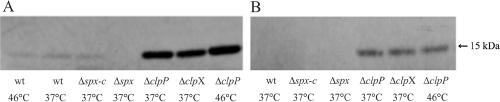
Multiple cellular processes in Staphylococcus aureus are influenced by the ClpP proteases (11, 12, 13). While ClpP influences some of these processes by its ability to degrade nonnative proteins, its role in other processes is likely to be associated with targeted degradation of specific protein substrates. As the Spx protein is highly conserved between S. aureus and B. subtilis (79% identical residues, including the N-terminal CXXC motif), Spx is also a likely candidate for a substrate of the ClpXP protease in S. aureus. To test this notion, we used an antibody raised against B. subtilis Spx to detect the homologous protein in S. aureus. Protein extracts were prepared from cells in the mid-exponential growth phase and from cells at the transition to stationary phase either growing at 37°C or heat treated at 46°C for 40 min prior to sampling. In wild-type cells isolated from the exponential growth phase, the antibody recognized a protein of the expected size (15 kDa). This protein was absent in cells in which the spx gene had been deleted (Fig. 1A) (construction of strain 8325-4Δspx is described in Materials and Methods), demonstrating that the antibody specifically recognizes Spx of S. aureus. Furthermore, the level of Spx returned to the wild-type level in an S. aureus 8325-4Δspx mutant strain complemented with a chromosomally integrated copy of spx (8325-4Δspx-c; for construction, see Materials and Methods) (Fig. 1A). The Western blot revealed that the amount of Spx protein did not change in heat-shocked wild-type cells, suggesting that the Spx level is not subject to temperature-dependent regulation. However, more Spx protein was detected in cells in the exponential growth phase than in cells in transition to stationary phase (compare panel A with panel B in Fig. 1), suggesting growth phase-dependent regulation of the Spx level.
FIG. 1.
Spx accumulates in cells lacking the ClpXP protease. Proteins were prepared from cells in exponential phase (A) (OD600 of 0.5 ± 0.1) either growing at 37°C or shifted to 46°C for 40 min prior to sampling or from cells in the transition to stationary growth phase (B) (OD600 of 2.5 ± 0.1). Proteins were separated on a NuPAGE 4 to 12% bis-Tris gel (Invitrogen) and subsequently blotted onto a polyvinylidene difluoride membrane and probed with a polyclonal antibody directed against Spx from B. subtilis (kindly provided by P. Zuber). (A) Lane 1, 8325-4 (wt) at 46°C; lane 2, 8325-4 at 37°C; lane 3, 8325-4Δspx-c at 37°C, lane 4, 8325-4Δspx at 37°C; lane 5, 8325ΔclpP at 37°C; lane 6, 8325ΔclpX at 37°C; lane 7, 8325ΔclpP at 46°C. (B) Lane 1, 8325-4 at 37°C; lane 2, 8325-4Δspx-c at 37°C; lane 3, 8325-4Δspx at 37°C; lane 4, 8325ΔclpP at 37°C; lane 5, 8325ΔclpX at 37°C; lane 6, 8325ΔclpP at 46°C.
Most notably, the amount of Spx protein was far higher in clpP and clpX mutant cells than in wild-type cells, strongly indicating that Spx is a substrate of the ClpXP protease in S. aureus.
Transcription of spx is reduced in cells lacking ClpX or ClpP.
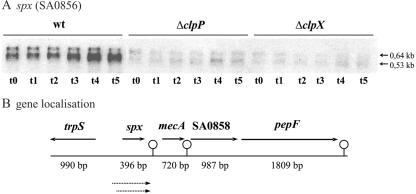
To examine if the accumulation of Spx protein observed in the clpXP mutant cells is due to increased transcription, we determined the amount of spx transcript in wild-type and mutant cells by Northern blot analysis (Fig. 2A). In wild-type cells, the spx-specific probe detected two transcripts of 0.64 and 0.53 kb, respectively (Fig. 2A). Inspection of the genome sequence showed that the spx gene is followed by a putative rho-independent terminator (ΔG = −14.2 kcal/mol) (Fig. 2B). Thus, the sizes of the detected transcripts suggest that spx is transcribed monocistronically from two separate promoters situated approximately 50 and 150 bp upstream of the translation initiation codon. Most interestingly, the Northern blot revealed that the amount of both spx transcripts was significantly reduced (about fivefold) in clpP and clpX mutant cells. Hence, Spx accumulates in the clpX and clpP mutant cells despite the fact that transcription of the gene is severely reduced, supporting the possibility that Spx is degraded by ClpXP in wild-type cells. The significant decrease in the amount of spx transcript in cells with high Spx levels may suggests that spx transcription is subjected to autoregulation.
FIG. 2.
Transcription of spx is reduced in cells lacking ClpP or ClpX. (A) Total RNA was prepared from S. aureus 8325-4 (wt), 8325-4ΔclpX, and 8325-4ΔclpP grown in TSB at 37°C under vigorous shaking in exponential growth phase (t0). To analyze transcription after a shift to anaerobic conditions, samples were taken in exponential growth phase (at an OD600 of 0.5) and 15 min (t1), 30 min (t2), 60 min (t3), 120 min (t4), and 180 min (t5) following a shift to anaerobic conditions. Ten micrograms of RNA was separated on a denaturing agarose gel and transferred onto a positively charged nylon membrane. The membrane was hybridized with a digoxigenin-labeled probe specific for spx. The sizes of relevant transcripts are indicated by arrows. (B) The chromosomal gene localization and the putative transcripts are illustrated schematically.
In wild-type cells, the amount of spx transcript increased slightly if cells were shifted from aerobic to anaerobic growth conditions, and this increase was not observed in cells lacking ClpXP (Fig. 2A). In contrast, the amount of spx transcript was not affected by high temperatures or by the presence of 1 M NaCl or 2 mM diamide (data not shown).
8325-4 has reduced levels of the alternative sigma factor σB due to a defect in rsbU, encoding an activator of σB. To examine if any of the two spx transcripts originate from a σB-dependent promoter, we additionally examined the expression of spx in SH1000, a rsbU+ derivative of 8325-4 (21). As the same amount of the two spx transcripts was present in both strains, we conclude that transcription of spx occurs independently of σB (data not shown). In support of this, −10 elements that resemble the consensus for σ70-dependent promoters were identified 50 bp upstream (TATAAC) and 100 bp upstream (TATAAT) of the spx start codon.
Inactivation of spx impairs growth under both stress and nonstress conditions.
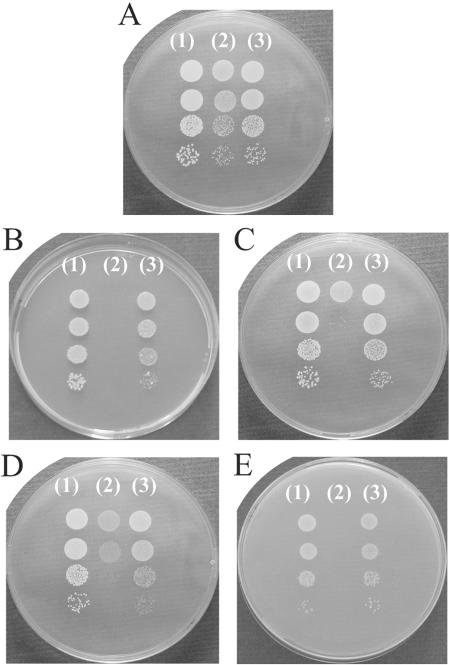
Mutants carrying the spx deletion were obtained only with very low frequency, and they formed colonies of reduced size under normal growth conditions (Fig. 3A). In liquid cultures (TSB at 37°C with vigorous shaking), the wild-type strain grew with a doubling time of 35 min; in comparison, deletion of spx increased the doubling time to approximately 48 min. When the spx mutant was complemented by a chromosomally integrated copy of spx, the growth defect was alleviated, confirming that Spx is important for growth of nonstressed S. aureus cells (Fig. 3A and data not shown).
FIG. 3.
The absence of Spx impairs growth under both stressed and nonstressed conditions. S. aureus 8325-4 (1), 8325-4Δspx (2), and 8325-4Δspx-c (3) were grown exponentially in TSB at 37°C up to an OD600 of 0.2. The cultures were then serially diluted (10−1-, 10−2-, 10−3-, and 10−4-fold), and 10 μl of each dilution was spotted onto TSA plates. The plates were incubated at 37°C (A, B, and E), 42°C (C), or 30°C (D) for 22 h. Growth was examined in the presence of 0.5 mM diamide (B) or in the presence of 2.2 M NaCl (E).
In Bacillus, Spx is required to induce transcription of genes encoding proteins involved in maintaining thiol homeostasis. Accordingly, the spx mutant strain is hypersensitive to diamide, a thiol-specific oxidant that promotes disulfide bond formation (40). Likewise, we observed that the S. aureus spx mutant could not grow in the presence of 0.5 mM diamide (Fig. 3B). However, in contrast to the B. subtilis spx mutant, the staphylococcal spx mutant was hypersensitive to a wide range of stress conditions, including high and low temperatures (Fig. 3C and D), high osmolarity, 2.2 M NaCl (Fig. 3E), and hydrogen peroxide (data not shown). In all cases, the stress-sensitive phenotypes could be complemented with a chromosomally integrated copy of spx. Hence, Spx fulfills an important role in general stress protection in S. aureus.
In B. subtilis, many developmental phenotypes associated with mutations in clpP or clpX are due to the accumulation of Spx, and consequently, these phenotypes are suppressed by a concomitant mutation in spx (36). In order to examine whether any of the many phenotypes described for clpP mutant cells in S. aureus are due to the accumulation of Spx, we attempted to construct a clpP spx double mutant. However, all attempts failed, and we therefore speculate that the double mutant will not be viable.
Interestingly, inactivation of the trmA gene, encoding one of seven Spx orthologous in Lactococcus lactis, alleviated the temperature-sensitive phenotype of clpP mutant cells and conferred increased heat tolerance to wild-type cells (10). Thus, despite the fact that TrmA and Spx have more than 50% identity, inactivation of the corresponding genes leads to very distinct phenotypes. L. lactis encodes a trmA paralogue that is more closely related to Spx on the sequence level than is TrmA, and it is conceivable that this protein is the functional homologue of Spx in L. lactis.
In the absence of Spx, transcription of trxB is severely reduced under both stress and nonstress conditions.
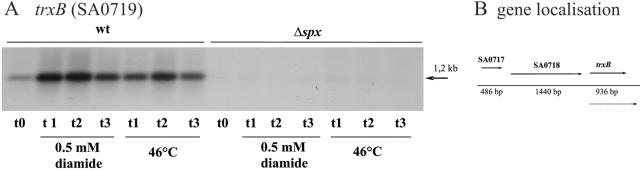
The diamide sensitivity of the spx mutant prompted us to investigate whether Spx is required for the induction of genes encoding proteins involved in maintaining thiol homeostasis. As a candidate gene, we examined the expression of trxB (encoding thioredoxin reductase), which was recently shown to be strongly induced by the addition of diamide in S. aureus (47). RNA was extracted from wild-type and mutant cells before and after the addition of diamide. Notably, the addition of diamide immediately stopped the growth of the spx mutant; however, the cells remained viable for at least 1 h (data not shown). To see if trxB transcription was affected by other types of stress, we additionally extracted RNA from heat-stressed cells. The Northern blot confirmed that the addition of diamide to the growth medium induced transcription of trxB three- to fivefold in wild-type cells (Fig. 4). Similarly, trxB expression was stimulated by shifting the wild-type culture from 37°C to 46°C (Fig. 4). Interestingly, the absence of Spx not only eliminated transcriptional induction of trxB by diamide but also significantly decreased the basal level of trxB transcription (Fig. 4). Moreover, heat induction of trxB transcription was abolished in cells that did not produce Spx (Fig. 4). We conclude that Spx contributes to the transcriptional regulation of trxB under both stressed and nonstressed conditions.
FIG. 4.
trxB transcription is severely reduced in cells lacking Spx. RNA was extracted from wild-type (wt) or spx mutant (Δspx) cells either growing exponentially in TSB at 37°C (t0) or 3 (t1), 8 (t2), and 15 (t3) minutes following the addition of 0.5 mM diamide or following a shift to 46°C. Northern blot analysis was performed using internal radioactively labeled probes of trxB.
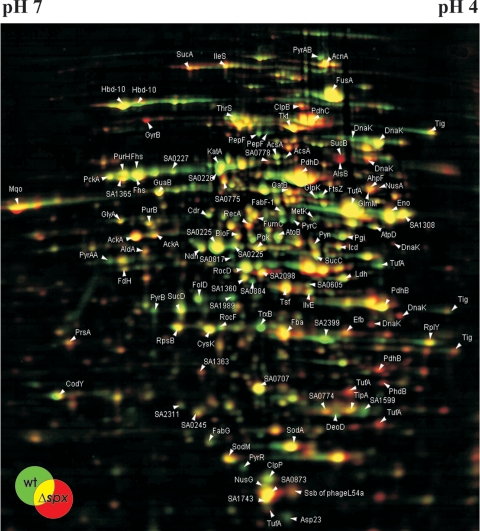
Two-dimensional gel electrophoresis indicates that Spx is a global regulator in S. aureus.
To examine whether the inactivation of spx impacted the overall protein expression, proteins extracted from wild-type and spx mutant cells (in either exponential, postexponential, or stationary growth phase) were analyzed by two-dimensional protein gel electrophoresis. As an example, the proteomes of the wild-type and the spx mutant strains in stationary growth phase are shown in Fig. 5. Gels from at least three independent experiments were compared, and proteins that were consistently present in higher or lower amounts in the spx mutant cells are listed in Table 1. In all growth phases, the absence of Spx resulted in major changes in the proteome. Most notably, inactivation of Spx changed the expression of enzymes of the pyrimidine and purine ribonucleotide biosynthesis pathways both positively and negatively. Similarly, expression of genes encoding proteins involved in purine and pyrimidine or amino acid biosynthesis is affected by the level of Spx in B. subtilis, but only negatively (40). Other proteins affected by the absence of Spx in S. aureus included proteins involved in DNA metabolism (GyrB and RecA) and the cell division protein FtsZ (Fig. 5). Moreover, the proteome analysis confirmed that the amount of TrxB was reduced in the spx mutant (Fig. 5). We also observed that the amount of several members of the HrcA heat shock regulon, including GroEL, GroES, GrpE, and DnaK, did not reach the wild-type level in the spx mutant cells (Fig. 5 and data not shown). While the reason for this finding is currently unknown, it could be related to the appearance of protein fragments of DnaK and GroEL in the spx mutant (Fig. 5).
FIG. 5.
Dual-channel image of a 2D gel showing the difference in intracellular protein patterns between 8325-4 (wild type) and 8325-4Δspx during stationary growth phase. Two hundred fifty micrograms of protein extract, isolated from wild-type and mutant cells grown in TSB at 37°C, was separated by two-dimensional gel electrophoresis and stained with colloidal Coomassie blue stain. Upon analysis with Delta2D software (Deocodon GmbH, Greifswald, Germany), proteins present in higher amounts in the spx mutant than in wild-type cells appear red, while proteins present in relatively lower amounts in the spx mutant cells appear green. Proteins identified by mass spectrometry are indicated by arrows. Gels from at least three independent experiments were compared, and proteins that were consistently present in lower or higher amounts in the spx mutant cells are summarized in Table 1.
TABLE 1.
Staphylococcal cytoplasmic proteins from stationary growth phase affected by an spx mutation
| Protein | Locus in S. aureus N315 | Function |
|---|---|---|
| Induced by a yjbD mutation | ||
| DnaK fragments | SA1409 | Protein fate; protein folding and stabilization |
| GyrBb | SA0005 | DNA metabolism; DNA replication, recombination, and repair |
| PdhB fragments | SA0944 | Energy metabolism; pyruvate dehydrogenase |
| RecA | SA1128 | DNA metabolism; DNA replication, recombination, and repair |
| TufA fragments | SA0506 | Protein synthesis; translation factors |
| Reduced by a yjbD mutation | ||
| Asp23 | SA1984 | Cellular processes; adaptation to atypical conditions |
| FtsZ | SA1029 | Cellular processes; cell division |
| PyrAAa | SA1045 | Pyrimidine ribonucleotide biosynthesis |
| PyrABa,b | SA1046 | Pyrimidine ribonucleotide biosynthesis |
| PyrBa,b | SA1043 | Pyrimidine ribonucleotide biosynthesis |
| PyrCa,b | SA1044 | Pyrimidine ribonucleotide biosynthesis |
| PyrRa | SA1041 | Pyrimidine ribonucleotide biosynthesis |
| TrxBa,b | SA0719 | Energy metabolism; electron transport |
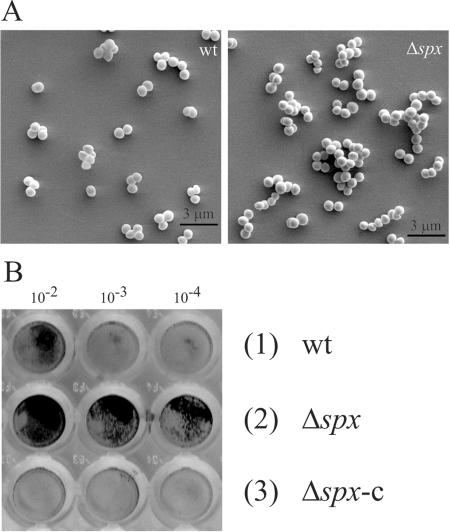
Biofilm formation is enhanced in the absence of Spx.
When we examined growth of the spx mutant strain, we observed that cells in overnight cultures had a tendency to clump and to adhere to the (glass) sides of test tubes (data not shown). Therefore, we analyzed the clustering of wild-type and mutant cells by light and scanning electron microscopy (Fig. 6A). Interestingly, the pictures revealed that the spx mutant in the transition state started to form larger clumps of attached cells than did the wild type. To determine if the spx mutation also influences the ability of S. aureus to bind to abiotic surfaces, the strains were grown in TSB overnight, diluted, and allowed to adhere to polystyrene microtiter wells. Adherence was assessed by visual inspection of the dry, stained biofilms (Fig. 6B). As described previously, the wild-type strain showed a weak or no tendency to adhere to polystyrene when grown in the absence of glucose or other biofilm-promoting additives (5). However, interestingly, the spx mutant strain showed strong adherence to the flat bottom of microtiter plates under the same conditions (Fig. 6B). Complementation of the spx mutant strain with an intact copy of the spx gene returned biofilm formation nearly to the wild-type level (Fig. 6B, lanes 3 and 6); thus, we conclude that Spx inactivation induces biofilm formation in S. aureus.
FIG. 6.
Cells lacking Spx are aggregative and prone to biofilm formation. (A) Visualization of cell-to-cell attachment by SEM. Cultures of S. aureus 8325-4 (wt) (1) and 8325-4Δspx (Δspx) (2) were grown in sterile filtered TSB medium at 37°C. Sampling occurred in the transient phase, between exponential and stationary growth phase, at an OD600 of 3.0. Cells were subsequently analyzed by SEM at 5,000-fold resolution. (B) Attachment to abiotic surfaces. S. aureus 8325-4 (1), 8325-4Δspx (2), and 8325-4Δspx-c (3) were grown exponentially in TSB medium at 37°C up to an OD600 of 0.2. The cultures were then serially diluted (10−1-, 10−2-, 10−3-, and 10−4-fold) in TSB medium, and 100 μl of 10−2-, 10−3-, and 10−4-fold dilutions were transferred into wells of a flat-bottom microtiter plate and incubated for 24 h at 37°C. Attached bacteria were stained with crystal violet as described in Materials and Methods.
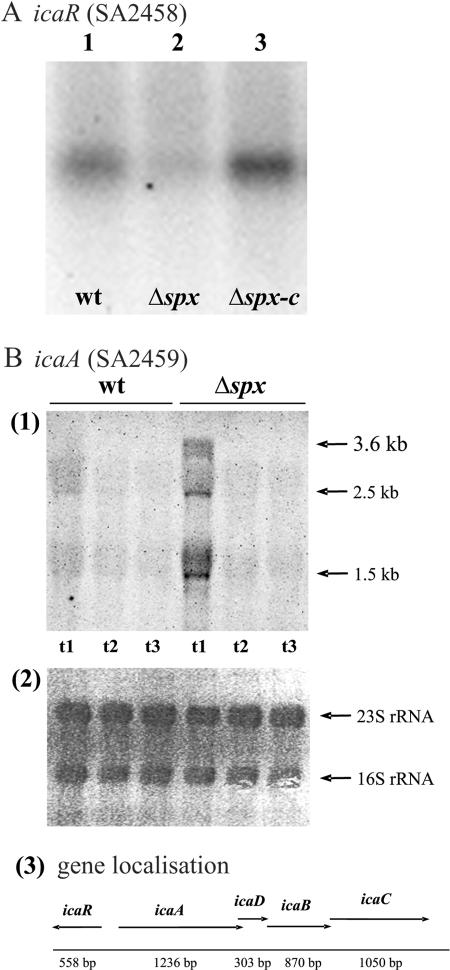
Transcription of the icaADBC operon is elevated in the absence of Spx.
Several genetic loci contribute to the ability of S. aureus to form biofilms (6, 18, 32, 48). Among the best-characterized loci is ica, which enables S. aureus to synthesize the polysaccharide intercellular adhesin (PIA), composed of linear β-1,6-linked glucosaminylglycans (5). The ica locus consists of the icaABCD operon encoding proteins involved in the structural synthesis of PIA as well as the divergently transcribed icaR, which was recently shown to encode a negative regulator of the icaABCD genes (17, 24, 25). Recently, it was proposed that IcaR plays a role in the suppression of PIA production in the absence of glucose (25). Thus, as cells devoid of Spx are characterized by forming biofilms in medium without added glucose, we examined the expression of icaR and the icaADBC operon by Northern blot analysis. Interestingly, the amount of icaR transcript was greatly reduced, while the amount of icaA transcript was substantially increased, in cells lacking Spx (Fig. 7A and B, respectively). These data indicate that the augmented capacity of the spx mutant to form biofilms is due to Spx modulating the expression of icaR, resulting in increased transcription of icaABCD.
FIG. 7.
icaR transcription is reduced, while icaADBC is increased, in cells devoid of Spx activity. (A) Total RNA was prepared from (1) S. aureus 8325-4 (wt), (2) 8325-4Δspx (Δspx), and (3) 8325-4Δspx-c (Δspx-c) grown exponentially in TSB at 37°C until an OD600 of 0.5 was reached. Twelve micrograms of RNA was separated on an agarose gel, transferred onto a positively charged nylon membrane, and probed with an icaR-specific radiolabeled probe. (B) Total RNA was prepared from S. aureus 8325-4 and 8325-4Δspx grown exponentially in TSB medium at 37°C. Sampling occurred in exponential phase at an OD600 of 0.5 (t1), in transition phase at an OD600 of 3.0 (t2), and 2 h after the onset of stationary phase (t3). Ten micrograms of RNA was separated on an agarose gel, transferred onto a positively charged nylon membrane, and probed with an icaA-specific digoxigenin-labeled probe. (1) Transcripts detected by the icaA probe. The sizes of the detected transcripts are indicated by arrows. (2) A picture of the methylene blue-stained membrane (staining rRNA). (3) The chromosomal region of the ica locus is depicted schematically based on the sequence of S. aureus 8325-4.
DISCUSSION
In B. subtilis, Spx is a global regulator that controls the transcription of target genes, negatively or positively, by a unique mechanism that requires direct interaction with the α subunit of RNA polymerase but not with DNA. Orthologs of Spx are highly conserved in most low-G+C-content gram-positive bacteria, including many pathogens such as Staphylococcus, Listeria, Enterococcus, and Streptococcus (55).
Here, we show that the inactivation of Spx has a profound negative effect on growth of S. aureus under both stressful and nonstressful conditions, demonstrating that the activity of Spx is important for the general well-being of staphylococci. Similarly, Spx seems to be of vital importance to L. monocytogenes, as attempts to delete the spx gene (denoted yjbD) in this organism failed (2). In contrast, a Bacillus spx mutant strain was reported to be sensitive only to thiol-oxidizing conditions, a phenotype linked to the role of Spx as a positive regulator of genes encoding proteins involved in thiol homeostasis (40, 41). This role of Spx is also conserved in staphylococci, as we demonstrate that transcription of trxB, encoding thioredoxin reductase, was almost abolished in cells lacking Spx. Thioredoxin reductase together with thioredoxin, TrxA, form part of the cell redox-regulating system (1). Thioredoxin is a small, abundant protein that contains a pair of redox-active cysteines that serve to maintain the intracellular thiol-disulfide balance and also provides reducing power to key reductive enzymes such as ribonucleotide reductases. Thioredoxin reductase, TrxB, is required to regenerate thioredoxin in its reduced form. Interestingly, trxB was recently reported to be essential for growth in S. aureus (47). Thus, the growth defects of the S. aureus spx mutant could be linked to very low levels of TrxB. Moreover, we showed that trxB transcription was stimulated by both heat and diamide stress, indicating that increased levels of TrxB are required not only in disulfide-stressed cells but also in otherwise stressed cells. Interestingly, both thioredoxin and thioredoxin reductase were recently shown to possess genuine chaperone activity towards proteins not containing cysteines (28). Under conditions of stress, proteins tend to denature, and increased levels of TrxB and TrxA may in general help stressed cells to maintain the native and reduced state of cellular proteins. An increased requirement for TrxB during stress may explain why the S. aureus spx mutant was hypersensitive to many types of stress.
While the dramatic reduction of trxB expression could account for the general growth deficiency of the staphylococcal spx mutant, this phenotype was not observed for the spx mutant of B. subtilis, in spite of the fact that the TrxA/TrxB system is essential for cell viability in both organisms (45, 47). We hypothesize that transcription of the trx genes is more dependent on Spx activity in S. aureus than in B. subtilis, as the absence of Spx in staphylococcal cells reduced trxB transcription to undetectable levels under all growth conditions, while transcription of trxB (and trxA) in nonstressed B. subtilis cells appeared to be unaffected by the absence of Spx (41). This could be explained by the finding that in B. subtilis, trxA is transcribed from two promoters, one utilizing the general stress factor σB and the other utilizing the vegetative σA factor (+Spx) (45). In contrast, σB had no affect on the transcription of trxA and trxB in S. aureus (47). We propose that the resulting differences in TrxA and TrxB levels could explain the phenotypic differences between the S. aureus and B. subtilis spx mutants.
This work was performed using strain 8325-4, one of the best-characterized S. aureus laboratory strains. This strain, however, is predicted to have reduced activity of the alternative sigma factor σB due to a natural deletion in rsbU, encoding an activator of SigB (30). Mutations in the SigB pathway are common among S. aureus isolates, including clinical isolates, indicating that SigB is not required for S. aureus to persist in the environment or for mediating at least certain infections (27). In B. subtilis, SigB-mediated transcription is important for general stress protection (20). This raises the possibility that some of the stress-related phenotypes observed in the spx mutant are synergistic outcomes of the absence of Spx and the defect in SigB regulon control. Although we cannot entirely exclude this possibility, we would like to emphasize that there are no indications in the literature that SigB is important for growth of S. aureus under the examined stress conditions. Moreover, under the conditions used, growth of 8325-4 is not stimulated further by introducing an intact copy of the rsbU gene (12).
Cells devoid of Spx exhibited an aggregative phenotype in the transition to stationary phase when grown under standard growth conditions. Accordingly, inspection of the cells by light or scanning electron microscopy demonstrated that the spx mutant cells formed large clumps of attached cells. Notably, mutants in clpX and clpP that have high intracellular levels of Spx are conversely seen mostly as single cells (12). Thus, the level of Spx seems to impact cell-cell adhesion. Moreover, the absence of Spx substantially increased the ability of the strain to form biofilms on abiotic surfaces. The genetic basis of biofilm formation in S. aureus is multifactorial, and much remains to be explored (17). However, the glucosamine-based PIA plays a major role in cell-cell adhesion (5, 24, 25). Very interestingly, we found that while the spx deletion enhanced transcription of the icaABCD operon (encoding the enzymes necessary for PIA synthesis), it reduced transcription of icaR, encoding a repressor of the structural ica genes. Thus, we propose that Spx reduces PIA-dependent biofilm formation by enhancing transcription of icaR. This hypothesis is strengthened by the finding that the amount of icaR transcript is significantly induced in cells having substantially increased levels of Spx due to the inactivation of the ClpXP protease (data not shown). Consistent with this finding, the transcription of icaABCD was reduced two- to ninefold upon inactivation of clpP in the closely related strain 8325 (Knut Ohlsen, personal communication). Curiously, clpX and clpP mutations oppositely influence the ability of S. aureus to form biofilms, as the clpP mutant cells are more prone, while the clpX mutant cells are less prone, to biofilm formation (12). However, it was recently shown that S. aureus is capable of forming a PIA-independent biofilm (46). Thus, we speculate that ClpP contributes positively to PIA-based biofilm formation by degrading Spx while at the same time reducing PIA-independent biofilm formation. To directly examine the significance of PIA for the biofilms made by the clpP and spx mutant strains, we would like to determine the amount of PIA produced by the mutant strains by performing a PIA dot blot. However, at present, we have not succeeded in obtaining PIA antibodies.
Environmental conditions affect the biofilm formation capacity of S. aureus, indicating the existence of positive and negative regulators of this process. Under the growth conditions we have used, the addition of glucose or NaCl to the growth medium is normally required for the wild-type strain to form a biofilm. However, this requirement was bypassed by the absence of Spx. Interestingly, it was recently shown that the glucose-mediated increase in biofilm formation is at least partly due to the alleviation of IcaR repression of the structural ica genes (25). Thus, our data suggest that Spx may impact biofilm formation by modulating the expression of icaR in response to glucose. Interestingly, the existence of a regulatory protein that regulates ica transcription in response to the redox status of the cell was recently predicted to be present in Staphylococcus epidermidis (50). We speculate that Spx could fulfill this role in staphylococci, and this issue will be a subject for future investigations.
Acknowledgments
We are grateful to Peter Zuber (Oregon Health and Science University) for providing the Spx antibody and to Chia Y. Lee (University of Kansas Medical Center) for providing us with plasmids pYL112Δ19 and pCL25. We thank Knut Ohlsen (Universität Würzburg) for sharing information for publication. Furthermore, we are very grateful to Renate Hanschke and Hartmut Fischer for their cooperation and generation of the SEM pictures and to Christel G. Buerholt for valuable technical assistance.
This research was supported by grants from the DFG (Graduiertenkolleg) and from the BMBF (Pathogenomic Network) to M. Hecker and by research funds from the Danish Technical Research Council and from the LMC to H. Ingmer. Finally, D. Frees was supported by a grant from the Danish Research Council on Technology and Production.
REFERENCES
- 1.Åslund, F., and J. Beckwith. 1999. The thioredoxin superfamily: redundancy, specificity, and gray-area genomics. J. Bacteriol. 181:1375-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borezee, E., T. Msadek, L. Durant, and P. Berche. 2000. Identification in Listeria monocytogenes of MecA, a homologue of the Bacillus subtilis competence regulatory protein. J. Bacteriol. 182:5931-5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Büttner, K., J. Bernhardt, C. Scharf, R. Schmid, U. Mader, C. Eymann, H. Antelmann, A. Völker, U. Völker, and M. Hecker. 2001. A comprehensive two-dimensional map of cytosolic proteins of Bacillus subtilis. Electrophoresis 22:2908-2935. [DOI] [PubMed] [Google Scholar]
- 4.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause for persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 5.Cramton, S. E., C. Gerke, N. F. Schnell, W. W. Nichols, and F. Götz. 1999. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 67:5427-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cucarella, C., C. Solano, J. Valle, B. Amorena, I. Lasa, and J. R. Penades. 2001. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 183:2888-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derre, I., G. Rapoport, K. Devine, M. Rose, and T. Msadek. 1999. ClpE, a novel type of HSP100 ATPase, is part of the CtsR heat shock regulon of Bacillus subtilis. Mol. Microbiol. 32:581-593. [DOI] [PubMed] [Google Scholar]
- 8.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duwat, P., S. D. Ehrlich, and A. Gruss. 1999. Effects of metabolic flux on stress response pathways in Lactococcus lactis. Mol. Microbiol. 31:845-858. [DOI] [PubMed] [Google Scholar]
- 10.Frees, D., P. Varmanen, and H. Ingmer. 2001. Inactivation of a gene that is highly conserved in gram-positive bacteria stimulates degradation of non-native proteins and concomitantly increases stress tolerance in Lactococcus lactis. Mol. Microbiol. 41:93-103. [DOI] [PubMed] [Google Scholar]
- 11.Frees, D., S. N. Qazi, P. J. Hill, and H. Ingmer. 2003. Alternative roles of ClpX and ClpP in Staphylococcus aureus stress tolerance and virulence. Mol. Microbiol. 48:1565-1578. [DOI] [PubMed] [Google Scholar]
- 12.Frees, D., A. Chastanet, S. Qazi, K. Sørensen, P. Hill, T. Msadek, and H. Ingmer. 2004. Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus. Mol. Microbiol. 54:1445-1462. [DOI] [PubMed] [Google Scholar]
- 13.Frees, D., K. Sørensen, and H. Ingmer. 2005. Global virulence regulation in Staphylococcus aureus: pinpointing the roles of ClpP and ClpX in the sar/agr regulatory network. Infect. Immun. 73:8100-8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerth, U., E. Krüger, I. Derré, T. Msadek, and M. Hecker. 1998. Stress induction of the Bacillus subtilis clpP gene encoding the proteolytic component of the Clp protease and involvement of ClpP and ClpX in stress tolerance. Mol. Microbiol. 28:787-802. [DOI] [PubMed] [Google Scholar]
- 15.Gerth, U., J. Kirstein, J. Mostertz, T. Waldminghaus, M. Miethke, H. Kock, and M. Hecker. 2004. Fine-tuning in regulation of Clp protein content in Bacillus subtilis. J. Bacteriol. 186:179-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gertz, S., S. Engelmann, R. Schmid, K. Ohlsen, J. Hacker, and M. Hecker. 1999. Regulation of sigmaB-dependent transcription of sigB and asp23 in two different Staphylococcus aureus strains. Mol. Gen. Genet. 261:558-566. [DOI] [PubMed] [Google Scholar]
- 17.Götz, F. 2002. Staphylococcus and biofilms. Mol. Microbiol. 43:1367-1378. [DOI] [PubMed] [Google Scholar]
- 18.Gross, M., S. E. Cramton, F. Götz, and A. Peschel. 2001. Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infect. Immun. 69:3423-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holmes, D. S., and M. Quigley. 1981. A rapid boiling method for the preparation of bacterial plasmids. Anal. Biochem. 114:193-197. [DOI] [PubMed] [Google Scholar]
- 20.Höper, D., U. Völker, and M. Hecker. 2005. Comprehensive characterization of the contribution of individual SigB-dependent general stress genes to stress resistance of Bacillus subtilis. J. Bacteriol. 187:2810-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horsburgh, M. J., J. L. Aish, I. J. Shaw, J. K. Lithgow, and S. J. Foster. 2002. σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoskins, J. R., M. Pak, M. R. Maurizi, and S. Wickner. 1998. The role of the ClpA chaperone in proteolysis by ClpAP. Proc. Natl. Acad. Sci. USA 95:12135-12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoskins, J. R., A. K. Singh, M. R. Maurizi, and S. Wickner. 2000. Protein binding and unfolding by the chaperone ClpA and degradation by the protease ClpAP. Proc. Natl. Acad. Sci. USA 97:8892-8897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jefferson, K. K., S. E. Cramton, F. Götz, and G. B. Pier. 2003. Identification of a 5-nucleotide sequence that controls expression of the ica locus in Staphylococcus aureus and characterization of the DNA-binding properties of IcaR. Mol. Microbiol. 48:889-899. [DOI] [PubMed] [Google Scholar]
- 25.Jefferson, K. K., D. B. Pier, D. A. Goldmann, and G. B. Pier. 2004. The teicoplanin-associated locus regulator (TcaR) and the intercellular adhesin locus regulator (IcaR) are transcriptional inhibitors of the ica locus in Staphylococcus aureus. J. Bacteriol. 186:2449-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jongebloed, W. L., and D. Kalicharan. 1994. Tannic-acid/arginine/osmiumtetroxide fixation of rat tissue by the microwave procedure, observed by SEM. Beitr. Elektronenmikroskop Direktabb. Oberfl. 27:243-252. [Google Scholar]
- 27.Karlsson-Kanth, A., K. Tegmark-Wisell, S. Arvidson, and J. Oscarsson. Natural human isolates of Staphylococcus aureus selected for high production of proteases and α-hemolysin are σB deficient. Int. J. Med. Microbiol., in press. [DOI] [PubMed]
- 28.Kern, R., A. Malki, A. Holmgren, and G. Richarme. 2003. Chaperone properties of Escherichia coli thioredoxin and thioredoxin reductase. Biochem. J. 371:965-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kreiswirth, B. N., S. Löfdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 30.Kullik, I., P. Giachino, and T. Fuchs. 1998. Deletion of the alternative sigma factor σB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J. Bacteriol. 180:4814-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, C. Y., S. L. Buranen, and Z.-H. Ye. 1991. Construction of single-copy integration vectors for Staphylococcus aureus. Gene 103:101-105. [DOI] [PubMed] [Google Scholar]
- 31a.Leichert, L. I. O., C. Scharf, and M. Hecker. 2003. Global characterization of disulfide stress in Bacillus subtilis. J. Bacteriol. 185:1967-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim, Y., M. Jana, T. T. Luong, and C. Y. Lee. 2004. Control of glucose- and NaCl-induced biofilm formation by rbf in Staphylococcus aureus. J. Bacteriol. 186:722-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McNamara, P. J., and J. J. Iandolo. 1998. Genetic instability of the global regulator agr explains the phenotype of the xpr mutation in Staphylococcus aureus KS9051. J. Bacteriol. 180:2609-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Msadek, T., F. Kunst, and G. Rapoport. 1994. MecB of Bacillus subtilis, a member of the ClpC ATPase family, is a pleiotropic regulator controlling competence gene expression and survival at high temperature. Proc. Natl. Acad. Sci. USA 91:5788-5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Msadek, T., V. Dartois, F. Kunst, M. L. Herbaud, F. Denizot, and G. Rapoport. 1998. ClpP of Bacillus subtilis is required for competence development, motility, degradative enzyme synthesis, growth at high temperature and sporulation. Mol. Microbiol. 27:899-914. [DOI] [PubMed] [Google Scholar]
- 36.Nakano, M. M., F. Hajarizadeh, Y. Zhu, and P. Zuber. 2001. Loss-of-function mutations in spx result in ClpX- and ClpP-independent competence development of Bacillus subtilis. Mol. Microbiol. 42:383-394. [DOI] [PubMed] [Google Scholar]
- 37.Nakano, M. M., S. Nakano, and P. Zuber. 2002. Spx (YjbD), a negative effector of competence in Bacillus subtilis, enhances ClpC-MecA-ComK interaction. Mol. Microbiol. 44:1341-1349. [DOI] [PubMed] [Google Scholar]
- 38.Nakano, S., G. Zheng, M. M. Nakano, and P. Zuber. 2002. Multiple pathways of Spx (YjbD) proteolysis in Bacillus subtilis. J. Bacteriol. 184:3664-3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakano, S., M. M. Nakano, Y. Zhang, M. Leelakriangsak, and P. Zuber. 2003. A regulatory protein that interferes with activator-stimulated transcription in bacteria. Proc. Natl. Acad. Sci. USA 100:4233-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakano, S., E. Kuster-Schock, A. D. Grossman, and P. Zuber. 2003. Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 100:13603-13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakano, S., K. N. Erwin, M. Ralle, and P. Zuber. 2005. Redox-sensitive transcriptional control by a thiol/disulphide switch in the global regulator, Spx. Mol. Microbiol. 55:498-510. [DOI] [PubMed] [Google Scholar]
- 42.Novick, R. P. 1967. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology 33:155-166. [DOI] [PubMed] [Google Scholar]
- 43.Opdyke, J. A., J. R. Scott, and C. P. Moran, Jr. 2003. Expression of the secondary sigma factor σX in Streptococcus pyogenes is restricted at two levels. J. Bacteriol. 185:4291-4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schäferkordt, S., and T. Chakraborty. 1995. Vector plasmid for insertional mutagenesis and directional cloning in Listeria spp. BioTechniques 19:720-725. [PubMed] [Google Scholar]
- 45.Scharf, C., S. Riethdorf, H. Ernst, S. Engelmann, U. Völker, and M. Hecker. 1998. Thioredoxin is an essential protein induced by multiple stresses in Bacillus subtilis. J. Bacteriol. 180:1869-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toledo-Arana, A., N. Merino, M. Vergara-Irigaray, M. Débarbouillé, J. R. Penadés, and I. Lasa. 2005. Staphylococcus aureus develops an alternative, ica-independent biofilm in the absence of the arlRS two-component system. J. Bacteriol. 187:5318-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uziel, O., I. Borovok, R. Schreiber, G. Cohen, and Y. Aharonowitz. 2004. Transcriptional regulation of the Staphylococcus aureus thioredoxin and thioredoxin reductase genes in response to oxygen and disulfide stress. J. Bacteriol. 186:326-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valle, J., A. Toledo-Arana, C. Berasain, J.-M. Ghigo, B. Amorena, J. R. Penades, and I. Lasa. 2003. SarA and not σB is essential for biofilm formation by Staphylococcus aureus. Mol. Microbiol. 48:1075-1087. [DOI] [PubMed] [Google Scholar]
- 49.Vaudaux, P. E., P. Francois, F. D. Lew, and F. A. Waldvogel. 2000. Host factors predisposing to and influencing therapy of foreign body infections, p. 1-27. In A. L. Bisno and F. A. Waldvogel (ed.), Infections associated with indwelling medical devices, 3rd ed. American Society for Microbiology, Washington, D.C.
- 50.Vuong, C., J. B. Kidder, E. R. Jacobson, M. Otto, R. A. Proctor, and G. A. Somerville. 2005. Staphylococcus epidermidis polysaccharide intercellular adhesin production significantly increases during tricarboxylic acid cycle stress. J. Bacteriol. 187:2967-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, J., J. A. Hartling, and J. M. Flanagan. 1997. The structure of ClpP at 2.3 Å resolution suggests a model for ATP-dependent proteolysis. Cell 91:447-456. [DOI] [PubMed] [Google Scholar]
- 52.Wawrzynow, A., D. Wojtkowiak, J. Marszalek, B. Banecki, M. Jonsen, B. Graves, C. Georgopolous, and M. Zylicz. 1995. The ClpX heat shock protein of Escherichia coli, the ATP-dependent substrate specificity component of the ClpP-ClpX protease, is a novel molecular chaperone. EMBO J. 14:1867-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wetzstein, M., U. Völker, J. Dedio, S. Löbau, U. Zuber, M. Schiesswohl, C. Herget, M. Hecker, and W. Schumann. 1992. Cloning, sequencing, and molecular analysis of the dnaK locus from Bacillus subtilis. J. Bacteriol. 174:3300-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wickner, S., S. Gottesmann, D. Skowyra, J. Hoskins, K. McKenny, and M. R. Maurizi. 1994. A molecular chaperone, ClpA, functions like DnaK and DnaJ. Proc. Natl. Acad. Sci. USA 91:12218-12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zuber, P. 2004. Spx-RNA polymerase interaction and global transcriptional control during oxidative stress. J. Bacteriol. 186:1911-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]