Abstract
Type IV fimbriae are expressed by several bacterial pathogens and are essential for virulence in Dichelobacter nodosus, which causes ovine footrot. We have identified a two-component signal transduction system (PilR/S) and an alternative sigma factor (σ54) that were shown by insertional inactivation to be required for the regulation of fimbrial biogenesis in D. nodosus. Western blots showed that in both pilR and rpoN mutants, fimbrial subunit production was significantly reduced by a process that was shown to occur at a PilR- and σ54-dependent promoter. The mutants lacked surface fimbriae, which were shown to be required for the adherence of D. nodosus cells to tissue culture monolayers. The reduction in fimbrial subunit production in these mutants also resulted in a concomitant loss of the ability to secrete extracellular proteases. A maltose binding protein-PilR fusion protein was purified and was shown to bind specifically to a region located 234 to 594 bp upstream of the fimA transcriptional start point. To determine additional targets of PilR and σ54, genome-wide transcriptional profiling was performed using a whole-genome oligonucleotide microarray. The results indicated that PilR and σ54 regulated genes other than fimA; these genes appear to encode surface-exposed proteins whose role in virulence is unknown. In conclusion, this study represents a significant advancement in our understanding of how the ability of D. nodosus to cause ovine footrot is regulated, as we have shown that the biogenesis of type IV fimbriae in D. nodosus is regulated by a σ54-dependent PilR/S system that also indirectly controls protease secretion.
Dichelobacter nodosus is a fastidious gram-negative anaerobe that is the causative agent of footrot in sheep. Ovine footrot is a debilitating disease that is characterized by necrotic separation of the hoof from the underlying soft tissue and leads to significant economic losses as a result of lameness, loss of body condition, and reduced wool growth (51). The virulence of D. nodosus isolates varies and depends on factors such as the production of type IV fimbriae and extracellular proteases and the presence of two genomic regions preferentially associated with virulent isolates (3).
Type IV fimbriae are expressed by a broad range of organisms and are characterized by sequence similarity of the fimbrial subunit, a polar location, and the presence of N-methylated phenylalanine or methionine as the N-terminal residue (13). The type IV fimbriae of D. nodosus are required for a flagellum-independent form of motility termed twitching motility, natural competence, and extracellular protease secretion and have been shown to be essential for virulence (31). D. nodosus isolates are classified into 10 serogroups based on agglutination to antibodies against these fimbriae (12, 20). Little is known about the regulation of fimbrial biogenesis in D. nodosus, but preliminary evidence suggests that it involves a regulatory system based on σ54-dependent promoters (22, 30). The regulation of type IV fimbriae in Pseudomonas aeruginosa and Myxococcus xanthus has been shown to involve σ54 and NtrC family response regulators, and given the mechanistic similarities of type IV fimbrial biogenesis, similar regulatory systems are likely to exist in other species (21, 25, 36, 60). However, in some bacteria that express type IV fimbriae, there are no functional homologues of the P. aeruginosa and M. xanthus regulatory systems (9, 34).
Two-component signal transduction systems are able to sense and respond to environmental or growth phase stimuli by a histidine-aspartate phosphorelay using a membrane-bound sensor histidine kinase and a cytoplasmic response regulator. Response regulators are transcriptional regulators composed of a conserved N-terminal receiver domain that is the site of phosphorylation and a C-terminal DNA binding domain. The NtrC family of response regulators possesses an additional central domain, which is responsible for ATP hydrolysis, oligomerization, and interaction with σ54 (7). NtrC-like regulators belong to the AAA+ (ATPases associated with a variety of cellular activities) family of proteins and interact with σ54-RNA polymerase (RNAP) to regulate diverse functions such as nitrate utilization, xylene degradation, and flagellar synthesis (7, 56). The association of σ54 with core RNAP and its subsequent binding to the promoter produce an activator-dependent, closed transcriptional complex. NtrC regulators bind to regions of DNA upstream of the σ54-dependent promoter and, through ATP hydrolysis, convert σ54-RNAP into an open transcriptional complex, thereby allowing transcription to occur (7, 38, 61, 64).
The overall aim of this study was to determine how the production of fimbriae was regulated in D. nodosus. In this report, we describe the successful identification and genetic analysis of a σ54-dependent PilRS system and show that it is responsible for the transcriptional regulation of fimbrial subunit synthesis and therefore fimbrial biogenesis in D. nodosus. We have also shown that the fimbriae of D. nodosus are required for adherence and that protease secretion is linked to the levels of the fimbrial subunit. In addition to fimbrial biogenesis, this system appears to concomitantly regulate additional, previously unidentified, putative virulence factors.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Bacterial strains and plasmids are listed in Table 1. Escherichia coli and P. aeruginosa strains were grown on 2× YT medium (47) supplemented with 100 μg ampicillin/ml or 150 μg erythromycin/ml for E. coli strains and 250 μg carbenicillin/ml for P. aeruginosa strains. D. nodosus strains were grown on Eugon (Difco) yeast extract (EYE) agar containing 5% defibrinated horse blood (Equicell) and supplemented with 1 μg erythromycin/ml for selection of transformants or in Eugon (Difco) broth with yeast extract in an anaerobic chamber as described previously (40).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | F−endA1 hsdR17(rk− mk−) thi-1 λ−recA1 gyrA96 relA1 phoA supE44 deoR φ80dlacZΔM15 Δ(lacZYA argF)U169 | Invitrogen |
| C43(DE3) | F′ ompT hsdSB(rB mB) gal dcm (DE3) (C43) | 37 |
| D. nodosus | ||
| VCS1703A | Serogroup G, transformable, virulent strain | J. Egerton, University of Sydney |
| JIR3727 | VCS1703A fimAΩ tet(M) | 31 |
| JIR3772 | VCS1703A pilRΩ erm(B) (pilR1) | Natural transformation |
| JIR3773 | VCS1703A pilRΩ erm(B) (pilR2) | Natural transformation |
| JIR3887 | VCS1703A rpoNΩ erm(B) (rpoN1) | Natural transformation |
| JIR3888 | VCS1703A rpoNΩ erm(B) (rpoN2) | Natural transformation |
| P. aeruginosa | ||
| PAK | Wild type | M. Hobbs and J. Mattick, University of Queensland |
| R94 | PAK pilRΩ Tn5G | 23 |
| Plasmids | ||
| pBluescript SK(+) | AprlacZ′ cloning vector | Stratagene |
| pUCP18 | AprE. coli-P. aeruginosa shuttle vector | 50 |
| pET22b(+) | Apr C-terminal His6 expression vector | Novagen |
| pT7Blue-3 | Apr cloning vector | Novagen |
| pMal-c2 | Apr N-terminal malE maltose binding protein expression vector | New England Biolabs |
| pJIR2586 | pBluescript SK(+) containing the erm(B) gene inserted into the ClaI site of pilR | Recombinant |
| pJIR2596 | pBluescript SK(+) containing the erm(B) gene inserted into the EcoRI site of rpoN | Recombinant |
| pJIR2764 | pUCP18 EcoRI/PstIΩ 4.5-kb PCR product containing D. nodosus pilR/S genes and upstream region | Recombinant |
| pJIR2776 | pT7Blue-3 EcoRVΩ 729-bp fimA promoter region | Recombinant |
| pJIR2824 | pMal-c2 EcoRI/XbaI pilR+ | Recombinant |
Molecular techniques.
Unless otherwise stated, molecular techniques were performed as described elsewhere previously (40, 47). Chemically competent P. aeruginosa cells were prepared as previously described (16) by using nutrient yeast broth.
Mutant construction and analysis.
Suicide vectors for pilR (pJIR2586) and rpoN (pJIR2596) were constructed by amplifying the pilR (primers 18635 [GGGGTACCTTAATGCTCAATGCCGTTGCC] and 19397 [GGAATTCATAATGATCGCTCGTGCTCGC]) and rpoN (primers 19606 [GTTTCCCAGGCATACTTTAGG] and 19760 [GGGGTACCTATTCGGCACATTATTCAACA]) genes by PCR and cloning each product into the Asp718/EcoRI and Asp718/BamHI sites of pBluescript SK(+), respectively. Unique internal restriction sites in pilR (ClaI) and rpoN (EcoRI) were then used to insert an erythromycin resistance cassette, erm(B).
Mutant strains were constructed by inserting the suicide vectors into D. nodosus VCS1703A by natural transformation (31). Two independently isolated mutants were obtained for each gene and analyzed by PCR using 16S rRNA gene primers to confirm that the strains were D. nodosus and using erm(B) primers to detect the presence of the erm(B) gene (40). To confirm that each gene was insertionally inactivated, a combination of erm(B) primers and gene-specific primers was used. To confirm that the antibiotic-resistant strains were derived from the wild-type strain, PCR-restriction fragment length polymorphism analysis of the outer membrane protein-encoding gene omp1 was performed (19).
Phenotypic analysis.
Qualitative protease (40) and elastase (52) assays, quantitative azocaseinase assays (31), twitching motility assays (31), and transmission electron microscopy (31) were performed as previously described. Slide agglutination assays were performed using a 10-μl aliquot of serogroup G fimbrial antibodies (1:10 dilution; John Egerton, University of Sydney) mixed with a loopful of cells from a 2-day-old EYE agar culture.
Adhesion assay.
Semiconfluent CHO-K1 cells, grown in 24-well tissue culture plates, were washed three times with prewarmed (37°C) phosphate-buffered saline (PBS) before the addition of 500 μl of prewarmed α-minimal essential medium (1 × HEPES buffer [25.9 mM HEPES, 11.5 mM NaOH, and 0.00005% phenol red], 8% fetal calf serum; Gibco) containing 0.5% mannose. D. nodosus cells were removed from EYE agar cultures grown for 48 h using 1.5 ml of prewarmed PBS. Fifty microliters of bacterial culture (turbidity at 600 nm of 0.4) was added to each cell, and the mixture was allowed to incubate for 6 h at 37°C in the anaerobic chamber. After incubation, the cells were washed four times with prewarmed PBS, and the wells were resuspended in 100 μl of 0.1% (wt/vol, in PBS) digitonin and allowed to sit for 5 min to lyse the CHO-K1 cells. The digitonin solution was then removed, serially diluted, subcultured onto EYE blood agar, and incubated for 5 days at 37°C in the anaerobic chamber. Viable counts were then compared to viable counts done on the original cultures. Adhesion ratios were then calculated relative to wild-type levels. Adhesion assays were performed in triplicate on three separate occasions.
RNA extraction and analysis.
RNA was extracted from D. nodosus cells grown in EYE broth using TRIzol (Invitrogen) according to the manufacturer's instructions. To determine if pilS and pilR were cotranscribed, reverse transcriptase (RT) PCR was performed using primers 25964 (CGGGGTTAGGTTTATTTTTAG) and 26714 (GCAGCAACAAATAACCCATCC) as described elsewhere previously (31). Quantitative real-time RT-PCR (QRT-PCR) was performed with an ABI PRISM 7700 sequence detector as described previously (40) using primers JRP2044 (ACATCGCTCGTACCCAGGTT) and JRP2045 (AAGTTGTCAGCGATACGGATTTTTA) for fimA, JRP2042 (CGGAATGACTGGGCGTAAA) and JRP2043 (CTAGGTTGAGCCCAGGGATTT) for 16S rRNA, JRP2724 (CGCATCAGTTAATTTCCGTAGGT) and JRP2725 (ACTCCTTGGAATAGCCATTTGC) for gdn1292, JRP2728 (GCAATCCGGACCTTTGTTGA) and JRP2729 (CGCCGCTTTATGATTTTTCG) for gdn1329, JRP2730 (ACGATTCACGGCGAAGATATG) and JRP2731 (CCAAGCTTTCAATGCGATCA) for gdn1330, and JRP2732 (CGCCGCCCGAGGAA) and JRP2733 (GATGCGTTCGGCTTTCATTT) for gdn1331. The data were standardized to the control 16S rRNA gene levels, and Student's t tests were performed to determine significance (P < 0.05).
Primer extension reactions on the fimA promoter region were performed using 50 μg of RNA and primer 22796 (TCATGAGTTCGATTAAGGTGA) with the Primer Extension System-AMV Reverse Transcription (Promega) according to the manufacturer's instructions. Reaction mixtures were ethanol precipitated overnight and run on 8% acrylamide sequencing gels alongside a sequencing ladder generated with primer 22796 and a pJIR2776 template using [α-35S]dATP and the T7 sequencing kit (USB) according to the manufacturer's instructions.
Microarray hybridization and analysis.
Microarrays consisted of 2,317 custom 70-mer oligonucleotides (designed to reduce cross-hybridization) spotted in triplicate on Ultra-GAPS slides (Corning) using a QArray Mini (Genetix) arrayer, representing 1,343 open reading frames (ORFs) determined from the unpublished D. nodosus genome sequence (http://www.tigr.org). Labeled cDNA was synthesized using amino-allyl dUTP at a 3:2 ratio with dTTP. Random hexamers (6 μg) were annealed to 5 μg of RNA in RT reaction mixtures that contained 20 U of RNasin (Promega); 10 mM dithiothreitol (DTT); 400 U Superscript II (Invitrogen); 500 μM dATP, dCTP, and dGTP; 200 μM dTTP; and 300 μM amino-allyl dUTP. Reaction mixtures were allowed to incubate overnight at 42°C. RNA was hydrolyzed at 65°C for 15 min in the presence of 200 mM NaOH and 100 mM EDTA before neutralization in 300 mM Tris-HCl, pH 7.4. Reaction mixtures were purified with Microcon columns (Millipore) and dried using a Speedvac SVC (Savant). The amino-allyl-labeled cDNA was coupled to monoreactive Cy3 or Cy5 dye (Amersham Biosciences) for 2 h at room temperature in the dark in 50 mM sodium bicarbonate buffer (pH 9) and 50% dimethyl sulfoxide before purification using Microcon columns and drying under vacuum. Typically, 15 μg of labeled cDNA (Cy3 and Cy5 cDNA probes) was hybridized to the slides.
Slides were prepared for hybridization by incubation at 42°C for 45 min in a solution containing 25% formamide, 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% sodium dodecyl sulfate (SDS), 10 mg/ml bovine serum albumin, and 1 mg/ml salmon sperm DNA under a coverslip in a humidified chamber. The coverslip was removed in double-distilled water before drying by centrifugation for 10 min at 1,000 × g. Hybridizations were carried out overnight at 42°C under a coverslip in humidified CMT-hybridization chambers (Corning) in the presence of a solution containing 25% formamide, 5× SSC, 0.1% SDS, and 1 mg/ml salmon sperm DNA. Probes were heated at 95°C for 5 min, cooled slightly, and then applied to the slides. After hybridization, the slides were washed once in 2× SSC-0.1% SDS at 42°C for 5 min, once in 0.1× SSC-0.1% SDS for 10 min at room temperature with mixing, and four times for 30 s each in 0.1× SSC and were then rinsed once in double-distilled water, dried by centrifugation, and scanned.
Slides were scanned using a GMS 418 microarray scanner (Genetic Microsystems), and the Cy3 and Cy5 channels were combined and quantified using ImaGene version 5.1 software (Biodiscovery). A minimum of three slides from separate biological replicates, including dye swaps, were used in the data analysis. Expression values for each gene were determined using BASE v1.2.9 software (46). Low-intensity spots were removed before global median ratio and lowess normalizations (63) were performed, and ratios (n-fold) were determined. The statistical significance of the derived ratios was determined using a Wilcoxon signed-rank test. Genes that were consistently twofold up- or down-regulated across the majority of slides, with a P value of <0.005, were designated differentially expressed.
Protein expression and purification.
To express PilR as a maltose binding protein (MBP) fusion, the pilR gene was cloned into pMAL-c2 using primers 25321 (GGAATTCAATCACGCGTTAGTTGTTG) and 25322 (GCTCTAGATCAATCAATTCCTAATTTTTTTAA), which contained EcoRI and XbaI sites, respectively. The resultant plasmid, pJIR2824, was used to transform E. coli C43 cells to ampicillin resistance. The cells were grown in 500 ml of 2× YT containing ampicillin to a turbidity at 600 nm of 0.5 and then induced with 0.3 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The cultures were grown for an additional 3 h before pelleting at 6,000 × g for 10 min at 4°C. The pellets were washed in Tris buffer (20 mM Tris-HCl, pH 7.8, 250 mM NaCl) and lysed using a French press. After centrifugation at 12,000 × g for 30 min at 4°C, the supernatant was mixed with amylose resin (NEB) equilibrated in the same buffer. The resin mixture was applied to a Poly-Prep chromatography column (Bio-Rad) and washed with 20 ml of Tris buffer. PilR was eluted in 2-ml fractions using Tris buffer containing 10 mM maltose. Elutions containing high concentrations of protein were then pooled and subjected to dialysis overnight at 4°C in dialysis buffer (20 mM Tris-HCl, pH 7.8, 250 mM NaCl, 50% glycerol) using dialysis tubing (Spectra/Por; Spectrum Laboratories) with a 12,000- to 14,000-Da cutoff.
Gel filtration analysis.
Proteins (2 to 10 mg) were separated on a Superdex 200 16/60 column (Amersham Biosciences) that was equilibrated in a solution containing 20 mM Tris-HCl, pH 7.8, 300 mM NaCl, and 1 mM DTT. Proteins were eluted at a flow rate of 0.4 ml/min, and fractions of 1 ml were collected and analyzed by 12% SDS-polyacrylamide gel electrophoresis (PAGE). Molecular sizes were determined by calibrating the column with standard proteins (gel filtration calibration kit [Bio-Rad] and gel filtration high-molecular-weight markers [Amersham Biosciences]).
Immunoblot analysis.
Samples to be separated by SDS-PAGE were isolated from either whole-cell lysates (31) or surface fimbriae. Surface fimbriae were isolated from vortexed suspensions of 2-day-old EYE blood agar cultures. Samples were resolved on 12% polyacrylamide gels and transferred onto nitrocellulose membranes (PROTRAN; Schleicher & Schuell). Blots were blocked with 5% (wt/vol) skim milk powder, washed in Tris-buffered saline (5 mM Tris-HCl, pH 7.4, 15 mM NaCl) containing 0.05% Tween 20, and probed with polyclonal rabbit anti-FimA antibodies (dilution, 1:1,000; John Egerton, University of Sydney) or polyclonal rabbit anti-P. aeruginosa PilR antibodies (1:3,000; Jessica Boyd, Harvard University) (5). The blots were then washed and probed with sheep anti-rabbit antibodies (1:2,000) conjugated to horseradish peroxidase (Chemicon) before detection using the Western Lightning chemiluminescence reagent (Perkin-Elmer) according to the manufacturer's instructions.
Gel mobility shift assays.
DNA for gel shift reactions (amplified using primers 22795 [GGCGCCGTAAAATTGCTGTGA] and 22797 [GCCTTATTATTAGCAGCGTTA] for fimA and 24915 [GGAATTCCATATGCAGGCGCGGGAAAATAAAAT] and 24935 [CCGCTCGAGTCCCCCTGATGCAGATGATT] for pilS) was labeled using terminal transferase (Roche) using [α-32P]dATP (Perkin-Elmer) according to the manufacturer's instructions for 3′ end labeling with radioactive nucleotides. Labeling reaction mixtures were then purified with the QIAquick PCR purification kit (QIAGEN) according to the manufacturer's instructions before counting on a TriCarb 2100TR Liquid Scintillation Analyser (Packard). Gel shift reactions were carried out in 20-μl volumes containing binding buffer (100 mM HEPES, pH 7.6, 50 mM ammonium sulfate, 5 mM DTT, 1% [vol/vol] Tween 20, 150 mM KCl, 5 mM MgCl2), 1 μg poly(dI-dC) (Amersham Biosciences), 4,000 cpm of DNA (0.05 to 0.5 fmol) and the desired concentrations of protein (15 to 1,500 nM). Reaction mixtures were incubated for 15 min at room temperature, and 5 μl of loading buffer (0.25× TBE [1× TBE contains 89 mM Tris-HCl, pH 8.0, 89 mM boric acid, 2 mM EDTA], 60% glycerol, 0.2% [wt/vol] bromophenol blue) was then added before the samples were loaded onto the gel. Samples were loaded onto 4% acrylamide gels containing 0.25× TBE and run for 3 h at 200 V.
Microarray accession numbers.
Microarray data have been deposited at the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) under the accession numbers GSE3465 (rpoN) and GSE3466 (pilR).
RESULTS
The D. nodosus genome contains pilR and rpoN homologues.
Little is known about the regulation of virulence factors in D. nodosus, with a fur homologue being the only regulatory gene that has been mutated and analyzed (40). To identify genes whose products may be involved in the regulation of fimbrial biogenesis in D. nodosus, the unpublished genome sequence (http://www.tigr.org) was analyzed using standard bioinformatic tools. Three genes with a potential coordinated role in fimbrial gene regulation were identified, pilR, pilS, and rpoN.
PilR was identified based on its 55% identity to the PilR regulator of pilin production in P. aeruginosa (26). PilR was a putative 528-amino-acid protein with a predicted size of 58.8 kDa, 83 residues larger than its P. aeruginosa homologue. These 83 residues were localized to a single C-terminal region. PilR belonged to the NtrC family of response regulators (53) and had significant identity (40%, over 75% of the protein) to NtrC from Klebsiella pneumoniae (15). NtrC-type regulators contain three domains, a response regulator receiver domain (pfam00072), a σ54 interaction/ATPase domain (pfam00158) that contains a Walker A motif, and a DNA binding domain, all of which were present in PilR from D. nodosus. Only one other putative response regulator was identified with the domain hallmarks of NtrC family proteins, but it did not display similarity to PilR. The NtrC ATPase domain contains a motif, GAFTGA (present as GSFTGA in D. nodosus PilR), that is unique to this class of activators and is thought to be required for interaction with the σ54 subunit of RNAP (57, 64). The putative DNA binding domain of D. nodosus PilR, which in NtrC-like activators is required for dimerization, was shown to contain a helix-turn-helix motif by using established algorithms (14, 42).
The gene encoding the putative sensor of this two-component system, pilS, was located upstream of pilR. PilS had 26% identity to PilS from P. aeruginosa (21), was 581 residues in size, and was predicted to have six transmembrane domains. Similar identity (26%) was also observed with the PilS protein of Xanthomonas axonopodis, which is not involved in fimbrial synthesis (62). PilS contained the conserved histidine residue that is the site of phosphorylation (pfam00512) and GXGL, DXGXG, N, and F motifs, which are conserved in sensor histidine kinases and are thought to form a nucleotide binding surface (41, 54). The region containing the putative site of phosphorylation was highly conserved in D. nodosus and P. aeruginosa, with more variation occurring in the N-terminal sensor region of the protein. Only 9 bp separated the stop and start codons of pilS and pilR, respectively, which indicated that the two genes may represent an operon. RT-PCR using a primer internal to pilS and a second primer located 677 bp into the coding region of pilR confirmed that pilS and pilR were cotranscribed (data not shown).
The alternative σ factor, σ54 (encoded by rpoN), is associated with the regulation of pilin production in P. aeruginosa and flagellar synthesis in numerous organisms (7, 25, 56). The product of rpoN from D. nodosus was predicted to be a 469-amino-acid protein, 53.8 kDa in size, and was 35% identical to σ54 factors of P. aeruginosa and E. coli. The D. nodosus protein was predicted to contain all the domains consistent with its assignment as σ54: an activator interaction domain (pfam00309) that is involved in conformational changes necessary for open complex formation (11), a central core RNA polymerase binding domain (pfam04963) (7), and a C-terminal region required for DNA binding (pfam04552) (7). The DNA binding domain contained a helix-turn-helix motif (14) that in enteric species has been shown to be involved in binding to the −12 promoter element (59). A second motif associated with DNA binding, the RpoN box, is associated with interactions at the −24 promoter region (8) and was conserved in 9 of 10 motif residues in the D. nodosus σ54 protein.
pilR and rpoN are required for fimbrial production.
Since type IV fimbriae are an essential D. nodosus virulence factor, it was important to determine if pilR and rpoN were involved in fimbrial biogenesis. Therefore, insertionally inactivated pilR and rpoN mutants were constructed by allelic exchange. Suicide vectors were constructed and introduced into D. nodosus by natural transformation, and erythromycin-resistant colonies were selected. PCR and restriction fragment length polymorphism analysis confirmed that the genes had been disrupted by double-crossover events (data not shown). Two independently derived mutants were obtained for each gene. Since their phenotypic properties were the same, detailed analysis focused on JIR3772 (pilR1) and JIR3887 (rpoN1).
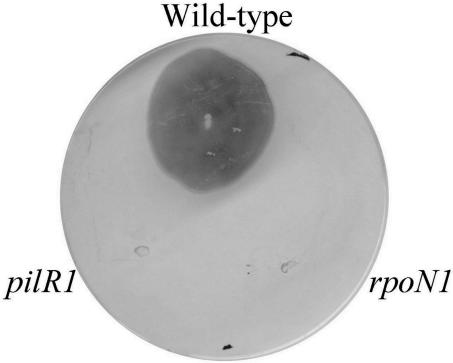
Both types of mutants had an altered colony morphology. The colonies were convex, differing from the usual spreading phenotype that is a result of their twitching motility. This altered phenotype was consistent with a loss of fimbriae, which was first confirmed by a slide agglutination assay using fimbria-specific antiserum. Also consistent with a loss of surface fimbriae was the observation that the mutants were no longer able to undertake twitching motility, as determined by using a subsurface stab assay of agar medium (Fig. 1) (31). Electron microscopy showed that no surface fimbriae were present in the mutants (Fig. 2B), unlike the wild-type strain, which had numerous polar fimbriae (Fig. 2A). This observation was confirmed by Western immunoblotting of sheared surface fimbriae using fimbrial antiserum (Fig. 2C). Since the fimbriae of D. nodosus are essential for natural transformation (31), the loss of surface fimbriae made the pilR and rpoN mutants no longer transformable. Therefore, for technical reasons, it was not possible to complement these mutants, as there are no other genetic exchange methods available for D. nodosus.
FIG. 1.
Twitching motility analysis of pilR and rpoN mutants. Strains were stab inoculated into 1% EYE agar plates and incubated for 7 days, and the agar plates were then compressed and stained with Coomassie blue.
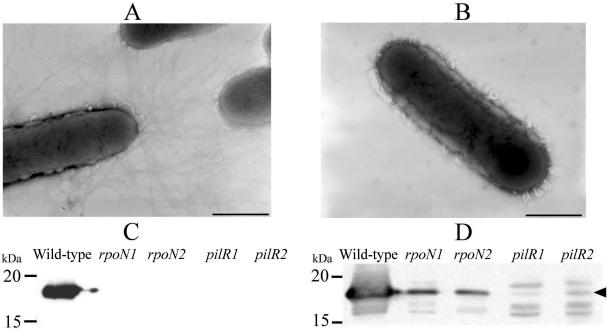
FIG. 2.
Absence of fimbriae in pilR and rpoN mutants. The presence of surface fimbriae was observed using electron microscopy. (A and B) Wild-type cells showing the presence of fimbriae (A) and the pilR mutant lacking surface fimbriae (B). Note that the rpoN mutants appeared similar to those shown in B. Bars equal 1 μm. The presence of surface fimbriae was also determined using Western immunoblotting with fimbrial antisera on sheared fimbriae (C) and whole-cell lysates (D).
Western immunoblotting was also performed on whole-cell lysates to determine if the fimbrial subunit FimA was still being produced even though surface fimbriae were absent. The results (Fig. 2D) showed that FimA levels were significantly reduced in both the pilR and rpoN mutants, with almost negligible levels present in the pilR mutants. Determination of the fimA transcript levels by QRT-PCR showed that the loss of fimbriae in the pilR and rpoN mutants was due to a significant reduction in the level of fimA (pilR/pilR+ transcript ratio of 0.00078 ± 0.00043; rpoN/rpoN+ transcript ratio of 0.012 ± 0.0081).
D. nodosus pilR does not complement a P. aeruginosa pilR mutant.
Previous studies (30) showed that the D. nodosus prepilin peptidase gene, fimP, can complement its homologous P. aeruginosa pilD mutant. To determine if the sequence identity between the PilR proteins of D. nodosus and P. aeruginosa was sufficient to enable genetic complementation, the pilSR region of D. nodosus, including 900 bp upstream of pilS, was cloned into the shuttle vector pUCP18 and introduced into P. aeruginosa pilR mutant R94 (21). The results showed that the pilR mutant was not complemented by the D. nodosus pilSR operon since the recombinant strain did not produce fimbriae, nor was it able to undergo twitching motility. RT-PCR was used to confirm that the D. nodosus pilR gene was being transcribed. Western immunoblotting using antisera against P. aeruginosa PilR revealed a cross-reacting band, approximately 60 kDa in size, which was consistent with the size of D. nodosus PilR. This band was observed in the complemented strain but was absent in the wild-type and pilR P. aeruginosa strains (data not shown). These data indicate that sequence similarity does not necessarily correlate with the ability to carry out functional complementation in these different bacterial species. Alternatively, the sequence variation between the PilS homologues may mean that the environmental signal required to activate the D. nodosus PilS protein was not present.
D. nodosus fimbriae are required for adherence.
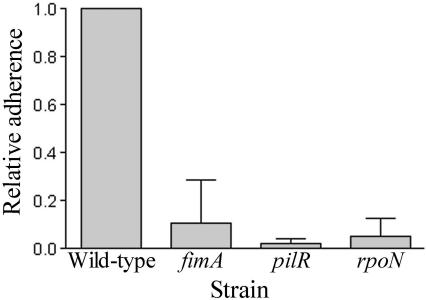
Type IV fimbriae are involved in many biological functions, including the attachment to various surfaces and epithelial cells (36, 55). The type IV fimbriae of D. nodosus are essential for virulence (31), but their role in attachment is yet to be determined. To determine if the type IV fimbriae were required for adherence, an assay was established using Chinese hamster ovary epithelial cells (CHO-K1). The adherence capabilities of the wild-type strain, the pilR and rpoN mutants, and a fimA mutant (31) were determined in an adherence assay undertaken for 6 h in an anaerobic chamber. The fimA mutant was included as a control for any pleiotropic effects caused by the pilR or rpoN mutations. The CHO-K1 cells were examined microscopically to confirm that they were not affected by the anaerobic conditions. The results showed that wild-type D. nodosus cells were adherent and that this adherence was dependent on the presence of the type IV fimbriae (Fig. 3). The pilR, rpoN, and fimA mutants, which all lacked surface fimbriae, showed a decreased (≥90% reduction) ability to adhere compared to the wild-type strain.
FIG. 3.
Fimbriae mediate adherence to epithelial cells. D. nodosus cells were incubated with CHO-K1 cell monolayers for 6 h anaerobically and washed, and viable counts were determined. Levels of adherent bacteria are shown relative to the wild-type level of adherence.
Extracellular protease activity correlates with levels of the fimbrial subunit.
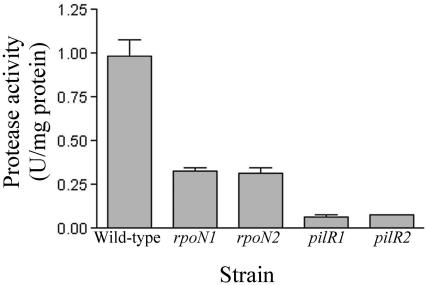
Type IV fimbriae play a role in protein secretion, and it has been postulated that the fimbrial extension and retraction reactions involved in twitching motility may act to pump proteases out of the cell (48). As mutants lacking fimbriae in both D. nodosus (31) and P. aeruginosa (35) have reduced extracellular protease activity, this property was examined with the pilR and rpoN mutants. Qualitative plate assays detecting elastase (52) and caseinase activity revealed an absence of elastase activity and a significant reduction in caseinase activity in both types of mutant. This reduction in activity was quantitated using culture supernatants and an azocasein substrate (Fig. 4). Extracellular protease activity levels were significantly reduced (P < 0.05) in all of the pilR and rpoN mutants compared to that of the wild type, consistent with their loss of fimbriae (31). Extracellular protease activity levels were also different in the pilR mutant compared to that of the rpoN mutant (P < 0.05). Protease secretion by the pilR mutants was less than that by the rpoN mutants (Fig. 4), commensurate with the different levels of fimbrial subunit produced by the two types of mutant (Fig. 2D). RNA dot blots that were carried out using separate probes specific for the three known D. nodosus extracellular protease genes (aprV2, aprV5, and bprV) did not reveal any significant differences, indicating that the reduction in extracellular protease levels was not due to direct regulatory effects on the transcription of the protease genes by PilR and σ54. Note that no extracellular protease activity was observed in a triple aprV2 aprV5 bprV mutant (R. M. Kennan, D. Parker, D. W. Wong, and J. I. Rood, unpublished results).
FIG. 4.
Reduced extracellular protease activity in pilR and rpoN mutants. Culture supernatants from wild-type, rpoN, and pilR strains were assessed for protease activity using azocasein as the substrate.
fimA is transcribed from a PilR- and σ54-dependent promoter.
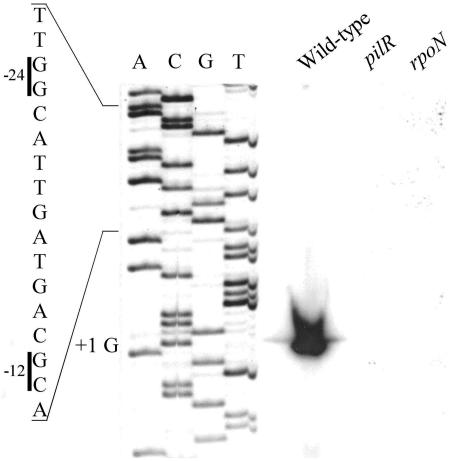
The transcriptional start point of fimA has been determined previously from strains of two different serogroups (serogroups A and H) (22). This work showed the presence of a major transcript starting 63 bp upstream of the ATG start codon and two less intense bands 45 and 46 bp upstream. Primer extension analysis of our type G wild-type strain indicated the presence of one major product that corresponded to a start point 63 bp upstream of ATG (Fig. 5), indicating that the same start site is used in this strain. No additional products were observed. Upstream of the start site, we identified a σ54-dependent promoter with the conserved −12 GC and −24 GG elements. The position of the −12 element was 13 bp upstream of the start site, which occurs most frequently (27%) among σ54-dependent promoters (2). The guanine start site is also the most common: 64% of the starting bases are purines (2). In lieu of this putative σ54-dependent promoter (22), we went on to determine if transcription from this start site was dependent on σ54 and PilR. Primer extension analysis was repeated on RNA isolated from the rpoN and pilR mutants. The results revealed an inability to obtain extension products from these mutants (Fig. 5), even upon longer exposure, which indicated that the fimA promoter was dependent on both σ54 and PilR.
FIG. 5.
fimA is transcribed from a PilR- and σ54-dependent promoter. Primer extension reactions were performed on 50 μg of RNA from wild-type, pilR1, and rpoN1 strains using a primer 42 bp downstream of the translational start site. Shown is the σ54 promoter region with the −12 and −24 elements indicated along with the transcriptional start point. Product sizes were determined by comparison to a sequencing ladder generated using the same primer (lanes A, C, G, and T).
PilR binds to a region upstream of the fimA promoter.
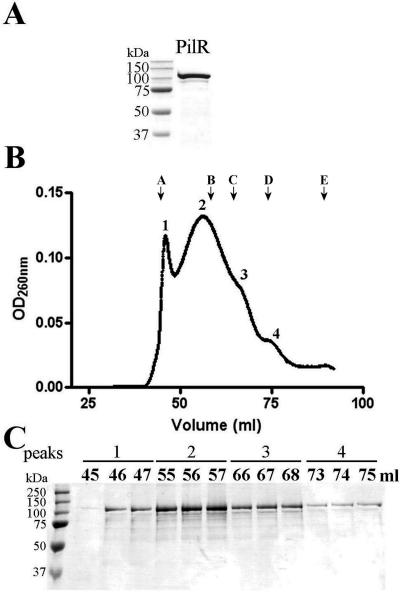
To investigate the role of PilR in the regulation of fimbrial biogenesis in D. nodosus further, PilR was overproduced and purified. PilR was initially expressed with a hexahistidine tag, but this fusion protein was insoluble. To increase its solubility, PilR was produced with an N-terminal MBP tag. Upon induction, this construct was overexpressed, and the soluble fusion protein was purified using amylose affinity chromatography to >90% purity (Fig. 6A).
FIG. 6.
Purification and gel filtration analysis of PilR. (A) PilR purification. PilR was purified as an MBP-tagged fusion using affinity chromatography. Samples were run on SDS-PAGE gels and stained with Coomassie blue. (B) Gel filtration elution profile of MBP-PilR. MBP-PilR was separated on a Superdex 200 gel filtration column, and peaks 1 to 4 correspond to the aggregated, octameric, tetrameric, and dimeric forms, respectively, based on comparisons to standard proteins (see Materials and Methods). Elution volumes of standards are indicated as follows: A, blue dextran (2 MDa); B, thyroglobin (670 kDa); C, ferritin (440 kDa); D, gamma globulin (158 kDa); E, ovalbumin (44 kDa). OD260nm, optical density at 260 nm. (C) SDS-PAGE analysis of selected fractions corresponding to the different peaks (1 to 4) from the gel filtration elution profile and stained with Coomassie blue. Markers are Precision Plus protein standards (Bio-Rad).
NtrC-like proteins are typically present as dimers in solution (32) and are able to oligomerize into tetramers and bind at tandem recognition sites upon phosphorylation (61). To determine the multimeric state of nonphosphorylated PilR, the fusion protein was applied to a Superdex 200 16/60 gel filtration column. The elution profile indicated that there were four reproducible and distinct peaks (Fig. 6B) that all gave a dominant protein band corresponding to the size of MBP-PilR when analyzed by SDS-PAGE (Fig. 6C). When these peaks were compared to gel filtration standards, the peaks corresponded to an aggregate at the void volume (Fig. 6B, peak 1), an octamer (Fig. 6B, peak 2), a tetramer (Fig. 6B, peak 3), and a dimer (Fig. 6B, peak 4). Most of the protein was present in the higher multimeric states as well as in the aggregate. The presence of these multiple forms is consistent with the need for NtrC-like transcriptional activators to have the ability to oligomerize.
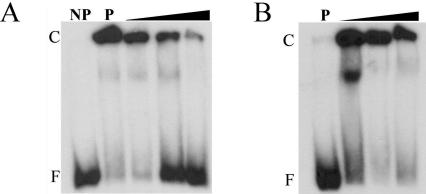
To identify the site at which PilR binds to the fimA promoter region, gel mobility shift assays were carried out. NtrC proteins typically bind from 100 bp (29) to 1 kb (38) upstream of the target promoter. Two 32P-labeled DNA fragments were used as targets: a 370-bp fragment encompassing the transcriptional start site and promoter region and a 359-bp fragment that was upstream of the promoter and corresponded to a region 234 to 593 bp upstream of the fimA transcriptional start site. These DNA fragments were incubated with various concentrations of PilR (15 to 1,500 nM) and separated on 4% polyacrylamide gels. A concentration-dependent shift of DNA was observed with the 359-bp fragment but not with the 370-bp promoter-containing fragment (data not shown). Binding appeared to be specific for PilR, since purified MBP did not bind (data not shown). To confirm that binding was specific for the target site, PilR (150 nM) was incubated with the 359-bp fragment and various levels (10 to 10,000 times) of specific and nonspecific (pilS coding sequence) competitor DNA. When competed with nonspecific DNA, no significant changes were observed (Fig. 7B). However, the addition of specific unlabeled DNA resulted in a consistent decrease in protein-DNA complexes and an increase in free, unbound DNA, confirming the specificity of the binding reaction (Fig. 7A). PilR was also shown to be able to bind, with slightly reduced affinity, to a 217-bp fragment that corresponded to a region 376 to 595 bp upstream of the transcriptional start site (data not shown). Further attempts to reduce the size of the PilR target site did not lead to the formation of PilR-DNA complexes.
FIG. 7.
DNA binding of PilR to the fimA promoter region. (A) A 359-bp 32P-labeled DNA fragment located upstream of the fimA promoter was incubated with PilR and increasing concentrations (0.005, 0.05, and 0.5 fmol) of unlabeled competitor DNA and analyzed using gel mobility shift analysis on 4% polyacrylamide gels. Lanes are as follows: NP, no protein; P, 150 nM PilR and no competitor DNA and increasing concentrations (10 to 10,000 times) of specific competitor DNA, compared to labeled DNA. The locations of free DNA (F) and protein-DNA complexes (C) are indicated. (B) Gel mobility shift analysis of PilR and the fimA promoter region incubated with increasing concentrations of nonspecific unlabeled competitor DNA.
Global transcriptional profiling of D. nodosus pilR and rpoN mutants.
To determine the full complement of genes regulated by PilR and σ54 in D. nodosus, the transcriptome of D. nodosus pilR and rpoN mutants was compared to that of the wild-type strain. To profile the transcription of D. nodosus genes, 70-mer oligonucleotide microarrays were constructed using 2,317 gene-specific probes, representing 1,343 ORFs predicted from the genome sequence. The ORFs were determined from the unpublished D. nodosus genome sequence (http://www.tigr.org), and, where possible, two gene probes per ORF were synthesized.
Microarray analysis was carried out for at least three biological replicates (including dye swaps) using a conservative selection criterion of a more-than-twofold difference with a P value of <0.005. Genes that were less-than-twofold regulated were included if they were part of a highly regulated operon. The observation that there was a more-than-fourfold decrease in fimA transcript in both mutants (Table 2) agreed with our previous observations that fimA transcription was significantly reduced. The microarray data were further validated by performing QRT-PCR on RNA from the wild-type and the pilR and rpoN mutants by using four of the genes differentially expressed in both strains as targets. The QRT-PCR results confirmed the microarray data, showing significantly reduced transcript levels. As expected, QRT-PCR was a more sensitive method than microarray analysis for quantifying expression levels.
TABLE 2.
Differentially expressed genes in the pilR and rpoN mutants
| Gene IDa | Putative function |
pilR/WT expression ratio
|
rpoN/WT expression ratio
|
σ54 promoter distance (bp)e | Evidence and/or function | ||
|---|---|---|---|---|---|---|---|
| Microarrayb | QRT-PCRc | Microarray | QRT-PCR | ||||
| gdn1074 | FimA | 0.23 | 0.00078 ± 0.00043 | 0.15 | 0.012 ± 0.0081 | 75 | FimA D. nodosus |
| gdn1291 | Outer membrane protein | 0.29 | NTd | 0.39 | NT | 383 | Signal sequence, outer membrane OprF of Pseudomonas stutzeri, 37% identity |
| gdn1292 | Outer membrane RTX protein | 0.58 (P < 0.15) | 0.12 ± 0.06 | 0.39 | 0.07 ± 0.02 | PSI-BLAST: RTX toxin, leukotoxins | |
| gdn1328 | Lipoprotein | 0.58 (P < 0.007) | NT | 0.64 | NT | 43 | Lipoprotein signal sequence with 61% identity to NMA1549 |
| gdn1329 | Unknown | 0.57 (P < 0.006) | 0.12 ± 0.03 | 0.46 | 0.09 ± 0.04 | Conserved hypothetical protein | |
| gdn1330 | Inner membrane protein | 0.31 | 0.12 ± 0.05 | 0.44 | 0.11 ± 0.02 | COG1585 membrane protein implicated in regulation of membrane protease activity | |
| gdn1331 | Inner membrane protease | 0.39 | 0.16 ± 0.04 | 0.63 | 0.19 ± 0.11 | COG0330 HflC membrane protease subunit | |
Numbers refer to the annotation at the time of this writing.
All expression ratios presented are significant (P < 0.005) unless otherwise stated.
QRT-PCR ratios are significant (P < 0.02).
NT, not tested.
Distance of the potential σ54 binding site from the ATG of the first gene in the operon.
NtrC family response regulators require σ54 for transcriptional activation to occur (7). Therefore, since any genes regulated by PilR should also be differentially expressed in an rpoN background, we used this property as a filter to determine the genes of interest. In addition to fimA, only two transcriptional units were identified as being coregulated (gdn1291-2 and gdn1328-31) (Table 2). The operon containing gdn1328-31 was observed as being down-regulated in both the pilR and rpoN mutants compared to the wild-type. The proteins encoded by this operon were largely hypothetical; however, bioinformatic analysis suggested that they may be involved in protein turnover, with the products of gdn1331 and gdn1330 having domains associated with membrane protease activity and the regulation of membrane protease activity, respectively. The second operon regulated by both PilR and σ54 had potential virulence functions. The products encoded by gdn1292 and gdn1291 were similar to RTX (repeats in toxin) proteins and their accessory proteins, respectively. The product of gdn1291 showed similarity to outer membrane proteins with OmpA domains. This protein may play a role similar to that of the FrpD protein of Neisseria meningitidis, which is involved in an interaction with the RTX protein FrpC of N. meningitidis (43). PSI-BLAST (1) analysis indicated that the 300-kDa product of gdn1292 had similarity to numerous RTX toxins. RTX toxins are cytotoxic proteins with a characteristic repeated motif (GGXGXDX[L/I/V/W/Y/F], where X is any amino acid) (33). Since the putative RTX protein identified here has only two such motifs, less than other family members described to date (33), further studies are required to determine if the GDN1292 protein is a functional RTX protein.
The genes that were observed to be down-regulated in the rpoN and pilR strains were likely to be directly regulated by σ54. Subsequently, we attempted to identify potential binding sites for σ54 by searching the promoter regions of the three transcriptional units for matches to the σ54 binding consensus (2). σ54 binding sites were identified in all three operons (Table 2). These sites had, as a minimum, the correct spacing and the highly conserved −24 GG and −12 GC elements.
There were also some genes that were differentially expressed in only one of the mutants. Two genes that were only down-regulated by PilR encoded two characterized Omp1 outer membrane proteins of D. nodosus. A gene with potential stress response functions was also observed to be up-regulated in the rpoN mutant, suggesting a potential role for σ54 in the reaction of D. nodosus to physiological stress. The putative protein product of this gene had some domain similarity to OstA-like proteins. In E. coli, OstA or Imp is σE regulated and functions as an outer membrane protein involved in envelope biogenesis and envelope remodeling and may be involved in responding to periplasmic stress (6). Further studies would be required to determine the function of the D. nodosus OstA-like protein.
In D. nodosus, the rpoN gene was located at the end of the putative operon encoding the OstA-like protein. Interestingly, rpoN transcription was shown to be increased over twofold in the rpoN mutant. The position of the two microarray oligonucleotides, 5′ and 3′ of the erm(B) promoter, excluded the explanation that the altered transcription levels resulted solely from readthrough from the erm(B) promoter. The increase in transcription (also observed by RT-PCR) suggested that the cell was compensating for the loss of σ54 protein by up-regulating the transcription of this operon.
DISCUSSION
Type IV fimbriae are present in many microorganisms and are required for diverse biological functions, including virulence, cell adhesion, natural transformation, and twitching motility (36). Previous studies suggested that the type IV fimbriae of D. nodosus are expressed from a σ54-dependent promoter (22, 30). In this study, we have provided direct evidence that the fimA fimbrial subunit gene is regulated at the transcriptional level by a σ54-dependent two-component signal transduction system that requires the PilR response regulator. We also show that the type IV fimbriae of D. nodosus are required for adherence, that the levels of subunit expression correlate with extracellular protease activity, and that two other operons are regulated by this system. This study represents a significant advancement in our understanding of how the ability of D. nodosus to cause ovine footrot is regulated.
In P. aeruginosa and M. xanthus, there is a definite link between the regulation of type IV fimbrial biogenesis and the presence of NtrC-like response regulators and σ54 (21, 25, 26, 60). By constructing chromosomal mutants by allelic exchange and then carrying out functional assays and transcriptional analysis, we have shown that this association extends to the regulation of type IV fimbriae in D. nodosus. By contrast, although there is an rpoN gene in the pathogenic neisseriae, it is not functional, and no biological activity has been attributed to a regulator that has similarity to PilR (9, 34).
In D. nodosus, a basal level of fimA transcription occurred even in the absence of PilR or σ54, and some fimbrial subunit was still produced in the mutants. The rpoN mutant produced more of the fimbrial subunit and the fimA transcript than did the pilR mutant (Fig. 2D). In the rpoN mutant, a low level of fimA transcription may occur by σ70-based transcription, analogous to that observed in Neisseria gonorrhoeae and Neisseria meningitidis (10, 18). In the pilR mutant, there may be a basal level of fimA transcription that occurs in a PilR-independent manner. Although an additional transcriptional start site was not detected in these studies, there is evidence for an alternative minor promoter in this system (22).
PilR was shown to bind specifically to a region 234 to 593 bp upstream of the fimA start site. This binding region is much further upstream than that observed for P. aeruginosa PilR, which bound to the region from positions −120 to −80 (29) and to NtrC-like regulators in general. However, studies with NtrC have shown that it is able to activate transcription when its promoter is some distance from the transcriptional start site (38, 44). The significant distance of the PilR binding region from the fimA transcriptional start site may connote a role for the integration host factor (IHF) in our system. We have identified a potential IHF binding site upstream of fimA (positions −58 to −46) as well as genes encoding IHF in the D. nodosus genome (G. S. Myers, D. Parker, R. M. Kennan, J. G. Songer, I. T. Paulsen, and J. I. Rood, unpublished data). IHF may act to bend the DNA to allow the interaction between PilR and σ54-RNAP. It is proposed that σ54 binds to core RNA polymerase and that the resultant holoenzyme then binds to the promoter region. At this stage, transient interactions between σ54-RNAP and PilR may occur, a process facilitated through intrinsic DNA curvature or DNA bending induced by factors such as IHF. Phosphorylation of PilR may induce the conformational change necessary for the hydrolysis of ATP and the conversion of σ54-RNAP into an open transcriptional complex, allowing transcription to occur. Phosphorylation in similar systems normally induces activator oligomerization and DNA binding (44, 58, 61). Phosphorylation may also alter the monomer/oligomer ratio, leading to a higher proportion of larger multimers.
One function of type IV fimbriae is to mediate adherence to various surfaces and cell types (36, 55). Although type IV fimbriae are essential for virulence in D. nodosus (31), their precise role in the disease process is yet to be determined. This study has shown for the first time that the fimbriae of D. nodosus are essential for adherence. A requirement for type IV fimbriae in adherence to epithelial cells also has been demonstrated in P. aeruginosa, N. gonorrhoeae, and Moraxella bovis (24, 27, 45). Loss of adherence was observed in both the pilR and rpoN mutants at levels similar to those of the fimA mutant. However, in both mutant strains, some fimbrial subunit was still being produced, albeit at highly reduced levels. In M. xanthus, a threshold level of the subunit protein was required before functional fimbriae were formed (28). This result agrees well with our observations and implies that in D. nodosus, a threshold level of fimbrial subunits is also required before fimbriae are produced. In vivo, it is likely that the fimbriae adhere to the epithelium of the interdigital skin, enabling the cells to penetrate into the skin-horn junction by twitching motility and to grow and produce extracellular subtilisin-like proteases that contribute to tissue destruction (3).
There is clearly an association between type IV fimbriae and extracellular protease secretion. It has been observed that inactivation of the fimbrial subunit in D. nodosus (31, 35) and P. aeruginosa (31, 35) leads to a loss of extracellular protease activity. Previous hypotheses have suggested that as some type II secretion proteins share similarity to fimbrial subunits, the type IV fimbriae may act as a secretion portal, expelling proteases through the motive force of fimbrial extension and retraction (39, 48, 49). In this study, analysis of the pilR and rpoN mutants showed that the loss of extracellular protease activity was proportional to the levels of the fimbrial subunit, which suggests that the fimbrial subunit interacts with components of the type IV fimbrial apparatus to provide a type II secretion pathway through which the proteases are secreted.
To determine additional regulatory targets of PilR and σ54, the unpublished D. nodosus genome sequence was utilized to construct an oligonucleotide microarray and to carry out global transcriptional profiling of the pilR and rpoN mutants. These experiments represent the first whole-genome transcriptomic study of a type IV fimbrial regulator such as PilR. It appears that this regulatory system, in addition to regulating a known virulence factor, the type IV fimbriae, may also coordinate the regulation of additional putative virulence factors. Reduced expression of genes encoding a putative RTX protein and its outer membrane accessory protein was observed in the pilR and rpoN mutants. The RTX family includes cytotoxic proteins, hemolysins, leukotoxins, lipases, and metalloproteases (33). The RTX toxins are calcium binding proteins that act on various cells types, are involved in cell lysis, and have been shown to be necessary for virulence in some organisms (4, 33). However, these proteins are not always involved in virulence. For example, the RTX protein of N. meningitidis was not required for virulence, and based on this result, it was suggested that it does not have toxic activity (17). Since there is no evidence that D. nodosus exhibits any hemolytic activity, further studies are required to determine if the putative RTX protein of D. nodosus is an RTX toxin. The outer membrane protein whose expression was differentially regulated with the putative RTX protein is likely to anchor the protein to the cell surface (43). The other putative operon that was down-regulated in the pilR and rpoN mutants appeared to encode a hypothetical membrane-bound protein turnover system whose role in virulence is unknown.
In summary, we were able to use genomic data to identify the genes involved in the regulation of type IV fimbriae in D. nodosus. This system regulates the fimbrial subunit gene fimA at the level of transcription using a PilR transcriptional activator and a σ54-dependent promoter, with PilR binding specifically to a region upstream of this promoter. The fimbriae of D. nodosus were shown to be required for adherence, and extracellular protease secretion was correlated with the levels of the fimbrial subunit. Finally, using global transcriptional profiling, for the first time in D. nodosus, we were able to show that PilR and σ54 regulate additional genes, including two putative operons that may encode other virulence factors.
Acknowledgments
We thank Khim Hoe for assistance with electron microscopy, Catherine Ryan for Pseudomonas strains and PilR antisera, Ben Howden for help with microarray analysis, and Denisse Leyton for assistance with cell culture.
D.P. was the recipient of an Australian Postgraduate Award, a Monash Faculty of Medicine, Nursing, and Health Sciences Postgraduate Excellence Award, and a Monash Postgraduate Publication Award. This work was supported by grants from the Australian Research Council and Initiative for Future Agriculture and Food Systems grant no. 2001-52100-11445 from the USDA Cooperative State Research, Education, and Extension Service.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrios, H., B. Valderrama, and E. Morett. 1999. Compilation and analysis of σ54-dependent promoter sequences. Nucleic Acids Res. 27:4305-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Billington, S. J., J. L. Johnston, and J. I. Rood. 1996. Virulence regions and virulence factors of the ovine footrot pathogen, Dichelobacter nodosus. FEMS Microbiol. Lett. 145:147-156. [DOI] [PubMed] [Google Scholar]
- 4.Boekema, B. K., E. M. Kamp, M. A. Smits, H. E. Smith, and N. Stockhofe-Zurwieden. 2004. Both ApxI and ApxII of Actinobacillus pleuropneumoniae serotype 1 are necessary for full virulence. Vet. Microbiol. 100:17-23. [DOI] [PubMed] [Google Scholar]
- 5.Boyd, J. M., and S. Lory. 1996. Dual function of PilS during transcriptional activation of the Pseudomonas aeruginosa pilin subunit gene. J. Bacteriol. 178:831-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun, M., and T. J. Silhavy. 2002. Imp/OstA is required for cell envelope biogenesis in Escherichia coli. Mol. Microbiol. 45:1289-1302. [DOI] [PubMed] [Google Scholar]
- 7.Buck, M., M. T. Gallegos, D. J. Studholme, Y. Guo, and J. D. Gralla. 2000. The bacterial enhancer-dependent σ54 (σN) transcription factor. J. Bacteriol. 182:4129-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burrows, P. C., K. Severinov, A. Ishihama, M. Buck, and S. R. Wigneshweraraj. 2003. Mapping σ54 polymerase interactions at the −24 consensus promoter element. J. Biol. Chem. 278:29728-29743. [DOI] [PubMed] [Google Scholar]
- 9.Carrick, C. S., J. A. Fyfe, and J. K. Davies. 2000. The genome of Neisseria gonorrhoeae retains the remnants of a two-component regulatory system that once controlled piliation. FEMS Microbiol. Lett. 186:197-201. [DOI] [PubMed] [Google Scholar]
- 10.Carrick, C. S., J. A. Fyfe, and J. K. Davies. 1997. The normally silent sigma54 promoters upstream of the pilE genes of both Neisseria gonorrhoeae and Neisseria meningitidis are functional when transferred to Pseudomonas aeruginosa. Gene 198:89-97. [DOI] [PubMed] [Google Scholar]
- 11.Chaney, M., R. Grande, S. R. Wigneshweraraj, W. Cannon, P. Casaz, M. T. Gallegos, J. Schumacher, S. Jones, S. Elderkin, A. E. Dago, E. Morett, and M. Buck. 2001. Binding of transcriptional activators to sigma 54 in the presence of the transition state analog ADP-aluminium fluoride: insights into activator mechanochemical action. Genes Dev. 15:2282-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Claxton, P. D. 1989. Antigenic classification of Bacteroides nodosus, p. 155-166. In J. R. Egerton, W. K. Yong, and G. G. Riffkin (ed.), Footrot and foot abscess of ruminants. CRC Press, Inc., Boca Raton, Fla.
- 13.Craig, L., M. E. Pique, and J. A. Tainer. 2004. Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2:363-378. [DOI] [PubMed] [Google Scholar]
- 14.Dodd, I., and J. B. Egan. 1990. Improved detection of helix-turn-helix DNA binding motifs in protein sequences. Nucleic Acids Res. 18:5019-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drummond, M., P. Whitty, and J. Wootton. 1986. Sequence and domain relationships of ntrC and nifA from Klebsiella pneumoniae: homologies to other regulatory proteins. EMBO J. 5:441-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elleman, T. C., P. A. Hoyne, D. J. Stewart, N. M. McKern, and J. E. Peterson. 1986. Expression of pili from Bacteroides nodosus in Pseudomonas aeruginosa. J. Bacteriol. 168:574-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forman, S., I. Linhartova, R. Osicka, X. Nassif, P. Sebo, and V. Pelicic. 2003. Neisseria meningitidis RTX proteins are not required for virulence in infant rats. Infect. Immun. 71:2253-2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fyfe, J. A., C. S. Carrick, and J. K. Davies. 1995. The pilE gene of Neisseria gonorrhoeae MS11 is transcribed from a σ70 promoter during growth in vitro. J. Bacteriol. 177:3781-3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghimire, S. C., and J. R. Egerton. 1999. PCR-RFLP of outer membrane proteins gene of Dichelobacter nodosus: a new tool in the epidemiology of footrot. Epidemiol. Infect. 122:521-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghimire, S. C., J. R. Egerton, O. P. Dhungyel, and H. D. Joshi. 1998. Identification and characterisation of serogroup M among Nepalese isolates of Dichelobacter nodosus, the transmitting agent of footrot in small ruminants. Vet. Microbiol. 62:217-233. [DOI] [PubMed] [Google Scholar]
- 21.Hobbs, M., E. S. Collie, P. D. Free, S. P. Livingston, and J. S. Mattick. 1993. PilS and PilR, a two-component transcriptional regulatory system controlling expression of type 4 fimbriae in Pseudomonas aeruginosa. Mol. Microbiol. 7:669-682. [DOI] [PubMed] [Google Scholar]
- 22.Hobbs, M., B. P. Dalrymple, P. T. Cox, S. P. Livingstone, S. F. Delaney, and J. S. Mattick. 1991. Organization of the fimbrial gene region of Bacteroides nodosus: class I and class II strains. Mol. Microbiol. 5:543-560. [DOI] [PubMed] [Google Scholar]
- 23.Hobbs, M., and J. S. Mattick. 1993. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol. Microbiol. 10:233-243. [DOI] [PubMed] [Google Scholar]
- 24.Irvin, R. T., P. Doig, K. K. Lee, P. A. Sastry, W. Paranchych, T. Todd, and R. S. Hodges. 1989. Characterization of the Pseudomonas aeruginosa pilus adhesin: confirmation that the pilin structural subunit protein contains a human epithelial cell-binding domain. Infect. Immun. 57:3720-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishimoto, K. S., and S. Lory. 1989. Formation of pilin in Pseudomonas aeruginosa requires the alternative sigma factor (RpoN) of RNA polymerase. Proc. Natl. Acad. Sci. USA 86:1954-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishimoto, K. S., and S. Lory. 1992. Identification of pilR, which encodes a transcriptional activator of the Pseudomonas aeruginosa pilin gene. J. Bacteriol. 174:3514-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jackman, S. H., and R. F. Rosenbusch. 1984. In vitro adherence of Moraxella bovis to intact corneal epithelium. Curr. Eye Res. 3:1107-1112. [DOI] [PubMed] [Google Scholar]
- 28.Jelsbk, L., and D. Kaiser. 2005. Regulating pilin expression reveals a threshold for S motility in Myxococcus xanthus. J. Bacteriol. 187:2105-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin, S., K. S. Ishimoto, and S. Lory. 1994. PilR, a transcriptional regulator of piliation in Pseudomonas aeruginosa, binds to a cis-acting sequence upstream of the pilin gene promoter. Mol. Microbiol. 14:1049-1057. [DOI] [PubMed] [Google Scholar]
- 30.Johnston, J. L., S. J. Billington, V. Haring, and J. I. Rood. 1998. Complementation analysis of the Dichelobacter nodosus fimN, fimO, and fimP genes in Pseudomonas aeruginosa and transcriptional analysis of the fimNOP gene region. Infect. Immun. 66:297-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kennan, R. M., O. P. Dhungyel, R. J. Whittington, J. R. Egerton, and J. I. Rood. 2001. The type IV fimbrial subunit gene (fimA) of Dichelobacter nodosus is essential for virulence, protease secretion, and natural competence. J. Bacteriol. 183:4451-4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klose, K. E., A. K. North, K. M. Stedman, and S. Kustu. 1994. The major dimerization determinants of the nitrogen regulatory protein NTRC from enteric bacteria lie in its carboxy-terminal domain. J. Mol. Biol. 241:233-245. [DOI] [PubMed] [Google Scholar]
- 33.Lally, E. T., R. B. Hill, I. R. Kieba, and J. Korostoff. 1999. The interaction between RTX toxins and target cells. Trends Microbiol. 7:356-361. [DOI] [PubMed] [Google Scholar]
- 34.Laskos, L., J. P. Dillard, H. S. Seifert, J. A. Fyfe, and J. K. Davies. 1998. The pathogenic neisseriae contain an inactive rpoN gene and do not utilize the pilE σ54 promoter. Gene 208:95-102. [DOI] [PubMed] [Google Scholar]
- 35.Lu, H. M., S. T. Motley, and S. Lory. 1997. Interactions of the components of the general secretion pathway: role of Pseudomonas aeruginosa type IV pilin subunits in complex formation and extracellular protein secretion. Mol. Microbiol. 25:247-259. [DOI] [PubMed] [Google Scholar]
- 36.Mattick, J. S. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56:289-314. [DOI] [PubMed] [Google Scholar]
- 37.Miroux, B., and J. E. Walker. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260:289-298. [DOI] [PubMed] [Google Scholar]
- 38.Ninfa, A. J., L. J. Reitzer, and B. Magasanik. 1987. Initiation of transcription at the bacterial glnAp2 promoter by purified E. coli components is facilitated by enhancers. Cell 50:1039-1046. [DOI] [PubMed] [Google Scholar]
- 39.Nunn, D. 1999. Bacterial type II protein export and pilus biogenesis: more than just homologies? Trends Cell Biol. 9:402-408. [DOI] [PubMed] [Google Scholar]
- 40.Parker, D., R. M. Kennan, G. S. Myers, I. Paulsen, and J. I. Rood. 2005. Identification of a Dichelobacter nodosus ferric uptake regulator and determination of its regulatory targets. J. Bacteriol. 187:366-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parkinson, J. S., and E. C. Kofoid. 1992. Communication modules in bacterial signalling proteins. Annu. Rev. Genet. 26:71-112. [DOI] [PubMed] [Google Scholar]
- 42.Pelton, J. G., S. Kustu, and D. E. Wemmer. 1999. Solution structure of the DNA-binding domain of NtrC with three alanine substitutions. J. Mol. Biol. 292:1095-1110. [DOI] [PubMed] [Google Scholar]
- 43.Prochazkova, K., R. Osicka, I. Linhartova, P. Halada, M. Sulc, and P. Sebo. 2005. The Neisseria meningitidis outer membrane lipoprotein FrpD binds the RTX protein FrpC. J. Biol. Chem. 280:3251-3258. [DOI] [PubMed] [Google Scholar]
- 44.Rippe, K., M. Guthold, P. H. von Hippel, and C. Bustamante. 1997. Transcriptional activation via DNA-looping: visualization of intermediates in the activation pathway of E. coli RNA polymerase · σ54 holoenzyme by scanning force microscopy. J. Mol. Biol. 270:125-138. [DOI] [PubMed] [Google Scholar]
- 45.Rudel, T., J. P. M. van Putten, C. P. Gibbs, R. Haas, and T. F. Meyer. 1992. Interaction of two variable proteins (PilE and PilC) required for pilus-mediated adherence of Neisseria gonorrhoeae to human epithelial cells. Mol. Microbiol. 6:3439-3450. [DOI] [PubMed] [Google Scholar]
- 46.Saal, L. H., C. Troein, J. Vallon-Christersson, S. Gruyberger, A. Borg, and C. Peterson. 2003. BioArray Software Environment: a platform for comprehensive management and analysis of microarray data. Genome Biol. 3:software0003. [DOI] [PMC free article] [PubMed]
- 47.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 48.Sandkvist, M. 2001. Biology of type II secretion. Mol. Microbiol. 40:271-283. [DOI] [PubMed] [Google Scholar]
- 49.Sauvonnet, N., G. Vignon, A. P. Pugsley, and P. Gounon. 2000. Pilus formation and protein secretion by the same machinery in Escherichia coli. EMBO J. 19:2221-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schweizer, H. P. 1991. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene 97:109-112. [DOI] [PubMed] [Google Scholar]
- 51.Stewart, D. J. 1989. Footrot of sheep, p. 5-45. In J. R. Egerton, W. K. Yong, and G. G. Riffkin (ed.), Footrot and foot abscess of ruminants. CRC Press, Boca Raton, Fla.
- 52.Stewart, D. J. 1979. The role of elastase in the differentiation of Bacteroides nodosus infections in sheep and cattle. Res. Vet. Sci. 27:99-105. [PubMed] [Google Scholar]
- 53.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 54.Stock, J. B., M. G. Surette, M. Levit, and P. Park. 1995. Two-component signal transduction systems: structure-function relationships and mechanisms of catalysis, p. 25-51. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. ASM Press, Washington, D.C.
- 55.Strom, M. S., and S. Lory. 1993. Structure-function and biogenesis of the type IV pili. Annu. Rev. Microbiol. 47:565-596. [DOI] [PubMed] [Google Scholar]
- 56.Studholme, D. J., and M. Buck. 2000. The biology of enhancer-dependent transcriptional regulation in bacteria: insights from genome sequences. FEMS Microbiol. Lett. 186:1-9. [DOI] [PubMed] [Google Scholar]
- 57.Studholme, D. J., and R. Dixon. 2003. Domain architectures of σ54-dependent transcriptional activators. J. Bacteriol. 185:1757-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weiss, D. S., J. Batut, K. E. Klose, J. Keener, and S. Kustu. 1991. The phosphorylated form of the enhancer-binding protein NtrC has an ATPase activity that is essential for activation of transcription. Cell 67:155-167. [DOI] [PubMed] [Google Scholar]
- 59.Wigneshweraraj, S. R., M. K. Chaney, A. Ishihama, and M. Buck. 2001. Regulatory sequences in sigma 54 localise near the start of DNA melting. J. Mol. Biol. 306:681-701. [DOI] [PubMed] [Google Scholar]
- 60.Wu, S. S., and D. Kaiser. 1997. Regulation of expression of the pilA gene in Myxococcus xanthus. J. Bacteriol. 179:7748-7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wyman, C., I. Rombel, A. K. North, C. Bustamante, and S. Kustu. 1997. Unusual oligomerization required for activity of NtrC, a bacterial enhancer-binding protein. Science 275:1658-1661. [DOI] [PubMed] [Google Scholar]
- 62.Yang, Y. C., C. P. Chou, T. T. Kuo, S. H. Lin, and M. K. Yang. 2004. PilR enhances the sensitivity of Xanthomonas axonopodis pv. citri to the infection of filamentous bacteriophage Cf. Curr. Microbiol. 48:251-261. [DOI] [PubMed] [Google Scholar]
- 63.Yang, Y. H., S. Dudoit, P. Luu, D. M. Lin, V. Peng, J. Ngai, and T. P. Speed. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, X., M. Chaney, S. R. Wigneshweraraj, J. Schumacher, P. Bordes, W. Cannon, and M. Buck. 2002. Mechanochemical ATPases and transcriptional activation. Mol. Microbiol. 45:895-903. [DOI] [PubMed] [Google Scholar]