Abstract
Rapid turnover of actin structures is required for dynamic remodeling of the cytoskeleton and cell morphogenesis, but the mechanisms driving actin disassembly are poorly defined. Cofilin plays a central role in promoting actin turnover by severing/depolymerizing filaments. Here, we analyze the in vivo function of a ubiquitous actin-interacting protein, Aip1, suggested to work with cofilin. We provide the first demonstration that Aip1 promotes actin turnover in living cells. Further, we reveal an unanticipated role for Aip1 and cofilin in promoting rapid turnover of yeast actin cables, dynamic structures that are decorated and stabilized by tropomyosin. Through systematic mutagenesis of Aip1 surfaces, we identify two well-separated F-actin–binding sites, one of which contributes to actin filament binding and disassembly specifically in the presence of cofilin. We also observe a close correlation between mutations disrupting capping of severed filaments in vitro and reducing rates of actin turnover in vivo. We propose a model for balanced regulation of actin cable turnover, in which Aip1 and cofilin function together to “prune” tropomyosin-decorated cables along their lengths. Consistent with this model, deletion of AIP1 rescues the temperature-sensitive growth and loss of actin cable defects of tpm1Δ mutants.
INTRODUCTION
Dynamic remodeling of the actin cytoskeleton is required for key processes such as endocytosis, cell migration, polarized growth, and cytokinesis. Actin networks are rapidly formed or expanded by de novo actin nucleation and/or severing and uncapping of existing filaments to amplify the number of free barbed ends for elongation. Subsequently, actin networks are rapidly depolymerized through the combined actions of filament severing, barbed end capping, and acceleration of subunit dissociation from filament pointed ends; “turnover” refers to the process of recycling actin subunits as they assemble into filaments, disassemble, and subsequently are restored to an ATP-bound form for new rounds of assembly. The rate of actin filament turnover in vivo is ∼100-fold faster than that of filaments assembled from purified actin in vitro (Zigmond, 1993), indicating that cells express factors that strongly promote turnover. One such protein is ADF/cofilin (Bamburg, 1999), which severs filaments and promotes dissociation of subunits from filament pointed ends, accelerating actin disassembly in vitro and in vivo (Carlier et al., 1997; Lappalainen and Drubin, 1997; Maciver et al., 1998; Hotulainen et al., 2005). Significantly, cofilin severing can induce actin assembly rather than disassembly, depending on the cellular conditions. Severing increases the number of free barbed ends of filaments, and if these ends remain uncapped and actin monomers are available, they undergo rapid growth (Chan et al., 2000; DesMarais et al., 2005). Thus, whether cofilin severing results in filament assembly or disassembly in vivo depends on whether the newly generated barbed ends are capped. It is not yet clear what cellular factors cap the barbed ends of cofilin-severed filaments. The ubiquitous CapZ family of capping proteins may contribute, but they have not been observed on some actin structures known to undergo rapid turnover in vivo (e.g., actin cables, stress fibers, and filopodia). Further, capping protein may not always be active at sites where cofilin is active. Thus, other factors are likely to participate in capping of cofilin-severed filaments.
One highly conserved component of the cytoskeleton that is thought to function with cofilin is actin interacting protein 1 (Aip1; reviewed in Ono, 2003). Aip1 was first identified in Saccharomyces cerevisiae, where it interacts with actin and cofilin by the two-hybrid assay and colocalizes with cofilin on actin patches (Amberg et al., 1995; Rodal et al., 1999). Deletion of the yeast AIP1 gene is synthetic lethal with cof1 alleles, and aip1Δ cells have thickened actin cables aberrantly decorated with cofilin (Rodal et al., 1999), suggesting that Aip1 function is linked tightly to cofilin. However, the cellular mechanism by which Aip1 functions has remained elusive, due in part to its biochemical activities being complex. Purified Aip1 alone has negligible effects on actin, but in the presence of cofilin it induces net disassembly of actin filaments (Aizawa et al., 1999; Okada et al., 1999; Rodal et al., 1999; Mohri et al., 2003). Two recent in vitro studies suggest that filament disassembly results from Aip1 capping the barbed ends of cofilin-severed filaments (Okada et al., 2002; Balcer et al., 2003). A separate study showed that Aip1 enhances filament severing (Ono et al., 2004). Thus, at least two different biochemical activities of Aip1 may be important for its in vivo function. Further, it has been suggested that a physiological function of Aip1 is to promote actin turnover, based on its in vitro activities and genetic interactions with cofilin. However, this function has never been demonstrated in living cells, and it remains unclear which specific actin structures Aip1 may target for disassembly.
Yeast cells contain two prominent actin structures, cables and patches, both of which are rapidly assembled and disassembled. Cables and patches are required for two different cellular functions, polarized growth and endocytosis, respectively. Further, they are nucleated by different mechanisms and decorated by distinct sets of actin-binding proteins (Pruyne and Bretscher, 2000; Pruyne et al., 2004). Cables are nucleated by formins and decorated by tropomyosins, a cable-specific marker, whereas patches are nucleated by the Arp2/3 complex and decorated with cofilin and other patch-specific markers. Tropomyosin and cofilin compete for F-actin binding in vitro (Bamburg, 1999), suggesting their decoration of filaments may be mutually exclusive, and this is consistent with their separate detection in vivo on patches and cables. Actin patches and cables are highly dynamic structures, disappearing <60 s after treatment of cells with latrunculin A (Lat-A; Ayscough et al., 1997; Karpova et al., 1998; Pruyne et al., 1998; Yang and Pon, 2002). The rapid turnover of patches depends on cofilin (Lappalainen and Drubin, 1997), but almost nothing is known about how cable turnover is regulated. Only four actin binding proteins (Tpm1, Tpm2, Sac6, Abp140) have been localized to cables, and each of these stabilizes and/or bundles filaments in vitro (Liu and Bretscher, 1989; Pruyne et al., 1998; Drubin et al., 1988; Asakura et al., 1998; Yang and Pon, 2002). The failure to detect severing/destabilizing factors on cables has left it unclear how rapid cable disassembly is achieved.
Here, we provide the first direct evidence that Aip1 accelerates actin turnover in vivo, demonstrating that Aip1 and cofilin promote rapid turnover of actin patches and cables. The cellular mechanism of Aip1 function is dissected through systematic mutagenesis of its conserved surface residues, performing parallel in vivo analysis of aip1 alleles and biochemical analysis of mutant Aip1 proteins. We define two actin-binding sites on Aip1 required for its ability to cap the barbed ends of cofilin-severed filaments in vitro. Further, we observe a close correlation between loss of capping activity in vitro and loss of rapid actin turnover in vivo. These data suggest that one critical function of Aip1 in living cells is to promote actin turnover by capping severed filaments.
MATERIALS AND METHODS
Strains and Plasmids
Strains and their genotypes are listed in Supplementary Table S1. We constructed an integration plasmid pBG363 that carries the AIP1 open reading frame (ORF) plus 379-base pair upstream and 516-base pair downstream sequence, with a KanMX gene inserted 79 base pairs downstream of the AIP1 stop codon. Mutant aip1 alleles were generated by site-directed mutagenesis (Quickchange, Stratagene, Cedar Creek, TX) using pBG363 as the template. N-aip1 (aa 1-326) was generated by introducing an early stop codon, and C-aip1 (Δ14–319) was generated by internal deletion. Alleles were integrated at the AIP1 locus by replacing aip1Δ::LEU2 in yeast strain DBY6529 with aip1-KanMX. Integration at the correct locus was confirmed by PCR. Plasmids for purifying GST-Aip1 overexpressed in yeast were constructed by gap repair in the yeast strain BGY697 using the pEG(KT) vector (Mitchell et al., 1993). Transformants were tested for galactose-induced GST-Aip1 expression by immunoblotting with anti-Aip1 antibodies. Plasmids were recovered from the transformants and digested with specific restriction enzymes to confirm the presence of mutations. For two-hybrid analyses, wild-type and mutant AIP1 alleles were amplified by PCR and cloned into the NcoI and BamHI sites of pGBKT7 (BD Biosciences, Palo Alto, CA). All constructs described above were sequenced. pGAD-ACT1 was constructed by excising the ACT1 ORF with NdeI and BamHI from pACTII-ACT1 (Amberg et al., 1995) and subcloning the insert into pGADT7 (BD Biosciences). Further details of constructions are available upon request.
Immunoblotting and Fluorescence Microscopy of Cells
Polyclonal chicken antibodies (Aves Labs, Tigard, OR) were raised against yeast Aip1 and cofilin, purified as described (Lappalainen et al., 1997; Rodal et al., 1999). Aip1 antibodies were affinity-purified. A rabbit anti-yeast actin antibody was affinity purified, and immunostaining was performed as described (Pringle et al., 1991). For monitoring the time courses of actin patch and cable turnover, cells were grown at 30°C to OD600 = 0.5 and then treated with 20–50 μM Lat-A as specified in figure legends. At different time points after treatment with Lat-A, aliquots of cells were removed and immediately fixed in 4% formaldehyde at 30°C for 1 h. Cells were washed and then stained with either Alexa-fluor 488 phalloidin (Molecular Probes, Eugene, OR) to visualize filamentous actin in patches or anti-actin antibodies to visualize actin cables. Cells were imaged on a Zeiss Axioskop2 mot plus (Carl Zeiss, Thornwood, NY). Images were captured using a Hamamatsu IEEE1394 digital CCD camera (Hamamatsu Photonics, Bridgewater, NJ) and analyzed using Openlab software (Improvision, Lexington, MA).
Yeast Two-Hybrid Interactions
Y187 strains carrying pGBKT7-aip1 allele plasmids were crossed to Y190 strains carrying either pGADT7-ACT1 or pACT II-COF1. Resulting diploid strains carrying both plasmids were selected on −Trp/−Leu/−His plates. Y187 strains carrying pDAb7-ACT1 or empty pGBKT7 vector were included as positive and negative controls, respectively. Serial dilutions of diploid cells from each cross were analyzed for growth on −Trp/−Leu/−His plates containing 0–10 mM 3-amino-1,2,4-triazole (3-AT; Sigma, St. Louis, MO).
Biochemical Analyses
Wild-type and mutant Aip1 proteins were expressed in yeast as GST-fusions and purified as described (Rodal et al., 1999). Yeast actin (Goode, 2002), cofilin (Lappalainen et al., 1997), and profilin (Eads et al., 1998) were purified as described. Rabbit skeletal muscle actin (RMA) was purified as described (Spudich and Watt, 1971) and gel filtered. For all of the different assays in Figure 4, purified yeast actin or RMA was assembled into filaments in F-buffer (50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 0.2 mM CaCl2, 0.2 mM ATP, 0.2 mM DTT, 10 mM Tris, pH 7.5) and then incubated with Aip1 and/or cofilin as described in the figure legends. Reactions were centrifuged at 436,000 × g for 20 min in a TLA-100 rotor (Beckman Instruments, Fullerton, CA) to pellet filaments. Proteins in the supernatants, and pellets were examined by SDS-PAGE, scanned, and analyzed with NIH image software v1.63.
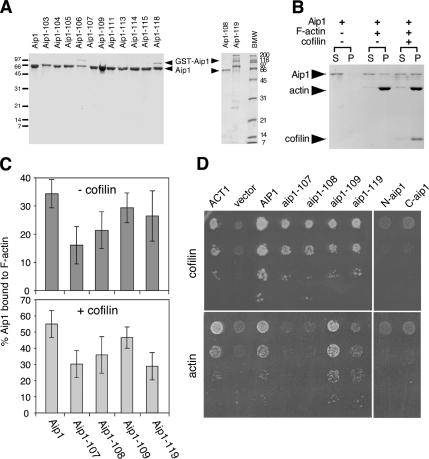
Figure 4.
Actin- and cofilin-interaction of purified wild-type and mutant Aip1 proteins. (A) Coomassie-stained gel of purified proteins. Lower arrow points to Aip1 lacking GST tag; upper arrow points to residual uncleaved GST-Aip1. Note that Aip1-119 has retarded migration compared with wild-type Aip1. (B) Cosedimentation of 0.5 μM wild-type Aip1 with 2 μM rabbit skeletal muscle F-actin (RMA) in the presence and absence of 3 μM cofilin; supernatants (S) and pellets (P) were analyzed on Coomassie-stained gels. (C) Comparison of wild-type and mutant Aip1 cosedimentation with RMA, with conditions same as above, in the presence (bottom panel) and absence (top panel) of cofilin (n = 3). Error bars, SD. (D) Two-hybrid interactions of wild-type and mutant Aip1 with cofilin (top) and actin (bottom). Cultures of diploid strains carrying Aip1-DBD bait plasmids and cofilin-AD or actin-AD prey plasmids were serially diluted and plated on triple selective media and 10 mM 3-AT.
The design and rationale of the filament capping assay used is described in Balcer et al. (2003) and herein (see Results and legend for Figure 5C). Because one recent study has challenged whether this assay measures Aip1-dependent capping (Clark et al., 2006), below we provide detailed commentary on the assay and its requirements. In brief, this is a modified filament sedimentation assay, in which barbed end capping is indicated by a shift of actin to the supernatant fraction after incubation of preformed yeast actin filaments (4 μM) with reaction ingredients (6 μM yeast profilin, 0.4 μM yeast cofilin, and 0.1 μM yeast Aip1). Profilin is included to restrict repolymerization of released monomers to barbed ends of filaments, because profilin blocks addition of monomers to pointed ends. As a result, when barbed ends are capped, actin subunits steadily dissociate from the pointed ends of filaments and accumulate as a profilin-bound monomer pool (supernatant fraction). The assay was developed by Balcer et al. (2003) to compare the effects of Cof1, Cof1-22, and Cof1-19 in Aip1-dependent capping. That study demonstrated that Cof1-19 (a mutant with no obvious defects in filament severing/disassembly, but impaired in Aip1 interactions and showing the same thickened cable phenotype as aip1Δ mutants) is defective in capping. In contrast, Cof1-22 (a mutant with clear defects in filament severing/disassembly, but that does not exhibit the thickened cable phenotype) exhibited normal Aip1-dependent capping. These findings indicated that the assay depends on productive interactions between Aip1 and cofilin to cap barbed ends and does not depend on efficient severing/depolymerization by cofilin. Further, it was shown that capping protein can functionally replace Aip1, providing further demonstration that what is being measured is capping, rather than a different activity of Aip1. In contrast, Clark et al. (2006) report profilin- and cofilin-dependent disassembly of filaments even in the absence of Aip1. To explain these data, they suggest a new mechanism in which cofilin caps the barbed ends of filaments to block profilin–actin addition. However, no independent verification of this mechanism was provided. At minimum, such capping by cofilin alone could have been tested independently in a filament elongation assay, as we showed previously for Aip1-cofilin capping (Okada et al., 2002).
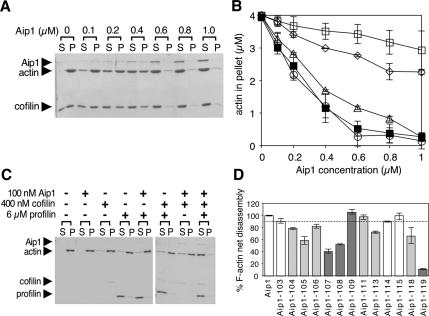
Figure 5.
Activities of purified wild-type and mutant Aip1 proteins. (A) Concentration-dependent effects of wild-type Aip1 on actin filament net disassembly. Preassembled yeast F-actin, 4 μM, was incubated at 25°C for 20 min in the presence or absence of 6 μM cofilin and the indicated concentrations of Aip1. The reactions were centrifuged, and the pellets and supernatants were analyzed on Coomassie-stained gels. (B) Concentration-dependent effects of wild-type and mutant Aip1 proteins on actin filament net disassembly. Reactions were carried out as in A. Results from three independent experiments were quantified by densitometry and graphed: wild-type Aip1 (■), Aip1-107 (◇), Aip1-108 (▵), Aip1-109 (○) Aip1-119 (□). Error bars, SD. (C) Cofilin-dependent capping and net disassembly of actin filaments by Aip1. Preassembled yeast F-actin, 4 μM, was incubated for 4 h at 25°C with and without 100 nM Aip1 and/or 400 nM cofilin and/or 6 μM profilin and then centrifuged, and the pellets and supernatants were analyzed on Coomassie-stained gels. (D) Effects of wild-type and mutant Aip1 proteins on cofilin-dependent capping and net disassembly of actin filaments with reactions performed as in C. F-actin levels in the pellet were quantified by densitometry and graphed as a percentage of F-actin in the pellet for wild-type Aip1. n = 2 for Aip1, Aip1-103, Aip1-104, Aip1-106, Aip1-111, Aip1-113, Aip1-114, Aip1-115, and Aip1-118. n = 3 for Aip1-105 Aip1-107, Aip1-108, Aip1-109, and Aip1-119. Error bars, SD. The dotted line separates effects of Aip1 proteins with pseudowild-type phenotypes in vivo (white columns) versus those with moderate or severe (light and dark gray columns, respectively) phenotypes in vivo (see Table 1).
We also point out that there are number of technical issues that might explain why they failed to detect Aip1-dependent capping using our assay. Both the nucleotide state of actin and the purity and precise concentrations of cofilin and profilin used are critical. First, we consider the nucleotide state of actin. Throughout the assay, actin monomers must be bound primarily to ATP, not ADP, because profilin has 5–10-fold weaker affinity for ADP-actin than ATP-actin (Vinson et al., 1998). ADP-actin also has a higher critical concentration for actin assembly (5 μM) than ATP-actin (0.1 μM; Pollard, 1984). Thus, if the nucleotide composition of the actin monomer pool shifts toward ADP during the course of the assay, one would predict a shift of actin to the supernatant independent of capping, as they observed. Cofilin and profilin synergize to promote rapid turnover of filaments, which involves hydrolysis of nucleotide bound to actin (Didry et al., 1998). Therefore, limiting concentrations of ATP can lead to the accumulation of ADP-actin monomers. We note that the G-actin buffer used by Clark et al. (2006) contains only 0.05 μM ATP, orders of magnitude lower than the ATP concentration in standard G-buffer recipes (0.1–0.2 mM ATP). In the presence of cofilin and profilin, these low concentrations of ATP would be rapidly converted to ADP and thus explain the observed cofilin- and profilin-dependent shift of actin to the supernatant. Second, we consider the purity of proteins used, which is also critical. Their study used yeast actin that may not be sufficiently purified. Specifically, after actin is eluted from the DNase1 affinity column, it should be purified further by ion exchange chromatography to separate actin from contaminants (including endogenous cofilin) that can affect its properties (Goode, 2002). Clark et al. (2006) did not carry out this step. It is also difficult to gauge the purity of their profilin, cofilin, and Aip1, because they show limited areas of the gels insufficient to evaluate contamination.
RESULTS
Cofilin and Aip1 Promote the Rapid Disassembly of Yeast Actin Cables
Actin disassembly-promoting factors such as cofilin have not been detected on actin cables in wild-type cells. However, aip1Δ and cof1-19 cells accumulate aberrantly thick actin cables decorated with cofilin (Rodal et al., 1999). This led us to explore the possible roles of cofilin and Aip1 in regulating cable dynamics. We first tested the role of cofilin, encoded by the essential gene COF1 (Iida et al., 1993; Moon et al., 1993). We used two mutant alleles of cofilin, cof1-19 and cof1-22. Cof1-19 has normal actin filament–severing activity in vitro (Ojala et al., 2001) but disrupts Aip1 capping and net disassembly of cofilin-decorated filaments (Balcer et al., 2003). Further, cof1-19 disrupts localization of Aip1 to patches, indicating that cofilin-Aip1 interactions are required to maintain Aip1 localization (Rodal et al., 1999). Interestingly, Cof1-19 localizes to both patches and cables (whereas wild-type cofilin is only detected on patches). Thus, normal functional interactions between cofilin and Aip1 may result in a highly dynamic decoration of cables by cofilin (i.e., short-lived), not detected in wild-type cells, but detected in cof1-19 cells where the interaction is impaired. Cof1-22 has partially impaired actin filament disassembly activity in vitro and greatly reduced rates of patch turnover in vivo (Lappalainen et al., 1997). Cof1-22 does not disrupt Aip1 capping of filaments (Balcer et al., 2003) and consequently does not accumulate thickened cables or stably decorate cables.
To investigate the role of cofilin in cable turnover, we treated wild-type, cof1-19, and cof1-22 cells with Lat-A. Lat-A is an actin monomer sequestering agent that blocks new actin assembly, thus allowing measurement of actin turnover rate, which correlates with the rate of disappearance of F-actin structures. The actual half-life of patches and cables is likely to be <20 s, as measured in the presence of high concentrations of Lat-A (Yang and Pon, 2002; Kaksonen et al., 2003, 2005). However, lower concentrations of Lat-A can be used to compare strains for relative rates of actin turnover. In fact, in the presence of higher concentrations of Lat-A, actin structures disappear so rapidly that mutant effects (e.g., 2–5-fold reduced rates of turnover) can be difficult to assess. For this reason, we used concentrations of Lat-A optimized for detecting differences between wild-type and mutant strains (20 μM for cable turnover and 50 μM for patch turnover).
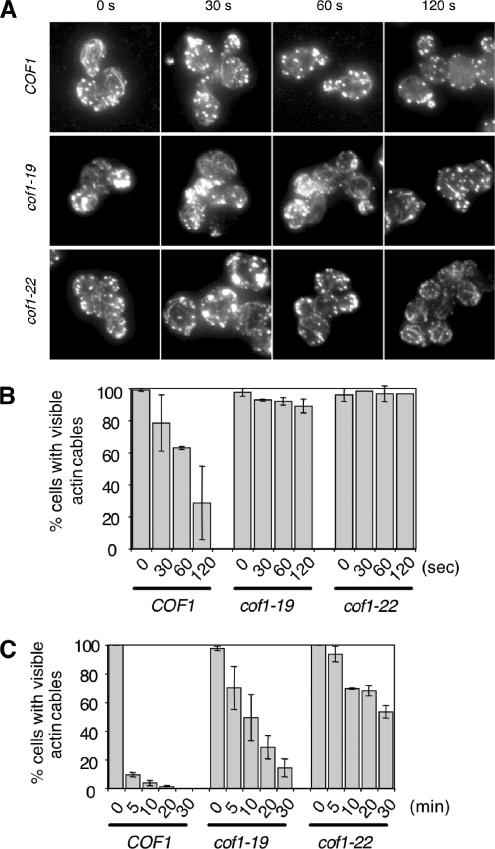
After cells were treated with 20 μM Lat A, cable disappearance was examined at different time points by immunofluorescence microscopy using anti-actin antibodies (representative staining in Figure 1A). Quantified results (Figure 1, B and C) show that cof1-22 markedly reduces the rate of cable disassembly, increasing by ∼20-fold the time in Lat A required for 50% of cells to lose visible cable staining (wild-type cells, 1 min; cof1-22 cells, 20 min). Thus, the filament severing and disassembly activity of cofilin is required for rapid turnover of cables. cof1-19 cells also showed significantly reduced rates of cable turnover, ∼10-fold slower than wild-type cells, indicating that rapid cable turnover also requires functional interactions between cofilin and Aip1. Consistent with this view, similar cable turnover defects were observed for aip1Δ cells (see below). These observations led us to dissect the cofilin-dependent function of Aip1 in promoting actin turnover.
Figure 1.
Reduced rates of actin cable turnover in cof1 mutant cells. Wild-type (COF1) and mutant (cof1-19, cof1-22) yeast cells were incubated with 20 μM Latrunculin A (Lat-A). Samples of cells were removed at the indicated time points, fixed, stained with anti-actin antibodies, and scored for visible actin cables. (A) Representative cell staining. (B and C) Percent cells with visible cables remaining after Lat-A treatment for shorter (B) or longer (C) incubation periods. More than 200 cells were counted for each time point, and the columns are an average of two independent experiments. Error bars, SD.
Identification of aip1 Alleles with Actin Cable Defects
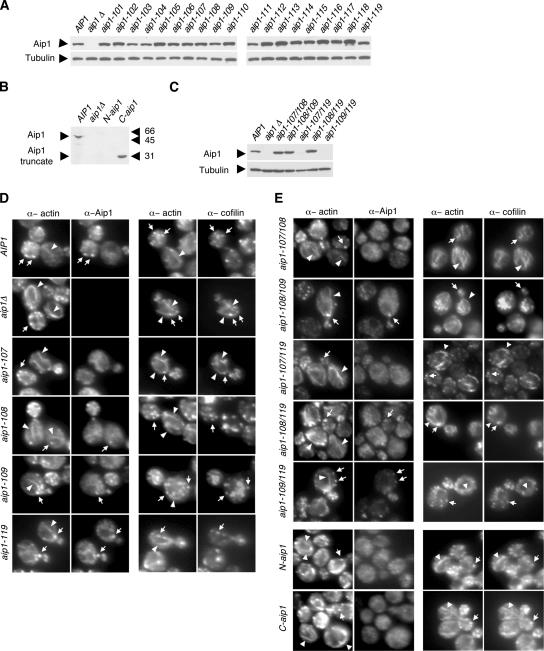
The cellular function of Aip1 has remained undefined, in part because point mutations have not been introduced into Aip1 to disrupt its specific molecular interactions and activities. Therefore, we performed a systematic mutagenesis of Aip1 surface residues. Using an alignment of Aip1 sequences from diverse organisms (Supplementary Figure S1) and the crystal structures of S. cerevisiae Aip1 (Voetgli et al., 2003) and Caenorhabditis elegans UNC-78/Aip1 (Mohri et al., 2004), we targeted all residues that are both conserved and solvent exposed. The Aip1 structure resembles an open clamshell and consists of two β-propellers, each formed by seven WD-blades, connected at an angle of ∼110°. A total of 31 surface residues were mutated within 18 different alleles (Table 1 and Supplementary Figure S1). The residues mutated are found primarily on one face of Aip1, designated as the “front” face (Figure 2), distributed between the N- and C-terminal lobes. The “back” face of Aip1 is not well conserved. In addition, we generated one substitution allele at a yeast-specific insertion located in the C-terminal lobe (aip1-119) and two truncation alleles that express the N- and C-terminal propellers of Aip1 alone: N-aip1 (Δ326–615) and C-aip1 (Δ14–319). All 21 resulting alleles were integrated at the AIP1 locus. Immunoblotting demonstrated that mutant protein levels are similar to Aip1 levels in wild-type cells (Figure 3A). Expression of N-aip1 was detected by immunofluorescence (cytoplasmic localization, Figure 3E), but not by immunoblotting (Figure 3B).
Table 1.
Data compilation for aip1 mutants
| Allele | Mutations | Aip1 on actin patches | Thickened actin cables | Cofilin on cables | Genetic interaction with cof1-22 | Two-hybrid interactions |
F-actin capping activity (% WT) | |
|---|---|---|---|---|---|---|---|---|
| With actin | With cofilin | |||||||
| AIP1 | + | − | − | − | + | + | 100 | |
| aip1Δ | Full deletion (Δ1–615) | − | C | + | SLa | |||
| N-Aip1 | Truncation (Δ326−615) | − | C | + | SL | − | − | ND |
| C-Aip1 | Truncation (Δ14−319) | − | C | + | SL | − | − | ND |
| N-terminal propeller | ||||||||
| aip1-101 | R18A | + | − | − | − | ND | + | ND |
| aip1-102 | V108A, K109A | + | s | − | − | ND | + | ND |
| aip1-103 | E127A | + | − | − | − | ND | ND | 88 |
| aip1-104 | E136A, R138A, D139A | + | s | − | − | ND | + | 73 |
| aip1-105 | F141A | + | s | − | ND | ND | + | 59 |
| aip1-106 | R172A | + | s | − | SS | ND | + | 84 |
| aip1-107 | K194A, F195A | − | C | + | SS | − | + | 36 |
| aip1-108 | K204A, F208A | + | C | − | SS | − | + | 53 |
| aip1-109 | D217A | + | C | − | SS | + | + | 106 |
| aip1-110 | D235A, K237A | + | − | − | − | ND | + | 11 |
| aip1-111 | D263A | + | s | − | ND | ND | + | 76 |
| C-terminal propeller | ||||||||
| aip1-112 | K340A | + | − | − | SS | ND | + | ND |
| aip1-113 | D356A | + | − | − | − | ND | ND | 89 |
| aip1-114 | D393A, K396A | + | − | − | ND | ND | + | 95 |
| aip1-115 | E465A | + | − | − | − | ND | + | 104 |
| aip1-116 | Y502A | + | − | − | − | ND | + | ND |
| aip1-117 | K511A | + | s | − | ND | ND | + | ND |
| aip1-118 | K533A | + | s | − | ND | ND | + | 66 |
| aip1-119 | 548–554Ab | + | C | − | SS | w | + | ND |
| Double mutant | ||||||||
| aip1-107/108 | − | C | + | SL | ND | ND | ND | |
| aip1-108/109 | + | C | − | SS | ND | ND | ND | |
| aip1-107/119 | − | C | + | SL | ND | ND | ND | |
| aip1-109/119 | + | C | − | SL (66%)/ | ND | ND | ND | |
| SS (34%) | ||||||||
+, yes; −, no; C, clearly thickened actin cables; s, subtly thickened actin cables; SL, double mutants that are synthetic lethal; SS, double mutants showing synthetic sick growth at 25°C; ND, not determined; w, weakened interaction; WT, wild type.
a Data from Rodal et al. (1999) and Balcer et al. (2003).
b aip1-119, 548-DDDIDDE/AAAIAAA-554.
Figure 2.
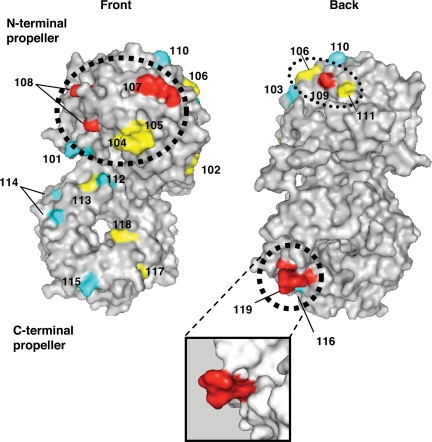
Surfaces on Aip1 required for its in vivo function. Mutated residues were modeled on the crystal structure of S. cerevisiae Aip1 (1PI6) with mutant allele numbers indicated (Table 1). The majority of conserved surface residues map to the “front” face of Aip1 (see Supplementary Figure S1 for sequence alignment). The color scheme for the mutated residues is based on the severity of mutant phenotypes (Table 1): Cyan, pseudowild type; yellow, moderately impaired; red, severely impaired. The two F-actin-binding sites identified (Figure 4) are circled by thick dotted lines. One additional surface that is important for Aip1 in vivo function (disrupted by aip1-109) is circled by a thin dotted line. The inset shows an enlarged view from a different angle of the site mutated in aip1-119. Note that the residues mutated in aip1-119 (rendered in red) protrude from the surface of the β-propeller. Molecular graphics were generated using PyMOL software (DeLano Scientific, South San Francisco, CA).
Figure 3.
Actin cytoskeleton organization and Aip1 and cofilin localization in wild-type and mutant aip1 cells. (A–C) Aip1 protein levels in wild-type and mutant aip1 cells. Whole cell extracts from integrated wild-type and mutant strains were immunoblotted with anti-Aip1 and anti-tubulin antibodies. (D and E) Cells with the designated genotypes were double-labeled with anti-actin and anti-Aip1 antibodies (left panels) or anti-actin and anti-cofilin antibodies (right panels). Arrowheads point to actin cables; arrows point to actin patches.
Each aip1 strain was examined for actin organization and localization of cofilin and Aip1. Four alleles showed thickened actin cables, similar to the aip1Δ mutations: aip1-107, aip1-108, aip1-109, and aip1-119 (Figure 3D). The remaining aip1 alleles did not show striking phenotypes compared with wild-type cells (Table 1). The thickened cables observed in specific aip1 alleles as well as in aip1Δ and cof1-19 are not actin bars, because they are detected by Alexa-488 phalloidin (Supplementary Figure S2). Of the four alleles that showed actin cable defects, clear decoration of thickened cables by cofilin was observed only for aip1-107 (Figure 3D, right panels). In addition, the severity of these defects correlated with loss of Aip1 localization from actin patches (Figure 3D, left panels). aip1-107 showed a complete loss of Aip1 from patches (cytoplasm only), whereas aip1-108, aip1-109, and aip1-119 caused partial mislocalization of Aip1. These data suggest that thickened cables arise from partial loss of Aip1 function, but detectable cofilin-decoration on cables only occurs with more severe loss of Aip1 function. Consistent with this hypothesis, N-aip1 and C-aip1 alleles, which behave genetically like null mutants, showed thick cables with cofilin decoration similar to aip1Δ (Figure 3E). Further, biochemical data suggest that Aip1-107 is more severely impaired than Aip1-108 or Aip1-109 (see below).
Genetic Interactions of aip1 Alleles with cof1-22
The complete loss of AIP1 function (aip1Δ) is synthetic lethal with cof1-22 (Rodal et al., 1999). Similarly, the complete loss of SRV2 function is synthetic lethal with cof1-22 (Balcer et al., 2003). This suggests that when the essential function of COF1 in promoting actin turnover is partially impaired by the cof1-22 mutation, cell viability becomes dependent on the activities of cofilin cofactors Aip1 and Srv2 (Rodal et al., 1999; Balcer et al., 2003). In these cases, synthetic lethality does not reflect two independent and parallel pathways leading to one essential function, but rather the functionally linked contributions of multiple actin-binding proteins driving actin turnover. We tested genetic interactions of our aip1 alleles with cof1-22 to evaluate loss of function. The results are summarized in Table 1. Similar to an aip1Δ mutation, N-aip1 and C-aip1 were synthetic lethal with cof1-22, demonstrating that each half of Aip1 is required for its normal cellular function. No single aip1 allele carrying point mutations was synthetic lethal with cof1-22. However, a number of aip1 alleles displayed synthetic sick interactions with cof1-22; that is, the double mutants grew more slowly than either single mutant alone at 25°C (Supplementary Figure S3). These included the alleles described above with thickened cable phenotypes (aip1-107, aip1-108, aip1-109, aip1-119) and one allele with a moderate cable phenotype (aip1-106). This suggests that these mutations each cause partial but not complete loss of Aip1 function.
Highlighting the mutated residues on the S. cerevisiae Aip1 structure reveals three key functional surfaces (Figure 2, dotted circles). The largest functional surface we identified (thick dotted circle) is located on the front face of the N-terminal lobe of Aip1 and encompasses residues mutated in aip1-107 and aip1-108, as well as aip1-104 and aip1-105. The two other functional surfaces are located along the rim that separates the front and back faces, one on the rim of the N-terminal lobe, marked by aip1-109 (thin dotted circle), and the other on the rim of the C-terminal lobe, marked by aip1-119 (thick dotted circle).
Because no single allele with point mutations showed complete loss of Aip1 function, we next tested the additive effects of combining partial loss of function alleles in pairs on a single molecule: aip1-107/108, aip1-108/109, aip1-107/119, aip1-108/119, and aip1-109/119. Aip1 protein levels in aip1-107/108, aip1-108/109, and aip1-108/119 cells were similar to wild-type cells (Figure 3C). Aip1 expression in aip1-107/119 and aip1-109/119 cells was detected by immunofluorescence (Figure 3E, left panel), but not by immunoblotting (Figure 3B). Both of the double aip1 mutants that include aip1-107 showed cofilin decoration on thickened cables (Figure 3E, right panel) and were synthetic lethal with cof1-22 (Table 1), suggesting that they cause a more complete loss of Aip1 function than the single mutants from which they are derived. In contrast, the phenotypes of aip1-108/109 and aip1-108/119 double mutants were not significantly different from their respective single mutants in terms of cable thickness, Aip1 localization, and genetic interactions with cof1-22. Effects of the aip1-109/119 double mutant were intermediate, displaying a stronger genetic interaction with cof1-22 than the single mutants, but not complete synthetic lethality (Table 1). Together, these data suggest that aip1-107 represents a critical site for Aip1 function in vivo and that the other surfaces we identified as important (aip1-108, aip1-109, aip1-119) play supportive roles, in some cases additive (particularly when combined with aip1-107).
Biochemical Defects of Mutant Aip1 Proteins
To understand better the cause of the more severe mutant phenotypes, we investigated the underlying biochemical and molecular defects of aip1-107, -108, -109, -119, N-aip,1 and C-aip1. Wild-type and mutant Aip1 proteins were overexpressed in yeast as GST-fusion proteins under control of the GAL1 promoter and purified by glutathione affinity. GST tags were removed by thrombin digestion, and the released Aip1 proteins were purified further by ion exchange chromatography (Figure 4A). Purified wild-type and mutant Aip1 proteins were compared for 1) binding to actin filaments in the presence and absence of cofilin (Figure 4, B and C), 2) cofilin-dependent net disassembly of filaments (Figure 5, A and B), and 3) cofilin-dependent capping of filaments (Figure 5, C and D). Attempts to purify N-Aip1 and C-Aip1 were unsuccessful.
In most studies thus far, biochemical interactions of Aip1 with F-actin have been demonstrated using RMA. Aip1 exhibits cofilin-enhanced association with RMA F-actin and a moderate net disassembly activity on filaments, shifting a fraction of the actin at steady state into the supernatant (Aizawa et al., 1999; Okada et al., 1999; Mohri et al., 2003). By comparison, using S. cerevisiae actin, yeast Aip1 and cofilin induce almost a complete shift of actin into the supernatant fraction (Rodal et al., 1999; also Figure 5A). This represents the strongest net disassembly activity observed for Aip1 and precludes the use of yeast actin for testing cofilin-dependent association of Aip1 with F-actin by cosedimentation. Thus, the employment of different actins can uncouple Aip1’s cofilin-dependent functions in binding filaments versus promoting filament disassembly.
Using RMA, we compared F-actin association of wild-type and mutant Aip1 proteins in the presence and absence of cofilin. Wild-type Aip1 showed modest association with F-actin in the absence of cofilin (32% bound), with increased binding in the presence of cofilin (52% bound, Figure 4B). Aip1-107 showed weaker association than wild-type Aip1 both in the presence and absence of cofilin (Figure 4C). Aip1-108 showed a similar pattern, though slightly more modest defects. Despite having weaker overall associations with F-actin, Aip1-107 and Aip1-108 both showed improved F-actin association in the presence of cofilin, similar to wild-type Aip1. In contrast, Aip1-119 had relatively normal F-actin associations in the absence of cofilin (72% of wild type), but displayed only a negligible increase in binding in the presence of cofilin, making this mutant unique. Aip1-109 associated with F-actin, similar to wild-type Aip1 levels.
We also compared wild-type and mutant Aip1 proteins for two-hybrid interactions with actin and cofilin (Figure 4D). In biochemical assays using purified proteins, Aip1 binds specifically to actin filaments and not monomers (Okada et al., 1999; Rodal et al., 1999). However, Aip1 also interacts with actin by the two-hybrid assay, suggesting that the actin-AD fusion protein has a conformation that may partially mimic F-actin. Our results from two-hybrid analysis showed a similar pattern to our biochemical results above: aip1-107 had the most severe defects, failing to interact with actin; aip1-108 and aip1-119 showed intermediate weakened interactions with actin; and aip1-109 interacted normally with actin. On the other hand, none of these aip1 mutants were defective in two-hybrid interactions with cofilin. However, N-aip1 and C-aip1 mutants (the expression of which were verified by immunoblotting) failed to interact with cofilin, suggesting that cofilin binding requires contributions from each half of Aip1. An important conclusion we draw from these data is that the actin and cofilin interactions of Aip1 are separable, because Aip1 mutants defective in actin interactions (e.g., aip1-107) still interact normally with cofilin.
We next compared the ability of wild-type and mutant Aip1 proteins to induce net disassembly of actin filaments (assembled from purified yeast actin) in the presence of cofilin. In this assay, filaments disassemble into fragments, but do not necessarily depolymerize into monomers. High concentrations of Aip1 and cofilin cause the formation of very short fragments of F-actin that fail to sediment by high-speed ultracentrifugation and are not recognized as monomeric actin in DNase I–binding assays (Mohri et al., 2003). Formation of these short fragments depends on two distinct activities of cofilin and Aip1, severing and capping (Balcer et al., 2003). Disassembly, as measured by sedimentation, depends on high concentrations of cofilin, possibly because severing into very short fragments requires heavy decoration of filaments by cofilin. On the other hand, cofilin-dependent capping of filaments by Aip1 requires much lower concentrations of cofilin (Balcer et al., 2003; and below). As shown in Figure 5, A and B, wild-type Aip1 induces concentration-dependent net disassembly of filaments specifically in the presence of cofilin. Aip1-107 was significantly impaired in this activity, whereas Aip1-108 showed a subtle defect (Figure 5B). These results are in good agreement with differences in the severity of their in vivo phenotypes (Table 1) and their actin binding defects in biochemical and two-hybrid assays (Figure 4, C and D).
The data also suggest that direct interactions with F-actin are critical for Aip1-mediated filament disassembly in vitro and Aip1 function in vivo. Further, the analysis with Aip1-119 indicates that net filament disassembly requires cofilin-dependent association of Aip1 with filaments. Aip1-109 showed no defects in filament disassembly, consistent with its normal binding to actin.
Aip1 has been shown to cap the barbed ends of actin filaments in a cofilin-dependent manner (Okada et al., 2002; Balcer et al., 2003). Capping can be measured using a modified net disassembly assay that contains low concentrations of cofilin and Aip1 and high concentrations of F-actin and profilin. Low concentrations of cofilin and Aip1 are sufficient to sever and cap filaments but are not sufficient to induce net disassembly of filaments. Further addition of high concentrations of profilin blocks monomer addition to the pointed ends of filaments and thereby leads to filament net disassembly specifically when barbed ends are capped (by Aip1 or Cap1/2; Balcer et al., 2003). As expected, omission of Aip1 from the reaction ingredients restores filaments, because barbed ends are no longer capped (Figure 5C). Using this assay, we compared the capping activities of Aip1 mutant proteins with variable phenotypes (pseudowild type, moderately defective, or severely defective; Figure 2). As shown in Figure 5D, loss of capping activity in vitro correlates closely with loss of function in vivo (Table 1). However, there are two mutants (Aip1-109 and Aip1-111) that show defects in vivo but are not obviously impaired in capping in vitro. Because Aip1-111 defects in vivo are subtle, this may explain its lack of biochemical defects. On the other hand, Aip1-109 has strong in vivo defects. As mentioned above, Aip1-109 interacts normally with actin and cofilin in the two-hybrid assay and biochemical tests. Because this mutant is expressed at normal levels in vivo, the basis of its loss of function in vivo remains a mystery.
Aip1 Promotes Actin Patch and Cable Turnover In Vivo
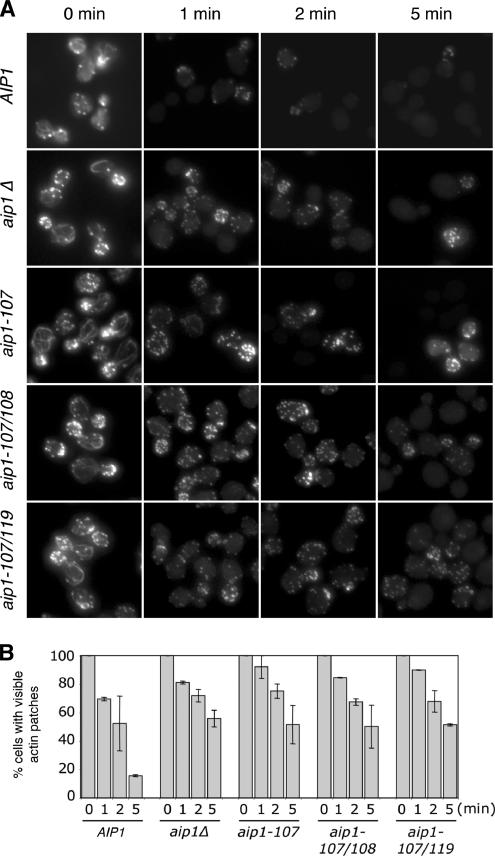
Although it has been postulated that Aip1 promotes actin disassembly based on in vitro activities and genetic interactions with cofilin, this has never been demonstrated in living cells. Therefore, we compared rates of actin turnover in vivo for wild-type and aip1 mutant strains, focusing first on patch turnover. Isogenic wild-type, aip1Δ, aip1-107, aip1-107/108, and aip1-107/119 cells were treated with 50 μM Lat-A to block new actin assembly. Samples were removed at different time points after Lat-A treatment, and cells were stained with fluorescently labeled phalloidin to visualize F-actin structures (Figure 6A). All of the aip1 mutant cells exhibited a delayed loss of F-actin staining compared with wild-type cells, indicating defects in actin turnover. In addition, three other alleles, aip1-108, aip1-109, and aip1-119, showed delayed actin turnover (Supplementary Figure S4). cof1-22 cells had an even more pronounced delay in patch turnover (unpublished data) as previously reported (Lappalainen and Drubin, 1997). To quantify these effects, we scored cells for visible F-actin structures at each time point (Figure 6B). The graphs reveal significant differences between wild-type and aip1 mutant cells after 5 min of Lat-A treatment. We also note that our scoring system provides a conservative measure of turnover defects, because cells scored as defective do not include those that have lost some or most of their actin staining (e.g., compare the actin staining of wild-type and aip1 mutant cells after Lat-A treatment in Figure 6A). Thus, mutant defects may be even more severe than suggested. Importantly, these data provide the first demonstration that Aip1 activity in vivo promotes turnover of a cellular actin structure.
Figure 6.
Defects in actin patch turnover in aip1 mutant cells. (A) Wild-type (AIP1) and mutant (aip1Δ, aip1-107, aip1-107/108, aip1-107/119) yeast cells were treated with 50 μM latrunculin A (Lat-A). Samples of cells were removed at the indicated time points, fixed, and stained with Alexa-488 phalloidin. (B) Cells from A were scored for visible actin patches and graphed. More than 200 cells were counted for each time point; columns are the average of two independent experiments. Error bars, SD.
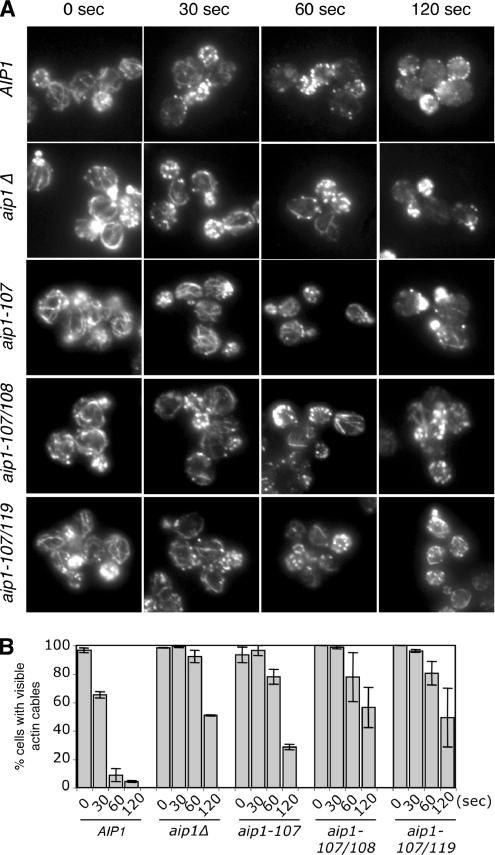
Given our new observations that cofilin promotes rapid turnover of cables (Figure 1) in addition to patches, we next tested Aip1 involvement in cable turnover. There are conflicting data in the literature as to whether patches or cables are more sensitive to Lat-A treatment. We determined that in our strain background cables are more sensitive to Lat-A treatment than patches. Therefore, we used a low concentration of Lat-A (20 μM) to visualize loss of cables compared with patches. Four mutant aip1 alleles were selected for these analyses based on the severity of their phenotypes and their strong genetic interactions with cof1-22: aip1Δ, aip1-107, aip1-107/108, and aip1-107/119. Each of these aip1 strains showed greatly reduced rates of cable turnover compared with the isogenic wild-type strain (Figure 7). These data, combined with those in Figures 1 and 6, demonstrate that Aip1 and cofilin contribute to the rapid turnover of both patches and cables. Further, our data show that specific defects in F-actin binding and capping by Aip1 lead to reduced turnover of these actin structures in vivo.
Figure 7.
Defects in actin cable turnover in aip1 mutant cells. (A) Wild-type (AIP1) and mutant (aip1Δ, aip1-107, aip1-107/108, aip1-107/119) yeast cells were treated with 20 μM latrunculin A (Lat-A). Samples of cells were removed at the indicated time points, fixed, and stained with anti-actin antibodies. (B) Cells from A were scored for visible actin cables and graphed. More than 200 cells were counted for each time point, and the columns were the average of two independent experiments. Defects in cable turnover were obvious in aip1 mutant cells at all stages of the cell cycle (unpublished data). These defects were quantified in small- and medium-budded cells, where cables were more prominent and visible than in larger budded cells. Error bars, SD.
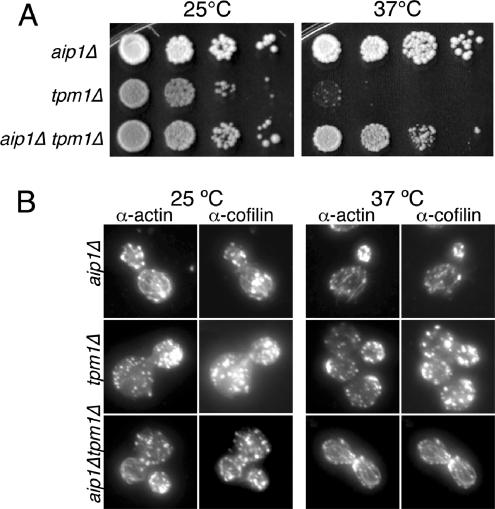
Deletion of AIP1 Rescues Loss of Actin Cables in tpm1 Mutants
The data above indicate that Aip1 and cofilin cooperatively disassemble actin filaments. In contrast, tropomyosin is known to decorate and stabilize cables (Liu and Bretscher, 1989; Pruyne et al., 1998). Thus these two sets of proteins may have an antagonistic relationship in regulating cable stability, with normal cable morphology resulting from their opposing activities. To test this hypothesis, we deleted the AIP1 gene in a tpm1Δ mutant background. tpm1Δ cells lack obvious actin cables and exhibit temperature-sensitive growth at 37°C (Liu and Bretscher, 1989, 1992). Deletion of AIP1 rescued both the temperature-sensitive growth and the actin cable defects of this strain (Figure 8, A and B). Similar results were obtained for aip1Δ tpm1-2 mutants (unpublished data). Further, the cables in aip1Δtpm1Δ cells were decorated with cofilin, which supports the view that cofilin and tropomyosin compete for binding to actin filaments. Also note that cap2Δ and cap1Δ mutations failed to rescue tpm1Δ phenotypes (unpublished data and Adams et al., 1993), suggesting that Aip1 plays a specific cellular role, distinct from capping protein.
Figure 8.
(A) Haploid strains were grown to log phase (OD600 = 0.5) and then cells were serially diluted, plated on YPD medium, and grown for 2 d at 25 or 37°C. (B) Cells were fixed and labeled with rabbit anti-actin and chicken anti-cofilin antibodies after growth at 25°C (left panel) or 37°C (right panel). Note the obvious loss of actin cables in tpm1Δ mutants and their restoration in aip1Δ tpm1Δ mutants.
DISCUSSION
We have taken a coupled genetic and biochemical approach to dissect Aip1 cellular function and made four important findings: 1) We provide the first in vivo demonstration that Aip1 promotes turnover of actin structures. Until now, an actin turnover function for Aip1 has only been inferred from its in vitro activities and genetic interactions with cofilin, but was never demonstrated in living cells. 2) By analyzing in parallel the cellular phenotypes and biochemical defects of aip1 alleles generated by systematic mutagenesis, we show that Aip1’s ability to promote actin disassembly in vitro and in vivo requires direct binding interactions with F-actin. This helps to define the Aip1 mechanism, because Aip1 interacts with both actin and cofilin, and until now it has been unclear whether one or both interactions are required for its function. Further, we mapped actin binding to two well-separated surfaces on Aip1, one in each major structural lobe of the protein. (3) We find a close correlation in aip1 alleles between loss of cofilin-dependent capping activity in vitro and loss of rapid actin turnover in vivo. This provides strong evidence that capping of severed filaments is important for Aip1 physiological function. (4) We provide the first mechanistic insights into how rapid turnover of yeast actin cables is maintained. Cables are comprised of overlapping cross-linked actin filaments that are decorated and stabilized by tropomyosin. However, they are also highly dynamic, undergoing rapid disassembly along their lengths rather than at one end (Yang and Pon, 2002). Until now, the mechanism by which cells maintain rapid turnover of cables has been elusive. We show that aip1 and cof1 mutations dramatically reduce rates of cable turnover (by 5- and 20-fold, respectively). This suggests that Aip1 and cofilin provide “cable destabilizing” activity that opposes the stabilizing activity of Tpm1 to maintain rapid cable turnover in vivo. In support of this model, tpm1Δ cells show a severe loss of actin cables and temperature-sensitive growth, and both defects are rescued by deletion of AIP1.
Mechanism of Aip1-mediated Actin Filament Disassembly
The dissection of Aip1 physiological function has been long awaited. A number of previous studies have defined the in vitro activities of Aip1 on actin (Rodal et al., 1999; Okada et al., 1999, 2002; Balcer et al., 2003; Mohri et al., 2004; Ono et al., 2004), but until now, there has been no mutational analysis to uncouple function and correlate loss of specific in vitro activities with loss of in vivo function. It has been suggested that Aip1, cofilin, and actin form at ternary complex (Rodal et al., 1999), but it has remained unclear which of these molecular interactions (Aip1-cofilin and/or Aip1-actin) contribute to cellular function. Further, there have been two distinct biochemical activities reported for Aip1: barbed end capping and enhanced filament severing. This has led to some confusion as to whether one or both of these activities are relevant in vivo. Two independent studies reported that Aip1 proteins from different species cap the barbed ends of actin filaments in a cofilin-dependent manner (Okada et al., 2002; Balcer et al., 2003). In the first study, capping activity was demonstrated by Aip1, decreasing the rate of filament elongation in the presence of cofilin, and by the preference of Aip1 to label barbed ends of filaments compared with sides of filaments, as analyzed by immunoelectron microscopy (Okada et al., 2002). In the second study, a modified F-actin sedimentation assay was used to demonstrate that Aip1 caps barbed ends of filaments (see Materials and Methods for details). Thus, by three independent in vitro assays, barbed end capping has been shown for Aip1. One group recently has challenged this view, however, based on a failure to detect a strong dependence on Aip1 for capping in a modified version of our sedimentation assay (Clark et al., 2006). Possible technical explanations for this discrepancy are discussed (see Materials and Methods).
Independently, another study used a light microscopy–based assay to suggest that Aip1 enhances cofilin-dependent severing of filaments (Ono et al., 2004). Although this group originally questioned capping by Aip1, shortly thereafter they confirmed that Aip1 caps barbed ends of filaments in vitro (Mohri et al., 2003, 2004). What remains to be determined is whether these two distinct activities, capping and enhanced severing, are related or perhaps even linked. It is possible that Aip1 might first assist cofilin in severing filaments and then cap the newly generated barbed ends to promote disassembly from pointed ends, i.e., an integrated severing/capping activity. In this manner, Aip1 and cofilin may combine their activities to perform a function similar to gelsolin. Indeed, arguments can be made that enhanced severing alone cannot explain Aip1 behavior. For instance, enhanced severing alone, without capping, would result in Aip1 increasing rates of filament elongation in the presence of cofilin. However, Aip1 reduces rates of filament elongation in the presence of cofilin, consistent with capping—possibly following enhanced severing events (Okada et al., 2002). Thus, we favor a unified mechanism, in which perhaps both reported activities operate together. However, we acknowledge that resolving more precisely the mechanism by which Aip1 and cofilin disassemble filaments will require new assays for measuring severing and capping, such as real time imaging of individual filaments by TIRF (total internal reflection fluorescence) microscopy.
Here, we have used mutagenesis to dissect Aip1 physiological function and demonstrated a close correlation between loss of barbed end capping activity in vitro and reduced actin turnover in vivo. Importantly, our analyses do not rule out possible contributions to Aip1 function made by enhanced severing. In fact, the mutants we show are defective in capping could also be defective in enhancing severing, a possibility that awaits future testing. We find that efficient capping by Aip1 requires two F-actin–binding sites, which we mapped to opposite ends of the Aip1 structure (Figure 2, thick dotted circles). The first actin-binding site is located on the N-terminal lobe of Aip1 and is evolutionarily conserved. This surface overlaps with an important functional site mapped by a recent biochemical study on C. elegans Aip1 (Mohri et al., 2004 and see Table 1). Both studies conclude that this general surface is critical for F-actin binding and disassembly in vitro. We extend these observations by demonstrating that this surface is required for Aip1 function in vivo. In another recent study on yeast Aip1, mutational analysis was used to identify residues proximal to this site as important for actin filament binding, although the residues mutated in that allele (aip1-15) were not conserved (Clark et al., 2006). Collectively, these studies show that the front side and edge of the N-terminal lobe of Aip1 are critical for actin binding and disassembly functions (Supplementary Figure S5).
In addition, we identify a second actin-binding site located in the C-terminal lobe of Aip1 and show that it is critical for function in vivo. This second F-actin–binding site is not conserved in other organisms, yet Aip1 homologues from other organisms cap actin filaments (Okada et al., 2002; Mohri et al., 2003). Thus, in other organisms a second actin-binding site may be functionally replaced by either stronger associations of the N-terminal lobe with F-actin and/or increased interactions of Aip1 with cofilin to enhance filament association. Alternatively, other Aip1 proteins may harbor a second actin-binding site in the C-terminal lobe that is not recognizable by sequence conservation. Such a site may have been overlooked for C. elegans Aip1 because only highly conserved residues were targeted for mutagenesis in that study (Mohri et al., 2004). We identified this second actin-binding site because we targeted for mutation the yeast-specific insertion in the C-terminal lobe. This actin-binding surface may extend beyond the insertion, as another study identified additional nonconserved residues in yeast Aip1, outside of the insertion, important for actin binding by the two-hybrid assay (Clark et al., 2006). Determining whether a second actin-binding site exists in Aip1 from other species will require mutational analysis of the nonconserved residues near this location in the C-terminal lobe.
How do the two separate actin-binding sites on Aip1 contribute to function? The binary nature of the Aip1 association with F-actin is a feature common to many capping proteins (e.g., CapZ, formins, and gelsolin). Such an arrangement can provide a mechanism to block two exposed actin subunits at the barbed end of a filament. Interestingly, we found distinct biochemical defects for mutations at the N-terminal actin-binding site (aip1-107) compared with the C-terminal actin-binding site (aip1-119). Aip1-107 was defective in actin filament binding both in the presence and absence of cofilin, whereas Aip1-119 was defective in actin filament binding specifically in the presence of cofilin. Given that Aip1-119 interacts normally with cofilin by the two-hybrid assay, this suggests that Aip1-119 may fail to interact with a site on F-actin that is exposed upon decoration with cofilin. Cofilin decoration alters the twist of actin filaments (McGough et al., 1997), and Aip1 functions to disassembly actin filaments specifically in the presence of cofilin. Thus, the second actin-binding site on Aip1 may play a critical role in disassembling filaments by recognizing a cofilin-induced conformational state of F-actin. Alternatively, cofilin interactions with Aip1 could alter Aip1 conformation to enable the C-terminal surface to interact with its cognate binding site on actin. Regardless of the mechanism, this site plays a critical role in cofilin-dependent capping and disassembly of filaments in vitro and in promoting rapid actin turnover in vivo.
Our analyses did not identify the cofilin-binding surface on Aip1. Because we did not identify any single conserved site that was critical for cofilin interactions by two-hybrid analysis, we speculate that cofilin may interact with a broad surface of Aip1 through multiple weak contacts. Indeed, Clark et al. (2006) propose a similar model and have used two-hybrid analysis and molecular dynamics simulations to map that cofilin interaction to the surface spanning the conserved front face of Aip1, where the two lobes are connected (Figure 2). This model is also consistent with our observation that each half of Aip1 is required for the cofilin interaction.
In Vivo Regulation of Yeast Actin Cable Turnover by Cofilin and Aip1
Our data shed new light on the mechanisms regulating yeast actin cable dynamics and have possibly broader reaching implications for understanding the rules governing turnover of tropomyosin-decorated actin structures in other organisms. Studies in live yeast cells have shown that actin cables grow from the bud tip and neck regions and stream rapidly toward the mother cell. On treatment of cells with Lat-A, cables disappear in seconds, suggesting they undergo very rapid turnover (Yang and Pon, 2002). Rapid disassembly suggests an active mechanism for destabilizing cables because the filaments in cables are decorated by stabilizing proteins, including the tropomyosins Tpm1 and Tpm2 and Sac6/fimbrin, but until now the mechanism has remained elusive.
Here, we found that mutations in cofilin and Aip1 reduce rates of cable disassembly by 20- and 5-fold, respectively. This result was unanticipated, because Aip1 and Cof1 are not readily detected on actin cables in wild-type yeast cells. However, our proposed mechanism (see below) offers a possible explanation for observation. On the basis of these functional data, we propose that cofilin and Aip1 “prune” cables along their lengths by a coordinated mechanism of severing and capping filaments (Figure 9). This model is consistent with observations made in cells fixed at different time points after Lat-A treatment to characterize cable disassembly, in which it was concluded that cables are comprised of short, overlapping filaments arranged in bundles that disassemble along their entire lengths (Karpova et al., 1998). It is also consistent with studies in live cells, which show (by following fiduciary marks) that cables do not disassemble at one end, but instead disassemble more uniformly (Yang and Pon, 2002).
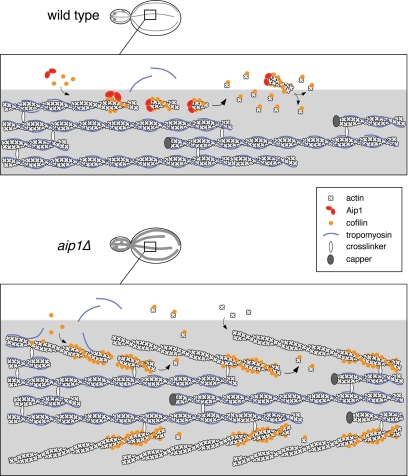
Figure 9.
A model for cofilin and Aip1 cellular function: coordinated pruning of tropomyosin-decorated actin cables. In this model, cofilin binds cooperatively to actin filaments in cables, helping to displace tropomyosin from a subset of filaments. The fate of cofilin-severed filaments differs greatly depending on whether Aip1 is available. In wild-type cells, Aip1 may assist cofilin in severing filaments and then rapidly caps the new barbed ends of filaments generated by severing. This leads to rapid net disassembly of those short filaments from their pointed ends. Thus cofilin-decorated filaments in actin cables are extremely short-lived and thus not easily detected in wild-type cells. However, in aip1Δ cells, cofilin severing generates uncapped barbed ends of filaments, which either reanneal or undergo rapid growth, leading to cable thickening. Further, in aip1Δ cells, cofilin-decorated of these cables can be detected by immunostaining because the filaments do not turnover as rapidly as in wild-type cells.
How do cofilin and Aip1 prune cables? Cofilin binds cooperatively to F-actin and competes for binding with tropomyosin in vitro (Bamburg, 1999). Therefore, cooperative binding of cofilin may displace tropomyosin rapidly from the sides of individual filaments in cables. Cofilin decoration also recruits Aip1 to filament sides, and Aip1 may enhance cofilin-dependent severing of filaments (Ono et al., 2004). After severing events, Aip1 caps the barbed ends of filaments, leading to the rapid disassembly of those filaments from their pointed ends (Figure 9, top row). The rapid nature of this process would explain why cofilin and Aip1 are not detected on cables by immunostaining in wild-type cells. Cables are faint to begin with, and if only a very small percentage of filaments are decorated with cofilin at any given time, it would be difficult to detect. On the other hand, in aip1Δ cells, cofilin severing generates uncapped barbed ends, which leads to filament growth and cable thickening (Figure 9, lower panel). Those cofilin-decorated filaments are slower to disassemble, which allows detection of cofilin on cables. Our model also explains why thickened cables decorated with cofilin are observed for cof1-19, a mutant that disrupts Aip1 function, and not cof1-22, a mutant defective in filament severing. Thus, cable thickening and stabilized cofilin decoration result specifically from loss of Aip1 capping, rather than from decreased severing/depolymerization activity and reduced rates of cable turnover. This cofilin-dependent severing/capping activity of Aip1 may be optimal for cable pruning and is distinct from the cellular function of conventional capping protein (Cap1/Cap2). cap2Δ cells do not exhibit thickened cables and in contrast to aip1Δ cells show diminished cable staining (Amatruda et al., 1990). Further, cap1Δ and cap2Δ mutations fail to restore actin cables in the tpm1Δ background (Adams et al., 1993 and our unpublished observations). Thus, Aip1 may have a specialized role in capping severed filaments, facilitated by its direct interactions with cofilin and/or a cofilin-induced conformation of F-actin.
Concluding Remarks
Given that the atomic structures and biochemical activities of cofilin and Aip1 are conserved across distant species (Ono, 2003), it seems likely that their combined mechanism of severing and capping to disassemble actin filaments demonstrated here in yeast extends to other organisms. Further, tropomyosin-decorated actin arrays may be a common target of their activity. In C. elegans, Aip1 and cofilin are required to maintain myofibrillar structure in body wall muscle, which is rich in tropomyosin-decorated filaments (Ono, 2001). In organisms ranging from fission yeast to mammals, cofilin is recruited during cytokinesis to the tropomyosin-decorated contractile ring (Abe et al., 1996; Nakano and Mabuchi, 2006). In addition, Aip1 and cofilin may prune the branched actin networks found in lammelipodia and trailing endocytic vesicles and pathogens. Aip1 is required for lammelapodia formation and cell motility (Konzok et al., 1999; Rogers et al., 2003). Thus, although Aip1 has not been specified as one of the core proteins required for formation of the motile actin comet tail behind Listeria (Loisel et al., 1999), it may be required for rapid turnover of the tail and sustained motility.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to A. Rodal, S. Almo, D. Amberg, S. Ono, and D. Sept for useful discussions and sharing data before publication and to members of the Goode laboratory for technical assistance. We also thank K. Daugherty, M. Gandhi, A. Goodman, C. Gould, J. Moseley, D. Nowakowski, and A. Rodal for critical reading of the manuscript. This research was supported by grants to B.G. from the American Cancer Society and National Institutes of Health (GM63691).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-02-0135) on April 12, 2006.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Abe H., Obinata T., Minamide L. S., Bamburg J. R. Xenopus laevis actin-depolymerizing factor/cofilin: a phosphorylation-regulated protein essential for development. J. Cell Biol. 1996;132:871–885. doi: 10.1083/jcb.132.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams A.E.M., Cooper J. A., Drubin D. G. Unexpected combinations of null mutations in genes encoding the actin cytoskeleton are lethal in yeast. Mol. Biol. Cell. 1993;4:459–468. doi: 10.1091/mbc.4.5.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizawa H., Katadae M., Maruya M., Sameshima M., Murakami-Murofushi K., Yahara I. Hyperosmotic stress-induced reorganization of actin bundles in Dictyostelium cells over-expressing cofilin. Genes Cells. 1999;4:311–324. doi: 10.1046/j.1365-2443.1999.00262.x. [DOI] [PubMed] [Google Scholar]
- Amatruda J. F., Cannon J. F., Tatchell K., Hug C., Cooper J. A. Disruption of the actin cytoskeleton in yeast capping protein mutants. Nature. 1990;344:352–354. doi: 10.1038/344352a0. [DOI] [PubMed] [Google Scholar]
- Amberg D. C., Basart E., Botstein D. Defining protein interactions with yeast actin in vivo. Nat. Struct. Biol. 1995;2:28–35. doi: 10.1038/nsb0195-28. [DOI] [PubMed] [Google Scholar]
- Asakura T. Isolation and characterization of a novel actin filament-binding protein from Saccharomyces cerevisiae. Oncogene. 1998;16:121–130. doi: 10.1038/sj.onc.1201487. [DOI] [PubMed] [Google Scholar]
- Ayscough K. R., Stryker J., Pokala N., Sanders M., Crews P., Drubin D. G. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J. Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balcer H. I., Goodman A., Rodal A. A., Smith E. M., Kugler J., Heuser J. H., Goode B. L. Coordinated regulation of actin filament turnover by a high molecular weight Srv2/CAP complex, cofilin, profilin, and Aip1. Curr. Biol. 2003;13:2159–2169. doi: 10.1016/j.cub.2003.11.051. [DOI] [PubMed] [Google Scholar]
- Bamburg J. R. Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu. Rev. Cell Dev. Biol. 1999;15:185–230. doi: 10.1146/annurev.cellbio.15.1.185. [DOI] [PubMed] [Google Scholar]
- Carlier M. F., Laurent V., Santolini J., Melki R., Didry D., Xia G. X., Hong Y., Chua N. H., Pantaloni D. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J. Cell Biol. 1997;136:1307–1322. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A. Y., Bailly M., Zebda N., Segall J. E., Condeelis J. S. Role of cofilin in epidermal growth factor-stimulated actin polymerization and lamellipod protrusion. J. Cell Biol. 2000;148:531–542. doi: 10.1083/jcb.148.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M. G., Teply J., Haarer B. K., Viggiano S. C., Sept D., Amberg D. C. A genetic dissection of Aip1p’s interactions leads to a model for Aip1p–cofilin cooperative activities. Mol. Biol. Cell. 2006;17:1971–1984. doi: 10.1091/mbc.E05-10-0956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesMarais V., Ghosh M., Eddy R., Condeelis J. S. Cofilin takes the lead. J. Cell Sci. 2005;118:19–26. doi: 10.1242/jcs.01631. [DOI] [PubMed] [Google Scholar]
- Didry D., Carlier M. F., Pantaloni D. Synergy between actin depolymerizing factor/cofilin and profilin in increasing actin filament turnover. J. Biol. Chem. 1998;273:25602–25611. doi: 10.1074/jbc.273.40.25602. [DOI] [PubMed] [Google Scholar]
- Drubin D. G., Miller K. G., Botstein D. Yeast actin-binding proteins: evidence for a role in morphogenesis. J. Cell Biol. 1988;107:2551–2561. doi: 10.1083/jcb.107.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eads J. C., Mahoney N. M., Vorobiev S., Bresnick A. R., Wen K. K., Rubenstein P. A., Haarer B. K., Almo S. C. Structure determination and characterization of Saccharomyces cerevisiae profilin. Biochemistry. 1998;37:11171–11181. doi: 10.1021/bi9720033. [DOI] [PubMed] [Google Scholar]
- Flick J. S., Johnston M. Two systems of glucose repression of the GAL1 promoter in Saccharomyces cerevisiae. Mol. Cell. Biol. 1990;10:4757–4769. doi: 10.1128/mcb.10.9.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode B. L. Purification of yeast actin and actin-associated proteins. Methods Enzymol. 2002;351:433–441. doi: 10.1016/s0076-6879(02)51862-0. [DOI] [PubMed] [Google Scholar]
- Hotulainen P., Paunola E., Vartiainen M. K., Lappalainen P. Actin-depolymerizing factor and cofilin-1 play overlapping roles in promoting rapid F-actin depolymerization in mammalian nonmuscle cells. Mol. Biol. Cell. 2005;16:649–664. doi: 10.1091/mbc.E04-07-0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida K., Moriyama K., Matsumoto S., Kawasaki H., Nishida E., Yahara I. Isolation of a yeast essential gene, COF1, that encodes a homologue of mammalian cofilin, a low-Mr actin-binding and depolymerizing protein. Gene. 1993;124:115–120. doi: 10.1016/0378-1119(93)90770-4. [DOI] [PubMed] [Google Scholar]
- Kaksonen M., Sun Y., Drubin D. G. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–487. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- Kaksonen M., Toret C. P., Drubin D. G. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell. 2005;123:305–320. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- Karpova T. S., McNally J. G., Moltz S. L., Cooper J. A. Assembly and function of the actin cytoskeleton of yeast: relationships between cables and patches. J. Cell Biol. 1998;142:1501–1517. doi: 10.1083/jcb.142.6.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konzok A., Weber I., Simmeth E., Hacker U., Maniak M., Muller-Taubenberger A. DAip1, a Dictyostelium homologue of the yeast actin-interacting protein 1, is involved in endocytosis, cytokinesis, and motility. J. Cell Biol. 1999;146:453–464. doi: 10.1083/jcb.146.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen P., Drubin D. G. Cofilin promotes rapid actin filament turnover in vivo. Nature. 1997;388:78–82. doi: 10.1038/40418. [DOI] [PubMed] [Google Scholar]
- Lappalainen P., Fedorof E. V., Almo S. C., Drubin D. G. Essential functions and actin-binding surfaces of yeast cofilin revealed by systematic mutagenesis. EMBO J. 1997;16:5520–5530. doi: 10.1093/emboj/16.18.5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. P., Bretscher A. Disruption of the single tropomyosin gene in yeast results in the disappearance of actin cables from the cytoskeleton. Cell. 1989;57:233–242. doi: 10.1016/0092-8674(89)90961-6. [DOI] [PubMed] [Google Scholar]
- Liu H. P., Bretscher A. Characterization of TPM1 disrupted yeast cells indicates an involvement of tropomyosin in directed vesicular transport. J. Cell Biol. 1992;118:285–299. doi: 10.1083/jcb.118.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loisel T. P., Boujemaa R., Pantaloni D., Carlier M. F. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 1999;401:613–616. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- Maciver S. K., Pope B. J., Whytock S., Weeds A. J. The effect of two actin depolymerizing factors (ADF/cofilins) on actin filament turnover : pH sensitivity of F-actin binding by human ADF, but not of Acanthamoeba actophorin. Eur. J. Biochem. 1998;256:388–397. doi: 10.1046/j.1432-1327.1998.2560388.x. [DOI] [PubMed] [Google Scholar]
- McGough A., Pope B., Chiu W., Weeds A. Cofilin changes the twist of F-actin: implications for actin filament dynamics and cellular function. J. Cell Biol. 1997;138:771–781. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell D. A., Marshall T. K., Deschenes R. J. Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast. 1993;9:715–722. doi: 10.1002/yea.320090705. [DOI] [PubMed] [Google Scholar]
- Mohri K., Ono S. Actin filament disassembling activity of Caenorhabditis elegans actin-interacting protein 1 (UNC-78) is dependent on filament binding by a specific ADF/cofilin isoform. J. Cell Sci. 2003;116:4107–4118. doi: 10.1242/jcs.00717. [DOI] [PubMed] [Google Scholar]
- Mohri K., Vorobiev S., Fedorov A. A., Almo S. C., Ono S. Identification of functional residues on Caenorhabditis elegans actin-interacting protein 1 (UNC-78) for disassembly of actin depolymerizing factor/cofilin-bound actin filaments. J. Biol. Chem. 2004;279:31697–31707. doi: 10.1074/jbc.M403351200. [DOI] [PubMed] [Google Scholar]
- Moon A. L., Janmey P. A., Louie K. A., Drubin D. G. Cofilin is an essential component of the yeast cortical cytoskeleton. J. Cell Biol. 1993;120:421–435. doi: 10.1083/jcb.120.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K., Mabuchi I. Actin-depolymerizing protein Adf1 is required for formation and maintenance of the contractile ring during cytokinesis in fission yeast. Mol. Biol. Cell. 2006;17:1933–1945. doi: 10.1091/mbc.E05-09-0900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojala P. J., Paavilainen V., Lappalainen P. Identification of yeast cofilin residues specific for actin monomer and PIP2 binding. Biochemistry. 2001;40:15562–15569. doi: 10.1021/bi0117697. [DOI] [PubMed] [Google Scholar]
- Okada K., Obinata T., Abe H. XAIP 1, a Xenopus homologue of yeast actin interacting protein 1 (AIP1), which induces disassembly of actin filaments cooperatively with ADF/cofilin family proteins. J. Cell Sci. 1999;112:1553–1565. doi: 10.1242/jcs.112.10.1553. [DOI] [PubMed] [Google Scholar]
- Okada K., Blanchoin L., Abe H., Chen H., Pollard T. D., Bamburg J. R. Xenopus actin interacting protein 1 (XAip1) enhances cofilin fragmentation of filaments by capping filament ends. J. Biol. Chem. 2002;277:43011–43016. doi: 10.1074/jbc.M203111200. [DOI] [PubMed] [Google Scholar]
- Ono S. The Caenorhabditis elegans unc-78 gene encodes a homologue of actin-interacting protein 1 required for organized assembly of muscle actin filaments. J. Cell Biol. 2001;152:1313–1319. doi: 10.1083/jcb.152.6.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S. Regulation of actin filament dynamics by actin depolymerizing factor/cofilin and actin-interacting protein 1, new blades for twisted filaments. Biochemistry. 2003;42:13363–13370. doi: 10.1021/bi034600x. [DOI] [PubMed] [Google Scholar]
- Ono S., Mohri K., Ono K. Microscopic evidence that actin-interacting protein 1 actively disassembles actin-depolymerizing factor/Cofilin-bound actin filaments. J. Biol. Chem. 2004;279:14207–14212. doi: 10.1074/jbc.M313418200. [DOI] [PubMed] [Google Scholar]
- Pollard T. D. Polymerization of ADP-actin. J. Cell Biol. 1984;99:769–777. doi: 10.1083/jcb.99.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle J. R., Adams A. E., Drubin D. G., Haarer B. K. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- Pruyne D. W., Schott D. H., Bretscher A. Tropomyosin-containing actin cables direct the Myo2p-dependent polarized delivery of secretory vesicles in budding yeast. J. Cell Biol. 1998;143:1931–1945. doi: 10.1083/jcb.143.7.1931. [DOI] [PubMed] [Google Scholar]
- Pruyne D., Bretscher A. Polarization of cell growth in yeast. J. Cell Sci. 2000;113:571–585. doi: 10.1242/jcs.113.4.571. [DOI] [PubMed] [Google Scholar]
- Pruyne D., Legesse-Miller A., Gao L., Dong Y., Bretscher A. Mechanisms of polarized growth and organelle segregation in yeast. Annu. Rev. Cell Dev. Biol. 2004;20:559–591. doi: 10.1146/annurev.cellbio.20.010403.103108. [DOI] [PubMed] [Google Scholar]
- Rodal A. A., Tetrault J., Lappalainen P., Drubin D. G., Amberg D. C. Aip1p interacts with cofilin to stimulate disassembly of actin filaments. J. Cell Biol. 1999;146:1251–1264. doi: 10.1083/jcb.145.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S. L., Wiedemann U., Stuurman N., Vale R. D. Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J. Cell Biol. 2003;162:1079–1088. doi: 10.1083/jcb.200303023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich J. A., Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the protelytic fragment of myosin. J. Biol. Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- Vinson V. K., De La Cruz E. M., Higgs H. N., Pollard T. D. Interactions of Acanthamoeba profilin with actin and nucleotides bound to actin. Biochemistry. 1998;37:10871–10880. doi: 10.1021/bi980093l. [DOI] [PubMed] [Google Scholar]
- Voegtli W. C., Madrona A. Y., Wilson D. K. The structure of Aip1p, a WD repeat protein that regulates cofilin-mediated actin depolymerization. J. Biol. Chem. 2003;278:34373–34379. doi: 10.1074/jbc.M302773200. [DOI] [PubMed] [Google Scholar]
- Yang H. C., Pon L. A. Actin cable dynamics in budding yeast. Proc. Natl. Acad. Sci. USA. 2002;99:751–756. doi: 10.1073/pnas.022462899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond S. H. Recent quantitative studies of actin filament turnover during cell locomotion. Cell Motil. Cytoskelet. 1993;25:309–316. doi: 10.1002/cm.970250402. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.