Abstract
Caveolin is the major protein component required for the formation of caveolae on the plasma membrane. Here we show that trafficking of Caenorhabditis elegans caveolin-1 (CAV-1) is dynamically regulated during development of the germ line and embryo. In oocytes a CAV-1-green fluorescent protein (GFP) fusion protein is found on the plasma membrane and in large vesicles (CAV-1 bodies). After ovulation and fertilization the CAV-1 bodies fuse with the plasma membrane in a manner reminiscent of cortical granule exocytosis as described in other species. Fusion of CAV-1 bodies with the plasma membrane appears to be regulated by the advancing cell cycle, and not fertilization per se, because fusion can proceed in spe-9 fertilization mutants but is blocked by RNA interference–mediated knockdown of an anaphase-promoting complex component (EMB-27). After exocytosis, most CAV-1-GFP is rapidly endocytosed and degraded within one cell cycle. CAV-1 bodies in oocytes appear to be produced by the Golgi apparatus in an ARF-1–dependent, clathrin-independent, mechanism. Conversely endocytosis and degradation of CAV-1-GFP in embryos requires clathrin, dynamin, and RAB-5. Our results demonstrate that the distribution of CAV-1 is highly dynamic during development and provides new insights into the sorting mechanisms that regulate CAV-1 localization.
INTRODUCTION
Caveolae are lipid-raft-enriched, flask-shaped invaginations present on the plasma membrane of many eukaryotic cell types (Anderson, 1998). Caveolae are enriched in glycosphingolipids and cholesterol and are thought to play a role in signaling, cholesterol homeostasis, clathrin-independent endocytosis, transcytosis, and potocytosis (Matveev et al., 2001; Parton and Richards, 2003). Caveolae are also implicated in internalization of some bacterial toxins, viruses, and bacteria into cells (Parton and Richards, 2003).
The major structural components of caveolae are caveolins, which are integral membrane proteins with carboxyl and amino termini located in the cytosol and a hydrophobic loop inserted into the membrane (Glenney and Soppet, 1992; Rothberg et al., 1992). Caveolins bind to cholesterol (Murata et al., 1995) and oligomerize to form filamentous structures that are thought to stabilize the membrane and to define the size and shape of caveolae (Fernandez et al., 2002). In mammalian cells, caveolin-1 and -2 are ubiquitously expressed, whereas caveolin-3 is specifically expressed in muscle tissues (Parton, 1996). Expression of caveolin-1 is sufficient to generate caveolae in cells previously lacking caveolae (Lipardi et al., 1998), and caveolin knockout mice show a remarkable loss of caveolae (Drab et al., 2001), indicating an essential role of caveolins in caveolar biogenesis. Caveolin knockout mice also show defects in diverse physiological processes (Le Lay and Kurzchalia, 2005). Previous studies in C. elegans found that CAV-1 is strongly expressed in the germ line and suggested that CAV-1 plays a role in meiotic progression as a negative regulator of Ras signaling (Scheel et al., 1999). Studies of caveolin in mammalian systems reached similar conclusions regarding its role in Ras signaling (Roy et al., 1999).
Caveolin-GFP fusions expressed in cultured cell lines have been extensively used to study caveolin dynamics in vivo (Pelkmans et al., 2001, 2002, 2004, 2005; Tagawa et al., 2005). Some groups studying caveolae in cultured cell lines have reported that caveolae are highly immobile structures that do not show a high turnover rate at the plasma membrane (Thomsen et al., 2002), whereas other groups indicate that a subset of surface caveolae are highly mobile under standard culture conditions (Pelkmans and Zerial, 2005). However, all groups agree that various stimuli can lead to greatly increased internalization of caveolae. Antibody cross-linking of MHC class I molecules or glycosyl phosphatidyl inositol–anchored proteins results in clustering of these molecules in caveolae and subsequent internalization of caveolae (Huet et al., 1980; Parton et al., 1994). Simian virus 40 (SV40) infection also induces a massive internalization of caveolae containing SV40 (Pelkmans et al., 2001). Recently, it has been reported that cell detachment from the extracellular matrix triggers internalization of lipid rafts and caveolin-1 from the plasma membrane to an intracellular compartment to down-regulate various signal transduction pathways (del Pozo et al., 2005). Furthermore, caveolins are redistributed from the plasma membrane to lipid bodies in early stages of liver regeneration after partial hepatectomy (Pol et al., 2004). These observations strongly suggest that the function and localization of caveolins are highly regulated in the cell. However, relatively little is known of caveolin trafficking in intact animal systems, and a genetically tractable system for the analysis of the molecular machinery that regulates the dynamic behavior of caveolins has been lacking.
To analyze caveolin dynamics, we have imaged CAV-1-GFP in living animals under a variety of experimental conditions. Here, we report that trafficking of C. elegans caveolin-1 is dynamically regulated during the development of the worm germ line and embryo. In the syncytial gonad CAV-1-GFP localizes to the plasma membrane and to punctate Golgi ministacks in the cytoplasm. As oocytes form, a significant fraction of the CAV-1-GFP in the cell begins to appear in large ring-like membrane compartments (CAV-1 bodies) in the cytoplasm that remain closely apposed to the Golgi. The formation of the CAV-1 body is clathrin-independent but ARF-1–dependent. Just after ovulation and fertilization, the large ring-like organelles positive for CAV-1-GFP apparently fuse with the plasma membrane. Shortly thereafter most of the CAV-1-GFP is internalized via clathrin-mediated endocytosis and is rapidly degraded. Our results demonstrate that the distribution of CAV-1 is highly dynamic during development, with at least two distinct mechanisms regulating CAV-1 localization at different developmental stages. Our results also suggest new potential functions for caveolin in development.
MATERIALS AND METHODS
General Methods and Strains
Methods for the handling and culturing of C. elegans were essentially those described by Brenner (1974). All strains were grown at 20°C unless otherwise stated. The wild-type parent for all strains was C. elegans var Bristol strain N2. Mutations used were LGIII, unc-119 (ed3) (Maduro and Pilgrim, 1995); LGIV, rme-2 (b1008) (Grant and Hirsh, 1999); LGI, spe-9 (hc52) (Singson et al., 1998); LGV, fog-2(q71) (Schedl and Kimble, 1988); and unmapped pwIs61 [GFP::cav-1, unc-119(+)] and pwIs281 [CAV-1-GFP, unc-119(+)] (this work). arf-1(ok796) was obtained from the C. elegans Gene Knockout Consortium.
Plasmids and Transgenic C. elegans
A genomic fragment containing the ORF in cav-1 was amplified by PCR and cloned into the Entry vector pDONR221 by Gateway recombinational cloning technology. The ORF of cav-1 was then cloned into pID3.01 (Pellettieri et al., 2003) to create an amino-terminal GFP fusion. pID3.01 utilizes pie-1 5′ and 3′ UTR sequences to drive expression of the transgene in the maternal germ line. To create a carboxy-terminal GFP fusion with CAV-1, a genomic fragment containing the ORF in cav-1 was amplified by PCR with cav-1Gw+ and cav-1Bgl2R [5′-GGGGACAACTTTGTACAAGAAAGTTGTTAagatctGACGCATGGAGCAGTAGTTTC-3′] and cloned into the Entry vector pDONR221, resulting pDONR221cav-1BglII. A DNA fragment encoding GFP was amplified using pID3.01 as a template and inserted into a BglII site of pDONR221cav-1BglII. The ORF of CAV-1-GFP was then cloned into pID2.01 (Pellettieri et al., 2003). GFP fusion expressing transgenic lines were created by the microparticle bombardment method as described previously (Praitis et al., 2001).
For RNA interference (RNAi) experiments, we cloned genomic DNA corresponding to F54C9.10, rab-27, rab-30, rab-33, rab-38, rabY4, rabY6, and chc-1 by PCR using purified N2 genomic DNA and inserting them into RNAi vector L4440. cDNAs corresponding to Y57G11C.13, Y116A8C.12, rab-3, rab-5, rab-7, rab-8, rab-11.1, rab-14, rab-18, rab-19, and rab-35 were prepared from EST clones provided by Yuji Kohara (National Institute of Genetics, Japan) and subcloned into L4440. All other feeding RNAi constructs were obtained from the Ahringer genomic RNAi library (Kamath and Ahringer, 2003).
Antibody Production
To generate antibodies directed against SQV-8, the nucleotides encoding amino acids 150–349 of SQV-8 were amplified from a cDNA library and inserted into pGEX6P-1 (GE Healthcare, Piscataway, NJ). The purified GST fusion protein was outsourced for injection into rabbits (Sigma Genosys), and affinity purification of anti-SQV-8 antibodies was performed as described previously, after removal of the GST by cleavage of the antigen with Precision protease (Precision Systems, Natick, MA; Audhya et al., 2005).
RNA-mediated Interference
RNAi experiments in this study were performed using the feeding method (Timmons et al., 2001). L4 larvae were placed on plates containing NGM agar with 5 mM IPTG and HT115 (DE3) bacteria carrying double-stranded RNA expression constructs and allowed to grow for 48 h at 20°C. P0 animals were transferred to a new plate and allowed to lay eggs for 1–2 h. Then, P0 animals were removed from the plate and observed by fluorescence microscopy. F1 progeny were further incubated for 4 d and observed by fluorescence microscopy.
Microscopy and Immunostaining
To observe live worms expressing transgenes, worms were mounted on agarose pads with 10 mM levamisole in M9 buffer. Fluorescence images were obtained using an Axiovert 200M (Carl Zeiss MicroImaging, Oberkochen, Germany) microscope equipped with a digital CCD camera (C4742-95-12ER, Hamamatsu Photonics, Hamamatsu, Japan) and deconvolved with AutoDeblur software (AutoQuant Imaging, Watervliet, NY). Confocal images were obtained using a Bio-Rad MRC-1024 confocal microscope system (Bio-Rad, Tokyo, Japan) and an Olympus confocal microscope system FV1000 (Olympus, Tokyo, Japan). Movies of the gonad were made using a spinning disk confocal microscope (Nikon Eclipse TE2000-E, Tokyo, Japan) equipped with a Hamamatsu Orca-ER CCD camera at 20°C using a Nikon 60×, 1.4 NA Planapo oil objective lens.
Immunostaining of dissected gonads was performed as described previously (Grant and Hirsh, 1999; Sato et al., 2005). Images of fixed worms stained with anti-SQV-8 antibody were acquired on a DeltaVision deconvolution Olympus IX70 microscope (Applied Precision, Issaquah, WA) equipped with a CoolSnap CCD camera (Roper Scientific, Tucson, AZ) at 20°C using a 100×, 1.35 NA Olympus U-Planapo oil objective lens.
RESULTS
CAV-1-GFP Displays Highly Dynamic Behavior during Development
To visualize caveolar trafficking in the context of a living animal, we expressed caveolin-GFP fusion proteins in transgenic nematodes. C. elegans has two caveolin genes, encoding the CAV-1 and CAV-2 proteins (Tang et al., 1997). Endogenous CAV-1 is known to be strongly expressed in the germ line of adult hermaphrodites and is thought to influence Ras/MAP-kinase–dependent progression through the pachytene-stage of the meiotic cell cycle (Scheel et al., 1999). To examine the dynamics of meiotic CAV-1 protein, we expressed CAV-1 with an amino- or carboxy-terminal GFP tag in the C. elegans germline using the germ line-specific pie-1 promoter (Pellettieri et al., 2003). We used both CAV-1-GFP fusions for the experiments presented here, but show the results for only one CAV-1-GFP reporter, because both fusions behaved identically in all cases.
The germ cells of adult hermaphrodites are contained within a U-shaped tubular gonad (McCarter et al., 1999; see Figure 7G). Germ cells in the most distal region of the gonad arm are in mitosis and enter meiosis as they move away from the distal tip. Oocytes first appear near the “bend” region of the gonad arm and are arrested in diakinesis of meiotic prophase I. The oocytes increase dramatically in size as they move from the bend proximally toward the spermatheca, but do not reenter meiosis until they receive signals from the overlying sheath and adjacent sperm cells to mature and ovulate. On receiving these signals, the first signs of maturation are nuclear envelope breakdown and cortical rearrangement. A mature oocyte in the proximal arm will then ovulate, entering the spermatheca for fertilization. After fertilization, embryos complete meiosis I and meiosis II and start zygotic development.
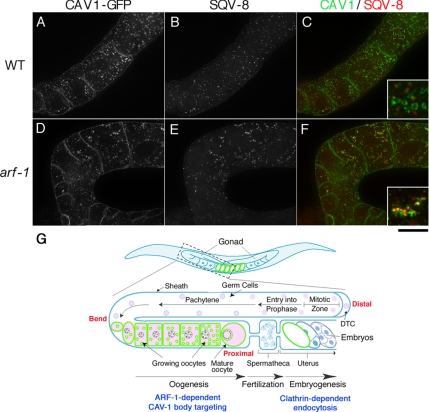
Figure 7.
ARF-1 is required for export of CAV-1-GFP from the Golgi. CAV-1-GFP–expressing wild-type (A–C) and arf-1(ok796) (D–F) strains were stained with an antibody against a Golgi membrane protein SQV-8. CAV-1-GFP failed to colocalize with SQV-8 in the wild-type strain (C, inset). Deletion of arf-1 caused a significant alteration in Golgi morphology and a loss of CAV-1 bodies (D–F). Colocalization of CAV-1-GFP with SQV-8 was often observed in arf-1 mutants (F, inset). Scale bar, 20 μm. The insets (bottom right) are 3× enlargements of the indicated regions in the merged panels. (G) Stylized drawing of one gonad arm connected to the spermatheca and the uterus. Schematic localization of CAV-1-GFP is also indicated with green. In growing oocytes, CAV-1-GFP is transported to the CAV-1 bodies in an ARF-1–dependent manner. After fertilization, CAV-1-GFP is internalized via clathrin-dependent endocytosis.
CAV-1-GFP expressed in the germ line displayed highly dynamic behavior during oocyte formation, ovulation, fertilization, and the first embryonic cell cycle. In mitotic and early meiotic cells of the distal germline CAV-1-GFP is mainly localized to the plasma membrane. As oocytes form in the bend region CAV-1-GFP additionally accumulates in small vesicles, and in larger oocytes appears in large ring-like structures deeper in the cytoplasm (Figure 1, A, C, and G).
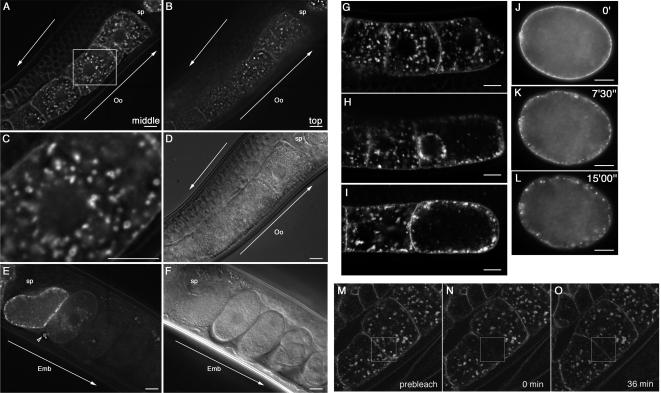
Figure 1.
Dynamic behavior of CAV-1-GFP during development. (A–D) Subcellular localization of CAV-1-GFP during oogenesis. CAV-1-GFP–labeled ring-like structures (CAV-1 bodies) and puncta. CAV-1-GFP puncta first appear in early oocytes in the bend region of gonad, near the plasma membrane and deeper in the cytoplasm (A and B). (C) An enlargement of the square indicated in A. (D) A Nomarski image of the oocytes. (E) Degradation of CAV-1-GFP after fertilization. After fertilization, CAV-1-GFP accumulates on the plasma membrane and then largely disappears before the first cell division of the embryo (E). CAV-1-GFP partitioned to the polar body of embryos remains highly fluorescent and does not disappear (arrowhead). (F) A Nomarski image of the embryos. (G–I) CAV-1-GFP changes localization dynamically, accumulating in the cortex and clustering around the nucleus (H). Perinuclear CAV-1-GFP is lost during nuclear breakdown (I). (J–L) Internalization of GFP-CAV-1 in the embryo. Plasma membrane localized GFP-CAV-1 (J, 0′) is then internalized (K and L). Oo, oocyte; emb, embryo; sp, spematheca. Arrows indicate the direction of maturation of oocytes and embryos. Scale bars, 10 μm. (M–O) CAV-1-GFP on the plasma membrane and in CAV-1 bodies was photobleached, and the recovery of fluorescence was followed as a function of time. From left to right, before (Prebleach), immediately after (0 min), and 36 min after bleaching are shown. The squares show the photobleached areas. Note that little fluorescence recovery occurred in CAV-1 bodies. Partial recovery occurred on the plasma membrane.
To determine if these CAV-1–positive vesicular structures define a unique compartment as is the case for mammalian caveosomes, we compared CAV-1-GFP localization with several membraneous organellar markers in oocytes (see Supplementary Material, Supplementary Figure S1). First we compared CAV-1-GFP with RME-2, the C. elegans yolk receptor (Grant and Hirsh, 1999). RME-2 is first expressed in very early oocytes in the bend region of the gonad where it appears primarily in the ER and Golgi. Soon thereafter it reaches the plasma membrane, and in large nearly full-grown oocytes is primarily found cycling through early endosomes and recycling endosomes (Grant and Hirsh, 1999; Grant et al., 2001; Sato et al., 2005). CAV-1-GFP partially colocalizes with RME-2 in early stage oocytes, probably in the ER and Golgi, but in larger oocytes did not overlap significantly with RME-2 in the endosomal system (Supplementary Figure S1, A and B). In addition CAV-1-GFP did not exhibit colocalization with a marker for early endosomes, EEA-1 (Supplementary Figure S1, C and D), or a marker for recycling endosomes, RME-1 (Supplementary Figure S1, E and F), at any stage of oogenesis. CAV-1-GFP–labeled organelles did not accumulate Lysotracker-Red, a marker for acidified organelles such as late endosomes and lysosomes, nor did they label with Nile Red, a marker for neutral lipid bodies (Supplementary Figure S1G). Thus at least in their unique labeling the CAV-1-GFP–labeled ring-like structures in large nearly full-grown oocytes are reminiscent of caveosomes (Pelkmans et al., 2001). Mammalian caveosomes are defined as caveolin-1–rich, intracellular organelles with a near neutral pH (Pelkmans et al., 2001). We refer to these CAV-1-GFP–labeled ring-like structures in C. elegans oocytes as CAV-1 bodies. The large CAV-1 bodies were relatively immobile, whereas the smaller CAV-1-GFP–labeled vesicles in the cytoplasm moved actively (Supplementary Video 1). CAV-1-GFP was also found in small, scattered spots on the plasma membrane (Figure 1B) that presumably represent individual caveolae or small caveolar vesicles beneath the plasma membrane.
Given the low mobility of large CAV-1 bodies, we sought to determine if these structures are still actively exchanging molecules with the rest of the cell or are quiescent. As a measure of molecular exchange we subjected the CAV-1-GFP–labeled CAV-1 bodies to fluorescence recovery after photobleaching (FRAP) analysis (Figure 1, M–O). Interestingly, fluorescence recovery in individual CAV-1 bodies was very slow, indicating that CAV-1-GFP associated with CAV-1 bodies in large, nearly full-grown oocytes has very little exchange with external pools. Significant recovery of fluorescence was noted on the plasma membrane, probably because of the lateral diffusion of CAV-1- GFP on the plasma membrane or influx of newly synthesized CAV-1-GFP to the plasma membrane.
CAV-1-GFP distribution changed rapidly during ovulation. The first change we observed in an oocyte about to ovulate was the clustering of all punctate CAV-1-GFP at the cortex and around the nucleus, clearing the cytoplasm in between (Figure 1H and Supplementary Video 2a). Shortly thereafter, accompanying nuclear breakdown, the perinuclear CAV-1-GFP signal was lost and the plasma membrane proximal CAV-1-GFP signal increased, with apparent fusion of the CAV-1 bodies with the plasma membrane (Figure 1I and Supplementary Video 2a). After fertilization, most CAV-1-GFP (Figure 1E), or GFP-CAV-1 (Figure 1, J–L, and Supplementary Video 2, a and b) was internalized and degraded in the one-cell stage embryo. CAV-1-GFP present in polar bodies persisted much longer and did not appear to be actively degraded (Figure 1E, arrowhead). These results indicate that CAV-1 localization and stability are tightly controlled in the germ line and embryo.
CAV-1-GFP Accumulates on the Plasma Membrane in Oocytes Lacking RME-2
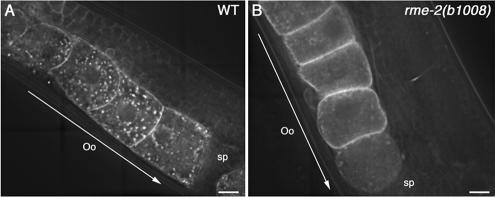
Endogenous CAV-1 was reported to be enriched in cholesterol-rich rafts in C. elegans, and cholesterol depletion disrupted the association of CAV-1 with glycosphingolipid-rich rafts (Scheel et al., 1999). The oocytes of C. elegans are enriched in cholesterol content relative to most other tissues as a consequence of their uptake of cholesterol-rich yolk particles (Matyash et al., 2001). To examine the effects of cholesterol depletion on CAV-1-GFP localization, we utilized rme-2 mutants, which lack the yolk receptor and thus fail in yolk uptake by oocytes (Grant and Hirsh, 1999). Because C. elegans does not possess the enzymes necessary for de novo sterol synthesis, all sterols enter the worm from their environment, most through ingestion and absorption by the intestine (Matyash et al., 2001). The bulk of the cholesterol entering oocytes is transported from the intestine via vitellogenins and RME-2 (Grant and Hirsh, 1999; Matyash et al., 2001). Thus, lack of RME-2 causes severe depletion of cholesterol in oocytes. As shown in Figure 2, rme-2(b1008) null mutant worms display aberrant accumulation of CAV-1-GFP on and near the plasma membrane accompanied by loss of intracellular CAV-1 bodies in the proximal oocytes (Figure 2B), suggesting that yolk uptake via RME-2 is important for correct localization of CAV-1-GFP in oocytes.
Figure 2.
CAV-1-GFP accumulates on the plasma membrane in rme-2 mutant oocytes. Subcellular localization of CAV-1-GFP in wild-type (A) and rme-2(b1008) (B) worms. CAV-1-GFP localizes to the CAV-1 body and the plasma membrane in oocytes of wild-type worms (A). In rme-2(b1008), CAV-1-GFP mainly localized to the plasma membrane, with the number and size of CAV-1-GFP labeled-CAV-1 bodies greatly reduced (B). Arrows indicate the direction of maturation of oocytes and embryos. Oo, oocyte; Sp, spematheca. Scale bars, 10 μm.
Fusion of CAV-1 Bodies with the Plasma Membrane Occurs after Anaphase
To better understand the molecular mechanisms controlling the trafficking of CAV-1 protein, we examined the effects of inactivation or depletion of proteins that regulate fertilization or cell-cycle changes associated with ovulation. In particular the timing of CAV-1 body fusion with the plasma membrane suggested that it might be an event triggered by fertilization. To test this hypothesis we examined CAV-1-GFP trafficking in spe-9 and fog-2 mutants. At the restrictive temperature of 25°C, spe-9(hc52) mutant sperm are morphologically normal and induce normal oocyte ovulation, but such sperm cannot fertilize the oocyte (Singson et al., 1998; Kadandale and Singson, 2004). fog-2(q71) mutants on the other hand develop as females devoid of sperm (Schedl and Kimble, 1988). Because major sperm proteins (MSPs) secreted by the sperm are important for ovulation, fog-2(q71) mutants show greatly reduced ovulation rates unless mated to males (Schedl and Kimble, 1988; Miller et al., 2001). Surprisingly, trafficking of CAV-1-GFP was nearly normal in both of these mutants (Figure 3, A–D), indicating that fertilization per se is not required for exocytosis of CAV-1 bodies or degradation of CAV-1-GFP after ovulation. The trafficking of CAV-1-GFP from CAV-1 bodies to the plasma membrane always accompanied ovulation, even in fog-2 mutant oocytes where ovulation is greatly delayed.
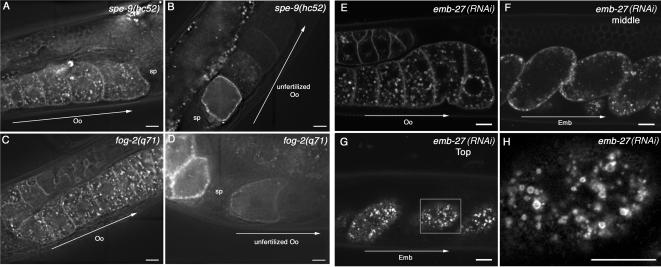
Figure 3.
Fusion of CAV-1 bodies with the plasma membrane occurs after anaphase. (A and B) CAV-1-GFP in a fertilization-defective mutant, spe-9(hc52). L4 hermaphrodites of spe-9(hc52)-expressing CAV-1-GFP were incubated at 25°C for 24 h and examined by fluorescence microscopy. CAV-1 body formation was normal in oocytes (A), and degradation of CAV-1-GFP occurred with normal kinetics in unfertilized eggs (B). (C and D) CAV-1-GFP in a sperm-deficient mutant, fog-2(q71). CAV-1 body formation was normal in oocytes (C), and the degradation of CAV-1-GFP occurred with normal kinetics in ovulated but unfertilized eggs (D) in fog-2(q71). (E-H) CAV-1-GFP in embryos defective in the metaphase-to-anaphase transition. L4 hermaphrodites expressing CAV-1-GFP were incubated at 20°C for 48 h. emb-27(RNAi) embryos are arrested in metaphase of meiosis I. In emb-27(RNAi) hermaphrodites expressing CAV-1-GFP, CAV-1 body formation appears normal in oocytes (E), but degradation of CAV-1-GFP in embryos was completely blocked (F). CAV-1 bodies were observed underneath the plasma membrane (G). (H) An enlargement of the square indicated in G. Oo, oocyte; Emb, embryo; Sp, spematheca. Arrows indicate the direction of maturation of oocytes and embryos. Scale bars, 10 μm.
Given the consistent timing of the fusion of CAV-1 bodies with the plasma membrane and the apparent fertilization independence of the event, we reasoned that perhaps it was the cell cycle that regulated the trafficking/fusion of CAV-1 at the plasma membrane. To test this hypothesis, we examined the effects of emb-27 RNAi on CAV-1-GFP trafficking. emb-27 encodes an ortholog of the cdc16p subunit of the anaphase-promoting complex that functions to target key cellular components for proteolytic destruction, advancing the cell cycle (Golden et al., 2000). RNAi of emb-27 results in accumulation of one-cell stage embryos arrested in metaphase of meiosis I (Golden et al., 2000). Such arrested embryos fail to transit to anaphase or to produce polar bodies. emb-27 RNAi did not interfere with CAV-1 body formation in oocytes (Figure 3E), but emb-27(RNAi) embryos arrested at the one-cell stage failed to degrade CAV-1-GFP (Figure 3F). CAV-1 bodies remained just beneath the plasma membrane in emb-27(RNAi) embryos (Figure 3, G and H), suggesting that fusion with the plasma membrane is inhibited until after metaphase.
CAV-1-GFP Is Degraded via Clathrin-mediated Endocytosis after Fertilization
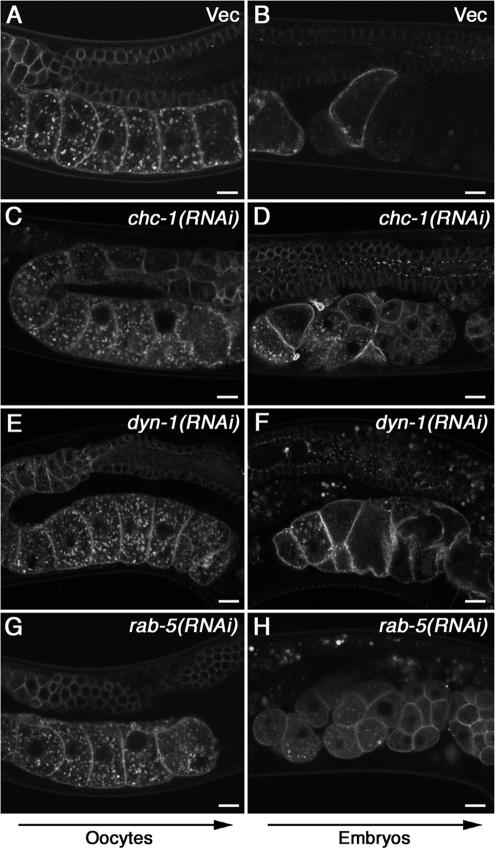
To gain insight into the mechanisms of both CAV-1 body formation in oocytes and CAV-1 down-regulation in embryos, we tested the importance of candidate trafficking regulators in either process. We began by depleting CAV-1-GFP animals of clathrin heavy chain (chc-1), dynamin (dyn-1), and rab-5 using RNAi conditions that we have previously established block endocytosis in oocytes (Grant and Hirsh, 1999; Sato et al., 2005). Clathrin is a major coat protein required for clathrin-mediated endocytosis by which most receptor-ligand complexes are internalized (Brodsky et al., 2001). Clathrin is also required for Golgi-to-endosome transport, a pathway utilized by newly synthesized lysosomal hydrolases (Bonifacino, 2004). Caveolar endocytosis is thought to be clathrin-independent. Dynamin is required for pinching off clathrin-coated vesicles and caveolae from the plasma membrane (Takei et al., 2005). The small GTPase RAB-5 is a key regulator of the early clathrin-mediated endocytic pathway and is also required for Simian Virus 40 infection via caveolar endocytosis in mammalian cells (Pelkmans et al., 2004). We did not observe any effect of RNAi-mediated depletion of chc-1, dyn-1, or rab-5 on CAV-1-GFP localization in oocytes (Figure 4, C, E, and G), suggesting that targeting of CAV-1-GFP to the CAV-1 bodies is a clathrin-, dynamin-, and RAB-5–independent process. In striking contrast we found that RNAi-mediated knockdown of chc-1, dyn-1, or rab-5 blocked degradation of CAV-1-GFP in embryos (Figure 4, D, F, and H). These results indicate that targeting of CAV-1-GFP to CAV-1 bodies in oocytes occurs in a clathrin- and RAB-5–independent-manner, but degradation of CAV-1-GFP in embryos is mediated by clathrin-dependent endocytosis.
Figure 4.
CAV-1-GFP is degraded via clathrin- and RAB-5–dependent endocytosis after fertilization. The subcellular localization of CAV-1-GFP was determined in chc-1(RNAi) (C and D), dyn-1(RNAi) (E and F), and rab-5(RNAi) worms (G and H) by a confocal microscopy. The localization of CAV-1-GFP was normal in mock RNAi-treated animals fed bacteria harboring the empty RNAi vector L4440 (A and B). In the chc-1(RNAi), dyn-1(RNAi), and rab-5(RNAi) worms, CAV-1 body formation appeared normal (C, E, and G), but degradation of CAV-1-GFP was blocked (D, F, and H). Arrows indicate the direction of maturation of oocytes and embryos. Scale bars, 10 μm.
ARF-1 Is Required for Targeting of CAV-1-GFP to CAV-1 Bodies in Oocytes
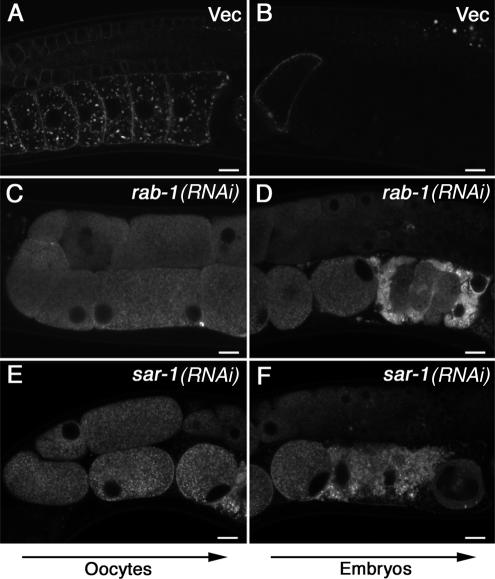
To gain additional insight into the mechanisms of CAV-1 trafficking we examined the effects of RNAi-mediated depletion of each C. elegans rab or sar/arf family GTPase on CAV-1-GFP localization in oocytes and embryos. Small GTPases of the Rab and Sar/Arf play pivotal roles in vesicular transport and each membrane transport step is thought to be regulated by at least one GTPase of this superfamily (Zerial and McBride, 2001; Nie et al., 2003). C. elegans has 1 sar, 10 arfs, and 29 rab genes. Among these genes, we found that RNAi of rab-1 or sar-1 caused severe accumulation of CAV-1-GFP in reticular structures, presumably the ER (Figure 5) and resulted in loss of CAV-1-GFP from other structures. These results indicate that CAV-1-GFP is transported through the early secretory pathway in the traditional manner and must exit the ER in a sar-1– and rab-1–dependent process before reaching the CAV-1 body and plasma membrane.
Figure 5.
CAV-1-GFP transport via the biosynthetic pathway. The subcellular localization of CAV-1-GFP was determined in rab-1(RNAi) (C and D) and sar-1(RNAi) (E and F) worms by confocal laser microscopy. The localization of CAV-1-GFP was normal in mock RNAi-treated animals fed bacteria harboring the empty RNAi vector L4440 (A and B). Arrows indicate the direction of maturation of oocytes and embryos. Scale bars, 10 μm.
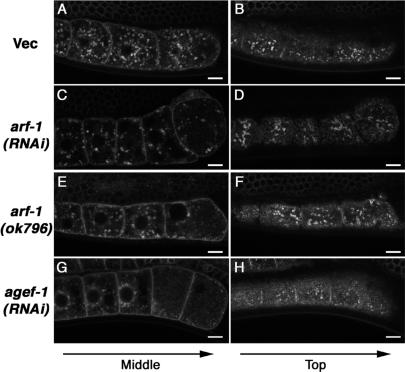
Interestingly, arf-1(RNAi) caused loss of normal CAV-1 bodies, which were replaced with fewer irregularly shaped CAV-1-GFP–positive vesicles in the cytoplasm (Figure 6C) and on or near the plasma membrane (Figure 6D). Furthermore, we obtained very similar results in arf-1(ok796) null mutants (Figure 6, E and F), confirming that ARF-1 is required for normal targeting of CAV-1-GFP to CAV-1 bodies. As a control for effects on general secretion, we also examined trafficking of GFP-tagged RME-2 in arf-1(RNAi) and arf-1(ok796) mutant animals (see Supplementary Material, Supplementary Figure S2, online). Most of the GFP-tagged RME-2 reached the plasma membrane and endosomes of these mutant oocytes, indicating fairly normal secretion under these conditions (Supplementary Figure S2, B and C). We did observe some extra RME-2-GFP puncta in the cytoplasm of oocytes lacking arf-1, however, perhaps indicating some delay in secretion in these strains (Supplementary Figure S2, B and C). We also found that RNAi of arf-3, a gene very similar in sequence to arf-1, caused a similar but weaker phenotype (our unpublished observation). RNAi of arf-3 in an arf-1(ok796) mutant background resulted in accumulation of CAV-1-GFP in reticular structures, presumably the ER (our unpublished observation), suggesting that ARF-1 and ARF-3 function redundantly in ER-Golgi transport and CAV-1 trafficking, but probably also have unique functions as indicated by their individual phenotypes.
Figure 6.
ARF-1–dependent transport of CAV-1-GFP to the CAV-1 body. CAV-1-GFP was normally localized to the CAV-1 body, and the plasma membrane in oocytes of mock RNAi-treated animals fed bacteria harboring the empty RNAi vector L4440 (A and B). In arf-1(RNAi) worms, the number of CAV-1 bodies was reduced (C) and large CAV-1-GFP–labeled aggregations were often observed (D, arrowheads). arf-1(ok796) displayed a very similar phenotype to arf-1(RNAi) (E and F). RNAi of agef-1, which encodes the predicted orthologue of a known Arf1 GEF, resulted in a severe defect in CAV-1 body formation (G and H). Scale bars, 10 μm.
To corroborate these findings, we further examined the effects of RNAi-mediated depletion of ARF guanine nucleotide exchange factor (GEF) homologues on CAV-1-GFP transport. The exchange of GDP for GTP in ARF family proteins is stimulated by GEFs containing a Sec7 domain (Jackson and Casanova, 2000). C. elegans has six genes encoding proteins with predicted Sec7 domains. Among these genes, only RNAi of Y6B3A.1 significantly blocked targeting of CAV-1-GFP to CAV-1 bodies (Figure 6, G and H). RNAi of Y6B3A.1 did not alter the localization of RME-2-GFP on the plasma membrane and endosomes (Supplementary Figure S2D). Y6B3A.1 encodes a homolog of mammalian BIG1 (brefeldin A–inhibited guanine nucleotide-exchange protein 1), which acts as a GEF for class I Arfs, predicted orthologues of C. elegans ARF-1 (Togawa et al., 1999). We named this gene agef-1, standing for Arf1 GEF homologue. Both Arf1 and BIG1 are known to function at the Golgi apparatus (Donaldson and Honda, 2005). Interestingly, agef-1 knockdown inhibited CAV-1 body formation more severely than did arf-1 disruption, suggesting that AGEF-1 might activate both ARF-1 and ARF-3 at the Golgi in C. elegans.
These results suggested that arf-1 and agef-1 function in CAV-1 body formation at the Golgi. We thus examined colocalization of CAV-1-GFP with the Golgi membrane protein SQV-8 (Herman and Horvitz, 1999). Although SQV-8 showed distinct localization from CAV-1-GFP in the large wild-type oocytes, SQV-8 was generally found directly adjacent to the CAV-1 bodies (Figure 7, A–C). Strikingly, deletion of arf-1 not only prevents normal CAV-1 body formation (Figure 7D) but also dramatically changed Golgi morphology (Figure 7E). Much of the CAV-1-GFP in arf-1 mutants was coincident with SQV-8 (Figure 7F), suggesting that ARF-1 is involved in export of CAV-1 from the Golgi to the CAV-1 bodies. Taken together, these results suggest that activation of class I Arfs by AGEF-1 is required for transport of caveolin from the Golgi to the CAV-1 bodies during oogenesis.
Time-lapse movies of CAV-1-GFP trafficking in the gonad of arf-1 mutants indicated that the abnormal CAV-1 bodies form by clustering of small cytoplasmic vesicles, like wild type, but in the absence of arf-1, clustering of small structures continues without reformation into distinct rings, consistent with CAV-1 entering Golgi ministacks but failing in the process of exiting (Supplementary Video 3). The defects in agef-1 RNAi animals were more severe, with apparent clustering of small CAV-1-GFP vesicles in the bend region of the gonad, but no apparent accumulation and no ring formation (Supplementary Video 4).
DISCUSSION
In this study we show that caveolin trafficking is highly regulated during development of the germ line and embryo in C. elegans. Caveolin-GFP dynamics have been studied in mammalian cultured cell lines (Pelkmans et al., 2001, 2002, 2004, 2005; Tagawa et al., 2005). However, few of the cultured cell results have been extended to the physiologically relevant context of a living animal. Our findings demonstrate that several distinct mechanisms regulate caveolin trafficking at different developmental stages in a living animal and suggest potential new functions for caveolin in development.
We found that CAV-1-GFP is first transported via the biosynthetic pathway from the ER to the Golgi and is then targeted to a novel membrane compartment, the CAV-1 body, during oocyte formation. We discovered that targeting of CAV-1-GFP to the CAV-1 bodies depends on the C. elegans homologues of Arf1 and its putative guanine nucleotide exchange factor BIG1. In arf-1 mutants CAV-1-GFP becomes trapped in a Golgi-associated structure, possibly an incomplete CAV-1 body that cannot detach from the TGN. In mammalian cells, Arf1 and BIG1 are localized to the Golgi (Stearns et al., 1990; Yamaji et al., 2000) and are required for the recruitment of cytosolic coat complexes to the Golgi membrane (Donaldson et al., 2005). Interestingly, newly assembled caveolar domains also first appear as structures budding from the Golgi in mammalian cells (Tagawa et al., 2005). Arf1 recruits the adaptor protein complex (AP1) and the monomeric Golgi-associated, γ-adaptin ear-containing, Arf-binding (GGA) proteins to the Golgi complex. Both of these coat proteins recognize the cytoplasmic tails of transmembrane cargo proteins and mediate their transport between the TGN and the lysosome in a clathrin-dependent manner (Robinson, 2004). Because targeting of CAV-1-GFP to CAV-1 bodies was not affected by knockdown of clathrin heavy chain, even under conditions stringent enough to redistribute clathrin light chain to the cytosol (our unpublished observation), it is likely that CAV-1 body formation is a novel clathrin-independent process. Strikingly, in rme-2 mutants defective in cholesterol-enriched yolk uptake, nearly all CAV-1-GFP accumulated on the plasma membrane concurrent with a loss of the CAV-1 bodies. One simple explanation would be that high cholesterol levels are required for correct sorting of CAV-1-GFP at the Golgi.
CAV-1 bodies normally appear to fuse with the plasma membrane after fertilization, suggesting that the CAV-1 body is a regulated secretory compartment derived from the TGN. The fusion of CAV-1 bodies with the plasma membrane is blocked by loss of EMB-27, a subunit of the anaphase-promoting complex required for the metaphase-to-anaphase transition of embryos after fertilization (Golden et al., 2000), indicating that fusion of CAV-1 bodies to the plasma membrane is tightly linked to progression of meiosis I. This process can proceed even if fertilization itself is blocked. In mammalian cells, cytoplasmic caveolar vesicles undergo kiss-and-run–like fusion with the plasma membrane (Pelkmans and Zerial, 2005), suggesting that caveolin-enriched vesicles in general undergo regulated targeting to the plasma membrane. The CAV-1 bodies are quite reminiscent of oocyte cortical granules observed in other animals (Fisher and Rebhun, 1983; Abbott and Ducibella, 2001). In animals, exocytosis of cortical granules located in the cortex of oocytes changes the extracellular environment to prevent additional spermatozoa from penetrating the newly fertilized egg. As such CAV-1 bodies may be a reservoir of cholesterol- and glycosphingolipid-enriched membranes or signaling molecules in the oocyte that are delivered to the plasma membrane to make oocytes resistant to polyspermy. Interestingly, electron microscopy indicates that C. elegans zygotes produce a transient “post-ovulation envelope” before eggshell formation (David Greenstein, personal communication). It is conceivable that CAV-1 bodies are involved in forming a postovulation envelope to block polyspermy. Because C. elegans embryos secrete their own eggshell, it is also possible that CAV-1 bodies contain components that contribute to eggshell formation (Grant and Sato, 2006).
Strikingly, CAV-1-GFP was internalized for degradation in a clathrin- and RAB-5–dependent manner. Because caveolins in cultured cells produce their own clathrin-independent endocytic pits and vesicles, this was unexpected. However, caveolins are not degraded by uptake into caveosomes, so redirection into the standard endocytosis pathway may be necessary for productive degradation. Regulated degradation of maternal proteins is under strict developmental control and is important for proper embryogenesis. For instance maternal proteins MEI-1 and MEI-2 are degraded in a ubiquitin-dependent reaction that is essential for transition of embryos from meiosis to mitosis (Srayko et al., 2000; Pellettieri et al., 2003). Mono-ubiquitination of membrane proteins is known to direct their endocytosis (Hicke and Dunn, 2003). In addition mono-ubiquitination is recognized by the ESCRT complex of the endosomes and directs transmembrane cargo into the internal vesicles of multivesicular bodies, leading to their degradation and preventing their recycling (Raiborg et al., 2003). Such ubiquitination of CAV-1 might be the means by which it is targeted for degradation.
In this study we reveal that trafficking of C. elegans caveolin-1 is dynamically regulated during the development of the germ line and embryo. We do note however that the CAV-1-GFP-fusion proteins examined here were driven by a heterologous promoter, and we were not able to test these fusions for function because no cav-1 mutants are currently available. Thus, it is not yet certain if our results completely recapitulate the expression and trafficking of the endogenous CAV-1 protein. Further studies will be required to determine the precise mechanisms that drive CAV-1 into this pathway.
Supplementary Material
ACKNOWLEDGMENTS
We thank Geraldine Seydoux for important reagents, the C. elegans Gene Knockout Consortium for the arf-1 knockout strain, and the C. elegans Genetic Center for many strains used in this study. We also thank Paul Andre Fonarev and Katsuya Sato for expert technical assistance. We thank Andrew Singson, David Greenstein, Willisa Liou, Akihiro Harada, and Reiko Harada for advice. This work was supported by a National Institutes of Health Grant GM67237 and MOD Grant 5-FY02-252 to B.G. B.G. also received support from the Chicago Community Trust Searle Scholars Program. This research was partially supported by the Ministry of Education, Science, Sports, and Culture, Grant-in-Aid for Young Scientists (A), 2005, Scientific Research on Priority Areas 2005, and Scientific Research (B), 2005. K.S. and M.S. were supported by Bioarchitect Research Projects II of RIKEN and a JSPS Postdoctoral Fellowship, respectively. A.A. is a Helen Hay Whitney Postdoctoral Fellow. K.O. is a Pew Scholar in the Biomedical Sciences supported by funding from the Ludwig Institute for Cancer Research.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-03-0211) on May 3, 2006.

 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Abbott A. L., Ducibella T. Calcium and the control of mammalian cortical granule exocytosis. Front Biosci. 2001;6:D792–D806. doi: 10.2741/abbott. [DOI] [PubMed] [Google Scholar]
- Anderson R. G. The caveolae membrane system. Annu. Rev. Biochem. 1998;67:199–225. doi: 10.1146/annurev.biochem.67.1.199. [DOI] [PubMed] [Google Scholar]
- Audhya A., Hyndman F., McLeod I. X., Maddox A. S., Yates J. R., 3rd, Desai A., Oegema K. A complex containing the Sm protein CAR-1 and the RNA helicase CGH-1 is required for embryonic cytokinesis in Caenorhabditis elegans. J. Cell Biol. 2005;171:267–279. doi: 10.1083/jcb.200506124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino J. S. The GGA proteins: adaptors on the move. Nat. Rev. Mol. Cell Biol. 2004;5:23–32. doi: 10.1038/nrm1279. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky F. M., Chen C. Y., Knuehl C., Towler M. C., Wakeham D. E. Biological basket weaving: formation and function of clathrin-coated vesicles. Annu. Rev. Cell Dev. Biol. 2001;17:517–568. doi: 10.1146/annurev.cellbio.17.1.517. [DOI] [PubMed] [Google Scholar]
- del Pozo M. A., Balasubramanian N., Alderson N. B., Kiosses W. B., Grande-Garcia A., Anderson R. G., Schwartz M. A. Phospho-caveolin-1 mediates integrin-regulated membrane domain internalization. Nat. Cell Biol. 2005;7:901–908. doi: 10.1038/ncb1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson J. G., Honda A. Localization and function of Arf family GTPases. Biochem. Soc. Trans. 2005;33:639–642. doi: 10.1042/BST0330639. [DOI] [PubMed] [Google Scholar]
- Donaldson J. G., Honda A., Weigert R. Multiple activities for Arf1 at the Golgi complex. Biochim. Biophys. Acta. 2005;1744:364–373. doi: 10.1016/j.bbamcr.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Drab M., et al. Loss of caveolae, vascular dysfunction, and pulmonary defects in caveolin-1 gene-disrupted mice. Science. 2001;293:2449–2452. doi: 10.1126/science.1062688. [DOI] [PubMed] [Google Scholar]
- Fernandez I., Ying Y., Albanesi J., Anderson R. G. Mechanism of caveolin filament assembly. Proc. Natl. Acad. Sci. USA. 2002;99:11193–11198. doi: 10.1073/pnas.172196599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher G. W., Rebhun L. I. Sea urchin egg cortical granule exocytosis is followed by a burst of membrane retrieval via uptake into coated vesicles. Dev. Biol. 1983;99:456–472. doi: 10.1016/0012-1606(83)90295-6. [DOI] [PubMed] [Google Scholar]
- Glenney J. R., Jr., Soppet D. Sequence and expression of caveolin, a protein component of caveolae plasma membrane domains phosphorylated on tyrosine in Rous sarcoma virus-transformed fibroblasts. Proc. Natl. Acad. Sci. USA. 1992;89:10517–10521. doi: 10.1073/pnas.89.21.10517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden A., Sadler P. L., Wallenfang M. R., Schumacher J. M., Hamill D. R., Bates G., Bowerman B., Seydoux G., Shakes D. C. Metaphase to anaphase (mat) transition-defective mutants in Caenorhabditis elegans. J. Cell Biol. 2000;151:1469–1482. doi: 10.1083/jcb.151.7.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B., Hirsh D. Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol. Biol. Cell. 1999;10:4311–4326. doi: 10.1091/mbc.10.12.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B., Zhang Y., Paupard M. C., Lin S. X., Hall D. H., Hirsh D. Evidence that RME-1, a conserved C. elegans EH-domain protein, functions in endocytic recycling. Nat. Cell Biol. 2001;3:573–579. doi: 10.1038/35078549. [DOI] [PubMed] [Google Scholar]
- Grant B. D., Sato M. WormBook, editor. Intracellular trafficking. 2006 doi: 10.1895/wormbook.1.77.1. The C. elegans Research Community, WormBook, doi/10.1895/wormbook. 1.77.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman T., Horvitz H. R. Three proteins involved in Caenorhabditis elegans vulval invagination are similar to components of a glycosylation pathway. Proc. Natl. Acad. Sci. USA. 1999;96:974–979. doi: 10.1073/pnas.96.3.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L., Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 2003;19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- Huet C., Ash J. F., Singer S. J. The antibody-induced clustering and endocytosis of HLA antigens on cultured human fibroblasts. Cell. 1980;21:429–438. doi: 10.1016/0092-8674(80)90479-1. [DOI] [PubMed] [Google Scholar]
- Jackson C. L., Casanova J. E. Turning on ARF: the Sec7 family of guanine-nucleotide-exchange factors. Trends Cell Biol. 2000;10:60–67. doi: 10.1016/s0962-8924(99)01699-2. [DOI] [PubMed] [Google Scholar]
- Kadandale P., Singson A. Oocyte production and sperm utilization patterns in semi-fertile strains of Caenorhabditis elegans. BMC Dev. Biol. 2004;4:3. doi: 10.1186/1471-213X-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R. S., Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Le Lay S., Kurzchalia T. V. Getting rid of caveolins: phenotypes of caveolin-deficient animals. Biochim. Biophys. Acta. 2005;1746:322–333. doi: 10.1016/j.bbamcr.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Lipardi C., Mora R., Colomer V., Paladino S., Nitsch L., Rodriguez-Boulan E., Zurzolo C. Caveolin transfection results in caveolae formation but not apical sorting of glycosylphosphatidylinositol (GPI)-anchored proteins in epithelial cells. J. Cell Biol. 1998;140:617–626. doi: 10.1083/jcb.140.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro M., Pilgrim D. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics. 1995;141:977–988. doi: 10.1093/genetics/141.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matveev S., Li X., Everson W., Smart E. J. caveolae and caveolin in vesicle-dependent and vesicle-independent trafficking. Adv. Drug Delivery Rev. 2001;49:237–250. doi: 10.1016/s0169-409x(01)00138-7. [DOI] [PubMed] [Google Scholar]
- Matyash V., Geier C., Henske A., Mukherjee S., Hirsh D., Thiele C., Grant B., Maxfield F. R., Kurzchalia T. V. Distribution and transport of cholesterol in Caenorhabditis elegans. Mol. Biol. Cell. 2001;12:1725–1736. doi: 10.1091/mbc.12.6.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter J., Bartlett B., Dang T., Schedl T. On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev. Biol. 1999;205:111–128. doi: 10.1006/dbio.1998.9109. [DOI] [PubMed] [Google Scholar]
- Miller M. A., Nguyen V. Q., Lee M. H., Kosinski M., Schedl T., Caprioli R. M., Greenstein D. A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation. Science. 2001;291:2144–2147. doi: 10.1126/science.1057586. [DOI] [PubMed] [Google Scholar]
- Murata M., Peranen J., Schreiner R., Wieland F., Kurzchalia T. V., Simons K. VIP21/caveolin is a cholesterol-binding protein. Proc. Natl. Acad. Sci. USA. 1995;92:10339–10343. doi: 10.1073/pnas.92.22.10339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z., Hirsch D. S., Randazzo P. A. Arf and its many interactors. Curr. Opin. Cell Biol. 2003;15:396–404. doi: 10.1016/s0955-0674(03)00071-1. [DOI] [PubMed] [Google Scholar]
- Parton R. G. Caveolae and caveolins. Curr. Opin. Cell Biol. 1996;8:542–548. doi: 10.1016/s0955-0674(96)80033-0. [DOI] [PubMed] [Google Scholar]
- Parton R. G., Joggerst B., Simons K. Regulated internalization of caveolae. J. Cell Biol. 1994;127:1199–1215. doi: 10.1083/jcb.127.5.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton R. G., Richards A. A. Lipid rafts and caveolae as portals for endocytosis: new insights and common mechanisms. Traffic. 2003;4:724–738. doi: 10.1034/j.1600-0854.2003.00128.x. [DOI] [PubMed] [Google Scholar]
- Pelkmans L., Burli T., Zerial M., Helenius A. Caveolin-stabilized membrane domains as multifunctional transport and sorting devices in endocytic membrane traffic. Cell. 2004;118:767–780. doi: 10.1016/j.cell.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Pelkmans L., Fava E., Grabner H., Hannus M., Habermann B., Krausz E., Zerial M. Genome-wide analysis of human kinases in clathrin- and caveolae/raft-mediated endocytosis. Nature. 2005;436:78–86. doi: 10.1038/nature03571. [DOI] [PubMed] [Google Scholar]
- Pelkmans L., Kartenbeck J., Helenius A. Caveolar endocytosis of simian virus 40 reveals a new two-step vesicular-transport pathway to the ER. Nat. Cell Biol. 2001;3:473–483. doi: 10.1038/35074539. [DOI] [PubMed] [Google Scholar]
- Pelkmans L., Puntener D., Helenius A. Local actin polymerization and dynamin recruitment in SV40-induced internalization of caveolae. Science. 2002;296:535–539. doi: 10.1126/science.1069784. [DOI] [PubMed] [Google Scholar]
- Pelkmans L., Zerial M. Kinase-regulated quantal assemblies and kiss-and-run recycling of caveolae. Nature. 2005;436:128–133. doi: 10.1038/nature03866. [DOI] [PubMed] [Google Scholar]
- Pellettieri J., Reinke V., Kim S. K., Seydoux G. Coordinate activation of maternal protein degradation during the egg-to-embryo transition in C. elegans. Dev. Cell. 2003;5:451–462. doi: 10.1016/s1534-5807(03)00231-4. [DOI] [PubMed] [Google Scholar]
- Pol A., Martin S., Fernandez M. A., Ferguson C., Carozzi A., Luetterforst R., Enrich C., Parton R. G. Dynamic and regulated association of caveolin with lipid bodies: modulation of lipid body motility and function by a dominant negative mutant. Mol. Biol. Cell. 2004;15:99–110. doi: 10.1091/mbc.E03-06-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis V., Casey E., Collar D., Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C., Rusten T. E., Stenmark H. Protein sorting into multivesicular endosomes. Curr. Opin. Cell Biol. 2003;15:446–455. doi: 10.1016/s0955-0674(03)00080-2. [DOI] [PubMed] [Google Scholar]
- Robinson M. S. Adaptable adaptors for coated vesicles. Trends Cell Biol. 2004;14:167–174. doi: 10.1016/j.tcb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Rothberg K. G., Heuser J. E., Donzell W. C., Ying Y. S., Glenney J. R., Anderson R. G. Caveolin, a protein component of caveolae membrane coats. Cell. 1992;68:673–682. doi: 10.1016/0092-8674(92)90143-z. [DOI] [PubMed] [Google Scholar]
- Roy S., Luetterforst R., Harding A., Apolloni A., Etheridge M., Stang E., Rolls B., Hancock J. F., Parton R. G. Dominant-negative caveolin inhibits H-Ras function by disrupting cholesterol-rich plasma membrane domains. Nat. Cell Biol. 1999;1:98–105. doi: 10.1038/10067. [DOI] [PubMed] [Google Scholar]
- Sato M., Sato K., Fonarev P., Huang C. J., Liou W., Grant B. D. Caenorhabditis elegans RME-6 is a novel regulator of RAB-5 at the clathrin-coated pit. Nat. Cell Biol. 2005;7:559–569. doi: 10.1038/ncb1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schedl T., Kimble J. fog-2, a germ-line-specific sex determination gene required for hermaphrodite spermatogenesis in Caenorhabditis elegans. Genetics. 1988;119:43–61. doi: 10.1093/genetics/119.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel J., Srinivasan J., Honnert U., Henske A., Kurzchalia T. V. Involvement of caveolin-1 in meiotic cell-cycle progression in Caenorhabditis elegans. Nat. Cell Biol. 1999;1:127–129. doi: 10.1038/10100. [DOI] [PubMed] [Google Scholar]
- Singson A., Mercer K. B., L’Hernault S. W. The C. elegans spe-9 gene encodes a sperm transmembrane protein that contains EGF-like repeats and is required for fertilization. Cell. 1998;93:71–79. doi: 10.1016/s0092-8674(00)81147-2. [DOI] [PubMed] [Google Scholar]
- Srayko M., Buster D. W., Bazirgan O. A., McNally F. J., Mains P. E. MEI-1/MEI-2 katanin-like microtubule severing activity is required for Caenorhabditis elegans meiosis. Genes Dev. 2000;14:1072–1084. [PMC free article] [PubMed] [Google Scholar]
- Stearns T., Willingham M. C., Botstein D., Kahn R. A. ADP-ribosylation factor is functionally and physically associated with the Golgi complex. Proc. Natl. Acad. Sci. USA. 1990;87:1238–1242. doi: 10.1073/pnas.87.3.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagawa A., Mezzacasa A., Hayer A., Longatti A., Pelkmans L., Helenius A. Assembly and trafficking of caveolar domains in the cell: caveolae as stable, cargo-triggered, vesicular transporters. J. Cell Biol. 2005;170:769–779. doi: 10.1083/jcb.200506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei K., Yoshida Y., Yamada H. Regulatory mechanisms of dynamin-dependent endocytosis. J Biochem. (Tokyo) 2005;137:243–247. doi: 10.1093/jb/mvi052. [DOI] [PubMed] [Google Scholar]
- Tang Z., Okamoto T., Boontrakulpoontawee P., Katada T., Otsuka A. J., Lisanti M. P. Identification, sequence, and expression of an invertebrate caveolin gene family from the nematode Caenorhabditis elegans. Implications for the molecular evolution of mammalian caveolin genes. J. Biol. Chem. 1997;272:2437–2445. doi: 10.1074/jbc.272.4.2437. [DOI] [PubMed] [Google Scholar]
- Thomsen P., Roepstorff K., Stahlhut M., van Deurs B. Caveolae are highly immobile plasma membrane microdomains, which are not involved in constitutive endocytic trafficking. Mol. Biol. Cell. 2002;13:238–250. doi: 10.1091/mbc.01-06-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L., Court D. L., Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Togawa A., Morinaga N., Ogasawara M., Moss J., Vaughan M. Purification and cloning of a brefeldin A-inhibited guanine nucleotide-exchange protein for ADP-ribosylation factors. J. Biol. Chem. 1999;274:12308–12315. doi: 10.1074/jbc.274.18.12308. [DOI] [PubMed] [Google Scholar]
- Yamaji R., Adamik R., Takeda K., Togawa A., Pacheco-Rodriguez G., Ferrans V. J., Moss J., Vaughan M. Identification and localization of two brefeldin A-inhibited guanine nucleotide-exchange proteins for ADP-ribosylation factors in a macromolecular complex. Proc. Natl. Acad. Sci. USA. 2000;97:2567–2572. doi: 10.1073/pnas.97.6.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerial M., McBride H. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.