Abstract
The yeast cell wall is an essential organelle that protects the cell from mechanical damage and antimicrobial peptides, participates in cell recognition and adhesion, and is important for the generation and maintenance of normal cell shape. We studied the localization of three covalently bound cell wall proteins in Saccharomyces cerevisiae. Tip1p was found only in mother cells, whereas Cwp2p was incorporated in small-to-medium–sized buds. When the promoter regions of TIP1 and CWP2 (responsible for transcription in early G1 and S/G2 phases, respectively) were exchanged, the localization patterns of Tip1p and Cwp2p were reversed, indicating that the localization of cell wall proteins can be completely determined by the timing of transcription during the cell cycle. The third protein, Cwp1p, was incorporated into the birth scar, where it remained for several generations. However, we could not detect any role of Cwp1p in strengthening the birth scar wall or any functional interaction with the proteins that mark the birth scar pole as a potential future budding site. Promoter-exchange experiments showed that expression in S/G2 phase is necessary but not sufficient for the normal localization of Cwp1p. Studies of mutants in which septum formation is perturbed indicate that the normal asymmetric localization of Cwp1p also depends on the normal timing of septum formation, composition of the septum, or both.
INTRODUCTION
The establishment and maintenance of asymmetry are of vital importance for the growth, differentiation, and morphogenesis of cells and organisms. For example, the yeast Saccharomyces cerevisiae grows asymmetrically, producing a bud that becomes the daughter cell (reviewed by Pringle et al., 1995; Lew and Reed, 1995; Pruyne and Bretscher 2000a,b). In the G1 phase of the cell cycle, a bud site is selected in a mating type-dependent manner. After the cell cycle-commitment point START, proteins required for bud initiation are recruited to this site, and a bud is formed in a process that depends largely on the delivery of vesicles containing new cell surface material by the polarized actin cytoskeleton. Growth occurs exclusively in the bud, first preferentially at the tip and later isotropically, until the time of cytokinesis. The actin cytoskeleton and cell surface growth are then redirected toward the mother-bud neck, where a chitinous primary septum is formed, followed by the deposition of secondary septa on both sides of the primary septum. Cells then separate by partial digestion of the primary septum, and a new cell cycle is initiated.
The cell wall of yeast is essential for growth and is involved in the establishment and maintenance of asymmetry, as illustrated by the loss of both buds and polarized localization of cytoskeletal proteins when the cell wall is removed (Reck-Peterson et al., 1999). The cell wall is comprised of glucans, mannoproteins, and a small amount of chitin (reviewed by Orlean, 1997; Smits et al., 2001; Klis et al., 2006). The mannoproteins can be divided into four classes: 1) noncovalently linked proteins, such as Bgl2p (Klebl and Tanner, 1989; Cappellaro et al., 1998); 2) proteins that are linked covalently to the cell wall through disulfide bridges (Orlean et al., 1986; Moukadiri et al., 1999); 3) PIR-proteins, which are linked via an alkali-sensitive bond to β-1,3-glucan (Kapteyn et al., 1999b; Castillo et al., 2003; Ecker et al., 2006); and 4) a set of ∼40 glycosylphosphatidylinositol (GPI)-dependent cell wall proteins (Caro et al., 1997; Hamada et al., 1998; Yin et al., 2005), which are covalently linked to β-1,3-glucan through β-1,6-glucan (Kapteyn et al., 1996; Kollár et al., 1997) and may or may not also be linked directly to β-1,3-glucan through an alkali-sensitive bond (Kapteyn et al., 2001). In general, the cell wall proteins are thought to protect the cell from the environment, and deletions of some cell wall protein genes with homologies to glycoside hydrolases produce the expected kinds of phenotypes (Yin et al., 2005). However, deletions of other genes encoding individual cell wall proteins, even those that are highly expressed, often have surprisingly little effect (van der Vaart et al., 1995; Hagen et al., 2004).
Cell wall proteins are processed through the secretory pathway, where they are O- and often also N-glycosylated and may receive a GPI anchor at their carboxy terminus. When GPI-dependent cell wall proteins arrive at the plasma membrane, the GPI anchor is thought to be processed, resulting in its cleavage and linkage of the protein to the β-1,6-glucan through a GPI remnant (Kollár et al., 1997; Lipke and Ovalle, 1998; Kapteyn et al., 1999a; Terashima et al., 2003).
Interestingly, more than half of the genes encoding cell wall proteins in S. cerevisiae are transcribed in a cell cycle-dependent manner (Caro et al., 1998; Spellman et al., 1998), compared with only 13% for the genome as a whole. This suggests that many cell wall proteins may be produced only when needed during the different stages of bud formation and cell division. Because of the rearrangements of the actin cytoskeleton during the cell cycle (see above), cell cycle-specific synthesis of a cell surface protein can also lead to its localized incorporation at discrete regions of the cell surface, as shown by the integral plasma membrane proteins Axl2p, Bud8p, and Bud9p (Lord et al., 2000; Schenkman et al., 2002). Consistent with this model, the GPI-dependent cell wall protein Crh1p seems to be synthesized in late G1 and then incorporated specifically at the presumptive bud site and in the surface of the emerging small bud (Spellman et al., 1998; Rodríguez-Peña et al., 2000). However, cell cycle-specific synthesis does not seem to be the only mechanism by which cells can achieve the localized incorporation of cell wall proteins. For example, the GPI-dependent cell wall protein Crh2p seems to be synthesized throughout the cell cycle and yet is localized primarily to a ring at the base of the bud by a mechanism that depends on a number of factors, including the septin ring at that location (Spellman et al., 1998; Rodríguez-Peña et al., 2000, 2002).
To investigate further the patterns and mechanisms of incorporation of GPI-dependent proteins into the cell wall, we tagged three different proteins with green fluorescent protein (GFP). All three proteins seemed to be incorporated into specific regions of the wall, and further investigation suggested that timed transcription during the cell cycle is involved in this localized incorporation. In addition, our studies suggest that incorporation of one of these proteins, Cwp1p, specifically into the birth scar also depends on the formation of a normal septum at the proper time.
MATERIALS AND METHODS
Strains, Growth Conditions, and Genetic Procedures
S. cerevisiae strains used in this study are listed in Table 1, and PCR primers are listed in Supplemental Table 1. Yeast cells were grown in YPD or appropriate selective media (Adams et al., 1996) at 28°C unless otherwise specified. Growth was routinely monitored by optical density (OD)600 measurements using spectrophotometers in their linear ranges (OD600 < 0.4; cultures were diluted before reading when necessary) and calibration curves relating OD600 to cell number. For mating, sporulation, tetrad dissection, and transformation, standard procedures were used (Adams et al., 1996). Molecular genetic procedures were also standard (Sambrook et al., 1989; Ausubel et al., 1995) except where noted. Strains containing complete deletions of the CWP1, CHS2, and SHE3 open reading frames (ORFs) in the YEF473 background were constructed by the PCR method (Baudin et al., 1993); plasmid pFA6a-His3MX6 (Longtine et al., 1998a) was used as template with primers LS137 and LS138 (CWP1), ML176 and ML177 (CHS2), or IC101 and IC102 (SHE3). In all cases, transformants were checked for the desired chromosomal insertions using PCR with other primer pairs including LS139 and LS144 (CWP1), ML176 and ML178 (CHS2), or IC103 and 5′TTEFCHK (SHE3).
Table 1.
Strains used in this study
| Strain | Relevant genotype | Source |
|---|---|---|
| FY833 | MATa his3-Δ200 leu2-Δ1 lys2Δ202 trp1-Δ63 ura3-52 | Winston et al. (1995) |
| JV96 | As FY833 except cwp1::LEU2 | Vossen et al. (1997) |
| YEF473 | MATa/α his3-Δ200/his3-Δ200 leu2-Δ1/leu2-Δ1 lys2-801/lys2-801 trp1-Δ63/trp1-Δ63 ura3-52/ura3-52 | Bi and Pringle (1996) |
| YEF473A | MATa his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 | Segregant from YEF473 |
| M-272 | As YEF473 except gin4-Δ9/gin4-Δ9 | Longtine et al. (1998b) |
| M-717 | As YEF473 except bni1Δ::HIS3/bni1Δ::HIS3 | M. Longtinea |
| ICY018 | As YEF473 except she3Δ::His3MX6/she3Δ::His3MX6 | See text |
| ICY028 | As YEF473 except chs2Δ::His3MX6/chs2Δ::His3MX6 | See text |
| KBY1012 | MATα his3 leu2 lys2 ura3-52 myo4::ura3 | Beach and Bloom (2001) |
| DDY172-2A | As YEF473A except chs4-Δ1 | DeMarini et al. (1997) |
| DDY181 | As YEF473 except CHS3/chs3-Δ1 | DeMarini et al. (1997) |
| DDY181-2D | As YEF473A except chs3-Δ1 | Segregant from DDY181 |
| KNY1048 | As YEF473 except chs6Δ::TRP1/chs6Δ::TRP1 bud7Δ::TRP1/bud7Δ::TRP1 ymr237wΔ::kanMX6/ymr237wΔ::kanMX6ykr027wΔ::His3MX6/ykr027wΔ::His3MX6 | K. Nakashimab |
| AM273 | As YEF473 except axl2Δ:: His3MX6 | A. McKenzieb |
| AM476 | As YEF473 except rax2Δ::His3MX6/rax2Δ::His3MX6 | A. McKenzieb |
| AM503 | As YEF473 except rax1Δ::kanMX6/rax1Δ::kanMX6 | A. McKenzieb |
| AM775 | As YEF473 except chs5Δ::TRP1/chs5Δ::TRP1 | A. McKenzieb |
| LSY252 | As YEF473A except cwp1Δ::His3MX6 | See text |
| LSY265 | As YEF473 except cwp1Δ::His3MX6/cwp1Δ::His3MX6 | See text |
| YHH415 | As YEF473 except bud8-Δ1/bud8-Δ1 | Harkins et al. (2001) |
| YHH615 | As YEF473 except bud9-Δ1/bud9-Δ1 | Harkins et al. (2001) |
a Constructed by mating strains YJZ426 and YJZ427 (Harkins et al., 2001).
b Nakashima, McKenzie, and Pringle, unpublished data. The strains have complete deletions of the indicated ORFs.
Plasmids
Multicopy plasmids expressing GFP-Cwp1p from the CWP1 promoter (pAR213) or GFP-Cwp2p from the CWP2 promoter (pAR205) were described previously (Ram et al., 1998a). Both constructs were subcloned into YCplac33 (Gietz and Sugino, 1988) using HindIII and BamHI for pAR213 and HindIII and EcoRI for pAR205, creating the low-copy plasmids pGS-GFP-CWP1-low and pGS-GFP-CWP2-lowR, respectively. The orientation of GFP-CWP2 relative to the vector sequences was then reversed (necessary for the promoter-exchange experiments described below) in two steps. First, the PstI-EcoRI fragment of pGS-GFP-CWP2-lowR was subcloned into the corresponding sites of pBluescript II KS. Second, the HindIII-PstI fragment of the resulting plasmid was subcloned into the corresponding sites of YCplac33, yielding pGS-GFP-CWP2-low.
To create a plasmid expressing GFP-tagged Tip1p, TIP1 was first cloned by gap repair. A linear gap-repair plasmid was created by PCR using primers GS2 and GS3 and plasmid YEplac181 (Gietz and Sugino, 1988) as template. The resulting fragment was transformed into strain FY833, selecting for Leu+. Plasmids were recovered into Escherichia coli, and the desired plasmid (pGS-TIP1) was identified by restriction analysis and PCR using primers GS22 and GS23. XhoI and KpnI restriction sites were then generated directly downstream of the TIP1 signal sequence in a two-step PCR reaction using Pfu polymerase (Fermentas, St.-Leon Rot, Germany) and plasmid pGS-TIP1 as template. Primer pairs GS6/GS7 and GS8/GS9 were used in the first reactions, and the combined purified products were used together with primers GS6 and GS8 in the second reaction. The final PCR product and plasmid pGS-TIP1 were digested with RsrII and XbaI and ligated, and the product was checked by restriction analysis. A XhoI-KpnI fragment containing the GFP (S65T) sequences from plasmid pREP4-GFP(S65T) (a derivative of plasmid pREP4; Maundrell, 1993; kindly supplied by Dr. F. Hochstenbach, Department of Medical Biochemistry, AMC, University of Amsterdam, Amsterdam, The Netherlands) was subcloned into the XhoI-KpnI sites to create pGS-GFP-TIP1-high. The HindIII-EcoRI fragment of pGS-GFP-TIP1-high was then subcloned into YCplac33 to create pGS-GFP-TIP1-low.
For promoter-exchange experiments, EagI (for CWP1 and TIP1) or NotI (for CWP2) sites were generated just upstream of the start codons using two-step Pfu-polymerase PCR reactions (see above) with plasmids pGS-GFP-CWP1-low, pGS-GFP-CWP2-low, and pGS-GFP-TIP1-low as templates. pGS-EagI-CWP1 was created using primer pairs GS24/GS25 and GS26/GS27 in the first reactions and primers GS24 and GS26 in the second reaction. The final product was digested with PstI and SalI and then cloned into PstI/SalI-digested pGS-GFP-CWP1-low. pGS-NotI-CWP2 was created using primer pairs GS56/GS29 and GS57/GS31 in the first reactions and primers GS56 and GS57 in the second reaction. The final product was digested with HindIII and KpnI and then cloned into HindIII/KpnI-digested pGS-GFP-CWP2-low. The resulting plasmid was again digested with KpnI, and the 1309-base pair KpnI fragment from pGS-GFP-CWP2-low was cloned into this site. The orientation of this last insertion was checked by restriction analysis. pGS-EagI-TIP1 was created using primer pairs GS32/GS33 and GS34/GS35 in the first reactions and primers GS32 and GS34 in the second reaction. The final product was digested with RsrII and KpnI and then cloned into RsrII/KpnI-digested pGS-GFP-TIP1-low. In all three cases, insertion of the restriction sites did not affect the localization patterns of the proteins (our unpublished data). Promoters were then exchanged by digesting the plasmids with either HindIII and EagI or HindIII and NotI and then ligating promoter and residual plasmid fragments from different origins. Products were checked by restriction analysis. In this way, six new plasmids were created: pCWP1-GFP-CWP2, pCWP1-GFP-TIP1, pCWP2-GFP-CWP1, pCWP2-GFP-TIP1, pTIP1-GFP-CWP1, and pTIP1-GFP-CWP2.
The MS2 tag was cloned into plasmid pAR213 (see above) using primers GS52 and GS53 and plasmid pIII/MS2-2 (Beach et al., 1999) as template. The PCR product was digested with BamHI and cloned into the unique BclI site just downstream of the CWP1 ORF, creating pGS-CWP1-MS2.
Cell Cycle Synchronization
Some cultures were synchronized by adding hydroxyurea (Sigma-Aldrich, St. Louis, MO) to an exponential phase culture (∼3 × 106 cells/ml) at 30°C to a final concentration of 200 μM. After a 3-h incubation, the culture was released by washing three times in medium prewarmed to 30°C and incubating further at that temperature. In addition, synchronization with α-factor was performed by washing cells from an exponential phase culture (∼1.5 × 106 cells/ml) at 30°C and resuspending them at 1.5 × 105 cells/ml in medium at 30°C containing 2 μg/ml α-factor (Sigma-Aldrich). After 3 h, >90% of the cells were unbudded, and the culture was released by washing the cells with water and resuspending them at 1.5 × 106 cells/ml in medium at 25°C.
Microscopy
For observations of GFP-tagged proteins by fluorescence microscopy, cells were harvested, washed once in phosphate-buffered saline (PBS), and kept on ice for at least 15 min. For chitin staining, calcofluor white (CFW) (Sigma-Aldrich) was added to a final concentration of 20 μg/ml. In most experiments, cells were observed and imaged using an Olympus BH-2 microscope equipped with a Hamamatsu (Ammersee, Germany) C5985 charge-coupled device camera and Object Image (http://simon.bio.uva.nl/object-image.html) 1.62n2 software. In some experiments, cells were observed and photographed on a Nikon Microphot SA microscope using an Apo 60X/1.40 numerical aperture oil immersion objective and Kodak T-Max 400 film.
Cell Fractionation, Cell Wall Digestion, and Western Blotting
Cells were harvested from an exponential phase culture (∼3 × 106 cells/ml). Proteins from the culture supernatant were precipitated using the sodium deoxycholate (DOC), trichloroacetic acid (TCA) method (Ozols, 1990). First, DOC was added to a final concentration of 200 μg/ml. After 10 min at 4°C, TCA was added to a final concentration of 6% (wt/vol), and the mixture was left overnight at 4°C. The precipitate was then pelleted at 10,000 × g and washed extensively with 80% (vol/vol) acetone. Meanwhile, the cells were washed and homogenized as described previously (Montijn et al., 1994; Kapteyn et al., 1995), and cell walls were spun down at 3000 × g. The resulting supernatant was precipitated with DOC and TCA as described above (“cytosolic fraction”). Meanwhile, the pellet from the 3000 × g spin was washed extensively with 1 M NaCl and then boiled twice for 10 min in 2% SDS, 100 mM EDTA, 40 mM β-mercaptoethanol, and 50 mM Tris-HCl, pH 7.8, to solubilize noncovalently linked cell wall proteins and proteins from the plasma membrane and other membranous compartments (Klis et al., 1998). The combined supernatants constitute the “SDS-soluble fraction.” The SDS-extracted cell walls were washed with 10 mM Tris-HCl, pH 7.8, and then treated overnight at 37°C with 0.8 U of recombinant Trichoderma harzianum endo-β-1,6-glucanase (Bom et al., 1998) per gram (wet weight) of cell walls to release the GPI-dependent cell wall proteins (Kapteyn et al., 1996).
Proteins were separated on 3–20% gradient polyacrylamide gels containing 0.1% SDS (Laemmli, 1970) and transferred electrophoretically onto Immobilon polyvinylidene difluoride membranes (Millipore, Billerica, MA) (Montijn et al., 1994). Glycoproteins were visualized by staining the membranes with peroxidase-labeled 1 μg/ml concanavalin A (Sigma-Aldrich) in PBS containing 3% (wt/vol) bovine serum albumin, 2.5 mM CaCl2, and 2.5 mM MnCl2 (Klis et al., 1998). Cwp1p was also visualized using polyclonal anti-Cwp1p antiserum (Shimoi et al., 1995), as described previously (Kapteyn et al., 1996). The blots were visualized using ECL Western blotting detection reagents (Amersham Europe, Braunschweig, Germany) according to the manufacturer's instructions.
RESULTS
Incorporation of Tip1p and Cwp2p into Specific Regions of the Cell Wall and Its Dependence on Their Times of Expression
Of genes encoding the ∼40 GPI-dependent cell wall proteins in S. cerevisiae, more than half are transcribed in a cell cycle-dependent manner (see Introduction). To begin examining the consequences of such expression patterns for protein localization and function, we generated GFP-fusion genes on plasmids for three such proteins: Tip1p, Cwp2p, and Cwp1p. The GFP sequences were inserted directly downstream of the signal peptide sequences so that the first amino acids of each mature fusion protein are from GFP. The resulting fusion proteins were extractable by β-1,6-glucanase but not by SDS, indicating that they were incorporated normally into the cell wall, and they were recognized by the lectin concanavalin A after separation by SDS-PAGE, indicating that they were glycosylated (our unpublished data). The genes encoding these proteins are expressed in different phases of the cell cycle: TIP1 is transcribed in late M and/or early G1 phase, well before bud emergence, whereas CWP1 and CWP2 are transcribed in S/G2 phase (Caro et al., 1998; Spellman et al., 1998).
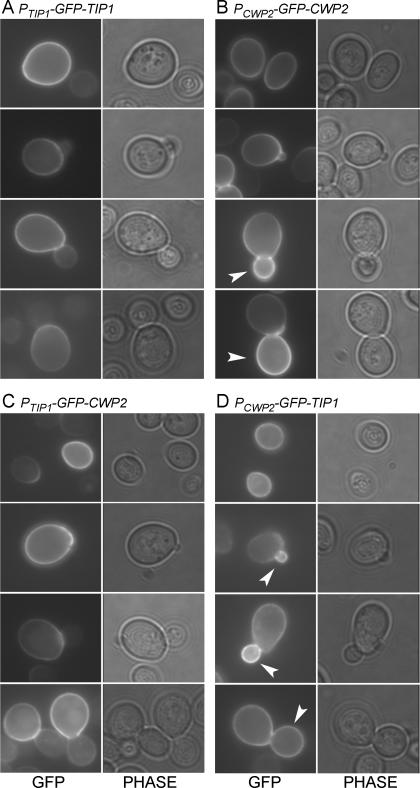
GFP-Tip1p showed little, if any, incorporation into buds: unbudded cells fluoresced at the cell surface, as did the mother portions of budded cells, but little fluorescence was seen in buds of any size (Figure 1A and Table 2). These observations suggest that Tip1p is incorporated into the cell wall predominantly or exclusively in unbudded, G1-phase cells, soon after TIP1 is transcribed.
Figure 1.
Localization of GFP-Tip1p and GFP-Cwp2p. Fusion proteins were expressed from low-copy plasmids in strain FY833. For each construct, >200 cells showing wall fluorescence were observed, and representative cells from four successive phases of the cell cycle are shown. (A) GFP-Tip1p expressed from its own promoter (plasmid pGS-GFP-TIP1-low). (B) GFP-Cwp2p expressed from its own promoter (plasmid pGS-GFP-CWP2-low). (C) GFP-Cwp2p expressed from the TIP1 promoter (plasmid pTIP1-GFP-CWP2). (D) GFP-Tip1p expressed from the CWP2 promoter (plasmid pCWP2-GFP-TIP1). Phase, phase-contrast images; arrowheads, buds with strong fluorescence.
Table 2.
Distribution of GFP fluorescence
| Promoter | GFP-fusion | Small-budded cellsa |
Large-budded cellsa |
||||
|---|---|---|---|---|---|---|---|
| b < m (%) | b = m (%) | b > m (%) | b < m (%) | b = m (%) | b > m (%) | ||
| TIP1 | Tip1p | 100 | 0 | 0 | 98.5 | 1b | 0.5b |
| Cwp2p | 100 | 0 | 0 | 98 | 2b | 0 | |
| Cwp1p | 100 | 0 | 0 | 99 | 1b | 0 | |
| CWP2 | Cwp2p | 19 | 31 | 50 | 14 | 71 | 15 |
| Tip1p | 40 | 25 | 35 | 21 | 67 | 12 | |
| CWP1 | Cwp2p | 30 | 21 | 49 | 8 | 67 | 25 |
| Tip1p | 32 | 16 | 52 | 11 | 57 | 32 | |
a From cultures of cells expressing each construct (see Figures 1 and 2), we counted ≥100 budded cells with bud diameter <50% that of the mother cell (small-budded cells) and ≥100 budded cells with bud diameter >50% that of the mother (large-budded cells). Bud wall fluorescence intensity was scored as lower than (b < m), equal to (b = m), or greater than (b > m) that of the mother.
b Examination of the CFW staining suggested that each of these cells had a septum and thus that each was actually a pair of unbudded cells.
Similarly, GFP-Cwp2p fluorescence was readily detectable in unbudded cells and in the mother portions of budded cells but was weak or undetectable in very small buds (Figure 1B). However, in contrast to GFP-Tip1p, GFP-Cwp2p signal was clearly present in most medium-sized and large buds and indeed was usually as strong or stronger there than in the corresponding mother cells (Figure 1B and Table 2). In a culture synchronized with α-factor, GFP-Cwp2p fluorescence was also clearly present in the growing buds observed after release and was almost always stronger there than in the mother cells (our unpublished observations). These observations suggest that Cwp2p is incorporated into the cell wall predominantly or exclusively during S/G2 phase, soon after CWP2 is transcribed. Further bud growth after the bulk of GFP-Cwp2p has been incorporated could explain why there is a decrease in the percentage of buds that fluoresce very strongly in large-budded cells compared with small-budded cells (Table 2).
The actin cytoskeleton is responsible for directing secretory vesicles to the sites of cell surface growth, and the apparent timing of Tip1p and Cwp2p incorporation correlates in each case with the polarization status of the actin cytoskeleton (see Introduction) at the time of gene expression. We therefore hypothesized that the timing of transcription might determine the observed patterns of incorporation. To test this, we exchanged the promoters of the two genes. Strikingly, this exchange essentially reversed the incorporation patterns of the two GFP-fusion proteins. When expressed from the TIP1 promoter, GFP-Cwp2p was no longer incorporated into growing buds but was instead found almost exclusively in unbudded cells and in the mother portions of budded cells (Figure 1C and Table 2). Conversely, GFP-Tip1p expressed from the CWP2 promoter was preferentially incorporated into medium-sized buds, which typically fluoresced more brightly than their mothers (Figure 1D and Table 2). Thus, the localizations of Tip1p and Cwp2p to specific regions of the wall seem to be determined largely or entirely by their times of expression during the cell cycle, as controlled by the 5′-upstream sequences.
Localization of Cwp1p to Birth Scars
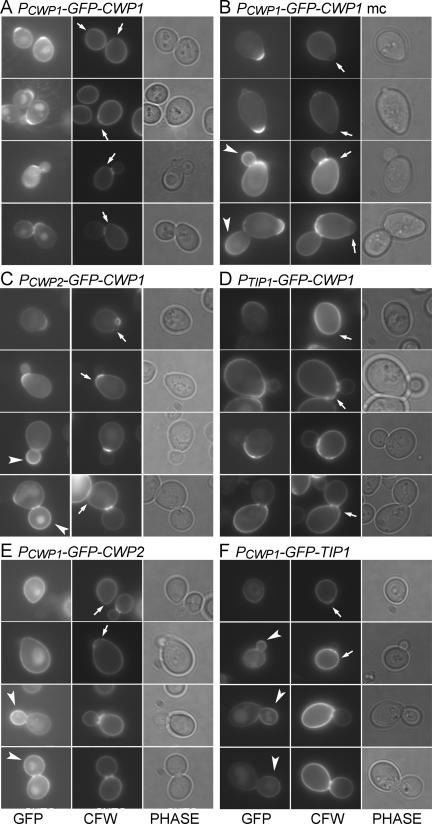
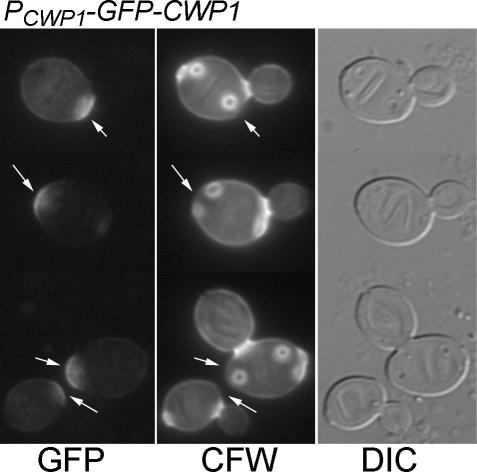
CWP1 is expressed concurrently with CWP2 (Caro et al., 1998; Spellman et al., 1998). However, the incorporation patterns of GFP-Cwp2p and GFP-Cwp1p were very different. When expressed under its own promoter from a low-copy vector, GFP-Cwp1p was incorporated mainly in the region of the birth scar (which is marked by a relatively low intensity of CFW staining and by the somewhat protuberant shape of the cell; Chant and Pringle, 1995) at one pole of the cell (Figure 2A). In many cases, some GFP fluorescence was also seen in the lateral walls of the cells, but the signal at the birth scar was much more intense, and lateral wall signal was never observed in the absence of birth scar signal. In addition, some intracellular GFP signal was often observed (Figure 2A). This signal probably corresponded to the vacuole (Kunze et al., 1999); its strength varied greatly between experiments (cf. Figure 2B). The signal at the birth scar was stable: cells with more than four bud scars were observed that still showed a clear GFP-Cwp1p signal at the birth scar (Figure 3).
Figure 2.
Localization of GFP-Cwp1p to the birth scar and its dependence on both promoter sequences and sequences within or downstream of the coding region. GFP-tagged proteins were expressed from low-copy (A and C–F) or high-copy (B) plasmids in strain FY833. For each construct, >200 cells showing wall fluorescence were observed, and representative cells from four successive phases of the cell cycle are shown. (A and B) GFP-Cwp1p expressed from its own promoter (plasmids pGS-GFP-CWP1-low and pAR213). (C) GFP-Cwp1p expressed from the CWP2 promoter (plasmid pCWP2-GFP-CWP1). (D) GFP-Cwp1p expressed from the TIP1 promoter (plasmid pTIP1-GFP-CWP1). (E) GFP-Tip1p expressed from the CWP1 promoter (plasmid pCWP1-GFP-TIP1). (F) GFP-Cwp2p expressed from the CWP1 promoter (plasmid pCWP1-GFP-CWP2). Phase, phase-contrast images; arrows, birth scars; arrowheads, buds with strong fluorescence.
Figure 3.
Stability of Cwp1p birth scar localization. Cells of strain LSY265 containing plasmid pGS-GFP-CWP1-low were grown at 30°C and stained with CFW, and cells were observed by DIC and fluorescence microscopy. Arrows, birth scars.
When the same GFP-Cwp1p construct was expressed from a high-copy vector, there was strong GFP fluorescence in medium-sized and large buds as well as strong signal at the birth scars (Figure 2B; Ram et al., 1998a). The bud fluorescence resembled that seen with GFP-Cwp2p (Figure 1B), suggesting that the mechanism for directing Cwp1p specifically to the birth scar is saturable and that excess protein is incorporated into the bud wall by the “default” pathway for S/G2-synthesized cell wall proteins (as seen normally with GFP-Cwp2p).
Dependence of Cwp1p Localization on Both Promoter and Other Sequences
To begin investigating the mechanisms of Cwp1p localization, we performed additional promoter-exchange experiments. When expressed from the CWP2 promoter, GFP-Cwp1p localized both to birth scars and to medium-sized and large buds (Figure 2C), much as when it was expressed from its own promoter on a high-copy plasmid (Figure 2B). These results seem consistent with the data indicating that the CWP1 and CWP2 promoters are expressed concurrently (see above) but with substantially stronger expression from the CWP2 promoter in the rich medium used here (Wodicka et al., 1997). Thus, upon expression from the CWP2 promoter, the mechanism for targeting Cwp1p to the birth scar would be saturated, and the excess protein would follow the default pathway to the surface of the bud.
In contrast, when GFP-Cwp1p was expressed from the TIP1 promoter, its localization resembled that of GFP-Tip1p (Figure 1A and Table 2) or of GFP-Cwp2p expressed from the TIP1 promoter (Figure 1C and Table 2); that is, fluorescence was seen almost exclusively in unbudded cells and in the mother portions of budded cells (Figure 2D and Table 2). Although this fluorescence sometimes seemed stronger around the birth and bud scars than elsewhere on the cell surface, it was much less localized than when GFP-CWP1 was expressed at its normal time in the cell cycle. These results suggest two conclusions. First, incorporation into the unbudded cell wall (and hence into the mother cell wall) seems to be a default pathway for cell wall proteins expressed in late M/early G1 phase, because all three proteins expressed from the TIP1 promoter followed this pattern (Table 2). Second, normal timing of expression (based on an appropriate promoter) seems to be critical for proper Cwp1p localization.
However, expression from the CWP1 promoter is not sufficient to target a protein to the birth scar: when either GFP-Tip1p or GFP-Cwp2p was expressed from this promoter, it was incorporated generally into growing buds (Figure 2, E and F, and Table 2), essentially as seen when either protein was expressed from the CWP2 promoter (see Figure 1, B and D). Thus, sequences downstream of the promoter, either in the coding region or in the 3′-untranslated region, also seem to be critical for normal Cwp1p localization, and proteins without such sequences seem to follow the default pathway for G2-synthesized cell wall proteins.
The Lag between CWP1 Transcription and Cwp1p Incorporation
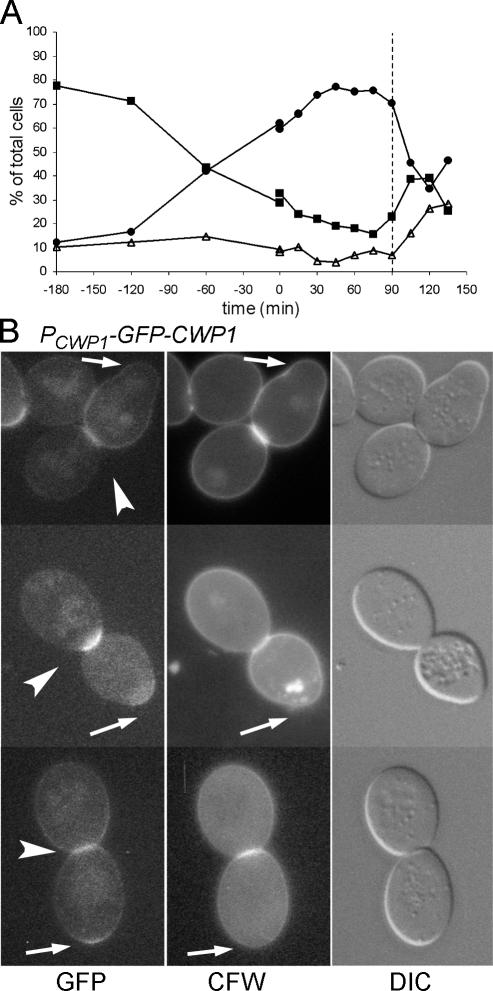
Although CWP1 is transcribed in late S to early G2 phase when the nucleus is in the mother cell, Cwp1p seems to be incorporated into the cell wall much later and specifically in the daughter cell, because the birth scar region of the wall is synthesized after cytokinesis as the secondary septum on the daughter side of the neck. This hypothesis was supported by observations on cells that had been synchronized at the DNA replication checkpoint using hydroxyurea. Even 90 min after release, when the cells were about to begin separating (Figure 4A), most cells did not show GFP-Cwp1p fluorescence at the neck (our unpublished data). The cells that did had already formed septa, as seen from the differential interference contrast (DIC) microscopy images and CFW staining (Figure 4B). Similarly, in an unsynchronized population of the same strain, 43 of 43 large-budded cells with GFP-Cwp1p fluorescence at the neck had completed septum formation as judged by DIC. In later samples of the synchronized population, most cells had separated, and most daughter cells did show fluorescence at the birth scar (our unpublished data). Thus, it seems that GFP-Cwp1p is indeed incorporated primarily soon after septum formation, just before cell separation in some cells, and probably during or after cell separation in others.
Figure 4.
Incorporation of GFP-Cwp1p into the secondary septum of the daughter cell after cytokinesis. Cells of strain LSY265 containing plasmid pGS-GFP-CWP1-low were synchronized by adding hydroxyurea at t = −180 min and washing it out at t = 0 (see Materials and Methods). (A) Synchronization shown as percentages of unbudded (■), small-budded (▵), and large-budded (●) cells. (B) Fluorescence and DIC images of large-budded cells from the 90-min sample (A, dashed line). Arrows, birth scars of the mother cells; arrowheads, GFP-Cwp1p on the daughter side of the necks.
These observations imply that there is a delay either in the translation of CWP1 mRNA or in the incorporation of Cwp1p into the wall. If the mRNA were translated immediately after transcription, Cwp1p should be detectable in an intracellular compartment where it would reside (in amounts similar to those finally incorporated) until incorporation into the wall. However, immunoblot analysis of asynchronously growing cultures showed that the Cwp1p in the cytosolic and SDS-soluble fractions together amounted to considerably <10% of the Cwp1p extractable from the cell wall using β-1,6-glucanase (Figure 5). It is therefore unlikely that the protein resides intracellularly, or even in the plasma membrane, for any significant period.
Figure 5.
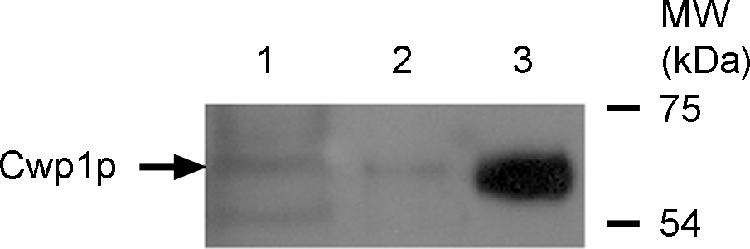
Abundance of Cwp1p in different cellular fractions. Proteins from strain FY833 were fractionated as described in Materials and Methods and analyzed by Western blotting using anti-Cwp1p antibodies. Lane 1, cytosolic fraction; lane 2, SDS-soluble fraction; lane 3, proteins released by β-1,6-glucanase digestion. Equal cell equivalents were loaded in each lane. The culture supernatant was also examined (see Materials and Methods) to determine whether Cwp1p was released from the cells (Ram et al., 1998b), but no Cwp1p was detected.
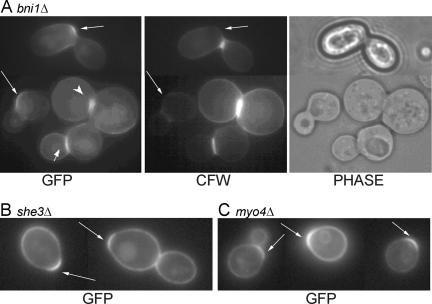
Although CWP1 is transcribed in the mother-cell nucleus before nuclear division, its product is found specifically in the daughter cell. A similar situation is known for the transcription factor Ash1p (Bobola et al., 1996; Sil and Herskowitz, 1996). In this case, the ASH1 mRNA is delivered specifically into the bud, and translation is inhibited while the mRNA is transported (Long et al., 1997; Takizawa et al., 1997). Several genes involved in this process have been identified, including BNI1, SHE3, and MYO4 (Jansen et al., 1996; Takizawa et al., 1997; Takizawa and Vale, 2000; Beach and Bloom, 2001), and it has been proposed that stem-loop structures present in the 3′-untranslated region of the mRNA are important for its localization (Chartrand et al., 1999, 2002; Gonzalez et al., 1999). Although the CWP1 mRNA also contains a large predicted stem-loop structure in its 3′-untranslated region (according to the Mfold 2.3 and 3.0 algorithms [Zuker et al., 1999; Mathews et al., 1999]), two types of experiments suggested that mRNA localization is not involved in the asymmetric distribution of Cwp1p. First, deletion of BNI1, SHE3, or MYO4 did not prevent the asymmetric localization of GFP-Cwp1p to the birth scar in many cells (Figure 6). Second, direct examination of CWP1 mRNA localization using the MS2 CP-GFP system (Beach et al., 1999) in strain JV96 did not detect any specific localization to the bud (our unpublished observations).
Figure 6.
Asymmetric localization of Cwp1p in mutants defective in mRNA localization. Strains (A) M-717 (bni1Δ/bni1Δ), (B) ICY018 (she3Δ/she3Δ), and (C) KBY1012 (myo4Δ/myo4Δ) were transformed with plasmid pGS-GFP-CWP1-low and examined for the localization of GFP-Cwp1p. Phase, phase-contrast images; long arrows, birth scars; short arrow, a septum with GFP-Cwp1p on the daughter side; arrowhead, GFP-Cwp1p localized on both sides of an incomplete septum.
Dependence of Cwp1p Localization on Normal Septum Formation
The incorporation of Cwp1p into the daughter cell wall soon after septum formation suggested that the two processes might be linked. To explore this possibility, we examined the localization of GFP-Cwp1p in several mutants in which septum formation is perturbed. Although mutants lacking the formin Bni1p are viable, bni1Δ/bni1Δ diploid cells often display defects such as the formation of incomplete, asymmetric, and/or misplaced septa and a delay in cell separation (Bi et al., 2000; Vallen et al., 2000). In a wild-type strain, unseparated cells that already had detectable GFP-Cwp1p signal at the daughter side of the septum were uncommon (see above), but such cells were frequently observed in a bni1Δ/bni1Δ strain (Figure 6A, short arrow), even when the septum was misplaced or otherwise abnormal, suggesting that Cwp1p incorporation followed closely upon septum formation even when cell separation was delayed. Moreover, in some cells in which the septum was incomplete and only partially traversed the neck, GFP-Cwp1p was present on both sides of the septum (Figure 6A, arrowhead), supporting the hypothesis that Cwp1p incorporation is linked both spatially and temporally to septum formation.
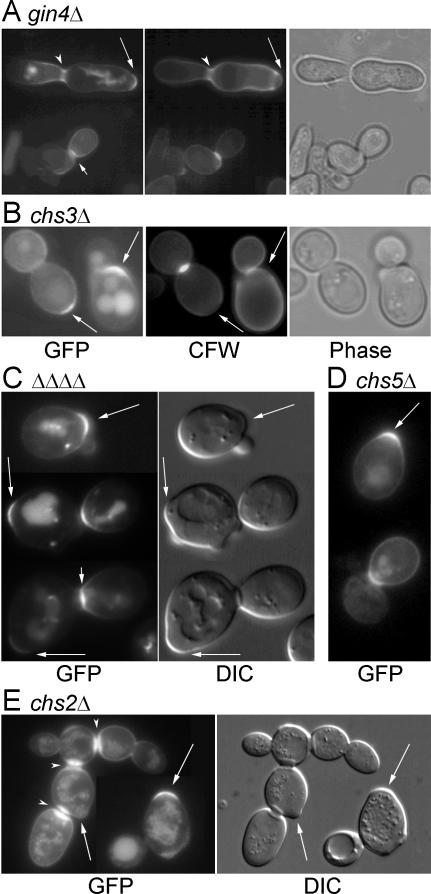
This hypothesis received further support from observations on mutant cells lacking Gin4p, a protein kinase that is involved in the organization of the ring of septin proteins at the neck, which in turn influences the pattern of septum formation (Longtine et al., 1996, 1998b, 2000; Gladfelter et al., 2001; Caviston et al., 2003). In a gin4Δ/gin4Δ diploid strain, many cells showed seemingly normal GFP-Cwp1p localization to the birth scar and to the daughter side of the neck in unseparated cells with relatively normal-looking septa (Figure 7A, long and short arrows). However, in many other cells, the neck region seemed elongated and relatively disorganized, and GFP-Cwp1p was observed in bars or patches that seemed to coincide with patches of increased CFW staining Figure 7A, arrowheads). These observations suggested that Cwp1p incorporation might be linked specifically to the deposition of septal chitin.
Figure 7.
Dependence of Cwp1p localization on normal septum formation. Strains (A) M-272 (gin4Δ/gin4Δ), (B) DDY181-2D (chs3-Δ1), (C), KNY1048 (chs6Δ/chs6Δ bud7Δ/bud7Δ ymr237wΔ/ymr237wΔ ykr027wΔ/ykr027wΔ), (D) AM775 (chs5Δ/chs5Δ), and (E) ICY028 (chs2Δ/chs2Δ) were transformed with plasmid pGS-GFP-CWP1-low and examined for the localization of GFP-Cwp1p. KNY1048 and ICY028 were grown at 30°C. Long arrows, birth scars; short arrows, fluorescent daughter sides of septa; arrowheads, GFP-Cwp1p spread through an elongated neck (A) or on both sides of an aberrant septum (E).
To investigate this possibility further, we first examined mutants defective in Chs3p, the catalytic subunit of chitin synthase III, or in one of its accessory proteins. Chitin synthase III is primarily responsible for the synthesis of the chitin ring at the incipient bud site and the chitin of the lateral cell wall (Shaw et al., 1991; Kollár et al., 1995; Cabib and Durán, 2005) as well as for increased chitin synthesis in response to various stresses (Valdivieso et al., 2000; Valdivia and Schekman, 2003), but it can also contribute to the formation of a remedial septum in the absence of CHS2 (Cabib and Schmidt, 2003). However, GFP-Cwp1p seemed to localize normally in a chs3Δ mutant (Figure 7B); in a mutant (DDY172-2A) with a deletion of CHS4, which encodes a protein necessary for the normal localization and activation of Chs3p (DeMarini et al., 1997; Trilla et al., 1997; Ono et al., 2000) (our unpublished data); and in a mutant with a deletion of CHS6, which encodes a protein involved in the trafficking of Chs3p through the secretory system (Ziman et al., 1998; Valdivia et al., 2002; Valdivia and Schekman, 2003) (our unpublished data).
Chs6p is one of a family of four proteins in S. cerevisiae. One of its homologues, Bud7p, is implicated in the vesicular transport of the bud site selection marker proteins Bud8p and Bud9p (Zahner et al., 1996; our unpublished data), and it seems likely that the other homologues, Ykr027p and Ymr237p, play similar roles in the transport of other specialized cargoes to specific sites at the cell surface. Thus, we also examined GFP-Cwp1p localization in a strain with deletions of these four genes as well as in a mutant with a deletion of CHS5, which encodes a protein that seems to function together with all four members of the Chs6p/Bud7p family (Valdivia et al., 2002; Sanchatjate and Schekman, personal communication; Trautwein and Spang, personal communication; Nakashima, McKenzie, and Pringle, unpublished data). However, in both cases, GFP-Cwp1p still seemed to localize normally to birth scars (Figure 7, C and D).
In contrast to the results with mutants lacking Chs3p or its accessory proteins, Cwp1p localization was strongly affected by an absence of chitin synthase II (Chs2p), the enzyme primarily responsible for synthesis of the primary septum (Shaw et al., 1991; Schmidt et al., 2002; Lesage et al., 2005). In a chs2 deletion strain, GFP-Cwp1p localization was rather variable from cell to cell. Nonetheless, many cells did show a well defined concentration of GFP-Cwp1p at the birth scar (Figure 7E). In addition, in the many necks that persist in the clumps of such a strain (reflecting its inefficient septum formation and cell separation; Shaw et al., 1991; Schmidt et al., 2002), GFP-Cwp1p was often highly concentrated. However, the fluorescence signal was typically bright on both sides of the neck (Figure 7E), instead of showing the normal restriction to the daughter side. These data suggest that the asymmetric localization of Cwp1p to the nascent birth scar requires the normal timing of septum formation, normal septum structure, or both.
Possible Functions of Cwp1p
Previous studies have reported that deletion of CWP1 is nonlethal and indeed produces no obvious phenotype other than a slightly increased sensitivity to cell surface-interactive chemicals (Shimoi et al., 1995; van der Vaart et al., 1995; Dielbandhoesing et al., 1998). We also found that haploid and diploid cwp1Δ cells did not differ significantly from wild type in growth rate, overall cell morphology, or appearance after staining chitin with CFW (our unpublished data). The localization of Cwp1p to the birth scar suggested that it might have a role in cell division and/or in bud site selection (which involves cortical marker proteins that are localized to the cell poles; Lord et al., 2000; Harkins et al., 2001; Kang et al., 2004). However, because the percentage of budded cells with septa visible by DIC microscopy did not differ between cwp1Δ and wild-type strains (31 and 32% for exponentially growing populations of strains LSY265 and YEF473, respectively), there seemed to be no delay in cell separation. Moreover, using several approaches, we were unable to obtain any evidence for a role of Cwp1p in bud-site selection or for interaction of Cwp1p with the bud site selection marker proteins. Haploid and diploid cwp1Δ cells displayed normal axial and bipolar budding; localization of the marker protein Bud9p to the birth scar was unaffected in cwp1Δ cells; and Cwp1p localization to the birth scar seemed to be independent of Bud9p and of the other marker proteins Bud8p, Rax2p, and Axl2p as well as of Bud7p and Rax1p, which seem to be involved in the trafficking of Bud8p, Bud9p, and Rax2p (Supplemental Figure 1).
DISCUSSION
Localized Incorporation of Yeast Cell Surface Proteins
Although studies of individual proteins and several large-scale surveys (Kumar et al., 2002; Huh et al., 2003) have described the localizations of many yeast proteins, cell surface proteins have mostly escaped attention because of the technical constraints imposed by their specific requirements for incorporation into the plasma membrane or cell wall. Thus, surprisingly little is known about the specific localizations of integral and GPI-anchored plasma membrane proteins, PIR proteins, and GPI-dependent cell wall proteins. Among the exceptions are the integral membrane proteins involved in bud site selection, which localize to the division sites and cell poles as noted above, and the enzymes that synthesize cell wall chitin and β-1,3-glucan, which localize dynamically to the bud neck or bud tip at appropriate times in the cell cycle (Chuang and Schekman, 1996; Drgonová et al., 1996; DeMarini et al., 1997; Utsugi et al., 2002). In addition, specific localizations have been reported for several GPI-dependent cell wall proteins, including the α-agglutinin Sag1p/Agα1p, which was found at the projection (shmoo) tip of Mata cells after stimulation with Mata mating pheromone (Wojciechowicz and Lipke, 1989), and the proteins Crh1p and Crh2p, which were found in small buds and the neck region (Crh1p) and all over the wall but preferentially in the neck region (Crh2p) (Rodríguez-Peña et al., 2000). At least in pheromone-treated cells, such localizations may reflect the distribution of lipid rafts enriched in the GPI-proteins (Bagnat and Simons, 2002).
In this study, we have demonstrated specific patterns of incorporation for three additional GPI-dependent cell wall proteins. We found Tip1p to be incorporated specifically in mother cells before bud emergence, Cwp2p to be incorporated specifically in small to medium-sized buds, and Cwp1p to be incorporated stably into the birth scar region. In the cases of Cwp1p and Cwp2p, these observations differ somewhat from those reported previously (Ram et al., 1998a), probably because the earlier study used high-copy plasmids that resulted in overexpression of the GFP-tagged proteins, whereas we have now expressed the proteins from their normal promoters on low-copy plasmids.
Mechanisms for Localized Incorporation of Cell Surface Proteins
In S. cerevisiae, the actin cytoskeleton is responsible for delivering secretory vesicles to specific areas of the cell surface; as actin organization changes during the cell cycle, the patterns of vesicle delivery show corresponding changes (Lew and Reed, 1995; Pringle et al., 1995; Pruyne and Bretscher, 2000b). Because plasma membrane and cell wall proteins travel to the cell surface in secretory vesicles, and because the traffic of such vesicles to the cell surface is normally rapid (Novick et al., 1981; Pastor et al., 1982; Novick and Schekman, 1983), it seems that in the absence of additional mechanisms, the synthesis of such a protein at a specific time in the cell cycle would automatically determine its incorporation into the membrane or wall in a specific pattern. Thus, the normal pulse of AXL2 expression in late G1 seems to allow the incorporation of Axl2p specifically at the presumptive bud site (at which it remains as the bud emerges); artificial expression of AXL2 in S/G2 led to the incorporation of Axl2p throughout the bud membrane (Lord et al., 2000). Similarly, the induction of α-agglutinin expression by Mata pheromone (Lipke et al., 1989), coinciding with the polarization of the actin cytoskeleton toward the pheromone source (Hašek et al., 1987), seems to be sufficient to explain its localized incorporation into the shmoo tip (Wojciechowicz and Lipke, 1989). This simple model also seems to explain the behavior of the cell wall proteins Tip1p and Cwp2p: when either of these proteins is expressed from the TIP1 promoter in late M/early G1 phase, it is incorporated specifically into the wall of the unbudded G1 cell and not into growing buds, but when either protein is expressed from the CWP2 or CWP1 promoter in S/early G2 phase, it is incorporated specifically into the wall of the growing bud. In each case, the site of incorporation coincides with the directionality of actin-based secretion at that phase of the cell cycle.
In contrast, the behavior of Cwp1p presents a more complicated picture. The normal expression of CWP1 in late S/early G2 seems to be critical for the normal pattern of Cwp1p incorporation, because expression from the TIP1 promoter in late M/early G1 causes Cwp1p to be incorporated specifically, and without apparent delay, into the wall of the unbudded cell, much as are Tip1p and Cwp2p when expressed at this time (although the Cwp1p pattern does show more heterogeneity than is evident with either of the other two proteins). However, when Cwp1p is expressed at normal levels from its own promoter in late S/early G2, it is incorporated with high specificity, and after a long delay, into the birth scar of the daughter cell. This behavior is strikingly different from that of Tip1p and Cwp2p when expressed at the same time, indicating that the CWP1-encoded mRNA and/or protein must contain (a) signal(s) that diverts Cwp1p from the general flow of secretory material produced around the time at which CWP1 is transcribed. Interestingly, the mechanism that produces this diversion seems to be saturable, because when CWP1 is overexpressed in late S/early G2 (by being expressed either from its own promoter on a high-copy plasmid or from the stronger CWP2 promoter), it is incorporated into the surface of the growing bud as well as at the birth scar.
An explanation for the behavior of Cwp1p must account for at least two unusual features, namely, the long delay between gene transcription and protein incorporation into the wall and the incorporation specifically into the daughter side of the septum. Both features might have a common explanation if the CWP1 mRNA, like the mRNAs of ASH1 (Chartrand et al., 2002; Gu et al., 2004) and certain other genes (Shepard et al., 2003; Gerber et al., 2004), were delayed in translation while being transported specifically into the bud, where it might be translated only at the time of cytokinesis, when the actin cytoskeleton would deliver it to the forming septum. This mechanism seemed the more likely because a large stem-loop structure, such as is thought to be critical for the transport of ASH1 mRNA (Chartrand et al., 1999, 2002; Gonzalez et al., 1999), is also predicted to be present in the 3′-untranslated region of the CWP1 mRNA. However, we could obtain no evidence to support this hypothesis. In particular, we could detect no enrichment of CWP1 mRNA in the bud using methods that have worked for other mRNAs, and we found that normal Cwp1p localization did not depend on any of several proteins that are essential for ASH1 mRNA transport. CWP1 mRNA was also not detected in other studies of the mRNAs that interact with known components of the mRNA-localization machinery (Shepard et al., 2003; Gerber et al., 2004). The localization of Cwp1p to the bud surface when CWP1 is transcribed at the normal time in the cell cycle but at higher-than-normal levels showed that CWP1 mRNA does have the potential to be translated at that time. However, because we did not detect any substantial pool of Cwp1p other than that covalently bound to the cell wall, we conclude that a delay in mRNA translation must account, at least in part, for the delay in Cwp1p incorporation into the wall. The mechanism of this translation delay seems to be novel and should be interesting to investigate further.
Another interesting feature of Cwp1p incorporation is its apparent dependence on the normal timing of septum formation and/or the normal structure of the septum. Most conspicuously in the chs2 mutant, but also in some cells of the bni1 and gin4 mutants, Cwp1p was found localized to both sides of the septal region rather than asymmetrically to the daughter side. This phenotype may simply reflect the inefficient septum formation in these mutants; thus, if Cwp1p is delivered to the neck before the septum is complete, it may become incorporated symmetrically along with other septum components rather than being confined to the daughter side. Moreover, the remedial septum in a chs2 mutant is formed differently from the normal primary and secondary septa in wild-type cells, which may also disturb the generation of asymmetry. Alternatively, the altered distribution of Cwp1p may reflect a specific association with other septal components whose distribution is altered in these mutants.
Functional Significance of Localized Incorporation of Cell Wall Proteins
What purposes are served by the localized incorporation of cell wall proteins? The cell cycle-regulated incorporation of the bulk of the cell wall mannoproteins into the wall (presumably of the bud) in G2/M phase has been shown to correlate with a decreased cell wall permeability at that stage (de Nobel et al., 1991), which may result from an increased cross-linking of the glucan network by some of the proteins. However, it is difficult to see the significance of the localized incorporation of Tip1p (in unbudded cells) and Cwp2p (in buds). Because every unbudded cell was once a bud and every bud soon becomes an unbudded cell, the initial asymmetry generated by this localized incorporation is only transient, and lasts less than one cell cycle. Because the phenotypes of tip1Δ and cwp2Δ mutants are very mild (van der Vaart et al., 1995; Dielbandhoesing et al., 1998), they also provide little guidance, and we are presently without a clear hypothesis as to the functions of these proteins or of their cell cycle-regulated and initially asymmetric incorporation.
In the case of Cwp1p, its localization primarily to the birth scar and persistence at that site suggest strongly that it should have a role in some specific function of that region of the cell wall. One well established function of the birth scar pole is that it houses both the transient marker used in the axial budding pattern of haploid cells and a persistent marker used in the bipolar budding pattern of diploid cells (see above). However, despite performing a seemingly full suite of tests, we were unable to detect any role of Cwp1p in either axial or bipolar budding or any functional interaction of Cwp1p with the proteins that serve as the bud site selection markers.
During the separation of mother and daughter cells, digestion of the chitinous primary septum proceeds from the daughter side (Colman-Lerner et al., 2001; Ríos Muñoz et al., 2003), with the result that the division site on the mother cell (the bud scar) retains both a thick chitin ring and a layer of chitin across the entire scar, whereas the division site on the daughter cell (the birth scar) contains a thinner or nonexistent chitin ring and little or no chitin in the wall within the ring (Streiblová and Beran, 1963; Beran et al., 1972; Molano et al., 1980; Roberts et al., 1983; Shaw et al., 1991; Chant and Pringle, 1995; Powell et al., 2003). Perhaps for this reason, the bud scar is a rigid structure that expands little or not at all as the cell grows during subsequent generations, whereas the birth scar is a more plastic structure that expands both during division and during the subsequent growth of the cell (Bartholomew and Mittwer, 1953; Streiblová and Beran, 1963; Bacon et al., 1966; Talens et al., 1973; Molano et al., 1980; Chant and Pringle, 1995; Powell et al., 2003). Thus, it seems plausible that the birth scar might be an intrinsically weaker structure than the bud scar and the rest of the cell wall, and thus in need of reinforcement from special birth scar-localized proteins such as Cwp1p, whose expression is known to be induced by cell wall stress (Ram et al., 1998b; Jung and Levin, 1999; Terashima et al., 2000), and that can cross-link the glucan network because of its multiple possible linkages (Kapteyn et al., 2001). However, cwp1Δ cells form septa at normal rates, have bud and birth scars of normal shape, and are hardly, if at all, more susceptible to environmental stress (van der Vaart et al., 1995; Dielbandhoesing et al., 1998; our unpublished data; see Results). Thus, we are presently without a good clue to the function of Cwp1p or of its specific localization to the birth scar.
It remains possible that the roles of Tip1p, Cwp2p, and Cwp1p are obscured because these proteins are functionally redundant with other proteins that have not yet been identified or tested for such possible redundancy. If further studies bring deeper insight into the specific roles of these proteins in the cell wall, this may also clarify the functional significance, if any, of their specifically timed, and thus localized, patterns of incorporation.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Klis laboratory, particularly H. de Nobel, for critical discussion of the manuscript. T. van Rij and M. J. Kedde helped greatly with the experimental work during their undergraduate research. We thank H. Sietsma for many useful discussions. We thank D. L. Beach and K. Bloom for assistance with the mRNA-localization studies and for the generous gift of strains and plasmids. G.J.S. much appreciated the warm reception in the Pringle laboratory, and we thank K. Nakashima, I.-C. Yu, and A. McKenzie III for the use of unpublished strains and information. This work was supported by the council for Earth and Life Sciences (ALW) from the Netherlands Organization for Scientific Research (NWO) and by National Institutes of Health Grant GM-31006 (to J.R.P.).
Abbreviations used:
- CFW
calcofluor white
- DOC
sodium deoxycholate
- GFP
green fluorescent protein
- GPI
glycosylphosphatidyl-inositol
- ORF
open reading frame
- TCA
trichloroacetic acid
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-08-0738) on May 3, 2006.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Adams A., Gottschling D., Kaiser C. A. Methods in Yeast Genetics. A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Struhl K., editors. Current Protocols in Molecular Biology. Hoboken, NJ: John Wiley & Sons; 1995. [Google Scholar]
- Bacon J.S.D., Davidson E. D., Jones D., Taylor I. F. The location of chitin in the yeast cell wall. Biochem. J. 1966;101:36C–38C. doi: 10.1042/bj1010036c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnat M., Simons K. Cell surface polarization during yeast mating. Proc. Natl. Acad. Sci. USA. 2002;99:14183–14188. doi: 10.1073/pnas.172517799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomew J. W., Mittwer T. Demonstration of yeast bud scars with the electron microscope. J. Bacteriol. 1953;65:272–275. doi: 10.1128/jb.65.3.272-275.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin A., Ozier-Kalogeropoulos O., Denouel A., Lacroute F., Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach D. L., Bloom K. ASH1 mRNA localization in three acts. Mol. Biol. Cell. 2001;12:2567–2577. doi: 10.1091/mbc.12.9.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach D. L., Salmon E. D., Bloom K. Localization and anchoring of mRNA in budding yeast. Curr. Biol. 1999;9:569–578. doi: 10.1016/s0960-9822(99)80260-7. [DOI] [PubMed] [Google Scholar]
- Beran K., Holan Z., Baldrián J. The chitin-glucan complex in Saccharomyces cerevisiae. I. IR and X-ray observations. Folia Microbiol. 1972;17:322–330. doi: 10.1007/BF02884098. [DOI] [PubMed] [Google Scholar]
- Bi E., Chiavetta J. B., Chen H., Chen G. C., Chan C. S., Pringle J. R. Identification of novel, evolutionarily conserved Cdc42p-interacting proteins and of redundant pathways linking Cdc24p and Cdc42p to actin polarization in yeast. Mol. Biol. Cell. 2000;11:773–793. doi: 10.1091/mbc.11.2.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E., Pringle J. R. ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:5264–5275. doi: 10.1128/mcb.16.10.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobola N., Jansen R.-P., Shin T. H., Nasmyth K. Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell. 1996;84:699–709. doi: 10.1016/s0092-8674(00)81048-x. [DOI] [PubMed] [Google Scholar]
- Bom I. J., Dielbandhoesing S. K., Harvey K. N., Oomes S. J., Klis F. M., Brul S. A new tool for studying the molecular architecture of the fungal cell wall: one-step purification of recombinant trichoderma β-(1–6)-glucanase expressed in Pichia pastoris. Biochim. Biophys. Acta. 1998;1425:419–424. doi: 10.1016/s0304-4165(98)00096-8. [DOI] [PubMed] [Google Scholar]
- Cabib E., Durán A. Synthase III-dependent chitin is bound to different acceptors depending on location on the cell wall of budding yeast. J. Biol. Chem. 2005;280:9170–9179. doi: 10.1074/jbc.M414005200. [DOI] [PubMed] [Google Scholar]
- Cabib E., Schmidt M. Chitin synthase III activity, but not the chitin ring, is required for remedial septa formation in budding yeast. FEMS Microbiol. Lett. 2003;224:299–305. doi: 10.1016/S0378-1097(03)00477-4. [DOI] [PubMed] [Google Scholar]
- Cappellaro C., Mrša V., Tanner W. New potential cell wall glucanases of Saccharomyces cerevisiae and their involvement in mating. J. Bacteriol. 1998;180:5030–5037. doi: 10.1128/jb.180.19.5030-5037.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caro L.H.P., Smits G. J., van Egmond P., Chapman J. W., Klis F. M. Transcription of multiple cell wall protein-encoding genes in Saccharomyces cerevisiae is differentially regulated during the cell cycle. FEMS Microbiol. Lett. 1998;161:345–349. doi: 10.1111/j.1574-6968.1998.tb12967.x. [DOI] [PubMed] [Google Scholar]
- Caro L.H.P., Tettelin H., Vossen J. H., Ram A.F.J., van den Ende H., Klis F. M. In silico identification of glycosyl-phosphatidylinositol-anchored plasma-membrane and cell wall proteins of Saccharomyces cerevisiae. Yeast. 1997;13:1477–1489. doi: 10.1002/(SICI)1097-0061(199712)13:15<1477::AID-YEA184>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Castillo L., Martinez A. I., Garcera A., Elorza M. V., Valentin E., Sentandreu R. Functional analysis of the cysteine residues and the repetitive sequence of Saccharomyces cerevisiae Pir4/Cis 3, the repetitive sequence is needed for binding to the cell wall β-1,3-glucan. Yeast. 2003;20:973–983. doi: 10.1002/yea.1016. [DOI] [PubMed] [Google Scholar]
- Caviston J. P., Longtine M., Pringle J. R., Bi E. The role of Cdc42p GTPase-activating proteins in assembly of the septin ring in yeast. Mol. Biol. Cell. 2003;14:4051–4066. doi: 10.1091/mbc.E03-04-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chant J., Pringle J. R. Patterns of bud-site selection in the yeast Saccharomyces cerevisiae. J. Cell Biol. 1995;129:751–765. doi: 10.1083/jcb.129.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartrand P., Meng X.-H., Huttelmaier S., Donato D., Singer R. H. Asymmetric sorting of Ash1p in yeast results from inhibition of translation by localization elements in the mRNA. Mol. Cell. 2002;10:1319–1330. doi: 10.1016/s1097-2765(02)00694-9. [DOI] [PubMed] [Google Scholar]
- Chartrand P., Meng X.-H., Singer R. H., Long R. M. Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr. Biol. 1999;9:333–336. doi: 10.1016/s0960-9822(99)80144-4. [DOI] [PubMed] [Google Scholar]
- Chuang J. S., Schekman R. W. Differential trafficking and timed localization of two chitin synthase proteins, Chs2p and Chs3p. J. Cell Biol. 1996;135:597–610. doi: 10.1083/jcb.135.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman-Lerner A., Chin T. E., Brent R. Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell. 2001;107:739–750. doi: 10.1016/s0092-8674(01)00596-7. [DOI] [PubMed] [Google Scholar]
- DeMarini D. J., Adams A.E.M., Fares H., De Virgilio C., Valle G., Chuang J. S., Pringle J. R. A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J. Cell Biol. 1997;139:75–93. doi: 10.1083/jcb.139.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nobel J. G., Klis F. M., Ram A., Van Unen H., Priem J., Munnik T., van den Ende H. Cyclic variations in the permeability of the cell wall of Saccharomyces cerevisiae. Yeast. 1991;7:589–598. doi: 10.1002/yea.320070606. [DOI] [PubMed] [Google Scholar]
- Dielbandhoesing S. K., Zhang H., Caro L.H.P., van der Vaart J. M., Klis F. M., Verrips C. T., Brul S. Specific cell wall proteins confer resistance to nisin upon yeast cells. Appl. Environ. Microbiol. 1998;64:4047–4052. doi: 10.1128/aem.64.10.4047-4052.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drgonová J., Drgon T., Tanaka K., Kollár R., Chen G.-C., Ford R. A., Chan C.S.M., Takai Y., Cabib E. Rho1p, a yeast protein at the interface between cell polarization and morphogenesis. Science. 1996;272:277–279. doi: 10.1126/science.272.5259.277. [DOI] [PubMed] [Google Scholar]
- Ecker M., Deutzmann R., Lehle L., Mrsa V., Tanner W. Pir-proteins of Saccharomyces cerevisiae are attached to β-1,3-glucan by a new protein-carbohydrate linkage. J. Biol. Chem. 2006;281:11523–11529. doi: 10.1074/jbc.M600314200. [DOI] [PubMed] [Google Scholar]
- Gerber A. P., Herschlag D., Brown P. O. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2:342–354. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R. D., Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Gladfelter A. S., Pringle J. R., Lew D. J. The septin cortex at the yeast mother-bud neck. Curr. Opin. Microbiol. 2001;4:681–689. doi: 10.1016/s1369-5274(01)00269-7. [DOI] [PubMed] [Google Scholar]
- Gonzalez I., Buonomo S.B.C., Nasmyth K., von Ahsen U. ASH1 mRNA localization in yeast involves multiple secondary structural elements and Ash1 protein translation. Curr. Biol. 1999;9:337–340. doi: 10.1016/s0960-9822(99)80145-6. [DOI] [PubMed] [Google Scholar]
- Gu W., Deng Y., Zenklusen D., Singer R. H. A new yeast PUF family protein, Puf6p, represses ASH1 mRNA translation and is required for its localization. Genes Dev. 2004;18:1452–1465. doi: 10.1101/gad.1189004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen I., et al. Sed1p and Srl1p are required to compensate for cell wall instability in Saccharomyces cerevisiae mutants defective in multiple GPI-anchored mannoproteins. Mol. Microbiol. 2004;52:1413–1425. doi: 10.1111/j.1365-2958.2004.04064.x. [DOI] [PubMed] [Google Scholar]
- Hamada K., Fukuchi S., Arisawa M., Baba M., Kitada K. Screening for glycosylphosphatidylinositol (GPI)-dependent cell wall proteins in Saccharomyces cerevisiae. Mol. Gen. Genet. 1998;258:53–59. doi: 10.1007/s004380050706. [DOI] [PubMed] [Google Scholar]
- Harkins H. A., Pagé N., Schenkman L. R., De Virgilio C., Shaw S., Bussey H., Pringle J. R. Bud8p and Bud9p, proteins that may mark the sites for bipolar budding in yeast. Mol. Biol. Cell. 2001;12:2497–2518. doi: 10.1091/mbc.12.8.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hašek J., Rupeš I., Svobodová J., Streiblová E. Tubulin and actin topology during zygote formation of Saccharomyces cerevisiae. J. Gen. Microbiol. 1987;133:3355–3363. doi: 10.1099/00221287-133-12-3355. [DOI] [PubMed] [Google Scholar]
- Huh W.-K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O’Shea E. K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Jansen R.-P., Dowzer C., Michaelis C., Galova M., Nasmyth K. Mother cell-specific HO expression in budding yeast depends on the unconventional myosin Myo4p and other cytoplasmic proteins. Cell. 1996;84:687–697. doi: 10.1016/s0092-8674(00)81047-8. [DOI] [PubMed] [Google Scholar]
- Jung U. S., Levin D. E. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signalling pathway. Mol. Microbiol. 1999;34:1049–1057. doi: 10.1046/j.1365-2958.1999.01667.x. [DOI] [PubMed] [Google Scholar]
- Kang P. J., Angerman E., Nakashima K., Pringle J. R., Park H.-O. Interactions among Rax1p, Rax2p, Bud8p, and Bud9p in marking cortical sites for bipolar bud-site selection in yeast. Mol. Biol. Cell. 2004;15:5145–5157. doi: 10.1091/mbc.E04-07-0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapteyn J. C., Montijn R. C., Vink E., de la Cruz J., Llobell A., Douwes J. E., Shimoi H., Lipke P. N., Klis F. M. Retention of Saccharomyces cerevisiae cell wall proteins through a phosphodiester-linked β-1,3-/β-1,6-glucan heteropolymer. Glycobiology. 1996;6:337–345. doi: 10.1093/glycob/6.3.337. [DOI] [PubMed] [Google Scholar]
- Kapteyn J. C., Montijn R. C., Dijkgraaf G.J.P., van den Ende H., Klis F. M. Covalent association of β-1,3-glucan with β-1,6-glucosylated mannoproteins in cell walls of Candida albicans. J. Bacteriol. 1995;177:3788–3792. doi: 10.1128/jb.177.13.3788-3792.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapteyn J. C., ter Riet B., Vink E., Blad S., de Nobel H., van den Ende H., Klis F. M. Low external pH induces HOG1-dependent changes in the organization of the Saccharomyces cerevisiae cell wall. Mol. Microbiol. 2001;39:469–479. doi: 10.1046/j.1365-2958.2001.02242.x. [DOI] [PubMed] [Google Scholar]
- Kapteyn J. C., van den Ende H., Klis F. M. The contribution of cell wall proteins to the organization of the yeast cell wall. Biochim. Biophys. Acta. 1999a;1426:373–383. doi: 10.1016/s0304-4165(98)00137-8. [DOI] [PubMed] [Google Scholar]
- Kapteyn J. C., van Egmond P., Sievi E., van den Ende H., Makarow M., Klis F. M. The contribution of the O-glycosylated protein Pir2p/Hsp150 to the construction of the yeast cell wall in wild-type cells and β-1,6-glucan-deficient mutants. Mol. Microbiol. 1999b;31:1835–1844. doi: 10.1046/j.1365-2958.1999.01320.x. [DOI] [PubMed] [Google Scholar]
- Klebl F., Tanner W. Molecular cloning of a cell wall exo-β-1,3-glucanase from Saccharomyces cerevisiae. J. Bacteriol. 1989;171:6259–6264. doi: 10.1128/jb.171.11.6259-6264.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klis F. M., Boorsma A., de Groot P. W. Cell wall construction in Saccharomyces cerevisiae. Yeast. 2006;23:185–202. doi: 10.1002/yea.1349. [DOI] [PubMed] [Google Scholar]
- Klis F. M., Ram A.F.J., Montijn R. C., Kapteyn J. C., Caro L.H.P., Vossen J. H., van Berkel M.A.A., Brekelmans S.S.C., van den Ende H. Posttranslational modifications of secretory proteins. Methods Microbiol. 1998;26:223–238. [Google Scholar]
- Kollár R., Petráková E., Ashwell G., Robbins P. W., Cabib E. Architecture of the yeast cell wall. The linkage between chitin and β(1→3)-glucan. J. Biol. Chem. 1995;270:1170–1178. doi: 10.1074/jbc.270.3.1170. [DOI] [PubMed] [Google Scholar]
- Kollár R., Reinhold B. B., Petráková E., Yeh H.J.C., Ashwell G., Drgonová J., Kapteyn J. C., Klis F. M., Cabib E. Architecture of the yeast cell wall. β(1→6)-glucan interconnects mannoprotein, β(1→3)-glucan, and chitin. J. Biol. Chem. 1997;272:17762–17775. doi: 10.1074/jbc.272.28.17762. [DOI] [PubMed] [Google Scholar]
- Kumar A., et al. Subcellular localization of the yeast proteome. Genes. Dev. 2002;16:707–719. doi: 10.1101/gad.970902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze I., Hensel G., Adler K., Bernard J., Neubohn B., Nilsson C., Stoltenburg R., Kohlwein S. D., Kunze G. The green fluorescent protein targets secretory proteins to the yeast vacuole. Biochim. Biophys. Acta. 1999;1410:287–298. doi: 10.1016/s0005-2728(99)00006-7. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lesage G., Shapiro J., Specht C. A., Sdicu A.-M., Ménard P., Hussein S., Tong A.H.Y., Boone C., Bussey H. An interactional network of genes involved in chitin synthesis in Saccharomyces cerevisiae. BMC Genet. 2005;6:8. doi: 10.1186/1471-2156-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew D. J., Reed S. I. Cell cycle control of morphogenesis in budding yeast. Curr. Opin. Genet. Dev. 1995;5:17–23. doi: 10.1016/s0959-437x(95)90048-9. [DOI] [PubMed] [Google Scholar]
- Lipke P. N., Ovalle R. Cell wall architecture in yeast: new structure and new challenges. J. Bacteriol. 1998;180:3735–3740. doi: 10.1128/jb.180.15.3735-3740.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipke P. N., Wojciechowicz D., Kurjan J. AGα1 is the structural gene for the Saccharomyces cerevisiae α-agglutinin, a cell surface glycoprotein involved in cell-cell interactions during mating. Mol. Cell. Biol. 1989;9:3155–3165. doi: 10.1128/mcb.9.8.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long R. M., Singer R. H., Meng X., Gonzalez I., Nasmyth K., Jansen R.-P. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science. 1997;277:383–387. doi: 10.1126/science.277.5324.383. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., DeMarini D. J., Valencik M. L., Al-Awar O. S., Fares H., De Virgilio C., Pringle J. R. The septins: roles in cytokinesis and other processes. Curr. Opin. Cell Biol. 1996;8:106–119. doi: 10.1016/s0955-0674(96)80054-8. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., Fares H., Pringle J. R. Role of the yeast Gin4p protein kinase in septin assembly and the relationship between septin assembly and septin function. J. Cell Biol. 1998b;143:719–736. doi: 10.1083/jcb.143.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., III, DeMarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998a;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., Theesfeld C. L., McMillan J. N., Weaver E., Pringle J. R., Lew D. J. Septin-dependent assembly of a cell cycle-regulatory module in Saccharomyces cerevisiae. Mol. Cell. Biol. 2000;20:4049–4061. doi: 10.1128/mcb.20.11.4049-4061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord M., Yang M. C., Mischke M., Chant J. Cell cycle programs of gene expression control morphogenetic protein localization. J. Cell Biol. 2000;151:1501–1512. doi: 10.1083/jcb.151.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews D. H., Sabina J., Zuker M., Turner D. H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- Molano J., Bowers B., Cabib E. Distribution of chitin in the yeast cell wall. An ultrastructural and chemical study. J. Cell Biol. 1980;85:199–212. doi: 10.1083/jcb.85.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montijn R. C., van Rinsum J., van Schagen F. A., Klis F. M. Glucomannoproteins in the cell wall of Saccharomyces cerevisiae contain a novel type of carbohydrate side chain. J. Biol. Chem. 1994;269:19338–19342. [PubMed] [Google Scholar]
- Moukadiri I., Jaafar L., Zueco J. Identification of two mannoproteins released from cell walls of a Saccharomyces cerevisiae mnn1 mnn9 double mutant by reducing agents. J. Bacteriol. 1999;181:4741–4745. doi: 10.1128/jb.181.16.4741-4745.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P., Ferro S., Schekman R. Order of events in the yeast secretory pathway. Cell. 1981;25:461–469. doi: 10.1016/0092-8674(81)90064-7. [DOI] [PubMed] [Google Scholar]
- Novick P., Schekman R. Export of major cell surface proteins is blocked in yeast secretory mutants. J. Cell Biol. 1983;96:541–547. doi: 10.1083/jcb.96.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono N., Yabe T., Sudoh M., Nakajima T., Yamada-Okabe T., Arisawa M., Yamada-Okabe H. The yeast Chs4 protein stimulates the trypsin-sensitive activity of chitin synthase 3 through an apparent protein-protein interaction. Microbiology. 2000;146:385–391. doi: 10.1099/00221287-146-2-385. [DOI] [PubMed] [Google Scholar]
- Orlean P. Biogenesis of yeast wall and surface components. In: Pringle J. R., Broach J. R., Jones E. W., editors. The Molecular and Cellular Biology of the Yeast Saccharomyces. Cell Cycle and Cell Biology. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 229–362. [Google Scholar]
- Orlean P., Ammer H., Watzele M., Tanner W. Synthesis of an O-glycosylated cell surface protein induced in yeast by α-factor. Proc. Natl. Acad. Sci. USA. 1986;83:6263–6266. doi: 10.1073/pnas.83.17.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozols J. Amino acid analysis. Methods Enzymol. 1990;182:587–601. doi: 10.1016/0076-6879(90)82046-5. [DOI] [PubMed] [Google Scholar]
- Pastor F. I., Herrero E., Sentandreu R. Metabolism of Saccharomyces cerevisiae envelope mannoproteins. Arch. Microbiol. 1982;132:144–148. doi: 10.1007/BF00508720. [DOI] [PubMed] [Google Scholar]
- Powell C. D., Quain D. E., Smart K. A. Chitin scar breaks in aged Saccharomyces cerevisiae. Microbiology. 2003;149:3129–3137. doi: 10.1099/mic.0.25940-0. [DOI] [PubMed] [Google Scholar]
- Pringle J. R., Bi E., Harkins H. A., Zahner J. E., De Virgilio C., Chant J., Corrado K., Fares H. Establishment of cell polarity in yeast. Cold Spring Harb. Symp. Quant. Biol. 1995;60:729–743. doi: 10.1101/sqb.1995.060.01.079. [DOI] [PubMed] [Google Scholar]
- Pruyne D., Bretscher A. Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J. Cell Sci. 2000a;113:365–375. doi: 10.1242/jcs.113.3.365. [DOI] [PubMed] [Google Scholar]
- Pruyne D., Bretscher A. Polarization of cell growth in yeast. II. The role of the cortical actin cytoskeleton. J. Cell Sci. 2000b;113:571–585. doi: 10.1242/jcs.113.4.571. [DOI] [PubMed] [Google Scholar]
- Ram A.F.J., Kapteyn J. C., Montijn R. C., Caro L.H.P., Douwes J. E., Baginsky W., Mazur P., van den Ende H., Klis F. M. Loss of the plasma membrane-bound protein Gas1p in Saccharomyces cerevisiae results in the release of β1,3-glucan into the medium and induces a compensation mechanism to ensure cell wall integrity. J. Bacteriol. 1998b;180:1418–1424. doi: 10.1128/jb.180.6.1418-1424.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram A.F.J., van den Ende H., Klis F. M. Green fluorescent protein-cell wall fusion proteins are covalently incorporated into the cell wall of Saccharomyces cerevisiae. FEMS Microbiol. Lett. 1998a;162:249–255. doi: 10.1111/j.1574-6968.1998.tb13006.x. [DOI] [PubMed] [Google Scholar]
- Reck-Peterson S. L., Novick P. J., Mooseker M. S. The tail of a yeast class V myosin, Myo2p, functions as a localization domain. Mol. Biol. Cell. 1999;10:1001–1017. doi: 10.1091/mbc.10.4.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ríos Muñoz W., Irizarry Ramírez M., Rivera Molina F., González Crespo S., Rodríguez-Medina J. R. Myosin II is important for maintaining regulated secretion and asymmetric localization of chitinase 1 in the budding yeast Saccharomyces cerevisiae. Arch. Biochem. Biophys. 2003;409:411–413. doi: 10.1016/s0003-9861(02)00614-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. L., Bowers B., Slater M. L., Cabib E. Chitin synthesis and localization in cell division cycle mutants of Saccharomyces cerevisiae. Mol. Cell. Biol. 1983;3:922–930. doi: 10.1128/mcb.3.5.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Peña J. M., Cid V. J., Arroyo J., Nombela C. A novel family of cell wall-related proteins regulated differently during the yeast life cycle. Mol. Cell. Biol. 2000;20:3245–3255. doi: 10.1128/mcb.20.9.3245-3255.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Peña J. M., Rodriguez C., Alvarez A., Nombela C., Arroyo J. Mechanisms for targeting of the Saccharomyces cerevisiae GPI-anchored cell wall protein Crh2p to polarised growth sites. J. Cell Sci. 2002;115:2549–2558. doi: 10.1242/jcs.115.12.2549. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schenkman L. R., Caruso C., Pagé N., Pringle J. R. The role of cell cycle-regulated expression in the localization of spatial landmark proteins in yeast. J. Cell Biol. 2002;156:829–841. doi: 10.1083/jcb.200107041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M., Bowers B., Varma A., Roh D.-H., Cabib E. In budding yeast, contraction of the actomyosin ring and formation of the primary septum at cytokinesis depend on each other. J. Cell Sci. 2002;115:293–302. doi: 10.1242/jcs.115.2.293. [DOI] [PubMed] [Google Scholar]
- Shaw J. A., Mol P. C., Bowers B., Silverman S. J., Valdivieso M. H., Durán A., Cabib E. The function of chitin synthases 2 and 3 in the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 1991;114:111–123. doi: 10.1083/jcb.114.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard K. A., Gerber A. P., Jambhekar A., Takizawa P. A., Brown P. O., Herschlag D., DeRisi J. L., Vale R. D. Widespread cytoplasmic mRNA transport in yeast: identification of 22 bud-localized transcripts using DNA microarray analysis. Proc. Natl. Acad. Sci. USA. 2003;100:11429–11434. doi: 10.1073/pnas.2033246100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoi H., Iimura Y., Obata T. Molecular cloning of CWP1: a gene encoding a Saccharomyces cerevisiae cell wall protein solubilized with Rarobacter faecitabidus protease I. J. Biochem. 1995;118:302–311. doi: 10.1093/oxfordjournals.jbchem.a124907. [DOI] [PubMed] [Google Scholar]
- Sil A., Herskowitz I. Identification of asymmetrically localized determinant, Ash1p, required for lineage-specific transcription of the yeast HO gene. Cell. 1996;84:711–722. doi: 10.1016/s0092-8674(00)81049-1. [DOI] [PubMed] [Google Scholar]
- Smits G. J., van den Ende H., Klis F. M. Differential regulation of cell wall biogenesis during growth and development in yeast. Microbiology. 2001;147:781–794. doi: 10.1099/00221287-147-4-781. [DOI] [PubMed] [Google Scholar]
- Spellman P. T., Sherlock G., Zhang M. Q., Iyer V. R., Anders K., Eisen M. B., Brown P. O., Botstein D., Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streiblová E., Beran K. Demonstration of yeast scars by fluorescence microscopy. Exp. Cell Res. 1963;30:603–605. [Google Scholar]
- Takizawa P. A., Sil A., Swedlow J. R., Herskowitz I., Vale R. D. Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature. 1997;389:90–93. doi: 10.1038/38015. [DOI] [PubMed] [Google Scholar]
- Takizawa P. A., Vale R. D. The myosin motor, Myo4p, binds ASH1 mRNA via the adapter protein, She3p. Proc. Natl. Acad. Sci. USA. 2000;97:5273–5278. doi: 10.1073/pnas.080585897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talens L. T., Miranda M., Miller M. W. Electron micrography of bud formation in Metschnikowia krissii. J. Bacteriol. 1973;114:413–423. doi: 10.1128/jb.114.1.413-423.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima H., Hamada K., Kitada K. The localization change of Ybr078w/Ecm33, a yeast GPI-associated protein, from the plasma membrane to the cell wall, affecting the cellular function. FEMS Microbiol. Lett. 2003;218:175–180. doi: 10.1111/j.1574-6968.2003.tb11515.x. [DOI] [PubMed] [Google Scholar]
- Terashima H., Yabuki N., Arisawa M., Hamada K., Kitada K. Up-regulation of genes encoding glycosylphosphatidylinositol (GPI)-attached proteins in response to cell wall damage caused by disruption of FKS1 in Saccharomyces cerevisiae. Mol. Gen. Genet. 2000;264:64–74. doi: 10.1007/s004380000285. [DOI] [PubMed] [Google Scholar]
- Trilla J. A., Cos T., Duran A., Roncero C. Characterization of CHS4 (CAL2), a gene of Saccharomyces cerevisiae involved in chitin biosynthesis and allelic to SKT5 and. CSD4. Yeast. 1997;1 3:795–807. doi: 10.1002/(SICI)1097-0061(199707)13:9<795::AID-YEA139>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Utsugi T., Minemura M., Hirata A., Abe M., Watanabe D., Ohya Y. Movement of yeast 1,3-β-glucan synthase is essential for uniform cell wall synthesis. Genes Cells. 2002;7:1–9. doi: 10.1046/j.1356-9597.2001.00495.x. [DOI] [PubMed] [Google Scholar]
- Valdivia R. H., Baggott D., Chuang J. S., Schekman R. W. The yeast clathrin adaptor protein complex 1 is required for the efficient retention of a subset of late Golgi membrane proteins. Dev. Cell. 2002;2:283–294. doi: 10.1016/s1534-5807(02)00127-2. [DOI] [PubMed] [Google Scholar]
- Valdivia R. H., Schekman R. The yeasts Rho1p and Pkc1p regulate the transport of chitin synthase III (Chs3p) from internal stores to the plasma membrane. Proc. Natl. Acad. Sci. USA. 2003;100:10287–10292. doi: 10.1073/pnas.1834246100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivieso M.-H., Ferrario L., Vai M., Duran A., Popolo L. Chitin synthesis in a gas1 mutant of Saccharomyces cerevisiae. J. Bacteriol. 2000;182:4752–4757. doi: 10.1128/jb.182.17.4752-4757.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallen E. A., Caviston J., Bi E. Roles of Hof1p, Bni1p, Bnr1p, and Myo1p in cytokinesis in Saccharomyces cerevisiae. Mol. Biol. Cell. 2000;11:593–611. doi: 10.1091/mbc.11.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vaart J. M., Caro L.H.P., Chapman J. W., Klis F. M., Verrips C. T. Identification of three mannoproteins in the cell wall of Saccharomyces cerevisiae. J. Bacteriol. 1995;177:3104–3110. doi: 10.1128/jb.177.11.3104-3110.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vossen J. H., Müller W. H., Lipke P. N., Klis F. M. Restrictive glycosylphosphatidylinositol anchor synthesis in cwh6/gpi3 yeast cells causes aberrant biogenesis of cell wall proteins. J. Bacteriol. 1997;179:2202–2209. doi: 10.1128/jb.179.7.2202-2209.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F., Dollard C., Ricupero-Hovasse S. L. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- Wodicka L., Dong H., Mittmann M., Ho M.-H., Lockhart D. J. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat. Biotechnol. 1997;15:1359–1367. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]
- Wojciechowicz D., Lipke P. N. α-agglutinin expression in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 1989;161:46–51. doi: 10.1016/0006-291x(89)91557-x. [DOI] [PubMed] [Google Scholar]
- Yin Q. Y., de Groot P.W.J., Dekker H. L., de Jong L., Klis F. M., de Koster C. G. Comprehensive proteomic analysis of Saccharomyces cerevisiae cell walls: identification of proteins covalently attached via GPI remnants or mild-alkali sensitive linkages. J. Biol. Chem. 2005;280:20894–20901. doi: 10.1074/jbc.M500334200. [DOI] [PubMed] [Google Scholar]
- Zahner J. E., Harkins H. A., Pringle J. R. Genetic analysis of the bipolar pattern of bud site selection in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:1857–1870. doi: 10.1128/mcb.16.4.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziman M., Chuang J. S., Tsung M., Hamamoto S., Schekman R. Chs6p-dependent anterograde transport of Chs3p from the chitosome to the plasma membrane in Saccharomyces cerevisiae. Mol. Biol. Cell. 1998;9:1565–1576. doi: 10.1091/mbc.9.6.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M., Mathews D. H., Turner D. H. Algorithms and Thermodynamics for RNA Secondary Structure Prediction: A Practical Guide in RNA Biochemistry and Biotechnology. In: Barciszewski J., Clark B.F.C., editors. NATO ASI Series. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1999. pp. 11–43. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.