Abstract
AP-1 and Gga adaptors participate in clathrin-mediated protein transport between the trans-Golgi network and endosomes. Both adaptors contain homologous domains that act to recruit accessory proteins involved in clathrin-coated vesicle formation, but the spectrum of known adaptor-binding partners is limited. This study describes an evolutionarily conserved protein of Saccharomyces cerevisiae, Laa1p (Yjl207cp), that interacts and functions specifically with AP-1. Deletion of LAA1, when combined with a conditional mutation in clathrin heavy chain or deletion of GGA genes, accentuated growth defects and increased disruption of clathrin-dependent α-factor maturation and transport of carboxypeptidase Y to the vacuole. In contrast, such genetic interactions were not observed between deletions of LAA1 and AP-1 subunit genes. Laa1p preferentially interacted with AP-1 compared with Gga proteins by glutathione S-transferase-fusion affinity binding and coimmunoprecipitations. Localization of AP-1 and Laa1p, but not Gga proteins, was highly sensitive to brefeldin A, an inhibitor of ADP-ribosylation factor (Arf) activation. Importantly, deletion of LAA1 caused mislocalization of AP-1, especially in cells at high density (postdiauxic shift), but it did not affect Gga protein distribution. Our results identify Laa1p as a new determinant of AP-1 localization, suggesting a model in which Laa1p and Arf cooperate to direct stable association of AP-1 with appropriate intracellular membranes.
INTRODUCTION
Protein transport between membrane-bound organelles is an essential aspect of eukaryotic life. Clathrin-coated vesicles (CCV) are major players in intracellular transport, mediating traffic of proteins from the plasma membrane to endosomes (endocytosis) and between endosomes and the trans-Golgi network (TGN). Vesicle formation involves assembly of coat proteins that collect cargo proteins as well as promote invagination and scission of the membrane to release a fully coated transport vesicle. Once formed, the coats are rapidly shed to allow targeting and fusion with the appropriate target organelle (Kirchhausen, 2000; Brodsky et al., 2001; Mousavi et al., 2004).
Clathrin is a hexamer of heavy and light chain subunits assembled into a radial three-legged structure termed a triskelion (Kirchhausen, 2000; Owen et al., 2004; Wilbur et al., 2005). Triskelia associate to form polyhedral lattices that encase the vesicle. Because clathrin has no intrinsic lipid-binding ability, it is linked to membranes by adaptors that also bind lipids and/or the cytoplasmic domains of cargo proteins (Kirchhausen, 1999). Through these activities, adaptors are thought to be key factors in clathrin coat nucleation. In addition to clathrin and adaptors, an increasing number of coat-interacting proteins have been recognized as important contributors to the transport process (Slepnev and De Camilli, 2000; Mousavi et al., 2004; Owen, 2004; Perrais and Merrifield, 2005). Most commonly, these so-called accessory proteins are transiently recruited by adaptors and clathrin. All stages of vesicle formation and uncoating seem to involve accessory proteins (Slepnev and De Camilli, 2000). Currently, more is known about the identity and functions of accessory factors that act in endocytic CCV formation than those acting in CCV formation at the TGN and endosomes.
Two major types of adaptors that function in clathrin-mediated transport between the TGN and endosomes are the AP-1 complex and the Gga family of proteins (Hinners and Tooze, 2003). AP-1 is composed of two large subunits (β1 and γ), one medium subunit (μ1), and a small subunit (ς1). In mammalian cells, there are three other complexes with homology to AP-1 that function in different transport pathways: AP-2 is involved in clathrin-mediated endocytosis (Kirchhausen, 2002), AP-3 transports proteins from endosomes and possibly the TGN to lysosomes and lysosome-related organelles (Cowles et al., 1997; Stepp et al., 1997; Theos et al., 2005), and AP-4 has been implicated in sorting of cargo destined for lysosomes and the basolateral membrane in different cell types (Aguilar et al., 2001; Simmen et al., 2002). Whether AP-3 and AP-4 function together with clathrin is currently unclear (Simpson et al., 1996; Dell’Angelica et al., 1998; Vowels and Payne, 1998). In yeast, there are three AP complexes homologous to mammalian AP-1, AP-2, and AP-3 (Yeung et al., 1999).
Gga proteins are monomeric adaptors with C-terminal domains homologous to the C-terminal “ear” domain of the AP-1 γ subunit, so called because it protrudes from the core structure of the AP complex. Mammalian cells express three Gga proteins (Boman et al., 2000; Dell’Angelica et al., 2000; Hirst et al., 2000; Poussu et al., 2000; Takatsu et al., 2000) and yeast express two (Dell’Angelica et al., 2000; Hirst et al., 2000). Gga proteins share many activities with AP-1, including binding to cargo, clathrin, and accessory proteins (Bonifacino, 2004; Ghosh and Kornfeld, 2004).
The functional relationship between Gga proteins and AP-1 has not been clearly defined (Hinners and Tooze, 2003). Both adaptors are localized to the TGN and endosomes, although the extent of AP-1 and Gga colocalization has varied in different reports (Dell’Angelica et al., 2000; Hirst et al., 2000; Puertollano et al., 2001a; Doray et al., 2002), and both are dependent on the ADP-ribosylation factor (Arf)1 GTPase for membrane association in mammalian cells (Stamnes and Rothman, 1993; Dell’Angelica et al., 2000). Both yeast and mammalian Gga proteins physically interact with AP-1 (Costaguta et al., 2001; Doray et al., 2002; Bai et al., 2004), and studies of mannose-6-phosphate receptor sorting in mammalian fibroblasts suggest a sequential role for the two adaptors (Doray et al., 2002). In contrast, results from genetic studies in yeast and mammalian cells are consistent with Gga and AP-1 function in distinct pathways between the TGN and endosomes (Black and Pelham, 2000; Meyer et al., 2000; Costaguta et al., 2001; Puertollano et al., 2001b; Ha et al., 2003). Identification and characterization of factors specific for each adaptor could help clarify the relative roles of AP-1 and Gga proteins in TGN/endosome traffic.
The γ-ears of AP-1 and Gga proteins serve as docking sites for accessory factors in CCV biogenesis at the TGN and endosomes. Several putative accessory factors have been identified that bind γ-ears, but their function remains largely unknown (Duncan and Payne, 2003; Ghosh and Kornfeld, 2004). One protein that associates with γ-ears, albeit indirectly, is mammalian p200 (Lui et al., 2003). It is a large protein (∼200 kDa) that is highly conserved throughout eukaryotes, with homologues from humans to yeast. Most vertebrates contain two p200 paralogues, and at least one isoform is present in a complex with aftiphilin and γ-synergin, two γ-ear binding proteins (Hirst et al., 2005). The complex seems to be involved in AP-1–mediated transport, but the specific role of the complex, and p200 in particular, has not been determined. Furthermore, it remains uncertain whether any p200 isoforms provide function outside of the complex.
The yeast Saccharomyces cerevisiae encodes a homologue of p200, Yjl207cp, but no homologues to aftiphilin or γ-synergin, suggesting that Yjl207cp may provide a conserved function independent of the aftiphilin/γ-synergin complex. Here, we have examined the function of Yjl207cp, which we have named large AP-1 accessory (Laa1p), in clathrin-dependent transport. Our results indicate preferential association of Laa1p with AP-1 compared with Gga adaptors and a corresponding functional specificity for AP-1–mediated transport. Analysis of Laa1p-deficient cells reveals a role for Laa1p in AP-1 localization, mediated in part by the region of Laa1p most similar to p200.
MATERIALS AND METHODS
Plasmids
Plasmids were constructed using standard molecular biology techniques and verified by DNA sequencing (Sambrook et al., 1989). LAA1 was cloned by PCR amplification of two fragments from the genome of SEY6210 that correspond to nucleotides −413 to +3172 and +2948 to +6928, where +1 is the first nucleotide of the coding region. Elongase enzyme mix (Invitrogen, Carlsbad, CA) was used in the PCR reactions. The two LAA1 fragments were ligated into pBKS+ (Stratagene, La Jolla, CA) using genomic EcoRI and BamHI restriction sites. To remove the LAA1 highly conserved region (HCR), XmaI sites were introduced by QuikChange site-directed mutagenesis (Stratagene), creating the following mutations: A3940C and A3942G (T1314P), A3944G (D1315G), T4623C (silent), A4624C, and T4626G (T1542P), and T4628G (V1543G). LAA1 was then moved from pBKS+ into pRS315 (Sikorski and Hieter, 1989) by digestion with ApaI/SacII to create pRS315-LAA1, which was digested with XmaI and religated to remove nucleotides +3940 to +4623 (amino acids 1314–1541), creating pRS315-laa1ΔHCR. To create pRS315-laa1ΔHCR-13Myc and pRS315-laa1ΔHCR-GFP, a PacI site was created by QuikChange mutagenesis at the 3′ end of LAA1 (A6044T, T6046A, G6048T, C6049A) in pRS315-laa1ΔHCR, changing the ochre codon to leucine. Into that plasmid were ligated PacI-digested 13Myc and green fluorescent protein (GFP) tags that were PCR-amplified from pFA6a-13Myc-His3MX6 and pFA6a-GFP(S65T)-His3MX6, respectively (Longtine et al., 1998). The PacI sites at the 3′ end of the 13Myc and GFP tags were incorporated into the relevant primers.
Yeast Strains and Media
The yeast strains used in this study are listed in Table 1. Targeted gene disruption of LAA1 and tagging of genomic LAA1, clathrin heavy chain, β1, and GGA2 with GFP, monomeric red fluorescent protein (mRFP), and/or a myc epitope was accomplished using standard techniques (Baudin et al., 1993; Longtine et al., 1998). The plasmid pFA6a–mRFP–KanMX6 was used for integrations of mRFP (Huh et al., 2003). Gene disruption and tagging of all genes was verified by PCR amplification of the corresponding genomic loci. Strains that contain tagged or deleted LAA1 in combination with mutated or tagged clathrin, β1, or GGA genes were constructed by mating parental strains followed by sporulation and dissection of the resulting heterozygous diploid.
Table 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| SEY6210 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 | Robinson et al. (1988) |
| GPY404 | MATa ura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 | Yeung et al. (1999) |
| FY1679-28C | MATa ura3-52 trp1-Δ63 leu2Δ1 his3Δ300 | B. Dujon |
| GPY2983 | FY1679-28C LAA1-13Myc::His3MX6 | This study |
| GPY3011 | FY1679-28C LAA1-13Myc::His3MX6GAL-AUX1::TRP1 | This study |
| GPY2853 | SEY6210 LAA1-13Myc::His3MX6 | This study |
| GPY2736 | SEY6210 ENT5-13Myc::His3MX6 | Duncan et al. (2003) |
| GPY2867 | SEY6210 laa1Δ::HIS3 | This study |
| GPY2869 | GPY404 laa1Δ::HIS3 | This study |
| GPY1064 | SEY6210 chc1-ts::URA3 | Bensen et al. (2000) |
| GPY3347 | SEY6210 chc1-ts::URA3laa1Δ::HIS3 | This study |
| GPY1783-10A | SEY6210 apl2Δ::TRP1 | Yeung et al. (1999) |
| GPY2903 | SEY6210 apl2Δ::TRP1laa1Δ::HIS3 | This study |
| GPY2149 | SEY6210 gga2Δ::HIS3 | Costaguta et al. (2001) |
| GPY2957 | SEY6210 gga2Δ::HIS3laa1Δ::HIS3 | This study |
| GPY2420-3C | SEY6210 gga2Δ::HIS3 apl2Δ::URA3 | Costaguta et al. (2001) |
| GPY2385 | SEY6210 gga1Δ::TRP1 gga2Δ::HIS3 | Costaguta et al. (2001) |
| GPY2960 | SEY6210 gga1Δ::TRP1 gga2Δ::HIS3laa1Δ::HIS3 | This study |
| GPY2967 | SEY6210 LAA1-GFP(S65T)::TRP1 | This study |
| GPY3262 | GPY404 LAA1-GFP(S65T)::TRP1 | This study |
| GPY3116 | GPY404 LAA1-GFP(S65T)::TRP1 apl2Δ::TRP1 | This study |
| GPY3111 | GPY404 LAA1-GFP(S65T)::TRP1 gga1Δ::TRP1 gga2Δ::HIS3 | This study |
| GPY3018 | SEY6210 LAA1-GFP(S65T)::TRP1GGA2-mRFP::KanMX | This study |
| GPY3028 | SEY6210 LAA1-GFP(S65T)::TRP1CHC1-mRFP::KanMX | This study |
| GPY3034 | SEY6210 LAA1-GFP(S65T)::TRP1APL2-mRFP::KanMX | This study |
| GPY3086 | SEY6210 APL2-GFP(S65T)::His3MX6 | This study |
| GPY3085 | SEY6210 APL2-GFP(S65T)::His3MX6laa1Δ::HIS3 | This study |
| GPY3106 | GPY404 GGA2-mRFP::KanMX | This study |
| GPY3007 | GPY404 GGA2-mRFP::KanMXlaa1Δ::HIS3 | This study |
| GPY3109 | SEY6210 GGA2-mRFP::KanMX APL2-GFP(S65T)::His3MX6 | This study |
| GPY3942 | SEY6210 APL2-GFP(S65T)::His3MX6erg6Δ::HIS3 | This study |
| GPY3943 | SEY6210 GGA2-mRFP::KanMX erg6Δ::HIS3 | This study |
| GPY3944 | GPY404 LAA1-GFP(S65T)::TRP1 erg6Δ::HIS3 | This study |
SD complete medium is 0.67% yeast nitrogen base without amino acids (Difco, Detroit, MI) and 2% dextrose supplemented with 40 μg/ml adenine, 30 μg/ml leucine and lysine, and 20 μg/ml histidine, tryptophan, and uracil. SD-leu is SD complete without leucine. SDCAA complete is SD complete with 0.5% vitamin assay casamino acid mix (Difco). SDYE is SD complete with 2% Bacto yeast extract. YP is 1% Bacto yeast extract and 2% Bacto peptone. YPD is YP with 2% dextrose. YPGal is YP with 2% galactose. Cell densities in liquid culture were measured at 500 nm in 1-cm plastic cuvettes using a DU-62 spectrophotometer (Beckman Coulter, Fullerton, CA); 1 OD500 unit is equivalent to 2.3 × 107 cells.
Affinity Chromatography with Glutathione S-transferase (GST) Fusion Proteins
Bacterial strain BL21-CodonPlus(DE3) (Stratagene) carrying plasmid pGEX-4T-1 (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom), pGEX-KG-Apl4pΔN609 (Yeung and Payne, 2001), or pGEX-Gga2 (Duncan et al., 2003) was used to express GST alone and fused to the AP-1 and Gga2p γ-ears, respectively. GST proteins were bound to glutathione-Sepharose (GE Healthcare), and 50% bead suspensions were made in phosphate-buffered saline buffer (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, and 1.4 mM KH2PO4) + 1% Triton X-100. Extract from GPY2853 cells was prepared and subjected to affinity chromatography with equal amounts of each GST protein as described previously (Yeung et al., 1999), except the beads were resuspended in Laemmli sample buffer (LSB; 62 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, and 0.01% bromphenol blue) and incubated at 100°C for 2–3 min. Proteins in the samples were analyzed by SDS-PAGE and immunoblotting with an antibody against the myc epitope (Calbiochem/EMD Biosciences, Darmstadt, Germany).
Nondenaturing Immunoprecipitation
Cultures were grown to mid-logarithmic phase in YPD or SD-leu, and 5 × 108 cells were converted to spheroplasts, washed once with 1.25 M sorbitol, and lysed by resuspending in 1 ml of HEKG5 buffer (20 mM HEPES, pH 6.8, 50 mM KCl, 1 mM EDTA, 5% glycerol, and 1% Triton X-100) with protease inhibitors [1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride, 0.8 μM aprotinin, 20 μM leupeptin, 40 μM bestatin, 15 μM pepstatin A, and 14 μM E-64; Sigma-Aldrich, St. Louis, MO]. The lysate was cleared by centrifugation at 12,000 × g for 20 min at 4°C followed by incubation with protein A-Sepharose (GE Healthcare) or IgSorb (The Enzyme Center, Malden, MA) for 30–45 min and centrifugation at 15,000 × g for 30 s (protein A-Sepharose) or 20,000 × g for 20 min (IgSorb). The extracts were incubated with 2 μg of anti-myc antibody and protein A-Sepharose for 16 h at 4°C. The beads were washed three times with buffer, resuspended in LSB, and the samples were analyzed by SDS-PAGE and immunoblotting.
Size Exclusion Chromatography
GAL-AUX1 (GPY3011) cells were grown in YPGal to stationary phase, diluted 1:500 into YPD, and allowed to grow for at least 24 h to repress AUX1 and accumulate CCV. Wild-type (GPY2983) cells were grown in YPD. For each fractionation, 5 × 109 cells in mid-logarithmic phase were converted to spheroplasts and lysed by agitation with glass beads in 1 ml of buffer A (100 mM MES-NaOH, pH 6.5, 0.5 mM MgCl2, 1 mM EGTA, 0.2 mM dithiothreitol, and 10 mM NaN3) with protease inhibitors. The lysate was sedimented by centrifugation at 17,000 × g for 30 min at 4°C, and the supernatant was loaded onto a 2 × 100-cm Sephacryl S-1000 column (Sigma-Aldrich) preequilibrated with buffer A. Fractions of 3 ml were collected, and 1 ml of each was precipitated with 10% trichloroacetic acid in the presence of 100 μg/ml bovine serum albumin, resuspended in LSB, and analyzed by SDS-PAGE and immunoblotting. The presence of CCV in the shifted clathrin peak from GAL-AUX1 cells was previously established by electron microscopy (Pishvaee et al., 2000).
Metabolic Radiolabeling and Immunoprecipitation
For metabolic labeling of α-factor and carboxypeptidase Y (CPY), cells were grown to mid-logarithmic phase in SDYE or SD-leu at 24°C (α-factor) or 30°C (CPY). Radiolabeling and immunoprecipitation were performed as described previously (Yeung et al., 1999), except that labeling was not terminated before cells were processed for α-factor immunoprecipitation. Immunoprecipitates were analyzed by SDS-PAGE and autoradiography on PhosphorImager screens (GE Healthcare) for image capture and quantification.
Microscopy
Cells were grown in SDCAA complete, SD complete (erg6Δ strains), or SD-leu (for plasmid selection) at 30°C. For visualization of late stage cultures, cells were grown to a density of 0.9–1.0 × 108 cells/ml in SDCAA complete or 4.5 × 107 cells/ml in SD-leu. For visualization of cells in early logarithmic phase, late stage cultures were diluted to 1–3 × 106 cells/ml and allowed to grow for at least 4 h (two doublings) at 30°C. For microscopy, cultures were concentrated to 5 × 108 cells/ml by centrifugation at 10,000 × g for 30 s and resuspension in fresh medium. Concentrated cells were incubated at 24°C for 10 min and visualized by pipetting 1.5 μl onto a microscope slide and applying a coverslip. To visualize erg6Δ strains, log phase cultures were concentrated to 108 cells/ml by centrifugation at 1500 × g for 60 s and resuspension in fresh medium containing 75 μg/ml brefeldin A (BfA; Sigma-Aldrich) or 1.5% ethanol (BfA solvent). Immediately, 25 μl of the concentrated cell suspension was spotted onto a concanavalin A-treated coverslip, and the cells were allowed to adhere for 1–3 min. The coverslip was washed with medium containing ethanol or BfA to remove loose cells and placed onto a glass slide containing 1.5 μl of the same medium. BfA-treated cells were visualized 10–15 min after addition of the drug. All images were captured using a 100× objective and Zeiss Axiovert 200M microscope. Images for colocalization and BfA experiments were captured with a Hamamatsu Orca II camera; all other images were captured with an Orca ER.
For quantification of colocalization, the GFP and mRFP channels of each image were exported to 8-bit TIFF files with Axiovision LE version 4.3 software (Carl Zeiss, Jena, Germany) and analyzed using ImageJ version 1.35e software (National Institutes of Health, Bethesda, MD). For each channel, the most frequent pixel intensity (mode) was determined and background signal was defined as 1 intensity unit above the mode. Only bright puncta were considered for colocalization analysis by thresholding the images to an intensity value such that 1% of the pixels above background were displayed. Examination by eye confirmed that 1% was a reasonable cut-off to remove diffuse cell staining and display most puncta of all the proteins visualized. Colocalization for a protein was expressed as the percentage of pixels from the corresponding channel that overlap with the other channel from the same image. Three fields from two different slides were quantified and averaged for each strain visualized; standard deviations from the mean are reported in the text.
Multiple Sequence Alignment
Multiple sequence alignment was performed with the PILEUP and PRETTYBOX programs of the Wisconsin Package version 10.3 (Accelrys, San Diego, CA).
RESULTS
Genetic Interactions between Mutant Alleles of LAA1 and Genes Encoding Clathrin and Clathrin Adaptors
A previous whole-genome study identified S. cerevisiae open reading frame YJL207c (LAA1) as encoding a protein that colocalizes with clathrin (Huh et al., 2003). To determine whether LAA1 functions in a clathrin-dependent process, we tested for a genetic interaction between mutations in LAA1 and the clathrin heavy chain gene (CHC1) using an assay that exploits the slow-growth phenotype of cells that have disrupted clathrin function. A temperature-sensitive allele of CHC1, chc1-521 (chc1-ts), causes increasingly slower growth at temperatures above 24°C (Seeger and Payne, 1992b; Bensen et al., 2000). Deletions of genes whose products act in clathrin-dependent transport further debilitate growth of chc1-ts cells, whereas such mutations in CHC1 cells have little or no effect. Such synthetic growth defects have been observed with deletions of genes encoding a number of clathrin adaptors and accessory factors, including AP-1 and Gga proteins (Yeung et al., 1999; Bensen et al., 2000, 2001; Costaguta et al., 2001). Thus, a synthetic growth defect with chc1-ts provides a sensitive genetic indicator for function in a clathrin-dependent process.
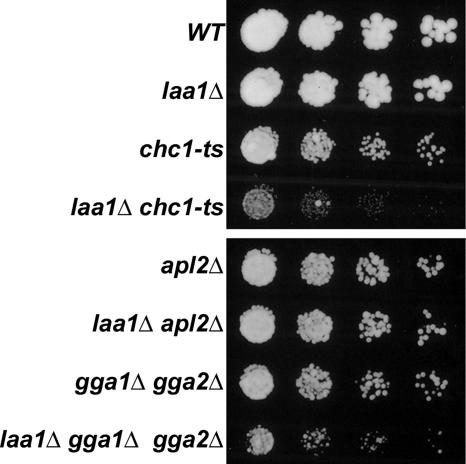
To test whether deletion of LAA1 (laa1Δ) causes synthetic growth defects when combined with chc1-ts, cultures of single and double mutant strains were diluted onto agar media and incubated at 30°C, a semipermissive temperature for chc1-ts (Figure 1). As observed previously, chc1-ts cells exhibited a mild growth defect at 30°C (Bensen et al., 2000). By comparison, laa1Δ cells grew as well as wild type. In contrast, the laa1Δ chc1-ts double mutant grew substantially slower than either single mutant, demonstrating a genetic interaction between LAA1 and CHC1.
Figure 1.
Genetic interactions of laa1Δ with chc1-ts and gga1Δ gga2Δ. Wild-type (SEY6210, WT), laa1Δ (GPY2867), chc1-ts (GPY1064), laa1Δ chc1-ts (GPY3347), apl2Δ (GPY1783-10A), laa1Δ apl2Δ (GPY2903), gga1Δ gga2Δ (GPY2385), and gga1Δ gga2Δ laa1Δ (GPY2960) cells were cultured in YPD at 30°C, spotted onto YPD agar in serial dilutions, and incubated at 30°C. Top, growth after 2 d; bottom, growth after 1 d.
We also used synthetic genetic analysis as an initial way to assess whether Laa1p preferentially acts in conjunction with AP-1 or Gga adaptors. Growth is robust when either AP-1– or Gga-mediated transport is abolished, but a deletion of both is near-lethal (Costaguta et al., 2001). Thus, cells deleted for either type of adaptor are sensitized to perturbations in the function of the other. We assessed whether laa1Δ affects the growth of cells lacking AP-1 function by introducing laa1Δ into apl2Δ cells, which harbor a deletion in the β1 gene that maximally disrupts AP-1 function (Yeung et al., 1999). The laa1Δ apl2Δ double mutant grew just as well as wild type, suggesting that laa1Δ does not affect Gga function (Figure 1). In contrast, laa1Δ enhanced the mild growth defect of a gga1Δ gga2Δ strain (Figure 1), consistent with selective function of Laa1p in AP-1–mediated traffic.
Synthetic Genetic Effects of laa1Δ on TGN–Endosome Traffic
To more directly evaluate the role of Laa1p in clathrin-dependent transport between the TGN and endosomes, we examined proteolytic maturation of the secreted pheromone α-factor. This peptide pheromone is synthesized as part of a larger polypeptide that is transported through the secretory pathway to the TGN where proteolytic maturation is initiated by the furin-like endoprotease Kex2p (Fuller et al., 1988). Kex2p localization to the TGN depends on clathrin; disrupting clathrin function results in Kex2p redistribution to the plasma membrane, thereby depleting the protease from the TGN (Payne and Schekman, 1989). Without adequate Kex2p at the TGN, α-factor precursor is not cleaved efficiently and is secreted as various immature forms that can be distinguished from mature α-factor by SDS-PAGE. Accordingly, secreted α-factor provides a convenient assay for the integrity of clathrin-dependent transport between the TGN and endosomes (Payne and Schekman, 1989).
Although clathrin mutations strongly affect Kex2p localization and α-factor maturation, single gene deletions of TGN/endosome clathrin adaptors such as AP-1 subunits or Gga2 proteins have little or no effect, most likely because of gene redundancy and/or alternative pathways (Yeung et al., 1999; Costaguta et al., 2001). However, such deletions cause α-factor maturation defects when combined with the chc1-ts allele at a temperature (24°C) where the chc1-ts allele is relatively innocuous on its own. Similarly, combination of gene deletions in the AP-1 β1 gene and Gga2p result in a marked α-factor maturation defect (Costaguta et al., 2001). Thus, in a manner analogous to the growth assays, synthetic genetic effects on α-factor maturation can be used to probe the relationships between Laa1p, clathrin, and adaptors in transport between the TGN and endosomes.
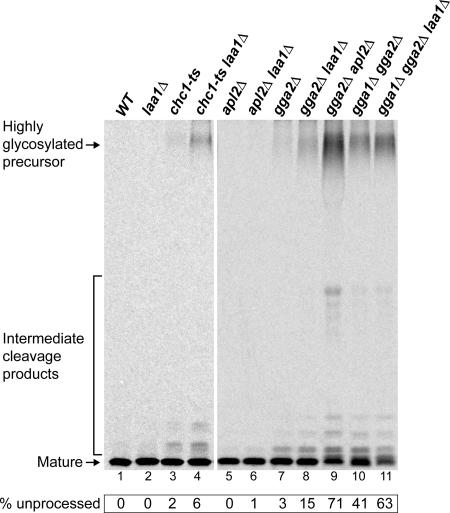
Proteolytic maturation of α-factor was assessed in strains with laa1Δ alone and in combination with mutations in genes encoding clathrin, the AP-1 β1 subunit (APL2), and Gga proteins. Cells were metabolically labeled with [35S]methionine for 45 min at 24°C, and secreted α-factor was immunoprecipitated from the culture supernatant and analyzed by SDS-PAGE (Figure 2). Like wild-type cells, laa1Δ and chc1-ts cells secreted mature α-factor almost exclusively (Figure 2, lanes 1–3). However, combining laa1Δ with chc1-ts significantly enhanced the level of secreted precursor (Figure 2, lane 4), consistent with a role for Laa1p in clathrin-mediated TGN/endosome traffic.
Figure 2.
Deletion of LAA1 enhances the α-factor processing defect of chc1-ts and GGA deletions. The same strains as in Figure 1, plus gga2Δ (GPY2149), gga2Δ laa1Δ (GPY2957), and gga2Δ apl2Δ (GPY2420-3C) strains, were metabolically labeled with [35S]methionine for 45 min at 24°C. The α-factor was immunoprecipitated from the culture supernatants and analyzed by SDS-PAGE and autoradiography. All samples were run on the same gel, and lanes were rearranged for presentation. Unprocessed α-factor was quantified by phosphorimaging analysis.
The laa1Δ mutation also exacerbated the α-factor maturation defect of gga2Δ cells, although not to the same extent as a full disruption of AP-1 (Figure 3, lanes 8 and 9). Importantly, even in a gga1Δ gga2Δ strain where Gga-dependent transport is abolished and α-factor maturation is significantly reduced (41% precursor), introduction of laa1Δ still exaggerated the maturation defect (63% precursor; Figure 2, lanes 10–11). In contrast, neither apl2Δ or laa1Δ apl2Δ cells secreted precursor α-factor (Figure 2, lanes 5 and 6), suggesting that Gga-mediated transport is fully functional in the absence of LAA1. Overall, the genetic interactions observed by α-factor maturation assays mirror those in the growth assays, providing functional evidence that Laa1p acts preferentially with AP-1 in TGN/endosome traffic.
Figure 3.
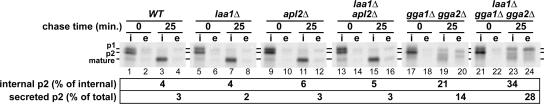
Deletion of LAA1 disrupts CPY trafficking in GGA-deficient cells. The strains used in Figure 1 (except chc1-ts strains) were metabolically labeled with [35S]methionine for 10 min at 30°C. Internal (i) and external (e) CPY was immunoprecipitated either immediately after labeling or after a 25-min chase. Immunoprecipitates were analyzed by SDS-PAGE and autoradiography and quantified by phosphorimaging. The p1 (endoplasmic reticulum), p2 (Golgi), and mature (vacuole) forms of CPY are indicated. The experiment was performed twice with similar results.
laa1Δ Disrupts Transport of CPY in gga1Δ gga2Δ Cells
As another way to investigate Laa1p function in TGN/endosome traffic, we examined transport of CPY, a vacuolar hydrolase. CPY is synthesized as an inactive precursor that is core glycosylated in the endoplasmic reticulum (p1 CPY). After transport to the Golgi, p1 CPY is glycosylated further to produce a higher molecular weight p2 form. In the TGN, Vps10p serves as a sorting receptor that directs p2 CPY into clathrin-coated vesicles destined for endosomes. Once delivered to endosomes, Vps10p releases its cargo and recycles back to the TGN for additional rounds of transport, whereas endosomal p2 CPY is delivered to the vacuole and proteolytically activated to the mature form (mCPY) (Conibear and Stevens, 1998). A sudden block in clathrin function with a temperature-sensitive mutant inhibits Vps10p sorting, resulting in redistribution of the receptor to the cell surface, and secretion of p2 CPY (Seeger and Payne, 1992a).
Clathrin-dependent transport of CPY depends primarily on Gga adaptors; deletion of GGA1 and GGA2, but not AP-1 subunit genes, causes substantial defects in p2 CPY transport and sorting (Dell’Angelica et al., 2000; Hirst et al., 2000). However, even in gga1Δ gga2Δ cells, a significant proportion of CPY is matured (Dell’Angelica et al., 2000; Hirst et al., 2001), suggesting that p2 CPY can reach the vacuole by Gga-independent routes, including an AP-1–mediated pathway (Black and Pelham, 2000). Thus, if Laa1p acts in AP-1–mediated transport, laa1Δ should affect CPY maturation in gga1Δ gga2Δ cells but not in apl2Δ cells.
Cells carrying laa1Δ alone or combined with deletions in either APL2 or both GGA genes were radiolabeled with [35S]methionine for 10 min, subjected to a 25-min chase period, and then CPY was immunoprecipitated from internal and extracellular fractions. No CPY sorting or maturation defect was apparent in laa1Δ or laa1Δ apl2Δ cells (Figure 3, lanes 5–8 and 13–16), as expected, because AP-1 is not required for CPY transport when Gga proteins are present. The gga1Δ gga2Δ strain secreted moderate levels of p2 CPY by 25 min (Figure 3, lane 20) and also accumulated internal p2 CPY (Figure 3, lane 19). Compared with the gga double mutant, the gga1Δ gga2Δ laa1Δ cells exhibited increases in both p2 CPY secretion (Figure 3, lane 24) and the relative level of internal p2 CPY (Figure 3, lane 23). These findings bolster the view that Laa1p functions specifically in AP-1–mediated traffic. The results also suggest that at least some of the residual CPY maturation in gga1Δ gga2Δ cells is due to transport through a Laa1p- and AP-1–dependent pathway.
Laa1p Physically Associates with AP-1
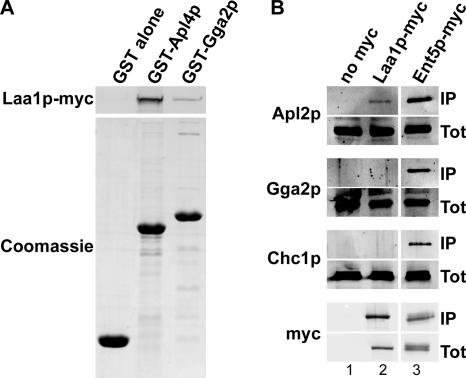
The mammalian homologue of Laa1p, p200, was first identified as a binding partner of the γ-ears of AP-1 and Gga proteins in affinity binding experiments using cell extracts and GST-γ-ear fusions (Lui et al., 2003). We also observed binding of Laa1p in cell extracts to GST fusions containing the AP-1 and Gga2p γ-ears (Figure 4A). More Laa1p bound to the AP-1 γ-ear, suggesting a higher affinity interaction (Figure 4A, compare lanes 2 and 3). To assess whether Laa1p associates with AP-1 and Ggas in vivo, we applied a coimmunoprecipitation strategy. A C-terminally myc epitope-tagged version of Laa1p expressed from the LAA1 chromosomal locus was immunoprecipitated from cell extracts with an anti-myc antibody. The precipitate was analyzed by immunoblotting for the presence of associated clathrin (Chc1p), AP-1 (Apl2p), and Gga2p, the more abundant of the two Gga proteins of yeast (Costaguta et al., 2001). For comparison, myc-tagged Ent5p, a protein known to interact directly with clathrin, AP-1, and Gga2p (Duncan et al., 2003), was subjected to the same analysis. As shown in Figure 4B, Apl2p, but not Gga2p or clathrin, was detected in the Laa1p-myc immunoprecipitate (Figure 4B, lane 2). The level of AP-1 that coimmunoprecipitated with Laa1p-myc was low compared with that with Ent5p-myc (Figure 4B, compare lanes 2 and 3), suggesting that the interaction between Laa1p and AP-1 is transient or indirect. The association of Laa1p with AP-1 and not with Gga2p is consistent with the genetic evidence for selective function of Laa1p in AP-1–mediated transport.
Figure 4.
Laa1p associates with AP-1. (A) Extract from cells expressing myc-tagged Laa1p (GPY2853) was subjected to affinity chromatography with GST alone and GST fusions to the γ-ear regions of AP-1 (Apl4p) and Gga2p. The amount of each pull-down analyzed in the Coomassie lanes corresponds to 25% of that analyzed in the Laa1p-myc lanes. (B) Extracts from cells expressing untagged proteins (SEY6210), myc-tagged Laa1p (GPY2853), or myc-tagged Ent5p (GPY2736) were subjected to immunoprecipitation with an anti-myc antibody. Immunoprecipitates were analyzed by SDS-PAGE and immunoblotting with antibodies against the β subunit of AP-1 (Apl2p), Gga2p, clathrin heavy chain (Chc1p), and myc. The amount of total extract in each Tot lane corresponds to 5% of the input extract used for immunoprecipitations analyzed in the IP lanes.
Laa1p Is Not a Stable Component of Clathrin-coated Vesicles
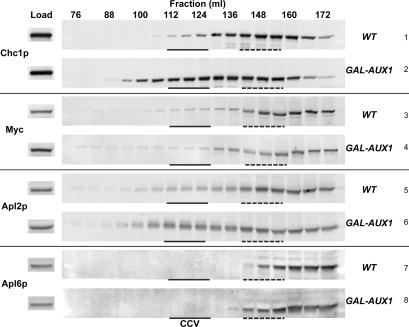
We tested whether Laa1p is a stable component of CCV by probing for the presence of Laa1p-myc in fractions of cell extracts enriched for CCV. For this purpose, we used a strain engineered to conditionally accumulate CCV. This strain expresses Aux1p/Swa2p under control of the galactose-inducible GAL1 promoter. Aux1p is an Hsp70 cochaperone that functions in CCV uncoating (Ungewickell et al., 1995; Gall et al., 2000; Pishvaee et al., 2000). Growth of GAL-AUX1 cells in glucose to repress Aux1p expression leads to inhibition of CCV uncoating and accumulation of CCV that cannot fuse with target organelles. CCV accumulation can be readily detected as a shift in clathrin distribution from soluble to vesicle-containing fractions by size exclusion chromatography. Coat-associated proteins and CCV cargo display a corresponding shift in distribution (Pishvaee et al., 2000). Thus, enrichment of a protein in CCV fractions from Aux1p-depleted cells compared with wild-type cells, can be used as a straightforward and reliable measure of CCV association.
Cell extracts from wild-type and Aux1p-depleted cells were subjected to medium-speed centrifugation to remove unbroken cells and large membranes and the resulting supernatant was applied to a Sephacryl S-1000 gel filtration column. Column fractions were analyzed by immunoblotting for clathrin heavy chain and Laa1p-myc. As controls, we also probed for the distribution of the β subunits of AP-1 as a CCV-associated marker and AP-3 (Apl6p) as a CCV-independent marker (Pishvaee et al., 2000). The majority of clathrin from the wild-type cell extract eluted with a peak at ∼148 ml, which corresponds to free triskelia (Figure 5, row 1). A dramatic shift in the clathrin elution pattern occurred upon repression of Aux1p, with a broad peak of clathrin present in CCV centered around 118 ml (Figure 5, row 2). As expected, Apl2p from Aux1p-depeleted cells exhibited a shift in elution profile to CCV-containing fractions (Figure 5, compare rows 5 and 6), whereas Apl6p did not fractionate with CCV in either wild-type or Aux1p-depleted extracts (Figure 5, rows 7 and 8). Analysis of Laa1p revealed little or no shift to CCV fractions from Aux1p-depleted cells in multiple experiments; a representative example is presented in Figure 5, rows 3 and 4. These results suggest that only a small fraction of Laa1p, if any, is present on CCV, either because Laa1p is not incorporated into fully formed vesicles or because the association is unstable. In this regard, Laa1p is similar to many other clathrin accessory factors (Slepnev and De Camilli, 2000).
Figure 5.
A minor population of Laa1p associates with CCV. LAA1-13Myc cells expressing AUX1/SWA2 from its endogenous promoter (GPY2983, WT) or a galactose-inducible promoter (GPY3011, GAL-AUX1) were grown for at least 24 h at 30°C in YPD, a repressive medium for GAL-AUX1. Extracts were fractionated by differential centrifugation, and medium-speed supernatants were applied to a Sephacryl S-1000 column. Proteins in each fraction were trichloroacetic acid (TCA)-precipitated and analyzed by SDS-PAGE and immunoblotting with antibodies against clathrin heavy chain (Chc1p), myc, and the β subunits of AP-1 (Apl2p) and AP-3 (Apl6p). CCV-containing fractions are indicated with a solid line, and soluble triskelion fractions are indicated with a dotted line. Each lane contains 25% of TCA precipitate (fractions) or 0.5% of total material loaded on column (load).
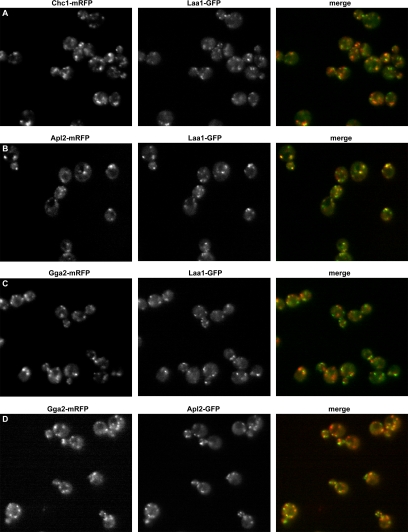
Laa1p Preferentially Localizes with AP-1
To compare localization of Laa1p with clathrin and TGN/endosome adaptors in living cells, we carried out epifluorescence microscopy of cells coexpressing functional forms of Laa1-GFP with mRFP-tagged Chc1p, Apl2p, or Gga2p. Relative localization of Apl2p-GFP and Gga2-mRFP was also examined. A subset of Laa1-GFP puncta (30 ± 9%) overlapped with Chc1p-mRFP (Figure 6A), confirming results from a previous whole genome localization study (Huh et al., 2003). Interestingly, Laa1-GFP colocalization with Apl2p-mRFP was more pronounced than with clathrin (50 ± 8%; Figure 6, A and B), suggesting that Laa1p is associated with AP-1 at times or locations where clathrin is not assembled. A substantial overlap of Apl2p and Gga2p (∼40% of each) was also observed (Figure 6D). Even so, Laa1p puncta were preferentially associated with AP-1 compared with Gga2p (50 ± 8 versus 26 ± 9%; Figure 6, B and C), in agreement with results from coimmunoprecipitations.
Figure 6.
Laa1p colocalization with clathrin, AP-1, and Gga2p. Cells expressing GFP-tagged Laa1p (Laa1-GFP) together with mRFP-tagged clathrin heavy chain (GPY3028, Chc1-mRFP) (A), AP-1 β1 subunit (GPY3034, Apl2-mRFP) (B), or Gga2p (GPY3018, Gga2-mRFP) (C) were grown to logarithmic phase and visualized by epifluorescence microscopy. (D) Cells expressing Gga2-mRFP and GFP-tagged β1 (Apl2-GFP) together (GPY3109) were visualized as described in A–C.
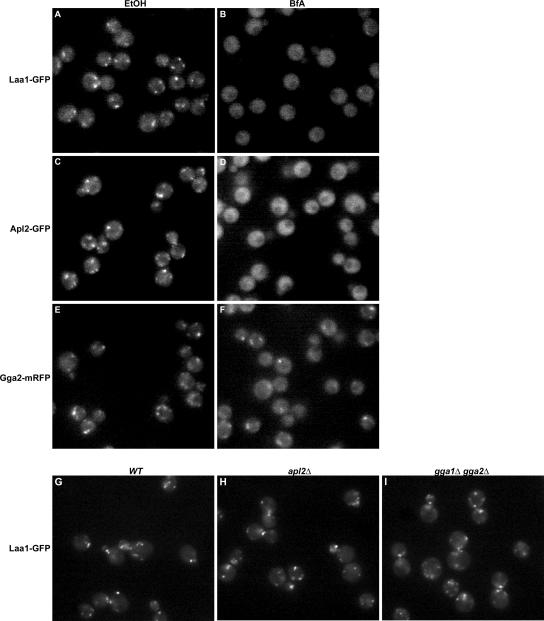
Laa1p and AP-1 Localization Is Brefeldin A Sensitive
In mammalian cells, clathrin adaptors such as AP-1 and Gga proteins rely on the activated form of the Arf1 GTPase for recruitment to TGN/endosome membranes. As a consequence, localization of these adaptors is highly sensitive to BfA, an inhibitor of Arf GTP exchange activity (Donaldson et al., 1992; Robinson and Kreis, 1992; Stamnes and Rothman, 1993; Dell’Angelica et al., 2000). To explore the relative sensitivities of yeast AP-1, Gga proteins, and Laa1p localization to BfA, we treated BfA-permeable (Graham et al., 1993; Shah and Klausner, 1993; Vogel et al., 1993) cells expressing GFP-tagged Apl2p, Gga2p, and Laa1p and assessed localization by fluorescence microscopy. BfA treatment caused extensive mislocalization of Apl2-GFP (Figure 7D). In contrast, Gga2-mRFP puncta were still observed in the presence of BfA (Figure 7F), consistent with previous observations that an interaction with Arf is dispensable for Gga localization in yeast (Boman et al., 2002). Laa1-GFP was almost completely diffuse in BfA-treated cells (Figure 7B), mirroring the effect of BfA on AP-1. These results implicate Arf in Laa1p localization and are also the first demonstration of the contribution of activated Arf to the localization of yeast AP-1.
Figure 7.
Laa1p localization depends on Arf but not AP-1 or Gga proteins. (A–F) Laa1p and AP-1 are sensitive to brefeldin A. Cells expressing GFP-tagged Laa1p (GPY3944, Laa1-GFP; A and B), Apl2p (GPY3942, Apl2-GFP; C and D), or mRFP-tagged Gga2p (GPY3943, Gga2-mRFP; E and F) were grown to log phase and then treated with 75 μg/ml BfA (B, D, and F) or without (ethanol, EtOH; A, C, and E). Cells were visualized by epifluorescence microscopy 10–15 min after addition of BfA. (G–I) Laa1p localization does not depend on AP-1 or Gga adaptors. Wild-type (GPY3262, WT, G), apl2Δ (GPY3116, H), and gga1Δgga2Δ (GPY3111, I) cells expressing GFP-tagged Laa1p were grown to early log phase and visualized.
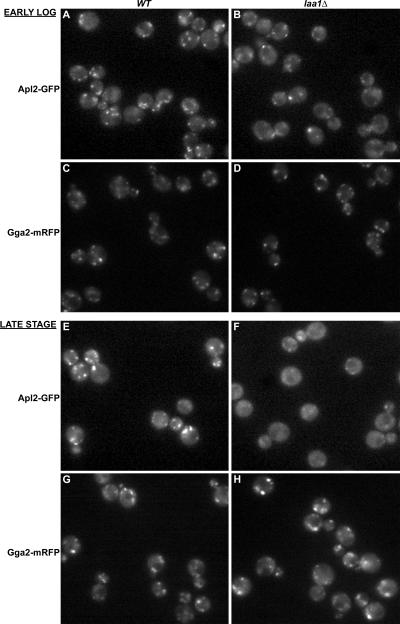
Laa1p Is Important for AP-1 Localization
Given the association of Laa1p with AP-1, we tested whether Laa1p localization depends on AP-1. However, Laa1-GFP distribution was unaltered in either apl2Δ (Figure 7H) or gga1Δ gga2Δ cells (Figure 7I). In a reciprocal experiment, Apl2p-GFP was examined in wild-type and laa1Δ cells. In cells growing in logarithmic phase, the intensity and frequency of Apl2p-GFP puncta was reduced in laa1Δ cells (Figure 8, A and B). Because the effect on AP-1 localization was not complete, we also examined five different pairs of LAA1/laa1Δ strains under the same growth conditions in blind experiments, and in each case the laa1Δ strain was identified by a defect in AP-1 localization. In contrast, Gga2-mRFP localization was unaffected in laa1Δ strains, providing evidence that the effect on AP-1 was specific.
Figure 8.
AP-1 is mislocalized in laa1Δ cells. Wild-type (A, C, E, and G) and laa1Δ (B, D, F, and H) cells expressing GFP-tagged β1 (Apl2-GFP; A and B and E and F) or mRFP-tagged Gga2p (Gga2-mRFP; C and D and G and H) were grown to a late stage (E–H) and visualized by epifluorescence microscopy. The late stage cultures were diluted and grown to early log phase before visualizing again (A–D).
Interestingly, we noticed that Apl2p-GFP mislocalization was more pronounced in laa1Δ cells at a late growth phase where cells shift energy production from fermentative metabolism to respiration (diauxic shift) (Figure 8, E and F). At this phase of growth, Apl2p-GFP puncta in wild-type cells were generally brighter than in cells in logarithmic growth (compare Figure 8, A to E). However, in laa1Δ cells Apl2p-GFP was largely diffuse (Figure 8F), whereas Gga2-mRFP was unperturbed (Figure 8, G and H). At both logarithmic and late growth phases, the overall levels of Apl2p-GFP were equal in wild-type and laa1Δ cells as assessed by immunoblotting (our unpublished data). Together, these findings indicate a specific role for Laa1p in AP-1 localization.
The HCR of Laa1p Is Important for Localization
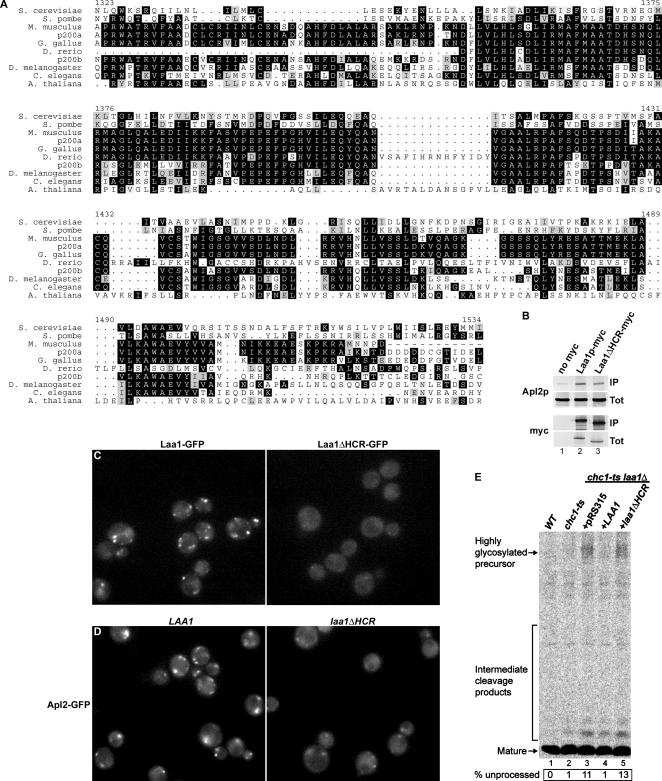
Evolutionary conservation of Laa1p is especially striking in an ∼200-amino acid segment of the protein, which we have termed the HCR (Figure 9A). S. cerevisiae Laa1p averages 45.7% similarity/25.1% identity to the two isoforms of human p200 in the HCR, compared with 25.3% similarity/10.1% identity outside the region. To investigate whether this high degree of conservation reflects a critical role for the HCR in Laa1p function, we determined whether deletion of the HCR affects interaction with AP-1 or Laa1p localization. To monitor AP-1 interaction by coimmunoprecipitation, a myc-tagged mutant lacking the HCR was generated (Laa1ΔHCRp-myc). By immunoblotting, the mutant was expressed at the same levels as wild-type, myc-tagged Laa1p (Figure 9B, myc, Tot). When Laa1p-myc or Laa1ΔHCRp-myc were immunoprecipitated, associated AP-1 was detected at nearly the same levels (Figure 9B, compare Apl2p in lanes 2 and 3), indicating that the HCR is dispensable for AP-1 interaction. Laa1ΔHCRp was also tagged at the C terminus with GFP to allow localization studies in living cells. Although Laa1ΔHCR-GFP was expressed at the same levels as Laa1-GFP by immunoblotting (our unpublished data), no fluorescent puncta like those typical of wild-type Laa1-GFP were observed (Figure 9C). We also fused the HCR to GFP to determine whether HCR alone is sufficient for localization. HCR-GFP was expressed but did not localize to puncta (our unpublished data), suggesting that the HCR is necessary but not sufficient for localization of Laa1p.
Figure 9.
The HCR of Laa1p is important for Laa1p localization and function. (A) Alignment of the Laa1p HCR with homologues from various organisms; numbering corresponds to S. cerevisiae Laa1p residues. (B) Laa1ΔHCRp associates with AP-1. Extracts from cells expressing untagged proteins (SEY6210 + pRS315), myc-tagged Laa1p (GPY2853 + pRS315), or myc-tagged Laa1p missing the HCR (Laa1ΔHCRp-myc, GPY2867 + pRS315-laa1ΔHCR-13Myc) were subjected to immunoprecipitation and analysis as in Figure 4B. (C) Laa1ΔHCRp is mislocalized. Cells expressing GFP-tagged Laa1ΔHCRp (Laa1ΔHCR-GFP, GPY2867 + pRS315-laa1ΔHCR-GFP) were grown and visualized as in Figure 7, G–I. (D) AP-1 is mislocalized in cells lacking the HCR of Laa1p. Cells expressing GFP-tagged β1 (Apl2p) and either Laa1ΔHCRp-myc (GPY3085 + pRS315-laa1ΔHCR-13Myc, laa1ΔHCR) or full-length Laa1p (GPY3086 + pRS315, WT) were analyzed as in Figure 8. Cells at a late stage of growth are shown. (E) laa1ΔHCR causes a synthetic α-factor processing defect with chc1-ts. Wild-type (SEY6210 + pRS315) and chc1-ts (GPY1064 + pRS315) cells, as well as chc1-ts laa1Δ cells carrying either full-length LAA1 (GPY3347 + pRS315-LAA1) or laa1ΔHCR (GPY3347 + pRS315-laa1ΔHCR) on plasmids were analyzed as described in Figure 2.
As independent tests of the importance of the HCR, we monitored AP-1 localization and α-factor maturation in cells expressing an untagged form of Laa1ΔHCRp. AP-1 localization was assayed using Apl2-GFP in cells grown to a late phase of growth where the localization defect in laa1Δ cells is most apparent. Apl2-GFP was mislocalized in laa1ΔHCR cells (Figure 9D) to an extent similar to that in laa1Δ cells (Figure 8F). Thus, even though Laa1ΔHCRp can associate with AP-1, it is defective in localizing AP-1. Further supporting the importance of the HCR, a synthetic defect in α-factor maturation was observed in chc1-ts laa1ΔHCR cells (Figure 9E, compare lanes 4 and 5). These experiments define the highly conserved region as a critical determinant for Laa1p localization, and, consequently, for Laa1p-mediated localization of AP-1.
DISCUSSION
Paradigms for clathrin-mediated vesicle formation have become more complex with the identification of multiple adaptors and accessory proteins, many conserved from yeast to humans, that participate together with clathrin and AP-2 in endocytic vesicle formation (Mousavi et al., 2004). It has been unclear whether CCV formation at the TGN and endosomes involves a similar level of complexity and evolutionary conservation (Traub, 2005). To address these issues, we have focused on identifying and characterizing TGN/endosome adaptors and accessory proteins in yeast. Like mammalian cells, yeast express two types of adaptors involved in TGN/endosome CCV formation, AP-1 and Gga proteins. Here, we identify yeast Laa1p as an evolutionarily conserved accessory factor specific for AP-1, but not Gga adaptors. Our data reveal that Laa1p plays an important role in AP-1 localization and AP-1–mediated protein traffic in yeast.
We favor the idea that Laa1p-mediated localization of AP-1 involves recruitment of cytoplasmic AP-1 to membranes. However, we have not observed differences in AP-1 fractionation in extracts of wild-type and laa1Δ cells by differential centrifugation or CCV chromatography (our unpublished data). The differences between in vivo and in vitro assays are probably due to nonphysiological effects that occur upon cell lysis and fractionation, particularly given our finding that even in live laa1Δ cells AP-1 localization is sensitive to changes that occur in response to different growth phases. An alternative explanation for AP-1 mislocalization in laa1Δ cells is that AP-1 is recruited to membranes, but those membranes are grossly perturbed by the absence of Laa1p. This possibility seems unlikely because there is substantial overlap of AP-1 and Gga proteins in wild-type cells, and while AP-1 is mislocalized in laa1Δ cells, Gga2p localization is unaffected.
The role for Laa1p in AP-1 localization defined by our studies seemingly contrasts with results from analysis of the human Laa1p homologues, p200a and p200b. One p200 isoform, p200a, was identified as a component of a complex also containing two γ-ear-binding proteins, aftiphilin and γ-synergin (Hirst et al., 2005). Depletion of aftiphilin by small interfering RNA (siRNA) reduced overall levels of the complex, but it did not alter AP-1 localization. Instead, siRNA-mediated depletion of AP-1 abolished perinuclear concentration of aftiphilin and presumably of its binding partners, although p200 was not directly examined. Several explanations can account for the apparent discrepancy between the findings in yeast and mammalian cells. Probably most reasonable is the existence of a second mammalian p200 isoform, p200b. Presumptive p200b depletion by siRNA did not affect stability of aftiphilin or γ-synergin whereas depletion of p200a reduced levels of both binding partners (Hirst et al., 2005), raising the possibility that p200b functions independently of the aftiphilin complex. However, even though depletion of p200b did not alter stability of the aftiphilin complex, there was an effect on AP-1–mediated TGN/endosome traffic comparable with that observed with p200a depletion, a result consistent with an aftiphilin-independent role for p200b in AP-1–dependent transport (Hirst et al., 2005). Alternatively, either or both p200 isoforms could function both in the context of an aftiphilin complex and also separately. In support of this scenario, a proportion of p200a did not fractionate with aftiphilin and γ-synergin by gel filtration chromatography (Hirst et al., 2005). Given these considerations we propose that the function of Laa1p in AP-1 localization revealed by our studies in yeast reflects a fundamental function that is conserved in mammalian p200 that accounts for the stronger evolutionary conservation of this protein family compared with other members of the aftiphilin complex. Our finding that the most highly conserved region of Laa1p is critical for AP-1 localization (Figure 9D) comports with this hypothesis.
Mammalian p200a seems to bind the AP-1 γ-ear domain indirectly by virtue of its association with aftiphilin and γ-synergin, each of which has multiple γ-ear-binding motifs matching a general consensus of (D/E)nΦXXΦ (Duncan and Payne, 2003; Mattera et al., 2004). Like the aftiphilin complex, Laa1p recognizes the AP-1 γ-ear domain as revealed by GST-fusion affinity binding (Figure 4A). However, because yeast lacks obvious homologues of aftiphilin and γ-synergin it is presently unclear whether AP-1 binding by Laa1p is direct or indirect. Additionally, the absence of aftiphilin and γ-synergin may explain the unstable association we observed between Laa1p and yeast CCV (Figure 5). The interaction between Laa1p and AP-1 detected by coimmunoprecipitation was weak compared with Ent5p, which is known to directly bind γ-ears through multiple (D/E)nΦXXΦ motifs (Duncan et al., 2003). This could reflect indirect binding of Laa1p to the γ-ear or weak direct binding through one or more of the several (D/E)nΦXXΦmotifs present in Laa1p. Current efforts to identify Laa1p interaction partners may help distinguish between these possibilities as will mutagenesis of the putative γ-ear interaction motifs.
The residual AP-1 localization and function in Laa1p-deficient cells (Figure 8) indicates that Laa1p is likely to be one of several factors that contribute to AP-1 localization, including Arf, phosphoinositides, and cargo proteins (Robinson, 2004). The parallel sensitivities of Laa1p and AP-1 to brefeldin A (Figure 7) suggest that Laa1p may act with Arf to localize AP-1. If this is the case, interaction of Laa1p with the AP-1 γ-ear and Arf with the AP-1 core as described for mammalian Arf1 (Austin et al., 2000) could provide multivalent interactions to promote cooperative clathrin coat nucleation or foster coat stability. The highly conserved domain of Laa1p is necessary for localization, not interaction with AP-1 (Figure 9), making it a candidate region for interaction with Arf, either directly or indirectly. Preliminary two-hybrid analysis has not revealed interaction of an HCR-containing region of Laa1p with Arf1p (our unpublished data), but further experiments will be needed to fully test the possibility of Arf1 binding.
Unexpectedly, we observed that AP-1 displayed greater reliance on Laa1p for localization in late stages of growth when cell densities near saturation (Figure 8). We can only speculate on the basis of this effect. Perhaps metabolic remodeling caused by the change from fermentative to respiratory growth increases the importance of Laa1p relative to other factors involved in AP-1 recruitment to the TGN/endosomes. Regardless of the cause, this observation hints at regulatory mechanisms that affect AP-1–mediated trafficking in response to the growth and metabolic status of the cell. It is intriguing to consider that such regulatory pathways may also account for the observation that an Arf1 guanine nucleotide exchange factor, Gcs1p, is necessary at low temperatures for transition of stationary cells to active growth (Drebot et al., 1987; Poon et al., 1996).
The relationship between AP-1 and Gga-mediated pathways between the TGN and endosomes has not been well defined (Hinners and Tooze, 2003). AP-1 has been implicated in both forward traffic from the TGN to endosomes and retrieval from endosomes to the TGN. Gga proteins seem to function primarily in the forward route, but whether that route involves AP-1 or is parallel to an AP-1–mediated pathway is unclear. Studies of mannose-6-phosphate receptor sorting in mammalian cells suggest a sequential action of Gga proteins and AP-1 (Doray et al., 2002). Although a similar situation could exist in yeast, the results of our genetic interaction studies are most consistent with models invoking parallel AP-1 and Gga-mediated pathways (Black and Pelham, 2000; Costaguta et al., 2001; Ha et al., 2003). If AP-1 and Gga proteins act in an obligatory sequential manner then inactivation of both adaptors should not be significantly worse than inactivation of individual adaptors. However, as previously reported, deletion of GGA genes and the AP-1 β subunit gene resulted in a near-lethal synthetic growth defect (Costaguta et al., 2001).
Here, we report that deletion of LAA1 also causes synthetic growth defects with gga mutations. Because the laa1Δ gga mutant cells are viable, we could also assess synthetic effects on specific protein trafficking pathways. Synthetic effects on both α-factor maturation (Figure 2) and CPY sorting (Figure 3) were observed when laa1Δ was combined with gga1Δ gga2Δ but not with apl2Δ. These results suggest that Laa1p, an AP-1-specific accessory factor, functions in parallel to Gga proteins in both Kex2p and Vps10p traffic between the TGN and endosomes. Because both Kex2p and Vps10p cycle between the TGN and endosomes, the synthetic defects could in principal be due to effects of laa1Δ on AP-1–mediated retrieval from endosomes. However, using an assay to monitor AP-1–mediated retrieval of chitin synthase III (Valdivia et al., 2002), laa1Δ did not cause defects in retrieval, whereas apl2Δ did (our unpublished data). Thus, our data are most consistent with a model in which AP-1 and Gga adaptors provide parallel functions in TGN to endosome traffic, providing support for similar conclusions based primarily on trafficking of chimeric reporter proteins (Black and Pelham, 2000; Ha et al., 2003). Whether these parallel pathways represent separate routes from the TGN to early (AP-1) and late (Gga) endosomes, parallel pathways to the same endosome, or even parallel function in the same vesicles, remains to be established.
In summary, our study defines Laa1p as an AP-1–specific accessory protein necessary for optimal AP-1 localization. Laa1p joins a small group of AP-1 accessory factors, including members of the aftiphilin complex (Hirst et al., 2005) and γ-BAR, a protein that is necessary for AP-1 localization in mammalian cells (Neubrand et al., 2005). Of these accessories, only Laa1p is conserved between yeast and mammals; aftiphilin is conserved in multicellular organisms, and γ-synergin and γ-BAR seem restricted to mammals. This pattern of conservation offers a preliminary indication that an expanded constellation of AP-1 accessory factors is present in metazoa compared with yeast. Based on this idea, we suggest that Laa1p and its homologues represent a core component of the mechanism responsible for AP-1 localization and AP-1–mediated traffic in eukaryotic cells; in multicellular organisms, the core Laa1p role is integrated into a more complex localization mechanism involving factors such as γ-BAR and is also diversified by incorporation into the aftiphilin complex.
ACKNOWLEDGMENTS
We are grateful to Alex van der Bliek for help with microscopy and members of the van der Bliek and Payne laboratories for helpful discussions. We thank Giancarlo Costaguta, Santiago Di Pietro, Mara Duncan, Todd Lorenz, and Alex van der Bliek for comments on this manuscript. G.E.F. was supported by National Institutes of Health predoctoral fellowship GM-72119 and training grant GM-07185. This work was supported by National Institutes of Health Grants GM-39040 and GM-67911 (to G.S.P.).
Abbreviations used:
- BfA
brefeldin A
- CCV
clathrin-coated vesicle
- CPY
carboxypeptidase Y
- HCR
highly conserved region
- TGN
trans-Golgi network.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06.02-0096) on May 10, 2006.
REFERENCES
- Aguilar R. C., Boehm M., Gorshkova I., Crouch R. J., Tomita K., Saito T., Ohno H., Bonifacino J. S. Signal-binding specificity of the mu4 subunit of the adaptor protein complex AP-4. J. Biol. Chem. 2001;276:13145–13152. doi: 10.1074/jbc.M010591200. [DOI] [PubMed] [Google Scholar]
- Austin C., Hinners I., Tooze S. A. Direct and GTP-dependent interaction of ADP-ribosylation factor 1 with clathrin adaptor protein AP-1 on immature secretory granules. J. Biol. Chem. 2000;275:21862–21869. doi: 10.1074/jbc.M908875199. [DOI] [PubMed] [Google Scholar]
- Bai H., Doray B., Kornfeld S. GGA1 interacts with the adaptor protein AP-1 through a WNSF sequence in its hinge region. J. Biol. Chem. 2004;279:17411–17417. doi: 10.1074/jbc.M401158200. [DOI] [PubMed] [Google Scholar]
- Baudin A., Ozier-Kalogeropoulos O., Denouel A., Lacroute F., Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensen E. S., Costaguta G., Payne G. S. Synthetic genetic interactions with temperature-sensitive clathrin in Saccharomyces cerevisiae. Roles for synaptojanin-like Inp53p and dynamin-related Vps1p in clathrin-dependent protein sorting at the trans-Golgi network. Genetics. 2000;154:83–97. doi: 10.1093/genetics/154.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensen E. S., Yeung B. G., Payne G. S. Ric1p and the Ypt6p GTPase function in a common pathway required for localization of trans-Golgi network membrane proteins. Mol. Biol. Cell. 2001;12:13–26. doi: 10.1091/mbc.12.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black M. W., Pelham H. R. A selective transport route from Golgi to late endosomes that requires the yeast GGA proteins. J. Cell Biol. 2000;151:587–600. doi: 10.1083/jcb.151.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boman A. L., Salo P. D., Hauglund M. J., Strand N. L., Rensink S. J., Zhdankina O. ADP-ribosylation factor (ARF) interaction is not sufficient for yeast GGA protein function or localization. Mol. Biol. Cell. 2002;13:3078–3095. doi: 10.1091/mbc.E02-02-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boman A. L., Zhang C., Zhu X., Kahn R. A. A family of ADP-ribosylation factor effectors that can alter membrane transport through the trans-Golgi. Mol. Biol. Cell. 2000;11:1241–1255. doi: 10.1091/mbc.11.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino J. S. The GGA proteins: adaptors on the move. Nat. Rev. Mol. Cell. Biol. 2004;5:23–32. doi: 10.1038/nrm1279. [DOI] [PubMed] [Google Scholar]
- Brodsky F. M., Chen C. Y., Knuehl C., Towler M. C., Wakeham D. E. Biological basket weaving: formation and function of clathrin-coated vesicles. Annu. Rev. Cell Dev. Biol. 2001;17:517–568. doi: 10.1146/annurev.cellbio.17.1.517. [DOI] [PubMed] [Google Scholar]
- Conibear E., Stevens T. H. Multiple sorting pathways between the late Golgi and the vacuole in yeast. Biochim. Biophys. Acta. 1998;1404:211–230. doi: 10.1016/s0167-4889(98)00058-5. [DOI] [PubMed] [Google Scholar]
- Costaguta G., Stefan C. J., Bensen E. S., Emr S. D., Payne G. S. Yeast Gga coat proteins function with clathrin in Golgi to endosome transport. Mol. Biol. Cell. 2001;12:1885–1896. doi: 10.1091/mbc.12.6.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles C. R., Odorizzi G., Payne G. S., Emr S. D. The AP-3 adaptor complex is essential for cargo-selective transport to the yeast vacuole. Cell. 1997;91:109–118. doi: 10.1016/s0092-8674(01)80013-1. [DOI] [PubMed] [Google Scholar]
- Dell’Angelica E. C., Klumperman J., Stoorvogel W., Bonifacino J. S. Association of the AP-3 adaptor complex with clathrin. Science. 1998;280:431–434. doi: 10.1126/science.280.5362.431. [DOI] [PubMed] [Google Scholar]
- Dell’Angelica E. C., Puertollano R., Mullins C., Aguilar R. C., Vargas J. D., Hartnell L. M., Bonifacino J. S. GGAs: a family of ADP ribosylation factor-binding proteins related to adaptors and associated with the Golgi complex. J. Cell Biol. 2000;149:81–94. doi: 10.1083/jcb.149.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson J. G., Finazzi D., Klausner R. D. Brefeldin A inhibits Golgi membrane-catalysed exchange of guanine nucleotide onto ARF protein. Nature. 1992;360:350–352. doi: 10.1038/360350a0. [DOI] [PubMed] [Google Scholar]
- Doray B., Ghosh P., Griffith J., Geuze H. J., Kornfeld S. Cooperation of GGAs and AP-1 in packaging MPRs at the trans-Golgi network. Science. 2002;297:1700–1703. doi: 10.1126/science.1075327. [DOI] [PubMed] [Google Scholar]
- Drebot M. A., Johnston G. C., Singer R. A. A yeast mutant conditionally defective only for reentry into the mitotic cell cycle from stationary phase. Proc. Natl. Acad. Sci. USA. 1987;84:7948–7952. doi: 10.1073/pnas.84.22.7948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan M. C., Costaguta G., Payne G. S. Yeast epsin-related proteins required for Golgi-endosome traffic define a gamma-adaptin ear-binding motif. Nat. Cell Biol. 2003;5:77–81. doi: 10.1038/ncb901. [DOI] [PubMed] [Google Scholar]
- Duncan M. C., Payne G. S. ENTH/ANTH domains expand to the Golgi. Trends Cell Biol. 2003;13:211–215. doi: 10.1016/s0962-8924(03)00076-x. [DOI] [PubMed] [Google Scholar]
- Fuller R. S., Sterne R. E., Thorner J. Enzymes required for yeast prohormone processing. Annu. Rev. Physiol. 1988;50:345–362. doi: 10.1146/annurev.ph.50.030188.002021. [DOI] [PubMed] [Google Scholar]
- Gall W. E., Higginbotham M. A., Chen C., Ingram M. F., Cyr D. M., Graham T. R. The auxilin-like phosphoprotein Swa2p is required for clathrin function in yeast. Curr. Biol. 2000;10:1349–1358. doi: 10.1016/s0960-9822(00)00771-5. [DOI] [PubMed] [Google Scholar]
- Ghosh P., Kornfeld S. The GGA proteins: key players in protein sorting at the trans-Golgi network. Eur. J. Cell Biol. 2004;83:257–262. doi: 10.1078/0171-9335-00374. [DOI] [PubMed] [Google Scholar]
- Graham T. R., Scott P. A., Emr S. D. Brefeldin A reversibly blocks early but not late protein transport steps in the yeast secretory pathway. EMBO J. 1993;12:869–877. doi: 10.1002/j.1460-2075.1993.tb05727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha S. A., Torabinejad J., DeWald D. B., Wenk M. R., Lucast L., De Camilli P., Newitt R. A., Aebersold R., Nothwehr S. F. The synaptojanin-like protein Inp53/Sjl3 functions with clathrin in a yeast TGN-to-endosome pathway distinct from the GGA protein-dependent pathway. Mol. Biol. Cell. 2003;14:1319–1333. doi: 10.1091/mbc.E02-10-0686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinners I., Tooze S. A. Changing directions: clathrin-mediated transport between the Golgi and endosomes. J. Cell Sci. 2003;116:763–771. doi: 10.1242/jcs.00270. [DOI] [PubMed] [Google Scholar]
- Hirst J., Borner G. H., Harbour M., Robinson M. S. The aftiphilin/p200/gamma-synergin complex. Mol. Biol. Cell. 2005;16:2554–2565. doi: 10.1091/mbc.E04-12-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J., Lindsay M. R., Robinson M. S. GGAs: roles of the different domains and comparison with AP-1 and clathrin. Mol. Biol. Cell. 2001;12:3573–3588. doi: 10.1091/mbc.12.11.3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J., Lui W. W., Bright N. A., Totty N., Seaman M. N., Robinson M. S. A family of proteins with gamma-adaptin and VHS domains that facilitate trafficking between the trans-Golgi network and the vacuole/lysosome. J. Cell Biol. 2000;149:67–80. doi: 10.1083/jcb.149.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O’Shea E. K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Kirchhausen T. Adaptors for clathrin-mediated traffic. Annu. Rev. Cell Dev. Biol. 1999;15:705–732. doi: 10.1146/annurev.cellbio.15.1.705. [DOI] [PubMed] [Google Scholar]
- Kirchhausen T. Clathrin. Annu. Rev. Biochem. 2000;69:699–727. doi: 10.1146/annurev.biochem.69.1.699. [DOI] [PubMed] [Google Scholar]
- Kirchhausen T. Clathrin adaptors really adapt. Cell. 2002;109:413–416. doi: 10.1016/s0092-8674(02)00751-1. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lui W. W., Collins B. M., Hirst J., Motley A., Millar C., Schu P., Owen D. J., Robinson M. S. Binding partners for the COOH-terminal appendage domains of the GGAs and gamma-adaptin. Mol. Biol. Cell. 2003;14:2385–2398. doi: 10.1091/mbc.E02-11-0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattera R., Ritter B., Sidhu S. S., McPherson P. S., Bonifacino J. S. Definition of the consensus motif recognized by gamma-adaptin ear domains. J. Biol. Chem. 2004;279:8018–8028. doi: 10.1074/jbc.M311873200. [DOI] [PubMed] [Google Scholar]
- Meyer C., Zizioli D., Lausmann S., Eskelinen E. L., Hamann J., Saftig P., von Figura K., Schu P. mu1A-adaptin-deficient mice: lethality, loss of AP-1 binding and rerouting of mannose 6-phosphate receptors. EMBO J. 2000;19:2193–2203. doi: 10.1093/emboj/19.10.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi S. A., Malerod L., Berg T., Kjeken R. Clathrin-dependent endocytosis. Biochem. J. 2004;377:1–16. doi: 10.1042/BJ20031000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubrand V. E., Will R. D., Mobius W., Poustka A., Wiemann S., Schu P., Dotti C. G., Pepperkok R., Simpson J. C. Gamma-BAR, a novel AP-1-interacting protein involved in post-Golgi trafficking. EMBO J. 2005;24:1122–1133. doi: 10.1038/sj.emboj.7600600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen D. J. Linking endocytic cargo to clathrin: structural and functional insights into coated vesicle formation. Biochem. Soc. Trans. 2004;32:1–14. doi: 10.1042/bst0320001. [DOI] [PubMed] [Google Scholar]
- Owen D. J., Collins B. M., Evans P. R. Adaptors for clathrin coats: structure and function. Annu. Rev. Cell Dev. Biol. 2004;20:153–191. doi: 10.1146/annurev.cellbio.20.010403.104543. [DOI] [PubMed] [Google Scholar]
- Payne G. S., Schekman R. Clathrin: a role in the intracellular retention of a Golgi membrane protein. Science. 1989;245:1358–1365. doi: 10.1126/science.2675311. [DOI] [PubMed] [Google Scholar]
- Perrais D., Merrifield C. J. Dynamics of endocytic vesicle creation. Dev. Cell. 2005;9:581–592. doi: 10.1016/j.devcel.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Pishvaee B., Costaguta G., Yeung B. G., Ryazantsev S., Greener T., Greene L. E., Eisenberg E., McCaffery J. M., Payne G. S. A yeast DNA J protein required for uncoating of clathrin-coated vesicles in vivo. Nat. Cell Biol. 2000;2:958–963. doi: 10.1038/35046619. [DOI] [PubMed] [Google Scholar]
- Poon P. P., Wang X., Rotman M., Huber I., Cukierman E., Cassel D., Singer R. A., Johnston G. C. Saccharomyces cerevisiae Gcs1 is an ADP-ribosylation factor GTPase-activating protein. Proc. Natl. Acad. Sci. USA. 1996;93:10074–10077. doi: 10.1073/pnas.93.19.10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poussu A., Lohi O., Lehto V. P. Vear, a novel Golgi-associated protein with VHS and gamma-adaptin “ear” domains. J. Biol. Chem. 2000;275:7176–7183. doi: 10.1074/jbc.275.10.7176. [DOI] [PubMed] [Google Scholar]
- Puertollano R., Aguilar R. C., Gorshkova I., Crouch R. J., Bonifacino J. S. Sorting of mannose 6-phosphate receptors mediated by the GGAs. Science. 2001a;292:1712–1716. doi: 10.1126/science.1060750. [DOI] [PubMed] [Google Scholar]
- Puertollano R., Randazzo P. A., Presley J. F., Hartnell L. M., Bonifacino J. S. The GGAs promote ARF-dependent recruitment of clathrin to the TGN. Cell. 2001b;105:93–102. doi: 10.1016/s0092-8674(01)00299-9. [DOI] [PubMed] [Google Scholar]
- Robinson M. S. Adaptable adaptors for coated vesicles. Trends Cell Biol. 2004;14:167–174. doi: 10.1016/j.tcb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Robinson M. S., Kreis T. E. Recruitment of coat proteins onto Golgi membranes in intact and permeabilized cells: effects of brefeldin A and G protein activators. Cell. 1992;69:129–138. doi: 10.1016/0092-8674(92)90124-u. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Maniatis T., Fritsch E. F. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Seeger M., Payne G. S. A role for clathrin in the sorting of vacuolar proteins in the Golgi complex of yeast. EMBO J. 1992a;11:2811–2818. doi: 10.1002/j.1460-2075.1992.tb05348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger M., Payne G. S. Selective and immediate effects of clathrin heavy chain mutations on Golgi membrane protein retention in Saccharomyces cerevisiae. J. Cell Biol. 1992b;118:531–540. doi: 10.1083/jcb.118.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah N., Klausner R. D. Brefeldin A reversibly inhibits secretion in Saccharomyces cerevisiae. J. Biol. Chem. 1993;268:5345–5348. [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmen T., Honing S., Icking A., Tikkanen R., Hunziker W. AP-4 binds basolateral signals and participates in basolateral sorting in epithelial MDCK cells. Nat. Cell Biol. 2002;4:154–159. doi: 10.1038/ncb745. [DOI] [PubMed] [Google Scholar]
- Simpson F., Bright N. A., West M. A., Newman L. S., Darnell R. B., Robinson M. S. A novel adaptor-related protein complex. J. Cell Biol. 1996;133:749–760. doi: 10.1083/jcb.133.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slepnev V. I., De Camilli P. Accessory factors in clathrin-dependent synaptic vesicle endocytosis. Nat. Rev. Neurosci. 2000;1:161–172. doi: 10.1038/35044540. [DOI] [PubMed] [Google Scholar]
- Stamnes M. A., Rothman J. E. The binding of AP-1 clathrin adaptor particles to Golgi membranes requires ADP-ribosylation factor, a small GTP-binding protein. Cell. 1993;73:999–1005. doi: 10.1016/0092-8674(93)90277-w. [DOI] [PubMed] [Google Scholar]
- Stepp J. D., Huang K., Lemmon S. K. The yeast adaptor protein complex, AP-3, is essential for the efficient delivery of alkaline phosphatase by the alternate pathway to the vacuole. J. Cell Biol. 1997;139:1761–1774. doi: 10.1083/jcb.139.7.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsu H., Yoshino K., Nakayama K. Adaptor gamma ear homology domain conserved in gamma-adaptin and GGA proteins that interact with gamma-synergin. Biochem. Biophys. Res. Commun. 2000;271:719–725. doi: 10.1006/bbrc.2000.2700. [DOI] [PubMed] [Google Scholar]
- Theos A. C., et al. Functions of adaptor protein (AP)-3 and AP-1 in tyrosinase sorting from endosomes to melanosomes. Mol. Biol. Cell. 2005;16:5356–5372. doi: 10.1091/mbc.E05-07-0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub L. M. Common principles in clathrin-mediated sorting at the Golgi and the plasma membrane. Biochim. Biophys. Acta. 2005;1744:415–437. doi: 10.1016/j.bbamcr.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Ungewickell E., Ungewickell H., Holstein S. E., Lindner R., Prasad K., Barouch W., Martin B., Greene L. E., Eisenberg E. Role of auxilin in uncoating clathrin-coated vesicles. Nature. 1995;378:632–635. doi: 10.1038/378632a0. [DOI] [PubMed] [Google Scholar]
- Valdivia R. H., Baggott D., Chuang J. S., Schekman R. W. The yeast clathrin adaptor protein complex 1 is required for the efficient retention of a subset of late Golgi membrane proteins. Dev. Cell. 2002;2:283–294. doi: 10.1016/s1534-5807(02)00127-2. [DOI] [PubMed] [Google Scholar]
- Vogel J. P., Lee J. N., Kirsch D. R., Rose M. D., Sztul E. S. Brefeldin A causes a defect in secretion in Saccharomyces cerevisiae. J. Biol. Chem. 1993;268:3040–3043. [PubMed] [Google Scholar]
- Vowels J. J., Payne G. S. A dileucine-like sorting signal directs transport into an AP-3-dependent, clathrin-independent pathway to the yeast vacuole. EMBO J. 1998;17:2482–2493. doi: 10.1093/emboj/17.9.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbur J. D., Hwang P. K., Brodsky F. M. New faces of the familiar clathrin lattice. Traffic. 2005;6:346–350. doi: 10.1111/j.1600-0854.2005.00277.x. [DOI] [PubMed] [Google Scholar]
- Yeung B. G., Payne G. S. Clathrin interactions with C-terminal regions of the yeast AP-1 beta and gamma subunits are important for AP-1 association with clathrin coats. Traffic. 2001;2:565–576. doi: 10.1034/j.1600-0854.2001.20806.x. [DOI] [PubMed] [Google Scholar]
- Yeung B. G., Phan H. L., Payne G. S. Adaptor complex-independent clathrin function in yeast. Mol. Biol. Cell. 1999;10:3643–3659. doi: 10.1091/mbc.10.11.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]