Abstract
GE82832, a secondary metabolite produced by Streptosporangium cinnabarinum (strain GE82832), has been identified as a translational inhibitor by in vitro screening of a library of natural products. Secondary functional tests specific for individual steps of the translational pathway demonstrated that translocation is the specific target of GE82832. Chemical probing in situ demonstrated that this antibiotic protects bases A1324 and A1333 and exposes C1336 of 16S rRNA, thereby indicating that its binding site is located on the head of the 30S ribosomal subunit. The ribosomal location of GE82832, near ribosomal protein S13 and G1338, two elements of the small subunit that are part of or close to the B1a intrasubunit bridge, suggests that translocation inhibition results from an altered dynamics of 30S–50S ribosomal subunit interaction.
Keywords: antibacterial agents, translational inhibitors, translocation, 30S ribosomal subunit
INTRODUCTION
Not all potential antibiotic targets within the translational machinery have been fully exploited. Therefore, in spite of the large number of antibiotics known to interfere with translation and of mutations conferring resistance to them, the search for new inhibitors of the bacterial translational apparatus can still be fruitful. One way of coping with the increasing number of bacterial pathogens displaying resistance to the available antibiotics is to develop new molecules with anti-infective properties that interfere with the function of nonexploited and/or underexploited molecular targets (Gale et al. 1981; Cundliffe 1990; Walsh 2003; Wilson 2004). The recent acquisition of detailed maps of the spatial organization of the ribosomal subunits complexed with known antibiotics at atomic resolution (Brodersen et al. 2000; Carter et al. 2000; Ogle et al. 2001; Pioletti et al. 2001; Vicens and Westhof 2002, 2003; Schlunzen et al. 2003) can ultimately allow the rational design of new inhibitors or of structural variants of known molecules.
A recent high-throughput screening, devised to identify translational inhibitors in a library of microbial products, yielded some novel and interesting molecules (e.g., Brandi et al. 2006a,b). Among these novel inhibitors identified is GE82832, a secondary metabolite of Streptosporangium cinnabarinum (strain GE82832) (L. Cavaletti, unpubl.). GE82832 is a complex of two bioactive isomeric substances (A and B), each with MW 1286 containing an aromatic and a peptidic part whose structure is being elucidated (M. Sette and A. Fabbretti, in prep.) GE82832 is very effective in vivo against Bacillus subtilis and fairly active against enterococci, including vancomycin-resistant strains and streptococci, being also able to cure an experimental septicemia caused in mice by Streptococcus pyogenes (D. Losi and G. Candiani, unpubl.).
These characteristics, which indicate that GE82832 might be a novel and promising antibiotic, have prompted us to undertake an in-depth characterization of its mechanism of action. The results presented in this article demonstrate that this antibiotic is capable of blocking translocation already at the earliest steps of translation by selectively binding to the head region of the 30S ribosomal subunit.
RESULTS
As described in the introduction, GE82832 was initially identified as an antibiotic capable of inhibiting mRNA translation in vitro in a primary HTS test. Thus, before beginning the characterization of the mechanism of action of GE82832, it was necessary to ascertain that its target is the translational apparatus also in vivo and that its microbiological activity is due to the inhibition of this function. That this is indeed the case is clearly demonstrated by its effect on the in vivo incorporation of specific precursors of DNA (thymidine), RNA (uracil), protein (methionine), and cell wall (N-acetylglucosamine). As seen from the results shown in Figure 1A, GE82832, at a concentration (3 μg/mL) 12 times higher than its minimum inhibitory concentration (MIC) (not shown), produces a substantial inhibition of protein synthesis in B. subtilis while the synthesis of RNA, DNA, and peptidoglycan are not significantly affected. Almost identical results were obtained when GE82832 was given to the cells at concentrations ≥300 times the MIC (not shown), indicating that even at very high dosages this antibiotic does not interfere with functions other than translation.
FIGURE 1.
In vivo and in vitro translation inhibition by GE82832. (A) Effect of GE82832 (3 μg/mL) on the in vivo incorporation of [3H] thymidine (□), [3H] uridine (○), [35S] methionine (▪), and [3H] N-acetylglucosamine (▵) by B. subtilis cells. (B) Comparison of the effect of increasing concentrations of GE82832 on the translation of cspAmRNA (•), p4hupBmRNA (▪), and 027mRNA (▴,▿) in a crude cell-free extract (closed symbols) or in a partially purified system (open symbol) derived from E. coli. For comparison, the effect of increasing concentrations of tetracycline on the 027mRNA-dependent translation in the E. coli crude extract is also presented (♦). (C) Effect of increasing concentrations of GE82832 (▪) and pactamycin (▴) on 027mRNA-directed translation in a Saccharomyces cerevisiae cell-free extract (Brandi et al. 2006b). Further experimental details are given in Materials and Methods.
In light of these results, analysis of the mechanism of action of GE82832 was started by testing its inhibitory activity in secondary in vitro assays in which crude Escherichia coli cell-free extracts were programmed with four different mRNAs, namely the model 027mRNA and three natural templates cspA, P4hupB, and Renilla luciferase (rluc) mRNAs.
As seen from the results shown in Figure 1B, GE82832 (Factor B) inhibits translation up to >80% with an estimated IC50 ≅ 3 μg/mL for all mRNAs, thereby displaying an activity somewhat higher than that of the well-characterized A-site inhibitor tetracycline (Gale et al. 1981; Cundliffe 1990; Walsh 2003; Wilson 2004) whose effect was tested in parallel as a positive control (Fig. 1B). On the other hand, confirming the results of the primary screening tests (not shown) GE82832 did not inhibit eukaryotic translation. In fact, GE82832 proved to be completely inactive in poly(U)-dependent polyphenylalane synthesis in cell-free extracts of both Candida albicans and rat hepatocytes (not shown) and, unlike pactamycin, in a 027mRNA-programmed yeast cell-free system (Fig. 1C).
To test the possible occurrence of GE82832-induced miscoding, the time course of luciferase mRNA translation was analyzed in the presence of increasing concentrations of this antibiotic. In fact, amino acid misincorporation during the synthesis of luciferase (311 amino acids) would be expected to reduce the enzymatic activity, thereby diminishing the level of luminescence without affecting the overall amount of protein synthesized. As seen in Figure 2A, under the conditions of our experiment and in the absence of any inhibitor, the luciferase activity appears after a fairly long (≅6 min) lag that can be attributed to the low initiation and elongation rates characteristic of translation at low temperature (Farewell and Neidhardt 1998; Brandi et al. 2006a) and to the time required for the correct folding of the de novo synthesized enzyme before its activity can be expressed (Fedorov and Baldwin 1995; Kolb et al. 2000). As seen from Figure 2A,C, the level of luciferase synthesized and the incorporation of radioactive precursors in the protein diminish in parallel with increasing concentrations of GE82832 (Fig. 2C) indicating that GE82832 inhibits translation without causing misreading. Furthermore, the results (Fig. 2A) show that the antibiotic has no effect on the kinetics of appearance of the luciferase activity and does not affect the length of the lag (Fig. 2B), indicating that it does not affect the rates of the events responsible for existence of the lag. This behavior is clearly different from that found for GE81112, another antibiotic recently discovered and characterized as a powerful inhibitor of translation initiation, which slows down initiation through a competition with fMet-tRNA for P-site binding, causing lengthening of the lag of luciferase appearance (Brandi et al. 2006a). These findings indicate that GE81112 and GE82832 hit different targets within the translational apparatus, both inhibiting early yet different steps of protein synthesis; unlike GE81112, GE82832 is not an inhibitor of translation initiation (see below), in spite of the fact that both antibiotics were identified by the same HTS test.
FIGURE 2.
Effect of GE82832 on luciferase synthesis. (A) Time course of Renilla luciferase production at 20°C with E. coli S30 extracts programmed with lucmRNA in the presence of the indicated concentrations (micromolar) of GE82832. (B) Effect of increasing concentrations of GE82832 on the length of the lag preceding the expression of 50% of the luciferase activity. (C) Differential inhibition by GE82832 of cspAmRNA (▪) and lucmRNA (▴,▵) translation measured as enzymatic luciferase activity (▴) and incorporation of a radioactive precursor ([35S] methionine) in an acid-insoluble peptide (▵). (D) Inhibition by GE82832 of cspAmRNA translation at 20°C (▴) and 37°C (▪). Further experimental details are given in Materials and Methods.
Comparison of the results shown in Figures 1B and 2C revealed a substantial difference in the GE82832 concentrations required to inhibit the two systems. In fact, the IC50 value estimated from the inhibition of luciferase synthesis (≅30 μg/mL; Fig. 2A) is about 10-fold higher than that found with the other three mRNAs (a comparison between inhibition of cspA and luc mRNA translation is shown in Fig. 2C). To explain this discrepancy we tested the influence of two parameters (monovalent cation and incubation temperature) that distinguish luciferase synthesis from the other translational systems. In fact, while in vitro mRNA translation is routinely carried out at 37°C in the presence of NH4 +, the synthesis and assay of luciferase activity must be performed at 20°C in the presence of K+ ions, since higher temperatures and presence of NH4 are inhibitory. After ruling out that the activity of GE82832 is influenced by the monovalent cation (not shown), the influence of temperature on GE82832 inhibition was tested comparing the inhibition of cspA mRNA translation at 37°C and 20°C. As seen from Figure 2D, the IC50 of GE82832 is ≅8-fold lower at the higher temperature. This result indicates that the lower inhibition by GE82832 seen with the luciferase mRNA is likely due to the temperature difference at which this and the other mRNAs are translated. This finding is also important insofar as it indicates that the translational step inhibited by GE82832 is markedly T-sensitive (Farewell and Neidhardt 1998).
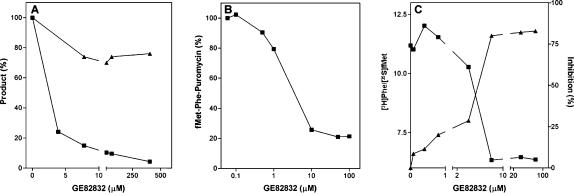
To identify the molecular target of GE82832 and clarify its mechanism of action, its effect on the individual steps of the translation pathway was tested. The first series of these experiments showed that GE82832 does not affect the initiation phase of translation since mRNA-dependent binding of fMet-tRNA to either 30S or 70S ribosomes was inhibited ≤10% and ≤20%, respectively (Fig. 3A). In addition, the formation of fMet-puromycin was not inhibited (inhibition ≤10%) by GE82832, even at >100 μM (Fig. 3B). Furthermore, unlike tetracycline, which inhibits Phe-tRNA binding to the A-site, both in the absence of EF-Tu and in its presence, albeit with lower efficiency (Fig. 3C), GE82832 did not inhibit A-site binding of Phe-tRNA in the presence of EF-Tu and caused only a modest inhibition (at most ≅45% at GE82832 concentrations >10 μM) in the absence of EF-Tu (Fig. 3C). Finally, formation of the initiation dipeptide fMet-Phe was found to be little inhibited (<20%) by GE82832 at concentrations up to about 400 μM (Fig. 4A). Taken together, these results indicate that GE82832 does not inhibit any of the steps involved in the translation initiation pathway from fMet-tRNA binding to the ribosomes, its adjustment in the P-site, the binding of the second aminoacyl-tRNA in the A-site to the formation of the first peptide bond.
FIGURE 3.
Effect of GE82832 on individual steps of the initiation pathway. (A) Effect of increasing concentrations of GE82832 (closed symbols) on the fMet-tRNA binding of to 30S (▪) and 70S (▴) ribosomes programmed with 027mRNA; (B) formation of fMet-puromycin (▪); (C) Phe-tRNA binding to the A-site in the presence (▴,▵) or absence (▪,□) of EF-Tu as a function of increasing concentrations of GE82832 (▵,□) or of tetracycline (▴,▪) taken as a positive control.
FIGURE 4.
Effect of GE82832 on translocation. Translocation activity is measured, as a function of the GE82832 concentrations indicated in the abscissa, from the level of initiation dipeptide (fMet-Phe) (▴) and initiation tripeptide (fMet-Phe-Leu) formed (▪) by ribosomes programmed with 012 mRNA (A), the level of fMet-Phe-puromycin formed by ribosomes programmed with 027 mRNA (B), and by the [3H]Phe/[35S]Met ratio (▪) compared to overall translation inhibition (▴) (C). In panel A, the amount of fMet-Phe and fMet-Phe-Leu formed (100% activity) corresponds to ≅55% and ≅38% of fMet-tRNA bound, respectively. In panel B, the amount of fMet-Phe-puromycin formed corresponds to 30% of fMet-tRNA bound. Further details are given in Materials and Methods.
On the other hand, the results presented in Figure 4A indicate that initiation tripeptide (fMet-Phe-Leu) formation is strongly inhibited by GE82832 with a reduction of >75% at 3 μM and suggest that translocation is the translational step inhibited by this antibiotic. This premise is fully supported by the finding that also the fMet-Phe-tRNA translocation necessary to yield fMet-Phe-puromycin is severely inhibited between 1 and 10 μM GE82832 (Fig. 4B). Also consistent with these data and with the conclusion that GE82832 is a translocation inhibitor is the finding that the size of the peptide product synthesized is progressively reduced in the presence of increasing concentrations of this antibiotic, with an inhibitory efficiency roughly corresponding to that displayed in the overall inhibition of translation (Fig. 4C).
The molecular target of GE82832 was investigated by testing the residual translational activity of ribosomal subunits preincubated with increasing amounts of the antibiotic and then pelleted by centrifugation. As seen in Figure 5A, the 30S ribosomal subunits pelleted after incubation even with the lowest concentration of GE82832 remain almost completely inhibited while the 50S subunits processed in the same way are fully active, some inhibition (≅40%) being seen only with subunits preincubated with the highest concentration (≅90 μM) of GE82832. Thus, this experiment demonstrated that GE82832 binds strongly and almost quantitatively to the 30S ribosomal subunit while the binding to the 50S subunit is either negligible or very unstable.
FIGURE 5.
Identification of the GE82832 target. (A) Residual translational activity expressed by 30S (▪) and 50S (▴) ribosomal subunits pelleted by ultracentrifugation after preincubation in the presence of the concentrations of GE82832 indicated in the abscissa. The translational activity of these ribosomal subunits is determined in the presence of a stoichiometric amount of control (untreated) complementary subunits as described in more detail in Materials and Methods. (B) Primer extension analysis of the 16S rRNA bases whose reactivity with DMS is affected by GE82832 (1, 10, and 100 μM). Lanes indicated K and C represent samples corresponding to 16S rRNA extracted from untreated 30S and from 30S treated with DMS in the absence of GE82832, respectively. (C) Localization of the bases whose reactivity is affected by GE82832 within the three-dimensional structure of the “head” region of the 30S ribosomal subunit (Vila-Sanjurjo et al. 2003; PDB file 1PNX) in which the bases protected (red) and exposed (blue) by GE82832 are shown together with ribosomal protein S13 (green). The localization within the 30S ribosomal subunits of the bases protected (red) or exposed (blue) by GE82832 is shown in panel D together with the localization of protein S13 (green and yellow). The structural elements of the 30S ribosomal subunit (i.e., part of r-protein S13 and base G1338) and of the 50S subunit (nucleotides C881, A882, and C883 of helix 38 of 23S rRNA) involved in intrasubunit bridging (Gao et al. 2003; PDB file 1PNX, 1PNY by Vila-Sanjurjo et al. 2003) are indicated in yellow on the background of protein S13 (green) and helix 38 (blue).
In agreement with the above findings, chemical probing experiments have shown that GE82832 protects bases A1333 and A1324 of 16S rRNA from DMS modification while moderately exposing C1336 (Fig. 5B). These results place the binding site of this antibiotic on the subunit interface side of the particle head, near ribosomal protein S13 (Fig. 5C,D). This localization can account for the biological activity of GE82832, which, as mentioned above, is an inhibitor of translocation (see Discussion).
DISCUSSION
The results presented in this article demonstrate that the newly discovered antibiotic GE82832, a secondary metabolite of S. cinnabarinum, is a powerful and selective inhibitor of bacterial protein synthesis. In fact, GE82832 was found to be inactive against the yeast translational apparatus but to inhibit translation in B. subtilis in vivo and in E. coli in vitro. These data indicate that this antibiotic is active on ribosomes of both g positive and g negative bacteria but not on those of eukarya and can therefore be considered a selective prokaryotic inhibitor. Moreover, these findings suggest that the failure of GE82832 to inhibit the growth of E. coli (not shown) is likely due to the existence of a permeability barrier in this organism.
The data presented here have also shown that translocation is the step of protein synthesis that is selectively inhibited by GE82832. In fact, while it has been shown that GE82832 is ineffective in blocking tRNA binding to P- and A-sites and peptidyl transferase activity and all the steps of the initiation pathway up to initiation dipeptide formation, GE82832 was found to severely inhibit the first translocation event, which allows the formation of the second peptide bond yielding a tripeptide and to cause a drastic reduction in the size of the translational product.
Translocation consists of the coordinated movement of the mRNA and tRNAs on the ribosome. This movement results in the advancement of the mRNA by one codon and in the shift of the peptidyl-tRNA from the A- to the P-site and of the deacylated tRNA from the P- to the E-site. Although basic translocation can occur also in the absence of exogenous factors and can therefore be regarded as an intrinsic property of the ribosome (Gavrilova et al. 1976), in nature elongation factor G (EF-G) catalyzes this process, increasing its rate by three to four orders of magnitude (Cukras et al. 2003). The translocation reaction is accompanied by conformational rearrangements of the ribosome. In particular, conformational changes of the 30S ribosomal subunit (Frank and Agrawal 2000; Stark et al. 2000; Peske et al. 2004), notably of elements of the head (Agrawal et al. 1999, Wilson 2004), occur during EF-G-dependent tRNA–mRNA translocation. The movement of the head may be involved, directly or indirectly, in tRNA displacement by affecting the structure or orientation of structural elements (intersubunit bridges) of the 30S subunit that interact with the 50S subunit (Agrawal et al. 2000; van Loock et al. 2000).
Several translocation inhibitors, having different targets and different mechanisms of action, are known; some target the activity of EF-G either directly or as a result of structural changes that they induce within the ribosomal structure; others interfere with the ribosomal dynamics required for translocation. Fusidic acid, which inhibits the dissociation of EF-G, and thiostrepton, which affects the 50S structure, belong to the first group, while viomycin (Vio), hygromycin B (HygB), paromomycin (Par), spectinomycin (Spc), and perhaps oxazolidinones (Oxo) are among the best-characterized translocation inhibitors of the second type. The localization of GE82823 on the head of the small ribosomal subunit suggests that GE82832 may also belong to this second class of antibiotics if an alteration of the dynamics of the intersubunit movement is at the root of its effect on translocation (see below). However, the properties of GE82832 are clearly different from those of the other inhibitors mentioned above whose mechanism of action is briefly summarized here below.
The tuberactinomycin Vio causes structural changes within the ribosome similar to those induced by miscoding agents and reinforces the interactions between aa-tRNA and the A-site after the codon–anticodon recognition step. Thus, translocation inhibition by Vio is attributed to the stabilization of tRNA binding in the A-site (Luehrmann 1980; Peske et al. 2004), which hinders the tRNA–mRNA movement, while the other partial reactions of translocation, including the unlocking rearrangement of the ribosome that precedes tRNA–mRNA movement (Savelsbergh et al. 2003), are not affected. Like Vio, HygB, Par, and Spc do not inhibit the partial reactions that precedes tRNA–mRNA movement (Savelsbergh et al. 2003) but impair the tRNA–mRNA movement itself. However, the tRNA–mRNA movement is inhibited in different ways by these antibiotics. In the case of HygB and Par the inhibition seems to be due to the stabilization of tRNA binding in the A-site, while Spc seems to have a direct inhibitory effect on the movement, probably due to a reduced conformational flexibility of the 30S subunit, caused by its binding to this subunit (Peske et al. 2004).
The above mentioned antibiotics target different ribosomal sites. In fact, Vio binds to the U913 and G914 of the 50S subunit (Moazed and Noller 1987; Yamada et al. 1980) while HygB and Spc bind to different sites of the 30S subunit. Spc binds to the minor groove at one end of helix 34 (H34), where it contacts C1064 and C1192 (Carter et al. 2000), and HygB interacts with bases that contact the tRNAs in both A- and P-sites, although its binding does not seem to induce any significant alterations in the structure of 16 S rRNA in this region. Conversely, HygB seems to cause subtle long-range effects on the 30S subunits (Brodersen et al. 2000). The nucleotides of 16S rRNA in contact with HygB are 1490–1500 and 1400–1410, located in both strands at the top of helix 44 (H44), which has been implicated in the subunit's movements that accompany translocation (Agrawal et al. 2000; van Loock et al. 2000), and disruption of the G1491:C1409 base pair, adjacent to the HygB binding site, confers resistance to this antibiotic (De Stasio and Dahlberg 1990).
Oxazolidinones are a class of synthetic antibacterial agents that strongly decrease the length of nascent peptide chains formed and the rate of elongation and are therefore regarded as possible tRNA translocation inhibitors (Matassova et al. 1999). Oxazolidinones target the rRNAs of both 30S and 50S subunits protecting from DMS reaction A864, in the central domain of 16S rRNA and bases (U2113, A2114, U2118, A2119 and C2153) belonging to domain V of the 23S rRNA, which are located near the binding site of protein L1 close to the 3′ end of E-site-bound tRNA (Matassova et al. 1999).
Unlike with these translocation inhibitors, the ribosomal target of GE82832 has been localized on the head of the 30S subunit since this antibiotic protects A1333 and A1324 and exposes C1336 belonging to helix 42 (H42) of 16S rRNA. These results place the binding site of GE82832 very close to ribosomal protein S13 and to G1338, two components of the 30S subunit that have been implicated in 30S–50S interaction and translocation (Merryman et al. 1999). Indeed, ribosomal protein S13 is responsible (through its residues 102–115) for the formation of an intersubunit bridge (B1a) with helix H38 (nucleotides 881–883) of 23S rRNA (Gao et al. 2003) and has been reported to be a control element in the translocation of the mRNA–tRNA complex (Cukras et al. 2003). The role played by S13 in translocation, in conjunction with S12, has been studied by a number of different approaches making use of ribosomes lacking S13. Since these ribosomes display an increased rate of factor-independent translocation, it has been suggested that S13 is involved in the maintenance of the pretranslocation state and is involved in the more ancient rRNA- and tRNA-driven movements of translocation (Cukras et al. 2003). Nucleotide G1338, on the other hand, was found to be moderately protected by the presence of the 50S ribosomal subunit, with which it was reported to form part of a bridge (B1a) (Merryman et al. 1999) whose existence depends upon the functional state of translocating ribosomes, being broken in the 70S bearing EF-G-GTP (Gao et al. 2003).
Taken together, our results indicate that GE82832 is a new translocation inhibitor and suggest that its mechanism of action consists of blocking translocation by interfering with the concerted movement of the two ribosomal subunits required for translocation.
MATERIALS AND METHODS
Buffers
Buffer A: 20 mM Tris-HCl (pH 7.7), 7 mM MgAc, 80 mM NH4Cl, 0.1 mM DTT.
Buffer B: 10 mM Tris-HCl (pH 7.7), 15 mM NH4Cl, 180 mM KCl, 0.1 mM DTT.
Buffer C: 20 mM Tris-HCl (pH 7.7), 10 mM MgAc, 80 mM NH4Cl, 0.1 mM DTT.
Buffer D: 50 mM Tris-HCl (pH 7.5), 100 mM NH4Cl, 30 mM KCl, 7 mM MgCl2.
Assessment of in vivo activity of GE82832
B. subtilis ATCC 6633 cells were grown at 37°C in Spizizen's medium (1.4% K2HPO4, 0.6% KH2PO4, 0.2% (NH4)2SO4, 0.1% trisodium citrate dehydrate, 0.02% MgSO4(H2O)7, 0.5% D-glucose supplemented with 0.1% casamino acids) to A600nm = 0.2 when (time = 0) the culture was divided into four identical aliquots and each of them received one of the following precursors: [3H] thymidine, [3H] uridine, [14C] phenylalanine, or [3H] N-acetylglucosamine. After 10 min, each radiolabeled culture was split in two aliquots, one of which was exposed to GE82832 (3 μg/mL) and the other to DMSO (final concentration 1.25%). At 10-min intervals, 50 μL samples of each culture were withdrawn, mixed with 50 μL of 2% SDS, and the hot acid (TCA)-insoluble radioactivity present in 50 μL of the resulting cell extracts was determined by liquid scintillation counting.
fMet-tRNA binding to 30S and 70S ribosomes
Each reaction mixture contained, in 40 μL of BufferA, 0.5 mM GTP, 30 pmol of 30S ribosomal subunits (or an equimolar mixture of 30S and 50S subunits), and the indicated amounts of GE82832. After 5 min incubation, 45 pmol each of either IF1, IF2, and IF3 were added. The binding reaction was triggered by the addition of 45 pmol each of 022 mRNA and f[35S]Met-tRNA. After 10 min incubation at 37°C, the amount of f[35S]Met-tRNA bound in either a 30S or a 70S initiation complex was determined by filtering 30 μL of each reaction mixture through a nitrocellulose disc.
fMet-puromycin formation
Primary mixtures containing 30 pmol of 30S ribosomal subunits, the indicated amounts of GE82832, 0.5 mM GTP, 45 pmol of 022 mRNA, 45 pmol of f[35S]Met-tRNA, and 45 pmol each of IF1, IF2, and IF3 were incubated for 10 min at 37°C in 100 μL of Buffer A to allow the formation of 30S initiation complexes. A 10-μL aliquot was used to determine the amount of 30S initiation complex formed. The remaining mixture was supplemented with a stoichiometric equivalent (25 pmol) of 50S ribosomal subunits. Puromycin (final concentration 1 mM) was then added and incubation continued at 37°C for 45 sec. The reaction was stopped by addition of 500 μL of 1 M (NH4)HCO3 (pH 9.0), and the f[35S]Met-puromycin formed was extracted with 1 mL of ethyl acetate. After vigorous vortex mixing for 1 min, the amount of f[35S]Met-puromycin present in 0.5 mL of the ethyl acetate phase was determined with a liquid scintillation counter.
fMet-Phe-puromycin formation
Primary mixtures containing 30 pmol of 30S ribosomal subunits, the indicated amounts of GE82832, 0.5 mM GTP, 45 pmol of 027 mRNA, 45 pmol of f[35S]Met-tRNA, and 45 pmol each of IF1, IF2, and IF3 were incubated for 10 min at 37°C in 100 μL of Buffer A to allow the formation of 30S initiation complexes. A 10-μL aliquot was used to determine the amount of 30S initiation complex formed. The remaining mixture (90 μL) was supplemented with a stoichiometric equivalent (25 pmol) of 50S ribosomal subunits and EF-Tu-GTP-[3H]Phe-tRNA ternary complex in 10 μL buffer D containing 1 mM GTP. After 5 min incubation at 37°C, 20 μL of Buffer D containing 6 mM puromycin and 25 pmol EF-G were added and the resulting 120-μL mixture was incubated at 37°C for 2 sec. The reaction was stopped by addition of 500 μL of 1 M (NH4)HCO3 (pH 9.0) and the f[35S]Met-[3H]Phe-puromycin formed was extracted with 1 mL of ethyl acetate. After vigorous vortex mixing for 1 min, the amount of f[35S]Met-[3H]Phe-puromycin present in 0.5 mL of the ethyl acetate phase was determined with a liquid scintillation counter.
In vitro translation
Translation in vitro was performed as previously described (La Teana et al. 1993). Each reaction mixture (30 μL of buffer C) contained 1 mM ATP, 0.5 mM GTP,10 mM phosphoenolpyruvate, 25 μg/mL pyruvate kinase, 0.12 mM citrovorum factor (Serva), 150 μg of total E. coli MRE600 tRNA, 30 pmol of high-salt-washed 70S ribosomes, 45 pmol 027mRNA, 20 μM [14C]-phenylalanine (460 mCi/mmol; Amersham), 30 pmol each IF1, IF2, and IF3, and the indicated amounts of inhibitors. In addition, the reaction mixtures contained 200 μM each amino acid (except phenylalanine) and an optimized amount of post-ribosomal S100 fraction. After 15 min incubation at 37°C, the hot TCA-insoluble radioactivity present in 20 μL of the reaction mixture was determined by liquid scintillation counting.
Luciferase synthesis
Each reaction mixture (50 μL of Buffer B) contained 0.4 mM GTP, 2 mM ATP, 10 mM PEP, 0.025 μg/μL PK, 2 μM Coelenterazine, 0.2 mM amino acids mix, 0.12 mM Citrovorum, 1 μg/μL E. coli MRE600 total tRNA (Sigma), 15 pmol of Renilla Luc mRNA, 12 μL S30 corresponding to ∼15 pmol 70S, and GE82832 as indicated. The luminescence produced in each luciferase reaction was recorded at 2-min interval using a 1450 Microbeta (Wallac).
Chemical probing and primer extension analysis
The modification reactions were carried out according to Moazed et al. (1986) using dimethylsulphate (DMS). After modification, ribosomal RNA was extracted (Moazed et al. 1986) and the modified residues identified by primer extension analysis.
Two picomoles of the selected primer, 5′-32P labeled with T4 kinase, were incubated with 2 μg of the modified rRNA for 3 min at 70°C in 10 μL of 50 mM Tris-HCl (pH 8.3), 40 mM KCl and then slowly cooled to 40°C. After addition of 2 U of AMV Reverse Transcriptase (Roche), 2 μL of each annealing reaction were mixed with 2 μL of a extension mixture (100 mM Tris-HCl at pH 8.3, 80 mM KCl, 12 mM Mg2Cl, 4 mM of each deoxynucleotide triphosphate) and incubated for 30 min at 45°C. The extension reaction was stopped by addition of 2 μL formamide/dye buffer, and 2.5 μL were loaded on a 10% sequencing gel.
Initiation dipeptide and tripeptide formation
To facilitate the HPLC analysis of the tripeptide product, a modified form of 022mRNA called 012mRNA was used for these experiments. In this mRNA the third triplet ACG of 022mRNA coding for Thr was changed into TTG, coding for Leu.
For these experiments, 30S initiation complexes were prepared incubating (10 min at 37°C) 0.3 μM 30S subunits, 0.45 μM each of IF1, IF2, and IF3, 0.45 μM f[35S]Met-tRNA, and 0.9 μM 012 mRNA in buffer D containing 1 mM GTP. The EF-Tu-GTP-Phe-tRNA ternary complex was prepared in buffer D containing 1 mM GTP, EF-Tu (0.3 μM final concentration), 3 mM phosphoenol-pyruvate and 0.25 μg/mL pyruvate kinase by incubation for 10 min at 37°C. The EF-Tu-GTP-Leu-tRNA ternary complex was prepared following the same procedure. To form the initiation dipeptide, 30S initiation complexes were mixed with an equal volume (40 μL) of a mixture containing the EF-Tu-GTP-Phe-tRNA ternary complex and 50S subunits (0.3 μM final concentration). After 5 min at 37°C formation of the fMet-Phe-Leu tripeptide was triggered by adding a mixture of EF-Tu-GTP-Leu-tRNA ternary complex and factor EF-G. After 5 min at 37°C the reaction was quenched with an equal volume of 0.5 M KOH followed by incubation at 37°C for 15 min, neutralization with acetic acid, and centrifugation at 12,000 rpm for 5 min. Analysis of the dipeptides and tripeptides was carried out by HPLC on the reverse phase (LiChrosorb RP-8, 5 μM-Merck) column with a linear (0%–65%) acetonitrile gradient in 0.1% TFA. The radioactivity present in the individual chromatographic fractions was then determined by liquid scintillation counting.
ACKNOWLEDGMENTS
The financial support of the EC grant (Contract QLRT-2001-00892 “Ribosome inhibitors”) to C.O.G. and S.D. is gratefully acknowledged. We are also grateful to Prof. Cynthia L. Pon for her invaluable discussions and kind and patient help in the preparation of this manuscript.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.61206.
REFERENCES
- Agrawal R.K., Heagle A.B., Penczek P., Grassucci R.A., Frank J. EF-G-dependent GTP hydrolysis induces translocation accompanied by large conformational changes in the 70S ribosome. Nat. Struct. Biol. 1999;6:643–647. doi: 10.1038/10695. [DOI] [PubMed] [Google Scholar]
- Agrawal R.K., Spahn C.M., Penczek P., Grassucci R.A., Nierhaus K.H., Frank J. Visualization of tRNA movements on the Escherichia coli 70S ribosome during the elongation cycle. J. Cell Biol. 2000;150:447–460. doi: 10.1083/jcb.150.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandi L., Fabbretti A., La Teana A., Abbondi M., Losi D., Donadio S., Gualerzi C.O. Specific, efficient, and selective inhibition of prokaryotic translation initiation by a novel peptide antibiotic. Proc. Natl. Acad. Sci. 2006a;103:39–44. doi: 10.1073/pnas.0507740102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandi L., Lazzarini A., Cavaletti L., Abbondi M., Corti E., Ciciliato I., Gastaldo L., Marazzi A., Feroggio M., Fabbretti A., et al. Novel tetrapeptide inhibitors of bacterial protein synthesis produced by a Streptomyces sp. Biochemistry. 2006b;45:3692–3702. doi: 10.1021/bi052540k. [DOI] [PubMed] [Google Scholar]
- Brodersen D.E., Clemons W.M., Carter A.P., Morgan-Warren R.J., Wimberly B.T., Ramakrishnan V. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell. 2000;103:1143–1154. doi: 10.1016/s0092-8674(00)00216-6. [DOI] [PubMed] [Google Scholar]
- Carter A.P., Clemons W.M., Brodersen D.E., Morgan-Warren R.J., Wimberly B.T., Ramakrishnan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- Cukras A.R., Southworth D.R., Brunelle J.L., Culver G.M., Green R.2003Ribosomal proteins S12 and S13 function as control elements for translocation of the mRNA–tRNA complex Mol. Cell 12321–328.s [DOI] [PubMed] [Google Scholar]
- Cundliffe E. Recognition sites for antibiotics within rRNA. In: Hill W.E., et al., editors. The ribosome. Structure, function & evolution. ASM Press; Washigton, DC: 1990. pp. 479–490. [Google Scholar]
- De Stasio E.A., Dahlberg A.E. Effects of mutagenesis of a conserved base-paired site near the decoding region of Escherichia coli 16S ribosomal RNA. J. Mol. Biol. 1990;212:127–133. doi: 10.1016/0022-2836(90)90309-A. [DOI] [PubMed] [Google Scholar]
- Farewell A., Neidhardt F.C. Effect of temperature on in vivo protein synthetic capacity in Escherichia coli . J. Bacteriol. 1998;180:4704–4710. doi: 10.1128/jb.180.17.4704-4710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov A.N., Baldwin T.O. Contribution of cotranslational folding to the rate of formation of native protein structure. Proc. Natl. Acad. Sci. 1995;92:1227–1231. doi: 10.1073/pnas.92.4.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J., Agrawal R.K. A ratchet-like intersubunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- Gale E.F., Cundliffe E., Reynolds P.E., Richmond M.H., Waring M.J. The molecular basis of antibiotic action. Wiley; London: 1981. Antibiotic inhibitors of ribosome function; pp. 402–547. [Google Scholar]
- Gao H., Sengupta J., Valle M., Korostelev A., Eswar N., Stagg S.M., Van Roey P., Agrawal R.K., Harvey S.C., Sali A., et al. Study of the structural dynamics of the E. coli 70S ribosome using real-space refinement. Cell. 2003;113:789–801. doi: 10.1016/s0092-8674(03)00427-6. [DOI] [PubMed] [Google Scholar]
- Gavrilova L.P., Kostiashkina V.E., Rutkevitch N.M., Spirin A.S. Factor-free (“non-enzymic”) and factor-dependent systems of translation of polyuridylic acid by Escherichia coli ribosomes. J. Mol. Biol. 1976;101:537–552. doi: 10.1016/0022-2836(76)90243-6. [DOI] [PubMed] [Google Scholar]
- Kolb V.A., Makeyev E.V., Spirin A.S. Co-translational folding of an eukaryotic multidomain protein in a prokaryotic translation system. J. Biol. Chem. 2000;275:16597–16601. doi: 10.1074/jbc.M002030200. [DOI] [PubMed] [Google Scholar]
- La Teana A., Pon C.L., Gualerzi C.O. Translation of mRNAs with degenerate initiation triplet AUU displays high initiation factor 2 dependence and is subject to initiation factor 3 repression. Proc. Natl. Acad. Sci. 1993;90:4161–4165. doi: 10.1073/pnas.90.9.4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luehrmann R. Dinucleotide codon–anticodon interaction as a minimum requirement for ribosomal aa-tRNA binding: Stabilisation by viomycin of aa-tRNA in the A site. Nucleic Acids Res. 1980;58:13–24. doi: 10.1093/nar/8.23.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matassova N.B., Rodnina M.V., Endermann R., Kroll H.P., Pleiss U., Wild H., Wintermeyer W. Ribosomal RNA is the target for oxazolidinones, a novel class of translational inhibitors. RNA. 1999;5:939–946. doi: 10.1017/s1355838299990210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merryman C., Moazed D., McWhirter J., Noller H.F. Nucleotides in 16S rRNA Protected by the association of 30S and 50S ribosomal subunits. J. Mol. Biol. 1999;285:97–105. doi: 10.1006/jmbi.1998.2242. [DOI] [PubMed] [Google Scholar]
- Moazed D., Noller H.F. Chloramphenicol, erythromycin, carbomycin and vernamycin B protect overlapping sites in the peptidyl transferase region of 23S ribosomal RNA. Biochimie. 1987;69:879–884. doi: 10.1016/0300-9084(87)90215-x. [DOI] [PubMed] [Google Scholar]
- Moazed D., Stern S., Noller H.F. Rapid chemical probing of conformation in 16 S ribosomal RNA and 30S ribosomal subunits using primer extension. J. Mol. Biol. 1986;187:399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- Ogle J.M., Brodersen D.E., Clemons W.M., Jr., Tarry M.J., Carter A.P., Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- Peske F., Savelsbergh A.K., Rodnina M.V., Wintermeyer W. Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA-mRNA translocation. J. Mol. Biol. 2004;343:1183–1194. doi: 10.1016/j.jmb.2004.08.097. [DOI] [PubMed] [Google Scholar]
- Pioletti M., Schlunzen F., Harms J., Zarivach R., Gluhmann M., Avila H., Bashan A., Bartels H., Auerbach T., Jacobi C., et al. Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J. 2001;20:1829–1839. doi: 10.1093/emboj/20.8.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savelsbergh A., Katunin V.I., Mohr D., Peske F., Rodnina M.V., Wintermeyer W. An elongation factor G-induced ribosome rearrangement precedes tRNA–mRNA translocation. Mol. Cell. 2003;11:1517–1523. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- Schlunzen F., Harms J.M., Franceschi F., Hansen H.A., Bartels H., Zarivach R., Yonath A. Structural basis for the antibiotic activity of ketolides and azalides. Structure (Camb.) 2003;11:329–338. doi: 10.1016/s0969-2126(03)00022-4. [DOI] [PubMed] [Google Scholar]
- Stark H., Rodnina M.V., Wieden H.-J., van Heel M., Wintermeyer W. Large-scale movement of elongation factor G and extensive conformational change of the ribosome during translocation. Cell. 2000;100:301–309. doi: 10.1016/s0092-8674(00)80666-2. [DOI] [PubMed] [Google Scholar]
- van Loock M.S., Agrawal R.K., Gabashvili I.S., Qi L., Frank J., Harvey S.C. Movement of the decoding region of the 16 S ribosomal RNA accompanies tRNA translocation. J. Mol. Biol. 2000;304:507–515. doi: 10.1006/jmbi.2000.4213. [DOI] [PubMed] [Google Scholar]
- Vicens Q., Westhof E. Crystal structure of a complex between the aminoglycoside tobramycin and an oligonucleotide containing the ribosomal decoding a site. Chem. Biol. 2002;9:747–755. doi: 10.1016/s1074-5521(02)00153-9. [DOI] [PubMed] [Google Scholar]
- Vicens Q., Westhof E. Molecular recognition of aminoglycoside antibiotics by ribosomal RNA and resistance enzymes: An analysis of x-ray crystal structures. Biopolymers. 2003;70:42–57. doi: 10.1002/bip.10414. [DOI] [PubMed] [Google Scholar]
- Vila-Sanjurjo A., Ridgeway W.K., Seymaner V., Zhang W., Santoso S., Yu K., Cate J.H.D. X-ray crystal structures of the WT and a hyper-accurate ribosome from Escherichia coli . Proc. Natl. Acad. Sci. 2003;100:8682–8687. doi: 10.1073/pnas.1133380100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C. Antibiotics. Actions, origins, resistance. ASM Press; Washington, DC: 2003. New Looks at targets; pp. 237–270. [Google Scholar]
- Wilson D.N. Antibiotics and the inhibition of ribosome function. In: Nierhaus K., Wilson D.N., editors. Protein synthesis and ribosome structure. Wiley-VSH Verlag; Weinheim, Germany: 2004. pp. 449–519. [Google Scholar]
- Yamada T., Teshima T., Shiba T., Nierhaus K.H. The translocation inhibitor tuberactinomycin binds to nucleic acids and blocks the in vitro assembly of 50S subunits. Nucleic Acids Res. 1980;8:5767–5777. doi: 10.1093/nar/8.23.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]