Abstract
Mammalian genomic imprinting is regulated by imprinting control regions (ICRs) that are usually associated with tandem arrays of transcription factor binding sites. In this study, the sequence features derived from a tandem array of YY1 binding sites of Peg3-DMR (differentially methylated region) led us to identify three additional clustered YY1 binding sites, which are also localized within the DMRs of Xist, Tsix, and Nespas. These regions have been shown to play a critical role as ICRs for the regulation of surrounding genes. These ICRs have maintained a tandem array of YY1 binding sites during mammalian evolution. The in vivo binding of YY1 to these regions is allele specific and only to the unmethylated active alleles. Promoter/enhancer assays suggest that a tandem array of YY1 binding sites function as a potential orientation-dependent enhancer. Insulator assays revealed that the enhancer-blocking activity is detected only in the YY1 binding sites of Peg3-DMR but not in the YY1 binding sites of other DMRs. Overall, our identification of three additional clustered YY1 binding sites in imprinted domains suggests a significant role for YY1 in mammalian genomic imprinting.
A subset of mammalian genes are subject to an unusual dosage control, genomic imprinting, by which one of two alleles of the genes are repressed in a parental-origin-specific manner. The imprinted genes are clustered in specific regions of chromosomes, and each imprinted domain is usually controlled by small genomic regions, termed imprinting control regions (ICRs) (Brannan and Bartolomei 1999; Spahn and Barlow 2003). These ICRs are usually located in CpG-rich regions near the promoters of imprinted genes and methylated differentially between two parental alleles. Surveys of the known ICRs indicated that these regions often show tandem repeat sequence structure and have evolved rapidly without any significant sequence conservation (Constancia et al. 1998; Reik and Walter 1998). Careful examinations of these ICRs revealed that the core sequences of these tandem repeats, ranging from 10 to 40 base pair (bp) in length, are conserved among different species and that these conserved core sequences usually turn out to be transcription factor binding sites. Known transcription factors binding to these tandem repeat regions include CTCF for the ICR of H19/Igf2-imprinted domain and YY1 for the DMR of Peg3 (Bell and Felsenfeld 2000; Hark et al. 2000; Kim et al. 2003). In the H19/Igf2-imprinted domain, CTCF has been shown to function as an enhancer-blocker for controlling allele-specific expression of H19 and Igf2 (Schoenherr et al. 2003; Fedoriw et al. 2004). However, the in vivo functions of YY1 for the Peg3-imprinted domain are currently unknown.
YY1 is a Gli-Kruppel-type zinc finger protein that controls the transcription of a large number of viral and cellular genes. YY1 can function as a repressor, activator, or transcriptional initiator depending on the sequence context of YY1-binding sites with respect to other regulator elements (Thomas and Seto 1999). The protein has a DNA-binding domain at the C terminus and other modulating domains at the N terminus displaying repression, activation, and protein–protein interaction activities. YY1 interacts with several key proteins, including TBP, TAFs, TFIIB and Sp1 (Lee et al. 1993; Seto et al. 1993; Usheva and Shenk 1994; Chiang and Roeder 1995; Austen et al. 1997). Other studies have also indicated that YY1 recruits histone-modifying enzymes including p300, HDACs, and PRMT1 to control transcription (Lee et al. 1995; Yang et al. 1996; Rezai-Zadeh et al. 2003). YY1 is evolutionarily well conserved throughout all vertebrate lineages and at least two genes similar to vertebrate YY1 are found in fly genomes. One of these YY1 homologs is known to be involved in the Polycomb complex-mediated repression mechanism (Brown et al. 1998). In vertebrates, several studies also support the potential connection of YY1 to this heritable silencing mechanism (Satijn et al. 2001; Caretti et al. 2004). YY1 is also thought to be involved in the formation of mammalian DNA repeat families based on the frequent detection of YY1 binding motifs in many families of different origin (Oei et al. 2004; Khan et al. 2006).
In contrast with known YY1 functions in cellular and viral genes, the function of YY1 in the Peg3-imprinted domain is expected to be very unique based on the localization of a tandem array of YY1 binding motifs within a genomic region undergoing an unusual epigenetic modification, allele-specific methylation. To gain insights for the functions of this tandem array of YY1 binding sites, we have sought to identify more regions with similar clustered YY1 binding sites in the current study. With newly implemented strategies, we have identified 35 human and 21 mouse genomic regions with a tandem array of clustered YY1 binding sites. We have characterized some of these potential regulatory regions, including clustered YY1 binding sites located within Xist, Tsix, and Gnas loci, in terms of their in vivo binding to YY1 as well as potential roles in transcription and imprinting. The locations of clustered YY1 binding sites coincide with the ICRs of these domains, suggesting a significant functional role of these clustered YY1 binding sites in imprinting regulation.
Results
Identification of clustered YY1 binding sites in ICRs
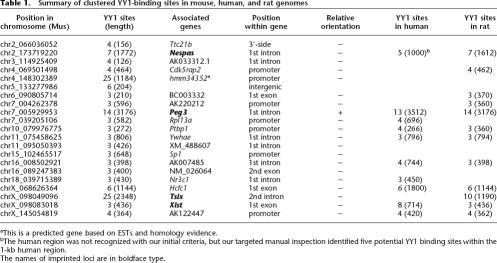
Two unique features observed from the clustered YY1 binding sites of Peg3-DMR were used as searching criteria for finding potential new regulatory regions. First, the YY1 binding sites of Peg3-DMR are all positioned in the same orientation. Second, YY1 binding sites appear to be in an evolutionarily dynamic state, constant decay and regeneration, based on the observation that half of the YY1 binding sites within Peg3-DMR have intact CpG sites while the remaining half have the mutated version of CpG sites (CpA or TpG) (Kim et al. 2003). We have developed a Perl-based script that can identify clustered YY1 binding sites exhibiting these two features. This program was designed to first identify any genomic region that has more than two YY1 binding sites (CGCCATnTT; n indicates any base at that position) with the same orientation within a 2-kb interval. The initial pool of the selected genomic regions was further tested for the presence of a less stringent binding site (CCATnTT) without the CpG dinucleotide or a minor form of YY1 binding sites (ACATnTT) (Yant et al. 1995). If the initial pool of the selected genomic regions had more than three YY1 binding sites at a density of one site per 300-bp genomic region, these genomic regions were selected for further investigation. With these initial settings, we found 35 human and 21 mouse genomic regions that have tandem arrays of YY1 binding sites (Table 1 and Supplemental material).
Table 1.
Summary of clustered YY1-binding sites in mouse, human, and rat genomes
aThis is a predicted gene based on ESTs and homology evidence.
bThe human region was not recognized with our initial criteria, but our targeted manual inspection identified five potential YY1 binding sites within the 1-kb human region.
The names of imprinted loci are in boldface type.
Detailed examination of the mouse and human sets derived several interesting observations (Table 1). First, the majority of these clustered YY1 binding sites (17/21) are derived from the first intron, exon, or promoter regions of individual genes, indicating the high success rate of our approach for finding potential regulatory regions. This high success rate is partly due to the CpG-containing binding motif of YY1, since the promoters of mammalian genes tend to be associated with CpG-rich sequences. Second, almost all of the clustered YY1 binding sites (19/20) show one particular orientation of YY1 binding sites relative to the transcriptional direction of their associated genes, whereas only one clustered YY1 binding site shows the other orientation. A recent survey has also reported a similar orientation bias of YY1 binding sites relative to transcriptional direction, but the reason for this bias is not well understood to date (Schug et al. 2005). Third, although these genomic regions have been independently identified because of their unusual high densities of YY1 binding sites, half of these binding sites appear to represent the orthologous genomic regions of different species: nine regions between human and mouse and twelve regions between mouse and rat. Most of these evolutionarily conserved YY1 binding sites show marginal sequence conservation between different species, ranging from 65% to 82% sequence identity. However, two YY1 binding sites located in the Peg3 and Xist locus show almost no sequence conservation beyond their YY1 binding sites, indicating that a tandem array of YY1 binding sites is the only evolutionarily selected feature in these two regions (Table 1).
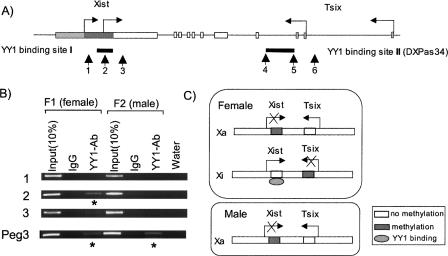
Our screening criteria have been designed to identify genomic regions in a dynamic state of CpG methylation, decay, and regeneration. Some of the clustered YY1 binding sites are indeed derived from the regions that are subject to constant DNA methylation as part of the epigenetic regulation of their associated genes. Besides one known cluster in Peg3-DMR, three more clustered YY1 binding sites have been identified from such genomic regions, including the differentially methylated region of Nespas located in the Gnas-imprinted domain (Fig. 1D), a small CpG island located 1 kb downstream of the Xist transcription start site, and a 1-kb tandem repeat region located in the second intron of Tsix, which is also known as DXPas34 (DNA segment from chromosome X, Pasteur Institute 34; a Sequence Tag Site marker) (Fig. 1A). The two clustered YY1 binding sites found in the Xist/Tsix locus are also localized within the CpG islands exhibiting differential methylation patterns between two parental alleles (Norris et al. 1994; Boumil et al. 2006) (Fig. 3C, see below). The 1.6-kb, Nespas-DMR harboring clustered YY1 binding sites has recently been shown to be the primary ICR for the Gnas-imprinted domain (Williamson et al. 2006).
Figure 1.
Three clustered YY1 binding sites located in the Xist/Tsix and Nespas loci. (A) The genomic structure of the Xist/Tsix locus. The arrows indicate the direction of Xist and Tsix transcription, and each gene has two different transcription start sites. The arrowheads indicate the positions of two clustered YY1 binding sites. (B,C) The conservation patterns of two clustered YY1 binding sites, the Xist cluster and the Tsix cluster. The two divergent motifs found in the Tsix clusters are marked: one with an underline and the other without an underline. (D) The genomic structure of the Gnas-imprinted domain and the conservation patterns of the clustered YY1 binding site located in the first intron of Nespas. The nucleotide bases are in boldface type if the sequences of each binding motif are identical to the consensus sequence of known YY1 binding sites.
Figure 3.
Allele-specific binding of YY1 to the Xist and Tsix clusters. A) The in vivo YY1 binding to each cluster was tested with three primer sets, and the positions of these are indicated with arrows with numbers. (B) Female-specific binding of YY1 to the Xist cluster. Two mouse brain tissues from female (F1) and male (F2) were used for our ChIP experiments. Our ChIP analyses included two control DNAs: 10% of the Input DNAs from F1 (lane 1) and F2 (lane 4) and the immunoprecipitated DNA with preimmune serum (lanes 2 and 5). Our ChIP analyses used the Peg3 locus as an internal control for female- and male-derived ChIP DNAs as shown in the bottom. Asterisks indicate the regions enriched by YY1-Ab. (C) Schematic representation of allele-specific YY1 binding to the Xist locus based on the results of B. YY1 binds to the Xist cluster located in the Xi of female, which is unmethylated. Because of the repeat structure of the Tsix cluster, however, our ChIP analyses could not confirm the in vivo YY1 binding to this locus. The methylation status of Tsix reflects the gametic difference between two sexes before the onset of random XCI (X chromosome inactivation). For the imprinted XCI in female, Xa is from oocytes (maternal) while Xi is from sperm (paternal).
Evolutionary conservation of the YY1 binding sites in the Xist/Tsix and Nespas locus
We analyzed the evolutionary conservation patterns of the clustered YY1 binding sites identified from the Xist/Tsix and Nespas loci (Fig. 1). The clustered YY1 binding site of mouse Xist has three potential YY1 binding motifs in the 400-bp genomic region, and the sequences of these three motifs are identical to the consensus sequence of known YY1 binding sites (CGCCATnTT). In contrast, the same region of other mammals with similar length has more YY1 binding motifs: eight in human, seven in cow (Fig. 1B), and eight in both horse and rabbit (GenBank accession nos. U50911 and U50910). The sequences of most YY1 binding motifs found in these species are also identical to the consensus sequence of known YY1 binding sites. The consensus sequence of known YY1 binding sites (CGCCATnTT) shows no base preference at the seventh position, which is also well reflected in each of potential YY1 binding motifs located in the Xist cluster, showing all possible base choices at that position (Fig. 1B). This position-specific, selective constraint on the potential YY1 binding motifs strongly suggests that these motifs have been selected for YY1 binding during evolution.
The clustered YY1 binding site located in the second intron of Tsix encompasses a 1.1-kb tandem repeat region that is made of 32 reiterations of a core sequence, 34 or 35 bp in length (Fig. 1C). Our inspection of this core sequence identified two similar, but different, types of motifs, AGACATTTT and AGGCATTTT. This region was initially identified because of the similarity of the first motif (AGACATTTT) to the minor form of known YY1 binding sites (ACATnTT). The evolutionary conservation of these two motifs has been assessed through comparing the orthologous regions of mouse and rat, but not through mouse and other mammals, because the orthologous sequence of mouse Tsix is absent in other mammals. The potential YY1 binding region of rat Tsix does not show any tandem repeat sequence structure, yet the two motifs appear to be the most obvious sequences that have been selected in this region (Fig. 1C).
The clustered YY1 binding site located in the first intron of mouse Nespas harbors seven potential YY1 binding motifs in a 1.7 kb-genomic region. Three out of these seven potential motifs still contain a CpG dinucleotide within their binding motifs, which is also true for the orthologous region of rat Nespas (Fig. 1D). In contrast, the orthologous human region was not initially identified as a clustered YY1 binding site with our searching criteria. Our targeted examination of the human region, however, identified five potential YY1 binding motifs with two of them showing one base difference to the minimal consensus sequence used for this search. Also, none of these sites are associated with the CpG dinucleotide. Further examination of this region in other mammals, such as dogs and cows, revealed that the orthologous regions of other mammals contain much lower numbers of YY1 binding motifs, one or two motifs (data not shown). Therefore, the evolutionary conservation of the clustered YY1 binding site of Nespas appears to be limited to rodents and possibly primates. In sum, our comparative analyses revealed a rapidly evolving pattern in the clustered YY1 binding sites of Xist/Tsix and Nespas. The only selected feature of these regions appears to be retention of multiple YY1 binding motifs.
Two divergent YY1 binding motifs in the Tsix cluster
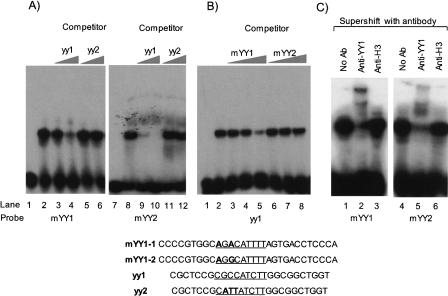
The two motifs identified from the Tsix cluster were further analyzed in terms of their binding capability to YY1 using gel shift assays (Fig. 2). For these experiments, we used four sets of duplex oligonucleotides. The two duplex probes, mYY1–1 and mYY1–2, contain the first (AGACATTTT) and second (AGGCA TTTT) motifs, respectively. These two probes were competed against two commercially available sets that have been designed for the YY1 gel shift assay. The yy1 probe contains the consensus sequence of YY1 binding sites (CGC CATCTT) and thus allows YY1 binding with high affinity. In contrast, the yy2 probe has three base changes in the critical region of YY1 binding (CATTATCTT) and thus the YY1 protein cannot bind to the yy2 probe. The first motif, mYY1–1, was tested with nuclear extracts prepared from HeLa cells (Fig. 2A, lanes 1–6). One protein complex binds to the mYY1–1 probe (lane 2), and this binding was diminished when the yy1 probe was added for competition (lanes 3 and 4), but not with the yy2 probe (lanes 5 and 6). This suggests that the protein complex binding to mYY1–1 may be YY1. This was further confirmed with a supershift assay (Fig. 2C, lanes 1–3), in which the protein complex binding to mYY1–1 was mobility-shifted by anti-YY1 antibody. We repeated a similar set of experiments with the second probe, mYY1–2, which contains the second motif found in the Tsix cluster (Fig. 2A, lanes 7–12; Fig. 2C, lanes 4–6). With similar results, the mYY1–2 probe appears to be a less favorable binding site for YY1 than the mYY1–1 probe based on the detection of weaker competition of the mYY1–2 probe against the yy1 probe (Fig. 2A, lanes 9 and 10) than the mYY1–1 probe (Fig. 2A, lanes 3 and 4). This conclusion was further confirmed through a set of reciprocal competition assays (Fig. 2B). These several sets of gel shift assays demonstrate that the two evolutionarily conserved motifs found in the tandem repeat region of Tsix indeed have the capability to bind to YY1.
Figure 2.
DNA mobility shift assays of two divergent motifs of the Tsix cluster. (A) The two motifs found in the Tsix cluster, AGACATTTT (mYY1–1) and AGGCATTTT (mYY1–2), were labeled as probes for gel shift assays. Each of two commercially available YY1 oligonucleotides, yy1 and yy2, were used as a competitor at two different molar ratios relative to a given probe (10×, lanes 3 and 9; 100×, lane 4 and 10). (B) Another series of the reciprocal competition assays using yy1 as a labeled probe were also performed. In this assay, the molar ratios of mYY1 and mYY2 to the labeled yy1 probe are 10× (lanes 3 and 6), 25× (lanes 4 and 7), and 100× (lanes 5 and 8), respectively. (C) Supershift assays were performed using two polyclonal antibodies, anti-YY1 antibody, and anti-H3 antibody as an unrelated negative control. The sequences of the four duplex oligonucleotide probes are shown at the bottom. The two motifs of the Tsix locus are positioned in the middle of these probes and marked with underlines. The nucleotide positions differing from the consensus sequence of YY1 binding sites are also indicated with boldface types.
Allele-specific binding of YY1 to the Xist/Tsix, Peg3, and Nespas-imprinted domains
The in vivo binding of YY1 to the clustered YY1 binding sites was tested with chromatin immunoprecipitation (ChIP) assays using the neonatal brain tissues derived from two hybrid mice, F1 (female) and F2 (male). Cross-linked chromatins were immunoprecipitated with polyclonal YY1 antibodies. The precipitated DNAs were analyzed by PCR using multiple primer sets for each clustered YY1 binding site. We have repeated our ChIP experiments more than three times for any given region, starting from ChIP to PCR, and one representative set is shown in Figures 3 and 4. As expected, the enrichment by the YY1 antibody was detected in the Xist cluster but not in the surrounding regions (Fig. 3B). Furthermore, this enrichment was detected mainly in the female tissue, whereas the enrichment was detected equally in both sexes at the autosomal Peg3 locus. The Xist locus is active only in the inactive X (Xi) present in female mammals, whereas the Xist locus of the active X (Xa) present in both sexes is inactivated with methylation on the 5′-side CpG islands (Fig. 3C). It has also been shown that YY1 binding is methylation sensitive (Kim et al. 2003). Therefore, the female-specific enrichment by the YY1 antibody may be an indication that the YY1 binding to the Xist cluster is allele specific, only to the unmethylated, active Xist locus present in Xi. A similar set of ChIP assays were also performed on the second site, the Tsix cluster, with three primer sets (DXPas34; Fig. 3A). However, because of the repetitive nature of this region, we have not been able to amplify this region efficiently and selectively. Therefore, the in vivo YY1 binding to the Tsix locus could not be confirmed from this study.
Figure 4.
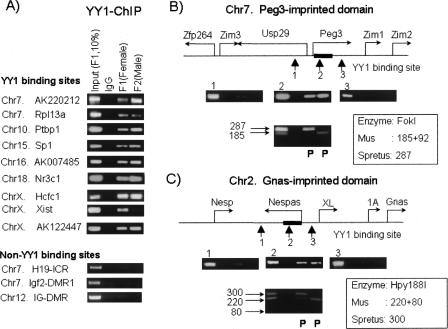
In vivo YY1 binding to other clustered regions. YY1 binding was tested on other clustered YY1 binding sites listed in Table 1. This series of ChIP analyses were also performed on three non-YY1 binding sites as negative control loci, including H19-ICR, Igf2-DMR1, and IG-DMR. The amplified PCR products of each cluster are shown in the following format: 10% of the Input DNA from F1 (lane 1), with pre-immune serum (lane 2), F1 brain (lane 3), and F2 brain (lane 4) with anti-YY1 antibody (A). We determined the allelic origin of the YY1-immunoprecipitated DNAs from the clustered site in Peg3-DMR (B) and Nespas-DMR (C). This analysis used sequence polymorphisms detected between the two parental species of our hybrid mice, F1 and F2. The restriction enzymes differentiating these sequence polymorphisms are shown with the estimated sizes of digested PCR products for each species. Two separate restriction enzyme digestions clearly demonstrated the presence of two alleles in the Input DNAs of F1 (lane 1). However, the YY1-immunoprecipitated DNAs at both Peg3-DMR and Nespas-DMR were mainly derived from the paternal allele (lanes 3 and 4).
Other clustered YY1 binding sites were also analyzed in terms of their in vivo binding to YY1 (Fig. 4A). Most of the clustered YY1 binding sites identified from our bioinformatic approach correspond to the in vivo binding sites of YY1, including the clusters in Rpl13a (ribosomal protein 13a), Ptbp1 (polypyrimidine tract binding protein 1), Sp1, Nr3C1 (glucocorticoid receptor), Hcfc1 (host cell factor C1), and three anonymous genes (AK220212, AK007485, and AK122447). In contrast, the same ChIP experiments did not show any specific enrichment at other YY1-unrelated loci, such as H19-ICR and Igf2-DMR1 of the H19/Igf2-domain and IG-DMR of the Gtl2/Dlk1-domain (Fig. 4A). The two clustered YY1 binding sites located in the Peg3-DMR and Nespas-DMR are also proven to be the in vivo binding sites of YY1 (Fig. 4B,C). Among three primer sets targeting the Nespas locus, only the primer set amplifying the clustered YY1 binding site showed detectable levels of enrichment by anti-YY1 antibody (Fig. 4C), which is also true for the Peg3 locus (Fig. 4B). Because these two clustered YY1 binding sites are located in differentially methylated regions, the allele-specific binding of YY1 was further tested at these two loci. For these experiments, we used two hybrid mice, F1 (C3H × M. spretus) and F2 (F1 × M. musculus). In F1 mice, the maternal allele is derived from M. musculus while the paternal is from M. spretus. In F2 mice, these two species’ alleles have been switched oppositely to two parental alleles. To differentiate the two parental alleles, sequence polymorphisms were first identified between two parental species, and subsequently these polymorphisms were visualized with restriction enzyme digestions. As shown in Figure 4, B and C, restriction enzyme digestions clearly demonstrated the presence of two alleles at the input DNAs, but only one dominant allele in the YY1-immunoprecipitated DNAs that were derived from both F1 and F2 tissues. This indicates that the immunoprecipitated DNA by the YY1 antibody was mainly derived from one of two parental alleles, in this case the paternal allele for both Nespas and Peg3.
Transcriptional activity of clustered YY1 binding sites
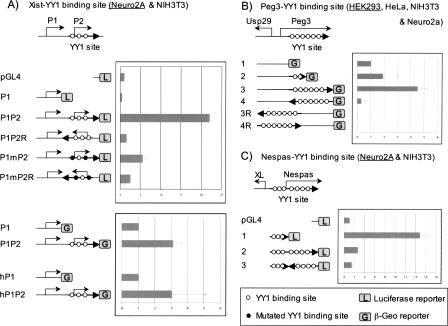
Because most of clustered YY1 binding sites are localized close to the promoter regions of the associated genes, we analyzed the potential transcriptional activity of these sites. For these experiments, we used the endogenous promoter regions that are associated with clustered YY1 binding sites to direct the transcription of two promoterless reporter genes, luciferase (pGL4, Promega) and IRES-β-Geo (Mountford et al. 1994). In this scheme, the transcriptional involvement of the YY1 binding sites was determined by comparing the activities of the two types of reporter constructs: one with and the other without the YY1 binding sites. We constructed a series of reporters with genomic fragments derived from mouse and human Xist, mouse Tsix, mouse Peg3, and mouse Nespas. These constructs were transfected into several different cell lines (Fig. 5).
Figure 5.
Transcriptional activity of the clustered YY1 binding sites. The transcriptional involvement of each clustered YY1 binding site was tested together with its endogenous promoter using pGL4 and/or IRES-β-Geo promoterless vector systems. (A) Transcriptional activity of YY1 binding sites of mouse and human Xist. The schematic diagram represents the genomic layout of two promoters with three YY1 binding sites of mouse Xist. Five constructs with one internal control (pGL4) were used for the analyses of the two promoters of mouse Xist, P1 and P2. The values shown in the graph on the right represent the averaged fold difference with standard deviation compared to that of the internal control. Each construct was analyzed more than three times using different cell lines shown inside a parenthesis. Because the overall patterns of data were similar among different cell lines, the graph shows only one representative data set from one of these cell lines, the name of which is underlined. The four constructs on the bottom represent the constructs of mouse and human Xist using another vector system, IRES-β-Geo. Two other YY1 binding sites were also analyzed in a similar way: the YY1 binding sites of mouse Peg3 (B) and mouse Nespas (C). In the case of constructs derived from IRES-β-Geo, the empty control vector does not yield any activity at all, and thus the vector construct containing each promoter without YY1 binding sites was used as an internal control. According to the results of Student’s t-test, most data points meet the statistical significance (P < 0.05) except a few constructs in each given cell line, as shown in construct 2 of Peg3 in HEK293 and construct hP1P2 of hXist in Neuro2a. However, the data of these constructs in other cell lines are consistent with the overall patterns shown above with much higher statistical significance.
As shown in Figure 5A, both human and mouse Xist contain two promoters and the second promoter region overlaps with the clustered YY1 binding site of Xist. The activity of the first promoter of mouse Xist (construct P1) was minimal with the pGL4 reporter system in both Neuro2a and NIH3T3 cells, even lower than that of the “empty” internal control (construct pGL4), indicating no significant activity of this promoter with this reporter system. However, when the second promoter (P2) of mouse Xist containing three YY1 binding sites was combined with the first promoter (P1), a genomic context similar to the endogenous one (construct P1P2), this construct yielded 22-fold higher activity than the control vector. This increased activity was detected only in the forward direction, indicating orientation-dependent increase of transcriptional activity by the P2 promoter. Mutations on two of the three YY1 binding sites (CGCCAT.TT to CAT TAT.TT) dramatically decreased the transcriptional activity in the forward direction (construct P1mP2), confirming the direct involvement of YY1 in the promoter activity of Xist. Another series of assays using the reporter system IRES-β-Geo also derived a similar conclusion that the inclusion of the YY1 binding sites increases the overall promoter activity of both mouse and human Xist (constructs with β-Geo in Fig. 5A). A similar pattern of results was also observed from the YY1 binding sites of mouse Tsix, boosting the transcriptional activity of promoters (data not shown).
The 2.5-kb clustered YY1 binding site of mouse Peg3 was also analyzed using constructs derived from the IRES-β-Geo system (Fig. 5B). The promoter region of Peg3 is bidirectional in vivo, directing the transcription of both Peg3 and Usp29, but only the constructs transcribing the reporter in the Peg3 direction showed transcriptional activity (constructs 1–4). The constructs in the Usp29 direction showed no activity (constructs 3R and 4R). The promoter without the YY1 binding sites (construct 1) yielded some levels of transcriptional activity, but the inclusion of the YY1 binding sites resulted in the further increase of this activity. The constructs containing the 1.2-kb and 2.5-kb YY1 binding sites yielded twofold and fourfold more activity, respectively (constructs 2 and 3). When the 2.5-kb genomic region of multiple YY1 binding sites was positioned in the reverse orientation (construct 4), the construct yielded very minimal levels of transcriptional activity, suggesting that the orientation of YY1 binding sites is critical for transcriptional activity.
As compared with other YY1 sites, the YY1 binding motifs identified from Nespas are scattered evenly throughout the entire 1.7-kb genomic interval spanning from the promoter to first intron. The promoter region with three YY1 binding motifs (construct 1) yielded relatively high transcriptional activity in both NIH3T3 and Neuro2A cell lines. However, the inclusion of the 1-kb first intron with the remaining four YY1 binding motifs (construct 2) resulted in a dramatic decrease of the promoter activity. Also, positioning the same region in an opposite orientation relative to that of the promoter further decreased the transcriptional activity of the Nespas promoter (construct 3). It remains to be investigated further, but the boosting effect observed consistently from other YY1 binding sites suggests that this decrease may be due to the presence of unknown repressors located within this interval but not due to the addition of the four YY1 binding motifs. Overall, our series of promoter assays on YY1 binding sites suggest that genomic regions with multiple YY1 binding sites function as potential enhancers with orientation dependency.
Insulator activity of clustered YY1 binding sites
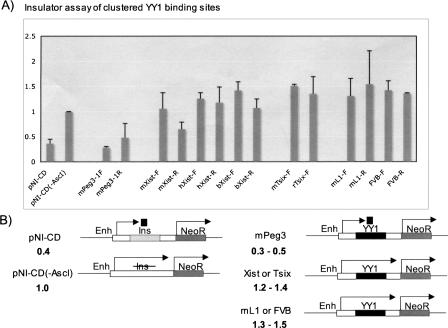
Since our initial observation on the clustered YY1 binding sites of Peg3-DMR revealed the presence of enhancer-blocking activity in the region (Kim et al. 2003), a series of similar insulator assays were performed to determine if the clustered YY1 binding sites identified in this study also have enhancer-blocking activity (Fig. 6). For these experiments, we used a cell line-based assay system utilizing a construct, pNI-CD, which contains the Neomycin resistance gene (neoR) as a reporter under the control of the erythroid-specific enhancer and promoter with a testing DNA fragment being positioned between this enhancer and promoter (Chung et al. 1997; Bell et al. 1999) (Fig. 6B).
Figure 6.
Insulator assay of the clustered YY1 binding sites. For insulator assays, the testing DNA fragments were positioned between an erythroid-specific enhancer (Enh) and a promoter directing the transcription of a reporter, neoR, as shown in B. This test used two control vectors: pNI-CD(-AscI) without any insulator and pNI-CD with a chicken insulator. The genomic fragments containing clustered YY1 binding sites were subcloned into the AscI site of the pNI-CD vector with two different orientations, forward (-F) and reverse (-R). The DNA fragments for the clustered sites of Xist and Tsix were derived from mouse (m), human (h), cow (b), and rat (r). Each fragment was tested more than three times, and the averaged values with standard errors are shown in A. The overall patterns observed in this series of experiments are summarized schematically in B with averaged values shown below the name of each construct.
Consistent with the human Peg3-DMR (Kim et al. 2003), the clustered YY1 binding sites of mouse Peg3-DMR also showed similar enhancer-blocking activity with the forward direction being more obvious, suggesting the evolutionary conservation of this activity in both species (mPeg3-1F). However, the clustered YY1 binding sites derived from both Xist and Tsix showed an unexpected, opposite outcome. The clustered YY1 binding sites of Xist and Tsix from mouse (mXist and mTsix), human (hXist), bovine (bXist), and rat (rTsix) all yielded much higher numbers of surviving colonies than the control construct without an insulator, pNI-CD(-AscI). This increase was observed in both orientations of the clustered YY1 binding sites. This is very unusual because any given DNA fragment without insulator activity, such as λ DNA fragments, usually yields either similar or slightly lower numbers of surviving colonies than the pNI-CD(-AscI) construct (Bell and Felsenfeld 2000; Hark et al. 2000). As an independent measure to resolve the two different outcomes between the YY1 clusters of Peg3-DMR versus the Xist/Tsix locus, we tested two unrelated DNA fragments containing a tandem array of YY1 binding sites: one 1.2-kb DNA fragment derived from one type of mouse LINE (mL1) and an 800-bp DNA fragment derived from the last intron of human fibrulin 1 gene (FVB). Both fragments also showed a similar result, the increased numbers of surviving colonies, suggesting that multiple YY1 binding sites be responsible for the observed increase. This is quite different from the enhancer-blocking activity observed from the YY1 sites of Peg3-DMR (Kim et al. 2003). It remains to be studied further in the future, but these results suggest that clustered YY1 binding sites may not function as an enhancer-blocker, and that the insulator activity observed from the clustered YY1 binding sites of Peg3-DMR may be a locus-specific activity.
Discussion
The clustered YY1 binding site of the Peg3-DMR region led us to identify three additional clustered YY1 binding sites that are localized in genomic regions undergoing similar epigenetic regulation as Peg3, the DMRs of Xist, Tsix, and Nespas. These genomic regions have maintained a tandem array of YY1 sites throughout mammalian evolution. The YY1 binding to these regions except for Tsix is shown to be allele specific and only to the unmethylated, active allele of the regions. The promoter and insulator assays also suggest that these regions function as a potential transcriptional enhancer but not as an enhancer-blocker.
Our screening strategy with the features derived from the Peg3-YY1 binding sites has been successful for finding additional ICRs as clustered YY1 binding sites. Four out of 21 clustered YY1 binding sites in mouse turn out to be ICRs, which is an unusual enrichment given the sheer number difference between imprinted and nonimprinted genes (∼100–40,000 per each mammalian genome). An independent study also reported the unusual enrichment of a particular sequence motif in imprinted domains, Motif 13: GGCCTGCCCTCCATCTTAG, which also appears to contain the minimal core motif of YY1 binding sequences (Wang et al. 2004). This frequent occurrence of YY1 binding sites in imprinted domains may be related to the two unique features of YY1. First, YY1 binding to its CpG-containing binding site (CGCCAT.TT) is methylation sensitive and thus can be controlled by the CpG methylation (Kim et al. 2003). This controllable binary mode of the YY1 binding to DNAs is an ideal feature for ICRs, which are predicted to regulate allele-specific involvement of trans factors. This has been further demonstrated in vivo in the current study showing the allele-specific binding of YY1 to Xist, Nespas, and Peg3 Figs. 3 and 4). Second, in Drosophila, YY1 is one of DNA-binding proteins responsible for the targeting of the Polycomb complex (Brown et al. 1998). The stable and heritable repression mediated through the Polycomb complex is similar, in many ways, to that of genomic imprinting. In fact, mammalian Polycomb complexes have been shown to be involved in maintaining the repressed state of imprinted domains (Mager et al. 2003; Lewis et al. 2004; Umlauf et al. 2004). Several studies also suggest that mammalian YY1 plays a similar role as shown in Drosophila as a targeting (or recruiting) protein of the Polycomb complexes (Caretti et al. 2004; Srinivasan et al. 2005). Thus, the unusual enrichment of YY1 binding sites may be for the targeting of the Polycomb complexes to the imprinted domains. In a similar line, it is interesting to note that the clustered YY1 binding sites of Peg3, Xist, Tsix, and Nespas all inherit their DNA methylation as a gametic signal from the previous generation (Norris et al. 1994; Lucifero et al. 2002; Coombes et al. 2003; Boumil et al. 2006). This suggests that YY1 and mammalian Polycomb complexes may be also involved in establishing imprinting signals on these ICRs during gametogenesis.
Most clustered binding sites are localized very close to promoters or first introns of genes (Table 1), suggesting that multiple YY1 binding sites are closely related to some unknown aspects of transcription. Consistently, the multiple YY1 binding sites of Peg3, Xist, and Tsix appear to increase the transcriptional strength of their promoters (Fig. 5). This also agrees well with the results of recent studies on the Tsix promoter, revealing that the YY1 binding sites in Tsix, DXPas34, have enhancer-like functions (Debrand et al. 1999; Stavropoulos et al. 2005). This region does not function as an independent enhancer, but some other upstream enhancers require this region for their transcriptional activation. The multiple YY1 binding sites also show an unusual orientation bias relative to the gene’s transcriptional direction (Table 1). The reason for this bias is not well understood but may be related to earlier observations regarding YY1 functions (Natesan and Gilman 1993; Kim and Shapiro 1996). These studies indicated that YY1 has an unusual DNA-bending capability and also that this bending activity is required for the transcription of the associated genes. The potential involvement of YY1 in the bending of DNA or chromatin structure is well supported, particularly, by the multiplicity of YY1 binding sites. It is easily conceivable that multiple binding sites with one particular orientation may be necessary for the bending of relatively large-size DNAs or chromatin of promoter regions.
The tandem arrays of YY1 binding sites derived from Xist, Tsix, and two other genomic loci do not have enhancer-blocking activity. This differs from the multiple YY1 binding sites of Peg3-DMR. According to the results from our insulator assays (Fig. 6), the multiple YY1 binding sites tend to derive an opposite outcome as expected from typical enhancer-blockers, resulting in more surviving colonies than the control vector. In this colony assay system, the boosted colony number could be caused by either increased transcriptional strength and/or increased protection from becoming heterochromatin. It is currently unclear which of these may contribute to the increased colony survivability, nevertheless the ultimate functional contribution by these multiple YY1 binding sites appears to be boosting or maintaining transcription of nearby genes. This is also consistent with the potential enhancer activity detected in the multiple YY1 binding sites of Peg3, Xist, and Tsix (Stavropoulos et al. 2005) (Fig. 5). If the in vivo function of multiple YY1 binding sites is indeed a transcriptional enhancer, the potential imprinting control mechanism(s) mediated by these ICRs may be different from the current enhancer-blocking model suggested from the H19/Igf2 domain (Wolffe 2000; Bell et al. 2001). The simplest model would be similar to the original enhancer competition hypothesis (Bartolomei and Tilghman 1997; Brannan and Bartolomei 1999). The ICRs with multiple YY1 binding sites in an unmethylated allele provide dominant transcriptional strength to immediate neighbor genes because of their proximity and/or compatibility, whereas in a methylated inactive allele the surrounding genes located in farther genomic distances can be transcribed because of the inactive state of the dominant genes. It needs to be tested in the future, but the ICRs with a tandem array of YY1 binding sites are predicted to provide new paradigm(s) for imprinting regulatory mechanism(s).
Methods
Identification of clustered YY1 binding sites
To test and optimize our bioinformatics searches, we performed a series of pre-tests, including Position Weight Matrix-based searches using more generous and statistical methods. One of the main lessons from these pre-tests was that many genomic regions could be easily identified as potential clustered YY1 binding sites with the total number of positive hits per each genome ranging from several hundreds to thousands. In particular, many positive hits turned out to be associated with repetitive DNA elements in both human and mouse. Therefore, we decided to perform more stringent searches using masked genome sequences. We also decided to perform more focused searches targeting evolutionarily dynamic regions with constant decay and regeneration of the CpG dinucleotide. All the lessons learned from the pre-tests have been incorporated into the final version of our homemade Perl script. Using this script, we have analyzed the masked genome sequences of human (hg17), mouse (mm5), and rat (rn3). The identified genomic regions were mapped and annotated using the BLAST and MapViewer programs. The Perl script and the sequences of the identified clustered YY1 binding sites are available as Supplemental material (www.genome.org) and at (http://jookimlab.lsu.edu/JooKimLab/Data.html).
Gel shift assay
The Gel Shift Assay system (Promega) was used for our DNA mobility shift assays. About 4 μg of nuclear extracts were first mixed with binding buffer and unlabeled competitor probes with varying amounts (10–100 pmol) for 20 min at room temperature and later mixed with the P32-labeled duplex probes (1 pmol) for an additional 10 min at room temperature. The reaction mixtures were separated on 5% native polyacrylamide gel (acryl:bis = 37.5:1) in 0.5× Tris buffer at pH 8.0 for 2 hr, and the separated gels were exposed to X-ray films for 1 hr. For supershift assays, the polyclonal antibody raised against the entire portion of human YY1 was obtained from a commercial firm (catalog no. sc-1703; Santa Cruz Biotechnology). Another polyclonal antibody raised against histone H3 was also used as a negative control for supershift assays (catalog no. sc-10809; Santa Cruz Biotechnology). All the gel shift assays used nuclear extracts prepared from human HeLa cell lines (Promega). The following oligonucleotides are the probes for our gel shift assays: mYY1–1 (5′-CCCCGT GGCAGACATTTTAGTGACCTCCCA-3′, 5′- TGGGAGGTCAC TAAATGTCTGCCACGGGG-3′); MYY1–2 (5′-CCCCGTGGCAG GCATTTTAGTGACCTCCCA-3′, 5′-TGGGAGGTCACTAAAAT GCCTGCCACGGGG-3); yy1 (5′-CGCTCCGCGCCATCTTGGCG GCTGGT-3′, 5′-ACCAGCCGCCAAGATGGCGCGGAGCG-3′); yy2 (5′-CGCTCCGCATTATCTTGGCGGCTGGT-3′, 5′-ACCAG CCGCCAAGATAATGCGGAGCG-3′).
ChIP assay
Chromatin immunoprecipitations were performed according to the protocol provided by Upstate Biotechnology with some modification as described previously (Kim et al. 2003). Briefly, we used mouse brain tissues from neonatal F1 (C3H × Mus spretus) and F2 (F1 × Mus musculus) hybrid homogenized in 10 ml of PBS for ChIP assay. The samples were treated with formaldehyde to final concentration of 1% and incubated at 37°C for 10 min. Treated samples were sheared by sonication and immunoprecipiated with anti-YY1 antibody. Precipitated DNA and protein complexes were reverse cross-linked and purified through phenol/chloroform extraction. Purified DNA was used as templates for PCR amplification. The oligonucleotide sequences used for this study can be available upon request. PCR reactions were carried out for 36 cycles using standard PCR conditions. The resulting PCR products were analyzed by running on 1.6% agarose gel and staining with ethidium bromide.
Promoter/enhancer assay
The 5.1-kb IRES-β-Geo fragment was transferred from the pGT1.8IresBgeo vector (Mountford et al. 1994) into the BamHI site of pBluescript SK(-). Genomic fragments containing a tandem array of YY1 binding sites were amplified by PCR, and subcloned into the NotI site of the modified pBluscript with IRES-β-Geo. For the reporter assays with the pGL4 system, the promoter and YY1 binding sites of Xist were subcloned into the BglII and HindIII sites, respectively, whereas the promoter and first intron of Nespas were subcloned into XhoI and BglII sites. The locations of the tested DNA fragments within the GenBank accession nos. are as follows: Peg3 construct 1 (AF105262; 2110–3379), construct 2 (AF105262; 2110–4281), construct 3 (AF105262; 2110–5905), mouse Xist construct P1 (AJ421479; 105154–107233), construct P1P2 (AJ421479; 105154–108089), human Xist construct hP1 (AL353840; 37006–35218), hP1P2 (AL353840; 37006–34435), mouse Tsix construct 1 (AJ421479; 140222–141899), construct 2 (AJ421479; 138971–141899), and mouse Nespas construct 1 (AJ251761; 13753–12730), construct 2 (AJ251761;12731–11797).
For the promoter/enhancer assays, HEK 293, NIH 3T3, and HeLa cells were grown in DMEM medium (Gibco BRL) and Neuro2A cell was maintained in MEM medium (Gibco BRL). Media were supplemented with 10% fetal bovine serum and 1% antibiotic-antimycotic (Gibco BRL). All cell lines were grown at 37°C in a humidified incubator containing 5% CO2. The 2 × 105 cells were plated per one well of six-well plate. On the next day, cells were cotransfected with GeneJuice transfection reagent (Novagen) according to the manufacturer’s protocol. Briefly, transfection was performed with serum-free medium containing 3 μl of GeneJuice and 1 μg of DNA (0.9 μg β-Geo vector + 0.1μg pGL3 Control vector [Promega]) per well. Two days after transfection, the cells were harvested, washed with PBS, and treated with 200 μl of lysis buffer (0.25 M Tris-HCl at pH 7.8 + 0.1% NP40) for 30 min at 4°C and cellular debris was removed by centrifugation for 10 min. For the β-galactosidase assay, 30 μl of cell lysate was mixed with the same volume of 2× β-galactosidase assay buffer (Promega) in a 96-well flat-bottom clear plate. The plate was incubated at 37°C, monitored visually, and terminated with 90 μl of 1 M sodium carbonate. The absorbance was measured at 405 nm with Wallac 1420 multilabel counter VICTOR3 (PerkinElmer). To control for transfection efficiency in each well, β-galactosidase activity was normalized to luciferase activity. For the luciferase assay, 20 μl of cell lysate was combined with 100 μl of luciferase assay reagent in a 96-well flat-bottom white plate (Corning). Luminescence was measured with Wallac 1420 multilabel counter VICTOR3.
Insulator assay
DNA fragments of interest were cloned into the AscI site of pNI-CD (generous gift from Drs. Gary Felsenfeld and Adam West). Each fragment was cloned in both orientations. The locations of the tested DNA fragments within the GenBank accession nos. are as follows: mPeg31–1 (AF105262; 1984–2932), mXist (AJ421479; 105842–107855), hXist (M97168; 1186–1710), bXist (AF104906; 945–1644), mTsix (AJ421479; 138971–140246), rTsix (NW_048043.1; 2951903–2953005), and FVB (NT_086921.1; 23995819–23996257). The mL1 was amplified from the BAC clone, RPCI23-93H2, with the following two primers: mL1-a (5′-GGCCTAGTCAGCCATCATTGG-3′) and mL1-b (5′- TTACAGGGAAGGTGCGCAGA-3′). Constructs were transfected into K562 cells by electroporation at 200V, 1000 mF (double pulse) using a Bio-Rad Gene Pulser II. After a 10-min recovery on ice, cells were plated into RPMI supplemented with 10% FBS, 200 mM L-glut, and Pen/Strep. Twenty-four hours after transfection, cells were washed and resuspended in Improved MEM Zn++ option (Gibco BRL). Cells were plated into 0.3% soft agar with 1050 μg/mL Geneticin (Gibco BRL) and incubated at 37°C for 18–21 d.
Acknowledgments
We thank Drs. Prescott Deininger and David Donze for critical reading of our manuscript. This research was supported by National Institutes of Health R01 GM66225, National Science Foundation EPS-0346411, and the State of Louisiana Board of Regents Support Fund (J.K) and the U.S. Department of Energy under contract no. W-7405-ENG-48.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5091406
References
- Austen M., Luscher B., Luscher-Firzlaff J.M., Luscher B., Luscher-Firzlaff J.M., Luscher-Firzlaff J.M. Characterization of the transcriptional regulator YY1. The bipartite transactivation domain is independent of interaction with the TATA box-binding protein, transcription factor IIB, TAFII55, or cAMP-responsive element-binding protein (CPB)-binding protein. J. Biol. Chem. 1997;272:1709–1717. doi: 10.1074/jbc.272.3.1709. [DOI] [PubMed] [Google Scholar]
- Bartolomei M.S., Tilghman S.M., Tilghman S.M. Genomic imprinting in mammals. Annu. Rev. Genet. 1997;31:493–525. doi: 10.1146/annurev.genet.31.1.493. [DOI] [PubMed] [Google Scholar]
- Bell A.C., Felsenfeld G., Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- Bell A.C., West A.G., Felsenfeld G., West A.G., Felsenfeld G., Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- Bell A.C., West A.G., Felsenfeld G., West A.G., Felsenfeld G., Felsenfeld G. Insulators and boundaries: Versatile regulatory elements in the eukaryotic genome. Science. 2001;291:447–450. doi: 10.1126/science.291.5503.447. [DOI] [PubMed] [Google Scholar]
- Boumil R.M., Ogawa Y., Sun B.K., Hyunh K.D., Lee J.T., Ogawa Y., Sun B.K., Hyunh K.D., Lee J.T., Sun B.K., Hyunh K.D., Lee J.T., Hyunh K.D., Lee J.T., Lee J.T. Differential methylation of Xite and CTCF sites in Tsix mirrors the pattern of X-inactivation choice in mice. Mol. Cell. Biol. 2006;26:2109–2117. doi: 10.1128/MCB.26.6.2109-2117.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannan C.I., Bartolomei M.S., Bartolomei M.S. Mechanisms of genomic imprinting. Curr. Opin. Genet. Dev. 1999;9:164–170. doi: 10.1016/S0959-437X(99)80025-2. [DOI] [PubMed] [Google Scholar]
- Brown J.L., Mucci D., Whiteley M., Dirksen M.L., Kassis J.A., Mucci D., Whiteley M., Dirksen M.L., Kassis J.A., Whiteley M., Dirksen M.L., Kassis J.A., Dirksen M.L., Kassis J.A., Kassis J.A. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol. Cell. 1998;1:1057–1064. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- Caretti G., Di Padova M., Micales B., Lyons G.E., Sartorelli V., Di Padova M., Micales B., Lyons G.E., Sartorelli V., Micales B., Lyons G.E., Sartorelli V., Lyons G.E., Sartorelli V., Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes & Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C.M., Roeder B.G., Roeder B.G. Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators. Science. 1995;267:531–536. doi: 10.1126/science.7824954. [DOI] [PubMed] [Google Scholar]
- Chung J.H., Bell A.C., Felsenfeld G., Bell A.C., Felsenfeld G., Felsenfeld G. Characterization of the chicken β-globin insulator. Proc. Natl. Acad. Sci. 1997;94:575–580. doi: 10.1073/pnas.94.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constancia M., Pickard B., Kelsey G., Reik W., Pickard B., Kelsey G., Reik W., Kelsey G., Reik W., Reik W. Imprinting mechanisms. Genome Res. 1998;8:881–900. doi: 10.1101/gr.8.9.881. [DOI] [PubMed] [Google Scholar]
- Coombes C., Arnaud P., Gordon E., Dean W., Coar E.A., Williamson C.M., Feil R., Peters J., Kelsey G., Arnaud P., Gordon E., Dean W., Coar E.A., Williamson C.M., Feil R., Peters J., Kelsey G., Gordon E., Dean W., Coar E.A., Williamson C.M., Feil R., Peters J., Kelsey G., Dean W., Coar E.A., Williamson C.M., Feil R., Peters J., Kelsey G., Coar E.A., Williamson C.M., Feil R., Peters J., Kelsey G., Williamson C.M., Feil R., Peters J., Kelsey G., Feil R., Peters J., Kelsey G., Peters J., Kelsey G., Kelsey G. Epigenetic properties and identification of an imprint mark in the Nesp-Gnasxl domain of the mouse Gnas imprinted locus. Mol. Cell. Biol. 2003;23:5475–5488. doi: 10.1128/MCB.23.16.5475-5488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debrand E., Chureau C., Arnaud D., Avner P., Heard E., Chureau C., Arnaud D., Avner P., Heard E., Arnaud D., Avner P., Heard E., Avner P., Heard E., Heard E. Functional analysis of the DXPas34 locus, a 3′ regulator of Xist expression. Mol. Cell. Biol. 1999;19:8513–8525. doi: 10.1128/mcb.19.12.8513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoriw A.M., Stein P., Svoboda P., Schultz R.M., Bartolomei M.S., Stein P., Svoboda P., Schultz R.M., Bartolomei M.S., Svoboda P., Schultz R.M., Bartolomei M.S., Schultz R.M., Bartolomei M.S., Bartolomei M.S. Transgenic RNAi reveals essential function for CTCF in H19 gene imprinting. Science. 2004;303:238–240. doi: 10.1126/science.1090934. [DOI] [PubMed] [Google Scholar]
- Hark A.T., Schoenherr C.J., Katz D.J., Ingram R.S., Levorse J.M., Tilghman S.M., Schoenherr C.J., Katz D.J., Ingram R.S., Levorse J.M., Tilghman S.M., Katz D.J., Ingram R.S., Levorse J.M., Tilghman S.M., Ingram R.S., Levorse J.M., Tilghman S.M., Levorse J.M., Tilghman S.M., Tilghman S.M. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- Khan H., Smit A., Boissinot S., Smit A., Boissinot S., Boissinot S. Molecular evolution and temporal amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res. 2006;16:78–87. doi: 10.1101/gr.4001406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Shapiro D.J., Shapiro D.J. In simple synthetic promoters YY1-induced DNA bending is important in transcription activation and repression. Nucleic Acids Res. 1996;24:4341–4348. doi: 10.1093/nar/24.21.4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Kollhoff A., Bergmann A., Stubbs L., Kollhoff A., Bergmann A., Stubbs L., Bergmann A., Stubbs L., Stubbs L. Methylation-sensitive binding of transcription factor YY1 to an insulator sequence within the paternally expressed imprinted gene, Peg3. Hum. Mol. Genet. 2003;12:233–245. doi: 10.1093/hmg/ddg028. [DOI] [PubMed] [Google Scholar]
- Lee J.S., Galvin K.M., Shi Y., Galvin K.M., Shi Y., Shi Y. Evidence for physical interaction between the zinc-finger franscription factors YY1 and Sp1. Proc. Natl. Acad. Sci. 1993;90:6145–6149. doi: 10.1073/pnas.90.13.6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.S., Galvin K.M., See R.H., Eckner R., Livingston D., Moran E., Shi Y., Galvin K.M., See R.H., Eckner R., Livingston D., Moran E., Shi Y., See R.H., Eckner R., Livingston D., Moran E., Shi Y., Eckner R., Livingston D., Moran E., Shi Y., Livingston D., Moran E., Shi Y., Moran E., Shi Y., Shi Y. Relief of YY1 transcriptional repression by adenovirus E1A is mediated by E1A-associated protein p300. Genes & Dev. 1995;9:1188–1198. doi: 10.1101/gad.9.10.1188. [DOI] [PubMed] [Google Scholar]
- Lewis A., Mitsuya K., Umlauf D., Smith P., Dean W., Walter J., Higgins M., Feil R., Reik W., Mitsuya K., Umlauf D., Smith P., Dean W., Walter J., Higgins M., Feil R., Reik W., Umlauf D., Smith P., Dean W., Walter J., Higgins M., Feil R., Reik W., Smith P., Dean W., Walter J., Higgins M., Feil R., Reik W., Dean W., Walter J., Higgins M., Feil R., Reik W., Walter J., Higgins M., Feil R., Reik W., Higgins M., Feil R., Reik W., Feil R., Reik W., Reik W. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat. Genet. 2004;36:1291–1295. doi: 10.1038/ng1468. [DOI] [PubMed] [Google Scholar]
- Lucifero D., Mertineit C., Clarke H.J., Bestor T.H., Trasler J.M., Mertineit C., Clarke H.J., Bestor T.H., Trasler J.M., Clarke H.J., Bestor T.H., Trasler J.M., Bestor T.H., Trasler J.M., Trasler J.M. Methylation dynamics of imprinted genes in mouse germ cells. Genomics. 2002;79:530–538. doi: 10.1006/geno.2002.6732. [DOI] [PubMed] [Google Scholar]
- Mager J., Montgomery N.D., de Villena F.P., Magnuson T., Montgomery N.D., de Villena F.P., Magnuson T., de Villena F.P., Magnuson T., Magnuson T. Genome imprinting regulated by the mouse Polycomb group protein Eed. Nat. Genet. 2003;33:502–507. doi: 10.1038/ng1125. [DOI] [PubMed] [Google Scholar]
- Mountford P., Zevnik B., Duwel A., Nichols J., Li M., Dani C., Robertson M., Chambers I., Smith A., Zevnik B., Duwel A., Nichols J., Li M., Dani C., Robertson M., Chambers I., Smith A., Duwel A., Nichols J., Li M., Dani C., Robertson M., Chambers I., Smith A., Nichols J., Li M., Dani C., Robertson M., Chambers I., Smith A., Li M., Dani C., Robertson M., Chambers I., Smith A., Dani C., Robertson M., Chambers I., Smith A., Robertson M., Chambers I., Smith A., Chambers I., Smith A., Smith A. Dicistronic targeting constructs: Reporters and modifiers of mammalian gene expression. Proc. Natl. Acad. Sci. 1994;91:4303–4307. doi: 10.1073/pnas.91.10.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natesan S., Gilman M.Z., Gilman M.Z. DNA bending and orientation-dependent function of YY1 in the c-fos promoter. Genes & Dev. 1993;7:2497–2509. doi: 10.1101/gad.7.12b.2497. [DOI] [PubMed] [Google Scholar]
- Norris D.P., Patel D., Kay G.F., Penny G.D., Brockdorff N., Sheardown S.A., Rastan S., Patel D., Kay G.F., Penny G.D., Brockdorff N., Sheardown S.A., Rastan S., Kay G.F., Penny G.D., Brockdorff N., Sheardown S.A., Rastan S., Penny G.D., Brockdorff N., Sheardown S.A., Rastan S., Brockdorff N., Sheardown S.A., Rastan S., Sheardown S.A., Rastan S., Rastan S. Evidence that random and imprinted Xist expression is controlled by pre-emptive methylation. Cell. 1994;77:41–51. doi: 10.1016/0092-8674(94)90233-x. [DOI] [PubMed] [Google Scholar]
- Oei S.L., Babich V.S., Kazakov V.I., Usmanova N.M., Kropotov A.V., Tomilin N.V., Babich V.S., Kazakov V.I., Usmanova N.M., Kropotov A.V., Tomilin N.V., Kazakov V.I., Usmanova N.M., Kropotov A.V., Tomilin N.V., Usmanova N.M., Kropotov A.V., Tomilin N.V., Kropotov A.V., Tomilin N.V., Tomilin N.V. Clusters of regulatory signals for RNA polymerase II transcription associated with Alu family repeats and CpG islands in human promoters. Genomics. 2004;83:873–882. doi: 10.1016/j.ygeno.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Reik W., Walter J., Walter J. Imprinting mechanisms in mammals. Curr. Opin. Genet. Dev. 1998;8:154–164. doi: 10.1016/s0959-437x(98)80136-6. [DOI] [PubMed] [Google Scholar]
- Rezai-Zadeh N., Zhang X., Namour F., Fejer G., Wen Y.D., Yao Y.L., Gyory I., Wright K., Seto E., Zhang X., Namour F., Fejer G., Wen Y.D., Yao Y.L., Gyory I., Wright K., Seto E., Namour F., Fejer G., Wen Y.D., Yao Y.L., Gyory I., Wright K., Seto E., Fejer G., Wen Y.D., Yao Y.L., Gyory I., Wright K., Seto E., Wen Y.D., Yao Y.L., Gyory I., Wright K., Seto E., Yao Y.L., Gyory I., Wright K., Seto E., Gyory I., Wright K., Seto E., Wright K., Seto E., Seto E. Targeted recruitment of a histone H4-specific methyltransferase by the transcription factor YY1. Genes & Dev. 2003;17:1019–1029. doi: 10.1101/gad.1068003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satijn D.P.E., Hamer K.M., Blaawen J.D., Otte A.P., Hamer K.M., Blaawen J.D., Otte A.P., Blaawen J.D., Otte A.P., Otte A.P. The polycomb group protein EED interact with YY1, and both proteins induce neural tissue in Xenopus embryos. Mol. Cell Biol. 2001;21:1360–1369. doi: 10.1128/MCB.21.4.1360-1369.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenherr C.J., Levorse J.M., Tilghman S.M., Levorse J.M., Tilghman S.M., Tilghman S.M. CTCF maintains differential methylation at the Igf2/H19 locus. Nat. Genet. 2003;33:66–69. doi: 10.1038/ng1057. [DOI] [PubMed] [Google Scholar]
- Schug J., Schuller W.P., Kappen C., Salbaum J.M., Bucan M., Stoeckert C.J., Jr., Schuller W.P., Kappen C., Salbaum J.M., Bucan M., Stoeckert C.J., Jr., Kappen C., Salbaum J.M., Bucan M., Stoeckert C.J., Jr., Salbaum J.M., Bucan M., Stoeckert C.J., Jr., Bucan M., Stoeckert C.J., Jr., Stoeckert C.J., Jr. Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol. 2005;6:R33. doi: 10.1186/gb-2005-6-4-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto E., Lewis B., Shenk T., Lewis B., Shenk T., Shenk T. Interaction between transcription factors Sp1 and YY1. Nature. 1993;365:462–464. doi: 10.1038/365462a0. [DOI] [PubMed] [Google Scholar]
- Spahn L., Barlow D.P., Barlow D.P. An ICE pattern crystallizers. Nat. Genet. 2003;35:11–12. doi: 10.1038/ng0903-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan L., Pan X., Atchison M., Pan X., Atchison M., Atchison M. Transient requirements of YY1 expression for PcG transcriptional repression and phenotypic rescue. J. Cell. Biochem. 2005;96:689–699. doi: 10.1002/jcb.20562. [DOI] [PubMed] [Google Scholar]
- Stavropoulos N., Rowntree R.K., Lee J.T., Rowntree R.K., Lee J.T., Lee J.T. Identification of developmentally specific enhancers for Tsix in the regulation of X chromosome inactivation. Mol. Cell. Biol. 2005;25:2757–2769. doi: 10.1128/MCB.25.7.2757-2769.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M.J., Seto E., Seto E. Unlocking the mechanisms of transcription factor YY1: Are chromatin modifying enzymes the key? Gene. 1999;236:197–208. doi: 10.1016/s0378-1119(99)00261-9. [DOI] [PubMed] [Google Scholar]
- Umlauf D., Goto Y., Cao R., Cerqueira F., Wagschal A., Zhang Y., Feil R., Goto Y., Cao R., Cerqueira F., Wagschal A., Zhang Y., Feil R., Cao R., Cerqueira F., Wagschal A., Zhang Y., Feil R., Cerqueira F., Wagschal A., Zhang Y., Feil R., Wagschal A., Zhang Y., Feil R., Zhang Y., Feil R., Feil R. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat. Genet. 2004;36:1296–1300. doi: 10.1038/ng1467. [DOI] [PubMed] [Google Scholar]
- Usheva A., Shenk T., Shenk T. TATA-binding protein-independent initiation: YY1, TFIIB, and RNA polymerase II direct basal transcription on supercoiled template DNA. Cell. 1994;76:1115–1121. doi: 10.1016/0092-8674(94)90387-5. [DOI] [PubMed] [Google Scholar]
- Wang Z., Fan H., Yang H.H., Hu Y., Buetow K.H., Lee M.P., Fan H., Yang H.H., Hu Y., Buetow K.H., Lee M.P., Yang H.H., Hu Y., Buetow K.H., Lee M.P., Hu Y., Buetow K.H., Lee M.P., Buetow K.H., Lee M.P., Lee M.P. Comparative sequence analysis of imprinted genes between human and mouse to reveal imprinting signatures. Genomics. 2004;83:395–401. doi: 10.1016/j.ygeno.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Williamson C.M., Turner M.D., Ball S.T., Nottingham W.T., Glenister P., Fray M., Tymowska-Lalanne Z., Plagge A., Powles-Glover N., Kelsey G., Turner M.D., Ball S.T., Nottingham W.T., Glenister P., Fray M., Tymowska-Lalanne Z., Plagge A., Powles-Glover N., Kelsey G., Ball S.T., Nottingham W.T., Glenister P., Fray M., Tymowska-Lalanne Z., Plagge A., Powles-Glover N., Kelsey G., Nottingham W.T., Glenister P., Fray M., Tymowska-Lalanne Z., Plagge A., Powles-Glover N., Kelsey G., Glenister P., Fray M., Tymowska-Lalanne Z., Plagge A., Powles-Glover N., Kelsey G., Fray M., Tymowska-Lalanne Z., Plagge A., Powles-Glover N., Kelsey G., Tymowska-Lalanne Z., Plagge A., Powles-Glover N., Kelsey G., Plagge A., Powles-Glover N., Kelsey G., Powles-Glover N., Kelsey G., Kelsey G., et al. Identification of an imprinting control region affecting the expression of all transcripts in the Gnas cluster. Nat. Genet. 2006;38:350–355. doi: 10.1038/ng1731. [DOI] [PubMed] [Google Scholar]
- Wolffe A.P. Transcriptional control: Imprinting insulation. Curr. Biol. 2000;10:R463–R465. doi: 10.1016/s0960-9822(00)00534-0. [DOI] [PubMed] [Google Scholar]
- Yang W.M., Inouye C., Zeng Y., Bearss D., Seto E., Inouye C., Zeng Y., Bearss D., Seto E., Zeng Y., Bearss D., Seto E., Bearss D., Seto E., Seto E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc. Natl. Acad. Sci. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant S.R., Zhu W., Millinoff D., Slightom J.L., Goodman M., Gumucio D.L., Zhu W., Millinoff D., Slightom J.L., Goodman M., Gumucio D.L., Millinoff D., Slightom J.L., Goodman M., Gumucio D.L., Slightom J.L., Goodman M., Gumucio D.L., Goodman M., Gumucio D.L., Gumucio D.L. High affinity YY1 binding motifs: Identification of two core types (ACAT and CCAT) and distribution of potential binding sites within the human β globin cluster. Nucleic Acids Res. 1995;23:4457–4465. doi: 10.1093/nar/23.21.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]