Abstract
Type III secretion systems are utilized by a number of gram-negative bacterial pathogens to deliver virulence-associated proteins into host cells. Using a PCR-based approach, we identified homologs of type III secretion genes in the gram-negative bacterium Burkholderia cepacia, an important pulmonary pathogen in immunocompromised patients and patients with cystic fibrosis. One of the genes, designated bscN, encodes a member of a family of ATP-binding proteins believed to generate energy driving virulence protein secretion. Genetic dissection of the regions flanking the bscN gene revealed a locus consisting of at least 10 open reading frames, predicted to encode products with significant homology to known type III secretion proteins in other bacteria. A defined null mutation was generated in the bscN gene, and the null strain and wild-type parent strain were examined by use of a murine model of B. cepacia infection. Quantitative bacteriological analysis of the lungs and spleens of infected C57BL/6 mice revealed that the bscN null strain was attenuated in virulence compared to the parent strain, with significantly lower bacterial recovery from the lungs and spleens at 3 days postinfection. Moreover, histopathological changes, including an inflammatory cell infiltrate, were more pronounced in the lungs of mice infected with the wild-type parent strain than in those of mice infected with the isogenic bscN mutant. These results implicate type III secretion as an important determinant in the pathogenesis of B. cepacia.
Cystic fibrosis (CF) is the most common lethal autosomal recessive disease of Caucasians, affecting approximately 1 in 2,000 live births. The major cause of morbidity and mortality in CF patients is chronic bacterial infection of the lungs, leading to repeated and destructive pulmonary exacerbations (28). While Pseudomonas aeruginosa is the major pathogen in CF patients, over the last decade the gram-negative bacterium Burkholderia cepacia has also emerged as an important pulmonary pathogen in individuals with CF as well as other compromised individuals (18). B. cepacia acquisition in individuals with CF generally leads to chronic infection. In some cases, however, B. cepacia infection can also result in acute pulmonary deterioration, accompanied by necrotizing pneumonia and septicemia (14, 55). In approximately 25% of CF patients infected with B. cepacia, the clinical course is ultimately fatal (23, 29).
B. cepacia is actually a complex of bacteria belonging to at least seven genomic species, or genomovars (57). Four of these genomovars were recently designated distinct species: Burkholderia multivorans (genomovar II), Burkholderia stabilis (genomovar IV), Burkholderia vietnamiensis (genomovar V), and Burkholderia ambifaria (genomovar VI). Although strains representing many of the known genomovars of the B. cepacia complex have been associated with opportunistic infections in humans, strains belonging to genomovar III have been most commonly associated with severe and chronic infections in CF patients (32, 57).
Investigation into the pathogenesis of B. cepacia has provided growing evidence that the organism possesses mechanisms for adherence (42, 46), invasion (3, 33, 56), intracellular survival (33, 41), and modulation of the host immune response (22, 36, 37, 52). B. cepacia has been shown to adhere to human mucins and respiratory epithelial cells, and this adherence may promote colonization of the lungs (46, 47). Invasion and intracellular survival have been demonstrated by use of cultured respiratory epithelial cells and macrophages (3, 33, 41), and the recent examination of lung explants from infected CF patients revealed the presence of B. cepacia intracellularly in surface airway epithelial cells and luminal macrophages (43). The ability of B. cepacia exoproducts, including lipopolysaccharide, to elicit cytokine release from lung epithelial cells and human monocytes may also contribute to infection, through both the induction and the modulation of the host proinflammatory response (22, 37, 51, 52). Despite these new insights, the specific virulence determinants and corresponding genetic elements required for B. cepacia infection are largely unknown.
A wide range of gram-negative bacterial pathogens utilize a conserved secretory system, termed type III, to deliver virulence proteins directly into host cells (12). The type III system-dependent delivery of virulence proteins, referred to as effector proteins, into host cells has been associated with (i) invasion by Salmonella enterica serovar Typhimurium and Shigella flexneri (9, 34), (ii) lesion formation by enteropathogenic Escherichia coli (24), (iii) cytotoxicity caused by P. aeruginosa (16, 19), (iv) evasion of phagocytosis and the host immune response by Yersinia spp. and Bordetella bronchiseptica (11, 61), and (v) intracellular survival in S. enterica serovar Typhimurium (6, 20). Type III secretion systems are also utilized by phytopathogens to cause disease on susceptible plants, as well as to elicit the so-called hypersensitive response in resistant plants (1). Delivery of the bacterial type III effector proteins into host cells requires a secretory apparatus comprised of approximately 10 highly conserved proteins (21). This core secretory apparatus is also found in the flagellar type III secretion system, utilized for the export and assembly of the structural components of the bacterial flagellum (31).
In this study, we identified genes encoding core components of a type III secretion system in B. cepacia strain J2315, a clinical isolate associated with epidemic outbreaks and mortality in CF patients. One of these genes, designated bscN (Burkholderia secretion protein N), encodes a new member of a family of ATP-binding proteins that likely function in the generation of energy driving effector protein secretion. To examine the role of B. cepacia type III secretion in virulence, a defined null mutation was generated in bscN, and the null strain and wild-type parent strain were compared by use of a murine model of B. cepacia infection.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this work are listed in Table 1. E. coli strains were grown in Luria-Bertani (LB) broth (48) or on LB agar plates supplemented with ampicillin (100 μg/ml), tetracycline (12 μg/ml), or chloramphenicol (30 μg/ml) as necessary. B. cepacia strain J2315 is a CF clinical isolate belonging to genomovar III (17). Strain J2315 was grown with aeration at 37°C in LB broth or in M9 broth-0.3% Casamino Acids (48) supplemented with tetracycline (50 μg/ml) or chloramphenicol (30 μg/ml) or on LB agar plates supplemented with tetracycline (500 μg/ml), kanamycin (50 μg/ml), or chloramphenicol (300 μg/ml) as necessary. For swarm agar plate assays, LB agar plates containing 0.25% (wt/vol) agar were stab inoculated with overnight cultures of B. cepacia and incubated for 24 h at 37°C. For the mouse infection experiments, B. cepacia strains were grown on LB agar plates with the addition of 0.25% (wt/vol) hog gastric mucin (Sigma).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | supE44 lacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Bethesda Research Laboratories |
| S17-1 | Integrated RP4-2; Tc::Mu; Km::Tn7 | 53 |
| B. cepacia | ||
| J2315 | CF clinical isolate; genomovar III | 17 |
| CM56 | bscN::cat derivative of J2315 | This work |
| CM62 | CM56 with plasmid pCM23 | This work |
| BC7 | CF clinical isolate; genomovar III | 44 |
| K56-2 | CF clinical isolate; genomovar III | 13 |
| PC184 | CF clinical isolate; genomovar III | 30 |
| Plasmids | ||
| pCRII | TA cloning vector; Apr | Invitrogen |
| pBluescript SK II(+) | Cloning and single-stranded phagemid; Apr | Stratagene |
| pCAT1 | Source of cat cassette; Cmr | 56 |
| pNPTS138 | Derivative of pLITMUS38 cloning vector with nptI, RK2 oriT, and Bacillus subtilis sacB; Kmr | M. R. K. Alley |
| pMR4 | Broad-host-range vector; Tcr | C. Mohr and R. Roberts |
| pUCBM20 | Cloning vector; Apr | Boehringer Mannheim |
| pCMT2 | Tcr derivative of pNPTS138 | C. Mohr and M. Tomich |
| pCH7 | pLAFR5-derived cosmid containing 20-kb B. cepacia genomic insert encoding the type III secretion cluster | 56 |
| pCM20 | 1.7-kb PstI fragment carrying bscN in pBluescript SK II(+) | This work |
| pCM21 | 1.7-kb BamHI-EcoRV fragment carrying bscN subcloned from pCM20 into pUCBM20 | This work |
| pCM22 | pCM21 containing bscN inactivated with the cat cassette; Cmr Apr | This work |
| pCM23 | 1.7-kb PstI fragment encoding bscN subcloned from pCM20 into pMR4 | This work |
| pMT1 | 1.7-kb bscN-cat fragment in pCM22 cloned as an EcoRI fragment into pCMT2 | This work |
DNA manipulations.
DNA-modifying enzymes, including restriction endonucleases, T4 DNA polymerase, T4 DNA ligase, and T4 polynucleotide kinase, were obtained from Roche, New England Biolabs, and Invitrogen. Plasmid DNA was isolated by the boiling lysis method (48) or by using a QIAprep spin miniprep kit (Qiagen Inc.). Genomic DNA was extracted from B. cepacia by using a PureGene kit (Gentra). Southern blot and colony hybridizations were generally performed as described by Sambrook et al. (48) with Hybond N nitrocellulose membranes and probes labeled with [α-32P]dCTP (Amersham Pharmacia Biotech) by the random-primer method. Recombinant plasmids were introduced into E. coli and B. cepacia strain J2315 by electroporation with a Gene Pulser II (Bio-Rad).
DNA sequencing.
Nucleotide sequencing was performed at the Advanced Genetic Analysis Center at the University of Minnesota by using the dideoxy chain termination method and an ABI 1371A DNA sequencer (Applied Biosystems). The oligonucleotide primers used for sequencing were standard forward and reverse (T3 and T7) pBluescript primers or custom oligonucleotides synthesized at Integrated DNA Technologies. The nucleotide sequence was determined on both strands. Double-stranded sequences were aligned and assembled by using the EditSeq and SeqMan components of a demonstration version of the Lasergene sequence analysis software package (DNASTAR Inc.). Nucleotide and amino acid sequence searches and analyses were done with the BLASTX and BLASTP programs from the National Center for Biotechnology Information.
Identification of type III secretion genes in B. cepacia strain J2315.
Degenerate primers for regions conserved in the Yersinia enterocolitica YscN protein (59) and its homologs in other bacteria were used in PCRs with genomic DNA from B. cepacia strain J2315. The degenerate primers used were Ysc1 (5′-GARMGNGGNMGNGARGT-3′) and Ysc2 (5′-TRTGNCCRTCNARDAT-3′), where R is A or G; M is A or C; N is A, C, G, or T; and D is A, G, or T. After 35 cycles consisting of incubations at 95°C for 1 min, 40°C for 1 min, and 72°C for 1 min, the amplified products were gel purified, cloned into TA cloning vector pCRII (Invitrogen), and sequenced. One of the cloned PCR products (PCR1A) contained a partial open reading frame (ORF) that encoded a product with predicted amino acid sequence homology to YscN and its homologs. PCR1A was used to probe restriction digests of J2315 genomic DNA, and a cross-hybridizing 1.7-kb PstI fragment was identified and subcloned into pBluescript SK II(+), generating plasmid pCM20. The 1.7-kb fragment contains the entire bscN gene. To clone the regions flanking bscN, the 1.7-kb fragment in plasmid pCM21 was used to probe a cosmid library of B. cepacia strain J2315 (56). Subclones of cross-hybridizing cosmids were generated, and the DNA sequence of the B. cepacia type III secretion locus shown in Fig. 1 was determined on both strands.
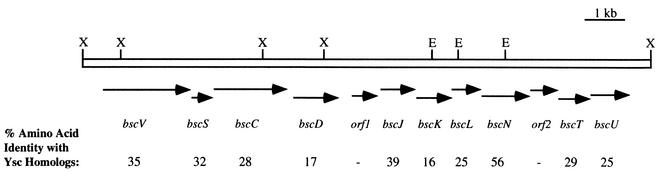
FIG. 1.
Type III secretion gene cluster of B. cepacia strain J2315. Arrows denote the orientations of ORFs corresponding to putative type III secretion genes in B. cepacia. The numbers below the ORFs indicate the percentages of amino acid identity of the products of the ORFs to corresponding homologs in Yersinia spp. The hyphens indicate that no significant homology to known type III secretion components was detected. Abbreviations: E, EcoRI; X, XhoI.
Generation of a bscN null mutant.
The construction of a bscN null mutant of B. cepacia strain J2315 is shown schematically in Fig. 2. The bscN gene was insertionally inactivated by allelic exchange mutagenesis with a chloramphenicol resistance cassette (cat) as a selectable marker. B. cepacia recombinants were selected on LB agar plates containing chloramphenicol (300 μg/ml) and kanamycin (50 μg/ml, to select against E. coli donor strain S17-1). Double-crossover events were confirmed by PCR and Southern analyses with bscN and cat as probes. The bscN null strain was designated CM56.
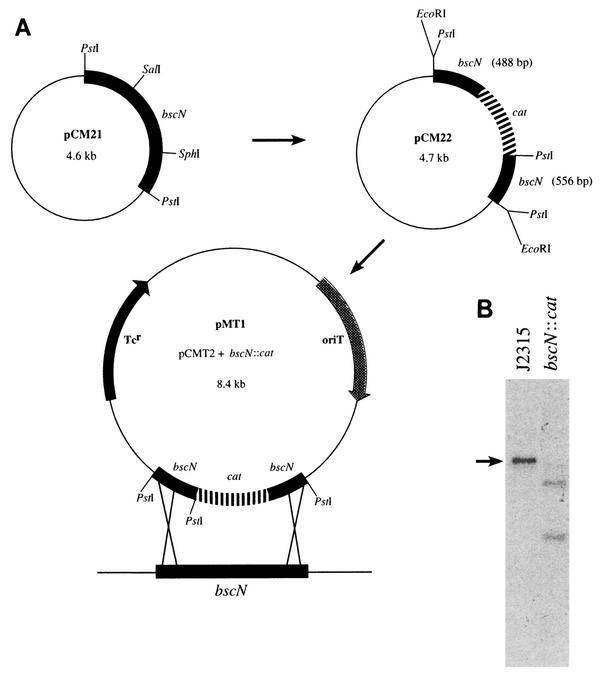
FIG. 2.
Construction of a bscN null mutant. (A) Strategy for the mutagenesis of bscN. Plasmid pCM21, carrying the full-length bscN gene, was digested with SalI-SphI and blunt ended with T4 polymerase, resulting in the removal of nucleotides 430 to 1213 of the bscN coding sequence. The deleted region was replaced with a chloramphenicol resistance cassette (cat) taken as a HincII fragment from pCAT1, generating plasmid pCM22. The 1.7-kb bscN::cat fragment in plasmid pCM22 was cloned as an EcoRI fragment into suicide vector pCMT2, generating plasmid pMT1. Plasmid pMT1 was delivered to B. cepacia by conjugation, and B. cepacia recombinants were selected as described in Materials and Methods. (B) Southern blot hybridization analysis of B. cepacia strain J2315 and the isogenic bscN mutant, confirming allelic exchange. Genomic DNAs were digested with PstI and hybridized with the 1.7-kb PstI fragment carrying bscN. The cat cassette contains a single PstI site. The arrow denotes the 1.7-kb PstI fragment carrying bscN in parent strain J2315.
Mouse agar bead model.
C57BL/6 mice were infected with B. cepacia by using the agar bead model essentially as described previously (5) but with the following modifications. Groups of mice were infected by intratracheal instillation of 109 bacteria embedded in agar beads. Mice infected with the bscN-complemented strain (CM62) were orally administered a low dose of tetracycline (100 μg/ml) in their drinking water in order to reduce plasmid loss during the course of the experiment. At 3 or 7 days postinfection, the animals were sacrificed; the lungs and spleens from three of the infected animals in each group were removed and homogenized in 0.5 ml of phosphate-buffered saline; and the number of B. cepacia CFU was determined by plating serial dilutions of the homogenates on LB agar. For histopathological examination, the lungs from two of the infected animals in each group were removed, inflated, and stored in 4% paraformaldehyde. Lung sections were embedded in paraffin, stained with hematoxylin and eosin, and examined by light microscopy. All animal experiments were carried out in accordance with National Institutes of Health guidelines and with protocols approved by the Animal Care and Use Committee at Children's Hospital and Regional Medical Center.
Quantitative invasion and adherence assays.
Invasion and adherence assays with cultured A549 respiratory epithelial cells were performed as described previously (56). All quantitative assays were performed by using triplicate wells with two samplings per well.
Statistical analysis.
The statistical significance of the observed differences in the mean log CFUs recovered from the animal infection experiments as well as the mean adherence and invasion frequencies in the A549 cell invasion and adherence assays was determined by calculating P values with the two-tailed Student t test for unpaired data sets.
Nucleotide sequence accession number.
The sequence of the B. cepacia type III secretion locus determined in this study has been deposited in GenBank under accession number AY166598.
RESULTS
Identification of a type III secretion system in B. cepacia strain J2315.
The ATP-binding proteins believed to provide energy for secretion are the central and most highly conserved components of type III systems. This family includes the YscN protein of Yersinia spp. and its homologs in other gram-negative bacterial pathogens (21). Homologs of YscN are also required for the export and assembly of the bacterial flagellum (31). In order to determine whether B. cepacia has a YscN homolog, degenerate primers based on conserved amino acid sequences found in the YscN protein and its homologs were generated and used in a PCR with genomic DNA from strain J2315, a CF clinical isolate of B. cepacia. Two products were amplified from a single reaction and designated PCR1A and PCR1B. Both products were cloned and sequenced. The translated amino acid sequence of PCR1A had the highest homology to the Yersinia sp. YscN protein and its homologs, while the translated amino acid sequence of the other PCR product (PCR1B) had the highest homology to S. enterica serovar Typhimurium flagellar protein FliI. FliI is the flagellar type III secretion homolog of YscN. The B. cepacia fliI gene, partially amplified to yield PCR1B, has been cloned and insertionally inactivated. The fliI null strain is nonmotile and blocked in flagellar biogenesis, confirming the role of fliI in flagellar export and assembly (56).
PCR1A, with translated amino acid sequence homology to YscN and its homologs, was used to probe restriction enzyme-digested chromosomal DNA of B. cepacia strain J2315. A 1.7-kb cross-hybridizing PstI fragment was identified and cloned. The PstI fragment carries a single ORF whose product has deduced amino acid sequence homology across its entire length to YscN (56% amino acid identity). A portion of the cloned region was identical in DNA sequence to the entire amplified product PCR1A, indicating that the corresponding gene had been cloned. We designated the gene bscN (Burkholderia secretion protein N), in keeping with the currently accepted nomenclature and the homology of its predicted product to known type III secretion ATP-binding proteins. The bscN gene encodes a predicted 52-kDa protein containing two conserved nucleotide-binding motifs, designated Walker boxes (58), suggesting that it can bind and hydrolyze ATP.
To further characterize the bscN locus, the 1.7-kb PstI fragment was used to probe a cosmid library of J2315 genomic DNA (56), and cross-hybridizing cosmids were identified. Subclones of these cosmids were generated and sequenced, revealing a locus containing multiple ORFs encoding products with predicted amino acid sequence homology to known type III secretion proteins. Homologs of the B. cepacia bscJ, bscT, and bscU genes shown in Fig. 1 encode both inner and outer membrane proteins involved in the formation of the channel through which type III effector proteins are secreted (39). Homologs of the bscK and bscL genes encode cytoplasmic components of type III secretion systems (39). The putative products of at least two ORFs (orf1 and orf2 in Fig. 1) within the B. cepacia type III secretion gene cluster have no significant homology to known type III secretion components, suggesting that they may encode functions specific to the type III secretion system in B. cepacia. No ORFs with homology to known type III secretion genes have been identified in the region immediately downstream of bscU (Fig. 1), suggesting that bscU may mark the end of the type III secretion gene cluster.
Generation of a bscN null strain.
To characterize further the role of type III secretion in B. cepacia, we generated a defined null mutation in bscN, encoding the putative energy-generating component of the secretory system. In order to insertionally inactivate bscN, an internal fragment of the cloned bscN gene (including both of the putative Walker boxes) was deleted and replaced with a chloramphenicol resistance cassette (Fig. 2A). The disrupted copy of bscN was delivered to B. cepacia strain J2315 via conjugation, and inactivation of the chromosomal copy of bscN was generated by allelic exchange. Southern blot hybridization analysis confirmed that the chromosomal copy of bscN had been disrupted via a double-crossover event (Fig. 2B). The bscN null strain was designated CM56.
We first tested whether the bscN disruption affected motility, since homologs of bscN are known to be involved in flagellar export and assembly. The bscN null strain was not defective in motility, as determined by a swarm agar plate assay (56) or by light microscopy, indicating that the bscN gene product is not involved in flagellar biogenesis in B. cepacia (data not shown). There was also no significant difference in the growth rates of the bscN null strain and the wild-type parent strain in either minimal (M9 broth) or rich (LB) medium.
Attenuated virulence of the bscN mutant.
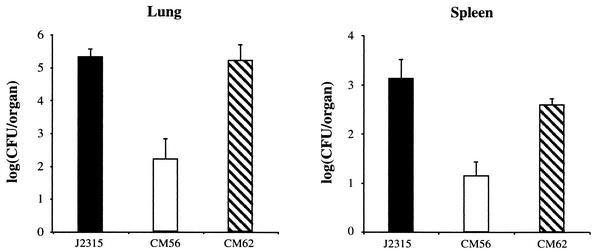
To examine the role of bscN in B. cepacia virulence, wild-type strain J2315 and type III secretion mutant strain CM56 were examined in a mouse agar bead model of infection. In the initial experiment with groups of three mice, a significant difference was seen between the number of CFU recovered from animals infected with wild-type strain J2315 and the number of CFU recovered from animals infected with strain CM56. An average of 3 × 105 CFU were recovered from the lungs of mice infected with wild-type B. cepacia, compared to an average of less than 5 × 102 CFU recovered from the lungs of mice infected with the bscN null strain (Fig. 3). A comparison of bacterial counts in the spleens of infected animals revealed a similar trend, with significantly fewer CFU recovered from mice infected with the type III secretion mutant strain CM56 than from mice infected with wild-type strain J2315 (Fig. 3).
FIG. 3.
Quantitative bacteriological analysis of lungs and spleens from C57BL/6 mice 72 h after infection with B. cepacia strain J2315 (wild type), CM56 (bscN::cat), or CM62 (CM56 complemented with bscN). Data shown are the means of log CFUs recovered from three animals. Error bars denote the standard errors of the mean. P values (determined by using the two-tailed Student t test for unpaired data sets) were 0.014 for lungs and 0.008 for spleens.
To demonstrate conclusively that the virulence defect of the bscN null strain was due to disruption of the type III secretion system and not to a polar effect of the bscN mutation, we complemented the bscN null strain by providing a wild-type copy of the bscN gene alone in trans on broad-host-range plasmid pMR4 (pCM23). This plasmid replicates at a low copy number in B. cepacia and encodes tetracycline resistance, allowing for its selection in the chloramphenicol-resistant background of CM56 (bscN::cat). Transcomplementation of the bscN mutant with pCM23 restored wild-type levels of CFU in the lungs and spleens of infected animals (Fig. 3), confirming that the virulence defect of strain CM56 was due to the disruption of bscN.
To confirm the findings of the mouse infection studies, we repeated the experiments by using groups of five animals and obtained similar results (data not shown). We also examined the lungs and spleens of infected animals for bacteria at 7 days postinfection. All of the mice infected with the bscN null strain had cleared the bacteria from both the lungs and the spleen. In contrast, none of the mice infected with the wild-type strain had completely cleared the bacteria from either the lungs or the spleen. Bacteria were also recoverable at 7 days from the lungs of all of the mice infected with the complemented strain, and only one mouse had cleared the complemented strain from the spleen. Recovery at 7 days postinfection, however, was variable and significantly lower than that at 72 h, suggesting that the bacteria were being cleared by the animals. Together, the data from the mouse infection studies support a role for type III secretion in the virulence of B. cepacia.
Histopathological analysis.
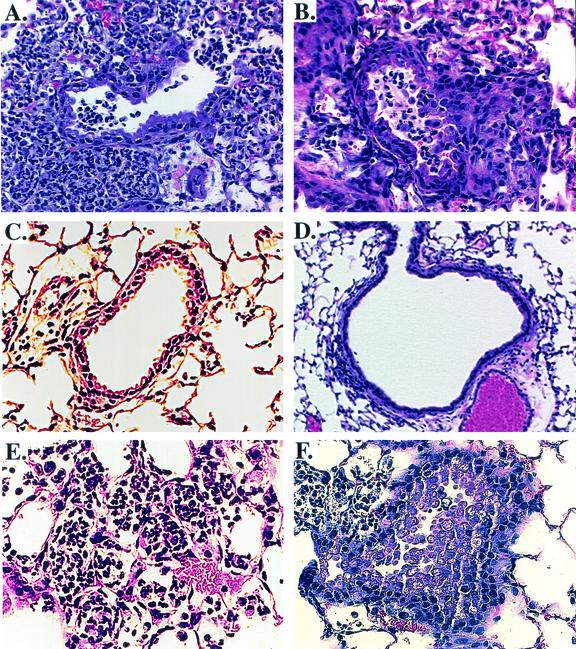
In order to determine the degree of pathologic changes and inflammatory responses associated with B. cepacia infection, lung sections from mice infected with either wild-type strain J2315 or the bscN null strain were examined for histopathological changes. The examination of lung sections from mice infected with wild-type strain J2315 revealed an extensive inflammatory cell infiltrate, comprised predominantly of macrophages and neutrophils (Fig. 4A and B). Other changes, including desquamated airway epithelium and interstitial inflammation, were also observed. In contrast, the pathologic changes in lung sections from mice infected with bscN mutant strain CM56 appeared less pronounced, with reduced parynchymal involvement and inflammatory cell infiltrate, as well as a more regular appearance of ciliated airway epithelial cells (Fig. 4C and D). The histopathological changes in lung sections from mice infected with complemented strain CM62 were similar to those in lung sections from mice infected with wild-type strain J2315 (Fig. 4E and F).
FIG. 4.
Comparison of lung histopathological findings in C57BL/6 mice following infection with B. cepacia. Shown are representative hematoxylin- and eosin-stained sections of mouse lung samples at 72 h after inoculation with B. cepacia strain J2315 (A and B), bscN null strain CM56 (C and D), or bscN-complemented strain CM62 (E and F). Sections A, B, E, and F show inflammatory cell infiltrate, airway blockage, and the appearance of irregular and desquamated airway epithelial cells. Sections C and D show minimal inflammatory cell infiltrate and airway blockage and a more regular appearance of ciliated airway epithelial cells. Magnification, ×150.
Examination of the bscN null strain with an in vitro invasion assay.
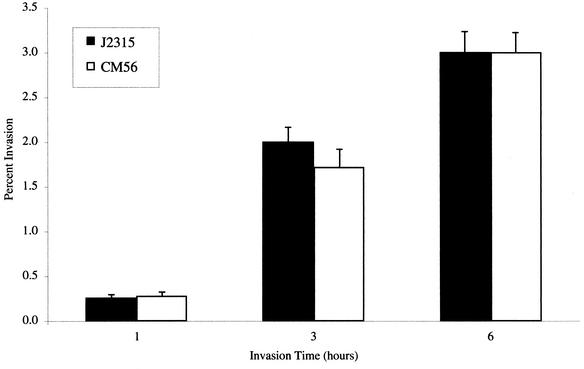
Several investigators have demonstrated the ability of B. cepacia to invade cultured human epithelial cells (3, 26, 33). The known role of type III secretion systems in the ability of other bacterial pathogens to invade host cells prompted us to examine whether the bscN null strain was defective in entry into cultured respiratory epithelial cells. Monolayers of human A549 respiratory epithelial cells were infected with either wild-type B. cepacia strain J2315 or bscN null strain CM56 at a multiplicity of infection of 10:1 for 1, 3, or 6 h, and the percentage of intracellular bacteria was determined at each time point. There was no significant difference between strain J2315 and the bscN null strain in their abilities to invade the cultured cell monolayers (Fig. 5). Quantitative adherence assays performed at 1 h postinfection also did not detect a significant difference between the wild-type strain and the type III secretion mutant strain (data not shown). Together, these results suggest that bscN is not required for adherence to or invasion of A549 respiratory epithelial cells under the conditions examined.
FIG. 5.
Invasion of A549 respiratory cell monolayers by B. cepacia strain J2315 (wild type) or mutant CM56 (bscN::cat). Invasion assays were performed as described previously (56). Values represent the percentages of the bacterial inoculum that survived 2 h of antibiotic treatment and are the means and standard deviations for six independent determinations from a total of three wells.
Distribution of type III secretion genes among epidemic strains of B. cepacia.
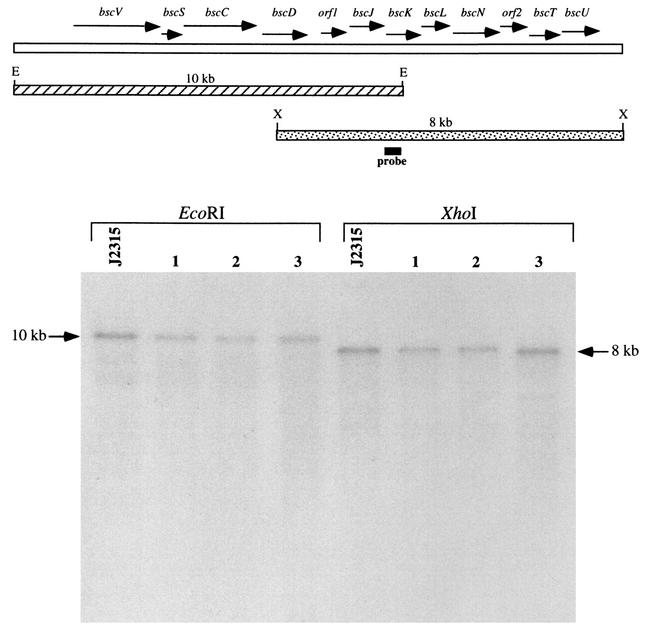
B. cepacia is a complex of bacteria constituting at least seven distinct genomic species, or genomovars (7, 8). While strains representing many of the known genomovars of the B. cepacia complex have been associated with opportunistic infections in humans, the majority of strains associated with morbidity and mortality belong to genomovar III. B. cepacia strain J2315 belongs to genomovar III and is a member of the virulent lineage known as electrophoretic type 12, which has been associated with epidemic spread among CF patients (25). In order to determine whether other strains of B. cepacia associated with epidemic spread also contain type III secretion genes, genomic DNA was isolated from three other genomovar III clinical isolates of B. cepacia and examined by Southern blot hybridization. In addition to strain J2315, which was isolated from a CF patient in the United Kingdom, CF isolates from Canada and the United States were examined. As a probe for these studies, a DNA fragment internal to the bscK gene (Fig. 1) was used, since homologs of bscK are not found in known flagellar type III secretion systems; this strategy thus avoided potential cross-hybridization with B. cepacia flagellar assembly components. DNA fragments corresponding to both the upstream and the downstream regions of the type III gene cluster hybridized in all three strains, and each of the three strains examined had a hybridization pattern identical to that of strain J2315 (Fig. 6). Identical hybridization patterns were also observed when probes specific to the bscV (also known as bcscV [38]) and bscN genes were used (data not shown). Together, these results indicate that each of the strains contains a type III secretion gene cluster.
FIG. 6.
Southern blot analysis of representative epidemic strains of B. cepacia genomovar III. Genomic DNA was digested with either EcoRI (E) or XhoI (X) and probed with a DNA fragment internal to the bscK gene. Lanes: J2315 (United Kingdom); 1, BC7 (Canada); 2, K56-2 (Canada); 3, PC184 (United States). Arrows denote the hybridizing 10-kb EcoRI and 8-kb XhoI bands.
DISCUSSION
Despite the known association of B. cepacia with chronic and sometimes fatal pulmonary infections in CF patients as well as other compromised patients, the specific virulence determinants utilized by B. cepacia during the course of infection are poorly understood. Given the known role of type III secretion in the pathogenesis of other gram-negative bacteria, we sought to determine whether B. cepacia may also contain type III secretion genes. We identified in a CF clinical isolate of B. cepacia a locus containing multiple type III secretion genes and provided evidence that the type III secretion locus is conserved in other clinical isolates of B. cepacia associated with epidemic spread. We insertionally inactivated one of these genes, designated bscN, encoding a putative ATP-binding protein required for protein export. Compared to the parent strain, the bscN null strain was attenuated in virulence in a murine model of infection, indicating that type III secretion plays an important role in the pathogenesis of B. cepacia.
At 3 days postinfection, significantly fewer bacteria were recovered from the lungs and spleens of mice infected with the bscN mutant strain than from those of mice infected with the parent strain. These findings suggest that bacteria lacking bscN are either less likely to establish infection or more likely to be cleared from infected organs. One hypothesis to explain the attenuated virulence of the bscN null strain is that type III secretion plays a role in modulation of the host immune response, allowing B. cepacia to evade host defense mechanisms and establish persistent infection. Histopathological changes in the lungs of mice infected with wild-type B. cepacia strain J2315 were characterized by an extensive inflammatory cell infiltrate, comprised predominantly of macrophages and neutrophils. Similar findings have been reported for B. cepacia in other animal models of infection (4, 45, 54). Using a mouse model of CF, Sajjan et al. (45) recently reported that B. cepacia infection was accompanied by infiltration of a mixed inflammatory cell population of macrophages and neutrophils into the lungs. Interestingly, however, the infiltrating immune cells appeared to be largely inactive, as determined by the reduced oxidant production and cell surface expression of the β2 integrin CD11b (45). It is conceivable that the inactivation of host macrophages and neutrophils through the secretion of type III effector molecules impairs the normal clearance of bacteria, resulting in the establishment of persistent B. cepacia infection. The significant reduction in the recovery of bacteria from mice infected with the type III secretion mutant may reflect improved clearance by fully activated immune cells in the lungs.
Invasion assays with cultured A549 respiratory epithelial cells did not reveal a significant difference between the wild-type and type III secretion mutant strains in their abilities to invade cultured respiratory epithelial cells. We cannot, however, completely discount a possible role for type III secretion in B. cepacia invasion of host tissues in vivo. It is possible that B. cepacia entry into cultured respiratory epithelial cells involves mechanisms distinct from those required for invasion in vivo or that cell culture conditions may not adequately mimic the lung environment, where additional host-specific signals may be required for full induction of the type III secretion system. An invasion defect due to disruption of the type III secretion system could explain the reduced extrapulmonary spread of B. cepacia to the spleens of mice infected with the bscN null strain. Primary cell cultures were recently used to investigate the invasion mechanisms of B. cepacia (42, 50) and may allow further dissection of the role of type III secretion in invasion under conditions more closely mimicking the in vivo environment.
Based on the known role of bscN homologs in the secretion of effector proteins by other bacterial pathogens, it is reasonable to suggest that the reduced virulence of the bscN null strain is due to a disruption in the delivery of B. cepacia virulence determinants into host cells. While the results of the present study clearly implicate type III secretion in the virulence of B. cepacia, the identity of the putative secreted effector(s) is unknown at this time. Unlike other components of the secretory apparatus, the effector proteins of bacterial type III secretion systems often share little or no homology, likely reflecting the diversity of their host cell targets (12). At least two ORFs that were identified within the type III secretion gene cluster appeared to be specific to B. cepacia, since they have no known homologs in other systems. These ORFs might encode effector proteins exported by the B. cepacia type III translocon. Alternatively, the genes encoding the type III effector proteins might not be linked to the genes encoding other components of the secretory apparatus, as has been reported for several type III effector genes in S. enterica serovar Typhimurium and B. bronchiseptica (35, 61).
We have compared the extracellular protein profiles of the B. cepacia wild-type and bscN mutant strains grown under several conditions known to stimulate type III secretion in other bacteria; these conditions include growth in minimal and tissue culture media, growth in the presence of Congo red, and growth under calcium limitation. Thus far, we have not detected differences between the two strains (data not shown). It is possible that the laboratory conditions tested do not adequately reproduce the environment in the animal model and human host. Similar difficulties have been encountered in the identification of substrates for the type III secretion systems of the respiratory pathogen Bordetella pertussis and the plant pathogen Xanthomonas campestris (15, 27). In the latter, identification of the effector proteins required specific inducing conditions as well as mutagenesis of a key regulatory gene, allowing for constitutive expression of the type III secretion genes (40). It is likely that similar approaches will be necessary to identify the substrate(s) for the B. cepacia type III secretion system.
We have identified a locus of B. cepacia type III secretion genes encoding predicted structural as well as energy-generating components of the secretory system. The entire arrangement of the B. cepacia type III secretion genes is not found in any other known bacterial type III secretion system, although the gene order bscJKLN is conserved in the type III secretion systems of the closely related respiratory pathogens B. bronchiseptica and B. pertussis (15, 60). Notably missing from the B. cepacia type III secretion gene cluster are genes encoding homologs of the Yersinia YscQ and YscR proteins. Members of these protein families are conserved in all known type III secretion systems, and their corresponding genes are typically found within clusters encoding homologs of the bsc genes described in this report. Parsons and coworkers (38) recently identified genes encoding predicted proteins with homology to the archetypal YscQ and YscR proteins in B. cepacia strain J2315. The corresponding genes, designated bcscQ and bcscR in reference 38, appear to be located outside of the gene cluster described in our study, suggesting a genetic organization distinct from those of all other known type III secretion systems. Although the B. cepacia bcscQ and bcscR genes have not been characterized, the possible presence of a YscQ homolog in B. cepacia is of particular interest, since SpaO and Spa33, the YscQ homologs in Salmonella spp. and Shigella spp., respectively, have been shown to be secreted via their respective type III secretion systems (10, 49).
It is worth noting that the B. cepacia type III secretion genes that we have identified are homologous not only to type III secretion components in other human pathogens but also to type III secretion components in several plant pathogens, including X. campestris and Ralstonia solanacearum. Recent studies have provided evidence that type III secretion genes may be widely distributed in the B. cepacia complex, including clinical as well as environmental isolates (38). Given the known role of B. cepacia as a phytopathogen (2), it is tempting to speculate that type III secretion can play a role in the establishment of commensal or pathogenic relationships between B. cepacia and plants and is also utilized during the course of infection in humans.
In summary, we have identified components of a type III secretion system in B. cepacia and have provided evidence that type III secretion is an important virulence determinant for this organism. Future studies will focus on further defining the role of B. cepacia type III secretion both in vitro and in vivo, including the identification of type III secretion molecules and characterization of their specific effects on host cells.
Acknowledgments
We thank Tim Leonard for technical assistance.
This work was supported by Minnesota Medical Foundation grant 13700, by grant MOHR0010 from the Cystic Fibrosis Foundation to C.D.M., and by NIH grant RO1 HL65898 to J.L.B.
Editor: V. J. DiRita
REFERENCES
- 1.Alfano, J. R., and A. Collmer. 1997. The type III (Hrp) secretion pathway of plant pathogenic bacteria: trafficking harpins, Avr proteins, and death. J. Bacteriol. 179:5655-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burkholder, W. H. 1950. Sour skin: a bacterial rot of onion bulbs. Phytopathology 40:115-117. [Google Scholar]
- 3.Burns, J. L., M. Jonas, E. Y. Chi, D. K. Clark, A. Berger, and A. Griffith. 1996. Invasion of respiratory epithelial cells by Burkholderia (Pseudomonas) cepacia. Infect. Immun. 64:4054-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiu, C.-H., A. Ostry, and D. P. Speert. 2001. Invasion of murine respiratory epithelial cells in vivo by Burkholderia cepacia. J. Med. Microbiol. 50:594-601. [DOI] [PubMed] [Google Scholar]
- 5.Cieri, M. V., N. Mayer-Hamblett, A. Griffith, and J. L. Burns. 2002. Correlation between an in vitro invasion assay and a murine model of Burkholderia cepacia lung infection. Infect. Immun. 70:1081-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cirillo, D. M., R. H. Valdivia, D. M. Monack, and S. Falkow. 1998. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol. Microbiol. 30:175-188. [DOI] [PubMed] [Google Scholar]
- 7.Coenye, T., J. J. LiPuma, D. Henry, B. Hoste, K. Vandemeulebroucke, M. Gillis, D. P. Speert, and P. Vandamme. 2001. Burkholderia cepacia genomovar VI, a new member of the Burkholderia cepacia complex isolated from cystic fibrosis patients. Int. J. Syst. E vol. Microbiol. 51:271-279. [DOI] [PubMed] [Google Scholar]
- 8.Coenye, T., P. Vandamme, J. R. W. Govan, and J. J. LiPuma. 2001. Taxonomy and identification of the Burkholderia cepacia complex. J. Clin. Microbiol. 39:3427-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collazo, C. M., and J. E. Galan. 1997. The invasion-associated type III system of Salmonella typhimurium directs the translocation of Sip proteins into the host cell. Mol. Microbiol. 24:747-756. [DOI] [PubMed] [Google Scholar]
- 10.Collazo, C. M., and J. E. Galan. 1996. Requirement for exported proteins in secretion through the invasion-associated type III system of Salmonella typhimurium. Infect. Immun. 64:3524-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cornelis, G., and H. Wolf-Watz. 1997. The Yersinia Yop virulon: a bacterial system for subverting eukaryotic cells. Mol. Microbiol. 23:861-867. [DOI] [PubMed] [Google Scholar]
- 12.Cornelis, G. R., and V. Van Gijsegem. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54:735-775. [DOI] [PubMed] [Google Scholar]
- 13.Darling, P., M. Chan, A. D. Cox, and P. A. Sokol. 1998. Siderophore production by cystic fibrosis isolates of Burkholderia cepacia. Infect. Immun. 66:874-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dobbin, C. J., R. Soni, T. Jelihovsky, and P. T. Bye. 2000. Cepacia syndrome occuring following prolonged colonisation with Burkholderia cepacia. Aust. N. Z. J. Med. 30:288-289. [DOI] [PubMed] [Google Scholar]
- 15.Fauconnier, A., A. Veithen, P. Gueirard, R. Antoine, L. Wacheul, C. Locht, A. Bollen, and E. Godfroid. 2001. Characterization of the type III secretion locus of Bordetella pertussis. Int. J. Med. Microbiol. 290:693-705. [DOI] [PubMed] [Google Scholar]
- 16.Fink-Barbancon, V., J. Goranson, L. Zhu, T. Sawa, J. P. Wiener-Kronish, M. J. Fleiszig, C. Wu, L. Mende-Mueller, and D. W. Frank. 1997. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol. Microbiol. 25:547-557. [DOI] [PubMed] [Google Scholar]
- 17.Govan, J. R., P. H. Brown, J. Maddison, C. J. Doherty, J. W. Nelson, M. Dodd, A. P. Greening, and A. K. Webb. 1993. Evidence for transmission of Pseudomonas cepacia by social contact in cystic fibrosis. Lancet 342:15-19. [DOI] [PubMed] [Google Scholar]
- 18.Govan, J. R. W., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hauser, A. R., P. J. Kang, and J. N. Engel. 1998. PepA, a secreted protein of Pseudomonas aeruginosa, is necessary for cytotoxicity and virulence. Mol. Microbiol. 27:807-818. [DOI] [PubMed] [Google Scholar]
- 20.Hensel, M., J. E. Shea, S. R. Waterman, R. Mundy, T. Nikolaus, G. Banks, A. Vazquez-Torres, C. Gleeson, F. C. Fang, and D. W. Holden. 1998. Genes encoding putative effector proteins of the typeIII secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol. Microbiol. 30:163-174. [DOI] [PubMed] [Google Scholar]
- 21.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hughes, J. E., J. Stewart, G. R. Barclay, and J. R. Govan. 1997. Priming of neutrophil respiratory burst activity by lipopolysaccharide from Burkholderia cepacia. Infect. Immun. 65:4281-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isles, A., I. Maclusky, M. Corey, R. Gold, C. Prober, P. Fleming, and H. Levison. 1984. Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. J. Pediatr. 104:206-210. [DOI] [PubMed] [Google Scholar]
- 24.Jarvis, K. G., J. A. Giron, A. E. Jerse, T. K. McDaniel, M. S. Donnenberg, and J. B. Kaper. 1995. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc. Natl. Acad. Sci. USA 92:7996-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson, W., S. Tyler, and K. Rozee. 1994. Linkage analysis of geographic and clinical clusters in Pseudomonas cepacia infections by multilocus enzyme electrophoresis and ribotyping. J. Clin. Microbiol. 32:924-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keig, P. M., E. Ingham, and K. G. Kerr. 2001. Invasion of human type II pneumocytes by Burkholderia cepacia. Microb. Pathog. 30:167-170. [DOI] [PubMed] [Google Scholar]
- 27.Kjemtrup, S., Z. Nimchuk, and J. L. Dangl. 2000. Effector proteins of phytopathogenic bacteria: bifunctional signals in virulence and host recognition. Curr. Opin. Microbiol. 3:73-78. [DOI] [PubMed] [Google Scholar]
- 28.Konstan, M. W., and M. Berger. 1993. Infection and inflammation of the lung in cystic fibrosis, p. 219-276. In P. B. Davis (ed.), Cystic fibrosis, vol. 64. Marcel Dekker, Inc., New York, N.Y. [Google Scholar]
- 29.LiPuma, J. J. 1998. Burkholderia cepacia. Management issues and new insights. Clin. Chest Med. 19:473-486. [DOI] [PubMed] [Google Scholar]
- 30.LiPuma, J. J., J. E. Mortensen, S. E. Dasen, T. D. Edlind, D. V. Schidlow, J. L. Burns, and T. L. Stull. 1988. Ribotype analysis of Pseudomonas cepacia from cystic fibrosis treatment centers. J. Pediatr. 113:859-862. [DOI] [PubMed] [Google Scholar]
- 31.Macnab, R. M. 1999. The bacterial flagellum: reversible rotary propellor and type III export apparatus. J. Bacteriol. 181:7149-7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahenthiralingam, E., P. Vandamme, M. E. Campbell, D. A. Henry, A. M. Gravelle, L. T. Wong, A. G. Davidson, P. G. Wilcox, B. Nakielna, and D. P. Speert. 2001. Infection with Burkholderia cepacia complex genomovars in patients with cystic fibrosis: virulent transmissible strains of genomovar III can replace Burkholderia multivorans. Clin. Infect. Dis. 33:1469-1475. [DOI] [PubMed] [Google Scholar]
- 33.Martin, D. W., and C. D. Mohr. 2000. Invasion and intracellular survival of Burkholderia cepacia. Infect. Immun. 68:24-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Menard, R., P. J. Sansonetti, and C. Parsot. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mirold, S., W. Rabsch, M. Rohde, S. Stender, H. Tschape, H. Russmann, E. Igwe, and W.-D. Hardt. 1999. Isolation of a temperate bacteriophage encoding the type III effector protein SopE from an epidemic Salmonella typhimurium strain. Proc. Natl. Acad. Sci. USA 96:9845-9850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mohr, C. D., M. Tomich, and C. A. Herfst. 2001. Cellular aspects of Burkholderia cepacia infection. Microbes Infect. 3:425-435. [DOI] [PubMed] [Google Scholar]
- 37.Palfreyman, R. W., M. L. Watson, C. Eden, and A. W. Smith. 1997. Induction of biologically active interleukin-8 from lung epithelial cells by Burkholderia (Pseudomonas) cepacia products. Infect. Immun. 65:617-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parsons, Y. N., K. J. Glendinning, V. Thornton, B. A. Hales, C. A. Hart, and C. Winstanley. 2001. A putative type III secretion gene cluster is widely distributed in the Burkholderia cepacia complex but absent from genomovar I. FEMS Microbiol. Lett. 203:103-108. [DOI] [PubMed] [Google Scholar]
- 39.Plano, G. V., J. B. Day, and F. Ferracci. 2001. Type III export: new uses for an old pathway. Mol. Microbiol. 40:284-293. [DOI] [PubMed] [Google Scholar]
- 40.Rossier, O., K. Wengelnik, K. Hahn, and U. Bonas. 1999. The Xanthomonas Hrp type III system secretes proteins from plant and mammalian bacterial pathogens. Proc. Natl. Acad. Sci. USA 96:9368-9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saini, L. S., S. B. Galsworthy, M. A. John, and M. A. Valvano. 1999. Intracellular survival of Burkholderia cepacia complex isolates in the presence of macrophage cell activation. Microbiology 145:3465-3475. [DOI] [PubMed] [Google Scholar]
- 42.Sajjan, U., C. Ackerley, and J. Forstner. 2002. Interaction of cblA/adhesin-positive Burkholderia cepacia with squamous epithelium. Cell. Microbiol. 4:73-86. [DOI] [PubMed] [Google Scholar]
- 43.Sajjan, U., M. Corey, A. Humar, E. Tullis, E. Cutz, C. Ackerley, and J. Forstner. 2001. Immunolocalisation of Burkholderia cepacia in the lungs of cystic fibrosis patients. J. Med. Microbiol. 50:535-546. [DOI] [PubMed] [Google Scholar]
- 44.Sajjan, U. S., L. Sun, R. Goldstein, and J. F. Forstner. 1995. Cable (Cbl) type II pili of cystic fibrosis-associated Burkholderia (Pseudomonas) cepacia: nucleotide sequence of the cblA major subunit pilin gene and novel morphology of the assembled appendage fibers. J. Bacteriol. 177:1030-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sajjan, U., G. Thanassoulis, V. Cherapanov, A. Lu, C. Sjolin, B. Steer, Y. J. Wu, O. D. Rotstein, G. Kent, C. McKerlie, J. Forstner, and G. P. Downey. 2001. Enhanced susceptibility to pulmonary infection with Burkholderia cepacia in Cftr−/− mice. Infect. Immun. 69:5138-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sajjan, U., Y. Wu, G. Kent, and J. Forstner. 2000. Preferential adherence of cable-piliated Burkholderia cepacia to respiratory epithelia of CF knockout mice and human cystic fibrosis lung explants. J. Med. Microbiol. 49:875-885. [DOI] [PubMed] [Google Scholar]
- 47.Sajjan, U. S., M. Corey, M. A. Karmali, and J. F. Forstner. 1992. Binding of Pseudomonas cepacia to normal human intestinal mucin and respiratory mucin from patients with cystic fibrosis. J. Clin. Investig. 89:648-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 49.Schuch, R., and A. T. Maurelli. 2001. Spa33, a cell surface-associated subunit of the Mxi-Spa type III secretory pathway of Shigella flexneri, regulates Ipa protein traffic. Infect. Immun. 69:2180-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwab, U., M. Leigh, C. Ribeiro, J. Yankaskas, K. Burns, P. Gilligan, P. Sokol, and R. Boucher. 2002. Patterns of epithelial cell invasion by different species of the Burkholderia cepacia complex in well-differentiated human airway epithelia. Infect. Immun. 70:4547-4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shaw, D., I. R. Poxton, and J. R. W. Govan. 1995. Biological activity of Burkholderia (Pseudomonas) cepacia lipopolysaccharide. FEMS Immunol. Med. Microbiol. 11:99-106. [DOI] [PubMed] [Google Scholar]
- 52.Shimomura, H., M. Matsuura, S. Saito, Y. Hirai, Y. Isshiki, and K. Kawahara. 2001. Lipopolysaccharide of Burkholderia cepacia and its unique character to stimulate murine macrophages with relative lack of interleukin-1β-inducing ability. Infect. Immun. 69:3663-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-790. [Google Scholar]
- 54.Sokol, P. A., P. Darling, D. E. Woods, E. Mahenthiralingam, and C. Kooi. 1999. Role of ornibactin biosynthesis in the virulence of Burkholderia cepacia: characterization of pvdA, the gene encoding l-ornithine N(5)-oxygenase. Infect. Immun. 67:4443-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tablan, O. C., W. J. Martone, C. F. Doershuk, R. C. Stern, M. J. Thomassen, J. D. Klinger, J. W. White, L. A. Carson, and W. R. Jarvis. 1987. Colonization of the respiratory tract with Pseudomonas cepacia in cystic fibrosis: risk factors and outcomes. Chest 91:527-532. [DOI] [PubMed] [Google Scholar]
- 56.Tomich, M., C. A. Herfst, J. W. Golden, and C. D. Mohr. 2002. Role of flagella in host cell invasion by Burkholderia cepacia. Infect. Immun. 70:1799-1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vandamme, P., B. Holmes, M. Vancanneyt, T. Coenye, B. Hoste, R. Coopman, H. Revets, S. Lauwers, M. Gillis, K. Kersters, and J. R. W. Govan. 1997. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int. J. Syst. Bacteriol. 47:1188-1200. [DOI] [PubMed] [Google Scholar]
- 58.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Woestyn, S., A. Allaoui, P. Wattiau, and G. R. Cornelis. 1994. YscN, the putative energizer of the Yersinia Yop secretion machinery. J. Bacteriol. 176:1561-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yuk, M. H., E. T. Harvill, and J. F. Miller. 1998. The BvgAS virulence control system regulates type III secretion in Bordetella bronchiseptica. Mol. Microbiol. 28:945-959. [DOI] [PubMed] [Google Scholar]
- 61.Yuk, M. H., E. T. Harvill, P. A. Cotter, and J. F. Miller. 2000. Modulation of host immune responses, induction of apoptosis and inhibition of NF-κB activation by the Bordetella type III secretion system. Mol. Microbiol. 35:991-1004. [DOI] [PubMed] [Google Scholar]