Abstract
Plasmodium falciparum apical membrane antigen 1 (AMA1) is a prime malaria vaccine candidate. Antigenic diversity within parasite populations is one of the main factors potentially limiting the efficacy of any asexual-stage vaccine, including one based on AMA1. The DNA coding for the most variable region of this antigen, domain I, was sequenced in 168 samples from the Wosera region of Papua New Guinea, including samples from symptomatic and asymptomatic infections. Neutrality tests applied to these sequences provided strong evidence of selective pressure operating on the sequence of ama1 domain I, consistent with AMA1 being a target of protective immunity. Similarly, a peculiar pattern of geographical diversity and the particular substitutions found were suggestive of strong constraints acting on the evolution of AMA1 at the population level, probably as a result of immune pressure. In addition, a strong imbalance between symptomatic and asymptomatic infections was detected in the frequency of particular residues at certain polymorphic positions, pointing to AMA1 as being one of the determinants of the morbidity associated with a particular strain. The information yielded by this study has implications for the design and assessment of AMA1-based vaccines and provides additional data supporting the importance of AMA1 as a malaria vaccine candidate.
Malaria is a major public health problem, resulting in approximately 300 to 500 million clinical cases and an estimated 1 to 3 million deaths each year (43). Development of a vaccine against Plasmodium falciparum, the parasite responsible for the most severe form of malaria, is an urgent priority, particularly because resistance to most traditionally used drugs is widespread. Identification of targets of naturally acquired protective immunity is an essential component of vaccine development.
The apical membrane antigen 1 (AMA1) is an 83-kDa type I integral membrane protein with an ectodomain organized in three domains stabilized by eight disulfide bonds (26). AMA1 is synthesized late during the development of parasite schizonts. Initially located in the merozoite apical organelles, it is processed to a 66-kDa form that relocates to the surfaces of mature merozoites (36, 38). The function of AMA1 is not well understood, but its stage specificity and location suggest that this protein is involved in the process of invasion of host red blood cells (RBCs). Such a role is also indicated by the finding that antibodies directed against AMA1 inhibit invasion of RBCs in vitro (25, 30). In good agreement, expression of P. chaubadi AMA1 in P. falciparum leads to more-efficient invasion of murine RBCs by the human parasite (48). Furthermore, the failure of “knockout” technology to generate parasites with the ama1 gene disrupted suggests that AMA1 plays a critical, perhaps essential role for asexual blood-stage growth (48).
Much evidence points to AMA1 as a leading candidate for inclusion in a vaccine directed against P. falciparum asexual blood stages. First, antibodies against AMA1 have been detected in populations exposed to malaria (46), and these antibodies have been shown to inhibit merozoite invasion in vitro (25); thus, they can be expected to be protective. Second, active immunization of mice and monkeys with AMA1 confers protection against rodent and simian malaria parasites, respectively (1, 6, 10, 11, 35).
Additional strong support for the importance of AMA1 as a vaccine candidate comes from the characteristics of its sequence. In contrast to most other blood-stage antigens, AMA1 is relatively well conserved among various Plasmodium species, consistent again with an essential role for this protein. AMA1 lacks the sequence repeats and size polymorphisms found in other malaria vaccine candidates. In P. falciparum and P. vivax, the limited diversity of this antigen results from point mutations that are concentrated in certain regions of the protein, most of them within domain I (14, 17, 29, 31, 37, 39, 47), and it has been suggested that these mutations are the consequence of diversifying selection by a protective immune response (14, 15, 39, 49).
Antigenic diversity is one of the main mechanisms used by malaria parasites to evade the host immune system, and it is believed to be one of the major difficulties confronting the development of an effective malaria vaccine. In a malaria vaccine trial conducted recently in Papua New Guinea, the 3D7-MSP2 component of the vaccine produced a shift in the parasites infecting vaccinated children toward parasites expressing MSP2 alleles from the alternative allelic family, FC27 (21). In mice, immunization with a recombinant protein containing the two epidermal growth factor (EGF)-like domains of MSP1 or with the AMA1 ectodomain conferred protection against homologous but not heterologous sporozoite challenge (10, 40), suggesting that protective immunity against these proteins has a high degree of strain specificity. Thus diversity in AMA1, though limited, could be a major obstacle to the efficacy of this antigen as a component of a malaria vaccine.
Here, the P. falciparum ama1 domain I from 168 infected blood samples was sequenced, and evidence for diversifying selection acting on this domain has been confirmed. In addition, differences have been detected between the sequences from symptomatic and asymptomatic infections, pointing to AMA1 as one of the determinants of the morbidity associated with a particular P. falciparum strain. The existence of other sets of published ama1 sequences from other locations allowed us to also study the geographical patterns of diversity of this gene.
MATERIALS AND METHODS
Study area.
The Wosera region (Eastern Sepik Province, Papua New Guinea) is an area of year-round high transmission of P. falciparum malaria, and P. vivax and P. malariae are also common there (19, 20). Transmission occurs mainly through members of the Anopheles punctulatus complex, and the number of P. falciparum infective bites per year is around 100 (23, 24).
Blood samples.
Finger-prick blood samples for this study were collected from 720 residents of the Wosera (Table 1). To obtain parasites from individuals with asymptomatic malaria infections, 628 samples were collected in two cross-sectional surveys from all individuals willing to take part in the study. In the first survey (June 2000), 367 samples were collected from three villages (Kitikum 1, Kitikum 2, and Numbunge) in North Wosera. In the second survey (November 2000), 261 samples were collected from three villages (Palgere, Wegior, and Patigo) in South Wosera. The longest distance between any two of the surveyed villages within North or South Wosera is approximately 3 km, and the longest distance between one of the villages in the North and one in the South is approximately 7 km.
TABLE 1.
Demographic description of samples
| Sample set | No. of AMA1 sequences obtained | No. of sequenced samples (% of total in sample set) in the following age group:
|
Mean age (95% confidence interval) in the following age groupa:
|
||
|---|---|---|---|---|---|
| <10 yr | ≥10 yr | <10 yr | ≥10 yr | ||
| North Wosera survey (n = 367) | 76 | 38 (50) | 38 (50) | 4.71 (3.96-5.46) | 30.37 (24.13-36.60) |
| South Wosera survey (n = 261) | 42 | 18 (42.9) | 24 (57.1) | 4.44 (3.31-5.58) | 22.37 (17.81-26.94) |
| Total asymptomatic infections (North + South) (n = 628) | 118 | 56 (47.5) | 62 (52.5) | 4.62 (4.02-5.23) | 27.27 (23.04-31.51) |
| Symptomatic infections (n = 66) | 44 | 28 (63.6) | 16 (36.4) | 5.57 (4.62-6.52) | 19.5 (12.37-26.63) |
| Others (n = 26) | 6 | 6 (100) | 0 | 3.5 (1.22-5.78) | |
| Total (n = 720) | 168 | 90 (53.6) | 78 (46.4) | 4.84 (4.35-5.34) | 25.68 (22.01-29.35) |
Results only for individuals from which an AMA1 sequence was obtained.
Samples from 66 symptomatic (clinical) infections were collected from individuals attending the Kunjingini (South Wosera) and Kaugia (North Wosera) health centers during the routine morbidity surveillance conducted as part of the malaria vaccine development program of the Papua New Guinea Institute of Medical Research. Samples were collected from individuals with a clinical diagnosis of malaria and a high-density P. falciparum infection (>3,000 parasites/μl for infants <1 year old and adults >19 years old, or >5,000 parasites/μl for individuals aged 1 to 19 years [20]). These symptomatic-malaria samples were collected at the health centers from patients who were residents either of the villages where the cross-sectional surveys were performed or of neighboring villages, between 1 month before and 1 month after the survey.
A small number of samples (“Others” in Table 1) collected from patients attending the health centers did not satisfy the criteria for symptomatic malaria. The information from these samples was not used in any comparison between symptomatic and asymptomatic infections.
Detailed epidemiological information for the full set of samples will be presented elsewhere (A. Cortés, M. Mellombo, A. Benet, and J. C. Reeder, unpublished data). Demographic data for the samples from which ama1 domain I sequences were obtained are presented in Table 1. The mean ages of symptomatic and asymptomatic individuals did not differ significantly in any age group (P > 0.05), but in individuals aged ≥10 years, the mean age was considerably higher in the asymptomatic group.
Ethical clearance for this study was obtained from the Papua New Guinea Medical Research Advisory Committee. All study participants or their parents or guardians gave informed consent.
MSP2 genotyping, ama1 nested PCR, and ama1 sequencing.
Isocode dipsticks for DNA isolation (Schleicher & Schuell) were prepared for all samples. In order to determine the multiplicity of infection, MSP2 genotyping was performed on all 720 samples as previously described (16) but using only HinfI and DdeI digestion. MSP2 genotyping results will be presented elsewhere (Cortés et al., unpublished).
Nested PCR was performed on all samples containing single P. falciparum infections (as determined by MSP2 genotyping) to amplify the region between nucleotides 325 and 955 of the ama1 coding sequence (31). The primers used were AMEX5 (5′-GAACCCGCACCACAAGAAC-3′) and AMEX3 (5′-TTGTTTAGGTTGATCCGAAGC3′) for the primary PCR and AMINT5 (5′-CCATGGACGGAATATATGGC-3′) and AMINT3 (5′-TTCCATCGACCCATAATCCG-3′) for the nested PCR. Both PCRs were carried out for 30 cycles with an annealing temperature of 55°C in a 50-μl volume with 1.5 mM MgCl2, 1.25 U of purified Taq polymerase (Gibco BRL), and standard concentrations of other reagents.
Positive ama1 PCR products were directly sequenced after purification in a 1.2% agarose gel by using a GFX PCR DNA and Gel Band Purification kit (Amersham Pharmacia Biotech). Automatic sequencing was carried out at the Australian Genome Research Facility in Brisbane by using the Big Dye Terminator system (Applied Biosystems) and an Applied Biosystems AB377 automatic DNA sequencer. All PCR products were sequenced by using primer AMINT3, and in all cases where uncertainties were detected, the complementary strand was sequenced by using primer AMINT5. Although direct sequencing of PCR products avoids sequence errors observed when an intermediate cloning step is used, PCR (starting from genomic DNA) and sequencing were repeated for all sequences containing mutations occurring in only one sample.
Sequences were obtained from direct sequencing of 163 ama1 PCR products. In addition, five ama1 nested PCR products from individuals 0 to 3 years old containing multiple P. falciparum infections were cloned in Escherichia coli and sequenced, to compensate for the low number of samples with single infections available from this age group (data not shown). Cloning was performed using the pGEM-T Easy vector (Promega), and four independent clones were sequenced for each of the five samples. Surprisingly, for each of the five samples a single ama1 sequence was obtained for all four clones, and these sequences were analyzed together with those from single infections.
Sequence analysis, population genetics, and statistical analysis.
Sequences were edited and aligned by using the BioEdit program (http://jwbrown.mbio.ncsu.edu/BioEdit/bioedit.html) (22). Pairs of sequences with the same haplotype were identified by using Arlequin software (version 2.0; Genetics and Biometry Laboratory, University of Geneva [http://anthropologie.unige.ch/arlequin/]) and were counted by using a text processor. Fixation indexes (Fst) were also determined by using Arlequin software. Other intrapopulation and interpopulation diversity indexes, as well as recombination estimations and neutrality tests, were calculated by using DnaSP software (version 3.53 [http://www.bio.ub.es/∼julio/DnaSP.html]) (42). Complex codons that were ignored by the program for the McDonald-Kreitman test were analyzed by eye according to the guidelines given in DnaSP 3.53. Phylogenetic trees were constructed by using BioEdit and MEGA software (version 1.02; The Pennsylvania State University [http://evolgen.biol.metro-u.ac.jp/MEGA]).
Stata (version 6.0 [http://www.stata.com]) was used for statistical analysis. The two-tailed Fisher exact test was used for all comparisons, and Bonferroni's correction for multiple testing was applied when appropriate.
The ama1 sequences from Nigeria that were used in many comparisons were obtained from blood samples from malaria patients attending a health center in Ibadam, southwest Nigeria (39).
Nucleotide sequence accession number.
Nucleotide sequences described in this report have been submitted to EMBL under accession no. AJ490528 to AJ490695.
RESULTS
Nucleotide and amino acid sequences.
The sequence for nucleotides 345 to 921 of the ama1 gene was determined for samples from 168 P. falciparum infections from the Wosera region of Papua New Guinea. This DNA codes for amino acids 116 to 307, which includes all the AMA1 domain I sequence (amino acids 149 to 302 [26]). Thirty-seven polymorphic nucleotide positions were found in this segment of the gene, and all of them fell within the domain I coding sequence. Consequently, only domain I was considered for further analysis. The polymorphic nucleotides generated a total of 27 haplotypes among the 168 sequences (Fig. 1 left). None of the haplotypes can be considered predominant, with frequencies ranging from 0.6 to 9.5%.
FIG. 1.
Nucleotide and amino acid polymorphisms in 168 AMA1 domain I sequences. Only polymorphic nucleotides (left) and amino acids (right) are shown. The number of occurrences of each particular haplotype is shown in parentheses after its designation on the left. Nucleotide or amino acid position numbers (31) are given above each polymorphic site. Asterisks above position numbers indicate trimorphic nucleotide positions; nucleotide positions that are part of the same codon are bracketed. The number of different amino acids that can be found at a position, when greater than two, is given in parentheses after the residue number. C1 through C6 indicate the positions of the six Cys residues. Sequences identical to those found in common laboratory strains are indicated on the left as follows: &, identical to 3D7 (only at the protein level, not at the DNA level, because of a synonymous substitution); #, identical to KF1916; $, identical to FCR-3. The three nonsynonymous mutations not reported previously are indicated by boldfaced underlined letters. The T590 and T730 mutations, which had been reported to occur only in the Papua New Guinea Highlands and nearby Western Papua, Indonesia, respectively, are indicated by italicized underlined letters. Numbers of conservative, semiconservative, and radical substitutions that can occur at each position, assigned according to the work of Tangri and collaborators (45), are given at the bottom. Since we cannot know the directionality of the mutations found, the average of the degree of similarity of each substitution in both directions was used to assign it to one of these categories.
Three nonsynonymous mutations not reported previously were identified (Fig. 1). Two of the new polymorphisms, G517 and T586, were found in only one sample each, and C672 was found in four samples. G517 was the only novel mutation occurring at a position that had not been previously described as polymorphic, but it produces an amino acid substitution at a site where substitutions had been described previously. The two other mutations, T586 and C672, extended the previously described dimorphisms in these positions to trimorphisms. The mutation C672 produces the same amino acid substitution (Met224→Ile224) as a different mutation found in Nigeria, T672 (39).
Two other mutations, T590 and T730 (Fig. 1), had been reported to occur only in the Papua New Guinea Highlands (34) (EMBL accession no. AJ487082 to AJ487086) or nearby Western Papua, Indonesia (12), respectively. T730 occurred in 13 of the Wosera samples, all of which had the same haplotype, WOS18, as the sample from Western Papua that contains this mutation. Similarly, the T590 mutation occurred in one sample from the Wosera, which had the same haplotype (WOS19) as the sequence from the Papua New Guinea Highlands that contains this mutation.
Translation of the 27 allelic domain I DNA sequences generated 27 different amino acid haplotypes containing a total of 29 polymorphic residues (Fig. 1 right). Of the 29 polymorphic positions, 11 accommodated at least one radical amino acid substitution and 24 accommodated at least one radical or one semiconservative substitution (Fig. 1 right).
Intrapopulation diversity analysis reveals lower haplotypic diversity in Papua New Guinea than in Nigeria.
Only the 51 ama1 domain I sequences from Nigeria (39) were used for most of the subsequent comparisons, because this is the only other large set of sequences from a single population without sequence uncertainties. These sequences from Nigeria were obtained from malaria patients attending hospitals, and consequently they are comparable to our 44 Wosera symptomatic-infection sequences.
The finding of 27 haplotypes out of 168 sequences (14 out of 44 sequences from symptomatic infections) indicates less diversity in the Wosera ama1 domain I sequences than in the Nigeria data set, where 35 domain I haplotypes were found in 51 sequences (39) (Table 2). In contrast, other measures of diversity were remarkably similar for the two populations. The average pairwise nucleotide variability per site (π) in the Wosera samples was 0.02605 (average number of pairwise nucleotide differences, 12.04), compared to 0.02679 in the Nigerian samples (average number of pairwise nucleotide differences, 12.38) (Table 2). The π values for sequences from symptomatic and asymptomatic Wosera infections were also very similar (Table 2). Furthermore, the numbers of segregating sites and mutations in the Wosera and the Nigeria samples were also similar (Table 2). Very similar values were also obtained for π and for the numbers of segregating sites and mutations in smaller sets of ama1 domain I sequences sampled from other locations (14).
TABLE 2.
Intrapopulation diversity indexes for AMA1 domain I
| Sample set | Indexa value
|
|||
|---|---|---|---|---|
| N | H | M (S) | π | |
| North Wosera | 76 | 23 | 43 (36) | 0.02568 |
| South Wosera | 42 | 19 | 40 (36) | 0.02579 |
| Total asymptomatic infections (North + South) | 118 | 26 | 44 (37) | 0.02573 |
| Wosera symptomatic infections | 44 | 14 | 37 (34) | 0.02634 |
| Total Wosera | 168 | 27 | 44 (37) | 0.02605 |
| Nigeriab | 51 | 35 | 42 (38) | 0.02679 |
N, number of sequences analyzed; H, number of haplotypes; M, number of mutations; S, number of segregating (polymorphic) sites; π, average pairwise nucleotide diversity per site.
Published sequences from Nigeria (39).
Interpopulation diversity analysis reveals that ama1 domain I sequences from different geographical locations have similar mutations and low Fst values but different haplotype repertoires.
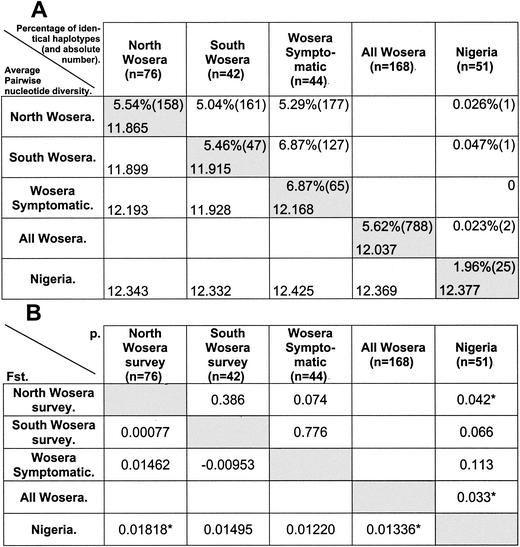
The percentage of sample pairs with the same haplotype (out of all the possible sample pairs) was drastically lower for comparisons between the Nigeria and Wosera samples than within each population (0.023% versus 1.96 and 5.62%, respectively [Fig. 2A ]). This result indicates that the repertoires of ama1 domain I haplotypes circulating in the two populations were very different, with only a slight overlap. On the other hand, the percentage of same-haplotype pairs was very similar for comparisons between North and South Wosera sequences and within each population (Fig. 2A), providing an appropriate control for the comparison method used.
FIG. 2.
Geographical differences in ama1 domain I sequences. (A) (Upper right section [upper values in shaded cells]) Percentage (and absolute number in brackets) of sample pairs with the same haplotype (self-pairs not considered). The percentage of pairs with the same haplotype was calculated as the absolute number divided by the number of possible sample pairs, which was calculated as the product of the number of samples in the two populations for interpopulation comparisons, and as ∑1 → n−1n (where n is the number of samples in the population) for intrapopulation comparisons. (Lower left section [lower values in shaded cells]) Average number of pairwise nucleotide differences. Shaded and unshaded cells contain, respectively, intrapopulation and interpopulation values. (B) (Lower left section) Pairwise Fst estimates determined from pairwise nucleotide diversity. (Upper right section) Respective P values. Asterisks indicate significant differences (P < 0.05). “Nigeria” refers to published sequences from Nigeria (39).
Despite the different repertoires of haplotypes found in the Wosera and Nigeria, most of the mutations were shared between the two populations. Of the 44 mutations detected in the Wosera region and the 42 detected in Nigeria, 37 were common to both populations while only 7 were specific to Wosera and 5 were specific to Nigeria. All mutations occurring in only one of the two populations had a very low prevalence (data not shown). Similarly, the vast majority of the mutations found in smaller sets of sequences from other locations were the same (12, 14, 29, 31, 37).
The close similarity between Wosera and Nigeria sequences was also indicated by the finding that average pairwise nucleotide diversity levels between and within populations were almost identical, again in sharp contrast to the differences in the haplotypes found. The interpopulation average number of pairwise nucleotide differences, π(xy), for the Wosera and Nigeria sequences was 12.37, very similar to the average pairwise diversity observed within each population (Fig. 2A). Consequently, the Fst value, which gives an estimation of the part of the diversity that can be attributed to differences between populations and the part that can be accounted for by diversity within populations, was very low, though significant (Fig. 2B), for the Wosera and Nigeria sequences (0.0133). Thus, at the level of nucleotide pairwise diversity, most of the diversity between ama1 alleles occurs within a population, and only a small part of it (around 1.3%) can be attributed to geographical differences.
No evidence of clustering of sequences from the same geographical origin was observed on phylogenetic trees constructed with Wosera and Nigeria sequences (data not shown) by using either distance methods (neighbor joining and unweighted pair group method with arithmetic means) or discrete character methods (maximum parsimony and maximum likelihood).
Neutrality tests provide evidence of natural selection acting on AMA1 sequences.
Of the 44 mutations occurring at 37 segregating sites in the Wosera domain I sequences, only one was a synonymous mutation (Table 3). In contrast, 4 of the 12 fixed differences between the Wosera sequences and the P. reichenowi AMA1 domain sequence (29) were synonymous. This difference was highly significant (P < 0.01) by the McDonald-Kreitman test (32) and indicates selection operating on the ama1 domain I sequence.
TABLE 3.
Neutrality tests on AMA1 domain I
| Sample set | Tajima's Db | Fu and Li's Dc,d | Fu and Li's Fc,d | McDonald-Kreitman testc,d
|
||
|---|---|---|---|---|---|---|
| Mutations within species synonymous, replacement | Fixed mutations between species synonymous, replacement | P (Fisher's exact) | ||||
| Asymptomatic infections (n = 118) | 1.373 | 1.180 | 1.523* | 1, 43 | 4, 8 | 0.0059*** |
| Symptomatic infections (n = 44) | 1.484 | 2.084*** | 2.255*** | 1, 36 | 4, 9 | 0.0131*** |
| Total (n = 168) | 1.670 | 1.122 | 1.643* | 1, 43 | 4, 8 | 0.0059*** |
| Nigeriaa (n = 51) | 1.110 | 1.694** | 1.780** | 3, 39 | 4, 8 | 0.0362** |
Published sequences from Nigeria (39).
Tajima's test was performed by considering all mutations and not only segregating sites.
P. reichenowi AMA1 domain I was used as an outgroup.
*, nonsignificant, 0.10 > P > 0.05; **, 0.05 > P > 0.02; ***, 0.02 > P > 0.005.
Analysis of the Wosera sequences by Tajima's D test of neutrality (44) gave positive values (Table 3), which might suggest departure from neutral evolution, but these values did not reach significance. This test is known to be conservative when applied to sequences subject to recombination events (44). Positive values of Tajima's D test result from the differential effect of natural selection on the number of segregating sites and the average number of nucleotide differences.
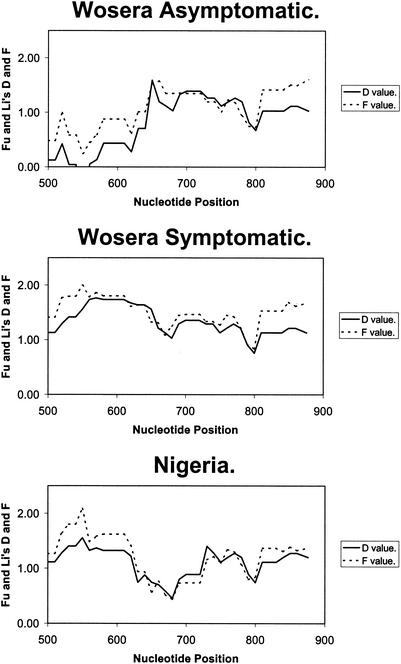
Stronger evidence of natural selection was obtained from analyses of the Wosera sequences with Fu and Li's D and F tests (18) by using the sequence of P. reichenowi ama1 (29) as an outgroup. Neither test reached significance when applied to sequences from asymptomatic infections, but both gave highly significant values for sequences from symptomatic infections (Table 3). When Fu and Li's tests were applied across domain I by using the sliding-window method, the highest values in symptomatic infections were found in the region between nucleotides 500 and 680 (middle points, 550 to 630), which codes for a region where the most polymorphic amino acid positions are clustered, whereas in asymptomatic infections this region had the lowest values (Fig. 3). The profile of the sliding-window plot for the symptomatic infections from Nigeria was found to be similar to that for the Wosera clinical infections (Fig. 3). Positive values of Fu and Li's D and F tests result from a deficit of mutations in external branches (identified as mutations occurring in only one haplotype and not present in the outgroup) and indicate departure from neutrality with the presence of unusually ancient alleles that are probably maintained by balancing selection (18). This test is also known to be conservative when applied to sequences subject to recombination events (18).
FIG. 3.
Sliding-window analysis of Fu and Li's D and F tests across AMA1 domain I by using a window size of 100 nucleotides and a step size of 10 nucleotides. The analysis was performed by using DnaSP 3.53 software (42). “Nigeria” refers to published sequences from Nigeria (39).
Particular residues at certain clustered polymorphic positions are associated with morbidity.
At some sites of amino acid substitutions there were strong imbalances in the frequency of particular residues between symptomatic and asymptomatic infections (Table 4). The frequencies were compared independently in two different age groups (<10 years and ≥10 years). The ≥10-year group roughly corresponds to individuals in whom naturally acquired immunity against clinical malaria is well developed, whereas in the <10-year group protective immunity against a broad range of antigenic variants is still developing.
TABLE 4.
Differences between symptomatic and asymptomatic infections and between age groups at certain polymorphic positions
| Position (amino acid) | Age group
|
Pa (asymptomatic infections of the <10-yr vs the ≥10-yr group) | |||||
|---|---|---|---|---|---|---|---|
| <10 yr
|
≥10 yr
|
||||||
| No. of sequences with the indicated amino acid in:
|
Pa | No. of sequences with the indicated amino acid in:
|
Pa | ||||
| Asymptomatic infections (n = 56) | Symptomatic infections (n = 28) | Asymptomatic infections (n = 62) | Symptomatic infections (n = 16) | ||||
| 187 | |||||||
| E | 8 | 17 | <0.001*** | 26 | 5 | 0.383 | 0.003* |
| K | 20 | 4 | 17 | 3 | |||
| N | 28 | 7 | 19 | 8 | |||
| 196 | |||||||
| D | 35 | 25 | 0.024 | 50 | 11 | 0.321 | 0.048 |
| N | 20 | 3 | 12 | 5 | |||
| Y | 1 | 0 | 0 | 0 | |||
| 197 | |||||||
| D | 6 | 11 | 0.006 | 20 | 5 | 0.838 | 0.024 |
| E | 2 | 3 | 5 | 0 | |||
| G | 19 | 3 | 12 | 5 | |||
| H | 2 | 0 | 2 | 0 | |||
| Q | 26 | 11 | 22 | 6 | |||
| R | 0 | 0 | 1 | 0 | |||
| V | 1 | 0 | 0 | 0 | |||
| 230 | |||||||
| E | 22 | 6 | 0.022 | 15 | 5 | 0.314 | 0.004 |
| K | 34 | 19 | 38 | 11 | |||
| Q | 0 | 3 | 9 | 0 | |||
| 242 | |||||||
| D | 39 | 9 | 0.002* | 34 | 8 | 0.784 | 0.129 |
| Y | 17 | 19 | 28 | 8 | |||
| 243 | |||||||
| E | 2 | 8 | <0.001** | 13 | 4 | 0.924 | 0.009 |
| K | 47 | 13 | 39 | 10 | |||
| N | 7 | 7 | 10 | 2 | |||
| 244 | |||||||
| D | 55 | 25 | 0.034 | 50 | 16 | 0.276 | 0.002* |
| N | 1 | 0 | 3 | 0 | |||
| Y | 0 | 3 | 9 | 0 | |||
| 245 | |||||||
| K | 54 | 22 | 0.015 | 55 | 16 | 0.334 | 0.168 |
| N | 2 | 6 | 7 | 0 | |||
| 283 | |||||||
| L | 18 | 5 | 0.202 | 12 | 9 | 0.009 | 0.140 |
| S | 38 | 23 | 50 | 7 | |||
| 285 | |||||||
| E | 10 | 3 | 0.529 | 8 | 7 | 0.010 | 0.609 |
| Q | 46 | 25 | 54 | 9 | |||
| 296 | |||||||
| D | 41 | 20 | 1.000 | 55 | 9 | 0.006 | 0.036 |
| H | 15 | 8 | 7 | 7 | |||
| 300 | |||||||
| E | 22 | 3 | 0.010 | 12 | 6 | 0.181 | 0.025 |
| K | 34 | 25 | 50 | 10 | |||
Fisher's exact test value (before correction). P values of <0.05 before correction for multiple testing are boldfaced. *, 0.10 > P > 0.05 after Bonferroni's correction for 29 tests; **, 0.05 > P > 0.01 after Bonferroni's correction; ***, P < 0.01 after Bonferroni's correction.
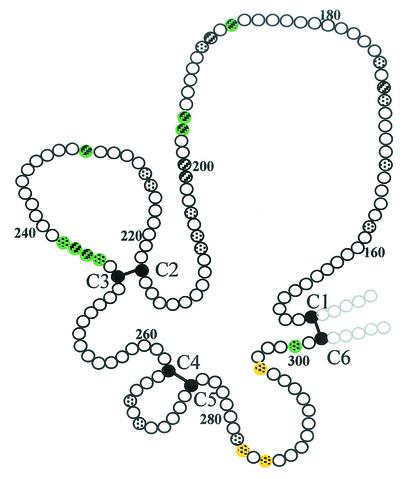
Large differences in the frequency of residues in any age group were found at 12 of the 29 polymorphic positions (Table 4). After Bonferroni's correction for multiple testing (29 comparisons), the difference remained significant in the younger age group at two of the positions (P < 0.01 at position 187; P < 0.05 at position 243) and almost reached significance at position 242 (0.1 > P > 0.05). Most of the residues where strong imbalances were observed in the younger age group are clustered in two different regions of domain I, whereas the positions at which imbalances were observed only in the older age group are clustered in another region (Fig. 4).
FIG. 4.
Schematic diagram of AMA1 domain I indicating the positions where strong imbalances in the frequencies of particular amino acids between symptomatic and asymptomatic infections were observed. Positions at which imbalances were observed in the <10-year and the ≥10-year age groups are colored green and yellow, respectively. Spotted circles, dimorphic positions; striped circles, positions with more than two possible residues. Solid circles, cysteine residues; solid bars, disulfide bonds. This figure was adapted and reprinted from reference 26 with permission of the publisher.
The frequency at which particular substitutions occurred was compared in asymptomatic infection sequences between the two age groups. Imbalances occurred at most of the positions where large differences between symptomatic and asymptomatic infections were observed (Table 4), consistent with an age-dependent selection against AMA1 variants containing certain residues. However, these differences did not reach significance after correction for multiple testing.
The most significant imbalance was found at position 187. There were three possible residues at this position: Asn, Lys, and Glu. In asymptomatic infections, Glu was the most common residue at this position (42%) in individuals 10 years old or older but was rather infrequent (14%) in the younger age group. The imbalance between age groups approached significance after correction for multiple testing (0.1 > P > 0.05). In sharp contrast, in individuals below the age of 10 years with symptomatic infections, Glu was the residue most frequently found at this position (61%; P < 0.01 after correction). This result suggests that for young children, parasites containing an AMA1 form with Glu187 are associated with high malaria morbidity but that this is not the case for older children or adults. A similar situation occurred at position 243, where, interestingly, the same three residues as in position 187 were found, and again the presence of Glu was associated with higher morbidity only in the younger age group (P < 0.05 after correction). At both positions 187 and 243, the presence of Glu was also associated with a higher geometric mean of parasitemia in individuals below the age of 10 years with asymptomatic infections (the number of parasites per microliter was 3,742 for Glu187, 1,233 for Asn187, and 1,514 for Lys187; it was 5,370 for Glu243, 2,303 for Asn243, and 1,389 for Lys243), but the wide range and large variability of parasitemias observed prevented the difference from reaching statistical significance.
DISCUSSION
This study of AMA1 diversity in a large number of symptomatic and asymptomatic P. falciparum infections from the Wosera region of Papua New Guinea provides a detailed analysis of the diversity of this malaria vaccine candidate in a single population. In order to analyze a large number of samples, we have restricted our analysis to domain I of AMA1, which is the most polymorphic region of this antigen (14, 31, 39) and appears to be a major target of anti-AMA1 protective antibodies (25). Multiple lines of evidence are presented indicating that polymorphisms in AMA1 domain I are the result of selective pressure exerted by protective immune responses. The inference that polymorphisms are located within the epitopic targets of protective immunity is fully consistent with previous reports of strain specificity of anti-AMA1 protective antibodies (10, 25, 30).
The results of the population genetics analyses on the Wosera samples reinforce the existing evidence for natural selection acting on the sequence of AMA1 domain I, which had been indicated by others from analyses of smaller numbers of samples (14, 15, 39, 49). The significance of the large excess of nonsynonymous versus synonymous substitutions in ama1 domain I was tested with the McDonald-Kreitman test, which would not be affected by the biased codon usage in P. falciparum because of the use of P. reichenowi as an outgroup (15). The highly significant values obtained with this test (P < 0.01) provide clear evidence of natural selection. The other neutrality tests used, Tajima's and Fu and Li's tests, indicated departure from neutrality, but most of the results were not significant. However, these tests would have underestimated the significance of a departure from neutrality because recombination in AMA1 was not taken into account (18, 44); it was difficult to obtain an accurate estimation of the recombination parameter because of the scarcity of synonymous substitution sites. Recombination in the ama1 gene is apparent from the decay of linkage disequilibrium with distance (39; data not shown for the sequences presented here), and using even a relatively low value for the recombination parameter would have resulted in both tests giving a significant result (39).
In contrast to the results with Tajima's test, Fu and Li's tests gave highly significant evidence of balancing selection acting on Wosera ama1 sequences from symptomatic but not asymptomatic infections, even without taking recombination into consideration. The difference between the two groups of sequences was most marked in the region between nucleotides 500 and 680. The relevance of this result, which indicates a greater deficit of mutations in external branches of the genealogy in this region of the sequence among parasites producing clinical disease, remains unclear. Nevertheless, it can be speculated that parasites carrying recent mutations in this region of AMA1 have compromised growth and as a consequence are less likely to produce clinical malaria.
Intrapopulation diversity indexes revealed greater haplotypic diversity for P. falciparum ama1 in Nigeria than in Papua New Guinea. This may reflect the higher transmission intensity in Africa, associated previously with increased diversity (2, 3, 27), but may also reflect the longer evolutionary time of P. falciparum in Africa (9) and the relative isolation of Papua New Guinea from other major malarious areas. More interesting results came from interpopulation diversity comparisons, which revealed that the ama1 domain I haplotype repertoires in Wosera and Nigeria overlap only slightly. The finding that the vast majority of the haplotypes are unique to one or the other of the populations suggests limited gene flow between distant populations, in agreement with previous reports (2, 9). In contrast to the haplotypes, most of the mutations were shared between Wosera and Nigeria sequences, and the average number of pairwise nucleotide differences was almost identical within and between the two populations. Consequently, the Fst value between the two populations was very low, consistent with other reports of low Fst values in Plasmodium genes under selective pressure (7, 8), and in contrast with the high Fst values observed in sequences experiencing neutral evolution (2, 9). It is noteworthy that no difference was observed between interpopulation and intrapopulation parameters for the populations of North and South Wosera, indicating that there was no heterogeneity in the parasite populations at this small regional level.
This peculiar geographical structure of diversity is suggestive of strong constraints acting on the evolution of ama1 at the population level. The striking similarity observed in the pairwise nucleotide comparison between distant populations with very different sets of haplotypes is likely to reflect the maintenance of similar balanced frequencies of the individual polymorphisms by the selection pressure of a protective immune response. The similar mutations and pairwise nucleotide differences between the two populations are clear imprints of natural selection. However, the lack of conservation at the haplotype level indicates that selection operates at the level of the individual polymorphic position (or small cluster of polymorphisms), and not at the level of the full domain.
The conservation of the observed mutations suggests that these particular substitutions are essential for immune evasion at the population level. If other mutations could play the same role in disrupting epitopic targets of effective immunity, different mutations would presumably be observed between populations that have evolved separately (as the different haplotype repertoires indicate). As many of the substitutions are radical, it is unlikely that the mutations observed would be the only mutations allowed by functional constraints.
Some of the amino acid replacements found in AMA1 domain I support this view. The Met→Ile replacement at codon 224 has been found in both Africa and Papua New Guinea but resulting from different nucleotide substitutions, indicating convergent evolution between the populations. Also, a relatively rare residue, Tyr, occurs at 5 of the 29 polymorphic positions and is always found at positions where an alternative residue is Asp. Tyr-Asp is a radical substitution and may be common in AMA1 because it can completely disrupt recognition of putative epitopes. Substitutions that involve change from a positive to a negative charge or vice versa, which have a drastic effect on recognition by antibodies, also occurred in a remarkably high number of polymorphic positions (nine) in AMA1 domain I.
Other mutations provide additional insights into the evolution of the ama1 gene. The T730 mutation found in the Wosera has been reported previously only in nearby Western Papua (12), and all the samples from both locations that had this mutation had the same ama1 domain I haplotype. These data suggest that this mutation has arisen recently, and although it has had time to spread among the Papua New Guinea and Western Papua populations to relatively high frequencies, apparently there has not been time for recombination or point mutations at other positions to generate new alleles bearing this mutation. A parallel situation occurs with the T590 mutation, which has also been detected in the Papua New Guinea Highlands (34).
Also consistent with AMA1 being a target of protective immunity was the finding of different AMA1 sequences associated with symptomatic and asymptomatic infections. It is not clear why certain P. falciparum infections cause clinical malaria while others remain asymptomatic, but there is increasing evidence that parasite factors are involved (33). One parasite factor appears to be the particular var gene expressed, because forms of P. falciparum EMP1 (the var gene product) that are poorly recognized by the infected individuals' immune systems are more likely to be associated with symptomatic infections (5). Although more controversial, there have also been reports that particular allelic families of the merozoite antigens MSP1 and MSP2 are associated with increased morbidity (4, 13, 28, 41). Here we report an association between clinical outcome and particular amino acid substitutions rather than particular alleles of AMA1.
A strong imbalance between symptomatic and asymptomatic infections was found in the distribution of residues at several clustered polymorphic positions. The most significant imbalances were observed in the younger age group. Residues Glu187 and Glu243 were strongly associated with clinical disease in children below the age of 10 years, but not in older individuals, and a parallel situation was observed at several other clustered positions. A possible explanation for these imbalances in children is that certain residues decrease the immunogenicity of AMA1, and consequently, longer exposure is necessary in order to develop an effective immune response against these forms of AMA1. In agreement with this explanation, a strong age dependence was observed in asymptomatic individuals in the distribution of residues at most of the positions at which imbalances between symptomatic and asymptomatic infections occurred. This finding suggests that there is an age-dependent selection against parasites with certain residues at particular AMA1 positions, consistent with age-dependent strain-specific immunity against these positions being effective in controlling parasite growth. The higher morbidity associated with the presence of certain residues in AMA1 could also be explained by improved parasite fitness, but the age dependence of the observed imbalances is not easily explained by changes in fitness, and the weight of the evidence indicates that these polymorphisms are selected by protective immune responses.
At this stage, we cannot determine whether the effect of certain residues on morbidity is a direct effect of these particular residues or an effect of their combination with other AMA1 residues with which they are linked. On the other hand, it is highly unlikely that the effect on morbidity is due to linkage with particular alleles at other loci, because the residues associated with clinical malaria appear in multiple AMA1 domain I haplotypes and thus cannot correspond to a few clonally transmitted parasites. Furthermore, linkage disequilibrium between different loci is usually low in areas with high malaria transmission (2).
The data presented here provide an extensive characterization of the diversity of the AMA1 domain I in the Wosera region of Papua New Guinea, where this antigen may be tested in phase I/IIb vaccine trials. These data provide a necessary baseline that will assist in interpretation of the outcome of such an intervention. Our data suggest that AMA1 domain I is a major target of naturally acquired protective immunity, in agreement with previous reports (25, 39), which favors its primacy as a malaria vaccine candidate and its inclusion in a subunit vaccine. On the other hand, our data strongly reinforce previous evidence (10, 25, 30, 39) that at least some of the epitopes for protective immunity in AMA1 are strain specific, which would hamper the efficacy of an AMA1-based vaccine containing only one form of the protein. Knowledge of the sequence diversity will contribute to the rational development of an AMA1-based vaccine intended to confer protection against a broad spectrum of challenging parasites, which is the final goal of any malaria vaccine. One possible strategy to achieve this aim would be to include a large number of forms of the protein, but such a vaccine would be difficult and costly to manufacture. A vaccine containing multiple allelic variants of polymorphic fragments of AMA1 that are considered likely targets of protective immunity would be less likely to be effective because important conformational epitopes may be lacking. Because of these considerations, it is likely that the first field testing of AMA1 will involve a vaccine containing one or, at the most, two allelic forms of the AMA1 ectodomain. However, detailed genotyping to determine which of the polymorphisms allow evasion of vaccine-induced protective immune responses, together with the knowledge of the sequences found in the population, might make possible the development of a future AMA1-based vaccine that, with a limited number of antigenic variants, would induce an immune response effective against most AMA1 variants. It will also be important to assess further the association between morbidity and AMA1 genotype, particularly in the context of interventions with vaccines containing AMA1.
Acknowledgments
This work was supported by the Australian Agency for International Development and the Australian Cooperative Research Centre for Vaccine Technology.
We are especially thankful to the Wosera communities for donating their blood for the malaria vaccine development program in which they are also involved. We are grateful to I. Betuela, L. Rare, and all the PNGIMR Maprik staff for their contributions to the field work, to L. Tavul for assistance in the lab, and to B. Genton for useful discussions.
Editor: J. M. Mansfield
REFERENCES
- 1.Anders, R. F., P. E. Crewther, S. Edwards, M. Margetts, M. L. Matthew, B. Pollock, and D. Pye. 1998. Immunisation with recombinant AMA-1 protects mice against infection with Plasmodium chabaudi. Vaccine 16:240-247. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, T. J., B. Haubold, J. T. Williams, J. G. Estrada-Franco, L. Richardson, R. Mollinedo, M. Bockarie, J. Mokili, S. Mharakurwa, N. French, J. Whitworth, I. D. Velez, A. H. Brockman, F. Nosten, M. U. Ferreira, and K. P. Day. 2000. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol. Biol. Evol. 17:1467-1482. [DOI] [PubMed] [Google Scholar]
- 3.Ariey, F., W. Chalvet, D. Hommel, C. Peneau, A. Hulin, O. Mercereau-Puijalon, J. B. Duchemin, J. L. Sarthou, J. M. Reynes, and T. Fandeur. 1999. Plasmodium falciparum parasites in French Guiana: limited genetic diversity and high selfing rate. Am. J. Trop. Med. Hyg. 61:978-985. [DOI] [PubMed] [Google Scholar]
- 4.Ariey, F., D. Hommel, C. Le Scanf, J. B. Duchemin, C. Peneau, A. Hulin, J. L. Sarthou, J. M. Reynes, T. Fandeur, and O. Mercereau-Puijalon. 2001. Association of severe malaria with a specific Plasmodium falciparum genotype in French Guiana. J. Infect. Dis. 184:237-241. [DOI] [PubMed] [Google Scholar]
- 5.Bull, P. C., B. S. Lowe, M. Kortok, C. S. Molyneux, C. I. Newbold, and K. Marsh. 1998. Parasite antigens on the infected red cell surface are targets for naturally acquired immunity to malaria. Nat. Med. 4:358-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins, W. E., D. Pye, P. E. Crewther, K. L. Vandenberg, G. G. Galland, A. J. Sulzer, D. J. Kemp, S. J. Edwards, R. L. Coppel, J. S. Sullivan, C. L. Morris, and R. F. Anders. 1994. Protective immunity induced in squirrel monkeys with recombinant apical membrane antigen-1 of Plasmodium fragile. Am. J. Trop. Med. Hyg. 51:711-719. [DOI] [PubMed] [Google Scholar]
- 7.Conway, D. J. 1997. Natural selection on polymorphic malaria antigens and the search for a vaccine. Parasitol. Today 13:26-29. [DOI] [PubMed] [Google Scholar]
- 8.Conway, D. J., D. R. Cavanagh, K. Tanabe, C. Roper, Z. S. Mikes, N. Sakihama, K. A. Bojang, A. M. Oduola, P. G. Kremsner, D. E. Arnot, B. M. Greenwood, and J. S. McBride. 2000. A principal target of human immunity to malaria identified by molecular population genetic and immunological analyses. Nat. Med. 6:689-692. [DOI] [PubMed] [Google Scholar]
- 9.Conway, D. J., C. Fanello, J. M. Lloyd, B. M. Al-Joubori, A. H. Baloch, S. D. Somanath, C. Roper, A. M. Oduola, B. Mulder, M. M. Povoa, B. Singh, and A. W. Thomas. 2000. Origin of Plasmodium falciparum malaria is traced by mitochondrial DNA. Mol. Biochem. Parasitol. 111:163-171. [DOI] [PubMed] [Google Scholar]
- 10.Crewther, P. E., M. L. Matthew, R. H. Flegg, and R. F. Anders. 1996. Protective immune responses to apical membrane antigen 1 of Plasmodium chabaudi involve recognition of strain-specific epitopes. Infect. Immun. 64:3310-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deans, J. A., A. M. Knight, W. C. Jean, A. P. Waters, S. Cohen, and G. H. Mitchell. 1988. Vaccination trials in rhesus monkeys with a minor, invariant, Plasmodium knowlesi 66 kD merozoite antigen. Parasite Immunol. 10:535-552. [DOI] [PubMed] [Google Scholar]
- 12.Eisen, D. P., A. Saul, D. J. Fryauff, J. C. Reeder, and R. L. Coppel. 2002. Alternations in Plasmodium falciparum genotypes during sequential infections suggest the presence of strain-specific immunity. Am. J. Trop. Med. Hyg. 67:8-16. [DOI] [PubMed] [Google Scholar]
- 13.Engelbrecht, F., I. Felger, B. Genton, M. Alpers, and H. P. Beck. 1995. Plasmodium falciparum: malaria morbidity is associated with specific merozoite surface antigen 2 genotypes. Exp. Parasitol. 81:90-96. [DOI] [PubMed] [Google Scholar]
- 14.Escalante, A. A., H. M. Grebert, S. C. Chaiyaroj, M. Magris, S. Biswas, B. L. Nahlen, and A. A. Lal. 2001. Polymorphism in the gene encoding the apical membrane antigen-1 (AMA-1) of Plasmodium falciparum. X. Asembo Bay Cohort Project. Mol. Biochem. Parasitol. 113:279-287. [DOI] [PubMed] [Google Scholar]
- 15.Escalante, A. A., A. A. Lal, and F. J. Ayala. 1998. Genetic polymorphism and natural selection in the malaria parasite Plasmodium falciparum. Genetics 149:189-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felger, I., L. Tavul, and H. P. Beck. 1993. Plasmodium falciparum: a rapid technique for genotyping the merozoite surface protein 2. Exp. Parasitol. 77:372-375. [DOI] [PubMed] [Google Scholar]
- 17.Figtree, M., C. J. Pasay, R. Slade, Q. Cheng, N. Cloonan, J. Walker, and A. Saul. 2000. Plasmodium vivax synonymous substitution frequencies, evolution and population structure deduced from diversity in AMA 1 and MSP 1 genes. Mol. Biochem. Parasitol. 108:53-66. [DOI] [PubMed] [Google Scholar]
- 18.Fu, Y. X., and W. H. Li. 1993. Statistical tests of neutrality of mutations. Genetics 133:693-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Genton, B., F. al-Yaman, H. P. Beck, J. Hii, S. Mellor, A. Narara, N. Gibson, T. Smith, and M. P. Alpers. 1995. The epidemiology of malaria in the Wosera area, East Sepik Province, Papua New Guinea, in preparation for vaccine trials. I. Malariometric indices and immunity. Ann. Trop. Med. Parasitol. 89:359-376. [DOI] [PubMed] [Google Scholar]
- 20.Genton, B., F. al-Yaman, H. P. Beck, J. Hii, S. Mellor, L. Rare, M. Ginny, T. Smith, and M. P. Alpers. 1995. The epidemiology of malaria in the Wosera area, East Sepik Province, Papua New Guinea, in preparation for vaccine trials. II. Mortality and morbidity. Ann. Trop. Med. Parasitol. 89:377-390. [DOI] [PubMed] [Google Scholar]
- 21.Genton, B., I. Betuela, I. Felger, F. Al-Yaman, R. F. Anders, A. Saul, L. Rare, M. Baisor, K. Lorry, G. V. Brown, D. Pye, D. O. Irving, T. A. Smith, H. P. Beck, and M. P. Alpers. 2002. A recombinant blood-stage malaria vaccine reduces Plasmodium falciparum density and exerts selective pressure on parasite populations in a phase 1-2b trial in Papua New Guinea. J. Infect. Dis. 185:820-827. [DOI] [PubMed] [Google Scholar]
- 22.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 23.Hii, J. L., T. Smith, A. Mai, E. Ibam, and M. P. Alpers. 2000. Comparison between anopheline mosquitoes (Diptera: Culicidae) caught using different methods in a malaria endemic area of Papua New Guinea. Bull. Entomol. Res. 90:211-219. [DOI] [PubMed] [Google Scholar]
- 24.Hii, J. L., T. Smith, P. Vounatsou, N. Alexander, A. Mai, E. Ibam, and M. P. Alpers. 2001. Area effects of bednet use in a malaria-endemic area in Papua New Guinea. Trans. R. Soc. Trop. Med. Hyg. 95:7-13. [DOI] [PubMed] [Google Scholar]
- 25.Hodder, A. N., P. E. Crewther, and R. F. Anders. 2001. Specificity of the protective antibody response to apical membrane antigen 1. Infect. Immun. 69:3286-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hodder, A. N., P. E. Crewther, M. L. Matthew, G. E. Reid, R. L. Moritz, R. J. Simpson, and R. F. Anders. 1996. The disulfide bond structure of Plasmodium apical membrane antigen-1. J. Biol. Chem. 271:29446-29452. [DOI] [PubMed] [Google Scholar]
- 27.Hoffmann, E. H., L. A. da Silveira, R. Tonhosolo, F. J. Pereira, W. L. Ribeiro, A. P. Tonon, F. Kawamoto, and M. U. Ferreira. 2001. Geographical patterns of allelic diversity in the Plasmodium falciparum malaria-vaccine candidate, merozoite surface protein-2. Ann. Trop. Med. Parasitol. 95:117-132. [DOI] [PubMed] [Google Scholar]
- 28.Jelinek, T., A. H. Kilian, A. Westermeier, S. Proll, G. Kabagambe, H. D. Nothdurft, F. von Sonnenburg, and T. Loscher. 1999. Population structure of recrudescent Plasmodium falciparum isolates from western Uganda. Trop. Med. Int. Health 4:476-480. [DOI] [PubMed] [Google Scholar]
- 29.Kocken, C. H., D. L. Narum, A. Massougbodji, B. Ayivi, M. A. Dubbeld, A. van der Wel, D. J. Conway, A. Sanni, and A. W. Thomas. 2000. Molecular characterisation of Plasmodium reichenowi apical membrane antigen-1 (AMA-1), comparison with P. falciparum AMA-1, and antibody-mediated inhibition of red cell invasion. Mol. Biochem. Parasitol. 109:147-156. [DOI] [PubMed] [Google Scholar]
- 30.Kocken, C. H., C. Withers-Martinez, M. A. Dubbeld, A. van der Wel, F. Hackett, A. Valderrama, M. J. Blackman, and A. W. Thomas. 2002. High-level expression of the malaria blood-stage vaccine candidate Plasmodium falciparum apical membrane antigen 1 and induction of antibodies that inhibit erythrocyte invasion. Infect. Immun. 70:4471-4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marshall, V. M., L. Zhang, R. F. Anders, and R. L. Coppel. 1996. Diversity of the vaccine candidate AMA-1 of Plasmodium falciparum. Mol. Biochem. Parasitol. 77:109-113. [DOI] [PubMed] [Google Scholar]
- 32.McDonald, J. H., and M. Kreitman. 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351:652-654. [DOI] [PubMed] [Google Scholar]
- 33.Miller, L. H., D. I. Baruch, K. Marsh, and O. K. Doumbo. 2002. The pathogenic basis of malaria. Nature 415:673-679. [DOI] [PubMed] [Google Scholar]
- 34.Mueller, I., J. Kaiok, J. C. Reeder, and A. Cortés. 2002. The population structure of Plasmodium falciparum and Plasmodium vivax during an epidemic of malaria in the Eastern Highlands of Papua New Guinea. Am. J. Trop. Med. Hyg. 67:459-464. [DOI] [PubMed] [Google Scholar]
- 35.Narum, D. L., S. A. Ogun, A. W. Thomas, and A. A. Holder. 2000. Immunization with parasite-derived apical membrane antigen 1 or passive immunization with a specific monoclonal antibody protects BALB/c mice against lethal Plasmodium yoelii yoelii YM blood-stage infection. Infect. Immun. 68:2899-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Narum, D. L., and A. W. Thomas. 1994. Differential localization of full-length and processed forms of PF83/AMA-1, an apical membrane antigen of Plasmodium falciparum merozoites. Mol. Biochem. Parasitol. 67:59-68. [DOI] [PubMed] [Google Scholar]
- 37.Oliveira, D. A., V. Udhayakumar, P. Bloland, Y. P. Shi, B. L. Nahlen, A. J. Oloo, W. E. Hawley, and A. A. Lal. 1996. Genetic conservation of the Plasmodium falciparum apical membrane antigen-1 (AMA-1). Mol. Biochem. Parasitol. 76:333-336. [DOI] [PubMed] [Google Scholar]
- 38.Peterson, M. G., V. M. Marshall, J. A. Smythe, P. E. Crewther, A. Lew, A. Silva, R. F. Anders, and D. J. Kemp. 1989. Integral membrane protein located in the apical complex of Plasmodium falciparum. Mol. Cell. Biol. 9:3151-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Polley, S. D., and D. J. Conway. 2001. Strong diversifying selection on domains of the Plasmodium falciparum apical membrane antigen 1 gene. Genetics 158:1505-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Renia, L., I. T. Ling, M. Marussig, F. Miltgen, A. A. Holder, and D. Mazier. 1997. Immunization with a recombinant C-terminal fragment of Plasmodium yoelii merozoite surface protein 1 protects mice against homologous but not heterologous P. yoelii sporozoite challenge. Infect. Immun. 65:4419-4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robert, F., F. Ntoumi, G. Angel, D. Candito, C. Rogier, T. Fandeur, J. L. Sarthou, and O. Mercereau-Puijalon. 1996. Extensive genetic diversity of Plasmodium falciparum isolates collected from patients with severe malaria in Dakar, Senegal. Trans. R. Soc. Trop. Med. Hyg. 90:704-711. [DOI] [PubMed] [Google Scholar]
- 42.Rozas, J., and R. Rozas. 1999. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174-175. [DOI] [PubMed] [Google Scholar]
- 43.Sachs, J., and P. Malaney. 2002. The economic and social burden of malaria. Nature 415:680-685. [DOI] [PubMed] [Google Scholar]
- 44.Tajima, F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tangri, S., G. Y. Ishioka, X. Huang, J. Sidney, S. Southwood, J. Fikes, and A. Sette. 2001. Structural features of peptide analogs of human histocompatibility leukocyte antigen class I epitopes that are more potent and immunogenic than wild-type peptide. J. Exp. Med. 194:833-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomas, A. W., J. F. Trape, C. Rogier, A. Goncalves, V. E. Rosario, and D. L. Narum. 1994. High prevalence of natural antibodies against Plasmodium falciparum 83-kilodalton apical membrane antigen (PF83/AMA-1) as detected by capture-enzyme-linked immunosorbent assay using full-length baculovirus recombinant PF83/AMA-1. Am. J. Trop. Med. Hyg. 51:730-740. [DOI] [PubMed] [Google Scholar]
- 47.Thomas, A. W., A. P. Waters, and D. Carr. 1990. Analysis of variation in PF83, an erythrocytic merozoite vaccine candidate antigen of Plasmodium falciparum. Mol. Biochem. Parasitol. 42:285-287. [DOI] [PubMed] [Google Scholar]
- 48.Triglia, T., J. Healer, S. R. Caruana, A. N. Hodder, R. F. Anders, B. S. Crabb, and A. F. Cowman. 2000. Apical membrane antigen 1 plays a central role in erythrocyte invasion by Plasmodium species. Mol. Microbiol. 38:706-718. [DOI] [PubMed] [Google Scholar]
- 49.Verra, F., and A. L. Hughes. 2000. Evidence for ancient balanced polymorphism at the apical membrane antigen-1 (AMA-1) locus of Plasmodium falciparum. Mol. Biochem. Parasitol. 105:149-153. [DOI] [PubMed] [Google Scholar]