Abstract
The Arabidopsis thaliana histone H2A gene HTA1 is essential for efficient transformation of Arabidopsis roots by Agrobacterium tumefaciens. Disruption of this gene in the rat5 mutant results in decreased transformation. In Arabidopsis, histone H2A proteins are encoded by a 13-member gene family. RNA encoded by these genes accumulates to differing levels in roots and whole plants; HTA1 transcripts accumulate to levels up to 1000-fold lower than do transcripts of other HTA genes. We examined the extent to which other HTA genes or cDNAs could compensate for loss of HTA1 activity when overexpressed in rat5 mutant plants. Overexpression of all tested HTA cDNAs restored transformation competence to the rat5 mutant. However, only the HTA1 gene, but not other HTA genes, could phenotypically complement rat5 mutant plants when expressed from their native promoters. Expression analysis of HTA promoters indicated that they had distinct but somewhat overlapping patterns of expression in mature plants. However, only the HTA1 promoter was induced by wounding or by Agrobacterium infection of root segments. Our data suggest that, with respect to Agrobacterium-mediated transformation, all tested histone H2A proteins are functionally redundant. However, this functional redundancy is not normally evidenced because of the different expression patterns of the HTA genes.
INTRODUCTION
Histones are highly conserved components of eukaryotic chromatin. They are classified into four subcategories of core histones (H2A, H2B, H3, and H4) and linker histones (H1). Core histones are among the most abundant proteins in eukaryotic organisms. The expression of many histone genes is coupled to the S phase of the cell cycle and correlates with DNA replication and cell proliferation (Sundas et al., 1993; Brandstadter et al., 1994; Kouchi et al., 1995; Kanazin et al., 1996; Reichheld et al., 1998; Zhao et al., 2000; Nelson et al., 2002). However, some histones are expressed in tissues with low proliferation and cell division activity (Waterborg and Robertson, 1996; van den Heuvel et al., 1999; Xu et al., 1999). This latter expression pattern is characteristic of replacement histones and may result from the natural turnover of histones in nondividing mature cells or in cells undergoing substantial chromatin reorganization.
For many years, histones were considered as relatively passive structural units of eukaryotic chromatin. During the past decade, however, an increasing number of reports has emphasized the roles of histone modification and histone variants in the regulation of gene expression (Paull et al., 2000; Rice and Allis, 2001; Verbsky and Richards, 2001; Ausio and Abbott, 2002; Fransz and de Jong, 2002; Redon et al., 2002). Histone acetylation and methylation in particular have been implicated in gene expression (Vongs et al., 1993; Hirochika et al., 2000; Beisel et al., 2002; Volpe et al., 2002). Minor histone variants may contribute to diverse nuclear functions by directing distinct or unique chromatin architectures (Brown, 2001; Ahmad and Henikoff, 2002; Smith, 2002; Talbert et al., 2002). These more recent findings have brought histones once again into the limelight as important mediators of cellular events.
For most species, histone genes are members of multigene families. In addition to histone modification, the nature of these histone multigene families has attracted considerable attention relating to the specific roles of individual histones in mediating divergent cellular responses. Several publications have focused on H2A variants. One such variant, H2AX, becomes phosphorylated following induction of double-strand DNA breaks and is associated with the recruitment of repair factors to damaged DNA (Andegeko et al., 2001; Celeste et al., 2002; Tsukuda et al., 2005). Another histone H2A variant, H2A.Z, was first identified by West and Bonner (1980) and is highly conserved throughout evolution. H2A.Z proteins are nonuniformly spread among chromatin (Leach et al., 2000), and they may confer properties distinct from other H2A variants upon the organism (Iouzalen et al., 1996; Jackson and Gorovsky, 2000; Santisteban et al., 2000; Fan et al., 2002). Recently, this histone variant has been implicated in preventing the spread of silent heterochromatin into euchromatic regions of the genome (Mizuguchi et al., 2004). Histone macroH2A1 may be involved in inactivation of X chromosomes and in transcriptional silencing (Costanzi et al., 2000).
Although several plant histone H2A genes have been identified and their expression patterns analyzed (Koning et al., 1991; Callard and Mazzolini, 1997; Huh et al., 1997; van den Heuvel et al., 1999; Xu et al., 1999), their functions have not yet been well defined. We have identified an Arabidopsis thaliana mutant, rat5, in which the histone H2A gene HTA1 is disrupted. This mutant is resistant to Agrobacterium tumefaciens–mediated root transformation caused by a deficiency in T-DNA integration into the plant genome (Mysore et al., 2000b). More recently, we showed that this Arabidopsis HTA1 gene is expressed in nondividing tissues and that its expression is highly correlated to root cells that are most susceptible to Agrobacterium-mediated transformation (Yi et al., 2002). The expression of HTA1 in cells that are not undergoing mitotic division suggests that histone H2A-1 is a replacement histone variant. As with the situation in most species, the Arabidopsis HTA genes constitute a multigene family. The fact that disruption of the HTA1 gene causes a rat phenotype indicates that expression of other histone H2A family members at normal levels does not compensate for loss of HTA1 function in roots. In this article, we investigated the expression levels and patterns of several Arabidopsis HTA genes and directly tested the degree of functional redundancy afforded by HTA family members with regard to complementation of the rat phenotype in rat5. Our results indicate that multiple histone HTA genes can, when overexpressed from a strong promoter, compensate for loss of HTA1 gene activity. However, when expressed from their native promoters, only HTA1 can complement rat5. These results suggest that the tested histone H2A proteins are functionally redundant with respect to Agrobacterium-mediated transformation but that the levels and/or patterns of expression from their native promoters are insufficient to compensate for loss of HTA1 function.
RESULTS
Histone H2A Genes in Arabidopsis
The Arabidopsis genome contains 13 histone H2A (HTA) genes, each of which encodes a protein of a different sequence (http://www.chromdb.org/). Individual histone H2A genes are dispersed among the five Arabidopsis chromosomes (Table 1) with substantial distance not only between H2A genes but also between H2A genes and other histone genes H2B, H3, H4, and H1. Comparison of the 13 Arabidopsis histone H2A amino acid sequences indicates that they can be categorized into four major groups, plus one unique member (Figures 1 and 2). These histones share very high sequence similarity among themselves and to histone H2A proteins in other species. Four histone H2As (H2A-1, -2, -10, and -13) display >92% amino acid sequence identity. We set these histones into group I. H2A-3 and H2A-5 are classified as group II; they contain a SQEF motif in their C-terminal regions. This sequence is characteristic of H2AX, a histone H2A variant involved in double-strand DNA break repair induced by γ radiation (Rogakou et al., 1998). H2A-3 and H2A-5 show ∼76% amino acid sequence identity with H2A-1. Three histone H2As, H2A-6, -7, and -12, show ∼57% amino acid sequence identity with H2A-1 and ∼70% among themselves. These members have a SPKK motif in their C-terminal regions. This motif is found in plant histone H2As that are cell cycle regulated (Huh et al., 1995). The fourth group includes H2A-8, -9, and -11. These histones share 80 to 90% intragroup amino acid sequence identity and ∼52% sequence identity with H2A-1. Group IV members show strong (∼80%) identity with histone H2A F/Z variants in other organisms.
Table 1.
Arabidopsis Histone H2As
| Gene | Gene ID | Protein Length (Amino Acids) | Number of cDNAsa | Number of EST Clonesa | Function or Subtype (Consensus Sequences) |
|---|---|---|---|---|---|
| HTA1 | At5g54640 | 132 | 2 | 11 | Agrobacterium transformation |
| HTA2 | At4g27230 | 131 | 2 | 22 | |
| HTA3 | At1g54690 | 142 | 2 | 5 | H2AX (SQEF) |
| HTA4 | At4g13570 | 119 | 0 | 0 | |
| HTA5 | At1g08880 | 143 | 2 | 33 | H2AX (SQEF) |
| HTA6 | At5g59870 | 151 | 1 | 17 | DNA binding motif (SPKK) |
| HTA7 | At5g27670 | 151 | 2 | 17 | DNA binding motif (SPKK) |
| HTA8 | At2g38810 | 137 | 0 (1)b | 5 | H2A.F/Z |
| HTA9 | At1g52740 | 135 | 3 | 70 | H2A.F/Z |
| HTA10 | At1g51060 | 133 | 2 | 30 | |
| HTA11 | At3g54560 | 137 | 1 | 12 | H2A.F/Z |
| HTA12 | At5g02560 | 153 | 0 (1)b | 7 | DNA binding motif (SPKK) |
| HTA13 | At3g20670 | 133 | 7 | 22 |
Data from The Arabidopsis Information Resource (www.arabidopsis.org).
Numbers inside parenthesis were amplified by RT-PCR based on longest EST information for this research.
Figure 1.
Multiple Sequence Alignment of Arabidopsis H2A Proteins.
The 13 Arabidopsis histone H2A amino acid sequences were compared and analyzed by ClustalX (version 1.81) multiple alignment. Darkly shaded amino acid residues indicate identity; lightly shaded residues indicate synonymous amino acids.
Figure 2.
Phylogenetic Reconstruction of Arabidopsis Histone H2A Proteins Based on Amino Acid Sequence Similarity.
The Arabidopsis histone H2A proteins were classified into four groups plus one unique member. The members marked with an asterisk were used for further analysis. The topology and branch length of the phylogram was produced by distance/neighbor-joining analyses performed with PAUP* software (v4.10b) and reviewed using TreeView software (version 1.6.6). The percentage of bootstrap replications supporting each branch is shown at the nodes of the tree.
Histone H2A-4 is quite different from the other Arabidopsis histone H2As. H2A-4 has only 35% amino acid sequence identity with the other histone H2A family members and contains only 119 amino acids (compared with other family members that are 130 to 150 amino acids in length). A full-length cDNA clone is not available, and we have been unable to generate a cDNA using gene-specific primers. It is possible that the gene encoding histone H2A-4 (HTA4) is a pseudogene.
Arabidopsis Histone H2A Genes Are Not Functionally Redundant with Regards to Agrobacterium-Mediated Root Transformation
Histone H2A-1 plays an important role in Agrobacterium-mediated transformation of Arabidopsis roots (Mysore et al., 2000b; Yi et al., 2002). The rat5 mutant contains two tandem copies of T-DNA inserted into the 3′ untranslated region of the HTA1 gene. This mutation results in a deficiency in T-DNA integration upon subsequent attempts to retransform the roots (Mysore et al., 2000b). In order to determine the extent of functional redundancy among the histone H2A genes, we individually introduced various HTA genes (HTA1, -2, -3, -4, -5, -6, -8, and -10), representing the four histone H2A clades, into the rat5 mutant using a flower dip procedure (Clough and Bent, 1998). Mysore et al. (2000a) previously showed that many rat mutants, including rat5, are transformable by this method of Agrobacterium-mediated transformation. For each introduced gene, we analyzed 11 to 29 lines for restoration of transformation proficiency to the rat5 mutant.
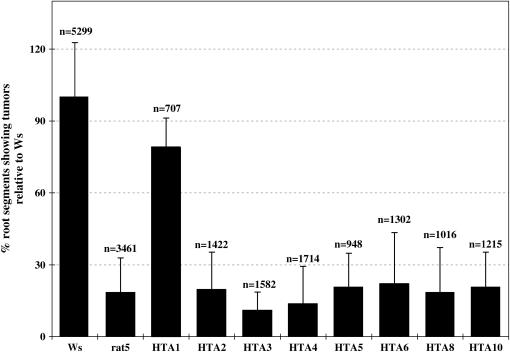
Figure 3 shows that compared with wild-type plants, roots of rat5 mutant plants transformed on average at only 18% frequency, and even those tumors that did form were very small compared with those incited on wild-type roots (Figure 5). Despite high amino acid sequence similarity among the introduced Arabidopsis histone H2As, none of the heterologous H2A gene family members could compensate for disruption of the HTA1 (RAT5) gene. As previously shown (Mysore et al., 2000b), introduction of a genomic copy of HTA1 into rat5 plants substantially increased their transformation competence; among the transgenic lines examined, transformation was restored, on average, to 79% that of wild-type plants. Thus, none of the other tested HTA genes were functionally redundant with HTA1. This makes sense in light of our ability to discern the rat phenotype in rat5 (HTA1) mutant plants despite the presence of intact copies of all other histone H2A genes.
Figure 3.
Restoration of rat5 Transformation Proficiency by Histone H2A Genomic DNA Clones.
Roots segments from rat5 transgenic plants containing one of the histone H2A genomic DNA clones were inoculated with A. tumefaciens A208. After 2 d, the root segments were transferred to hormone-free Murashige and Skoog (MS) medium containing timentin. After 1 month, the root segments were scored for tumor production. Note that only the HTA1 (RAT5) genomic clone complemented the rat5 mutant plant. n, number of root segments evaluated; Ws, Wassilewskija. Error bars represent standard deviation.
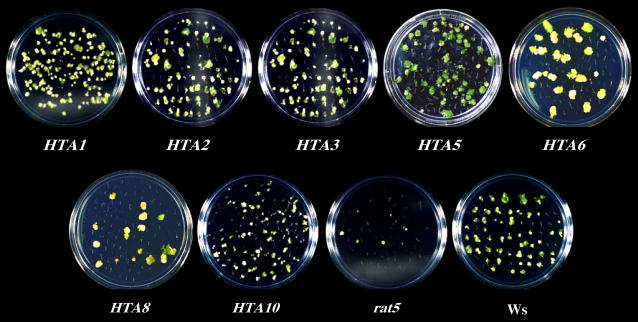
Figure 5.
Root Tumorigenesis Assays of rat5 Mutant Plants Containing Various Histone H2A cDNAs Controlled by a CaMV 35S Promoter.
Root segments from plants containing the indicated cDNAs, rat5 mutant plants, and Ws wild-type plants were inoculated with A. tumefaciens A208. After 2 d, the root segments were transferred to solidified MS medium containing timentin (to kill the bacteria) but lacking phytohormones. After 1 month, representative plates from each infection were photographed.
Heterologous Histone H2A cDNAs, under the Control of a Constitutive Cauliflower Mosaic Virus 35S Promoter, Can Restore Transformation Proficiency to the rat5 Mutant
It is possible that additional copies of HTA genes did not complement the rat5 mutant because these genes could not express to a sufficient extent in the root cells most susceptible to transformation by Agrobacterium (Yi et al., 2002). We therefore introduced into rat5 mutant plants cDNA copies of each of the genes investigated above (except HTA4, for which a cDNA was unavailable) and investigated the transformation efficiency of the derived transgenic lines. In these plants, the cDNAs were under the control of a strong, constitutive cauliflower mosaic virus (CaMV) 35S promoter. Figure 4 shows that, unlike the situation with genomic copies of the HTA genes, transgenic rat5 plants containing any of the tested HTA cDNAs were restored to transformation proficiency. In these experiments, root segments from rat5 mutant plants displayed a transformation efficiency only 7% that of wild-type plants. Introduction of the HTA1 cDNA into these plants restored transformation, on average, to 81% of the wild-type controls. Other HTA cDNAs restored transformation to lesser extents. Transgenic rat5 plants individually containing HTA groups I and II cDNA transgenes (HTA2, -3, -5, and -10) displayed a 50 to 70% restoration of transformation efficiency. Histones encoded by these cDNAs are closest in amino acid sequence to histone H2A-1 (Figures 1 and 2). Representatives of histone H2A groups III and IV, HTA6 and HTA8, restored transformation to rat5 mutant plants less efficiently (32 and 24%, respectively). Histones encoded by family members of groups III and IV are more distant in amino acid sequence to histone H2A-1 than are members of groups I and II.
Figure 4.
Restoration of rat5 Transformation Proficiency by Histone H2A cDNA Clones.
Roots segments from rat5 transgenic plants containing one of the histone H2A cDNA clones were inoculated with A. tumefaciens A208. After 2 d, the root segments were transferred to hormone-free MS medium containing timentin. After 1 month, the root segments were scored for tumor production. Note that all cDNA clones tested complemented the rat5 mutant plant. However, the HTA6 and HTA8 cDNAs did not complement as well as the other cDNAs tested. n, number of root segments evaluated.
Wild-type Arabidopsis ecotype Ws roots usually produce large green teratomata when infected by A. tumefaciens A208. The extent of tumorigenesis can be determined not only by the percentage of root segments that develop tumors but also by the morphology and color of the resulting tumors; greater susceptibility generally correlates with larger and greener tumors (Zhu et al., 2003). Figure 5 shows that rat5 lines individually transgenic for HTA1, -2, -3, -5, and -10 cDNAs produced large green teratomata when infected by A. tumefaciens A208, whereas rat5 lines transgenic for HTA6 or HTA8 produced less pigmented and more amorphous tumors. These tumor phenotypes additionally indicate that these transgenic lines are less susceptible to Agrobacterium-mediated transformation than are lines containing the other cDNAs. Taken together, our data indicate that, with regard to Agrobacterium-mediated transformation, all tested histone H2A proteins are (at least partially) functionally redundant. Our data further suggest that the lack of functional redundancy displayed by the tested HTA genes may result from insufficient levels of expression or from inappropriate patterns of expression in root cells most susceptible to transformation by Agrobacterium.
Histone H2A mRNAs Accumulate to Different Extents in Arabidopsis Plants
The ability of multiple HTA cDNAs, but not genomic DNAs, to complement the rat phenotype of the rat5 mutant suggests that the level of expression of the different HTA genes may also play a role in Agrobacterium-mediated transformation. We therefore measured the steady state levels of transcripts from eight HTA genes (HTA1, -2, -3, -4, -5, -6, -8, and -10) that were previously tested in the complementation assays described above. Because of the high level of sequence similarity among the histone H2A genes, we performed quantitative real-time RT-PCR analyses using primer sets specific for the individual HTA genes. In our initial analyses, we were able to amplify RT-PCR products of all tested HTA mRNAs except HTA4 and HTA8. As mentioned above, HTA4 may be a pseudogene. The amplification of HTA8 mRNA was weak and variable; we presume that the steady state level of HTA8 transcripts is very low. We analyzed the levels of other HTA transcripts from three independent RNA extractions of roots or whole plants grown in vitro. Their individual transcript levels were normalized to those of α-tubulin and glyceraldehydes-3-phosphate-dehydrogenase (GAPDH) and displayed as the number of molecules per microgram of total RNA.
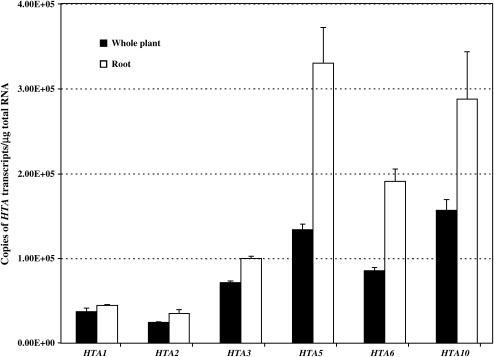
Figure 6 shows that HTA1 and HTA2 transcripts accumulated to only a low level in preparations of RNA from whole plants and root tissue. HTA3 and HTA5 transcripts were detected at a level 3 to 5 times higher than those of HTA1 and HTA2. HTA5, -6, and -10 showed a root preferential expression pattern. Considering that we use root tissues for transformation assays, it is interesting to note that HTA1 transcripts do not accumulate to high levels in roots. Despite this low level of HTA1 expression, disruption of HTA1 in the rat5 mutant resulted in recalcitrance of Arabidopsis to Agrobacterium-mediated transformation (Mysore et al., 2000b). These results indicate that insufficient levels of HTA gene expression in the cells undergoing transformation may be a reason for the nonredundant function of other HTA genes with regard to Agrobacterium-mediated transformation.
Figure 6.
Quantification of HTA Transcripts in Roots and Whole Arabidopsis Plants.
RNA was isolated from 3-week-old Arabidopsis roots or whole plants and used for reverse transcription. Equal amounts of the reverse transcription products were used as templates for quantitative real-time PCR using primers directed against each histone HTA, GAPDH, or α-tubulin gene. The level of expression of each HTA gene was determined after 30 cycles of reaction using Corbett Rotor gene software (version 1.4) and normalized to internal standards (GAPDH and α-tubulin). Three different RNA samples isolated from different batches of plant material were used for reverse transcription, and at least two PCR analyses were performed for each RNA sample. Closed bars, RNA from whole plants; open bars, RNA from roots. Error bars denote standard deviation.
Arabidopsis Histone H2A Promoters Display Different Patterns of Expression
Because our data suggested that the lack of functional redundancy among the various HTA genes may result from differential patterns of expression, we examined the expression of nine HTA promoters, plus the first encoded 10 amino acids of each H2A protein fused to a gusA gene, in transgenic Arabidopsis plants. Previous data indicated that β-glucuronidase (GUS) expression directed by a similar HTA1-gusA translational fusion construction paralleled expression of the HTA1 gene as determined by in situ RNA hybridization (Yi et al., 2002). For these experiments, we generated at least 10 lines of transgenic plants for each HTA promoter-gusA fusion construction and examined the resulting plants at weekly intervals after germination. Figures 7 and 8 indicate that in mature plants (i.e., plants that had just started to flower), the HTA promoters showed distinct but, in some cases, overlapping patterns of expression. These overlapping patterns of expression could be further delimited in some cases to distinguish between two promoters on the basis of tissue specificity (Figure 8).
Figure 7.
Expression of HTA Promoter-gusA Fusion Genes in Transgenic Arabidopsis Plants.
Transgenic plants containing each of the indicated HTA-gusA fusion genes were stained with X-gluc and photographed. Representative plants from representative transgenic lines are shown.
(A) Aerial parts of plant; insets show flower bolts.
(B) Plant roots; bottom section of each panel shows root tips.
Figure 8.
Transverse Sections of Roots of Transgenic Arabidopsis Plants Showing Expression of HTA Promoter-gusA Fusion Genes.
Approximately 20 transgenic lines for each promoter-gusA fusion were screened, and the roots were sectioned. Representative photomicrographs of the root apical, elongation, and maturation zones from representative transgenic lines are shown. Schematic diagrams of the root apical, elongation, and maturation zones are shown in the last panel. RC, root cap cells; Ep, epidermis; C, cortex; En, endodermis; P, pericycle; V, vasculature.
The HTA2, -3, -6, -8, and -13 promoters directed GUS expression predominantly in the meristems and actively dividing cells, including the root meristem and lateral root primordia (Figures 7B and 8) as well as the floral and leaf meristematic regions (Figure 7A). The HTA8 promoter could be distinguished from the other promoters by the specificity of GUS expression in the root tissues (Figure 8); expression was restricted to the vasculature in the maturation zone for the HTA8 promoter. HTA10 promoter-directed GUS expression was predominantly restricted to the root cap cells in the apical zone (Figure 8). This promoter expressed in nondividing regions of the plant, including the root elongation and maturation zones (Figures 7B and 8) and the leaf veins (Figure 7A). The HTA6 promoter displayed another pattern of expression. This promoter expressed strongly throughout the roots and leaves (Figures 7A and 7B). The HTA4 promoter expressed in a spotty manner at the margins of cauline and rosette leaves (Figure 7A). The promoter also expressed weakly in the root apical and elongation zones (Figures 7B and 8).
Although the expression patterns of the HTA1, -2, -3, -5, -6, -10, and -13 promoters looked similar, there were clear differences in several aspects. As shown previously, HTA1 expressed mainly in the root tip area, the elongation zone, and in lateral root primodia (Figure 7B; Yi et al., 2002). HTA1 showed weak expression in root cap and epidermal cells of the apical region, whereas HTA2, -3, -5, -6, -10, and -13 expression patterns were somewhat broader than that of HTA1. The HTA1 and -5 promoters did not express well in the floral bud, whereas the HTA2, -3, -10, and -13 promoters did (Figure 7A). The HTA5 and HTA10 promoter did not express as well in newly emerging leaves as did the HTA1 promoter.
Thus, although some of the HTA promoters showed overlapping patterns of expression, none of the promoters showed identical expression patterns (Table 2, Figures 7 and 8). In addition, we did not find any correlation between the various promoter activities and the extent of amino acid sequence similarity among the histone H2A proteins. Based upon these data, the tested Arabidopsis histone H2A genes are differentially regulated. It is likely that this differential regulation is responsible for the lack of functional redundancy among the histone H2A genes with regards to Agrobacterium-mediated genetic transformation.
Table 2.
Expression Patterns of Nine HTA Promoter-gusA Reporter Gene Fusions in Transgenic Arabidopsis Plants
| Gene | Root Apical Zone | Lateral Root Primordia | Root Elongation Zone | Root Maturation Zone | Leaves | Center of Rosette | Floral Bud |
|---|---|---|---|---|---|---|---|
| HTA1 | +/− | + | + | + | Expanded | + | + |
| HTA2 | + | _ | + | + | Veins | + | + |
| HTA3 | + | + | + | + | Young | + | + |
| HTA4 | +/− | − | − | − | Margin | − | +/− |
| HTA5 | + | + | + | + | Young | + | − |
| HTA6 | + | + | + | + | Expanded | + | +/− |
| HTA8 | + | + | +/− | + | Young | + | + |
| HTA10 | + | + | + | + | Veins | − | + |
| HTA13 | + | + | + | + | Expanded | + | + |
The +, −, and +/− indicate expression, no expression, and weak expression of the gusA reporter gene, respectively.
The Expression of Only HTA1 Is Increased in Response to Root Wounding
We previously demonstrated that the expression of HTA1 is induced by cutting Arabidopsis roots into small segments (Yi et al., 2002). Because of the importance of wounding in the transformation process (Baker et al., 1997), we examined the induction of different HTA promoter-gusA fusion constructions in wounded root segments. For this experiment, we cut the roots of transgenic plants containing one of seven different HTA-gusA fusion genes into 3- to 5-mm segments and assayed them for GUS activity immediately after cutting and 2 d later. We scored the percentage of cut ends that stained blue with 5-bromo-4-chloro-3-indolylglucuronide (X-gluc). Table 3 shows that, according to this assay, only HTA1 showed an increase in activity in response to wounding. All other tested HTA genes showed a decrease in expression. These data indicate that HTA genes respond differently to wounding and suggest that increased HTA1 expression in the region near the wound site may be important for Agrobacterium-mediated transformation.
Table 3.
Changes of Arabidopsis H2A Promoter Activity Near the Cut Site of Root Segments
| Freshly Cut
|
Incubated for 2 d on MS Medium
|
|||
|---|---|---|---|---|
| Gene | GUS Expression (%)a | Number of Root Segments Assayed | GUS Expression (%)a | Number of Root Segments Assayed |
| HTA1 | 16.4 | 354 | 34.6 | 399 |
| HTA2 | 51.0 | 776 | 17.3 | 1064 |
| HTA3 | 9.2 | 981 | 4.1 | 398 |
| HTA4 | 0.5 | 364 | 0.3 | 325 |
| HTA5 | 50.5 | 871 | 24.5 | 1191 |
| HTA6 | 49.1 | 793 | 29.7 | 1169 |
| HTA13 | 38.7 | 860 | 26.8 | 1086 |
Calculated as the percentage of root segments showing GUS activity near the cut site.
Response of HTA Genes to Infection of Root Segments by Agrobacterium
We had previously shown that high levels of GUS activity are directed by the HTA1 promoter after infection of root segments by Agrobacterium (Yi et al., 2002). We therefore investigated whether other HTA genes respond similarly to bacterial infection. We initially attempted to quantify various HTA mRNAs, using quantitative real-time RT-PCR, in cut root segments that were not inoculated or inoculated with either A. tumefaciens At793 (lacking a Ti-plasmid) or At804 (containing the disarmed Ti-plasmid pEHA105) plus the T-DNA binary vector pBISN1 (Narasimhulu et al., 1996). However, over a period of 36 h after inoculation, we could not detect any major changes in transcript abundance for any HTA gene, including HTA1 (data not shown). This lack of change in mRNA quantity is most likely explained by the fact that only a very few cells near the cut end of the root segment were infected by bacteria.
We therefore investigated expression of GUS activity directed by various HTA promoter-gusA fusions in cut root segments inoculated with A. tumefaciens A208 (a tumorigenic strain containing pTiT37). GUS activity, measured as the percentage of root segments staining blue with X-gluc, indicated the activity of the various HTA promoters. As a control, we examined root segments inoculated with the transfer-deficient strain A. tumefaciens A136 (which lacks a Ti-plasmid). Table 4 shows that the HTA1 promoter specifically responded to A. tumefaciens A208, a tumorigenic strain. More than five times as many root segments showed GUS activity when cocultivated with this strain than with A. tumefaciens A136, a nontumorigenic strain lacking a Ti-plasmid. No other HTA-gusA promoter fusion showed such an increase in activity after infection by A. tumefaciens A208. These experiments indicate that the various tested HTA promoters respond differently to infection by Agrobacterium and that the HTA1 promoter in particular is induced.
Table 4.
Changes of Arabidopsis HTA Promoter Activity Near the Cut Site of Root Segments 2 d after Cocultivation with Various Agrobacterium Strains
|
A. tumefaciers A208a
|
A. tumefaciens A136b
|
|||||
|---|---|---|---|---|---|---|
| Gene | GUS Expression (%)c | ± se | Number of Root Segments Assayed | GUS Expression (%)c | ± se | Number of Root Segments Assayed |
| HTA1 | 15.8 | 6.5 | 1040 | 2.8 | 0.1 | 1096 |
| HTA2 | 20.8 | 3.2 | 1179 | 16.4 | 2.4 | 873 |
| HTA3 | 4.8 | 0.7 | 1073 | 8.9 | 1.3 | 910 |
| HTA4 | 0.8 | 0.3 | 615 | 1.3 | 0.4 | 455 |
| HTA5 | 8.1 | 1.8 | 1378 | 5.9 | 0.3 | 1173 |
| HTA6 | 19.0 | 2.5 | 1615 | 17.9 | 1.5 | 799 |
| HTA8 | 6.1 | 0.7 | 655 | 8.3 | 1.4 | 495 |
| HTA10 | 3.4 | 0.9 | 520 | 10.1 | 0.0 | 355 |
| HTA13 | 20.7 | 2.4 | 1017 | 19.1 | 2.0 | 619 |
T-DNA and Vir protein transfer-competent strain.
T-DNA and Vir protein transfer-deficient strain.
Calculated as the percentage of root segments showing GUS activity near the cut site.
DISCUSSION
Arabidopsis Histone H2A Genes
Similar to the situation with other organisms, Arabidopsis histone H2A genes constitute a multigene family. However, unlike the arrangement of mammalian histone H2A genes (Albig and Doenecke, 1997), the 13 members of this family are not clustered in the genome but rather are dispersed among the five Arabidopsis chromosomes.
Mammalian H2A proteins are classified into four different variant families: core H2A, macroH2A, H2A.F/Z, and H2AX. Numerous groups have investigated the functions of each variant type (Iouzalen et al., 1996; Liu et al., 1996; Reichheld et al., 1998; Rogakou et al., 1998; Costanzi et al., 2000; Santisteban et al., 2000). However, little is currently known about the function of Arabidopsis histone H2A proteins. We compared Arabidopsis and mammalian histone H2A amino acid sequences with regard to classification into variant families. Arabidopsis H2A-8, -9, and -11 are similar to H2A.F/Z variants, whereas H2A-3 and H2A-5 show strong similarity to H2AX variants. Callard and Mazzolini (1997) reported that the expression of Arabidopsis H2AvAt (HTA11), a H2A.F/Z variant, is tightly related to cell proliferation. They suggested that expression of HTA11 might be associated with a switch from a quiescent to an actively replicating state. Arabidopsis H2A-3 and H2A-5 contain a conserved SQEF motif that is found in most H2AX variants in mammals, Drosophila, and yeast. The Ser residue of this motif becomes phosphorylated in response to double strand DNA breaks, meiotic recombination preceding synaptic cross-over, immune cell development, and apoptotic DNA fragmentation (Rogakou et al., 1998, 2000; Chen et al., 2000; Mahadevaiah et al., 2001; Tsukuda et al., 2005).
H2A-6, -7, and -12 contain a SPKK motif at their C termini. Huh et al. (1995) suggested that wheat (Triticum aestivum) H2A proteins can be divided into two groups based upon the existence of this SPKK motif, which is conserved in angiosperm H2As. The SPKK motif can bind to the DNA minor groove in A/T-rich regions (Suzuki, 1991). In sea urchins, histone H1 contains N-terminal SPKK repeats that are targets for the cdc2 kinase, and phosphorylation of these repeats is important for chromatin condensation during sperm development (Roth and Allis, 1992). The SPKK motif may be a unique feature of plant H2As because it is lacking in mammalian, insect, and yeast H2As.
Differential Expression of Histone H2A Members in Arabidopsis
In most organisms, core histone genes are expressed primarily during the S phase of the cell cycle (Tanimoto et al., 1993; Oswald et al., 1996; Downs et al., 2000). In addition, there are histones, called replacement histones, that are expressed constitutively in nondividing cells (Chaubet-Gigot et al., 2001). Replacement histones can be functionally redundant with core histones and can exchange with preexisting histones during development. We analyzed Arabidopsis histone H2A gene expression patterns using promoter-GUS fusions (Figures 7 and 8). Previous work validated that the expression pattern of a HTA1-gusA fusion gene using this technique was identical to that revealed by RNA in situ hybridization (Yi et al., 2002).
HTA2, -3, -6, -8, and -13 expression was predominantly in meristematic regions in a pattern similar to that of a cyclin gene, whose expression is directly related to the cell cycle (Ferreira et al., 1994; Yi et al., 2002). The other tested Arabidopsis histone H2A genes (HTA1, -4, -5, and -10) showed a broad range of expression patterns. Because they were expressed in cells not undergoing mitotic divisions, these HTA genes display some characteristics of replacement histones. It is notable that HTA13 is expressed mainly in nondividing tissues of the plant (Figure 7).
Based on amino acid sequence similarity, HTA8 is most closely related to known H2A.F/Z variants. Vertebrate H2A.F/Z variants are regarded as replacement histones that are used to replace core H2A proteins during portions of the cell cycle other than S phase (Wu et al., 1982; Dalton et al., 1989; Hatch and Bonner, 1990). However, Drosophila melanogaster H2A.F/Z variants are essential for viability and are expressed maximally during DNA replication (van Daal and Elgin, 1992). Arabidopsis HTA8 shows an expression pattern expected of a core histone in that its expression is restricted to meristematic cells. The expression of another Arabidopsis H2A.F/Z variant, cH2AvAt (HTA11), is tightly correlated with cell proliferation in cell suspension cultures (Callard and Mazzolini, 1997).
Although we grouped Arabidopsis histone H2A proteins on the basis of their primary protein structure, we did not find any relationship between histone H2A gene expression patterns and protein structural similarity. For example, H2A-3 and H2A-5 are classified as H2A.X variants, yet their expression patterns were quite disparate. Histone H2A-10 and H2A-13 also display strong similarity in amino acid sequence but differ substantially in their expression patterns (Figures 7 and 8, Table 2).
Functional Redundancy of Histone Genes
Although the rat5 mutant, which contains a disruption of the HTA1 gene, appears phenotypically normal, it is deficient in T-DNA integration (Mysore et al., 2000b) and thus is highly recalcitrant to Agrobacterium-mediated root transformation (Nam et al., 1999). Expression of the other HTA genes in this mutant is similar to that of wild-type plants (H.C. Yi, unpublished data). These findings suggest that the tested HTA genes are not functionally redundant with HTA1, at least with respect to Agrobacterium-mediated transformation. Indeed, none of the heterologous HTA genes tested could compensate for the loss of HTA1 when transformed into rat5 mutant plants (Figure 3). However, Arabidopsis histone H2A proteins display a high degree of structural similarity, and all tested Arabidopsis H2A cDNAs were functionally redundant with HTA1 when they were expressed constitutively (Figure 4). Taken together, these results suggest that histone H2A proteins are functionally redundant with histone H2A-1 with respect to Agrobacterium-mediated transformation. However, the expression patterns of these genes do not permit this redundancy to be displayed. Placement of the various HTA cDNAs under the regulation of the HTA1 promoter may shed more light on the role of cell-specific regulation of histone H2A expression in Agrobacterium-mediated transformation. These experiments are currently being conducted.
Functional redundancy of histone H2A genes differs among species. Takami et al. (1997) removed approximately half of the histone genes from a cultured chicken cell line. The results of these experiments indicated that some of the histone family members or variants are not completely redundant. In Tetrahymena thermophila, disruption of either of the two major histone H2A genes did not substantially affect viability of the organism. However, mutation of the HTA3 gene encoding a H2A.F/Z variant was not completely compensated by expression of the other major H2A genes (Liu et al., 1996).
Individual members of a multigene family could support disparate cellular metabolic activities and developmental responses. Histone H2A family members are abundant proteins that play important and sometimes different roles in constituting chromosomal structure (Redon et al., 2002). For example, histone H2AX may be involved in signaling double strand breaks that require repair (Paull et al., 2000), whereas histone H2A.Z is involved in marking telomeric heterochromatin and in preventing its spread into euchromatic regions of the chromosome (Meneghini et al., 2003; Mizuguchi et al., 2004). Particular histone variants may associate with various chromosomes or chromosomal regions. Histone macroH2A1 is found on inactive X chromosomes of female mice (Costanzi et al., 2000), whereas certain histone H3 variants are specifically associated with centromeric heterochromatin (Talbert et al., 2002).
As a first step in elucidating control of histone H2A gene expression, we used quantitative real-time RT-PCR to monitor the steady state level of transcripts encoded by six HTA genes (HTA1, -2, -3, -5, -6, and -10). Real-time PCR has been used by several research groups to estimate gene copy number and for quantification of transcript levels (Bustin, 2000; Kandasamy et al., 2002; Milligan et al., 2002; Toyooka et al., 2002). Because we used Arabidopsis roots to test for Agrobacterium-mediated transformation competence, we used this tissue for our RNA analysis and subsequently compared these results with those using RNA extracted from whole plants. The expression of many tested HTA genes in roots paralleled that of whole plants (Figure 6). However, HTA5, -6, and -10 showed enriched expression in root tissues. The quantitative real-time RT-PCR results correlated well with our HTA promoter-gusA fusion reporter gene expression data (Figures 7 and 8, Table 2). HTA1 is expressed at low levels and constitutes only 2 to 3% of total histone H2A gene expression. It is interesting to note that, despite the low level of expression, Arabidopsis HTA1 is essential for integration of Agrobacterium T-DNA and cannot be compensated for by normal levels of expression of the other HTA genes.
HTA1 also responds differently to wounding and to Agrobacterium infection than do the other investigated HTA genes (Tables 3 and 4). Thus, HTA1 shows a unique association with Agrobacterium-mediated plant transformation that is not shared by other Arabidopsis histone genes. We have searched for DNA sequence motifs in the HTA1 promoter that may confer wound-inducible expression using the database PLACE (http://www.dna.affrc.go.jp/PLACE/; Higo et al., 1999). A W-box motif, commonly found in the promoters of wound-inducible genes (Nishiuchi et al., 2004), exists in the HTA1 promoter at position −354. However, this motif also appears (sometimes in multiple places) in the promoters of HTA3, HTA4, HTA6, HTA8, and HTA9, promoters that are not wound inducible. Other DNA sequence motifs commonly found in wound-inducible promoters, such as a GT1 box, also appear in the HTA1 promoter, although a MYC box sometimes associated with wound-inducible promoters is absent from the HTA1 promoter. Thus, specific motifs causing wound inducibility of the HTA1 promoter remain to be established.
METHODS
Plant and Agrobacterium Growth
Arabidopsis thaliana seeds were surface sterilized and germinated in Petri dishes containing B5 medium (Gibco BRL) solidified with 7.5 g/L bactoagar (Difco). When appropriate, antibiotics were added either to kill Agrobacterium tumefaciens (100 μg/mL timentin) or for transgene selection (50 μg/mL kanamycin or 50 μg/mL hygromycin). The plates were incubated under a 16-h-light/8-h-dark photoperiod at 25°C for 1 week. Seedlings were individually transferred into baby food jars containing B5 medium and grown for 2 to 3 weeks for tumorigenesis assays. Alternatively, plants were grown in pots containing soil for harvesting seeds.
Agrobacterium was grown in YEP-rich or AB minimal medium (Lichtenstein and Draper, 1986) at 30°C. When appropriate, antibiotics were used at the following concentrations: 10 μg/mL rifampicin, 100 μg/mL kanamycin (on solidified medium), or 25 μg/mL (in liquid medium).
Cloning of Histone H2A Genomic DNAs and cDNAs
Arabidopsis histone H2A genomic DNAs were cloned by PCR amplification. PCR primers were designed to bind ∼2 kb upstream and 1 kb downstream of the coding sequence of individual histone H2A genes. The sequence of the primers used are: HTA1 forward, 5′-TGGGTCGTGGACATCAGATGGTTCGGA-3′; HTA1 reverse, 5′-GAGGCATAGACACTGTCACTCACTTGT-3′; HTA2 forward, 5′-GGCTATGGATCTTGAACAACCGGACCT-3′; HTA2 reverse, 5′-ACGAGAGCTCTAAACCGAATCGTCCCT-3′; HTA3 forward, 5′-GAGCAGTTTTTCGAGGAGGTGGTCTCC-3′; HTA3 reverse, 5′-TTGAACGATGTTCTGCGGTGGCTCGGC-3′; HTA4 forward, 5′-TGGGACTGACCTTAAAATTGTCACA-3′; HTA4 reverse, 5′-GTCTCGCTTCGTGAAATACAGTTGT-3′; HTA5 forward, 5′-ACCTGTCGACGACCAATAACTGCAGCA-3′; HTA5 reverse, 5′-AGCGAGCTAATTCAGGTGCGAACCAAC-3′; HTA6 forward, 5′-AGTAACAACCTGCCAAACCCGTCCCGC-3′; HTA6 reverse, 5′-GTCCATGGAAGCTAAGGCACTTGCAGC-3′; HTA8 forward, 5′-TCCGCAATCTTGGTCACATCAGTCA-3′; HTA8 reverse, 5′-ACCCTGCAAACAAGAAGAGTCAGGC-3′; HTA10 forward, 5′-TCGTCCCAAGCGTCACTCGAACCAA-3′; HTA10 reverse, 5′-ACAGAACCTTCCTTTCGCAATTGGT-3′; HTA13 forward, 5′-AGACAAGGCCAGAGGGGCAAGCCAT-3′; HTA13 reverse, 5′-GCCATGGCTTCTTCGACTCTCTTCA-3′.
For amplification accuracy and fidelity, AccuTaq LA DNA polymerase (Sigma-Aldrich) and Takara EX Taq DNA polymerase were used. PCR reactions were performed using the following cycling conditions: one cycle of denaturation at 98°C for 30 s; 35 cycles of denaturation at 94°C for 30 s, annealing at 65°C for 45 s, extension at 68°C for 5 min; one cycle of elongation at 68°C for 30 min. The annealing temperatures and times were occasionally modified depending on primer specificity. The AccuTaq reaction mixture contained 200 ng of Arabidopsis genomic DNA, 50 mM Tris-HCl, 15 mM ammonium sulfate, pH 9.3, 2.5 mM MgCl2, 0.1% Tween 20, 500 μM deoxynucleotide triphosphate (dNTP) mix, 400 nM of primers, and 5 units of Taq polymerase. The ExTaq reaction mixture contained 200 ng of Arabidopsis genomic DNA, 25 mM TAPS, pH 9.3, 50 mM KCl, 2 mM MgCl2, 1 mM 2-mercapoethanol, 200 μM dNTP mix, 1 μM primers, and 5 units of Takara ExTaq polymerase. Amplified PCR products were cloned into pBluescript II SK+ (Stratagene), pGEM-T (Promega), or pCR 2.1 (Invitrogen) vectors. Five histone H2A cDNAs that had not been isolated from an Arabiodopsis cDNA library in our laboratory were cloned by RT-PCR. Total RNA was isolated from Arabidopsis roots, and 1 to 2 μg of total RNA was used for reverse transcription using oligo(dT) as a primer (Promega). The reaction mixture was diluted 10 to 20 times, and 10 to 20 μL of the reverse transcription mixture was used for the PCR reaction. Five PCR primer sets for HTA1, -4, -6, -8, and -13 were designed based on Arabidopsis EST information. The sequences of the primers were as follows: HTA1 forward, 5′-CTCTTTGTGGGTTGTTGTTGTTGAA-3′; HTA1 reverse, 5′-GAAACAACATGTCGAAACAGAAACGG-3′; HTA4 forward, 5′-ACGGTCTCATTTTATGTTTCATGGT-3′; HTA4 reverse, 5′-GGTTGTTTTGTTGATGAGACTAGTG-3′; HTA6 forward, 5′-TCAGTAATCGATAACCGTAGCAATG-3′; HTA6 reverse, 5′-GACCAAAAGATTAGACGAAGCGTAT-3′; HTA8 forward, 5′-CTTTGATTTCGACGACGGATCTTGA-3′; HTA8 reverse, 5′-TAATAGAGACTTAGGACTCAGAGAG-3′; HTA13 forward, 5′-TTTCTATCTCTCTTCCCAAATCACA-3′; HTA13 reverse, 5′-CAACCACAACACAAATCCCTAATCT-3′.
RT reaction mixtures (10 or 20 μL) were added to PCR reaction mixtures containing 10 mM Tris-HCl, pH 8.3, 50 mM KCl, 2.5 mM MgCl2, 100 μM dNTP, and 3 units of high-fidelity Taq DNA polymerase (Boehringer Mannheim). PCR was performed using the following conditions: one cycle of denaturation at 95°C for 2 min; 35 cycles of denaturation at 94°C for 20 s, annealing at 58°C for 40 s, and extension at 72°C for 2.5 min; one cycle of elongation at 72°C for 15 min. The PCR products were cloned into pBS+ (Stratagene). All the cloned genomic and cDNA sequences were confirmed by analysis at the Purdue University Genomics Center.
T-DNA Binary Vector Construction and Plant Transformation for Complementation of the rat5 Mutant
For cDNA complementation analysis, histone H2A cDNAs (HTA1, -2, -3, -5, -6, -8, -10, and -13) were excised from plasmids using either SalI or SalI plus XbaI and ligated into the multicloning site of the T-DNA binary vector pS35-hpt behind the CaMV 35S promoter in the sense orientation. For genomic DNA complementation, histone H2A genomic DNA fragments (HTA1, -2, -3, -4, -5, -6, -8, -10, and -13) containing ∼2 kb of their native promoter sequence and 1 kb of downstream sequence were excised from plasmids using appropriate restriction sites: HTA1, -2, -5, and -6, XbaI and SalI; HTA3 and -4, HindIII and SpeI; HTA8 and -10, SmaI; HTA13, SalI and XhoI. The excised DNA fragments were cloned into the T-DNA binary vector pGPTV-hpt. The various constructions carrying histone H2A cDNAs or genes were introduced into A. tumefaciens GV3101 (Koncz and Schell, 1986) by electroporation and used for floral dip transformation (Clough and Bent, 1998) of rat5 mutant plants (Mysore et al., 2000a). Harvested seeds were spread on B5 medium containing kanamycin and hygromycin for selection of transgenic plants.
Tumorigenesis Assays and Phenotypic Analysis
Arabidopsis plants were grown axenically in baby food jars for 3 to 4 weeks at 25°C. Root segments were cut and infected with Agrobacterium as described previously (Nam et al., 1999). Root segments were separated onto solidified MS medium containing timentin but lacking phytohormones and scored 1 month later for the number and phenotype of the tumors.
Construction of Histone H2A Promoter-gusA Fusion Genes
Approximately 2 kb upstream of each histone H2A genomic DNA, plus sequences encoding the first 10 to 22 amino acids, were PCR amplified with SmaI linker sequences. The primer sequences were as follows: HTA1, 5′-CCCGGGGGATCCAAGAGTTTTTCCACGACCA-3′; HTA2, 5′-CCCGGGCTTGCTACTACGAGAAGTAGACTTC-3′; HTA3, 5′-CCCGGGACCTTTAGTTGTTCCACTGCCGGCG-3′; HTA4, 5′-CCCGGGATCTTTTAGTATATTCGTGTTGCAC-3′; HTA5, 5′-CCCGGGGGTTGTTCCGCTTCCTGCGCCTGTA-3′; HTA6, 5′-CCCGGGAGCTTTCTTCACTTTTCCGGTGGAT-3′, HTA8, 5′-CCCGGGTAGAAGCCCTTTCCCACCTTTACCA-3′, HTA10, 5′-CCCGGGAGATCCGAGTGTTTTACCACGACCC-3′; HTA13, 5′-CCCGGGGATCCGAGAGTTTTGCCGCGACCCG-3′. PCR reactions were performed using the same conditions as for genomic DNA PCR. Amplified products were cloned in the SmaI site of pBluescript II SK. Cloned DNA fragments were excised from the plasmid using appropriate restriction enzymes (HTA1, -2, and -6, XbaI and SmaI; HTA3, -4, -5, -10, and -13, SalI and SmaI; HTA8, SmaI) and cloned into the T-DNA binary vector pBI101.3 as a translational fusion with the gusA gene. The constructions were transferred into A. tumefaciens GV3101 and used to transform Arabidopsis ecotype Ws by a flower dip protocol (Clough and Bent, 1998). Transgenic plants were selected on medium containing kanamycin and used for analysis of promoter activity of each of the HTA genes.
Sequence Alignment and Phylogenetic Analyses
Protein sequences encoded by 13 histone H2A (HTA) genes were downloaded from the ChromDB database (http://www.chromdb.org/) and aligned using ClustalX 1.81 (Thompson et al., 1997). No manual adjustment was required for the alignment. The alignment was used for distance/neighbor-joining phylogenetic analyses performed in PAUP* v4.10b (Swofford, 2003) based on the total number of pairwise character differences. Confidence values for groupings in the tree were assessed by bootstrap resampling (Felsenstein, 1985) with 10,000 repetitions for distance/neighbor joining, and the tree was reviewed in TreeView 1.6.6 (Page, 1996).
Histochemical Analysis of GUS Activity
Detection of GUS expression in plant tissues was performed using X-gluc according to Jefferson et al. (1987) with modifications. Briefly, plant samples were harvested in 90% acetone incubated at room temperature for 20 min. The samples were rinsed once with staining solution (0.2% Triton X-100, 50 mM sodium phosphate buffer, pH 7.2, 2 mM potassium ferrocyanide, and 2 mM potassium ferricyanide). The staining solution was replaced by staining solution containing 2 mM X-gluc, and the plant samples were vacuum infiltrated on ice for 20 min. Following overnight incubation of the samples at 37°C, the plants were processed for sectioning by incubating them successively in 20, 35, and 50% ethanol at room temperature for 30 min each. Ethanol was replaced by FAA (5% formaldehyde, 10% glacial acetic acid, and 50% ethanol) for 30 min, and the ethanol series (80, 90, and 95%) was continued until the plants were finally in 100% ethanol. Plant tissue was infiltrated and embedded in JB-4 resin (Electron Microscopy Sciences) following the manufacturer's instructions. Tissues were sectioned at a thickness of 10 μm using a Sorvall JB-4 microtome and examined under a compound light microscope.
Quantitative Real-Time RT-PCR of HTA Transcripts
Total RNA isolated from roots or whole plants was used for reverse transcription and subsequent PCR amplification. Thirteen primer sets were designed, each with a melting temperature of 55 to 65°C. Each primer was >25 nucleotides and recognized gene-specific regions in the 5′ or 3′ untranslated regions of individual histone H2A genes. The sequences of the primers were as follows: HTA1 forward, 5′-CTCTTTGTGGGTTGTTGTTGTTGAAA-3′; HTA1 reverse, 5′-GAGAGACGTGTTCTATATCATTGTG-3′; HTA2 forward, 5′-CTATCTTGGGTAGTAGAGAGAAATG-3′; HTA2 reverse, 5′-AGCTTTCCCTATCTTCGTAATGAAC-3′; HTA3 forward, 5′-GCCTCTTCAAATTTCCCGATAAACA-3′; HTA3 reverse, 5′-GGAACAGAGAGCCATGTCTATGTTA-3′; HTA4 forward, 5′-ACGGTCTCATTTTATGTTTCATGGT-3′; HTA4 reverse, 5′-GGTTGTTTTGTTGATGAGACTAGTG-3′; HTA5 forward, 5′-TCTGTTCTAAATTTCGAAGAAGACG-3′; HTA5 reverse, 5′-CTGAACTAAGAAGTCTAAGAACCTC-3′; HTA6 forward, 5′-TCAGTAATCGATAACCGTAGCAATG-3′; HTA6 reverse, 5′-CTAGCAACGAAAACTCTAGCAGATT-3′; HTA7 forward, 5′-CTTCATCTAAGATCCGAATATAACC-3′; HTA7 reverse, 5′-CTTTTGGAGCTTTTGAACAATGGAG-3′; HTA8 forward, 5′-CTTTGATTTCGACGACGGATCTTGA-3′; HTA8 reverse, 5′-TAATAGAGACTTAGGACTCAGAGAG-3′; HTA9 forward, 5′-CAATCTCCAAGGATTTTACTGTGA-3′; HTA9 reverse, 5′-CAAAGCGGGTAACTAAAAAAGTCCT-3′; HTA10 forward, 5′-CTGTTTCCCTCTCATCTTTACACAA-3′; HTA10 reverse, 5′-CACAAGAGAGTGGATTTGGTTGATTA-3′; HTA11 forward, 5′-GTCTCAAGATCTAGAAGAAGGAAAC-3′; HTA11 reverse, 5′-GGAAAATGAGTCCAAGACGACAGAA-3′; HTA12 forward, 5′-TCAGAATCAAATTTCTCGTCGTGTC-3′; HTA12 reverse, 5′-CAGTAGGTTATACAGAGGAATGAAG-3′; HTA13 forward, 5′-TTTCTATCTCTCTTCCCAAATCACA-3′; HTA13 reverse, 5′-CAACCACAACACAAATCCCTAATCT-3′. The PCR reaction was performed using a Rotor-Gene real-time thermocycler (Corbett Research). The PCR reaction mixture contained 200 μM each dNTP, 2 mM MgCl2, 10 mM Tris-HCl, pH 9.0, 50 mM KCl, 0.1% Triton X-100, 0.24 μM of the gene-specific primer combination, 1× dilution of the fluorescent dye Sybr-Green I (Molecular Probes), and 2.5 units of Taq2000 DNA polymerase (Stratagene). As internal standards, GAPDH and α-tubulin genes were used. Real-time PCR was performed using the following cycle conditions: 95°C for 2 min; 30 to 32 cycles of 94°C for 20 s, 57°C for 45 s, and 72°C for 2 min; a final 5-min elongation at 72°C. After PCR amplification, the data were analyzed using Corbett Rotor gene software (version 4.4).
Accession Numbers
Designator numbers for the histone H2A genes are listed in Table 1.
Acknowledgments
This work was supported by grants from the National Science Foundation Plant Genome program (9975715 and 9975930), the Biotechnology Research and Development Corporation, the Corporation for Plant Biotechnology Research, and a P30 grant to the Purdue University Cancer Center. We thank the expertise of Debra Sherman of the Life Sciences Microscopy Facility, Purdue University for help with the microscopy.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with policy described in the Instructions for Authors (www.plantcell.org) is: Stanton B. Gelvin (gelvin@bilbo.bio.purdue.edu).
Open Access articles can be viewed online without a subscription.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.039719.
References
- Ahmad, K., and Henikoff, S. (2002). Histone H3 variants specify modes of chromatin assembly. Proc. Natl. Acad. Sci. USA 99 16477–16484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albig, W., and Doenecke, D. (1997). The human histone gene cluster at the D9S105 locus. Hum. Genet. 101 284–294. [DOI] [PubMed] [Google Scholar]
- Andegeko, Y., Moyal, L., Mittelman, L., Tsarfaty, I., Shiloh, Y., and Rotman, G. (2001). Nuclear retention of ATM at sites of DNA double strand breaks. J. Biol. Chem. 276 38224–38230. [DOI] [PubMed] [Google Scholar]
- Ausio, J., and Abbott, D.W. (2002). The many tales of a tail: Carboxyl-terminal tail heterogeneity specializes histone H2A variants for defined chromatin function. Biochemistry 41 5945–5949. [DOI] [PubMed] [Google Scholar]
- Baker, B., Zambryski, P., Staskawicz, B., and Dinesh-Kumar, S.P. (1997). Signaling in plant-microbe interactions. Science 276 726–733. [DOI] [PubMed] [Google Scholar]
- Beisel, C., Imhof, A., Greene, J., Kremmer, E., and Sauer, F. (2002). Histone methylation by the Drosophila epigenetic transcriptional regulator Ash1. Nature 419 857–862. [DOI] [PubMed] [Google Scholar]
- Brandstadter, J., Rossbach, C., and Theres, K. (1994). The pattern of histone H4 expression in tomato shoot apex changes during development. Planta 192 69–74. [DOI] [PubMed] [Google Scholar]
- Brown, D.T. (2001). Histone variants: Are they functionally heterogeneous? Genome Biol. 2 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin, S.A. (2000). Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 25 169–193. [DOI] [PubMed] [Google Scholar]
- Callard, D., and Mazzolini, L. (1997). Identification of proliferation-induced gene in Arabidopsis thaliana: Characterization of a new member of the highly evolutionarily conserved histone H2A.F/Z variant subfamily. Plant Physiol. 115 1385–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celeste, A., et al. (2002). Genomic instability in mice lacking histone H2AX. Science 296 922–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaubet-Gigot, N., Kapros, T., Flenet, M., Kahn, K., Gigot, C., and Waterborg, J.H. (2001). Tissue-dependent enhancement of transgene expression by introns of replacement histone H3 genes of Arabidopsis. Plant Mol. Biol. 45 17–30. [DOI] [PubMed] [Google Scholar]
- Chen, H.T., Bhandoola, A., Difilippantonio, M.J., Zhu, J., Brown, M.J., Tai, X., Rogakou, E.P., Brotz, T.M., Booner, W.M., Reid, T., and Nussenzweig, A. (2000). Response to RAG-mediated V(D)J cleavage by NBS1 and γH2AX. Science 290 1962–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Costanzi, C., Stein, P., Worrad, D.M., Schultz, R.M., and Pehrson, J.R. (2000). Histone macroH2A1 is concentrated in the inactive X chromosome of female preimplantation mouse embryos. Development 127 2283–2289. [DOI] [PubMed] [Google Scholar]
- Dalton, S., Robins, A.J., Harvey, R.P., and Wells, J.R.E. (1989). Transcription from the intron-containing chicken H2A.F gene is not S-phase regulated. Nucleic Acids Res. 16 10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs, J.A., Lowndes, N.F., and Jackson, S.P. (2000). A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 408 1001–1004. [DOI] [PubMed] [Google Scholar]
- Fan, J.Y., Gordon, F., Luger, K., Hansen, J.C., and Tremethick, D.J. (2002). The essential histone variant H2A.Z regulates the equilibrium between different chromatin conformational states. Nat. Struct. Biol. 9 172–176. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J. (1985). Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39 783–791. [DOI] [PubMed] [Google Scholar]
- Ferreira, P.C.G., Hemerly, A.S., de Almeida Engler, J., Van Montagu, M., Engler, G., and Inze, D. (1994). Developmental expression of the Arabidopsis cyclin gene cyc1At. Plant Cell 6 1763–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransz, F.F., and de Jong, J.H. (2002). Chromatin dynamics in plants. Curr. Opin. Plant Biol. 5 560–567. [DOI] [PubMed] [Google Scholar]
- Hatch, C.L., and Bonner, W.M. (1990). The human histone H2A.Z gene: Sequence and regulation. J. Biol. Chem. 265 15211–15218. [PubMed] [Google Scholar]
- Higo, K., Ugawa, Y., Iwamoto, M., and Korenaga, T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirochika, H., Okamoto, H., and Kakutani, T. (2000). Silencing of retrotransposons in Arabidopsis and reactivation by the ddm1 mutation. Plant Cell 12 357–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh, G.H., Matsuura, Y., Meshi, T., and Iwabuchi, M. (1995). Differential expression of the two types of histone H2A genes in wheat. Biochim. Biophys. Acta 1261 155–160. [DOI] [PubMed] [Google Scholar]
- Huh, G.H., Nakayama, T., Meshi, T., and Iwabuchi, M. (1997). Structural characteristics of two wheat histone H2A genes encoding distinct types of variants and functional differences in their promoter activity. Plant Mol. Biol. 33 791–802. [DOI] [PubMed] [Google Scholar]
- Iouzalen, N., Moreau, J., and Mechali, M. (1996). H2A.ZI, a new variant histone expressed during Xenopus early development exhibits several distinct features from the core histone H2A. Nucleic Acids Res. 24 3947–3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, J.D., and Gorovsky, M.A. (2000). Histone H2A.Z has a conserved function that is distinct from that of the major H2A sequence variants. Nucleic Acids Res. 28 3811–3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: β- Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazin, V., Blake, T., and Schoemaker, R.C. (1996). Organization of the histone H3 gene in soybean, barley and wheat. Mol. Gen. Genet. 250 137–147. [DOI] [PubMed] [Google Scholar]
- Kandasamy, M.K., McKinney, E.C., and Meagher, R.B. (2002). Plant profilin isovariants are distinctly regulated in vegetative and reproductive tissues. Cell Motil. Cytoskeleton 52 22–32. [DOI] [PubMed] [Google Scholar]
- Koncz, C., and Schell, J. (1986). The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204 383–396. [Google Scholar]
- Koning, A.J., Tanimoto, E.Y., Kiehne, K., Rost, T., and Comai, L. (1991). Cell-specific expression of plant histone H2A genes. Plant Cell 3 657–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouchi, H., Sekine, M., and Hata, S. (1995). Distinct classes of mitotic cyclins are differentially expressed in the soybean shoot apex during the cell cycle. Plant Cell 7 1143–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach, T.J., Mazzeo, M., Chotkowski, H.L., Madigan, J.P., Wotring, M.G., and Glaser, R.L. (2000). Histone H2A.Z is widely but nonrandomly distributed in chromosomes of Drosophila melanogaster. J. Biol. Chem. 275 23267–23272. [DOI] [PubMed] [Google Scholar]
- Lichtenstein, C., and Draper, J. (1986). Genetic engineering of plants. In DNA Cloning: A Practical Approach, Vol. 2, D.M. Glover, ed (Oxford, UK: IRL Press), pp. 67–119.
- Liu, X., Li, B., and Gorovsky, M.A. (1996). Essential and nonessential histone H2A variants in Tetrahymena themophila. Mol. Cell. Biol. 16 4305–4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevaiah, S.K., Turner, J.M., Baudat, F., Rogakou, E.P., de Boer, P., Blanco-Rodriguez, J., Jasin, M., Keeney, S., Bonner, W.M., and Burgoyne, P.S. (2001). Recombinational DNA double-strand breaks in mice precede synapsis. Nat. Genet. 27 271–276. [DOI] [PubMed] [Google Scholar]
- Meneghini, M.D., Wu, M., and Madhani, H.D. (2003). Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112 725–736. [DOI] [PubMed] [Google Scholar]
- Milligan, L., Forne, T., Antoine, E., Weber, M., Hemonnot, B., Dandolo, L., Brunel, C., and Cathala, G. (2002). Turnover of primary transcripts is a major step in the regulation of mouse H19 gene expression. EMBO Rep. 3 774–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi, G., Shen, X., Landry, J., Wu, W.H., Sen, S., and Wu, C. (2004). ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303 343–348. [DOI] [PubMed] [Google Scholar]
- Mysore, K.S., Kumar, C.T.R., and Gelvin, S.B. (2000. a). Arabidopsis ecotypes and mutants that are recalcitrant to Agrobacterium root transformation are susceptible to germ-line transformation. Plant J. 21 9–16. [DOI] [PubMed] [Google Scholar]
- Mysore, K.S., Nam, J., and Gelvin, S.B. (2000. b). An Arabidopsis histone H2A mutant is deficient in Agrobacterium T-DNA integration. Proc. Natl. Acad. Sci. USA 97 948–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam, J., Mysore, K.S., Zheng, C., Knue, M.K., Matthysse, A.G., and Gelvin, S.B. (1999). Identification of T-DNA tagged Arabidopsis mutants that are resistant to transformation by Agrobacterium. Mol. Gen. Genet. 261 429–438. [DOI] [PubMed] [Google Scholar]
- Narasimhulu, S.B., Deng, X.-B., Sarria, R., and Gelvin, S.B. (1996). Early transcription of Agrobacterium T-DNA genes in tobacco and maize. Plant Cell 8 873–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, D.M., Ye, X., Hall, C., Santos, H., Ma, T., Kao, G.D., Yen, T.J., Harper, J.W., and Adams, P.D. (2002). Coupling of DNA synthesis and histone synthesis in S phase independent of cyclin/cdk2 activity. Mol. Cell. Biol. 22 7459–7472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiuchi, T., Shinshi, H., and Suzuki, K. (2004). Rapid and transient activation of transcription of the ERF3 gene by wounding in tobacco leaves: Possible involvement of NtWRKYs and autorepression. J. Biol. Chem. 279 55355–55361. [DOI] [PubMed] [Google Scholar]
- Oswald, F., Dobner, T., and Lipp, M. (1996). The E2F transcription factor activates a replication-dependent human H2A gene in early S phase of the cell cycle. Mol. Cell. Biol. 16 1889–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, R.D.M. (1996). TREEVIEW: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12 357–358. [DOI] [PubMed] [Google Scholar]
- Paull, T.T., Rogakou, E.P., Yamazaki, V., Kirchgessner, C.U., Gellert, M., and Bonner, W.M. (2000). A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10 886–895. [DOI] [PubMed] [Google Scholar]
- Redon, C., Pilch, D., Rogakou, E., Sedelnikova, O., Newrock, K., and Bonner, W.M. (2002). Histone H2A variants H2AX and H2AZ. Curr. Opin. Genet. Dev. 12 162–169. [DOI] [PubMed] [Google Scholar]
- Reichheld, J.P., Gigot, C., and Gigot, N.C. (1998). Multilevel regulation of histone gene expression during the cell cycle in tobacco cells. Nucleic Acids Res. 26 3255–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, J.D., and Allis, C.D. (2001). Histone methylation versus histone acetylation: New insights into epigenetic regulation. Curr. Opin. Cell Biol. 13 263–273. [DOI] [PubMed] [Google Scholar]
- Rogakou, E.P., Nieves-Neira, W., Boon, C., Pommier, Y., and Bonner, W.M. (2000). Initiation of DNA fragmentation during apoptosis induces phosphorylation of H2AX histone at serine 139. J. Biol. Chem. 275 271–276. [DOI] [PubMed] [Google Scholar]
- Rogakou, E.P., Pilch, D.R., Orr, A.H., Ivanova, V.S., and Bonner, W.M. (1998). DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273 5858–5868. [DOI] [PubMed] [Google Scholar]
- Roth, S.Y., and Allis, C.D. (1992). Chromatin condensation: Does histone H1 dephosphorylation play a role? Trends Biochem. Sci. 17 93–98. [DOI] [PubMed] [Google Scholar]
- Santisteban, M.S., Kalashnikova, T., and Smith, M.M. (2000). Histone H2A.Z regulates transcription and is partially redundant with nucleosome remodeling complexes. Cell 103 411–422. [DOI] [PubMed] [Google Scholar]
- Smith, M.M. (2002). Centromeres and variant histones: What, where, when and why? Curr. Opin. Cell Biol. 14 279–285. [DOI] [PubMed] [Google Scholar]
- Sundas, A., Tandre, K., Kvarnheden, A., and Engstrom, P. (1993). cDNA sequence and expression of an intron-containing histone H2A gene from Norway spruce, Picea abies. Plant Mol. Biol. 21 595–605. [DOI] [PubMed] [Google Scholar]
- Suzuki, M. (1991). The DNA-binding motif, SPKK, and its variants. Nucleic Acids Mol. Biol. 5 126–140. [Google Scholar]
- Swofford, D.L. (2003). PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. (Sunderland, MA: Sinauer Associates).
- Takami, Y., Takeda, S., and Nakayama, T. (1997). An approximately half set of histone genes is enough for cell proliferation and a lack of several histone variants causes protein pattern changes in the DT40 chicken B cell line. J. Mol. Biol. 265 394–408. [DOI] [PubMed] [Google Scholar]
- Talbert, P.B., Masuelli, R., Tyagi, A.P., Comai, L., and Henikoff, S. (2002). Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell 14 1053–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto, E.Y., Rost, T.L., and Comai, L. (1993). DNA replication-dependent histone H2A mRNA expression in pea root tips. Plant Physiol. 103 1291–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka, K.O., Toyooka, S., Maitra, A., Feng, Q., Kiviat, N.C., Smith, A., Minna, J.D., Ashfaq, R., and Gazdar, A.F. (2002). Establishment and validation of real-time polymerase chain reaction method for CDH1 promoter methylation. Am. J. Pathol. 161 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukuda, T., Fleming, A.B., Nickoloff, J.A., and Osley, M.A. (2005). Chromatin remodeling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature 438 379–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Daal, A., and Elgin, S.C. (1992). A histone variant, H2Av, is essential in Drosophila melanogaster. Mol. Biol. Cell 3 593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel, K.J.P.T., van Esch, R.J., Barendse, G.W.M., and Wullems, G.J. (1999). Isolation and molecular characterization of gibberellin-regulated H1 and H2B histone cDNAs in the leaf of the gibberellin-deficient tomato. Plant Mol. Biol. 39 883–890. [DOI] [PubMed] [Google Scholar]
- Verbsky, M.L., and Richards, E.J. (2001). Chromatin remodeling in plants. Curr. Opin. Plant Biol. 4 494–500. [DOI] [PubMed] [Google Scholar]
- Volpe, T.A., Kidner, C., Hall, I.M., Teng, G., Grewal, S.I., and Martienssen, R.A. (2002). Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297 1833–1837. [DOI] [PubMed] [Google Scholar]
- Vongs, A., Kakutani, T., Martienssen, R.A., and Richards, E.J. (1993). Arabidopsis thaliana DNA methylation mutants. Science 260 1926–1928. [DOI] [PubMed] [Google Scholar]
- Waterborg, J.H., and Robertson, A.J. (1996). Common features of analogous replacement histone H3 genes in animal and plants. J. Mol. Evol. 43 194–206. [DOI] [PubMed] [Google Scholar]
- West, M.H., and Bonner, W.M. (1980). Histone 2A, a heteromorphous family of eight protein species. Biochemistry 19 3238–3245. [DOI] [PubMed] [Google Scholar]
- Wu, R.S., Tsai, S., and Bonner, W.M. (1982). Patterns of histone variant synthesis can distinguish G0 from G1 cells. Cell 31 367–374. [DOI] [PubMed] [Google Scholar]
- Xu, H., Swoboda, I., Bhalla, P.L., and Singh, M.B. (1999). Male gametic cell-specific expression of H2A and H3 histone genes. Plant Mol. Biol. 39 607–614. [DOI] [PubMed] [Google Scholar]
- Yi, H., Mysore, K.S., and Gelvin, S.B. (2002). Expression of the Arabidopsis histone H2A-1 gene correlates with susceptibility to Agrobacterium transformation. Plant J. 32 285–298. [DOI] [PubMed] [Google Scholar]
- Zhao, J., Kennedy, B.K., Lawrence, B.D., Barbie, D.A., Matera, G., Fletcher, J.A., and Harlow, E. (2000). NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 14 2283–2297. [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y., et al. (2003). Identification of Arabidopsis rat mutants. Plant Physiol 132 494–505. [DOI] [PMC free article] [PubMed] [Google Scholar]