Abstract
Both antibodies and T cells contribute to immunity against influenza virus infection. However, the generation of strong Th1 immunity is crucial for viral clearance. Interestingly, we found that human dendritic cells (DCs) infected with influenza A virus have lower allospecific Th1-cell stimulatory abilities than DCs activated by other stimuli, such as lipopolysaccharide and Newcastle disease virus infection. This weak stimulatory activity correlates with a suboptimal maturation of the DCs following infection with influenza A virus. We next investigated whether the influenza A virus NS1 protein could be responsible for the low levels of DC maturation after influenza virus infection. The NS1 protein is an important virulence factor associated with the suppression of innate immunity via the inhibition of type I interferon (IFN) production in infected cells. Using recombinant influenza and Newcastle disease viruses, with or without the NS1 gene from influenza virus, we found that the induction of a genetic program underlying DC maturation, migration, and T-cell stimulatory activity is specifically suppressed by the expression of the NS1 protein. Among the genes affected by NS1 are those coding for macrophage inflammatory protein 1β, interleukin-12 p35 (IL-12 p35), IL-23 p19, RANTES, IL-8, IFN-α/β, and CCR7. These results indicate that the influenza A virus NS1 protein is a bifunctional viral immunosuppressor which inhibits innate immunity by preventing type I IFN release and inhibits adaptive immunity by attenuating human DC maturation and the capacity of DCs to induce T-cell responses. Our observations also support the potential use of NS1 mutant influenza viruses as live attenuated influenza virus vaccines.
Influenza A virus is an important human pathogen that causes worldwide epidemics yearly and pandemics sporadically. Protection against reinfection relies on the presence of neutralizing antibodies to influenza virus in the host, while clearance of infection is mediated by cellular immunity. In order to clear influenza virus infection from the lungs, it is important to generate Th1 immunity against the virus (18). The optimal Th1 response consists of virus-specific gamma interferon (IFN-γ)-secreting CD4 T cells and cytotoxic CD8 T cells that lyse virus-infected cells (28). Dendritic cells (DCs), the most efficient antigen-presenting cells able to initiate primary immune responses (27), survey the body, and upon contact with particular pathogens, such as viruses or bacteria, undergo maturation and migrate to lymph nodes, where they present pathogen-specific antigens to T cells (7). The phenotypic changes that occur in maturation include the upregulation of major histocompatibility complex (MHC) class II and costimulatory molecules and the release of proinflammatory cytokines and chemokines that enhance DCs' ability to stimulate T cells, leading to the initiation of adaptive immune responses specific for the infecting pathogen (2, 3, 30). In addition to their critical role in initiating adaptive immune responses, DCs contribute to the antiviral innate immune system by secreting IFN-α/β, a powerful antiviral cytokine, in response to viral infection.
Employing human monocyte-derived DCs, we conducted a comprehensive analysis of human DC activation after influenza A virus infection by reverse transcription, followed by quantitative real-time PCR (qRT-PCR), using an extensive panel of genes associated with DC maturation and migration as well as genes involved in the IFN-α/β pathway. Additionally, we analyzed the functional maturation of human DCs by their ability to secrete cytokines and chemokines following exposure to virus, and we examined their T-cell stimulatory capacity. We also investigated the capacity of influenza A virus-infected human DCs to stimulate naïve CD4 T cells. Previous work from our group suggested a link between the activation of the IFN-α/β pathway and DC maturation (22). Thus, we hypothesized that the IFN-α/β antagonist protein of influenza A virus, the NS1 protein (16), might also be responsible for attenuation of DC maturation following influenza virus infection. Using recombinant viruses expressing or not expressing the NS1 protein, we assessed the impact of NS1 expression on human DCs exposed to viruses with respect to the ability to undergo functional maturation as well as to induce the IFN-α/β system. Our results demonstrate that the NS1 protein prevents not only the induction of IFN-α/β by human myeloid DCs but also the induction of a transcriptional program associated with DC maturation, resulting in suboptimal stimulation of T cells. In contrast, viruses lacking the NS1 gene are potent stimulators of human DCs and therefore might be potent immunogens to be used in live attenuated vaccine approaches.
MATERIALS AND METHODS
Viruses and cells.
Recombinant Newcastle disease viruses (NDVs) NDVB1 and NDVB1-NS1 were generated from the B1 Hitchner avian vaccine strain as previously described (34). Influenza virus DeltaNS1 was generated from influenza virus A/PR/8/34 (PR8) as previously described (10).
Influenza viruses PR8 (H1N1), A/Texas/91 (Texas) (H1N1), and A/Moscow/99 (Moscow) (H3N2) and recombinant NDVs were grown in 9-day-old embryonated chicken eggs (SPAFAS; Charles River Laboratories). The influenza virus DeltaNS1 was grown in 6-day-old embryonated chicken eggs (10). All influenza viruses were titrated on MDCK cells by detection of hemagglutination (HA) activity in the supernatants after 48 h of infection, as previously described (13). The PR8 and DeltaNS1 viruses were also titrated by immunoflourescence, using a monoclonal antibody, PY102, specific for the HA protein (obtained from Jerome L. Schulman). NDVs were titrated by immunofluorescence of Vero cells at 24 h postinfection, using the monoclonal antibody 7B1, which is specific for the NDV protein HN (5 μg/ml) (Mount Sinai Hybridoma Shared Research Facility). All virus infections were performed in infection medium (Dulbecco's modified Eagle's medium, 0.35% bovine serum albumin, 0.12% NaHCO3, 100 μg/ml penicillin-streptomycin). For influenza virus titrations, 2.5 μg/ml trypsin was included in the infection medium.
MDCK and Vero cells were grown in tissue culture medium (Dulbecco's modified Eagle's medium [Invitrogen] with 10% fetal calf serum [HyClone], 1 mM sodium pyruvate [Invitrogen], 2 mM l-glutamine [Invitrogen], and 50 μg/ml gentamicin [Invitrogen]). All cells were grown at 37°C in 7% CO2.
Isolation and culture of human DCs.
Peripheral blood mononuclear cells were isolated by Ficoll density gradient centrifugation (Histopaque; Sigma Aldrich) from buffy coats of healthy human donors (Mount Sinai Blood Donor Center and New York Blood Center). CD14+ cells were immunomagnetically purified using anti-human CD14 antibody-labeled magnetic beads and iron-based Midimacs LS columns (Miltenyi Biotec). After elution from the columns, cells were plated (0.7 × 106 cells/ml) in DC medium (RPMI [Invitrogen], 10% fetal calf serum [HyClone], 100 units/ml of penicillin, and 100 μg/ml streptomycin [Invitrogen]) supplemented with 500 U/ml human granulocyte-macrophage colony-stimulating factor (Peprotech) and 1,000 U/ml human interleukin-4 (IL-4; Peprotech) and incubated for 5 to 6 days at 37°C.
Infection and treatment of DCs.
After 5 to 6 days in culture, cells were infected with NDVs or influenza viruses at a multiplicity of infection (MOI) of 0.5 for 40 min in serum-free RPMI or treated with lipopolysaccharide (LPS; from Escherichia coli) at 100 ng/ml (Sigma Aldrich) or with 3,000 U/ml IFN-β (PBL). Cells were then plated in DC medium at 1 × 106 cells/ml for different time periods, depending on the experiment.
Capture ELISAs.
Capture enzyme-linked immunosorbent assays (ELISAs) for IL-1β, IL-6, tumor necrosis factor alpha (TNF-α), macrophage inflammatory protein 1β (MIP1β), IP10 (all from R & D Systems), IFN-α, and IFN-β (Biosource) were used according to the manufacturers' instructions to quantify the cytokines and chemokines in the DC supernatants. Plates were read in an ELISA reader from Biotek Instruments. ELISAs for RANTES and IL-8 (Upstate) were performed as part of a multiplex assay following the manufacturer's protocol. Plates were read in a Luminex plate reader, and data were analyzed using software from Applied Cytometry Systems.
Flow cytometry.
Cells were stained with fluorescein isothiocyanate-linked CD83 or HLA-DR and phycoerythrin-linked CD11c, CD80, or CD86 according to the manufacturer's instructions (Beckman Coulter or BD-Pharmingen), and the expression of each marker was determined by flow cytometry after gating on the lineage-specific marker, using an Epics XL Expo 32 or FC500 flow cytometer from Beckman Coulter. Data were analyzed using Flowjo software.
RNA extraction and generation of cDNAs from human DCs.
Samples of 1 × 106 to 2.5 × 106 DCs differentially treated according to the experimental protocol were pelleted, and RNAs were isolated and treated with DNase by using an Absolutely RNA RT-PCR miniprep kit (Stratagene). RNAs were quantified using a Nanodrop spectrophotometer (Nanodrop Technologies). The yields of RNA were approximately 50 to 100 μg/sample.
Quantitative real-time PCR.
qRT-PCR was performed by using a previously published SYBR green protocol with an ABI7900 HT thermal cycler (47). Each transcript in each sample was assayed two times, and the mean cycle threshold was used to calculate the x-fold change and control changes for each gene. Four housekeeping genes were used for global normalization in each experiment (actin, Rps11, glyceraldehyde-3-phosphate dehydrogenase, and tubulin genes) (17). Data validity by modeling of reaction efficiencies and analysis of measurement precision was determined as described previously (47). Primer sequences are listed in Table S1 in the supplemental material.
Alloantigen stimulation of naïve CD4 T cells and PBLs by DCs.
DCs that either were not treated or were infected with influenza virus or NDV were mixed together with allogeneic naive CD4 T cells (naïve CD4 isolation kit from Miltenyi Biotec) or PBLs from buffy coats and cultured for 4 days in 96-well plates at a ratio of 1:1 (106 DCs/ml to 106 PBLs/ml) or 1:5 (2 × 105 DCs/ml to 106 naïve CD4 T cells/ml). Supernatants from the cocultures were tested by ELISA for IFN-γ release at different times of culture (R&D Systems).
Topic-defined PIQOR immunology microarrays.
Samples of 1 × 106 to 3 × 106 DCs were infected with NDVB1 or NDVB1-NS1 or mock infected for 18 h as described above. Samples were shipped to Memorec (a Miltenyi Biotec company), and all the procedures for microarrays were performed according to their standard protocols. Briefly, RNAs were extracted from the cells (NucleoSpin RNA II; Macherey-Nagel) and amplified. Fluorescence-labeled probes were hybridized to topic-defined PIQOR immunology microarrays (human antisense) and subjected to overnight hybridization using a hybridization station.
RESULTS
Influenza virus-infected DCs induce lower levels of T-cell activation than do DCs treated with NDV or LPS.
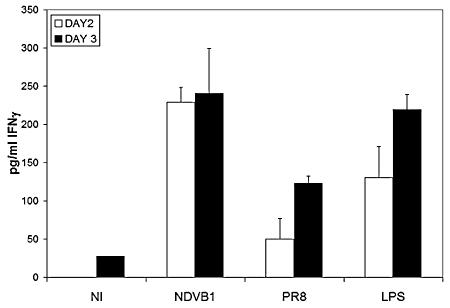
Mature DCs are much better stimulators of naïve T cells than are immature DCs. While T-cell proliferation can be associated with induction of immunity, the secretion of IFN-γ by T cells is the hallmark of Th1 immunity. When influenza virus-infected DCs were used to stimulate allogeneic purified naïve CD4 T cells, they induced considerably lower levels of IFN-γ release than did LPS-treated or NDV-infected DCs (Fig. 1). In contrast, all of the DCs induced comparable levels of proliferation of allogeneic naïve CD4 T cells (data not shown). Similar results were obtained with allogeneic PBLs (data not shown). Thus, there is impairment in the ability of human DCs infected with influenza viruses to prime T cells towards the Th1 phenotype required for effective antiviral adaptive immune responses.
FIG. 1.
Allospecific responses of purified CD4 T cells primed with differentially treated DCs. Human DCs were infected with PR8 influenza virus or Newcastle disease virus (NDVB1), left untreated (NI), or treated with 100 ng/ml of LPS for 45 min. Differentially treated DCs were then incubated with allogeneic naïve CD4 T cells for 3 days. Supernatants from the cocultures were harvested at different days of culture and tested by ELISA for the release of IFN-γ into the supernatants of the cocultures. Error bars are for triplicate samples. These results are representative of three independent experiments.
Influenza virus induces less maturation of human DCs than does NDV or LPS.
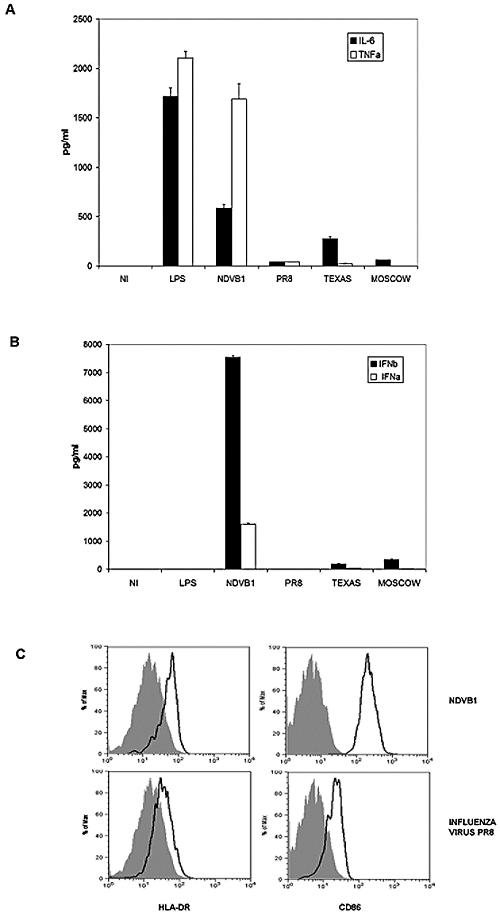
Myeloid DCs are present in the lungs, and after interaction with pathogens or their structural elements, they undergo a maturation process that culminates in their migration to the lymph nodes and the priming of naïve T cells to initiate the appropriate immune response. We tested the ability of influenza viruses, including PR8 (A/PR/8/34) and the more recent human isolates Texas (A/Texas/91) and Moscow (A/Moscow/99), to induce the maturation of human DCs in comparison to the response following infection with NDV (NDVB1 strain) or LPS treatment. An important aspect of virus-induced DC maturation is the release of proinflammatory cytokines, including IFN-α/β. Figure 2A shows the levels of TNF-α and IL-6, two representative proinflammatory cytokines, released into the culture supernatants of DCs treated with LPS or infected with either NDVB1 or influenza virus PR8, Texas, or Moscow. There was a significant reduction in the levels of TNF-α and IL-6 released by DCs infected with the influenza viruses compared to NDV- or LPS-treated cells, as measured by ELISA. Additionally, NDVB1 induced high levels of IFN-α and IFN-β release from DCs, while influenza viruses induced almost undetectable amounts (Fig. 2B). This was probably due to the expression by influenza viruses of the NS1 protein, a known antagonist of IFN-α/β production (16). In our experiments, LPS induced undetectable levels of IFN-α/β. DCs were also tested by flow cytometry for the upregulation of MHC class II and costimulatory molecules. Influenza virus induced less expression of MHC class II and costimulatory molecules on DCs than did NDVB1 (Fig. 2C).
FIG. 2.
Influenza virus and NDV induce different degrees of maturation in human DCs. Human DCs were infected on day 5 or 6 of culture with influenza virus (PR8, Texas, or Moscow) or NDV (NDVB1) at an MOI of 0.5. Supernatants from infected cells (at 18 h postinfection) were tested by ELISA for the release of (A) TNF-α and IL-6 and (B) IFN-α and IFN-β. (C) After infection, cells were incubated at 37°C for 18 h and stained for flow cytometry analysis of the expression of HLA-DR (left panels) or CD86 (right panels). Filled histograms represent uninfected cells, and open histograms represent infected cells (with NDVB1 [top panels] and influenza virus PR8 [bottom panels]). Concentrations are indicated in pg/ml. Error bars represent the standard errors of triplicate samples, and data are representative of at least three independent experiments with cells from different donors.
Virus infection induces a distinct transcriptional response in human myeloid DCs compared to that after IFN-β treatment.
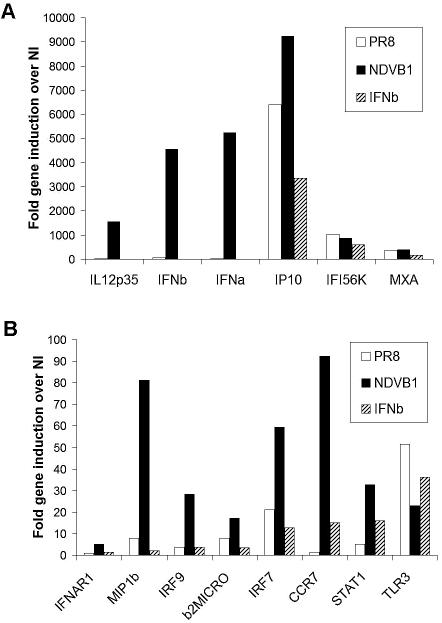
Previous data showed a correlation between the ability of a virus to induce high levels of IFN-α/β and its ability to trigger strong DC maturation (22). Since IFN-α/β released by the DCs could by itself be responsible for the enhanced maturation of NDV-infected DCs, we tested the effects of IFN-β treatment of human DCs compared to infection with NDVB1 (a virus without a gene product able to block IFN-α/β production in human DCs) or influenza virus PR8 (a virus with a known inhibitor of IFN-α/β production, NS1). Gene activation in DCs was determined by using qRT-PCR. Figure 3A shows genes upregulated at high levels (>100-fold induction), and Fig. 3B shows genes upregulated at moderate levels (up to 100-fold induction) compared with those in mock-infected DCs. Clearly, NDVB1 infection induced the upregulation of numerous genes in the DCs implicated in the generation of innate and adaptive immune responses compared to influenza virus PR8 infection and IFN-β treatment. NDVB1 infection induced the upregulation of IFN-β-responsive genes (such as those encoding IFI56K, MXA, IP-10, IRF7, STAT1, and CCR7) as well as genes that are not inducible by IFN-β (those encoding IL-12 p35, IFN-β, and MIP1β). Some genes, such as those for CCR7 and IRF7, are induced to moderate levels by IFN-β but to much higher levels by virus infection. Thus, the enhanced activation induced by NDVB1 was not solely the result of binding of released IFN-β.
FIG. 3.
Infection of human DCs with influenza virus PR8 resulted in low expression of genes involved in DC maturation compared with that in NDV infection. Human DCs were either infected with NDVB1 or influenza virus PR8 at an MOI of 0.5 or treated with 5,000 U/ml of IFN-β or 500 ng/ml of LPS. At different times after treatment, RNAs were isolated and used to generate cDNAs to test increases in specific gene expression. (A) Genes upregulated at high levels 24 h after treatment; (B) genes upregulated at moderate levels 24 h after treatment. The data are representative of at least three independent experiments with cells from different donors and are given as x-fold increases in gene expression over that in uninfected cells (NI).
The NS1 protein of influenza virus blocks the production of IFN-α/β and other proinflammatory cytokines by influenza virus-infected human DCs.
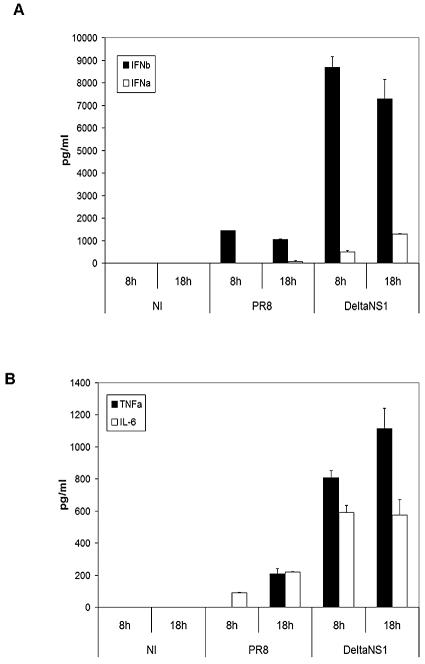
The NS1 protein of influenza virus has been shown to prevent the transcriptional activation of the IFN-β gene. An influenza virus lacking the NS1 protein, but otherwise identical to PR8 (DeltaNS1), was previously engineered by reverse genetics (16). DCs were infected with the PR8 and DeltaNS1 influenza viruses to study the role of the NS1 protein in their maturation. As expected, PR8 influenza virus, which expresses the NS1 protein, showed a dramatic reduction in IFN-α/β release from infected DCs compared to the same virus lacking the NS1 protein (DeltaNS1) (Fig. 4A). Additionally, PR8 virus-infected DCs showed a reduced release of the proinflammatory cytokines TNF-α and IL-6 (Fig. 4B) and less expression of surface MHC class II and costimulatory molecules (data not shown), suggesting that the NS1 protein inhibits DC maturation.
FIG. 4.
The NS1 protein of influenza virus abolishes the release of proinflammatory cytokines and IFN-α/β by human DCs after infection with influenza virus. Human DCs were infected with influenza virus PR8 or DeltaNS1 or mock infected. Supernatants from infected DCs were tested by ELISA for the release of (A) IFN-α (white bars) and IFN-β (black bars) or (B) TNF-α (black bars) and IL-6 (white bars) at different times after infection. Error bars are for triplicate samples. Data are representative of at least three independent experiments.
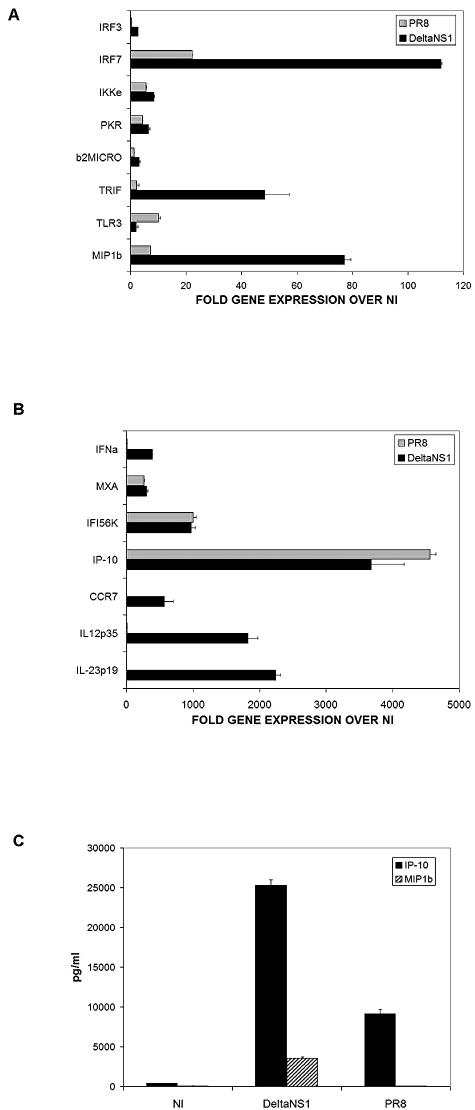
Inhibitory effect of NS1 protein on DCs is specific for genes important for DC maturation and migration.
We selected a panel of more than 60 genes involved in DC maturation, Toll-like receptor signaling, and the IFN-α/β pathway to be used in a high-throughput transcriptome qRT-PCR of RNAs isolated from differentially treated human DCs. This approach revealed that the inhibitory effect of the NS1 protein from influenza virus extended to several groups of genes, including cytokine, chemokine, chemokine receptor, and IFN-related genes. Some of these results are shown in Fig. 5. DCs infected with influenza viruses PR8 and DeltaNS1 were analyzed for their patterns of gene expression compared to that of uninfected DCs (Fig. 5A shows genes upregulated <150-fold, and Fig. 5B shows genes upregulated >150-fold over those in uninfected cells). Included are genes coding for cytokines involved in T-cell activation (IL-12 p35 and IL-23 p19), chemokines and chemokine receptors involved in DC migration (MIP1β and CCR7), and antiviral cytokine genes (IFN-α and IFN-β) that were highly affected by the presence of the NS1 protein. Several genes were moderately affected or not affected by the NS1 protein (Fig. 5A and B). The supernatants from DCs after infection were also tested by ELISA for the presence of the chemokines MIP1β and IP-10 (Fig. 5C), thus confirming the qRT-PCR data. The inhibition of the induction of MIP1β and CCR7 would be expected to diminish the capacity of the DCs to migrate to the lymph nodes and initiate antiviral (Th1) immunity (37).
FIG. 5.
The NS1 protein of influenza virus downregulates the expression of specific genes involved in DC maturation. Human DCs were infected with influenza viruses PR8 and DeltaNS1 for 18 h. (A and B) RNAs were isolated from the cells and used to generate cDNAs to perform qRT-PCR for genes involved in DC maturation. Values indicate changes in gene expression in DCs infected by viruses compared to that in uninfected DCs (NI). Error bars represent standard deviations for triplicate samples. Results are representative of three independent experiments with cells from three different donors. (C) At 18 h postinfection, supernatants from infected DCs were analyzed by ELISA for the presence of IP-10 and MIP1β. Error bars are for triplicate samples. Results are representative of three independent experiments with cells from three different donors.
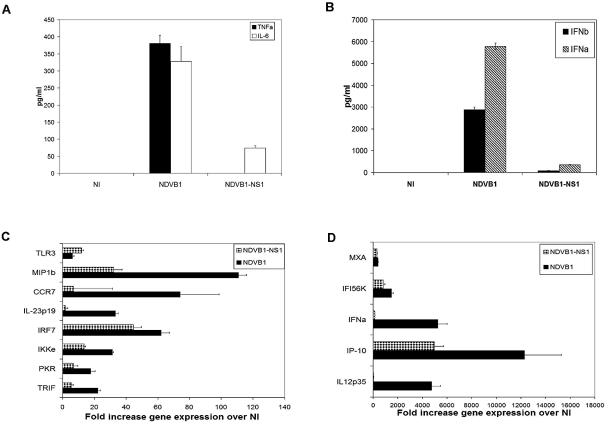
Human DC maturation induced by NDV infection is attenuated by the NS1 protein.
In order to further investigate the inhibitory effects of the NS1 protein on DC maturation, we used a chimeric NDV virus containing the NS1 gene of the influenza virus PR8, designated NDVB1-NS1 (34). This recombinant virus allowed us to study whether the effects of NS1 expression were specific for influenza virus or would function in another viral context. Human DCs were infected with the recombinant viruses NDVB1 and NDVB1-NS1, and their patterns of maturation were studied. Figure 6 shows the effects of NS1 in the context of NDV on cytokine production and the expression of genes related to DC maturation and function. Similar to data obtained from DCs infected with the influenza viruses PR8 and DeltaNS1, the maturation of DCs infected with the NDV virus expressing the NS1 protein of influenza virus (NDVB1-NS1) was clearly reduced. The presence of the NS1 protein had a dramatic inhibitory effect on the ability of DCs to express and secrete proinflammatory cytokines (Fig. 6A) and IFN-α/β (Fig. 6B and D) and on the expression of genes such as those for CCR7, MIP1β, IL-12 p35, and IL-23 p19, which are important in the initiation of antiviral immunity (5, 26, 44), (Fig. 6C and D). Overall, the NS1 protein showed the same inhibitory effect on IFN-α/β production and DC maturation regardless of the background virus.
FIG. 6.
The NS1 protein of influenza virus downregulates DC maturation when expressed in an NDV background. Human DCs were infected with NDVB1 and NDVB1-NS1 for 18 h. Supernatants from infected DCs were analyzed by ELISA for the presence of (A) TNF-α and IL-6 or (B) IFN-α and IFN-β. Error bars are for triplicate samples. Results are representative of three independent experiments with cells from three different donors. (C and D) RNAs were isolated from the cells and used to perform qRT-PCR for genes involved in DC maturation. Values indicate changes in gene expression in DCs infected by viruses compared to that in uninfected DCs (NI). Error bars represent standard deviations for triplicate samples. Results are representative of three independent experiments with cells from three different donors.
Viruses expressing the NS1 protein of influenza virus induce less chemokine production from DCs and are less efficient at activating DCs to prime T cells towards Th1 immunity.
Chemokines released from activated DCs are believed to play an important role in the migration of cell types involved in innate and adaptive immunity (37). The NS1 protein also showed an inhibitory effect on the ability of DCs to secrete chemokines such as RANTES, IL-8, and MIP1β. Thus, DCs infected with NDVB1-NS1 produced almost no RANTES, IL-8, or MIP1β relative to what was observed in response to NDVB1 (Fig. 7A).
FIG. 7.
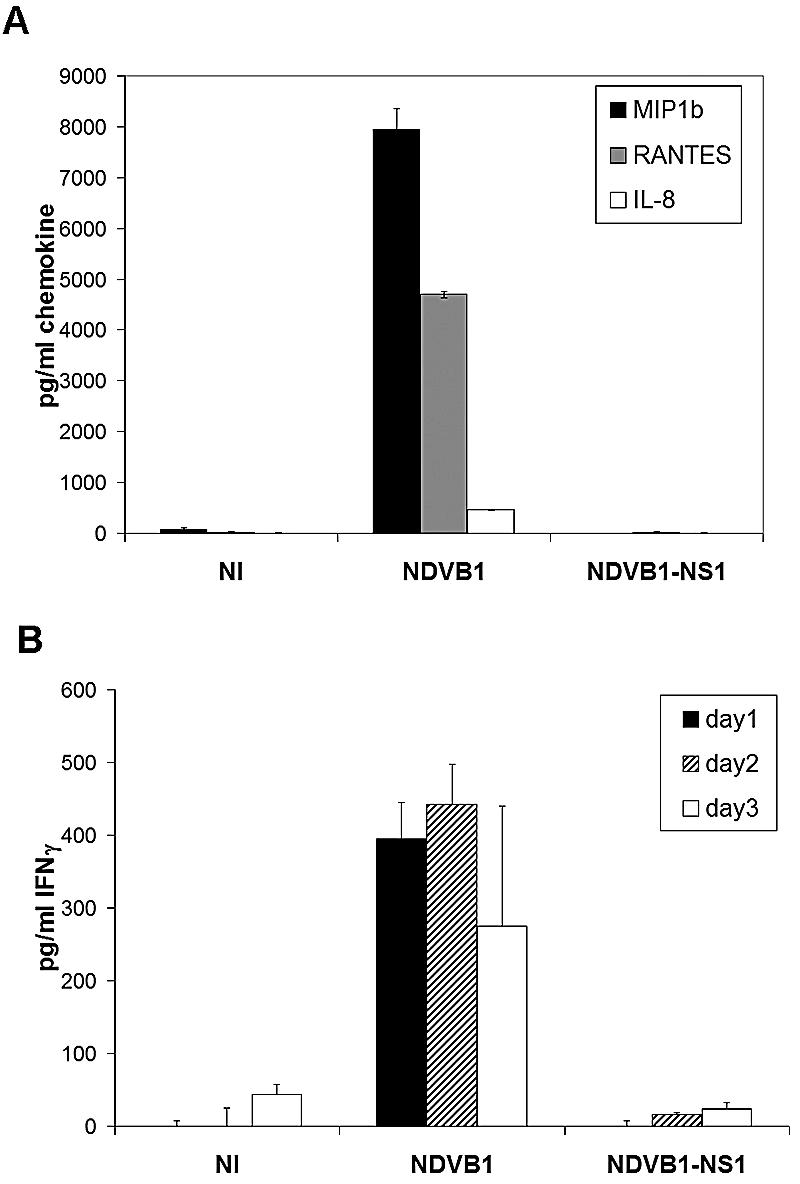
The NS1 protein of influenza virus has an inhibitory effect on chemokine secretion and T-cell priming by DCs. (A) Supernatants from DCs infected with NDVB1 or NDVB1-NS1 or not infected (NI) were tested by ELISA for the presence of MIP1β, RANTES, and IL-8. Error bars are for triplicate samples. Data are representative of at least three independent experiments. (B) Human DCs were infected with NDVB1 or NDVB1-NS1 at an MOI of 0.5 for 45 min and then incubated with peripheral blood mononuclear cells from an allogeneic donor at a ratio of 1:1 for 3 days. Supernatants were harvested every day and tested by ELISA for the release of IFN-γ. Results represent three independent experiments. Error bars are for triplicate samples in each experiment.
In order to investigate the functional significance of NS1 expression on the ability of DCs to induce allospecific stimulation of T cells, DCs were infected with NDV, with or without the NS1 protein of influenza virus, and subsequently cocultured with naïve PBLs. DCs infected with the NS1-expressing virus (NDVB1-NS1) were less efficient than DCs infected with virus lacking the NS1 protein (NDVB1) at priming allospecific lymphocytes for Th1 immunity, as determined by the release of IFN-γ (Fig. 7B).
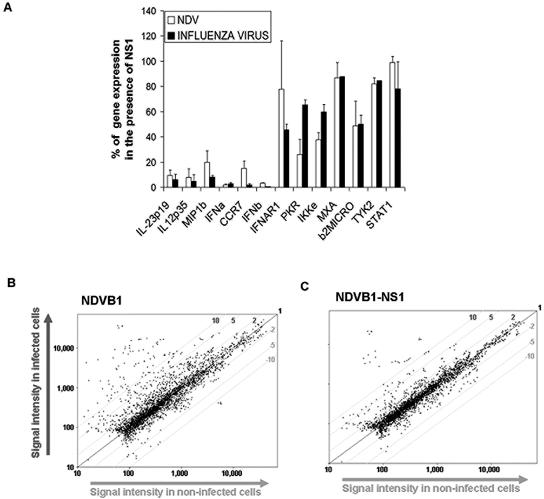
The inhibitory effect of the NS1 protein on DC maturation is gene specific and is not a global effect.
The effect of the NS1 protein on the maturation of DCs was not a global effect, since there were many genes upregulated or not affected by the presence of the NS1 protein in virus-infected human DCs (Fig. 8A). Additionally, we tested DCs infected with NDVB1 (Fig. 8B) and NDVB1-NS1 (Fig. 8C) relative to uninfected DCs by microarray analysis, using a panel of more than 1,000 genes (Memorec; Miltenyi Biotec). The results obtained by microarray analysis strongly support our qRT-PCR and ELISA data (data not shown) and indicate that the effect of the NS1 protein on DC gene expression is not a global effect, since many genes are not affected by its presence.
FIG. 8.
The inhibitory effect of the NS1 protein on DC maturation is not a global effect. (A) Human DCs were infected with influenza virus PR8 or DeltaNS1 or with NDVB1 or NDVB1-NS1 for 18 h. RNAs were isolated from the cells and used to perform qRT-PCR for genes involved in DC maturation. White bars represent effects of NDVB1-NS1 over those of NDVB1 (NDV), while black bars represent effects of PR8 influenza virus over those of DeltaNS1 (influenza virus). Values indicate percentages of gene expression by viruses expressing NS1 relative to that by the same viruses lacking NS1. Error bars represent standard deviations from three independent experiments with cells from three different donors. (B and C) Human DCs infected with NDVB1 and NDVB1-NS1 were analyzed by microarray analysis against uninfected DCs, using an immunology microarray panel of 1,070 genes (Memorec; Miltenyi Biotec). y axes show signal intensities in infected cells, and x axes show signal intensities in uninfected cells.
DISCUSSION
Influenza A virus is a negative-strand RNA virus that causes significant morbidity and mortality yearly in humans and other species (33). Protective neutralizing antibodies are able to prevent reinfection with the same strain, but due to the high rate of mutation of influenza viruses, the immunogenic epitopes in the HA and NA proteins change very rapidly, and neutralizing antibodies are not efficient at blocking reinfection with the mutated drifted viruses (reviewed in reference 46). Cellular immunity is crucial for clearance of the virus from the lungs, and both CD4 and CD8 T cells appear to play an important role. If viruses are able to avoid elements of innate immunity, such as the IFN-α/β response, then they have a great advantage, since adaptive immunity requires more time to generate. One efficient way to reduce the effects of the immune response on virus replication is to block both arms of immunity, i.e., IFN-α/β production and DC activation, interfering with T-cell priming by DCs. Although it is known that influenza A viruses induce T-cell responses in their hosts, it is not clear whether this induction is optimal or whether mechanisms are used by these viruses to prevent the onset of a very strong T-cell response that would otherwise limit viral replication.
Using a human in vitro system, we analyzed the ability of influenza A virus to induce DC activation and examined the CD4 T-cell priming capacity of the infected DCs as an indicator of their ability to initiate Th1 immunity. We observed that the PR8 influenza virus failed to induce a strong Th1 immune response in allospecific cultures of infected DCs and naïve CD4 T cells (Fig. 1). Influenza virus PR8, as well as more recent influenza virus isolates, failed to induce DC activation, as measured by the upregulation of costimulatory molecules, cytokine secretion, and the upregulation of the expression of several genes involved in the IFN-α/β response and in DC activation (Fig. 2, 3, 4, and 5).
Among the 11 proteins encoded by influenza virus, the NS1 protein has been shown to block the production of IFN-β in infected cells, including epithelial cells and DCs (22). This feature of influenza virus allows it to evade innate immunity. In our studies, we have analyzed the effects of the NS1 protein, not only on IFN-α/β production but also on virus-induced maturation of human myeloid DCs and their ability to prime CD4 T cells towards Th1 immunity. If the viruses used to infect DCs expressed influenza virus NS1 (influenza viruses PR8, Moscow, and Texas and NDVB1-NS1), the cells failed to mature fully in vitro (by upregulation of costimulatory molecules and release of proinflammatory cytokines) (Fig. 2, 4, 5, 6, and 7). The infection of DCs with viruses lacking the NS1 protein of influenza virus (NDVB1 and the influenza virus DeltaNS1) resulted in the induction of strong DC maturation, as shown by flow cytometry (Fig. 2C) and ELISA (Fig. 2A and B, 4A and B, and 6A and B). The inhibitory effect of the NS1 protein on DC maturation could be seen even more clearly at the level of gene transcription (Fig. 5A and B, 6C and D, and 8A; see Fig. S1 in the supplemental material). The genes affected by the NS1 protein included those encoding mediators of antiviral immunity, antiviral cytokines, inflammatory chemokines, and the chemokine receptors necessary for DC migration to lymph nodes. Recently, it was shown that influenza virus infection induces a very highly coordinated chemokine response in different subsets of human DCs (35), indicating the importance of the pattern of chemokine secretion by DCs in the generation of immunity to influenza virus. Importantly, the NS1 effect also alters the functionality of DCs by preventing the efficient activation of T cells. Thus, T cells proliferated normally but were insufficiently polarized to produce IFN-γ when incubated with DCs infected with an NS1-expressing virus (Fig. 1 and 7B).
This clear inhibitory effect by the influenza virus NS1 protein on human DC maturation and function is neither a global effect nor simply the result of inhibition of IFN-α/β release and signaling. The addition of antibodies to IFN-β at the time of infection of human DCs with the PR8 influenza virus affected the levels of expression of IFN-inducible genes, while IFN-independent genes were not affected (data not shown). There were numerous genes not affected or moderately affected by the presence of the NS1 protein in infected cells (Fig. 5A and B, 6C and D, and 8A and C). Additionally, this gene-specific effect was very reproducible among several donors and could be observed regardless of the background virus used to express NS1 (Fig. 8; see Fig. S2 in the supplemental material). The absolute values of gene expression between different donors could differ, but the effect of NS1 was maintained among multiple donors tested.
The NS1 protein of influenza virus has been described as mediating multiple effects on cells after virus infection. Predominantly, it has been shown to inhibit pathways involved in IFN-β production, such as the protein kinase R (PKR) pathway (6, 19, 24), and the activation of the transcription factors NF-κB, AP-1, IRF3, and IRF7 (39). Our hypothesis is that one or more of these elements involved in IFN-β production may also be the target for the inhibitory function of NS1 on the expression of genes involved in DC maturation. PKR has been reported to be important for the production of IFN-β in response to double-stranded RNA treatment (8); however, other groups have shown that it is not essential for IFN-β production or for DC activation after virus infection (39). IRF3 and IRF7, two molecules involved in IFN-β production, may be targets for the NS1 protein to downregulate gene transcription. However, we have observed that DCs genetically deficient in IRF3 mature normally to negative-strand RNA viruses (data not shown), and IRF7 synthesis in conventional DCs depends on IFN signaling, which is not essential for full DC maturation in response to RNA virus infection (22). Most likely, the inhibition of genes involved in DC maturation by NS1 is mediated by inhibition of transcription factors such as NF-κB and AP-1. These transcription factors, which were shown to be inhibited by the NS1 protein (25, 32, 45), participate in both IFN-β and cytokine production. Interestingly, we found that the chemokine IL-8, which is strongly dependent on NF-κB and AP-1 for its induction (20), is strongly inhibited by the NS1 protein in DCs (Fig. 7A; see Fig. S1 in the supplemental material). Although the effects of NS1 were gene specific, it is also possible that part of these effects are due to the proposed inhibition of gene expression by the NS1 protein through a block in cellular mRNA processing (29).
One of the most important elements of the antiviral innate immune system is IFN-α/β. Most cell types, including respiratory epithelial cells and DCs, synthesize IFN-α/β in response to virus infection (1, 15). Upon release from infected cells, IFN-α/β binds to its receptor, present in the secreting cells and in adjacent cells, thus establishing an antiviral state characterized by the production of numerous proteins that inhibit the ability of viruses to replicate (reviewed in references 1 and 15). Many viruses evade the IFN-α/β system through the expression of viral IFN antagonist proteins. In the case of influenza A virus, the NS1 protein was shown to be a potent IFN antagonist in epithelial cells through its N-terminal double-stranded RNA-binding domain, which prevents the induction of type I IFN synthesis during viral infection (9, 42). Our results demonstrate that influenza virus inhibits not only IFN-α/β release in human DCs but also maturation of DCs, and they suggest that the NS1 protein of influenza virus attenuates the initiation of adaptive immune responses in infected individuals. Recent reports have also shown low levels of IFN-α/β, TNF-α, and IFN-λ1,2,3 release by influenza virus-infected human DCs compared to those in Sendai virus infection and low levels of DC maturation induced by influenza virus (31). A number of reports have documented enhanced immunogenicities of viruses with mutated or truncated NS1, consistent with an inhibitory effect of this protein on adaptive immunity (12, 41, 43). Due to the attenuation properties of NS1 mutant viruses in their hosts (11, 36, 40, 41, 43) and to the enhanced ability of NS1 mutant viruses to induce human DC maturation (this report), we suggest that these viruses are good candidates to be considered as live attenuated vaccines against influenza. Thus, a lack of NS1 function results in mutant influenza viruses with more potent adjuvant properties in DCs. Further investigation will be required to demonstrate the vaccine efficacy of NS1 mutant viruses in humans as well as to determine whether our observations might also be generalized to other viruses that carry IFN-α/β antagonists. Interestingly, it has recently been shown that the IFN-α/β antagonist genes of respiratory syncytial viruses also appear to prevent optimal stimulation of human DCs (38).
Here we report an inhibitory effect of NS1 on elements from DCs, which are important for the initiation of Th1 immunity. This inhibitory effect on adaptive immunity may have been overlooked due to previous virus exposures in the human outbred population. This effect could be compensated for by the generation of a strong CD8 T-cell memory response after influenza virus infection that overcomes the weak CD4 T-cell response, as has been suggested for mouse models (48). On the other hand, the expected decrease in the adaptive immune response due to the NS1 effect could be compensated for by the release of type I IFN or other proinflammatory cytokines from plasmacytoid DCs (pDCs) in vivo, providing an adjuvant effect in trans on myeloid DCs, as recently suggested (4). While a direct role for pDCs in the induction of adaptive immune responses against viruses in vivo has not been documented, a number of studies have demonstrated a requirement for type I IFN for optimal maturation of myeloid DCs (14, 21, 23). Further experimentation will be required to evaluate the role of pDCs in the induction of immunity against influenza viruses.
In summary, using recombinant NDV and influenza viruses expressing the NS1 IFN antagonist of influenza virus, we have shown that the NS1 protein is able to attenuate a primary human cell system in both innate immunity and a critical element necessary for the initiation of adaptive immunity, DC maturation. This dual immune evasion strategy is likely to be utilized by other viral IFN antagonist proteins. Further characterization of this strategy, in which a single protein targets both innate and adaptive immunity, may prove to be extremely useful for the design of live virus vaccines or therapeutic agents with improved immune efficacy.
Supplementary Material
Acknowledgments
We thank Sharon Czelusniak for her invaluable help and proofreading of the manuscript.
This work was supported by an NIH training grant to A.F.-S. (1-T32-AI07605) and by an NIH U19 grant awarded to T.M.M., A.G.-S., and S.C.S. (U19 AI62623).
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Alcami, A., and U. H. Koszinowski. 2000. Viral mechanisms of immune evasion. Trends Microbiol. 8:410-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banchereau, J., F. Briere, C. Caux, J. Davoust, S. Lebecque, Y. J. Liu, B. Pulendran, and K. Palucka. 2000. Immunobiology of dendritic cells. Annu. Rev. Immunol. 18:767-811. [DOI] [PubMed] [Google Scholar]
- 3.Banchereau, J., and R. M. Steinman. 1998. Dendritic cells and the control of immunity. Nature 392:245-252. [DOI] [PubMed] [Google Scholar]
- 4.Barchet, W., A. Krug, M. Cella, C. Newby, J. A. Fischer, A. Dzionek, A. Pekosz, and M. Colonna. 2005. Dendritic cells respond to influenza virus through TLR7- and PKR-independent pathways. Eur. J. Immunol. 35:236-242. [DOI] [PubMed] [Google Scholar]
- 5.Belladonna, M. L., J. C. Renauld, R. Bianchi, C. Vacca, F. Fallarino, C. Orabona, M. C. Fioretti, U. Grohmann, and P. Puccetti. 2002. IL-23 and IL-12 have overlapping, but distinct, effects on murine dendritic cells. J. Immunol. 168:5448-5454. [DOI] [PubMed] [Google Scholar]
- 6.Bergmann, M., A. Garcia-Sastre, E. Carnero, H. Pehamberger, K. Wolff, P. Palese, and T. Muster. 2000. Influenza virus NS1 protein counteracts PKR-mediated inhibition of replication. J. Virol. 74:6203-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cella, M., F. Sallusto, and A. Lanzavecchia. 1997. Origin, maturation and antigen presenting function of dendritic cells. Curr. Opin. Immunol. 9:10-16. [DOI] [PubMed] [Google Scholar]
- 8.Diebold, S. S., M. Montoya, H. Unger, L. Alexopoulou, P. Roy, L. E. Haswell, A. Al-Shamkhani, R. Flavell, P. Borrow, and C. Reis e Sousa. 2003. Viral infection switches non-plasmacytoid dendritic cells into high interferon producers. Nature 424:324-328. [DOI] [PubMed] [Google Scholar]
- 9.Donelan, N. R., C. F. Basler, and A. Garcia-Sastre. 2003. A recombinant influenza A virus expressing an RNA-binding-defective NS1 protein induces high levels of beta interferon and is attenuated in mice. J. Virol. 77:13257-13266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egorov, A., S. Brandt, S. Sereinig, J. Romanova, B. Ferko, D. Katinger, A. Grassauer, G. Alexandrova, H. Katinger, and T. Muster. 1998. Transfectant influenza A viruses with long deletions in the NS1 protein grow efficiently in Vero cells. J. Virol. 72:6437-6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falcon, A. M., A. Fernandez-Sesma, Y. Nakaya, T. M. Moran, J. Ortin, and A. Garcia-Sastre. 2005. Attenuation and immunogenicity in mice of temperature-sensitive influenza viruses expressing truncated NS1 proteins. J. Gen. Virol. 86:2817-2821. [DOI] [PubMed] [Google Scholar]
- 12.Ferko, B., J. Stasakova, J. Romanova, C. Kittel, S. Sereinig, H. Katinger, and A. Egorov. 2004. Immunogenicity and protection efficacy of replication-deficient influenza A viruses with altered NS1 genes. J. Virol. 78:13037-13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernandez-Sesma, A., J. L. Schulman, and T. M. Moran. 1996. A bispecific antibody recognizing influenza A virus M2 protein redirects effector cells to inhibit virus replication in vitro. J. Virol. 70:4800-4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujii, S., K. Liu, C. Smith, A. J. Bonito, and R. M. Steinman. 2004. The linkage of innate to adaptive immunity via maturing dendritic cells in vivo requires CD40 ligation in addition to antigen presentation and CD80/86 costimulation. J. Exp. Med. 199:1607-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Sastre, A. 2002. Mechanisms of inhibition of the host interferon alpha/beta-mediated antiviral responses by viruses. Microbes Infect. 4:647-655. [DOI] [PubMed] [Google Scholar]
- 16.García-Sastre, A., A. Egorov, D. Matassov, S. Brandt, D. E. Levy, J. E. Durbin, P. Palese, and T. Muster. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324-330. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez-Maeso, J., T. Yuen, B. J. Ebersole, E. Wurmbach, A. Lira, M. Zhou, N. Weisstaub, R. Hen, J. A. Gingrich, and S. C. Sealfon. 2003. Transcriptome fingerprints distinguish hallucinogenic and nonhallucinogenic 5-hydroxytryptamine 2A receptor agonist effects in mouse somatosensory cortex. J. Neurosci. 23:8836-8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graham, M. B., V. L. Braciale, and T. J. Braciale. 1994. Influenza virus-specific CD4+ T helper type 2 T lymphocytes do not promote recovery from experimental virus infection. J. Exp. Med. 180:1273-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatada, E., S. Saito, and R. Fukuda. 1999. Mutant influenza viruses with a defective NS1 protein cannot block the activation of PKR in infected cells. J. Virol. 73:2425-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffmann, E., O. Dittrich-Breiholz, H. Holtmann, and M. Kracht. 2002. Multiple control of interleukin-8 gene expression. J. Leukoc. Biol. 72:847-855. [PubMed] [Google Scholar]
- 21.Honda, K., S. Sakaguchi, C. Nakajima, A. Watanabe, H. Yanai, M. Matsumoto, T. Ohteki, T. Kaisho, A. Takaoka, S. Akira, T. Seya, and T. Taniguchi. 2003. Selective contribution of IFN-alpha/beta signaling to the maturation of dendritic cells induced by double-stranded RNA or viral infection. Proc. Natl. Acad. Sci. USA 100:10872-10877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.López, C. B., A. García-Sastre, B. R. G. Williams, and T. M. Moran. 2003. Type I interferon induction pathway, but not released interferon, participates in the maturation of dendritic cells induced by negative-strand RNA viruses. J. Infect. Dis. 187:1126-1136. [DOI] [PubMed] [Google Scholar]
- 23.López, C. B., B. Moltedo, L. Alexopoulou, L. Bonifaz, R. Flavell, and T. M. Moran. 2004. Toll like receptor-independent induction of dendritic cell maturation and adaptive immunity by negative-strand RNA viruses. J. Immunol. 173:6882-6889. [DOI] [PubMed] [Google Scholar]
- 24.Lu, Y., M. Wambach, M. G. Katze, and R. M. Krug. 1995. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the eIF-2 translation initiation factor. Virology 214:222-228. [DOI] [PubMed] [Google Scholar]
- 25.Ludwig, S., X. Wang, C. Ehrhardt, H. Zheng, N. Donelan, O. Planz, S. Pleschka, A. García-Sastre, G. Heins, and T. Wolff. 2002. The influenza A virus NS1 protein inhibits activation of Jun N-terminal kinase and AP-1 transcription factors. J. Virol. 76:11166-11171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsui, M., O. Moriya, M. L. Belladonna, S. Kamiya, F. A. Lemonnier, T. Yoshimoto, and T. Akatsuka. 2004. Adjuvant activities of novel cytokines, interleukin-23 (IL-23) and IL-27, for induction of hepatitis C virus-specific cytotoxic T lymphocytes in HLA-A*0201 transgenic mice. J. Virol. 78:9093-9104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mellman, I., and R. M. Steinman. 2001. Dendritic cells: specialized and regulated antigen processing machines. Cell 106:255-258. [DOI] [PubMed] [Google Scholar]
- 28.Moran, T. M., H. Park, A. Fernandez-Sesma, and J. L. Schulman. 1999. Th2 responses to inactivated influenza virus can be converted to Th1 responses and facilitate recovery from heterosubtypic virus infection. J. Infect. Dis. 180:579-585. [DOI] [PubMed] [Google Scholar]
- 29.Nemeroff, M. E., S. M. Barabino, Y. Li, W. Keller, and R. M. Krug. 1998. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol. Cell 1:991-1000. [DOI] [PubMed] [Google Scholar]
- 30.O'Doherty, U., M. Peng, S. Gezelter, W. J. Swiggard, M. Betjes, N. Bhardwaj, and R. M. Steinman. 1994. Human blood contains two subsets of dendritic cells, one immunologically mature and the other immature. Immunology 82:487-493. [PMC free article] [PubMed] [Google Scholar]
- 31.Osterlund, P., V. Veckman, J. Siren, K. M. Klucher, J. Hiscott, S. Matikainen, and I. Julkunen. 2005. Gene expression and antiviral activity of alpha/beta interferons and interleukin-29 in virus-infected human myeloid dendritic cells. J. Virol. 79:9608-9617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ouaaz, F., J. Arron, Y. Zheng, Y. Choi, and A. A. Beg. 2002. Dendritic cell development and survival require distinct NF-kappaB subunits. Immunity 16:257-270. [DOI] [PubMed] [Google Scholar]
- 33.Palese, P., and A. Garcia-Sastre. 2002. Influenza vaccines: present and future. J. Clin. Investig. 110:9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park, M. S., A. Garcia-Sastre, J. F. Cros, C. F. Basler, and P. Palese. 2003. Newcastle disease virus V protein is a determinant of host range restriction. J. Virol. 77:9522-9532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piqueras, B., J. Connolly, H. Freitas, A. K. Palucka, and J. Banchereau. 2005. Upon viral exposure, myeloid and plasmacytoid dendritic cells produce three waves of distinct chemokines to recruit immune effectors. Blood 107:2613-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quinlivan, M., D. Zamarin, A. Garcia-Sastre, A. Cullinane, T. Chambers, and P. Palese. 2005. Attenuation of equine influenza viruses through truncations of the NS1 protein. J. Virol. 79:8431-8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sallusto, F., B. Palermo, D. Lenig, M. Miettinen, S. Matikainen, I. Julkunen, R. Forster, R. Burgstahler, M. Lipp, and A. Lanzavecchia. 1999. Distinct patterns and kinetics of chemokine production regulate dendritic cell function. Eur. J. Immunol. 29:1617-1625. [DOI] [PubMed] [Google Scholar]
- 38.Schlender, J., V. Hornung, S. Finke, M. Gunthner-Biller, S. Marozin, K. Brzozka, S. Moghim, S. Endres, G. Hartmann, and K. K. Conzelmann. 2005. Inhibition of Toll-like receptor 7- and 9-mediated alpha/beta interferon production in human plasmacytoid dendritic cells by respiratory syncytial virus and measles virus. J. Virol. 79:5507-5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith, E. J., I. Marié, A. Prakash, A. García-Sastre, and D. E. Levy. 2001. IRF3 and IRF7 phosphorylation in virus-infected cells does not require double-stranded RNA-dependent protein kinase R or IκB kinase but is blocked by vaccinia virus E3L protein. J. Biol. Chem. 276:8951-8957. [DOI] [PubMed] [Google Scholar]
- 40.Solorzano, A., R. J. Webby, K. M. Lager, B. H. Janke, A. Garcia-Sastre, and J. A. Richt. 2005. Mutations in the NS1 protein of swine influenza virus impair anti-interferon activity and confer attenuation in pigs. J. Virol. 79:7535-7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stasakova, J., B. Ferko, C. Kittel, S. Sereinig, J. Romanova, H. Katinger, and A. Egorov. 2005. Influenza A mutant viruses with altered NS1 protein function provoke caspase-1 activation in primary human macrophages, resulting in fast apoptosis and release of high levels of interleukins 1β and 18. J. Gen. Virol. 86:185-195. [DOI] [PubMed] [Google Scholar]
- 42.Talon, J., C. M. Horvath, R. Polley, C. F. Basler, T. Muster, P. Palese, and A. Garcia-Sastre. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 74:7989-7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Talon, J., M. Salvatore, R. E. O'Neill, Y. Nakaya, H. Zheng, T. Muster, A. Garcia-Sastre, and P. Palese. 2000. Influenza A and B viruses expressing altered NS1 proteins: a vaccine approach. Proc. Natl. Acad. Sci. USA 97:4309-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trinchieri, G. 2003. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat. Rev. Immunol. 3:133-146. [DOI] [PubMed] [Google Scholar]
- 45.Wang, X., M. Li, H. Zheng, T. Muster, P. Palese, A. A. Beg, and A. García-Sastre. 2000. Influenza A virus NS1 protein prevents activation of NF-κB and induction of alpha/beta interferon. J. Virol. 74:11566-11576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Webster, R. G., W. G. Laver, G. M. Air, and G. C. Schild. 1982. Molecular mechanisms of variation in influenza viruses. Nature 296:115-121. [DOI] [PubMed] [Google Scholar]
- 47.Yuen, T., E. Wurmbach, R. L. Pfeffer, B. J. Ebersole, and S. C. Sealfon. 2002. Accuracy and calibration of commercial oligonucleotide and custom cDNA microarrays. Nucleic Acids Res. 30:e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zammit, D. J., L. S. Cauley, Q. M. Pham, and L. Lefrancois. 2005. Dendritic cells maximize the memory CD8 T cell response to infection. Immunity 22:561-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.