Abstract
Dynamic telomere repositioning is a prominent feature of meiosis. Deletion of a telomere-associated protein, Ndj1, results in the failure of both attachment and clustering of telomeres at the nuclear envelope and delays several landmarks of meiosis I, such as pairing, synaptonemal complex formation, and timing of the meiosis I division. We explored the role of Ndj1 in meiotic recombination, which occurs through the formation and repair of programmed double-strand breaks. The ndj1Δ mutation allows for the formation of the first detectable strand invasion intermediate (i.e., single-end invasion) with wild-type kinetics; however, it confers a delay in the formation of the double-Holliday junction intermediate and both crossover and noncrossover products. These results challenge the widely held notion that clustering of telomeres in meiosis promotes the ability of homologous chromosomes to find one another in budding Saccharomyces cerevisiae. We propose that an Ndj1-dependent function is critical for stabilizing analogous strand invasion intermediates that exist in two separate branches of the bifurcated pathway, leading to either noncrossover or crossover formation. These findings provide a link between telomere dynamics and a distinct mechanistic step of meiotic recombination that follows the homology search.
Meiosis is a conserved cell division pathway that generates haploid gametes from diploid parents in sexual eukaryotes. In budding Saccharomyces cerevisiae and most eukaryotes, meiotic recombination produces crossovers (CRs) between homologous chromosomes. Together with sister chromatid cohesion, CRs hold homologs together prior to meiosis I disjunction to ensure proper chromosome segregation. Failure in this conserved process is a major cause of miscarriage and birth defects in humans (reviewed in references 21, 43, 45, and 64).
Meiotic recombination is initiated by programmed double-strand break (DSB) formation catalyzed by a topoisomerase-like protein, Spo11 (3, 30). The repair of DSBs can generate either CR or noncrossover (NCR) products. The subset of DSBs processed to CRs proceed primarily through a pathway (referred to here as the CR-I pathway) involving physically detectable intermediates: single-end invasion (SEI), followed by a double-Holliday junction (dHJ) with ligated DNA strands (26). It is thought that dHJs are resolved primarily as CRs (1, 26, 47). The CRs formed via the CR-I pathway are thought to be under crossover control and exhibit crossover interference. Their formation is dependent, to a large extent, on the ZMM group of proteins, comprising Zip1, Zip2, Zip3, Msh4-Msh5, and Mer3 (6, 53).
NCRs and a smaller subset of CRs (called class CR-II) are likely processed by a synthesis-dependent strand annealing pathway that is dependent in part on Mus81 and Mms4 in budding yeast (reviewed in references 13, 23, and 25). Both the CR-I and NCR-plus-CR-II pathways require Spo11-dependent DSB formation, resection of 5′ ends of the DSB, and presumably an unstable strand invasion intermediate promoted by the RecA homologs Dmc1 and Rad51 (60).
Multiple lines of evidence suggest that telomeres play a critical role(s) during meiotic recombination and synapsis in various sexual reproductive eukaryotes. In the fission yeast Schizosaccharomyces pombe, telomere-led, dynein-dependent, oscillatory movements are important for the efficiency of meiotic homolog pairing and recombination (8, 11, 16, 39, 62). Mutants defective in telomere-spindle pole body attachment and dynein heavy chain and light chain lead to drastic reductions in meiotic recombination (reviewed in references 12 and 24). In maize, a pam1 mutant fails to bring telomeres together into a clustered bouquet during zygotene and also exhibits defects in homologous synapsis (20). In mice, irregular telomere length results in decreased levels of synapsis and a modest reduction in CR recombination, as measured by the number of MLH1 foci (34).
Meiotic prophase is notable for the dynamic nature of chromosome mobility (12, 14, 24, 35, 64). It has long been postulated that the dynamic reorganization of chromosomes may facilitate meiotic recombination by aligning homologous chromosomes into close proximity (45, 64). In particular, the clustering of telomeres at the nuclear envelope at the “bouquet” stage has been postulated to reduce the homology search from three dimensions to two dimensions since allelic sequences would be placed at similar latitudes within the nuclear volume (45, 64). This view has been challenged by a recent study using Sordaria macrospora that shows presynaptic homolog coalignment is accomplished prior to the bouquet stage (51). Recent work by Peoples-Holst and Burgess (42) using Saccharomyces cerevisiae demonstrates that a mutation defective in attachment and clustering of telomeres at the nuclear envelope (ndj1Δ) is partially defective in forming close stable homolog connections. This defect, however, is dependent on the formation of meiotically induced DSBs, thus also arguing against an independent contribution of telomere organization in promoting stable interhomolog (IH) interactions.
Meiotic prophase is divided into several stages. DSB-induced recombination is initiated during early prophase (leptotene) and completed during pachytene when homologous chromosomes are fully synapsed. During the intervening zygotene stage, SEIs and synapsis initiation occur (6, 26). In budding yeast, dynamic reorganization of telomeres is concomitant with the events of meiotic recombination (57). Telomeres are found detached from the nuclear envelope following DNA replication and, by late leptotene, are found to be reassociated and dispersed around the nuclear envelope. At the leptotene-zygotene transition, telomeres cluster near the spindle pole body in the bouquet configuration. The meiotic telomere cluster repeatedly splits and reforms during zygotene (55). During pachytene, telomeres again appear dispersed around the nuclear envelope (57) and undergo actin-dependent oscillating movement over the entire nuclear periphery (55).
Ndj1 (also known as Tam1) is a meiosis-induced, telomere-associated protein in budding yeast (9, 10). In the ndj1Δ mutant, telomeres fail to retether to the nuclear envelope in early meiotic prophase, and therefore, the earliest step of telomere reorganization is impeded (56). Notably, several genes required for nuclear envelope-telomere interactions in mitotic cells (YKU70, YKU80, and SIR3) are not required for the nuclear envelope-telomere interaction in meiotic prophase (55, 58). To date, NDJ1 is the only gene identified to be involved in tethering telomeres with the nuclear envelope in meiotic prophase. Numerous events occurring during meiosis prophase I are delayed in the ndj1Δ mutant, including pairing, axial element formation, synaptonemal complex formation, and meiosis I division (9, 10). While no significant genetic effects on either allelic gene conversion or crossover recombination were found, the mutant does exhibit an increase in the frequency of precocious separation of sister chromatids and E0 (nonexchange) tetrads. Moreover, crossover interference is impaired (9). Homologous juxtaposition, meiosis I disjunction, and spore viability are also decreased in ndj1Δ cells. Ectopic recombination is increased in the ndj1Δ strain, suggesting that the homology search step of meiotic recombination in ndj1Δ cells is not impaired; however, compartmentalization of the nucleus may be less defined (18). NDJ1 also plays a role in homolog interaction in the absence of recombination initiation in a haploid-disome strain background (44). Recently, nonmeiotic roles for Ndj1 have been described that implicate it more directly in telomere biology, as it is required for telomere length regulation in both mitotic and meiotic cells (27). NDJ1 expression is also induced during a telomerase deletion response invoked in vegetatively dividing cells (40).
Meiotic defects exhibited by the ndj1Δ mutant have generally been attributed to defects in the formation of the bouquet, since telomeres fail to attach to the nuclear envelope. Here, we address the role of Ndj1 in meiotic recombination of budding yeast. First, we show that the delay in separation of chromosome masses at meiosis I (MI) in the ndj1Δ mutant is dependent on the formation of DSBs. We also show that the turnover of DSBs in the ndj1Δ mutant is delayed compared to that in the wild type. Unexpectedly, the timing of SEI formation in the ndj1Δ mutant is very close to that in the wild type while dHJ formation is delayed. Both CR and NCR formation is delayed, while NCRs are specifically decreased in the mutant compared to those in the wild type. These results imply that Ndj1 is not required for a genome-wide homology search but facilitates meiotic recombination primarily by promoting the transition between recombination intermediates at a step following the bifurcation of DSB repair into CR-I and NCR-plus-CR-II pathways. Our results provide insights into how disruption of meiotic telomere reorganization might influence specific steps of meiotic recombination along the lengths of chromosomes.
MATERIALS AND METHODS
Yeast strains.
All strains in this study are in the SK1 background (28) and are listed in Table 1. Kanamycin or hygromycin B resistant markers were used for all knockout strains generated in this study (19, 36).
TABLE 1.
Strains used in this study
| Strain | Genotype |
|---|---|
| NKY3230 (also known as SBY1903) | MATa/Matα ho::hisG/ho::hisG leu::hisG/leu2::hisG ura3/ura3 his4-X::LEU2-(NBam)-URA3/HIS4::LEU2-(NBam) |
| SBY2030 | NKY3230 except that it has ndj1Δ::hphB/ndj1Δ::hphB |
| SBY2249 | NKY3230 except that it has spo11Δ::kanMX/spo11Δ::kanMX |
| SBY2380 | NKY3230 except that it has spo11Δ::kanMX/spo11Δ::kanMX ndj1Δ::hphB/ndj1::hphB |
| SBY1765 | MATa/MATα ho::hisG/ho::hisG lys2/lys2 ura3D::hisG/ura3D::hisG leu2::hisG/leu2::hisG ade2D::hisG/ade2D::hisG |
| SBY1766 | SBY1765 except that it has ndj1Δ::kanMX/ndj1Δ::kanMX |
| SBY86 | MATa/MATα ho::LYS2/ho::LYS2 lys2/lys2 ura3/ura3 leu2::hisG/leu2::hisG trp1::hisG/trp1::hisG arg4-Nsp/arg4-Bgl his4-B::LEU2/his4-X(Bam)::LEU2-URA3 |
| SBY2118 | SBY86 except that it has ndj1Δ::hphB/ndj1::hphB |
| SBY1759 | ho::LYS2/ho::LYS2 lys2/lys2 ura3/ura3 leu2::hisG/leu2::hisG rad50-K181::URA3/rad50-K181::URA3 |
| SBY1795 | ho::hisG/ho::hisG lys2/lys2 ura3/ura3 leu2::hisG/leu2::hisG ade2D::hisG/ade2d::hisG rad50-k181::URA3/rad50-k181::URA3 ndj1Δ::kanMX/ndj1Δ::kanMX |
| SBY2368 | NKY3230 except that it has rad17Δ::kanMX/rad17Δ::kanMX |
| SBY2369 | NKY3230 except that it has rad17Δ::kanMX/rad17Δ::kanMX ndj1Δ::hphB/ndj1Δ::hphB |
| SBY2180 | NKY3230 except that both chromosome IIIs are circular chromosomes |
Sporulation conditions.
For meiotic division analysis and DNA analysis, yeast cells were synchronized and sporulated using the SPS (supplemented presporulation medium) method (1), except for the experiments whose results are shown in Fig. 2, which were done by the YPA method (41). The SPS and YPA methods differ by the composition of the presporulation medium (0.5% yeast extract, 1% yeast peptone, 0.17% yeast nitrogen base, 1% potassium acetate, 0.5% ammonium sulfate, and 1.02% potassium biphthalate [SPS]; 1% yeast extract, 2% yeast peptone, and 1% potassium acetate [YPA]). For all experiments, sporulation medium (1% potassium acetate, 0.02% raffinose, 0.009% amino acid drop-out powder) was used to induce the meiotic program. All cultures were incubated at 30°C except where otherwise noted.
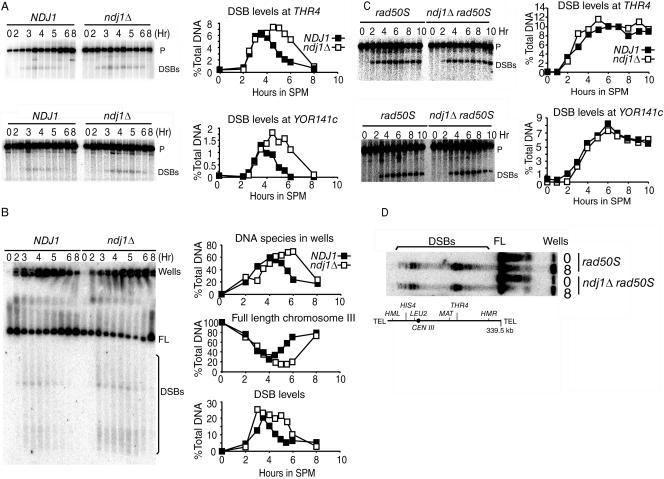
FIG. 2.
DSB turnover in the ndj1Δ mutant is delayed compared to that in NDJ1, but the timing and levels of DSB formation are not dramatically affected. Results in panels A and B are all from time course 6 (TC6). (A) DSB turnover of NDJ1 (SBY1765) and the ndj1Δ mutant (SBY1766) at THR4 (EcoRI digestion, THR4 open reading frame [ORF] probe) and YOR141C (HindIII digestion, YOR141C ORF probe). THR4 (YCR053W) localizes 100 kb away from the right end of chromosome III (total, 320 kb). YOR141C localizes 500 kb away from the right end of chromosome XV (total, 1,095 kb). P, parental DNA. (B) CHEF gel on chromosome III, probed with the CHA1 ORF from the left end of chromosome III. FL, full-length chromosome III; Wells, DNA species stuck in the wells of the CHEF gel. (C) DSB timing and levels in rad50S and ndj1Δ rad50S mutants at THR4 and YOR141C loci. (D) DSB levels in ndj1Δ rad50S and rad50S mutants on chromosome III, probed with the CHA1 ORF. For experiments whose results are shown in panels C and D, rad50S (SBY1759) and ndj1Δ rad50S (SBY1795) mutants were used.
Cytological analysis.
The number of chromosome masses was monitored by 4′,6′-diamidino-2-phenylindole (DAPI) staining of cells fixed in 50% ethanol. Two hundred cells were counted for each time point. Experiments were conducted in multiple independent trials, and similar results were obtained.
Molecular analysis.
Psoralen cross-linking, DNA preparation, one-dimensional gel, Southern blotting, and DNA probing were performed according to previous conditions (26, 48). Separation and analysis of chromosomes by contour-clamped homogeneous electric field (CHEF) electrophoresis were performed as described by Borde et al. (5). DNA species were quantified using a Storm phosphorimager and ImageQuant Software (Molecular Dynamics). Life spans of DSBs were calculated by an Excel macro kindly provided by N. Hunter (26). In our calculation, the maximum levels of DSBs at the HIS4::LEU2-(NBam)/his4-X::LEU2-(NBam)-URA3 hot spot in NDJ1 and ndj1Δ cells were set at 23.5%, according to Hunter and Kleckner (26). All experiments reported were carried out in at least two independent trials, and similar results were obtained.
RESULTS
The meiosis I delay conferred by the ndj1Δ mutation is linked to meiotic recombination.
We first sought to understand the nature of the delay in MI conferred by the ndj1Δ mutation. Several possible explanations could account for this phenotype. i) Ndj1 could be either directly or indirectly involved in meiotic recombination by any one of various mechanisms, as previously proposed (reviewed in reference 64). ii) Lack of Ndj1 could affect other cellular processes in a more general way. For example, expression of genes in the subtelomere region may be influenced by whether telomeres are attached to the nuclear envelope (17). Alternatively, since the transcription factor Rap1 has been shown to be mislocalized in the ndj1Δ mutant (10), a more general effect on gene expression may lead to a delay in the progression through meiosis. iii) The lack of telomere attachment to the nuclear envelope or a stage of meiotic telomere reorganization perturbed by the ndj1Δ mutation could activate a checkpoint unrelated to DNA recombination and slow the progression to meiosis I anaphase.
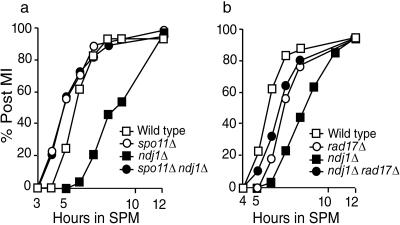
We addressed these models by comparing the kinetics of the first meiotic division in ndj1Δ mutant cells that do not initiate meiotic recombination with that in mutants that presumably initiate recombination normally (i.e., ndj1Δ spo11Δ cells with ndj1Δ cells). The timing of MI in ndj1Δ spo11Δ cells resembles that in spo11Δ cells (Fig. 1a). Chromosome masses separate faster in spo11Δ cells than in the wild type as previously reported (29). A catalytically inactive spo11-Y135F allele also suppressed ndj1Δ MI delay (data not shown). These results suggest that the delay of separation of chromosome masses at MI conferred by the ndj1Δ mutation depends upon the formation of DSBs. In this case, the accumulation of a DSB repair intermediate might induce a DNA damage checkpoint response (37). Consistent with this notion, we found that a DNA recombination checkpoint mutant, rad17Δ, rescues the meiotic delay conferred by the ndj1Δ mutation (Fig. 1b).
FIG. 1.
The meiosis I delay conferred by the ndj1Δ mutation is suppressed by the spo11Δ mutation. (a) Meiotic time course of SBY1903 (Wild type), SBY2030 (ndj1Δ), SBY2249 (spo11Δ), and SBY2380 (spo11Δ ndj1Δ). (b) Meiotic time course of SBY1903 (Wild type), SBY2030 (ndj1Δ), SBY2368 (rad17Δ), and SBY2369 (rad17Δ ndj1Δ). Separation of chromosome masses into two, three, or four foci was monitored by DAPI staining.
These results preclude the second and third models presented above; suppression of the ndj1Δ MI delay phenotype conferred by the spo11Δ mutation argues against an effect of the ndj1Δ mutation on gene expression, thereby causing a general delay or activating a nonrecombination checkpoint during meiosis in the ndj1Δ cells.
The ndj1Δ mutation delays the turnover of DSBs but does not change the total level of DSBs formed.
Since the MI delay conferred by the ndj1Δ mutation is dependent on the initiation of meiotic recombination, we next asked whether Ndj1 is required for the timely formation and/or processing of DSBs into recombination intermediates or products. Wild-type and ndj1Δ cultures were synchronized to enter into meiosis, and Southern blot analyses were carried out to measure DSB levels at several meiotic recombination hot spots (i.e., THR4, YOR141C, and HIS4::LEU2) at various subsequent time points. In all cases, ndj1Δ cells accumulated higher levels of DSBs than wild-type cells (Fig. 2A and 3A and B).
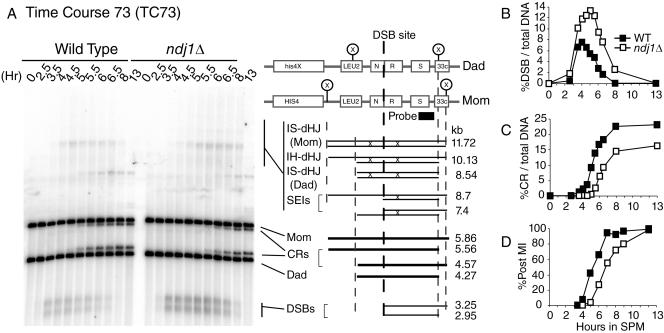
FIG. 3.
Meiotic recombination at HIS4::LEU2/his4-X::LEU2-URA3 hot spot in NDJ1 and the ndj1Δ mutant. Panels shown are from TC73 of meiosis of strains NKY3230 (Wild Type) and SBY2030 (ndj1Δ). TC73 samples were treated with psoralen. DNA isolated at various time points was digested with XhoI and separated on a 0.6% agarose gel. Southern blots were hybridized with probe A (3′ end of STE50 open reading frame). (A) One-dimensional gel analysis. The schematic of the HIS4::LEU2/his4-X::LEU2-URA3 hot spot was modified as described by Hunter and Kleckner (26). The joint molecule region contains both dHJs and SEIs. (B) Quantification of DSB levels from panel A. (C) Quantification of CR levels from panel A. (D) Meiosis I timing for TC73, monitored by DAPI staining. WT, wild type.
To test whether ndj1Δ mutants were either making greater overall levels of breaks or exhibiting delayed DSB turnover kinetics, the maximum levels of DSB formation were compared in ndj1Δ rad50S and NDJ1 rad50S strains. The rad50S mutation (rad50-K181) creates a block in the DSB repair pathway so that unprocessed meiotic DSBs accumulate prior to resection. Since the turnover of breaks occurs very slowly in the rad50S strain background, a close approximation of maximal levels of DSBs formed at recombination hot spots can be ascertained. Similar levels of breaks were formed in the rad50S strain background for both ndj1Δ and wild-type cells for the THR4 and YOR141C loci and for the major DSB hot spots resolved using CHEF gel analysis (Fig. 2C and D). Given that the ndj1Δ mutant appears to make the same total levels of breaks as the wild type, we interpret the increased levels of DSBs in a RAD50 background to suggest that the life spans of DSBs are increased in the ndj1Δ mutants compared to those in wild-type cells. In other words, the turnover of DSBs is delayed.
The timing of DSB formation was observed to be similar to that in the wild type (Fig. 2C and D) or slightly delayed (usually a slight delay [7 to 21 min] for large-sized cultures, i.e., >100-ml cultures) (data not shown). This effect could be due to a function for NDJ1 in telomere metabolism in vegetatively dividing cells (27). A gene required for nuclear reorganization in Caenorhabditis elegans appears to regulate the timing of DSB formation (2). It is formally possible that NDJ1 may play similar roles in budding yeast. Nevertheless, these possibilities were not distinguished by our experimental design.
DSB turnover rates at hot spots along the whole length of chromosome III were measured using CHEF gel electrophoresis (Fig. 2B). As with the single hot spot analysis, the turnover of meiotic DSB hot spots was also delayed in the ndj1Δ strain. There was no qualitative difference in timing or levels of breaks at the ends versus the middle regions of chromosome III.
A DNA species hybridizing to the chromosome III telomere proximal probe was absent in the well of the CHEF gel at 0 h but accumulated starting at 2 h in the wild type, reached a maximum at 4 h, and was significantly reduced by 8 h. In both wild-type and ndj1Δ strains, this DNA species was observed at an early time point (2 h), prior to the formation of DSBs, possibly indicating chromosomes undergoing DNA replication that cannot enter the gel due to large replication bubbles (22). If this interpretation is correct, then the kinetics of the appearance of this species in ndj1Δ and wild-type strains suggests that DNA replication timing is unaffected in the absence of Ndj1. We reasoned that the persistence of DNA molecules in wells at late time points (>5 h) likely represented joint molecules produced during recombination or late-replicating DNA. In ndj1Δ strains, this DNA species reached a maximum level at ≥6 h, suggesting that it persisted until late time points. This result suggests that ndj1Δ strains could be defective in joint molecule turnover in the DSB repair pathway of meiotic recombination. As expected for both accumulation of DSBs and the species in the wells, recovery of full-length chromosome III over time was also delayed in the ndj1Δ strain compared to that in the wild type (Fig. 2B).
Formation of the single-end invasion intermediate in the ndj1Δ mutant occurs with wild-type kinetics, while the formation of the double-Holliday junction intermediate is delayed.
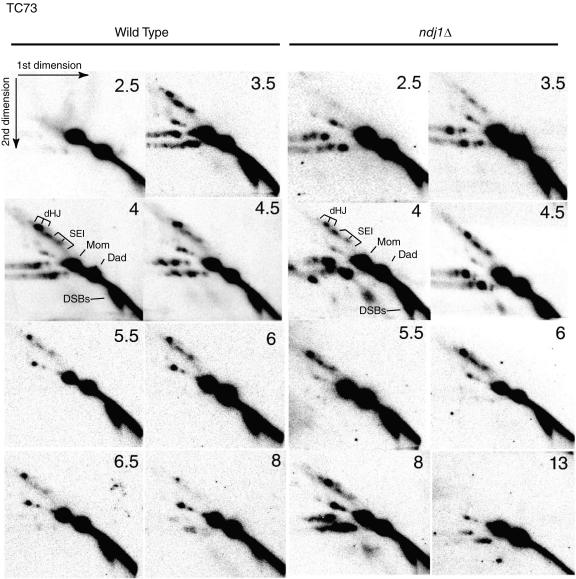
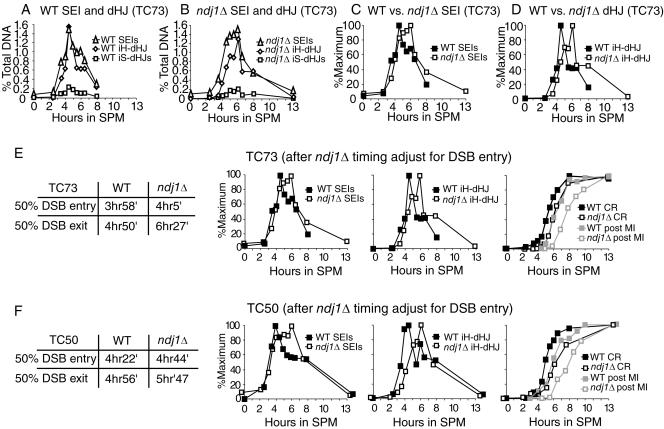
We next addressed the effect of the ndj1Δ mutation on the turnover of detectable intermediates of the CR-I pathway. When strains carrying the well-characterized HIS4::LEU2-(NBam)/his4-X::LEU2-(NBam)-URA3 hot spot (26) are used, a one-dimensional gel can separate a mixture of these intermediates (i.e., SEIs plus dHJs) from their parental strands (Fig. 3A, right), while a two-dimensional gel can separate SEIs, interhomolog dHJs, and intersister (IS) dHJs from one another (Fig. 4) (26, 49). We found that the maximum levels of SEIs and intersister and interhomolog dHJs in the wild type and the ndj1Δ mutant are similar (Fig. 5A and B).
FIG. 4.
Southern analysis of two-dimensional gels of meiotic recombination at HIS4::LEU2/his4-X::LEU2-URA3 hot spot in NDJ1 and the ndj1Δ mutant. Samples are from the same time course experiment (TC73) whose results are shown in Fig. 3.
FIG. 5.
Quantification and adjustment of two-dimensional gel results. (A to D) Two-dimensional gel quantification of TC73. (A) Quantification of joint molecules in NDJ1. (B) Quantification of joint molecules in the ndj1Δ mutant. (C) Normalized SEIs and dHJs of NDJ1 to their maximum levels. (D) Normalized SEIs and dHJs of the ndj1Δ mutant to their maximum levels. TC73 (E) and TC50 (F) entry of meiotic events of NDJ1 and the ndj1Δ mutant after adjustment of the ndj1Δ curves for the delay in DSB entry. DSB entry and exit timing at the HIS4::LEU2 recombination hot spot is shown. The SEI, dHJ, and CR curves of the ndj1Δ mutant were adjusted for the delay of DSB entry in the ndj1Δ mutant. All curves were normalized to the maximum level. In TC50, the maximum levels of SEIs, IH-dHJs, and CRs in the NDJ1 strain were 1.17%, 1.03%, and 20.62%, respectively; the maximum levels of SEIs, IH-dHJs, and CRs in the ndj1Δ mutant were 0.98%, 1.15%, and 17.08%, respectively. Entry and exit timing for these experiments of NDJ1 and ndj1Δ strains was calculated by an algorithm based on that described by Padmore et al. (26, 41). WT, wild type.
In ndj1Δ strains, both intersister and interhomolog dHJ formation was significantly delayed (Fig. 5D), while a slight delay in SEI formation was observed for two independent time course (TC) experiments (Fig. 5C, TC73; data not shown [TC50]). After adjustment for the delay in DSB entry, SEI formation in the ndj1Δ mutant was nearly identical to that in the wild type (Fig. 5E and F). SEIs in the ndj1Δ mutant reached 80 to 90% of their maximum level at the time the maximal level was attained in the wild-type strain (Fig. 5E and F). Notably, the SEI level in ndj1Δ strain persisted at a high level (i.e., >80% of its maximum signal) for about 2 h, while the SEI level in wild-type strain immediately decreased after reaching its peak. By contrast, the overall formation of dHJ molecules was significantly delayed in ndj1Δ strains compared to that in wild-type strains, with the peak of maximal dHJ formation delayed about 2 h.
The observed delay in DSB turnover is consistent with a delay in the transition from the SEI to the dHJ intermediate. The SEI intermediate is a three-armed structure that migrates slower than parental DNA fragments on a one-dimensional gel. The “fourth” arm represents the other side of the DSB that has not achieved a stable interaction with the homologous chromosomes. Thus, for every SEI detected, there should be a corresponding species detected at the position of the DSB.
Implicit in our interpretation is that “on-time” formation of the SEI intermediate in the ndj1Δ mutant represents IH interactions. It is possible that an early product formed between sister chromatids (IS-SEI) could confound this conclusion. We offer two arguments against this interpretation. (i) IS-dHJs do not form early, and their level is not increased in the ndj1Δ mutant (Fig. 5A and B). While an IS-SEI intermediate would not necessarily be converted to an IS-dHJ, the most parsimonious model would anticipate an early and increased level of an IS-dHJ intermediate in the ndj1Δ mutant compared with that in the wild type. (ii) A signal consistent with an IS-SEI intermediate is not observed in the ndj1Δ mutant. IS-SEI intermediates should migrate as 9.11- and 7.2-kb bands compared with 8.7 and 7.4 kb for the IH-SEI intermediates in the first dimension of separation. While these differences are not great, we found no indication of different-sized products in the ndj1Δ mutant at early versus late time points (3.5 versus 8 h and 4 versus 6 h) or between ndj1Δ and wild-type strains at an early time point (4 h) by analyzing superimposed images, using the IH-dHJ intermediate as a point of reference (data not shown).
Both crossover and noncrossover product formation is delayed in the ndj1Δ mutant compared with that in the wild type, but the level of noncrossover product is more severely affected.
CR products arising from the HIS4::LEU2-(NBam)/his4-X::LEU2-(NBam)-URA3 hot spot (TC73, psoralen-treated samples, 350-ml cultures for both the wild type and the ndj1Δ mutant) were delayed about 1.5 h. The final level of CR was reduced to about 70% of wild-type levels in the ndj1Δ mutant (Fig. 3C). The CR levels in NDJ1 and ndj1Δ cells were also determined from six independent small-sized (10 ml) cultures (13 h), without psoralen cross-linking treatment. The CR level (CR divided by total DNA) in the ndj1Δ mutant was found to be 88% of the wild-type level (wild type, 19.4 ± 0.7%; ndj1Δ mutant, 17.1 ± 0.5% [mean ± standard deviation]; P < 0.001 by t test) (data not shown).
While our physical analysis of CR levels showed a modest decrease in the ndj1Δ mutant compared to those in the wild type (70 to 90% of the wild type), a previous study showed that the ndj1Δ mutation conferred no significant effect on crossing over at any one of five genetic intervals (9). While these results may appear at odds with one another, it is possible that a subtle crossover phenotype may have been missed in the genetic analysis since only four-spore viable tetrads were analyzed and the strain under study gave 76% spore viability levels. Moreover, the frequency of E0 tetrads, in which chromosome III is nonrecombinant along its entire length, was elevated 20-fold in the ndj1Δ background (9). The modest defect in CR levels detected physically is therefore likely to be consistent with a high level of E0 tetrads and the levels of nondisjunction and spore inviability associated with the ndj1Δ mutant.
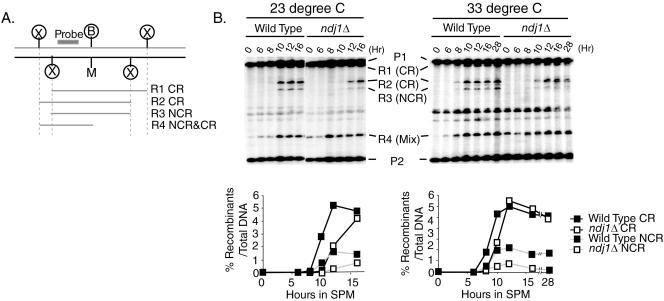
We have previously demonstrated a role for Ndj1 achievement of close, stable homolog juxtaposition during meiosis that functions through an SPO11-dependent pathway and is largely independent of ZMM function (i.e., the CR-I pathway) (42). From this phenotype, we inferred that Ndj1 contributes to close, stable homolog juxtaposition in part through the NCR-plus-CR-II pathway. Here, we directly addressed whether Ndj1 is involved in noncrossover recombination by using the his4-X::LEU2-URA3/his4-B::LEU2 test locus, in which gene conversion events associated with NCRs can be physically assessed by analysis of restriction-length polymorphisms built into the constructs (52). To make comparisons with published zmmΔ mutant phenotypes (6, 52), the ndj1Δ and wild-type CR/NCR phenotypes were examined at two temperatures (23°C and 33°C) (Fig. 6). Experiments presented here were performed as described by Borner et al. (6). We used R2 and R3 (Fig. 5) to represent a fraction of CRs and NCRs, respectively. Interestingly, the formation of both CR and NCR products was delayed at both temperatures. In the ndj1Δ strain, while a significant reduction in CRs was not discernible in this experiment—neither at 23°C nor at 33°C—a strong reduction in NCR levels was observed at both temperatures, particularly at 33°C, where the reduction was greater than 50% of wild-type levels for nearly all time points. The missing NCR events might be repaired from ectopic sequences (e.g., the leu2::hisG locus on chromosome III) or a sister chromatid after a delay. The first possibility is consistent with the previous finding that gene conversions in ndj1Δ meioses are unaffected for heteroalleles at allelic loci but are increased for ectopically located heteroalleles (18).
FIG. 6.
Both CR and NCR formation is delayed in the ndj1Δ mutant. NCRs were significantly reduced compared to CRs in the ndj1Δ mutant. Time course of meiosis of NDJ1 (SBY86) and ndj1Δ (SBY2118) strains. (A) Schematic of the his4-X::LEU2-URA3/his4-B::LEU2 hot spot (52). (B) Timing and levels of CR and NCR formation, probed with a PCR fragment of the 3′ end of the HIS4 open reading frame (1,715 bp to 2,133 bp). DNA samples were digested with XhoI and MluI. A background signal overlapped with the R4 band in this experiment.
Although fewer NCRs were detected in ndj1Δ than in wild-type strains, we cannot completely exclude the possibility that heteroduplex sequences remained in a portion of recombinants in the ndj1Δ mutant. Such hybrid DNA would be inaccessible to restriction digestion and thus would separate as parental-size bands in our assay.
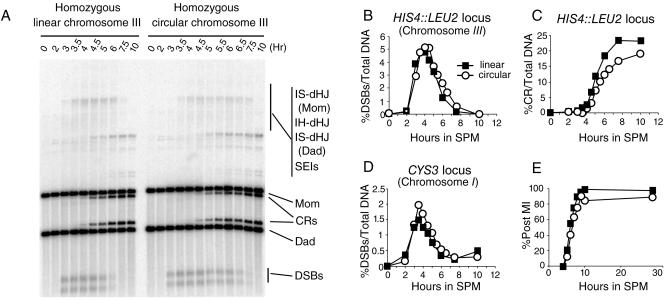
DSB turnover present on a circular version of chromosome III does not phenocopy the DSB turnover defect in the ndj1Δ mutant.
There are two general ways in which the ndj1Δ mutation could confer the mutant phenotypes we observe. First, the effect could act in a chromosome-autonomous fashion in which recombination is affected only on the chromosome on which a telomere is detached from the nuclear envelope. Alternatively, recombination could be affected in a global way, for example, through the general disruption of telomeres, the specific absence of the Ndj1 gene product, or any gene products whose activity is regulated by these factors.
To begin to address these possibilities, we asked whether the presence of telomere sequences on chromosome III would affect recombination at the HIS4::LEU2 hot spot in cis in an NDJ1 strain background. To this end, we constructed diploid strains homozygous for circular versions of chromosome III that lacked telomeric sequences and all sequences distal to the HML and HMR loci, including HMR. A time course study was carried out using large cultures with psoralen cross-linking; post-MI kinetics, DSB turnover, and CR formation at the HIS4::LEU2 locus on chromosome III were measured. As a control, DSB turnover at a hot spot near the CYS3 locus on chromosome I was assayed.
Notably, turnover times of DSBs at both the HIS4::LEU2 locus and the CYS3 locus were slightly increased (Fig. 7) and the post-MI kinetics was similar or slightly delayed for strains homozygous for a circular chromosome III compared with that for strains homozygous for a linear chromosome III (Fig. 7). Moreover, CR formation was slightly delayed and reduced (90% of linear chromosome levels) in strains containing circular chromosomes at the HIS4::LEU2 hot spot compared with that in strains containing the same locus on linear chromosomes. It should be noted that several genes are deleted in the circular chromosome, which could account for the delayed MI kinetics. Together, these results demonstrate that while the circular chromosomes are not entirely neutral during meiosis, they nonetheless do not confer a phenocopy of the ndj1Δ mutation for a DSB hot spot located in cis.
FIG. 7.
Meiotic recombination on circular and linear chromosomes. SBY1903 (wild type) and a strain derived from SBY1903 with homozygous circular chromosome III (SBY2180) were analyzed by time course studies carried out in parallel. Two additional late time points were analyzed for homozygous circular chromosome strains. (A) Southern blot analysis of HIS4::LEU2/his4-X::LEU2-URA3 hot spot. (B) Quantification of DSB turnover results shown in panel A. (C) The kinetics of crossover formation. (D) DSB turnover of the CYS3 hot spot on chromosome I (linear). Samples are from the same time course as that whose results are shown in panel A. Southern blot analysis is not shown. (E) Timing of meiosis I of SBY1903 and SBY2180.
DISCUSSION
The ndj1Δ mutant exhibits a novel recombination phenotype.
Study of meiotic recombination in the budding yeast Saccharomyces cerevisiae has defined many genetic requirements and physical intermediates of the double-strand break repair pathway. Analysis of the recombination hot spot HIS4::LEU2 has been of particular importance, since multiple studies involving a wide range of mutations in otherwise isogenic strains of SK1 in yeast have been compared to one another and to studies of the wild type. The analysis of physical intermediates and products in mutant backgrounds has allowed for an approximate ordering and assignment of recombination intermediates to one of two major pathways of DSB repair, branching from resected DSBs (possibly at the step of strand invasion): (i) a CR-I pathway arising via SEI and dHJ intermediates and (ii) an NCR-plus-CR-II pathway likely arising via the synthesis-dependent strand annealing pathway (see introduction).
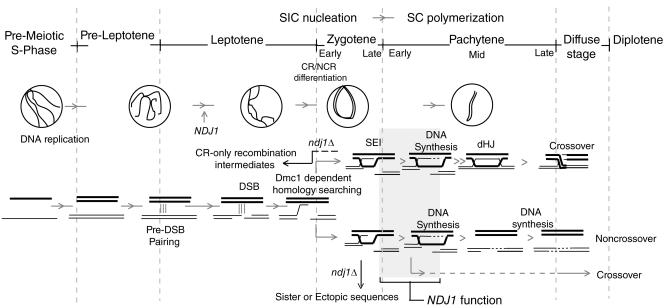
We have shown that the meiosis I delay conferred by the ndj1Δ mutation is fully suppressed by the spo11Δ mutation. Through physical analysis of two variants of the HIS4::LEU2 hot spot, we have observed a recombination phenotype in an ndj1Δ mutant that has not been previously described to date. Interpreting the defect conferred by the ndj1Δ mutation offers several challenges. First, while both CR and NCR recombinants are delayed in the ndj1Δ mutant compared to those in the wild type, NCRs are more-severely reduced than CRs. Second, while DSB turnover is delayed, the next physically detectable intermediate in this pathway, the SEI, appears on time in the mutant compared to that in the wild type, while the later intermediate, the dHJ, is delayed. Overall, these phenotypes suggest that the recombination defect conferred by the ndj1Δ mutation is due to a delay in the transition from SEIs to dHJs in the CR-I pathway and, most likely, an analogous intermediate in the NCR-plus-CR-II pathway. It is attractive to speculate that a process shared in common with both pathways could be attenuated in the ndj1Δ mutant. One such process could be a step related to DNA synthesis following strand invasion (Fig. 8; see below for more discussion). Extension of a 3′ strand invasion intermediate could act to stabilize such intermediates.
FIG. 8.
Ndj1 acts during early meiotic recombination to facilitate turnover of strand invasion intermediates into downstream intermediates. Timeline of events that happen during meiotic I prophase (1, 26, 41, 46, 64). We suggest that the NDJ1 function is required to promote recombination-dependent homolog interaction, possibly by facilitating the DNA synthesis step for both CR-I and NCR-plus-CR-II pathways of meiotic recombination (shaded area). SIC, synapsis initiation complex; SC, synaptonemal complex.
Because the DSB turnover defect in ndj1Δ strains was also observed at two other DSB hot spots and along the length of chromosome III, we suspect that the SEI-to-dHJ transition and the NCR-plus-CR-II pathway events are impaired across the genome.
Independent lines of evidence suggest early CR/NCR determination.
The strong reduction of NCRs at the HIS4::LEU2 locus in the ndj1Δ mutant is particularly interesting compared with that in other mutants analyzed using the same or a similar construct. For example, mutations in ZMM genes are temperature sensitive and result in a significant decrease in CR formation and increase in NCR formation compared to the wild type at high temperatures. By contrast, ndj1Δ phenotypes are not temperature sensitive and result in a substantial reduction in NCR products while CRs are only slightly affected. It has been suggested previously that the fate of each DSB is determined before strand invasion (4, 6, 26). The specific reduction of NCR exhibited by the ndj1Δ mutant in our study provides a new line of evidence that there exist two pools of DSBs: i) those that are committed to the CR-I pathway through the formation of SEIs and dHJs and ii) those that are destined for the NCR pathway.
The reduction in NCRs relative to CRs exhibited by the ndj1Δ mutant can be explained by an inherent instability of strand invasion intermediates so that they decompose to their respective DSB-like intermediates (Fig. 8). DSB CR-I intermediates would then remain committed to the SEI pathway, while DSB NCR-plus-CR-II intermediates would proceed into the synthesis-dependent strand annealing pathway using a either a homolog, a sister chromatid, or an ectopic sequence sharing DNA homology. In either case, the results are explained by the CR- and NCR-determined intermediates being distinguished at a very early stage (i.e., at or prior to strand invasion).
A major difference between zmmΔ and ndj1Δ mutants at high temperatures is that intermediates originally destined for the CR-I pathway can be shunted to the NCR pathway in zmmΔ mutants while in the ndj1Δ mutant, it appears as though the accumulated intermediate is not amenable to switching to the other branch, i.e., the NCR-plus-CR-II pathway. The supposition that Ndj1 also functions in the NCR pathway is further supported by data measuring close, stable homolog juxtaposition using Cre-loxP site-specific recombination. Double mutants containing an ndj1Δ and zmmΔ mutation give a more severe reduction in allelic interaction than either single mutant alone, suggesting that the two classes of genes function, at least in part, in different pathways (42).
Role of the bouquet in meiosis of S. cerevisiae.
It has been widely held that the bouquet plays a role in the early homology search by aligning allelic sequences in close juxtaposition in the space of the nucleus. For Sordaria, this view has been challenged by the observation that presynaptic coalignment of homologs takes place prior to bouquet formation (51, 63). Cytological evidence from various organisms also implies that the homology search has been accomplished before the bouquet stage (35, 64). In mice, short stretches of homologous chromosomes interact in regions where Rad51 foci are found during leptotene, at a time likely prior to bouquet formation (54).
Here, we show by eliminating the attachment of telomeres to the nuclear envelope (and thus, the formation of the bouquet) using the ndj1Δ mutant that the progression from SEI to dHJ intermediates of meiotic recombination is delayed compared with that using the wild type. By contrast, SEIs form at around the same time and levels in the ndj1Δ mutant as they do in the wild type. Therefore, early stages of homology recognition in budding yeast meiosis, as in Sordaria and mice, do not appear to require the formation of the bouquet.
How does NDJ1 affect meiotic recombination?
How does a mutation deleting a telomere protein affect recombination events tens or hundreds of kilobases away from telomeres? We propose here that stabilization of a strand invasion intermediate(s) may be defective in the ndj1Δ mutant. This might be achieved by one or more of several mechanisms. First, Ndj1, or a cofactor, could be directly involved in the biochemistry of meiotic recombination. While prominent accumulation of Ndj1 has been observed at telomeres by immunofluorescence analysis, it is possible that lower levels of Ndj1 protein function at sites of DSB repair. For example, Rap1 localizes predominantly at telomeres but is also found at interstitial sites when chromatin immunoprecipitation analysis is used (33).
Second, it is conceivable that detachment of telomeres from the nuclear envelope could cause mislocalization of one or more factors involved in the regulation or the biochemistry of homologous recombination. The composition of proteins at the telomere is dynamic during the cell cycle (32, 50). Some telomere proteins relocalize to DNA damage sites and presumably play roles in DNA damage repair (7, 61). Thus, telomeres could act as a repository for regulating free protein levels of damage response factors. Indeed, the raison d'être of a telomere is to promote elongation of the chromosome end by DNA replication. Polymerase δ, the same polymerase shown to be involved in gene conversion and crossing over during meiosis (38), is also required for telomerase-mediated telomere addition (i.e., lagging strand synthesis) (15). Perhaps a connection between telomere integrity and availability of these enzymes in the nucleus exists.
The failure to detect delayed CR formation on circular chromosomes indicates that the presence of telomeres in cis does not appear to promote homologous recombination at a locus on the same chromosome. Two possibilities could account for this observation: i) that Ndj1 regulates the nucleus-wide concentration or activity of a telomere-binding protein that participates in the SEI-to-dHJ transition (described above), perhaps through the combined function of the 120 remaining chromosome ends in the nucleus, or ii) that an element on the circular chromosome acting in cis (e.g., the centromere [31, 59] or sequence which directs the chromosome to the nuclear envelope) is redundant with Ndj1 function.
Third, Ndj1-dependent chromosome movement requiring telomere attachment to the nuclear envelope might select for strong interactions between homologous chromosomes. Alternatively, limiting Ndj1-dependent chromosome movement might facilitate the stabilization of a primer/template for a period of time sufficient to ensure the efficient repair of DSBs during meiosis. In this case, mobility of the circular chromosomes could be influenced by the bulk inertia imposed by the 30 remaining linear chromosomes. Future studies will be directed toward distinguishing between direct and indirect roles of Ndj1 and/or telomere dynamics in promoting meiotic recombination.
Acknowledgments
We thank Burgess Lab members, Elizabeth Blackburn, JoAnne Engebrecht, Neil Hunter, Nancy Kleckner, Scott Keeney, and Michael Lichten, for discussion. We especially thank Neil Hunter for strains and advice on protocols. We thank JoAnne Engebrecht, Joshua Chang Mell, and Scott Keeney for critical reading of the manuscript.
This work was supported by the American Cancer Society RSG-01-053-01-CCG award to S.M.B.
There are no conflicts of interest.
REFERENCES
- 1.Allers, T., and M. Lichten. 2001. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106:47-57. [DOI] [PubMed] [Google Scholar]
- 2.Alpi, A., P. Pasierbek, A. Gartner, and J. Loidl. 2003. Genetic and cytological characterization of the recombination protein RAD-51 in Caenorhabditis elegans. Chromosoma 112:6-16. [DOI] [PubMed] [Google Scholar]
- 3.Bergerat, A., B. de Massy, D. Gadelle, P.-C. Varoutas, A. Nicolas, and P. Forterre. 1997. An atypical topoisomerase II from archaea with implications for meiotic recombination. Nature 386:414-417. [DOI] [PubMed] [Google Scholar]
- 4.Bishop, D. K., and D. Zickler. 2004. Early decision: meiotic crossover interference prior to stable strand exchange and synapsis. Cell 117:9-15. [DOI] [PubMed] [Google Scholar]
- 5.Borde, V., T.-C. Wu, and M. Lichten. 1999. Use of a recombination reporter insert to define meiotic recombination domains on chromosome III of Saccharomyces cerevisiae. Mol. Cell. Biol. 19:4832-4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borner, G. V., N. Kleckner, and N. Hunter. 2004. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117:29-45. [DOI] [PubMed] [Google Scholar]
- 7.Bradshaw, P. S., D. J. Stavropoulos, and M. S. Meyn. 2005. Human telomeric protein TRF2 associates with genomic double-strand breaks as an early response to DNA damage. Nat. Genet. 37:193-197. [DOI] [PubMed] [Google Scholar]
- 8.Chikashige, Y., D.-Q. Ding, H. Funabiki, T. Haraguchi, S. Mashiko, M. Yanagida, and Y. Hiraoka. 1994. Telomere-led premeiotic chromosome movement in fission yeast. Science 264:270-273. [DOI] [PubMed] [Google Scholar]
- 9.Chua, P., and G. Roeder. 1997. Tam1, a telomere-associated meiotic protein, functions in chromosome synapsis and crossover interference. Genes Dev. 11:1786-1800. [DOI] [PubMed] [Google Scholar]
- 10.Conrad, M. N., A. M. Dominguez, and M. E. Dresser. 1997. Ndj1p, a meiotic telomere protein required for normal chromosome synapsis and segregation in yeast. Science 276:1252-1255. [DOI] [PubMed] [Google Scholar]
- 11.Cooper, J. P., Y. Watanabe, and P. Nurse. 1998. Fission yeast Taz1 protein is required for meiotic telomere clustering and recombination. Nature 392:828-831. [DOI] [PubMed] [Google Scholar]
- 12.Davis, L., and G. R. Smith. 2001. Meiotic recombination and chromosome segregation in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 98:8395-8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de los Santos, T., N. Hunter, C. Lee, B. Larkin, J. Loidl, and N. M. Hollingsworth. 2003. The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics 164:81-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dernburg, A. F., J. W. Sedat, W. Z. Cande, and H. W. Bass. 1995. The cytology of telomeres, p. 295-338. In E. H. Blackburn and C. W. Greider (ed.), Telomere. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Diede, S. J., and D. E. Gottschling. 1999. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell 99:723-733. [DOI] [PubMed] [Google Scholar]
- 16.Ding, D.-Q., A. Yamamoto, T. Haraguchi, and Y. Hiraoka. 2004. Dynamics of homologous chromosome pairing during meiotic prophase in fission yeast. Dev. Cell 6:329-341. [DOI] [PubMed] [Google Scholar]
- 17.Feuerbach, F., V. Galy, E. Trelles-Sticken, M. Fromont-Racine, A. Jacquier, E. Gilson, J.-C. Olivo-Marin, H. Scherthan, and U. Nehrbass. 2002. Nuclear architecture and spatial positioning help establish transcriptional states of telomeres in yeast. Nat. Cell Biol. 4:214-221. [DOI] [PubMed] [Google Scholar]
- 18.Goldman, A. S. H., and M. Lichten. 2000. Restriction of ectopic recombination by interhomolog interactions during Saccharomyces cerevisiae meiosis. Proc. Natl. Acad. Sci. USA 97:9537-9542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldstein, A. L., and H. J. McCusker. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15:1541-1553. [DOI] [PubMed] [Google Scholar]
- 20.Golubovskaya, I. N., L. C. Harper, W. P. Pawlowski, D. Schichnes, and W. Z. Cande. 2002. The pam1 gene is required for meiotic bouquet formation and efficient homologous synapsis in maize (Zea mays L.). Genetics 162:1979-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hassold, T., and P. Hunt. 2001. To err (meiotically) is human: the genesis of human aneuploidy. Nat. Rev. Genet. 2:280-291. [DOI] [PubMed] [Google Scholar]
- 22.Hennessy, K. M., A. Lee, E. Chen, and D. Botstein. 1991. A group of interacting yeast DNA replication genes. Genes Dev. 5:958-969. [DOI] [PubMed] [Google Scholar]
- 23.Heyer, W.-D., K. T. Ehmsen, and J. A. Solinger. 2003. Holliday junctions in the eukaryotic nucleus: resolution in sight? Trends Biochem. Sci. 28:548-557. [DOI] [PubMed] [Google Scholar]
- 24.Hiraoka, Y. 1998. Meiotic telomeres: a matchmaker for homologous chromosomes. Genes Cells 3:405-413. [DOI] [PubMed] [Google Scholar]
- 25.Hollingsworth, N. M., and S. J. Brill. 2004. The Mus81 solution to resolution: generating meiotic crossovers without Holliday junctions. Genes Dev. 18:117-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hunter, N., and N. Kleckner. 2001. The single-end invasion: an asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell 106:59-70. [DOI] [PubMed] [Google Scholar]
- 27.Joseph, I., D. Jia, and A. J. Lustig. 2005. Ndj1p-dependent epigenetic resetting of telomere size in yeast meiosis. Curr. Biol. 15:231-237. [DOI] [PubMed] [Google Scholar]
- 28.Kane, S. M., and R. Roth. 1974. Carbohydrate metabolism during ascospore development in yeast. J. Bacteriol. 118:8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kee, K., and S. Keeney. 2002. Functional interactions between SPO11 and REC102 during initiation of meiotic recombination in Saccharomyces cerevisiae. Genetics 160:111-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keeney, S., C. N. Giroux, and N. Kleckner. 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88:375-384. [DOI] [PubMed] [Google Scholar]
- 31.Kemp, B., R. M. Boumil, M. N. Stewart, and D. S. Dawson. 2004. A role for centromere pairing in meiotic chromosome segregation. Genes Dev. 18:1946-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laroche, T., S. G. Martin, M. Tsai-Pflugfelder, and S. M. Gasser. 2000. The dynamics of yeast telomeres and silencing proteins through the cell cycle. J. Struct. Biol. 129:159-174. [DOI] [PubMed] [Google Scholar]
- 33.Lieb, J. D., X. Liu, D. Botstein, and P. O. Brown. 2001. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat. Genet. 28:327-334. [DOI] [PubMed] [Google Scholar]
- 34.Liu, L., S. Franco, B. Spyropoulos, P. B. Moens, M. A. Blasco, and D. L. Keefe. 2004. Irregular telomeres impair meiotic synapsis and recombination in mice. Proc. Natl. Acad. Sci. USA 101:6496-6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loidl, J. 1990. The initiation of meiotic chromosome pairing: the cytological view. Genome 33:759-778. [DOI] [PubMed] [Google Scholar]
- 36.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 37.Lydall, D., Y. Nikolsky, D. Bishop, and T. Weinert. 1996. A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature 383:840-843. [DOI] [PubMed] [Google Scholar]
- 38.Maloisel, L., J. Bhargava, and G. S. Roeder. 2004. A role for DNA polymerase delta in gene conversion and crossing over during meiosis in Saccharomyces cerevisiae. Genetics 167:1133-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miki, F., K. Okazaki, M. Shimanuki, A. Yamamoto, Y. Hiraoka, and O. Niwa. 2002. The 14-kDa dynein light chain-family protein Dlc1 is required for regular oscillatory nuclear movement and efficient recombination during meiotic prophase in fission yeast. Mol. Biol. Cell 13:930-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nautiyal, S., J. L. DeRisi, and E. H. Blackburn. 2002. The genome-wide expression response to telomerase deletion in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 99:9316-9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Padmore, R., L. Cao, and N. Kleckner. 1991. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell 66:1239-1256. [DOI] [PubMed] [Google Scholar]
- 42.Peoples-Holst, T. L., and S. M. Burgess. 2005. Multiple branches of the meiotic recombination pathway contribute independently to homolog pairing and stable juxtaposition during meiosis in budding yeast. Genes Dev. 19:863-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petronczki, M., M. F. Siomos, and K. Nasmyth. 2003. Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell 112:423-440. [DOI] [PubMed] [Google Scholar]
- 44.Rockmill, B., and G. S. Roeder. 1998. Telomere-mediated chromosome pairing during meiosis in budding yeast. Genes Dev. 12:2574-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roeder, G. S. 1997. Meiotic chromosomes: it takes two to tango. Genes Dev. 11:2600-2621. [DOI] [PubMed] [Google Scholar]
- 46.Scherthan, H. 2001. A bouquet makes ends meet. Nat. Rev. Mol. Cell Biol. 2:621-627. [DOI] [PubMed] [Google Scholar]
- 47.Schwacha, A., and N. Kleckner. 1995. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell 83:783-791. [DOI] [PubMed] [Google Scholar]
- 48.Schwacha, A., and N. Kleckner. 1994. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell 76:51-63. [DOI] [PubMed] [Google Scholar]
- 49.Schwacha, A., and N. Kleckner. 1997. Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell 90:1123-1135. [DOI] [PubMed] [Google Scholar]
- 50.Smith, C. D., D. L. Smith, J. L. DeRisi, and E. H. Blackburn. 2003. Telomeric protein distributions and remodeling through the cell cycle in Saccharomyces cerevisiae. Mol. Biol. Cell 14:556-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Storlazzi, A., S. Tesse, S. Gargano, F. James, N. Kleckner, and D. Zickler. 2003. Meiotic double-strand breaks at the interface of chromosome movement, chromosome remodeling, and reductional division. Genes Dev. 17:2675-2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Storlazzi, A., L. Xu, L. Cao, and N. Kleckner. 1995. Crossover and noncrossover recombination during meiosis: timing and pathway relationships. Proc. Natl. Acad. Sci. USA 92:8512-8516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Storlazzi, A., L. Xu, A. Schwacha, and N. Kleckner. 1996. Synaptonemal complex (SC) component Zip1 plays a role in meiotic recombination independent of SC polymerization along the chromosomes. Proc. Natl. Acad. Sci. USA 93:9043-9048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tarsounas, M., T. Morita, R. E. Pearlman, and P. B. Moens. 1999. RAD51 and DMC1 form mixed complexes associated with mouse meiotic chromosome cores and synaptonemal complexes. J. Cell Biol. 147:207-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trelles-Sticken, E., C. Adelfalk, J. Loidl, and H. Scherthan. 2005. Meiotic telomere clustering requires actin for its formation and cohesin for its resolution. J. Cell Biol. 170:213-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trelles-Sticken, E., M. E. Dresser, and H. Scherthan. 2000. Meiotic telomere protein Ndj1p is required for meiosis-specific telomere distribution, bouquet formation and efficient homologue pairing. J. Cell Biol. 151:95-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trelles-Sticken, E., J. Loidl, and H. Scherthan. 1999. Bouquet formation in budding yeast: initiation of recombination is not required for meiotic telomere clustering. J. Cell Sci. 112:651-658. [DOI] [PubMed] [Google Scholar]
- 58.Trelles-Sticken, E., J. Loidl, and H. Scherthan. 2003. Increased ploidy and KAR3 and SIR3 disruption alter the dynamics of meiotic chromosomes and telomeres. J. Cell Sci. 116:2431-2442. [DOI] [PubMed] [Google Scholar]
- 59.Tsubouchi, T., and G. S. Roeder. 2005. A synaptonemal complex protein promotes homology-independent centromere coupling. Science 308:870-873. [DOI] [PubMed] [Google Scholar]
- 60.Villeneuve, A. M., and K. J. Hillers. 2001. Whence meiosis? Cell 106:647-650. [DOI] [PubMed] [Google Scholar]
- 61.Xu, L., and E. H. Blackburn. 2004. Human Rif1 protein binds aberrant telomeres and aligns along anaphase midzone microtubules. J. Cell Biol. 167:819-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamamoto, A., R. R. West, J. R. McIntosh, and Y. Hiraoka. 1999. A cytoplasmic dynein heavy chain is required for oscillatory nuclear movement of meiotic prophase and efficient meiotic recombination in fission yeast. J. Cell Biol. 145:1233-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zickler, D. 1977. Development of the synaptonemal complex and the “recombination nodules” during meiotic prophase in the seven bivalents of the fungus Sordaria macrospora Auersw. Chromosoma 61:289-316. [DOI] [PubMed] [Google Scholar]
- 64.Zickler, D., and N. Kleckner. 1998. The leptotene-zygotene transition of meiosis. Annu. Rev. Genet. 32:619-697. [DOI] [PubMed] [Google Scholar]