Abstract
The Nrf2 transcription factor is a key player in the cellular stress response through its regulation of cytoprotective genes. In this study we determined the role of Nrf2-mediated gene expression in keratinocytes for skin development, wound repair, and skin carcinogenesis. To overcome compensation by the related Nrf1 and Nrf3 proteins, we expressed a dominant-negative Nrf2 mutant (dnNrf2) in the epidermis of transgenic mice. The functionality of the transgene product was verified in vivo using mice doubly transgenic for dnNrf2 and an Nrf2-responsive reporter gene. Surprisingly, no abnormalities of the epidermis were observed in dnNrf2-transgenic mice, and even full-thickness skin wounds healed normally. However, the onset, incidence, and multiplicity of chemically induced skin papillomas were strikingly enhanced, whereas the progression to squamous cell carcinomas was unaltered. We provide evidence that the enhanced tumorigenesis results from reduced basal expression of cytoprotective Nrf target genes, leading to accumulation of oxidative damage and reduced carcinogen detoxification. Our results reveal a crucial role of Nrf-mediated gene expression in keratinocytes in the prevention of skin tumors and suggest that activation of Nrf2 in keratinocytes is a promising strategy to prevent carcinogenesis of this highly exposed organ.
Due to its accessibility, the skin is permanently exposed to harmful environmental influences, such as UV irradiation and noxious xenobiotics. Many of these induce the formation of reactive oxygen species (ROS), e.g., hydrogen peroxide, oxyradicals, or organic hydroperoxides. Endogenous ROS are generated in the course of common metabolic processes, and particularly large amounts are produced in wounded and inflamed tissues, where they are released by activated macrophages and neutrophils as a defense against microbial infection. Since excessive accumulation of ROS causes cell aging, severe cell damage, and even neoplastic transformation, cells need to develop strategies to protect themselves against these insults (4, 16, 48).
Of particular importance is the expression of ROS-detoxifying enzymes. Interestingly, many of them as well as other cytoprotective proteins are controlled by NF-E2-related factor 2 (Nrf2). The latter is a member of the “cap ‘n’ collar” family of transcription factors (40), which, among others, also includes the closely related proteins Nrf1 and Nrf3 (5, 6, 31). Together with their heteromeric interaction partners, the small Maf proteins, Nrf1, Nrf2, and Nrf3 bind to cis-acting elements in the promoters of target genes, called antioxidant response elements (AREs) or electrophile response elements (42). At least the binding of Nrf1 and Nrf2 activates the expression of these genes (34), which encode for example NAD(P)H:quinone oxidoreductase 1 (NQO1), several glutathione S-transferases (GSTs), γ-glutamylcysteine synthetase heavy subunit (γ-GCSh) and light subunit, heme oxygenase 1 (HO-1), and peroxiredoxin 1 (42).
Under normal conditions, Nrf2 is sequestered in the cytosol via binding to the actin-binding protein Keap1 (26), which also mediates its degradation through the ubiquitin-proteasome pathway (12, 32). Upon addition of electrophilic substances, which directly interact with Keap1 through Michael addition, and possibly through oxidation of Keap1 via ROS, Nrf2 becomes liberated and shuttles to the nucleus, where it activates its target genes. In addition, phosphorylation of Nrf2 by different kinases can also result in liberation from Keap1 (25, 43).
The important role of Nrf2 in the cellular stress response is reflected by the phenotype of Nrf2 knockout mice. Whereas young Nrf2 knockout animals have no obvious phenotype under normal laboratory conditions (7), older mice develop a severe autoimmune disease resembling systemic lupus erythematosus (36). Furthermore, even young Nrf2 null mice are highly susceptible to electrophilic and oxidative stress exerted by various chemicals (25).
Finally, our recent studies suggested a role of Nrf2 in tissue repair. Thus, it is strongly expressed in macrophages and keratinocytes of healing skin wounds. We showed that Nrf2 knockout mice have a prolonged inflammatory response after skin injury (3), most likely reflecting the important role of Nrf2 in macrophages (24). By contrast, no abnormalities in the epidermis were observed, possibly due to up-regulation of Nrf3 (3). To determine the effect of Nrf2 exclusively in keratinocytes and to overcome a possible compensatory effect exerted by Nrf3 and/or Nrf1, we generated and analyzed transgenic mice expressing a dominant-negative mutant of Nrf2 (dnNrf2) in basal keratinocytes of the epidermis. Our results reveal that Nrf-mediated gene expression in keratinocytes is dispensable for wound healing but crucial for skin tumor prevention.
MATERIALS AND METHODS
Plasmid construction.
The cDNA encoding dnNrf2 was amplified by PCR from cDNA of mouse skin wounds using oligonucleotides 5′-TCCCCCCGGGCCACCATGCGTGAATCCCAATG-3′ and 5′-CGGGATCCATCCTCCCGAAC-3′, subcloned, and completely sequenced. It was then inserted into an expression cassette, which includes a 2-kb human keratin 14 (K14) promoter followed by a 0.65-kb rabbit β-globin intron and a transcription termination/polyadenylation fragment (0.65 kb) of the human growth hormone gene (41).
Generation and identification of transgenic mice.
Fertilized eggs were obtained following superovulation and mating of FVB/N females. The 3.8-kb insert was separated from vector sequences, purified, and injected into the pronuclei of one-cell-stage embryos. Microinjected zygotes were transferred into the oviducts of pseudopregnant recipient females. Mouse tail DNA was analyzed for integration of the transgene by PCR using oligonucleotides specific for the rabbit β-globin intron fragment, since they do not cross-hybridize with chromosomal mouse DNA.
RNA isolation and RPA.
RNA isolation and RNase protection assay (RPA) were carried out as described previously (9, 53). The following templates were used: nucleotides (nt) 320 to 543 of the murine cyclooxygenase 2 (COX2) cDNA (accession no. NM_011198), nt 85 to 296 of the murine γ-GCSh cDNA (U85414), nt 160 to 436 of the murine GST-Ya cDNA (NM_008181), nt 659 to 898 of the NQO1 cDNA (NM_008706), nt 1816 to 1983 of the murine Nrf2 cDNA (BC026943), and fragments specific for murine interleukin 6 (IL-6) (3), murine HO-1 (19), and murine IL-1β (3). As a loading control the RNA was hybridized with a probe for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (nt 566 to 685 of the cDNA; NM_008084).
Histological analysis.
Back skin, tail skin (separated from the bone), or wound tissue was fixed overnight at 4°C in 4% paraformaldehyde in phosphate-buffered saline (PBS) and embedded in paraffin. Sections (6 μm) were stained with hematoxylin and eosin.
Wounding and preparation of wound tissues.
Mice 10 to 12 weeks old were anesthetized with a single intraperitoneal injection of ketamine-xylazine. For each time point of wound healing at least four mice were injured, and two independent experiments were performed. Four full-thickness excisional wounds (5-mm diameter, 3 to 4 mm apart) were generated on the shaved back, two on each side of the spinal cord, by excising skin and panniculus carnosus as described previously (52). The wounds were allowed to dry to form a scab. The complete wounds were isolated including 2 mm of the wound margins. Wounds used for RNA isolation were frozen in liquid nitrogen and stored at −80°C until further processing. For histological analysis bisected wounds were fixed overnight in 4% paraformaldehyde in PBS or in 95% ethanol-1% acetic acid, followed by paraffin embedding. Sections (6 μm) from the middle of the wound were stained with hematoxylin and eosin or used for immunofluorescence. Only littermates of the same sex were used for direct histological comparison. All experiments with animals were carried out with permission from the local veterinary authorities.
Topical application of tBHQ to hyperproliferative epidermis.
Mice 10 to 12 weeks old were anesthetized (see above), and the dorsal region was shaved and treated with a depilatory agent (Pilca). For the treatment of hyperproliferative epidermis with tert-butylhydroquinone (tBHQ) two full-thickness incisions (1 cm) were made at one anterior and one posterior dorsal site and the skin margins were closed with strips of a wound plaster (Fixomull; Beiersdorf, Hamburg, Germany). Four days after wounding, the skin area adjacent to the incisions was treated three times with a 2:1 mixture of tBHQ (100 mM dissolved in dimethyl sulfoxide [DMSO]) and hydrophilic cream within a period of 24 h. Two h after the last application the mice were killed, and the treated skin area was embedded in tissue freezing medium (Jung, Nussloch, Germany) without prior fixation. Twelve-μm frozen sections were used for human placental alkaline phosphatase (hPAP) staining using the nitroblue tetrazolium method as described below.
Immunofluorescence.
Ethanol-acetic acid-fixed paraffin sections (6 μm) from tail skin were incubated overnight at 4°C with the primary antibodies diluted in PBS containing 3% bovine serum albumin and 0.025% NP-40. After three 10-min washes with PBS-0.1% Tween 20, sections were incubated for 1 h with the secondary antibodies, washed again, mounted with Mowiol (Hoechst, Frankfurt, Germany), and photographed with a Zeiss Axioplan fluorescence microscope. We used a goat polyclonal antibody directed against pankeratin (Abcam, Cambridge, United Kingdom), a rabbit polyclonal anti-K14 antibody (BabCO, Richmond, CA), and a rabbit polyclonal anti-Nrf2 antibody (3). All secondary antibodies (coupled to Cy2 or Cy3) were from Jackson ImmunoResearch (West Grove, PA). To visualize cleaved caspase 3, methanol-fixed cryosections (7 μm) were used and treated as described above using a rabbit polyclonal antibody (Cell Signaling Technology, Beverly, MA).
Preparation of protein lysates and Western blot analysis.
Cultured cells were lysed in 20 mM Tris-HCl (pH 8.0), 1% (vol/vol) Triton X-100, 137 mM NaCl, 2 mM EDTA, 10% (vol/vol) glycerol, 0.5 mM aminoethylbenzenesulfonyl fluoride, and 0.15 U/ml aprotinin. Preparation of tissue lysates from skin was performed as described previously (53). Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to nitrocellulose filters. Antibody incubations were performed in 5% nonfat dry milk in TBS-T (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.05% Tween 20). The following antibodies were used: anti-β-actin (Sigma), anti-Cdk4 (Santa Cruz), anti-IκBα (Calbiochem), anti-RelA (Santa Cruz), anti-lamin A (Santa Cruz), anti-Nrf2 (3), anti-p42/44 mitogen-activated protein kinase (Cell Signaling Technology), anti-phospho-p42/44 mitogen-activated protein kinase (Cell Signaling Technology), and anti-phospho-Jun N-terminal protein kinase (anti-phospho-JNK; Cell Signaling Technology).
Oxyblot.
Oxidized proteins were detected using the Oxyblot assay kit (Chemicon, Temecula, CA) according to the manufacturer's instructions. The method is based on the detection of protein carbonyl groups derivatized with 2,4-dinitrophenylhydrazine to convert carbonyl groups to dinitrophenylhydrazone derivatives. After derivatization protein samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and subsequently analyzed by Western blotting using an antibody against dinitrophenylhydrazone.
hPAP histochemistry.
Frozen sections were fixed with 4% paraformaldehyde in PBS and subsequently incubated in TMN buffer (50 mM Tris, 5 mM MgCl2, 100 mM NaCl [pH 10]) at 65°C for 15 min to heat inactivate endogenous alkaline phosphatase. After being washed with PBS sections were stained for hPAP activity by replacing the TMN with a staining solution containing 1 mg/ml of nitroblue tetrazolium and 1 mg/ml 5-bromo-4-chloro-3-indolylphosphate (X-phosphate; Promega, Wisconsin) and incubating the mixture at 37°C for 30 to 45 min. In combination with immunofluorescence hPAP activity was visualized using the VECTOR Red alkaline phosphatase substrate kit (Vector Laboratories, Burlingame, CA) as recommended by the manufacturer.
Transient transfection of COS-1 cells.
COS-1 cells were transiently transfected using the calcium phosphate method (8). Briefly, 3 × 105 COS-1 cells in 60-mm dishes were transfected with 4 to 6 μg plasmid DNA in a calcium phosphate precipitate. After 5 h of incubation cells were washed once with 3 ml HEPES buffer (142 mM NaCl, 10 mM HEPES, 6.7 mM KCl, pH 7.3) and cultured for 2 days in Dulbecco modified Eagle medium-10% fetal calf serum before harvesting.
Establishment of primary mouse keratinocytes and tBHQ treatment.
Murine epidermal keratinocytes were isolated as described previously (3) with the exception that cells were seeded at a density of 5 × 104 cells per cm2 on collagen IV (2.5 μg per cm2)-coated dishes (17). Subsequently, cells were incubated for 20 min at 37°C, and the medium was replaced afterwards. For each experiment, keratinocytes were freshly isolated, grown to confluence in defined keratinocyte serum-free medium (Gibco, BRL) supplemented with 10 ng/ml of epidermal growth factor and 10−10 M cholera toxin, and rendered quiescent by incubation in defined keratinocyte serum-free basal medium without additives. Subsequently, the medium was replaced by the same medium containing tBHQ (Acros, New Jersey), and the cells were harvested for RNA isolation.
Two-stage skin carcinogenesis.
Female 7- to 9-week-old K14-dnNrf2 or wild-type mice were shaved, and 25 μg 7,12-dimethylbenz(a)anthracene (DMBA) dissolved in 200 μl acetone or acetone alone was applied topically to the shaved area on the back. One week later 5 μg 12-O-tetradecanoylphorbol-13-acetate (TPA) dissolved in 200 μl acetone or acetone alone was applied to the same site once a week for 20 weeks. Mice were weekly checked for occurrence and number of papillomas. To determine the rate of progression to squamous cell carcinomas, some of the animals were maintained for up to 15 weeks after the last TPA treatment. The number of papillomas and squamous cell carcinomas per mouse was determined, and the type of tumor was verified by histopathological analysis. To monitor ARE activation, ARE-hPAP mice were subjected to the same procedure, with the exception that at different time points after DMBA treatment and at different time points during promotion animals were sacrificed and analyzed for hPAP activity in the epidermis.
RESULTS
Generation of transgenic mice expressing dnNrf2 in the epidermis.
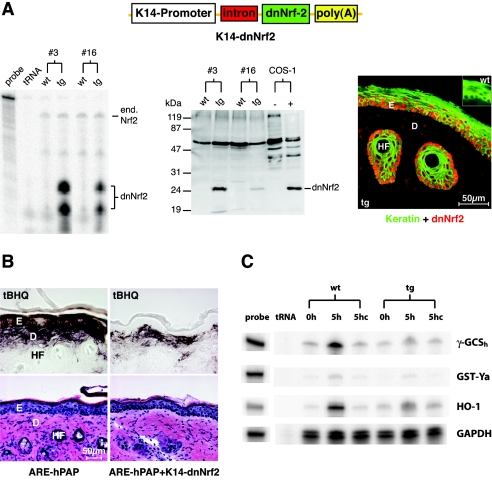
The analysis of Nrf2 knockout mice suggested a possible redundancy and/or compensation among the Nrf transcription factors (3). To circumvent this problem and to specifically determine the role of ARE-dependent transcription in keratinocytes, we expressed a dominant-negative mutant of Nrf2 (dnNrf2) (1) under the control of the keratin 14 (K14) promoter in the epidermis of transgenic mice (Fig. 1A) using a well-characterized expression cassette (41, 51). The K14 promoter targets transgenes to basal cells of stratified epithelia, particularly to the epidermis and to outer root sheath cells of the hair follicles (50). The dominant-negative mutant lacks both transactivation domains and the Keap1 binding domain but includes the DNA binding domain. Therefore, it is expected to continuously bind to AREs, thereby competing with endogenous Nrf transcription factors. This mutant had previously been characterized in vitro (1). In spite of the similarity between AREs and AP1 sites (42), AP1 responsiveness is not affected by dnNrf2 (data not shown). Two transgenic founder mice with different integration sites of the transgene were generated, which strongly overexpress the mutant compared to the endogenous Nrf2 as shown by RNase protection assay and Western blotting (Fig. 1A, left and middle panels). Immunofluorescence analysis revealed the expected expression and nuclear localization of dnNrf2 in the skin (Fig. 1A, right panel, red).
FIG. 1.
Expression and functionality of dnNrf2 in the skin of transgenic mice. (A) Left panel: 20 μg total cellular RNA from the skin of dnNrf2 transgenic mice (lines 3 and 16) and of wild-type littermates was analyzed by RNase protection assay for the expression of endogenous Nrf2 and dnNrf2 using a riboprobe that can distinguish between RNAs encoding the wild-type and the mutant proteins. One thousand counts per minute of the hybridization probe was loaded in the lane labeled “probe” and used as a size marker. Twenty micrograms of tRNA was used as a negative control. Middle panel: protein lysates from the skin of dnNrf2 transgenic mice (lines 3 and 16) and of wild-type littermates were analyzed by Western blotting for the presence of the dnNrf2 protein. Lysates from COS-1 cells transiently transfected with the transgene construct were used as a positive control. The sensitivity of the blot is insufficient to detect the endogenous Nrf2. Right panel: immunofluorescence analysis of tail skin of transgenic mice using antibodies against Nrf2 (red) and pankeratin (green). The transgene construct is shown above. (B) Alkaline phosphatase (upper panels) and hematoxylin and eosin (lower panels) staining of hyperproliferative back skin of ARE-hPAP (left panels) or ARE-hPAP/K14-dnNrf2 (right panels) double-transgenic mice topically treated with tBHQ. The staining in double-transgenic animals reveals the specific inhibition of Nrf2 action in the epidermis. D, dermis, E, epidermis, HF, hair follicle. (C) Primary keratinocytes from dnNrf2 transgenic mice and wild-type littermates were treated with tBHQ (50 μM) (5h) or DMSO (5hc) for 5 h. Ten μg total RNA was analyzed by RPA for the expression of γ-GCSh, GST-Ya, HO-1, and GAPDH.
To verify the dominant-negative effect of the transgene, we mated our K14-dnNrf2 mice with transgenic ARE reporter mice. The latter harbor a reporter construct in their genome that includes a 51-bp ARE-containing fragment of the rat NQO1 promoter upstream of an initiator element (Inr)-containing minimal promoter, followed by the hPAP cDNA (27). Therefore, they allow monitoring of ARE activation in vivo by staining for hPAP activity. Since Nrf2 is highly expressed in hyperproliferative keratinocytes (3, 46), we generated incisional wounds on the back of ARE transgenic animals and treated the hyperproliferative skin adjacent to the wound site with the electrophilic Nrf2-activator tBHQ or the solvent DMSO. There was a strong hPAP staining in keratinocytes of the hyperproliferative epidermis as well as in cells of the underlying dermis in single-transgenic ARE reporter mice, whereas the staining in double-transgenic mice (ARE-hPAP/K14-dnNrf2) was restricted to the mesenchyme (Fig. 1B). No staining was seen in any of these mice upon treatment with DMSO (data not shown). These results demonstrate that Nrf2 can be activated in keratinocytes of hyperproliferative skin and confirm the efficient blockade of ARE-dependent gene expression in K14-dnNrf2 mice.
To further determine the functionality of the transgene, we cultured primary keratinocytes from wild-type and dnNrf2-transgenic mice and treated them with tBHQ. The expression of known Nrf2 target genes (γ-GCSh, GST-Ya, and HO-1) in quiescent and tBHQ-treated primary keratinocytes was determined by RPA. A strong induction of these genes was observed in cells derived from wild-type mice but not in those from transgenic littermates (Fig. 1C).
dnNrf2 does not affect skin morphogenesis.
K14-dnNrf2 mice were macroscopically normal, and they had no defects in fertility. Histologically, no abnormalities in tail and back skin of these mice were detectable (data not shown), and the keratinocyte differentiation markers keratins 14, 10, and 6 were normally expressed in the epidermis (data not shown). These results reveal that Nrf-mediated gene expression in keratinocytes is dispensable for epidermal morphogenesis.
Normal wound healing in K14-dnNrf2 transgenic mice.
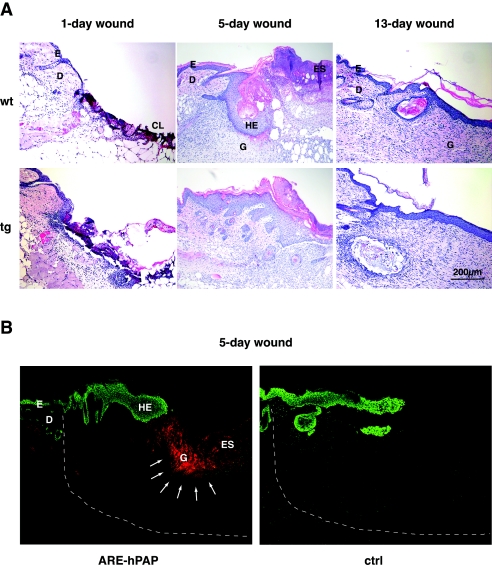
We next examined if our transgenic mice revealed phenotypic abnormalities under stress conditions. Therefore, we first subjected them to skin injury, since high levels of ROS are produced in wounded skin by inflammatory cells. However, no histological abnormalities were observed at any stage of the healing process of full-thickness excisional wounds (Fig. 2A). In particular, reepithelialization occurred normally, and 13 days after injury wounds in control and transgenic mice were fully reepithelialized. These results were unexpected, because Nrf2 is highly expressed in the hyperproliferative wound epithelium (3, 46). Since Nrf2 activity is regulated at the posttranscriptional level (25), we wondered whether the insult to keratinocytes in the hyperproliferative epithelium during the normal wound healing process is sufficient to activate Nrf2. For this purpose we performed wound healing studies in ARE reporter mice. Consistent with the lack of abnormalities in wound reepithelialization in K14-dnNrf2 mice, we could not see activation of the reporter in the wound epidermis (Fig. 2B). However, there was a strong induction of reporter activity in cells of the granulation tissue (Fig. 2B), which predominantly represent inflammatory cells (data not shown).
FIG. 2.
Normal wound healing in K14-dnNrf2 mice and ARE activation in murine skin wounds. (A) Full-thickness excisional wounds were made on the back of wild-type (wt) and K14-dnNrf2 transgenic (tg) mice. Sections from the middle of 1-day, 5-day, and 13-day wounds were stained with hematoxylin and eosin. (B) Wounds were made on the back of ARE reporter mice and wild-type littermates. Sections from the middle of 5-day wounds were stained for hPAP activity. Keratinocytes were visualized by keratin 14 staining (green). Note the hPAP staining (red) of cells in the granulation tissue (indicated by arrows) and the lack of hPAP activity in the hyperproliferative epithelium. CL, clot; D, dermis; E, epidermis; ES, eschar; HE, hyperproliferative epithelium; G, granulation tissue. The dashed white line indicates the border of the wound area.
Enhanced chemically induced skin carcinogenesis in K14-dnNrf2 mice.
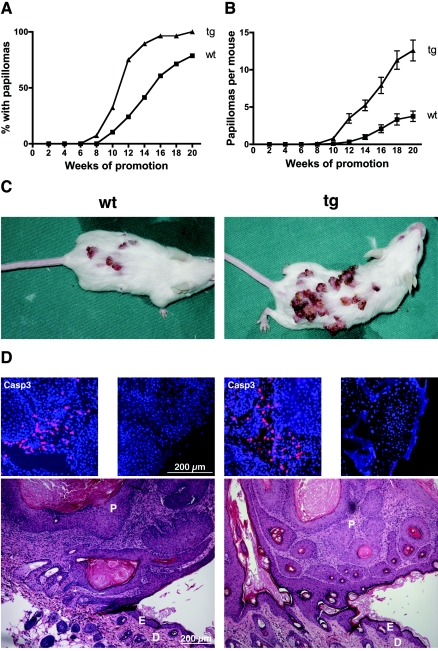
We subsequently analyzed the consequences of a blockade of Nrf-mediated gene expression in keratinocytes in a situation with abnormal and continuous hyperproliferation of the epidermis. For this purpose we used the well-characterized model of two-stage chemical carcinogenesis in mouse skin (14, 54). K14-dnNrf2 mice (n = 28) started to show papillomas already at 8 weeks after the first TPA treatment, whereas age- and sex-matched wild-type mice (n = 28) remained papilloma free up to this time point (Fig. 3A). At week 16 almost 100% of transgenic mice had developed papillomas, in comparison to only 60% of control mice (Fig. 3A). Furthermore, the number of papillomas per mouse was significantly higher in transgenic mice than in wild-type animals (Fig. 3B and C). Mice of both genotypes which were treated with only DMBA or TPA did not develop tumors, demonstrating that both a mutagenic insult and a proliferative stimulus are required (data not shown). No obvious histological differences were observed between the papillomas of wild-type and transgenic mice (Fig. 3D), and there were no differences in the number of apoptotic or proliferating cells between wild-type and transgenic mice—neither in the tumors nor in nontumorigenic skin after the first or last TPA treatment (Fig. 3D; data not shown).
FIG. 3.
Enhanced tumor susceptibility of K14-dnNrf2 transgenic mice. Back skin of transgenic mice and wild-type littermates was treated once topically with DMBA in acetone. Subsequently, the animals were treated weekly with TPA in acetone for 20 weeks. (A) Tumor incidence. The number of animals with papillomas was determined every week. The graph shows the percentages of animals with papillomas (n = 28 per genotype). (B) Tumor multiplicity. The number of papillomas per mouse was determined every week. The graph represents the averages of 28 animals per genotype. Error bars indicate the standard errors of the means. (C) Representative DMBA/TPA-treated wild-type or transgenic mice are shown. (D) Sections from papillomas from transgenic animals and wild-type littermates were stained with hematoxylin and eosin. Papillomas from both genotypes show characteristic cellular atypisms and inflammatory infiltrates. D, dermis; E, epidermis; P, papilloma. Stainings of tumor sections with an antibody against cleaved caspase 3 are shown above the histological pictures (left picture, middle of the tumor; right picture, edge of the tumor).
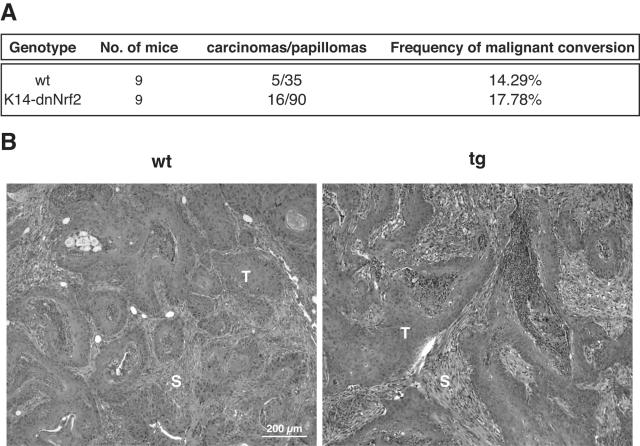
We also determined the rate of progression of papillomas to squamous cell carcinomas in nine wild-type animals and nine transgenic animals. Malignant conversion was observed in a subset of papillomas from mice of both genotypes, but the frequency of malignant conversion was not affected by the dominant-negative Nrf2 mutant (Fig. 4).
FIG. 4.
Inhibition of Nrf2-mediated gene expression does not affect the frequency of malignant conversion of skin tumors. Progression from papillomas to squamous cell carcinomas was monitored in nine wild-type and nine transgenic mice. (A) The ratio of carcinomas to papillomas at week 15 after the last TPA treatment as well as the frequency of malignant conversion is indicated. (B) Sections from squamous cell carcinomas of a wild-type mouse and a transgenic mouse were stained with hematoxylin and eosin. The tumors were identified as squamous cell carcinomas by a histopathologist. S, stroma; T, tumor cells.
To determine if the enhanced tumorigenesis in dnNrf2 transgenic mice results from reduced basal or inducible expression of Nrf target genes, we subjected ARE reporter mice to the two-stage skin carcinogenesis protocol. Activation of the reporter was determined 6, 12, 24, and 48 h after DMBA treatment and at different time points after TPA treatment (1 to 16 weeks). To our surprise, ARE reporter activation was not detectable at any time point (Fig. 5 and data not shown), although tBHQ treatment of hyperproliferative skin efficiently activated the reporter (Fig. 1B). This finding strongly suggests that the basal rather than the inducible expression of Nrf target genes is responsible for the tumor-preventive effect of Nrf transcription factors in the skin.
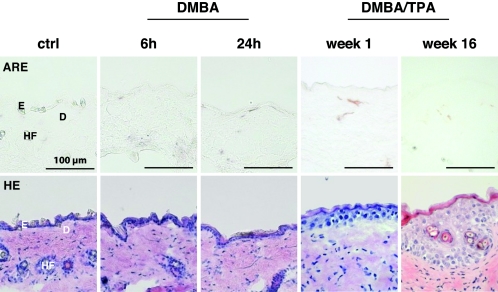
FIG. 5.
Lack of ARE activation by DMBA and TPA during the development of skin papillomas. ARE reporter mice were subjected to the two-stage skin carcinogenesis protocol. Sections from DMBA- and DMBA/TPA-treated skin were analyzed by alkaline phosphatase staining to determine ARE reporter activity (upper panels) or stained with hematoxylin and eosin (lower panels). D, dermis; E, epidermis; HF, hair follicle.
Although enhanced tumorigenesis is often a result of increased inflammation (11, 29), the inflammatory infiltrate and the expression of the proinflammatory cytokines IL-1β, IL-6, and tumor necrosis factor alpha (TNF-α) was rather reduced than enhanced in the papillomas of dnNrf2 transgenic mice (Fig. 6A; data not shown).
FIG. 6.
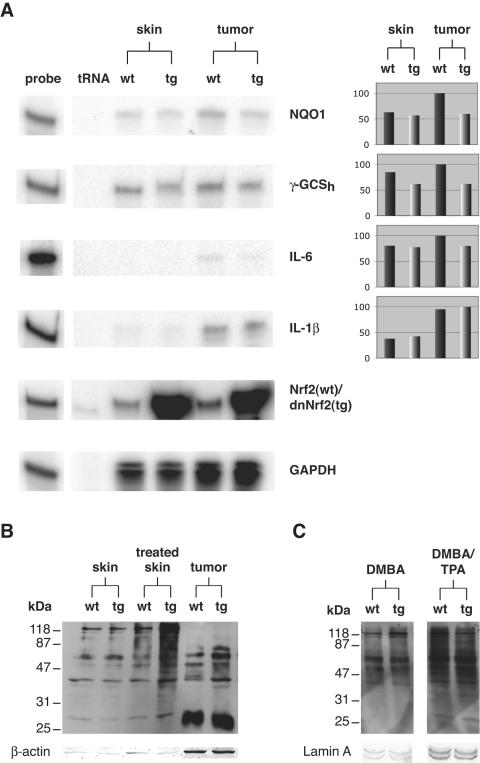
Reduced expression of cytoprotective target genes and enhanced oxidative stress in skin and papillomas of dnNrf2 transgenic mice. (A) Samples of 20 μg total RNA from nontumorigenic skin (pools of five biopsy samples per genotype) and from papillomas (pools of five biopsy samples per genotype) of wild-type and transgenic mice were analyzed by RPA for the presence of NQO1, γ-GCSh, IL-6, IL-1β, Nrf2, and GAPDH mRNAs. Twenty micrograms of tRNA was used as a negative control. One thousand cpm of the hybridization probes was loaded in the lanes labeled “probe” and used as a size marker. Densitometric quantification of each RNase protection assay result (normalized to GAPDH) is shown on the right side. The strongest signal was arbitrarily set as 100. (B) Thirteen micrograms of total protein from untreated skin, from nontumorigenic skin which was treated for 20 weeks with DMBA/TPA, and from papillomas of wild-type and transgenic animals was analyzed for the presence of oxidized proteins by Oxyblot analysis. (C) Twenty micrograms of total protein from DMBA-treated skin of wild-type and transgenic mice and 7.5 μg total protein from skin, which was treated once with DMBA and once with TPA were analyzed for the presence of oxidized proteins by Oxyblot analysis. Staining of the same membrane with an antibody directed against β-actin served as a loading control in panel B. Staining with an antibody against lamin A served as a loading control in panel C.
Blockage of the NF-κB signaling pathway with concomitant up-regulation of Cdk4 and enhanced JNK activity has been implicated in higher susceptibility to skin carcinogenesis (13, 38, 57, 58). However, Western blot analysis revealed that the levels of RelA, IκBα, Cdk4, and phospho-JNK were similar in wild-type and dnNrf2 transgenic mice, in both nontumorigenic and tumorigenic skin (see Fig. S1A in the supplemental material). Furthermore, expression of the NF-κB target gene cyclooxygenase 2 (cox-2) was unaltered in nontumorigenic and tumorigenic skin of our transgenic mice compared to control littermates as determined by RNase protection assay (see Fig. S1B in the supplemental material).
Therefore, the most likely mechanism underlying the enhanced tumorigenesis in K14-dnNrf2 transgenic mice is reduced basal expression of Nrf target genes. Indeed, expression of NQO1, which is involved in DMBA detoxification and in the prevention of ROS accumulation (44), and of γ-GCSh, the rate-limiting enzyme in glutathione biosynthesis, was reduced in DMBA/TPA-treated nontumorigenic skin as well as in papillomas of transgenic mice (Fig. 6A). Furthermore, we found higher levels of oxidized proteins in papillomas and adjacent nontumorigenic skin of our transgenic animals (Fig. 6B). By contrast, no obvious differences in the levels of oxidized proteins were observed in nontreated skin of mice from both genotypes (Fig. 6B, left lanes) as well as after the first DMBA or TPA treatment (Fig. 6C). Therefore, accumulation of oxidative damage in combination with reduced DMBA detoxification is most likely responsible for the increased tumor susceptibility of K14-dnNrf2 mice.
DISCUSSION
The cytoprotective transcription factor Nrf2 is strongly expressed in the hyperproliferative epithelium of skin wounds and in epidermal cancer (3, 46). In addition, Nrf1 and Nrf3 are also expressed in keratinocytes, in particular in Nrf2 knockout mice, suggesting functional redundancy/compensation (3). Therefore, we attempted to inhibit the activity of all Nrf transcription factors in the epidermis using a dominant-negative Nrf2 mutant. The latter was targeted to keratinocytes to specifically determine the role of Nrf transcription factors in this cell type.
Several findings confirm that the activity of Nrf transcription factors is efficiently inhibited in keratinocytes of K14-dnNrf2 transgenic mice: (i) the same mutant was shown to inhibit binding of Nrf transcription factors to the common ARE (1), (ii) expression of dnNrf2 was more than 10-fold higher than that of the endogenous Nrf2 (Fig. 1A), (iii) dnNrf2 was present in the nucleus of keratinocytes (Fig. 1A), (iv) tBHQ-mediated induction of Nrf2 target genes was inhibited in cultured keratinocytes of K14-dnNrf2 transgenic mice (Fig. 1C), and (v) mice doubly transgenic for dnNrf2 and an ARE-driven reporter gene failed to activate the reporter in the epidermis under conditions where single ARE reporter mice showed strong activation (Fig. 1B). In spite of the similarity between AREs and AP1 sites (42), AP1-mediated gene expression was not affected as demonstrated for HaCaT cells stably transfected with a dnNrf2 expression vector (data not shown). Furthermore, the phenotype seen in our transgenic mice was different from the phenotype of mice lacking c-jun or both c-jun and junB in the epidermis (35, 55, 56). In conclusion, Nrf-mediated gene expression is efficiently and specifically suppressed in keratinocytes of K14-dnNrf2 transgenic mice.
Surprisingly, dnNrf2-transgenic mice revealed no obvious phenotypic abnormalities, suggesting that Nrf2-mediated gene expression in keratinocytes is important only under stress conditions. To address this question, we performed wound healing studies in our transgenic mice, since Nrf2 and Nrf3 are highly expressed in the hyperproliferative wound epidermis (3) and since high amounts of ROS-producing inflammatory cells are present in the healing skin wound (10). To our surprise, however, wound reepithelialization proceeded normally in dnNrf2 transgenic mice (Fig. 2A), demonstrating that the strong expression of Nrf2 in the wound epidermis is obviously not a prerequisite for healing under normal laboratory conditions.
The normal reepithelialization seen in our transgenic mice and also in Nrf2 null mice (3) suggested that Nrf2 is not activated in keratinocytes of the wound epidermis (Fig. 2B). This hypothesis was proven through wounding of ARE reporter mice where no hPAP staining was observed in keratinocytes, even when the inflammatory response was enhanced by addition of neutrophil chemokines (data not shown). Consistent with this observation, we did not observe ARE activation in cultured keratinocytes from ARE reporter mice upon addition of hydrogen peroxide. By contrast, only electrophilic agents, which can directly couple to Keap1 via Michael addition, induced ARE-mediated gene expression in cultured keratinocytes. A similar result was obtained in vivo, where tBHQ and other agents that can undergo Michael addition, but not hydrogen peroxide, DMBA, or TPA, activated ARE-mediated gene expression in keratinocytes of hyperproliferative skin (Fig. 5 and data not shown). This is different from reports from other cells, where Nrf2 activation was achieved, e.g., by hydrogen peroxide, peroxynitrate, or ROS generated by particles of diesel exhaust (2, 23, 28). Interestingly, we found ARE activation in cells of the wound granulation tissue, which most likely represent inflammatory cells. These findings reveal cell-type-specific differences in ARE activation and indicate that Nrf2 is activated in keratinocytes only by electrophilic compounds. Thus, a major function of Nrf2 in the epidermis may be the protection from toxic environmental chemicals.
The most striking finding of our study was the enhanced onset, incidence, and multiplicity of skin tumors in K14-dnNrf2 transgenic mice (Fig. 3). This result is consistent with the observed susceptibility of Nrf2 knockout mice to acetaminophen-mediated liver cancer and indicates a general tumor-preventive role of Nrf2 (15). However, our study is the first demonstration that Nrf-mediated gene expression in epithelial cells is crucial for skin tumor prevention and that Nrf-mediated gene expression in stromal cells is not required for this protective effect. In spite of the enhanced formation of papillomas, progression to squamous cell carcinomas was not altered by dnNrf2 (Fig. 4), demonstrating that Nrf2-mediated gene expression protects from the development of skin tumors but not from malignant progression of existing benign tumors.
The enhanced skin carcinogenesis is obviously not due to excessive inflammation or to alterations in NF-κB signaling. No activation of the latter was observed, and expression of the NF-κB target gene cyclooxygenase 2 was also unaltered (see Fig. S1 in the supplemental material). This result confirms previous studies, which showed that enhanced levels of ROS, as seen in the papillomas and the adjacent nontumorigenic skin of our transgenic animals, do not result in NF-κB activation (20). However, our results strongly suggest that reduced expression of cytoprotective Nrf2 target genes is responsible for the phenotype. The latter encode proteins which are involved in the detoxification of the mutagen DMBA and/or of ROS. Indeed, we found reduced expression of NQO1, a DMBA-detoxifying enzyme (45, 49), in papillomas as well as in the adjacent nontumorigenic, DMBA/TPA-treated skin of transgenic mice (Fig. 6). This down-regulation is of obvious functional importance, since skin carcinogenesis was also enhanced in NQO1 knockout mice (39). However, the effect was less severe than in K14-dnNrf2 mice, demonstrating that other Nrf2 targets also contribute to the phenotype of our animals. One of them may be GST-π, another DMBA-detoxifying enzyme, which is important for skin tumor prevention (21). Indeed, mice lacking GST-π were also more susceptible to DMBA/TPA-induced carcinogenesis (21).
Finally, expression of enzymes involved in ROS detoxification was also reduced in DMBA/TPA-treated skin and in papillomas of our transgenic animals. The latter include γ-GCS, the rate-limiting enzyme in glutathione biosynthesis, as well as NQO1, which prevents accumulation of ROS through its ability to circumvent redox cycling of quinones (44). Thus, the reduced expression of these enzymes results in enhanced oxidative stress in the presence of DMBA and TPA as demonstrated by the elevated levels of oxidized proteins in the long-term DMBA/TPA-treated skin and in papillomas of our K14-dnNrf2 transgenic animals. The reduced detoxification of DMBA as well as of ROS is likely to enhance the rate of mutations in keratinocytes, including activating mutations in the ha-ras proto-oncogene. Furthermore, ROS accumulation during the promotion phase will speed up tumorigenesis. A comprehensive scheme of the proposed model of skin tumor prevention by Nrf transcription factors is shown in Fig. 7.
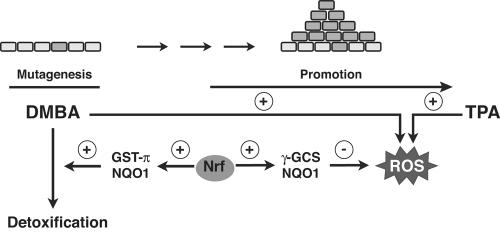
FIG. 7.
Model for skin tumor prevention by Nrf transcription factors. The carcinogen DMBA causes mutations in the DNA of keratinocytes, including a critical mutation in the ha-ras proto-oncogene (30). Detoxification of DMBA is at least in part achieved by the Nrf targets NQO1 and GST-π (21, 45). On the other hand, DMBA as well as TPA triggers the generation of ROS, which further enhance the rate of mutagenesis. In addition, ROS enforce the activity of TPA as a tumor-promoting agent (48). Nrf transcription factors allow ROS detoxification through positive regulation of ROS-detoxifying enzymes, such as γ-GCS and NQO1.
The contribution of the individual members of the Nrf family to the tumor-preventive effect remains to be determined. However, we recently mated ARE-hPAP reporter mice with Nrf2 knockout mice and observed that the lack of Nrf2 completely inhibited the tBHQ-induced activation of the reporter in keratinocytes and fibroblasts (see Fig. S2 in the supplemental material). This finding demonstrates that Nrf2 comprises the major ARE-binding activity upon activation by tBHQ. Nevertheless, it is still possible that in addition to Nrf2 the basal activities of the other family members also contribute to this effect or that other stimuli are required to activate these factors.
It has been suggested that the toxin-inducible expression of cytoprotective Nrf2 target genes is responsible for the tumor-suppressive effect (47). To our surprise, however, we did not observe ARE activation in our reporter mice, which were also subjected to the chemical carcinogenesis protocol (Fig. 5). Since Nrf2 also regulates the basal expression of several target genes (33), we propose that the long-term reduction in the basal levels of ROS-detoxifying enzymes results in a continuous increase in intracellular ROS as previously shown for the livers of Nrf2 knockout mice upon aging (36). Together with reduced detoxification of DMBA, the accumulation of oxidative damage is likely to result in enhanced tumorigenesis.
In summary, we identified a crucial role of Nrf-mediated gene expression in skin tumor prevention. This finding suggests that activation of Nrf2 in keratinocytes, e.g., by synthetic triterpenoids or avicins (18, 22, 37), could be used for the protection of this highly exposed organ from environmental carcinogenic insults.
Supplementary Material
Acknowledgments
We thank Yuet Wai Kan for providing Nrf2 knockout mice, Thomas Rülicke for generation of the transgenic mice, Moritz Durchdewald for help with the experiment shown in Fig. S2 in the supplemental material, and Christiane Born-Berclaz for excellent technical assistance.
This work was supported by the Swiss National Science Foundation (grants no. 31-61358.00 and 3100A0-109340/1 to S.W. and 3100AO-105459 to M.H. and D.H.), the AETAS foundation (to S.W.), a Boehringer-Ingelheim predoctoral fellowship (to U.A.D.K.), and a postdoctoral fellowship from the Deutsche Forschungsgemeinschaft (to C.S.).
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Alam, J., D. Stewart, C. Touchard, S. Boinapally, A. M. Choi, and J. L. Cook. 1999. Nrf2, a Cap′n'Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J. Biol. Chem. 274:26071-26078. [DOI] [PubMed] [Google Scholar]
- 2.Baulig, A., M. Garlatti, V. Bonvallot, A. Marchand, R. Barouki, F. Marano, and A. Baeza-Squiban. 2003. Involvement of reactive oxygen species in the metabolic pathways triggered by diesel exhaust particles in human airway epithelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 285:L671-L679. [DOI] [PubMed] [Google Scholar]
- 3.Braun, S., C. Hanselmann, M. G. Gassmann, U. auf dem Keller, C. Born-Berclaz, K. Chan, Y. W. Kan, and S. Werner. 2002. Nrf2 transcription factor, a novel target of keratinocyte growth factor action which regulates gene expression and inflammation in the healing skin wound. Mol. Cell. Biol. 22:5492-5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cerutti, P. A., and B. F. Trump. 1991. Inflammation and oxidative stress in carcinogenesis. Cancer Cells 3:1-7. [PubMed] [Google Scholar]
- 5.Chan, J. Y., X. L. Han, and Y. W. Kan. 1993. Cloning of Nrf1, an NF-E2-related transcription factor, by genetic selection in yeast. Proc. Natl. Acad. Sci. USA 90:11371-11375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan, J. Y., X. L. Han, and Y. W. Kan. 1993. Isolation of cDNA encoding the human NF-E2 protein. Proc. Natl. Acad. Sci. USA 90:11366-11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan, K., R. Lu, J. C. Chang, and Y. W. Kan. 1996. NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc. Natl. Acad. Sci. USA 93:13943-13948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, C., and H. Okayama. 1987. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7:2745-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 10.Clark, R. A. F. 1996. Wound repair. Overview and general considerations, p. 3-50. In R. A. F. Clark (ed.), The molecular and cellular biology of wound repair, 2nd ed. Plenum Press, New York, N.Y.
- 11.Coussens, L. M., and Z. Werb. 2002. Inflammation and cancer. Nature 420:860-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cullinan, S. B., J. D. Gordan, J. Jin, J. W. Harper, and J. A. Diehl. 2004. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol. Cell. Biol. 24:8477-8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dajee, M., M. Lazarov, J. Y. Zhang, T. Cai, C. L. Green, A. J. Russell, M. P. Marinkovich, S. Tao, Q. Lin, Y. Kubo, and P. A. Khavari. 2003. NF-κB blockade and oncogenic Ras trigger invasive human epidermal neoplasia. Nature 421:639-643. [DOI] [PubMed] [Google Scholar]
- 14.DiGiovanni, J. 1992. Multistage carcinogenesis in mouse skin. Pharmacol. Ther. 54:63-128. [DOI] [PubMed] [Google Scholar]
- 15.Enomoto, A., K. Itoh, E. Nagayoshi, J. Haruta, T. Kimura, T. O'Connor, T. Harada, and M. Yamamoto. 2001. High sensitivity of Nrf2 knockout mice to acetaminophen hepatotoxicity associated with decreased expression of ARE-regulated drug metabolizing enzymes and antioxidant genes. Toxicol. Sci. 59:169-177. [DOI] [PubMed] [Google Scholar]
- 16.Fridovich, I. 1978. The biology of oxygen radicals. Science 201:875-880. [DOI] [PubMed] [Google Scholar]
- 17.Hager, B., J. R. Bickenbach, and P. Fleckman. 1999. Long-term culture of murine epidermal keratinocytes. J. Investig. Dermatol. 112:971-976. [DOI] [PubMed] [Google Scholar]
- 18.Hanausek, M., P. Ganesh, Z. Walaszek, C. J. Arntzen, T. J. Slaga, and J. U. Gutterman. 2001. Avicins, a family of triterpenoid saponins from Acacia victoriae (Bentham), suppress H-ras mutations and aneuploidy in a murine skin carcinogenesis model. Proc. Natl. Acad. Sci. USA 98:11551-11556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanselmann, C., C. Mauch, and S. Werner. 2001. Haem oxygenase-1: a novel player in cutaneous wound repair and psoriasis? Biochem. J. 353:459-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayakawa, M., H. Miyashita, I. Sakamoto, M. Kitagawa, H. Tanaka, H. Yasuda, M. Karin, and K. Kikugawa. 2003. Evidence that reactive oxygen species do not mediate NF-κB activation. EMBO J. 22:3356-3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henderson, C. J., A. G. Smith, J. Ure, K. Brown, E. J. Bacon, and C. R. Wolf. 1998. Increased skin tumorigenesis in mice lacking pi class glutathione S-transferases. Proc. Natl. Acad. Sci. USA 95:5275-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hyer, M. L., R. Croxton, M. Krajewska, S. Krajewski, C. L. Kress, M. Lu, N. Suh, M. B. Sporn, V. L. Cryns, J. M. Zapata, and J. C. Reed. 2005. Synthetic triterpenoids cooperate with tumor necrosis factor-related apoptosis-inducing ligand to induce apoptosis of breast cancer cells. Cancer Res. 65:4799-4808. [DOI] [PubMed] [Google Scholar]
- 23.Ishii, T., K. Itoh, S. Takahashi, H. Sato, T. Yanagawa, Y. Katoh, S. Bannai, and M. Yamamoto. 2000. Transcription factor Nrf2 coordinately regulates a group of oxidative stress-inducible genes in macrophages. J. Biol. Chem. 275:16023-16029. [DOI] [PubMed] [Google Scholar]
- 24.Itoh, K., M. Mochizuki, Y. Ishii, T. Ishii, T. Shibata, Y. Kawamoto, V. Kelly, K. Sekizawa, K. Uchida, and M. Yamamoto. 2004. Transcription factor Nrf2 regulates inflammation by mediating the effect of 15-deoxy-Δ12,14-prostaglandin J2. Mol. Cell. Biol. 24:36-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Itoh, K., K. I. Tong, and M. Yamamoto. 2004. Molecular mechanism activating Nrf2-Keap1 pathway in regulation of adaptive response to electrophiles. Free Radic. Biol. Med. 36:1208-1213. [DOI] [PubMed] [Google Scholar]
- 26.Itoh, K., N. Wakabayashi, Y. Katoh, T. Ishii, K. Igarashi, J. D. Engel, and M. Yamamoto. 1999. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 13:76-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson, D. A., G. K. Andrews, W. Xu, and J. A. Johnson. 2002. Activation of the antioxidant response element in primary cortical neuronal cultures derived from transgenic reporter mice. J. Neurochem. 81:1233-1241. [DOI] [PubMed] [Google Scholar]
- 28.Kang, K. W., S. H. Choi, and S. G. Kim. 2002. Peroxynitrite activates NF-E2-related factor 2/antioxidant response element through the pathway of phosphatidylinositol 3-kinase: the role of nitric oxide synthase in rat glutathione S-transferase A2 induction. Nitric Oxide 7:244-253. [DOI] [PubMed] [Google Scholar]
- 29.Karin, M. 2005. Inflammation and cancer: the long reach of Ras. Nat. Med. 11:20-21. [DOI] [PubMed] [Google Scholar]
- 30.Kemp, C. J. 2005. Multistep skin cancer in mice as a model to study the evolution of cancer cells. Semin. Cancer Biol. 15:460-473. [DOI] [PubMed] [Google Scholar]
- 31.Kobayashi, A., E. Ito, T. Toki, K. Kogame, S. Takahashi, K. Igarashi, N. Hayashi, and M. Yamamoto. 1999. Molecular cloning and functional characterization of a new Cap ‘n’ collar family transcription factor Nrf3. J. Biol. Chem. 274:6443-6452. [DOI] [PubMed] [Google Scholar]
- 32.Kobayashi, A., M. I. Kang, H. Okawa, M. Ohtsuji, Y. Zenke, T. Chiba, K. Igarashi, and M. Yamamoto. 2004. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 24:7130-7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, J. M., M. J. Calkins, K. Chan, Y. W. Kan, and J. A. Johnson. 2003. Identification of the NF-E2-related factor-2-dependent genes conferring protection against oxidative stress in primary cortical astrocytes using oligonucleotide microarray analysis. J. Biol. Chem. 278:12029-12038. [DOI] [PubMed] [Google Scholar]
- 34.Leung, L., M. Kwong, S. Hou, C. Lee, and J. Y. Chan. 2003. Deficiency of the Nrf1 and Nrf2 transcription factors results in early embryonic lethality and severe oxidative stress. J. Biol. Chem. 278:48021-48029. [DOI] [PubMed] [Google Scholar]
- 35.Li, G., C. Gustafson-Brown, S. K. Hanks, K. Nason, J. M. Arbeit, K. Pogliano, R. M. Wisdom, and R. S. Johnson. 2003. c-Jun is essential for organization of the epidermal leading edge. Dev. Cell 4:865-877. [DOI] [PubMed] [Google Scholar]
- 36.Li, J., T. D. Stein, and J. A. Johnson. 2004. Genetic dissection of systemic autoimmune disease in Nrf2-deficient mice. Physiol. Genomics 18:261-272. [DOI] [PubMed] [Google Scholar]
- 37.Liby, K., T. Hock, M. M. Yore, N. Suh, A. E. Place, R. Risingsong, C. R. Williams, D. B. Royce, T. Honda, Y. Honda, G. W. Gribble, N. Hill-Kapturczak, A. Agarwal, and M. B. Sporn. 2005. The synthetic triterpenoids, CDDO and CDDO-imidazolide, are potent inducers of heme oxygenase-1 and Nrf2/ARE signaling. Cancer Res. 65:4789-4798. [DOI] [PubMed] [Google Scholar]
- 38.Lind, M. H., B. Rozell, R. P. Wallin, M. van Hogerlinden, H. G. Ljunggren, R. Toftgard, and I. Sur. 2004. Tumor necrosis factor receptor 1-mediated signaling is required for skin cancer development induced by NF-κB inhibition. Proc. Natl. Acad. Sci. USA 101:4972-4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Long, D. J., II, R. L. Waikel, X. J. Wang, D. R. Roop, and A. K. Jaiswal. 2001. NAD(P)H:quinone oxidoreductase 1 deficiency and increased susceptibility to 7,12-dimethylbenz[a]-anthracene-induced carcinogenesis in mouse skin. J. Natl. Cancer Inst. 93:1166-1170. [DOI] [PubMed] [Google Scholar]
- 40.Moi, P., K. Chan, I. Asunis, A. Cao, and Y. W. Kan. 1994. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc. Natl. Acad. Sci. USA 91:9926-9930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Munz, B., H. Smola, F. Engelhardt, K. Bleuel, M. Brauchle, I. Lein, L. W. Evans, D. Huylebroeck, R. Balling, and S. Werner. 1999. Overexpression of activin A in the skin of transgenic mice reveals new activities of activin in epidermal morphogenesis, dermal fibrosis and wound repair. EMBO J. 18:5205-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen, T., P. J. Sherratt, and C. B. Pickett. 2003. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu. Rev. Pharmacol. Toxicol. 43:233-260. [DOI] [PubMed] [Google Scholar]
- 43.Nguyen, T., C. S. Yang, and C. B. Pickett. 2004. The pathways and molecular mechanisms regulating Nrf2 activation in response to chemical stress. Free Radic. Biol. Med. 37:433-441. [DOI] [PubMed] [Google Scholar]
- 44.Nioi, P., and J. D. Hayes. 2004. Contribution of NAD(P)H:quinone oxidoreductase 1 to protection against carcinogenesis, and regulation of its gene by the Nrf2 basic-region leucine zipper and the arylhydrocarbon receptor basic helix-loop-helix transcription factors. Mutat. Res. 555:149-171. [DOI] [PubMed] [Google Scholar]
- 45.Park, J. H., S. Gopishetty, L. M. Szewczuk, A. B. Troxel, R. G. Harvey, and T. M. Penning. 2005. Formation of 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxo-dGuo) by PAH o-quinones: involvement of reactive oxygen species and copper(II)/copper(I) redox cycling. Chem. Res. Toxicol. 18:1026-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pedersen, T. X., C. Leethanakul, V. Patel, D. Mitola, L. R. Lund, K. Dano, M. Johnsen, J. S. Gutkind, and T. H. Bugge. 2003. Laser capture microdissection-based in vivo genomic profiling of wound keratinocytes identifies similarities and differences to squamous cell carcinoma. Oncogene 22:3964-3976. [DOI] [PubMed] [Google Scholar]
- 47.Ramos-Gomez, M., M. K. Kwak, P. M. Dolan, K. Itoh, M. Yamamoto, P. Talalay, and T. W. Kensler. 2001. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc. Natl. Acad. Sci. USA 98:3410-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sander, C. S., H. Chang, F. Hamm, P. Elsner, and J. J. Thiele. 2004. Role of oxidative stress and the antioxidant network in cutaneous carcinogenesis. Int. J. Dermatol. 43:326-335. [DOI] [PubMed] [Google Scholar]
- 49.Schor, N. A., K. Morgan, N. Epstein, and R. Knight. 1983. The influence of polycyclic hydrocarbons on the activity of NAD(P)H-dehydrogenating enzymes in rat thymus. A biochemical and histochemical study. Enzyme 29:167-174. [DOI] [PubMed] [Google Scholar]
- 50.Vassar, R., M. Rosenberg, S. Ross, A. Tyner, and E. Fuchs. 1989. Tissue-specific and differentiation-specific expression of a human K14 keratin gene in transgenic mice. Proc. Natl. Acad. Sci. USA 86:1563-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wankell, M., B. Munz, G. Hubner, W. Hans, E. Wolf, A. Goppelt, and S. Werner. 2001. Impaired wound healing in transgenic mice overexpressing the activin antagonist follistatin in the epidermis. EMBO J. 20:5361-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Werner, S., H. Smola, X. Liao, M. T. Longaker, T. Krieg, P. H. Hofschneider, and L. T. Williams. 1994. The function of KGF in morphogenesis of epithelium and reepithelialization of wounds. Science 266:819-822. [DOI] [PubMed] [Google Scholar]
- 53.Werner, S., W. Weinberg, X. Liao, K. G. Peters, M. Blessing, S. H. Yuspa, R. L. Weiner, and L. T. Williams. 1993. Targeted expression of a dominant-negative FGF receptor mutant in the epidermis of transgenic mice reveals a role of FGF in keratinocyte organization and differentiation. EMBO J. 12:2635-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuspa, S. H., A. A. Dlugosz, M. F. Denning, and A. B. Glick. 1996. Multistage carcinogenesis in the skin. J. Investig. Dermatol. Symp. Proc. 1:147-150. [PubMed] [Google Scholar]
- 55.Zenz, R., R. Eferl, L. Kenner, L. Florin, L. Hummerich, D. Mehic, H. Scheuch, P. Angel, E. Tschachler, and E. F. Wagner. 2005. Psoriasis-like skin disease and arthritis caused by inducible epidermal deletion of Jun proteins. Nature 437:369-375. [DOI] [PubMed] [Google Scholar]
- 56.Zenz, R., H. Scheuch, P. Martin, C. Frank, R. Eferl, L. Kenner, M. Sibilia, and E. F. Wagner. 2003. c-Jun regulates eyelid closure and skin tumor development through EGFR signaling. Dev. Cell 4:879-889. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, J. Y., C. L. Green, S. Tao, and P. A. Khavari. 2004. NF-κB RelA opposes epidermal proliferation driven by TNFR1 and JNK. Genes Dev. 18:17-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang, J. Y., S. Tao, R. Kimmel, and P. A. Khavari. 2005. CDK4 regulation by TNFR1 and JNK is required for NF-κB-mediated epidermal growth control. J. Cell Biol. 168:561-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.