Abstract
Vaccinia virus replicates in the cytoplasm of the host cell and encodes its own RNA polymerase and transcription factors. The proteins that target the poxvirus RNA polymerase to intermediate- and late-class promoters have not been identified. In this study, representatives of the intermediate and late promoters were characterized at the nucleotide level to identify essential motifs. Both intermediate and late viral promoters are shown to have an essential element suggestive of TATA boxes, which are potential targets for the TATA-binding protein (TBP). Several approaches were used to test for TBP requirement in vaccinia virus transcription, including overexpression of TBP, expression of a dominant negative mutant of TBP, RNA interference, and expression of adenovirus E1A protein, which inactivates TBP. In each case, the results support an essential role for TBP in vaccinia virus intermediate- and late-gene transcription. These findings indicate that poxviruses have integrated TBP as a central feature into an otherwise heterologous transcription system. A model for transcriptional switching, in which both intermediate and late promoter elements are targeted by TBP that recruits viral transcription factors to assemble a functional complex on their cognate promoters and a dysfunctional, repressed complex on the other class, is proposed.
Vaccinia virus is the prototypal member of the poxvirus family, which is comprised of a number of animal and human pathogens, including smallpox. These viruses have the largest DNA genomes of all animal viruses, typically encoding some 200 different proteins (reviewed in reference 23). They are unique among DNA viruses in their residence exclusively in the cytoplasm of the host cell, where they replicate DNA, synthesize mRNA, and assemble progeny virus. This apparent autonomy from the nucleus is possible because these viruses encode many of the proteins that function in nucleic acid biosynthesis, including a DNA polymerase, RNA polymerase, transcription factors, and a nearly complete repertoire of mRNA modification enzymes.
Vaccinia virus coordinates its progression through its replicative cycle by expressing individual proteins at specific times. The temporal regulation of gene expression is controlled at the level of transcriptional initiation (reviewed in reference 4). The multisubunit viral mRNA polymerase, which structurally resembles its cellular counterparts, is responsible for all mRNA synthesis. Virus-encoded transcription factors are required for transcription of the early, intermediate, and late classes of genes which are activated in that order. The factors required for activation of each class are products of the preceding class, establishing a cascade for gene activation. The early-class genes have a characteristic promoter element centered 20 nucleotides upstream of the transcriptional start site that is targeted by the virus-encoded protein ETF. Intermediate gene transcription in vitro has been reported to require the viral E4L gene product (28), the viral capping enzyme (41), and virus-encoded VITF-3 (30). A fourth factor called VITF-2 that stimulates transcription in vitro, was reported to be a cellular factor (29) recently identified as G3BP, a putative RNA binding protein, and/or p137 (15). Cellular transcription factor YY1 was reported to target a promoter element at the transcriptional start site in the I1L promoter, now known to be an intermediate-class promoter, suggesting that YY1 activates this promoter (6). Recent evidence, however, indicates that YY1 represses rather than activates the I1L promoter (unpublished results). Late transcription also requires three virus-encoded proteins for transcription in vivo and in vitro: the A1L, A2L, and G8R gene products (16). No role for any of the late factors in transcription is known. Cellular heterogeneous nuclear riboproteins A2/B1 and RBM3 were reported to stimulate transcription from a late vaccinia virus promoter in vitro and were reported to have affinity for poly(T) tracts (42). Some vaccinia virus late promoters have T-rich sequences (10), but these are not a universal feature of late promoters. Evidence supporting the activation of vaccinia virus transcription in virus-infected cells has not been reported.
None of the vaccinia virus-encoded intermediate and late transcription factors has been demonstrated to have promoter binding activity, raising the possibility that host proteins may fulfill that function. In this study, systematic analyses of the sequence requirements for a vaccinia virus intermediate and a vaccinia virus late promoter were performed. The results indicate that an essential element for both classes of promoters resembles a TATA box as found in RNA polymerase II promoters in nuclear genes. The TATA box is the target for the widely important cellular transcription factor TATA-binding protein (TBP; reviewed in references 7 and 25). TBP is utilized by RNA polymerases I, II, and III as an integral component of transcription factors selectivity factor I, TFIID, and TFIIIB, respectively. The incorporation of TBP into the seemingly independent vaccinia virus transcription system located in the cytoplasm of the cell was unexpected. Several approaches toward assessing the importance of TBP in vaccinia virus intermediate and late gene transcription are described.
MATERIALS AND METHODS
Cell culture, virus infection, and DNA transfections.
HeLa S-3 cells were grown in monolayers in 6 (9-cm2)- or 12 (3.7-cm2)-well trays in Dulbecco's modified Eagle medium supplemented with 10% bovine serum. Cells were infected with 10 PFU/cell vaccinia virus Western Reserve strain. Where indicated, hydroxyurea (HU) was included in the culture medium at 10 mM. Expression plasmids were constructed by ligating double-stranded synthetic oligonucleotides with the desired sequence in front of the Escherichia coli β-galactosidase gene. After 1 h of virus infection, plasmid transfections were performed in triplicate with 2 μg DNA/well, using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. TBP and adenovirus serotype 5 E1A cDNAs were gifts from Arnold Berk (University of California, Los Angeles) and Ourania Andrisani (Purdue University). Site-directed mutagenesis was performed as described by Tyagi et al. (36). Cells were harvested 16 to 18 h after transfections, and reporter enzyme assays for β-galactosidase were performed in duplicate or triplicate with o-nitrophenyl-β-d-galactoside as the substrate as described by Miller (22). Enzyme activity was normalized to protein content as determined with Bradford reagent (Bio-Rad).
RNA interference.
The RNA oligonucleotides UUGUAUCCACAGUGAAUCUdTdT (d is 2′ deoxy) and AGAUUCACUGUGGAUACAAdTdT were designed against TBP mRNA, and GGAGCGCACCAUCUUCUUCUU and GAAGAAGAUGGUGCGCUCCUU were designed against enhanced green fluorescent protein (GFP) (19). The siRNAs were purchased as annealed duplexes (Dharmacon). RNA transfections were performed with 20 μM RNA and 10 μl Oligofectamine transfection reagent (Invitrogen) according to the manufacturer's instructions. After 3 days, cells were infected with virus and transfected with reporter gene as described above.
Immunoblotting.
Total cell proteins were separated by electrophoresis on a 12% polyacrylamide gel and transferred onto nitrocellulose. Blots were probed with antibody directed against the N-terminal 12 amino acids of TBP (sc-204; Santa Cruz) and visualized by chemiluminescence with ECL reagents (Amersham). Antibody against β-actin was from Covance.
DNA binding assays.
DNA oligonucleotides of the indicated sequences were chemically synthesized (Integrated DNA Technologies), labeled at their 5′ ends with T4 kinase, and annealed to form duplex DNA. DNA binding reactions were performed 20 μl of a solution containing 60 mM KCl, 10 mM HEPES-KOH, pH 7.9, 5 mM MgCl2, 2 mM dithiothreitol, 8% glycerol, 100 ng bovine serum albumin, 100 ng poly(dG-dC), and 200 fmol radiolabeled DNA. Where indicated, 100 ng human TBP was added to the solution which was incubated for 30 min at 37°C. Protein-DNA complexes were resolved by electrophoresis in a 6% polyacrylamide gel in 25 mM Tris-HCl, pH 8.0, 190 mM glycine, and 10 mM EDTA (37). Gels were dried and analyzed by autoradiography and phosphorimage scanning.
RESULTS
Sequence requirements for vaccinia virus intermediate and late gene transcriptional promoters.
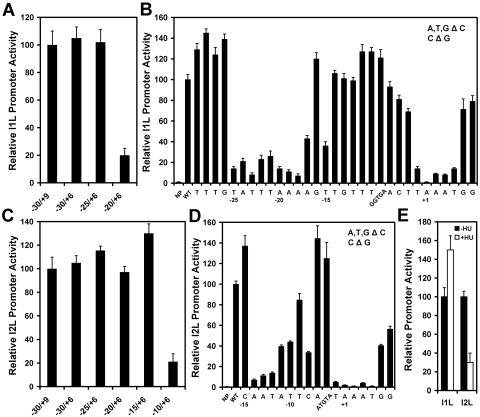
A systematic analysis of the DNA sequence requirements for a single vaccinia virus intermediate promoter has been reported. Baldick and Moss described two essential elements in the G8R promoter: a TAAAA sequence on the nontemplate strand at nucleotides −1 to +4 relative to the transcriptional start site, and another of the sequence TTTAAATAATT at −27 to −13 (2). In the current study, the vaccinia virus I1L promoter was selected for analysis because it has exceptionally high activity (20) and therefore would be expected to have optimized promoter elements. The promoter was linked to the E. coli β-galactosidase gene that served as a reporter for promoter activity and transfected into HeLa cells previously infected with vaccinia virus. The I1L promoter was originally reported to be a late promoter (31). This promoter is shown here to be enhanced in the presence of the DNA synthesis inhibitor HU in a transfection experiment (Fig. 1E), a hallmark of an intermediate promoter (16). A plasmid construct with 30 nucleotides upstream of the transcriptional start site was determined to retain full activity compared to one with 100 nucleotides upstream (not shown), and 6 nucleotides downstream were required for optimal activity (Fig. 1A). Therefore, the I1L promoter with nucleotides −30 to +6 served as the starting point for analysis of essential sequence elements. Initially, blocks of five nucleotides were replaced with C residues to scan across the promoter to rule out nonessential sequences (not shown). A segment spanning nucleotides −5 to −10 was determined to have little effect on promoter strength (Fig. 1A and B). Nucleotides in blocks found to be essential were replaced individually with C residues (or a G residue if originally a C). This analysis identified the sequence TATTTAAAAGT at nucleotides −29 to −14 as important for promoter activity as well as the sequence TAAATGG from −1 to +6 (Fig. 1B). Thus, the nucleotides identified here as important for the function of the I1L promoter are similar in location to those identified in the G8R promoter (2), they are similar in being very AT rich but have difference sequences.
FIG. 1.
Dissection of the vaccinia virus intermediate I1L and late I2L promoters. The I1L promoter (panels A and B) and I2L promoter (panel C and D) were linked to the β-galactosidase gene and transfected into HeLa cells that had been infected with vaccinia virus. Panels A and C show the activity of shortened promoters to determine the upstream and downstream boundaries of promoter elements. Panels B and D show activities of promoters with individual nucleotide substitutions. A, T, and G residues were replaced with C residues, and C residues were replaced with G residues. Bars labeled with five nucleotides are a block substitution. NP, no promoter; WT, wild-type sequence. Panel E shows the effect of HU in the culture medium on the activity of the I1L and I2L promoters. −HU, absence of HU; +HU, presence of HU. Error bars denote standard deviations.
An analysis of the sequence requirement for the L4R late promoter was reported (10). That study identified a sequence of TAAAT at nucleotides −1 to +4 on the nontemplate strand of the promoter as being essential for transcription. No other nucleotide at any of the five positions was tolerated. An oligo(T) tract at nucleotides −15 to −9 was found to be important as well, although not all nucleotides immediately upstream of the transcriptional start site were tested. The TAAAT motif was also shown to be essential for the F17R promoter (13). In the current study, the sequence requirements for the I2L promoter were determined systematically as described above for the I1L promoter. Reporter gene expression from the I2L promoter was reduced by HU (Fig. 1E), demonstrating that it is a late-class promoter. Fifteen nucleotides upstream of the transcriptional start and six nucleotides downstream were determined to be sufficient to retain full activity of the promoter (Fig. 1C). Nucleotide replacements identified one functional element at nucleotide positions −14 to −8 and another at nucleotide positions −1 to +6 (Fig. 1D). Therefore, both an intermediate and a late vaccinia virus promoter have two essential elements: one at the transcriptional start site and another upstream. Hereafter, the element at the start site for transcription hereafter is referred to as the initiator element, and the upstream element is referred to as the core element.
Interconversion of intermediate and late promoters.
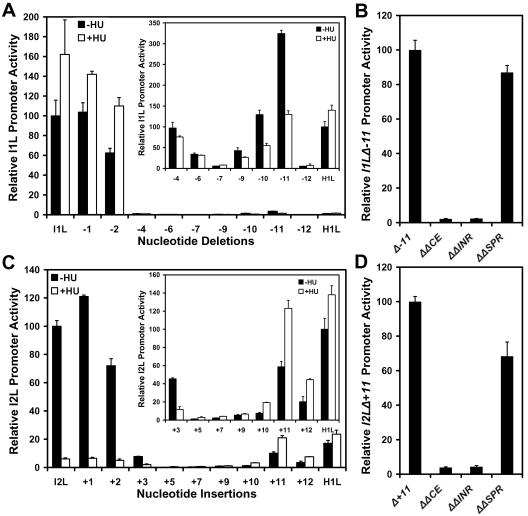
Comparison of the I1L promoter sequence with those of other vaccinia virus intermediate promoters and a comparison of that of the I2L promoter with those of other late promoters reveal similarities between the two promoter types. Both intermediate and late promoters have an initiator element containing the sequence TAAATGG at the transcriptional start site in which the two G residues are found in a subset of these promoters. This sequence is nearly invariant. The upstream core elements of both intermediate and late promoters are clearly AT-rich sequences; however, the sequences are highly variable and do not seem to conform to a consensus as such. The primary difference between intermediate and late promoter appears to be the length of the spacer between the initiator and core elements. To test this hypothesis, the number of nucleotides in the spacer region in intermediate and late promoters was varied by deletion or addition of nucleotides, respectively. At the same time, the effect of the DNA synthesis inhibitor (HU) on the promoters was monitored to discriminate between intermediate and late promoter behavior (16). Nucleotides were deleted incrementally from the spacer of the I1L promoter. The first few nucleotide deletions imposed a sharp reduction in promoter activity (Fig. 2A). As the number of deletions was increased, the activity of the promoter was restored partially, until optimal restoration occurred at 11 base pairs (bp) of deletion. The activity of the promoter with the 11-bp deletion appears to be low, but this is in comparison to the high activity of the native I1L promoter. The activity of the deleted promoter is actually quite comparable to other vaccinia virus promoters such as that for the H1L promoter (Fig. 2A, inset). Significantly, the levels of activity of the promoters with 9 to 11 bp deleted were inhibited by HU, demonstrating that they behaved as though they were late promoters. The activity of the promoter restored with the 11-bp deletion required both the initiator and the newly created late core elements (Fig. 2B). Replacement of nucleotides at the original position of the wild-type I1L promoter (−25 to −21) did not affect the activity of the restored promoter (Fig. 2B). These results indicate that transcription from the newly created late promoter was initiating within the initiator element and not elsewhere in plasmid construct. The complementary experiment was performed with the I2L promoter. Nucleotides were added incrementally up to 12 bp of additional sequence. Promoter activity actually increased with the addition of the first nucleotide, but declined with further nucleotide addition (Fig. 3C). Promoter activity was restored partially with the addition of 10 to 12 nucleotides and was optimal with an 11-bp addition. The restored promoters were stimulated by HU, demonstrating that they had the properties of an intermediate promoter. The activity of the promoter restored by the 11 bp addition also required both the initiator and newly created intermediate core element, but not the nucleotides at the −12 to −8 positions corresponding to the position of the original late core element (Fig. 2D). It was concluded that intermediate and late promoters could be interconverted by deletion and addition, respectively, of 11 nucleotides between the core and initiator promoter elements. The helical repeat of B-form DNA is 10.5 bp. The data presented here argue for two sites of protein interaction in vaccinia virus intermediate and late promoters that must reside on the same face of the DNA helix.
FIG. 2.
Effect of alteration of the number of nucleotides in the spacer between the core and initiator elements in the intermediate I1L promoter and late I2L promoter. Nucleotides were progressively deleted from spacer of the I1L promoter (panel A) or progressively added into the spacer of the I2L promoter (panel C). Reporter constructs were transfected into HeLa cells infected in the absence (−HU) or presence (+HU) of HU. The insets in panels A and C show activities in an expanded scale relative to that of the vaccinia virus H1L promoter, which is intermediate class. Panels B and D show the activity of the I1L 11-nucleotide spacer deletion and the I2L 11-nucleotide spacer addition, respectively. Shown are the effects of nucleotide replacements (Δ) in the initiator elements (ΔINR; +1), nucleotide replacements in the new core element in the converted promoters (ΔCE; −13 to −9 for the I1L promoter and −25 to −21 for the I2L promoter), and nucleotide replacement at the original location of the core element in the converted promoter (ΔSPR; −25 to −21 for the I1L promoter and −12 to −8 for the I2L promoter).
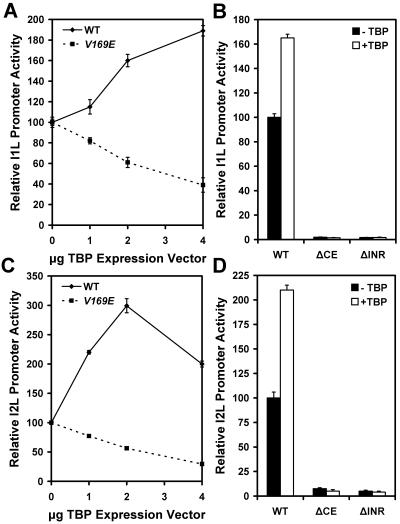
FIG. 3.
Effect of overexpression of TBP and a dominant negative mutant on the activity of vaccinia virus intermediate I1L and late I2L promoters. Vaccinia virus-infected HeLa cells were transfected with reporter vectors driven by the I1L (panels A and B) or the I2L promoter (panels C and D) and cotransfected with the indicated amount of TBP expression vector or V169E TBP mutant expression vector. Panels B and D show the effect of block substitutions in the core element (ΔCE) or initiator element (ΔINR) on the reporter gene activity when cotransfected with 3 μg empty expression vector (−TBP) or 3 μg TBP expression vector (+TBP). WT, wild-type sequence promoter.
An important clue to the identity of the protein(s) that target the vaccinia virus promoter elements came from their sequences. The initiator elements of intermediate and late promoters are nearly invariant in sequence. Almost all late promoters have the sequence TAAAT. Some intermediate initiator elements have the sequence TAAA (2), while others, such as I1L and I2L as shown here, have the sequence TAAATGG. The core elements of both intermediate and late promoters are very AT rich; however, there appears to be no consensus sequence. It is noted that the core sequence for the I1L promoter (TATTTAA) is an exact match to a sequence previously described as a high-affinity binding site for cellular TBP (14). This protein has preference for DNA having the first four nucleotides of this sequence but tolerates variation in the latter half. TBP tolerates sequence variation because it contacts DNA in the minor groove (17, 18, 34), and the stereochemistry of the minor groove of an AT pair is very similar to that of a TA pair. Thus, these properties make TBP an attractive candidate for the protein that targets vaccinia virus promoter core elements.
Impact of overexpression of TBP and a dominant negative mutant on vaccinia virus intermediate and late promoter activity.
Different approaches for perturbing the level of TBP in the cell were utilized to evaluate the importance of this protein in vaccinia virus transcription. First tested was the effect of overexpression of TBP on vaccinia virus promoter activity. Because vaccinia virus suppresses expression of host cell proteins, the TBP gene was placed under the control of the viral I1L promoter in an expression construct that was cotransfected with the vaccinia virus promoter-driven reporter gene into HeLa cells previously infected with vaccinia virus. Virus-infected cells were transfected with a fixed amount of I1L promoter-reporter plasmid and titrated with increasing amounts of TBP expression construct. The level of reporter enzyme activity increased with increasing TBP expression in a dose-responsive manner, approximately doubling with 4 μg of TBP expression vector (Fig. 3A). The activation response to TBP required a functional promoter. Replacement of five residues in the core element or five residues in the initiator element with C residues resulted in a promoter with very low activity that did not respond to TBP expression (Fig. 3B). As a control, the effect of expression of a TBP dominant negative mutant on reporter activity was examined. The V169E mutant was designed on the DNA binding surface of TBP, imparting a dominant negative effect on endogenous TBP (27, 40). The V169E TBP mutant was expressed behind the vaccinia virus I1L promoter and cotransfected with the reporter construct. Titration with increasing amounts of the mutant expression vector in a cotransfection experiment progressively decreased reporter enzyme levels (Fig. 3A), consistent with a dominant negative effect. If the V169E TBP down-regulates the I1L promoter, then it down-regulates its own synthesis, lessening the effect on reporter expression that otherwise might be observed. Similar experiments were performed with the vaccinia virus I2L late promoter-driven reporter gene. This promoter was about three times more active with 2 μg TBP expression vector (Fig. 3C). The I2L promoter was activated by titration with TBP more strongly than was observed with the I1L promoter. The ability of TBP expression to activate the I2L promoter was also dependent on both its core and initiator elements (Fig. 3D). The I2L promoter also was inactivated similarly by titration with the V169E TBP mutant. These results indicate that both the vaccinia virus intermediate I1L and late I2L promoters are responsive to TBP.
Effect of RNA interference knockdown of TBP on vaccinia virus promoter activity.
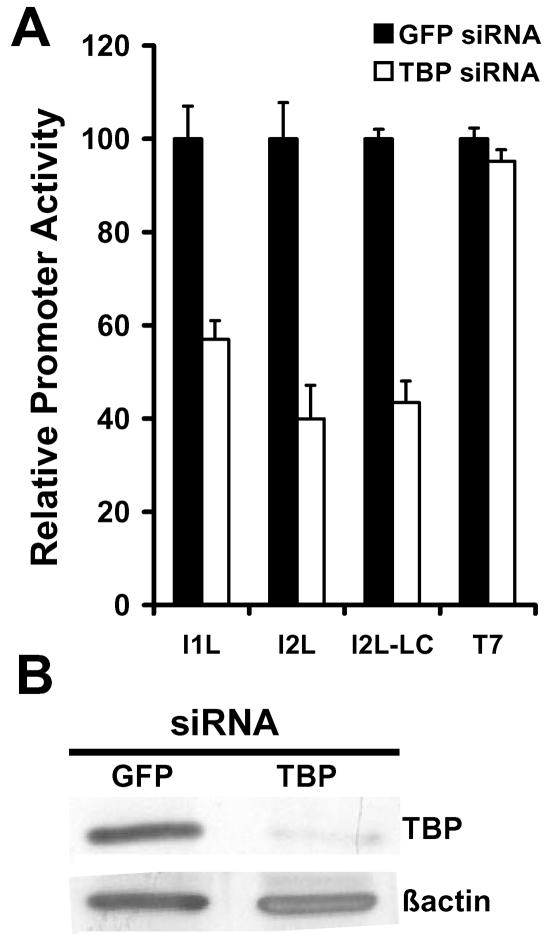
As an alternative approach toward evaluating the importance of TBP in vaccinia virus transcription, short interfering RNAs (siRNAs) were designed against the mRNA of TBP to reduce the steady-state levels of TBP in HeLa cells. siRNA directed against enhanced GFP, which is not expressed in HeLa cells, was used as a control. siRNA was transfected into cells and incubated for 3 days prior to infection with vaccinia virus and transfection with reporter construct. The siRNA treatment resulted in approximately 80% reduction in endogenous TBP levels (Fig. 4B). In TBP siRNA-treated cells the level of virus-directed reporter enzyme was reduced to 60% for the intermediate I1L-driven reporter and 40% for the late I2L-driven reporter relative to cells that were treated with control (GFP) siRNA. Thus, siRNA-mediated reduction in intracellular TBP levels reduced the activity of both the intermediate I1L and late I2L vaccinia virus promoters (Fig. 4A). It is possible that the reduced late promoter activity is a secondary effect of inhibition of transcription of intermediate genes which includes those for the late transcription factors A1L, A2L, and G8R. To circumvent the requirement for intermediate gene transcription in late promoter activation, TBP siRNA-treated cells were infected with vaccinia virus TF7-3 that expresses the T7 RNA polymerase in the presence of HU to block DNA synthesis and intermediate gene transcription. This virus expresses the T7 RNA polymerase from a tandem early/late promoter (11) and therefore would be active in HU-treated cells (16). Vectors directing the expression of the A1L, A2L, and G8R gene products from a T7 promoter were cotransfected with the I2L promoter reporter construct to allow transcription from late promoters (5, 16). Under these conditions, the I2L promoter was active and was similarly reduced to about 40% by pretreatment of the cells with TBP siRNA. The activity of the T7 promoter was unaffected by the siRNA, indicating that the reduction of late promoter activity is a direct result of TBP siRNA.
FIG. 4.
The effect of TBP siRNA on intermediate and late promoter activity. HeLa cells were transfected with siRNA directed against GFP or TBP mRNA, then infected with vaccinia virus and transfected with reporter gene driven by the indicated promoter. LC is promoter activity in cells infected with vaccinia virus T7-7, which expresses T7 RNA polymerase, and reporter gene construct was cotransfected with vectors expressing late transcription factors A1L, A2L, and G8R driven by a T7 promoter in the presence of HU. T7 shows the activity of reporter gene driven by a T7 promoter in the presence of HU. Promoter activity was normalized to that produced in extracts from GFP siRNA-treated cells. Panel E is an immunoblot showing the effect of siRNA directed against TBP and GFP on steady-state levels of TBP in cells prior to infection with vaccinia virus. A parallel blot was probed for β-actin as a loading control.
Inhibition of vaccinia virus transcription by adenovirus E1A protein.
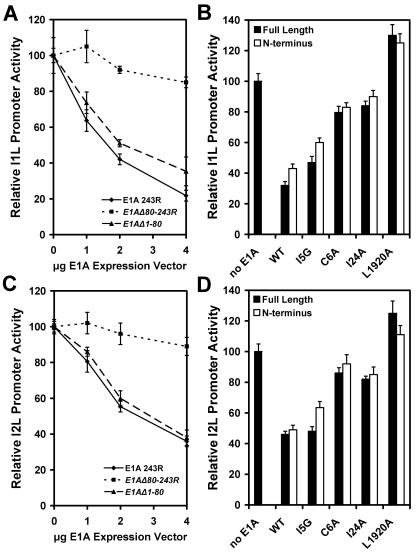
The E1A protein of adenovirus is a powerful probe for TBP function. The 243R form (also called 12S) of E1A that contains conserved regions 1 and 2 is well documented as being able to inhibit TBP function by sequestering it and interfering with DNA binding (32, 33). The TBP-inhibitory function localizes to the N-terminal 80 amino acids of E1A in conserved region 1, and several mutations that impair the inhibitory effect have been identified. Full-length E1A, truncated E1A, and E1A mutants were expressed under the control of the vaccinia virus I1L promoter by cotransfection into vaccinia virus-infected HeLa cells along with reporter genes driven by the vaccinia virus I1L promoter. Expression of full-length E1A 243R resulted in inhibition of the intermediate I1L promoter driven reporter (Fig. 5A). Titration with E1A expression vector showed the inhibition of the I1L promoter was responsive to the amount of E1A plasmid transfected into cells. Expression of E1A N-terminal 80 residues also resulted in inhibition of vaccinia virus promoter activity; however, the deletion of expression of E1A from the N-terminal 80 amino acids did not. These results demonstrate that the I1L promoter is sensitive to E1A, and it is the amino terminal portion of the protein that is responsible for the inhibition. The I2L promoter was similarly impaired by expression of E1A and its N-terminal 80 residues but not significantly affected by expression of residues 81 to 243 (Fig. 5B).
FIG. 5.
Effect of expression of adenovirus E1A protein on vaccinia virus intermediate I1L and late I2L promoter activities. Vaccinia virus-infected HeLa cells were transfected with reporter construct driven by the intermediate I1L promoter (panels A and B) or late I2L promoter (panels C and D). In panels A and C, cells were cotransfected with the indicated amount of vector directing the expression of full-length E1A 243R expression, E1A N-terminal residues 1 to 80 (E1AΔ1-80), or E1A C-terminal residues 80 to 243 (E1AΔ80-243R). In panels B and D, cells were cotransfected with 3 μg empty expression vector DNA (no E1A), 3 μg plasmid directing the expression of full-length E1A 243R (WT), or an E1A C6A, I5G, I24A, or L1920A mutant in the context of the full-length E1A 243R or truncated E1A (residues 1 to 80; N-terminal). Error brackets indicate standard deviations.
TBP is not the only known target for E1A. Several other cellular transcription factors have been described as interaction partners of E1A. Most significant among these for this discussion is CBP, whose site of interaction on E1A overlaps the site of interaction with TBP (3, 26). Extensive mutational analysis of E1A has identified amino acid residues specific to interaction with TBP and CBP, thus allowing discrimination between the two as the basis for repression of transcription. The L1920A double mutant is defective for interaction with both TBP and CBP, the C6A and I24A mutants are defective for interaction with TBP but not CBP, and the I5G mutant is defective for interaction with CBP but has nearly wild-type affinity for TBP (26). The expression of each of these mutant forms of E1A was placed under the control of the vaccinia virus I1L promoter and cotransfected with the I1L reporter construct. The mutants were expressed as full-length 243R proteins or in the context of the N-terminal 80 residue protein. Cotransfection with the I5G mutant E1A protein resulted in about 50% repression of the I1L and I2L promoter activities (Fig. 5B and D). Expression of the E1A C6A and I24A mutants suppressed reporter activity by about 10%, and the L1920A mutant actually activated promoter activity modestly. The E1A mutations produced the same effect in the context of the truncated form of E1A. These results indicate that TBP is responsible for the inhibition of vaccinia virus intermediate and late promoters by adenovirus E1A.
Localization of TBP binding in vaccinia virus promoters.
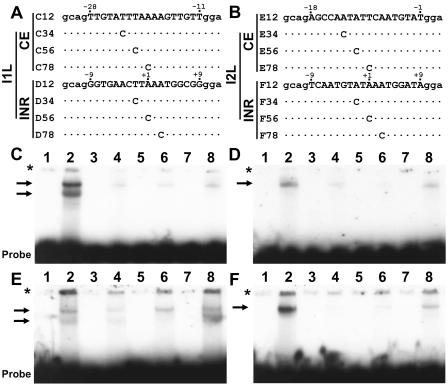
The I1L promoter core element has a nucleotide sequence previously shown to bind TBP (14). DNA electrophoretic mobility shift analysis was used to study the interaction of TBP with the vaccinia virus I1L and I2L promoters. Incubation of TBP with intact promoters containing both elements resulted in apparent aggregation of the DNA probe, as surmised because of the inability of the protein-DNA complexes to enter the gel wells despite the inclusion of poly(dG-dC) in the binding reactions (data not shown). To address the interaction of TBP with vaccinia virus promoter elements, oligonucleotides with individual core and initiation elements with relatively GC-rich sequences flanking them were incubated with TBP. The core elements of both the I1L and I2L promoters produced two gel-shifted species with TBP (Fig. 6). The initiator elements of both promoters produced a single shifted complex. The sequence requirements for each of the four elements were analyzed by introducing single-C nucleotide substitutions spanning the elements. Nucleotide substitutions at positions −23, −21, and −19 in the I1L core element impaired interaction with TBP, as did nucleotide replacements at positions −13 and −11 in the I2L core element. The substitution at nucleotide position −9 had minor effect on TBP binding, consistent with its minor effect on transcription. Interaction with TBP was also impaired by nucleotide replacements at positions −2, +1, and +3 in the initiator elements of both promoters. The effect of nucleotide alterations in the I1L and I2L core elements are consistent with the importance of these nucleotides in the function of both promoters in reporter gene experiments (Fig. 1). The inhibitory effect of alteration of the −2 position on TBP binding to the initiator elements is not in agreement with lack of importance of this nucleotide for promoter activity. These results indicate that the TBP binding to the initiator elements is due to fortuitous sites of interaction of TBP with the AT-rich promoter elements, and the essential sites of interaction of TBP with vaccinia virus promoters are the upstream core elements.
FIG. 6.
Localization of TBP binding in vaccinia virus intermediate I1L and late I2L promoters. Panels A and B show sequences of oligonucleotides used for the DNA binding assays of the I1L and I2L promoters, respectively. The oligonucleotides contain native promoter elements (uppercase nucleotides) and nonnative ends (lowercase nucleotides). Letters and numbers to the left of each oligonucleotide correspond to the panel and lane numbers in the gels shown below each. The positions of C nucleotide replacements are indicated. Numbers above each sequence indicate nucleotide positions relative to the start site for transcription. Recombinant TBP was incubated with oligonucleotides containing the core and initiator elements of the I1L promoter (panels C and D, respectively) and the core and initiator elements of the I2L promoter (panels E and F, respectively). Binding mixtures were subjected to electrophoresis on a polyacrylamide gel to resolve protein-DNA complexes, indicated by arrows. Odd-numbered lanes are oligonucleotide alone, and even-numbered lanes are oligonucleotides incubated with TBP. Asterisks indicate the top of the gel.
TBP domains required for activation of vaccinia virus promoters.
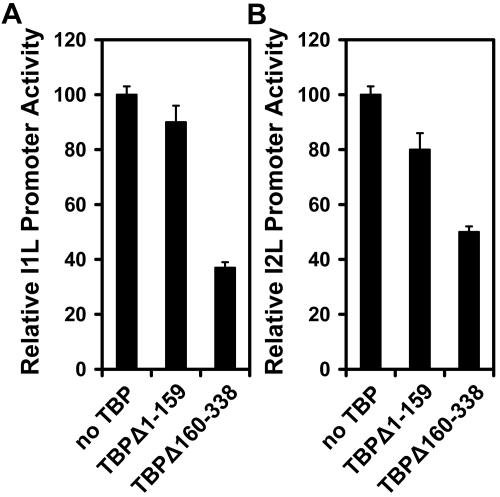
The C-terminal domain of TBP is a quasisymmetrical saddle-shaped structure that contacts the TATA box DNA (9). The N-terminal domain is believed to be required for response to activated transcription by nuclear RNA polymerases and may modulate DNA binding. The two halves of TBP were expressed from the vaccinia virus I1L promoter, and their effects on the I1L and I2L promoters were evaluated in reporter gene experiments. Expression of the N-terminal DNA binding domain of TBP had a modest effect on vaccinia virus promoter activity. The levels of activity were reduced by 10 and 20% for the I1L and I2L promoters, respectively (Fig. 7). Expression of the C-terminal domain resulted in greater inhibition of both promoters. I1L promoter activity was reduced to about 35% of its normal activity, and I2L was reduced to about 45%. It was concluded that both the N- and C-terminal domains of TBP are essential for its effect on vaccinia virus intermediate and late promoters.
FIG. 7.
Effect of expression of N- and C-terminal TBP truncations on the activity of the I1L and I2L promoters. HeLa cells were infected with vaccinia virus and transfected with 2 μg reporter vector DNA driven by the I1L (panel A) or I2L (panel B) promoter and cotransfected with 3 μg N-terminal TBP residues 1 to 159, C-terminal TBP residues 160 to 338, or empty expression vector.
DISCUSSION
In this study, the definition of functional elements in vaccinia virus intermediate and late promoters was used to identify proteins that target the elements. The sequence of the core element of the I1L promoter previously was described as a high-affinity binding site for TBP (14). The ability to convert the I1L promoter into a late-class promoter through deletion of spacer nucleotides strongly suggests that the same protein can target the core elements of both intermediate and late vaccinia virus promoters. When used with tests for the importance of TBP, an intermediate and a late promoter responded similarly. Both were activated by overexpression of TBP, both were inactivated by expression of a dominant negative mutant TBP, and both were impaired by knockdown of TBP with siRNA. The intermediate and late promoters were impaired by the adenovirus E1A expression and its isolated repression domain but not by E1A mutants that do not interact with TBP. Evidence for interaction of TBP with the core and initiator elements for both an intermediate and late promoters was also presented.
The identification of vaccinia virus intermediate and late promoter elements by sequence alignments has been largely uninformative. Typically, sequences are aligned by the TAAA sequence in the initiator (2, 10, 13); however, no consensus sequence upstream of the initiator can be derived from comparison of multiple promoters. The results presented here demonstrate that the intermediate- and late-class core and initiator elements require a narrow window of nucleotide spacer between them for optimal activity. While changing the number of nucleotides in the respective spacers reduced promoter activity, in no case was activity undetectable. It is quite possible that many natural vaccinia virus promoters have less than optimal spacing between their promoter elements as a mechanism of attenuating transcription to moderate protein expression. Indeed, the activity of the I2L promoter actually increased moderately by the addition of one nucleotide to the spacer between the core and initiator elements (Fig. 2D). If some promoters have less than optimal spacing between the functional elements, aligning sequences to identify promoter elements would be difficult.
DNA binding assays shown here indicate that TBP can form a reasonably stable complex with both the core and initiator element of intermediate and late vaccinia virus promoters. The interaction of TBP with the core elements was predictable, but interaction with the initiator elements was unexpected. There are good correlations between nucleotides in the core elements in both promoters required for promoter activity and interaction with TBP. The sequences of the core elements are consistent with the sequence specificity of TBP. Considerable variation in the core elements is observed through sequence comparison of vaccinia virus promoters, and TBP will interact with a variety of sequences that are mostly AT in base composition (14). The sequences of vaccinia virus initiator elements, on the other hand, are quite invariant. The sequence TAAAT is nearly universal, and any nucleotide replacement is tolerated weakly, if at all (2, 10). Thus, the conservation of sequence in initiator elements is not consistent with the DNA binding properties of TBP. Replacement of the −2 nucleotide in both promoters prevented interaction with TBP but had little effect on promoter activity. Thus, the interaction of TBP with initiator elements appears to be fortuitous and not functionally significant.
Several prior reports describe key information that supports the involvement of TBP in vaccinia virus postreplicative transcription. First, vaccinia virus intermediate and late gene transcription were reported to be inhibited by the antibiotic distamycin A (5). An antibiotic with potent antiviral activity against vaccinia virus (5, 21, 39). Distamycin A is a DNA minor groove ligand that targets AT-rich sequences (1) such as those in the functional elements of vaccinia virus transcriptional promoters. TBP contacts TATA box DNA exclusively in the helix minor groove (17, 18), and its binding to DNA is highly sensitive to distamycin A (8). Sequence-specific DNA binding proteins that contact DNA in the minor groove are very atypical (12). TBP's interaction with vaccinia virus promoter DNA is the likely explanation for the virus' sensitivity to distamycin A. A second line of support is a recent report showing that TBP accumulates abundantly in the cytoplasm of vaccinia virus-infected cells despite its normal location in the nucleus (24). Although TBP is a nuclear transcription factor, vaccinia virus has ample opportunity to take advantage of it as a transcriptional activator. Third, expression of adenovirus E1A protein was reported to suppress vaccinia virus protein expression when expressed by a recombinant vaccinia virus (35). The viral proteins whose expression was inhibited were identified by pulse-labeling experiments at 6 to 18 h postinfection, indicating that they are late proteins. The target for E1A-mediated suppression of vaccinia virus protein synthesis is likely TBP.
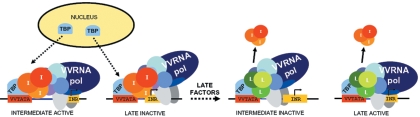
The functional interaction of TBP with both intermediate and late vaccinia virus promoter elements suggests a model for the regulatory switch from intermediate to late gene transcription that occurs during virus infection (Fig. 8). After DNA replication begins, both intermediate and late promoters would be expected to be occupied by TBP, but only intermediate promoters would be active because the architecture of the intermediate factors assembled on short late promoters is not conducive to assembly of a functional preinitiation complex. As the late transcription factors accumulate, they would be predicted to be more effective competitors for interaction with TBP and may effectively displace intermediate factors from preinitiation complexes on both types of promoters to activate late transcription with a concomitant inactivation of intermediate transcription.
FIG. 8.
Model for activation of vaccinia virus intermediate transcription and the switch from intermediate and late transcription. Following entry into the cell, the DNA is amplified by the viral replication machinery. The amplified DNA draws transcription factors such as TBP from the nucleus into the cytoplasm to activate intermediate gene transcription. TBP is capable of targeting both intermediate (I) and late promoters (L) but activates only the intermediate promoters initially because intermediate transcription factors assemble a dysfunctional preinitiation complex on the short late promoter. As late transcription factors accumulate, they displace intermediate factors to activate late promoters and inactivate intermediate transcription by formation of a dysfunctional complex at intermediate promoters. VV, vaccinia virus.
If TBP plays an essential role in vaccinia virus transcription, why does a virus of such high complexity not encode a TBP itself? The even more complex chlorella viruses encode a protein with a sequence consistent with a functional TBP (38). The answer may lie in the linkage between gene expression and DNA replication. Like with most viruses, the switch between the early and late phases of vaccinia virus gene regulation is inextricably linked to genome replication. Early gene transcription does not cease, and intermediate gene transcription does not begin until DNA replication initiates. Vaccinia virus has been shown to recruit host nuclear proteins, including TBP, into cytoplasmic replication complexes (24). The accumulation of these proteins in the cytoplasm requires viral DNA replication and a functional DNA binding domain in the nuclear protein. It is possible that TBP will remain restrained in the nucleus during the course of virus infection until a critical mass of DNA accumulates in the cytoplasm to pull transcription factors such as TBP into the cytoplasm to associate with viral replication complexes. Once pulled into the cytoplasm, the host factor would activate viral intermediate gene transcription, thereby inducing the switch (Fig. 8). This might also explain why intermediate promoters are active on transfected plasmids when DNA replication is blocked, but those in the viral genome are not. The transfected DNA may compensate for a need to amplify DNA to pull proteins from the nucleus. Not only are transfected intermediate promoters resistant to the effects of inhibition of DNA synthesis, they are stimulated by it, likely because intermediate genes including those for the late transcription factors are not activated, avoiding a competition between intermediate and late promoters for cellular activators such as TBP.
Acknowledgments
We are most grateful to Arnold Berk and Carol Eng for sharing numerous reagents used in this study and to Ourania Andrisani for an E1A clone. We are also grateful to Stewart Shuman for critical commentary on the manuscript.
REFERENCES
- 1.Abu-Daya, A., P. M. Brown, and K. R. Fox. 1995. DNA sequence specificity of several A-T-selective minor groove binding ligands. Nucleic Acids Res. 23:3385-3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldick, C. J., Jr., J. G. Keck, and B. Moss. 1992. Mutational analysis of the core, spacer, and initiator regions of vaccinia virus intermediate-class promoters. J. Virol. 66:4710-4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyd, J. M., P. M. Loewenstein, Q. Tang, L. Yu, and M. Green. 2002. Adenovirus E1A N-terminal amino acid sequence requirements for repression of transcription in vitro and in vivo correlate with those required for E1A interference with TBP-TATA complex formation. J. Virol. 76:1461-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broyles, S. S. 2003. Vaccinia virus transcription. J. Gen. Virol. 84:2293-2303. [DOI] [PubMed] [Google Scholar]
- 5.Broyles, S. S., M. Kremer, and B. A. Knutson. 2004. Antiviral activity of distamycin A against vaccinia virus is the result of inhibition of postreplicative mRNA synthesis. J. Virol. 78:2137-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broyles, S. S., X. Liu, M. Zhu, and M. Kremer. 1999. Transcription factor YY1 is a vaccinia virus late promoter activator. J. Biol. Chem. 274:35662-35667. [DOI] [PubMed] [Google Scholar]
- 7.Burley, S. K., and R. G. Roeder. 1996. Biochemistry and structural biology of transcription factor IID (TFIID). Annu. Rev. Biochem. 65:769-799. [DOI] [PubMed] [Google Scholar]
- 8.Chiang, S.-Y., J. Welch, F. J. Rauscher III, and T. A. Beerman. 1994. Effects of minor groove binding drugs on the interaction of TATA box binding protein and TFIIA with DNA. Biochemistry 33:7033-7040. [DOI] [PubMed] [Google Scholar]
- 9.Davidson, I. 2003. The genetics of TBP and TBP-related factors. Trends Biochem. Sci. 28:391-398. [DOI] [PubMed] [Google Scholar]
- 10.Davison, A. J., and B. Moss. 1989. Structure of vaccinia virus late promoters. J. Mol. Biol. 210:771-784. [DOI] [PubMed] [Google Scholar]
- 11.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garvie, C. W., and C. Wolberger. 2001. Recognition of specific DNA sequences. Mol. Cell 8:937-946. [DOI] [PubMed] [Google Scholar]
- 13.Hanggi, M., W. Bannwarth, and H. G. Stunnenberg. 1986. Conserved TAAAT motif in vaccinia virus late promoters: overlapping TATA box and site of transcription initiation. EMBO J. 5:1071-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoopes, B. C., J. F. LeBlanc, and D. K. Hawley. 1998. Contributions of the TATA box sequence to rate-limiting steps in transcription initiation by RNA polymerase II. J. Mol. Biol. 277:1015-1031. [DOI] [PubMed] [Google Scholar]
- 15.Katsafanas, G. C., and B. Moss. 2004. Vaccinia virus intermediate stage transcription is complemented by Ras-GTPase-activating protein SH3 domain-binding protein (G3BP) and cytoplasmic activation/proliferation-associated protein (p137) individually or as a heterodimer. J. Biol. Chem. 279:52210-52217. [DOI] [PubMed] [Google Scholar]
- 16.Keck, J. G., C. J. Baldick, Jr., and B. Moss. 1990. Role of DNA replication in vaccinia virus gene expression: a naked template is required for transcription of three late trans-activator genes. Cell 61:801-809. [DOI] [PubMed] [Google Scholar]
- 17.Kim, J. L., D. B. Nikolov, and S. K. Burley. 1993. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature 365:520-527. [DOI] [PubMed] [Google Scholar]
- 18.Kim, Y., J. H. Geiger, S. Hahn, and P. Sigler. 1993. Crystal structure of a yeast TBP/TATA-box complex. Nature 365:512-520. [DOI] [PubMed] [Google Scholar]
- 19.Leirdall, M., and M. Sioud. 2002. Gene silencing in mammalian cells by preformed small RNA duplexes. Biochem. Biophys. Res. Commun. 295:744-748. [DOI] [PubMed] [Google Scholar]
- 20.Liu, X., M. Kremer, and S. S. Broyles. 2004. A natural vaccinia virus promoter with exceptional capacity to direct protein synthesis. J. Virol. Methods 122:141-145. [DOI] [PubMed] [Google Scholar]
- 21.Lown, J. W., K. Krowicki, J. Balzarini, R. A. Newman, and E. De Clercq. 1989. Novel linked antiviral and antitumor agents related to netropsin and distamycin: synthesis and biological evaluation. J. Med. Chem. 32:2368-2375. [DOI] [PubMed] [Google Scholar]
- 22.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 23.Moss, B. 2001. Poxviruses and their replication, p. 2849-2884. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott, Williams, and Wilkins, Philadelphia, Pa.
- 24.Oh, J., and S. S. Broyles. 2005. Host cell nuclear proteins are recruited to cytoplasmic vaccinia virus replication complexes. J. Virol. 79:12852-12860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pugh, B. F. 2000. Control of gene expression through regulation of the TATA-binding protein. Gene 255:1-14. [DOI] [PubMed] [Google Scholar]
- 26.Rasti, M., R. J. Grand, J. S. Mymryk, P. H. Gallimore, and A. S. Turnell. 2005. Recruitment of CBP/p300, TATA-binding protein, and S8 to distinct regions at the N terminus of adenovirus E1A. J. Virol. 79:5594-5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reddy, P., and S. Hahn. 1991. Dominant negative mutations in yeast TFIID define a bipartite DNA-binding domain. Cell 65:349-357. [DOI] [PubMed] [Google Scholar]
- 28.Rosales, R., N. Harris, B. Y. Ahn, and B. Moss. 1994. Purification and identification of a vaccinia virus-encoded intermediate stage promoter-specific transcription factor that has homology to eukaryotic transcription factor SII (TFIIS) and an additional role as a viral RNA polymerase subunit. J. Biol. Chem. 269:14260-14267. [PubMed] [Google Scholar]
- 29.Rosales, R., G. Sutter, and B. Moss. 1994. A cellular factor is required for transcription of vaccinia viral intermediate-stage genes. Proc. Natl. Acad. Sci. USA 91:3794-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanz, P., and B. Moss. 1999. Identification of a transcription factor, encoded by two vaccinia virus early genes, that regulates the intermediate stage of viral gene expression. Proc. Natl. Acad. Sci. USA 96:2692-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmitt, J. F., and H. G. Stunnenberg. 1988. Sequence and transcriptional analysis of the vaccinia virus HindIII I fragment. J. Virol. 62:1889-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song, C. Z., P. M. Loewenstein, K. Toth, and M. Green. 1995. Transcription factor TFIID is a direct functional target of the adenovirus E1A transcription-repression domain. Proc. Nat. Acad. USA 92:10330-10333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song, C. Z., P. M. Loewenstein, K. Toth, Q. Tang, A. Nishikawa, and M. Green. 1997. The adenovirus E1A repression domain disrupts the interaction between the TATA binding protein and the TATA box in a manner reversible by TFIIB. Mol. Cell. Biol. 17:2186-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Starr, D. B., and D. K. Hawley. 1991. TFIID binds in the minor groove of the TATA box. Cell 67:1231-1240. [DOI] [PubMed] [Google Scholar]
- 35.Strauss, D., O. Elroy-Stein, and R. Ehrlich. 1997. Adenovirus E1A interferes with expression of vaccinia viral genes. Gene 184:279-284. [DOI] [PubMed] [Google Scholar]
- 36.Tyagi, R., R. Lai, and R. G. Duggleby. 2004. A new approach to ‘megaprimer’ polymerase chain reaction mutagenesis without an intermediate gel purification step. BMC Biotechnol. 4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vanathi, P., A. K. Mishra, and P. Bharagava. 2003. Regulation of actvity of the yeast TATA-binding protein through intra-molecular interactions. J. Biosci. 28:313-421. [DOI] [PubMed] [Google Scholar]
- 38.Van Etten, J. L. 2003. Unusual life style of giant chlorella viruses. Annu. Rev. Genet. 37:153-159. [DOI] [PubMed] [Google Scholar]
- 39.Verini, M. A., and M. Ghione. 1964. Activity of distamycin A on vaccinia infection in culture. Chemotherapy 9:144-157. [DOI] [PubMed] [Google Scholar]
- 40.Veschambre, P., P. Simard, and P. Jalinot. 1995. Evidence for functional interaction between the HIV-1 Tat transactivator and the TATA box binding protein in vivo. J. Mol. Biol. 250:169-180. [DOI] [PubMed] [Google Scholar]
- 41.Vos, J. C., M. Sasker, and H. G. Stunnenberg. 1991. Vaccinia virus capping enzyme is a transcription initiation factor. EMBO J. 10:2553-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wright, C. F., B. W. Oswald, and S. Dellis. 2001. Vaccinia virus late transcription is activated in vitro by cellular heterogeneous nuclear ribonucleoproteins. J. Biol. Chem. 276:40680-40686. [DOI] [PubMed] [Google Scholar]