Abstract
Erythroid Krüppel-like factor (EKLF) is an erythroid zinc finger protein identified by its interaction with a CACCC sequence in the β-globin promoter, where it establishes local chromatin structure permitting β-globin gene transcription. We sought to identify other EKLF target genes and determine the chromatin status of these genes in the presence and absence of EKLF. We identified alpha hemoglobin-stabilizing protein (AHSP) by subtractive hybridization and demonstrated a 95 to 99.9% reduction in AHSP mRNA and the absence of AHSP in EKLF-deficient cells. Chromatin at the AHSP promoter from EKLF-deficient cells lacked a DNase I hypersensitive site and exhibited histone hypoacetylation across the locus compared to hyperacetylation of wild-type chromatin. Wild-type chromatin demonstrated a peak of EKLF binding over a promoter region CACCC box that differs from the EKLF consensus by a nucleotide. In mobility shift assays, the AHSP promoter CACCC site bound EKLF in a manner comparable to the β-globin promoter CACCC site, indicating a broader recognition sequence for the EKLF consensus binding site. The AHSP promoter was transactivated by EKLF in K562 cells, which lack EKLF. These results support the hypothesis that EKLF acts as a transcription factor and a chromatin modulator for the AHSP and β-globin genes and indicate that EKLF may play similar roles for other erythroid genes.
EKLF (erythroid Krüppel-like factor, or KLF1) is one of several CACCC-binding regulatory proteins active in erythroid cells (2, 3, 5, 6, 17, 48, 74). It is the founding member of the mammalian Krüppel subfamily of transcription factors with 3 C2H2-type zinc fingers at the COOH terminus (7, 8, 35, 55). Its roles include β-globin gene activation, participation in the switch to adult β-globin, and coordination of erythroid cell proliferation and hemoglobinization (11, 18, 31, 39, 56, 65, 69). EKLF binds to the consensus sequence CCNCNCCCN in the β-globin gene promoter proximal CACCC box (22, 48), where it establishes local chromatin structure, as evidenced by DNase I hypersensitive site (HS) formation and chromatin immunoprecipitation (ChIP) studies, and directs high-level β-globin transcription. Mutations of this consensus sequence in the β-globin promoter lead to the phenotype of β-thalassemia (40, 52, 53).
The β-thalassemia syndromes are marked by clinical heterogeneity, and numerous modifier genes have been proposed (66). Recently, α-hemoglobin stabilizing protein (AHSP, also known as erythroid differentiation-related factor and erythroid-associated factor) has been proposed as a β-thalassemia modifier gene (19, 37, 38, 46, 68). AHSP binds and stabilizes free α-hemoglobin, inhibiting the production of reactive oxygen species from α-hemoglobin and preventing the precipitation of unstable, cytotoxic free α-globin chains. Free α-globin chains aggregate in erythroid precursors, damaging the membrane and triggering cell death. When β-thalassemic mice were bred to AHSP-deficient mice, AHSP deficiency worsened the thalassemic phenotype, leading to the suggestion that AHSP could be a modifier gene in human β-thalassemia syndromes (38).
EKLF-deficient mice die at embryonic day 14.5 (E14.5) to E15 from severe anemia due to defective definitive erythropoiesis (44, 50, 57). There is a marked decrease in β-globin mRNA and protein levels in EKLF-deficient erythroid cells. In addition, there are alterations in chromatin configuration at the β-globin gene promoter, including loss of a DNase I hypersensitive site at the proximal CACCC box as well as diminution of another HS site over 50 kb away in the β-globin locus control region (HS3) (26, 69). Large amounts of iron accumulate in the reticuloendothelial system of EKLF-deficient mice, consistent with ineffective erythropoiesis and hemolysis. These observations led to the suggestion that the fatal anemia experienced by these EKLF-deficient mice was due entirely to deficient β-globin expression.
Several observations have suggested that there are additional erythroid cell defects in EKLF-deficient mice. Fetal liver-derived circulating erythroid cells in EKLF-deficient embryos exhibit a greater degree of hemolysis than typically occurs in β-thalassemia major (50, 57). Disruption of the βmajor and βminor genes in mice leads to anemia and death, but unlike the EKLF-deficient embryos, erythroid morphology in these animals closely mirrors that seen in human patients with β-thalassemia major (16, 61, 63, 71). Finally, in rescue experiments with EKLF-deficient mice, overexpression of a human γ-globin transgene improved globin chain imbalance, but hemolysis persisted and survival was not improved (44, 49, 58).
We hypothesized that the expression of erythroid genes other than β-globin was also affected by EKLF deficiency, contributing to the anemia experienced by EKLF-deficient mice. Subtractive hybridization with fetal liver RNA of wild-type and EKLF-deficient mice identified several potential EKLF target genes, including AHSP. Levels of AHSP mRNA were reduced 95 to 99.9%, and AHSP was undetectable in the EKLF-deficient fetal liver. In the AHSP promoter region, chromatin from a wild-type fetal liver demonstrated a DNase I hypersensitive site that was absent in the EKLF-deficient fetal liver. ChIP analyses identified two regions of histone hyperacetylation in wild-type chromatin, one corresponding to the location of the hypersensitive site and the other in the 3′ end of the gene. In EKLF-deficient chromatin, histones across the AHSP locus were hypoacetylated. Two regions of EKLF binding were found in wild-type chromatin that corresponded to the regions of histone hyperacetylation, one at the hypersensitive site, peaking over an AHSP promoter CACCC box, and the other in the 3′ end of the gene. In mobility shift assays, the AHSP promoter CACCC box, which differs from the EKLF consensus by 1 nucleotide, bound EKLF in a manner comparable to the β-globin promoter CACCC box. Additional studies demonstrated that this nucleotide was not critical for EKLF binding. When combined with recent studies of finger one of the closely related transcription factor Sp1, these data indicate a broader recognition sequence for the EKLF consensus binding site. In K562 cells, the AHSP promoter was transactivated by EKLF. These results demonstrate that EKLF acts as a transcription factor and a chromatin modulator for both β-globin and AHSP and may perform the same function for other erythroid genes as well.
MATERIALS AND METHODS
Identification of EKLF target genes by cDNA subtraction.
To identify differentially expressed genes, in wild-type and EKLF-deficient erythroid cells, we used a cDNA subtraction technique. The subtraction utilized fetal livers obtained from mice rendered null for the EKLF by gene targeting (57). EKLF-deficient embryos were initially identified by pallor and confirmed by genotyping (57). Ten micrograms of total RNA isolated from E13.5 wild-type and EKLF-deficient fetal livers was reverse transcribed with a modified oligo(dT) primer, 5′-TTTTGTACAAGCTT30N1N-3′, followed by coupling with (dC) tailing using a reverse transcriptase with terminal transferase activity (PowerScript; Clontech). Differential subtraction was performed as described by the manufacturer (PCR-Select cDNA Subtraction; BD Clontech). Nucleotide sequence analysis of differentially expressed clones was performed.
Analysis of differentially expressed genes.
A dual riboprobe containing sequences for both exon 2 of the murine α-globin gene and the last exon of the gene of interest was created for use in RNase protection assay (RPA) quantitation. This riboprobe ensures that both sequences are labeled to equal specific activity, allowing direct comparison of mRNA levels of target genes and α-globin mRNA levels. Quantitative, real-time PCR confirmation of differential mRNA expression was performed using primers for individual target genes that amplify the penultimate exon to the 3′ untranslated region (UTR) or the 5′ UTR to exon 2 (to decrease amplification of potentially homologous genes and detect genomic DNA contamination) and an internal control, β-actin, and SYBR green for detection using an iCycler instrument (Bio-Rad) (Table 1). Reverse transcription (RT)-PCR was performed with Thermus thermophilus polymerase (BD Clontech), a polymerase with reverse transcriptase and polymerase activities. Values obtained for target gene expression were normalized to β-actin and were expressed relative to the expression in control samples. For calculations, the 2−ΔΔCT formula was used, with ΔΔCT = (CT, target − CT, GAPDH)experimental sample − (CT, target − CT, GAPDH)control samples, where CT is cycle threshold.
TABLE 1.
Primers used in quantitative RT-PCR
| Gene | Primer sequence (5′-3′) | Location |
|---|---|---|
| AHSP | CGAGGGTTCACCCAGTCATGAACCACAATC | 5′ UTR |
| ATTGTGGATGAGGCGGGCTC | Exon 2 | |
| Protein 4.9 | CTCCGGGTGTGGACCGCATGAGG | 3′ UTR |
| CAGCAGAGGAAGAGGACACTGTA | Penultimate exon | |
| Rh30 | GTTGTCTCCTAAGTGTCAGAGTGTGGAGGGCTCCC | 3′ UTR |
| CATCCCAAAGTATTAATAGTCTCTGTCATGAAACC | Penultimate exon | |
| Aquaporin 1 | GGTGCCCTGGCAGTGCTCATCTA | 3′ UTR |
| GGAGTGACTTTGGTCAGCTTGTC | Penultimate exon | |
| p55 | GTTCATTGCACCTACTGACCAGG | 3′ UTR |
| CTAGTTGGAGCAGCCCTGTTTGT | Penultimate exon | |
| Protein 4.2 | GGAAGTAGATTGTGACATGTTCC | 3′ UTR |
| GAGTTCCTGGACCAACAGCACTA | Penultimate exon | |
| ERMAP | GCCCGATGTCAGACCACCCTGCCTG | 5′ UTR |
| CAGGGACTTGGTCTCTCCATATGAAC | Exon 2 | |
| β-Actin | GTGGGCCGCTCTAGGCACCA | |
| CGGTTGGCCTTAGGGTTCAGGGGGG | ||
| α-Globin | GGAAGATTGGTGGCCATGGTG | |
| TGACCTGGGCAGAGCCGTGGC |
Western blot analysis was performed using wild-type and EKLF-deficient E13.5 total fetal liver proteins. Blots were probed with either a rat anti-mouse AHSP monoclonal antibody (37) or an anti-actin antibody (sc-1616; Santa Cruz). Recombinant glutathione S-transferase (GST)-AHSP, a gift from Mitchell Weiss, was prepared as described previously (25).
DNase I hypersensitive site mapping.
DNase I HS mapping was performed as described previously (45, 70), with minor modifications, with 107 murine fetal liver cell nuclei. For HS mapping, DNA from embryonic stem cells was digested with BamHI for Southern blot analysis using a 211-bp fragment containing exons 1 and 2 of the AHSP cDNA as a probe. For fine mapping, the migration of the band generated by DNase I and BamHI enzyme digestion was compared with the migration of bands generated by the digestion of high-molecular-weight embryonic stem cell DNA digested with BamHI and AflIII, BlnI, HindIII, NsiI, or SmlI, respectively.
Quantitative ChIP assay.
ChIP analysis of diacetylated histone H3 and tetraacetylated histone H4 was performed with antibodies from Upstate Biotechnology (06-599 and 06-866; Lake Placid, NY) as described previously (33). After elution and extraction, immunoprecipitated DNA was analyzed by quantitative real-time PCR (iCycler; Bio-Rad) as described previously using primers in Table 2 (33). Signals of test genes were normalized to a region from the murine α-globin promoter with a region from the keratin gene promoter utilized as a nonerythroid internal control. SYBR green fluorescence in 25-μl PCR mixtures was determined, and the amount of product was determined relative to a standard curve generated from a titration of input chromatin. Amplification of a single amplification product was confirmed by dissociation curve analysis and acrylamide gel electrophoresis. Samples from at least three independent immunoprecipitations were analyzed. Parallel controls for each experiment included samples of no chromatin, no antibody, preimmune serum, and nonimmune rabbit immunoglobulin G.
TABLE 2.
Primers used for ChIP analysis of the murine AHSP locus
| Primer no. | Location in AF485327 (bp) | Sequence (5′-3′) |
|---|---|---|
| 1 | 187 | GTGGCTCTGCTCTCCTCTCTATAAAG |
| 287 | GCTTGTTTGAAAAGCCAGAAAGC | |
| 2 | 975 | CACAGGTTGTAACTGTGAGATCTTGG |
| 1088 | TCTCACCCTGACTCTATCTGGTATGTAGTAG | |
| 3 | 1303 | AAAGGATACTTATGTGGGTCCAGG |
| 1581 | CCCTCCTCTGGTGTGTCTGAG | |
| 4 | 2022 | CCTTTGCCTCCTTACCCAGC |
| 2126 | GGTAATGGGCACTTTTGCGC | |
| 5 | 2260 | CTAACTCCAGGGAAGCCTCACC |
| 2401 | TTTGTGTGTCTTCTGCACTAAGCG | |
| 6 | 2486 | AGCGGGACTTGAAAGGTTTAGG |
| 2600 | TTCCCTCACGCCTGAATACC | |
| 7 | 2856 | GAGCCTCTCGGAGACCATCC |
| 3000 | CCAGTCATGAACCACAATCACC | |
| 8 | 3162 | ACTGCCCTCCTCCTCATAACTTAAAGGG |
| 3260 | CAACATCTTGGGAGAACGGTC | |
| 9 | 3472 | TCGGAGACTAAAGAGGATTCGG |
| 3613 | CAACATCTTGGGAGAACGGTC | |
| 10 | 4390 | CTGTAGAAACAACAGCGGGAGTG |
| 4536 | CTGGGCTGCTCTCAGAATCAG | |
| 11 | 5298 | CTCAGCCATCCTCCTCGAAC |
| 5445 | AGTGGATTGAACCCATTTCACAG |
ChIP analysis of EKLF binding in wild-type fetal liver cells was performed with chromatin immunoprecipitated from mice with a hemagglutinin (HA)-EKLF-tandem affinity purification (TAP)-tagged knock-in allele (77) using an HA antibody (sc-7392; Santa Cruz). These mice have an HA tag knocked into the 5′ end of the EKLF gene, producing a functional EKLF protein with an HA tag at the NH2 terminus of the protein. ChIP was performed as described previously (13), except chromatin DNA from 1 million cells was used in each reaction. Quantitative PCR amplification was performed as described above.
EMSA.
Recombinant GST-EKLF fusion protein was prepared using a GST-EKLF plasmid (clone C10) as described previously (48). Binding reactions, electrophoresis, and autoradiography were carried out as described previously (70). Oligonucleotide probes are shown in Table 3. Unlabeled competitor oligonucleotides were added at various molar excesses as described previously (22). EKLF antibody 6B3 was obtained from James Bieker. Quantitative electrophoretic mobility shift analyses (EMSA) was performed as described (27, 34, 73). Specifically, the fraction of free DNA, D/Dt, was determined by measuring the ratio of free DNA signal analyzed at each protein concentration at the DNA signal in a control lane containing no protein. The fraction of DNA in complex with protein, PD/Dt, was derived from the relationship PD/Dt = 1 − D/Dt. To derive the equilibrium dissociation constant (KD) with standard error, the data were fit to the rearranged mass action equation, PD/Dt = 1/(1 + KDP), using nonlinear least-square analyses. Multiple analyses (three or more) were performed with the same range of protein concentrations to provide mean and standard error values for each point.
TABLE 3.
Electrophoretic mobility shift assay probes
| Probe | Sequence (5′-3′) |
|---|---|
| AHSP 3′ flanking EKLF consensus site 1 | AGGATTCGGGGGGGGGGGGGAGGCGGGA |
| TCCCGCCTCCCCCCCCCCCCCGAATCCT | |
| AHSP 3′ flanking EKLF consensus site 2 | AGCACATACCCCCACCCCCACGGCCA |
| TGGCCGTGGGGGTGGGGGTATGTGCT | |
| AHSP 3′ flanking EKLF consensus site 3 | CCACCCCACCCCACCCCCACAGAAT |
| ATTCTGTGGGGGTGGGGTGGGGTGG | |
| AHSP 3′ flanking EKLF consensus site 4 | TGCTTGCCCACACCCGCCAGTTTC |
| GAAACTGGCGGGTGTGGGCAAGCA | |
| AHSP promoter CACCC box | ACCTTCTCCACCCTAGGATG |
| CATCCTAGGGTGGAGAAGGT | |
| β-Globin promoter CACCC box (wild type) | TAGAGCCACACCCTGGTAAG |
| CTTACCAGGGTGTGGCTCTA | |
| β-Globin promoter CACCC box (C to T) | TAGAGCTACACCCTGGTAAG |
| CTTACCAGGGTGTAGCTCTA | |
| β-Globin promoter CACCC box (C to A) | TAGAGCAACACCCTGGTAAG |
| CTTACCAGGGTGTTGCTCTA | |
| β-Globin promoter CACCC box (C to G) | TAGAGCGACACCCTGGTAAG |
| CTTACCAGGGTGTCGCTCTA | |
| γ-Globin promoter CACCC box | TGGCTAAACTCCACCCATGGGTTG |
| CCAGAAGCGAGTGTGTGGAACTGCT |
Transactivation analyses.
HS2-β-globin gene promoter-luc and HS2-γ-globin gene promoter-luc plasmids were used as positive and negative controls, respectively, in transactivation assays (4, 32). The β-globin gene promoter was removed from the HS2-β-globin-luc plasmid and replaced with either a promoter fragment −170/+269 from the human AHSP gene (21) or a β-globin promoter with a thalassemia-associated CACCC box mutation known to perturb EKLF binding (22). Integrity of all test plasmids was confirmed by sequencing. Transient K562 cell (ATCC, CCL 243) transfections were performed as described previously with 20 μg of test plasmid, 10 μg of an EKLF expression plasmid, (48), and 0.5 μg of pCMVβ, a mammalian reporter plasmid expressing β-galactosidase driven by the human cytomegalovirus immediate-early gene promoter (Clontech) as described previously (70, 74). At least two preparations of each plasmid were tested in triplicate.
RESULTS
β-Globin and alpha-hemoglobin stabilizing protein expression are decreased in EKLF-deficient fetal liver.
To identify potential EKLF target genes, subtractive hybridization was performed with total RNA isolated from day 13.5 fetal livers of wild-type (WT) and EKLF-deficient mice (78). This method has successfully been applied to a large number of applications, including gene expression in cancer, development, and hematopoiesis (28, 30, 62). In the subtraction utilizing wild-type RNA as the tester population and EKLF-deficient fetal liver RNA as the driver, ∼175 differentially expressed clones were identified and subjected to sequence analysis. Validating the subtractive approach, the most abundant clone isolated corresponded to β-globin (n = 122). The second most abundant clone isolated was AHSP (n = 22). No other single clone except G protein Gi2a (n = 8) was represented by more than 5 clones. Results of subtraction were compared to results of differential gene expression in EKLF-deficient cells identified by analysis of a noncommercial microarray (20). Both techniques identified β-globin, AHSP, and the membrane protein gene protein 4.9 (3 clones in the subtraction). Compared to the array, subtraction did not identify hemogen, hemoglobin Z, any enzymes involved in heme biosynthesis, and several membrane-associated genes, such as those for Kell, CD24a, Icam4, and Kcnn4. It did identify other membrane-associated proteins, such as aquaporin, Rh30, p55, protein 4.2, and ERMAP.
When subtraction was performed using EKLF-deficient RNA as the tester population and wild-type fetal liver RNA as the driver, only 12 clones were obtained. No clone was represented more than once, even though EKLF has been shown to have a repressor function (14, 15).
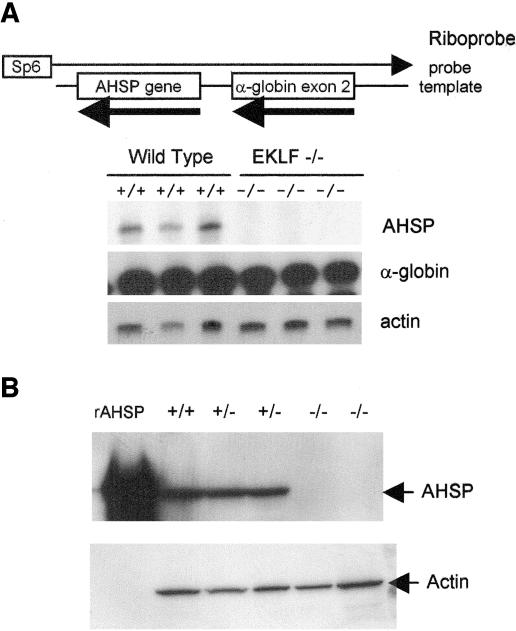
Reduced expression of potential target genes in EKLF-deficient fetal liver RNA was confirmed using either quantitative, real-time RT-PCR (Table 4) or RPA. The most frequently isolated clone after β-globin, AHSP, had nearly undetectable mRNA levels in EKLF-deficient RNA (Table 4; Fig. 1A). Western blot analysis with a rat anti-mouse monoclonal antibody against AHSP demonstrated that there was no AHSP in day 13.5 fetal livers from EKLF-deficient mice (Fig. 1B). Other genes with significantly decreased mRNA levels in EKLF-deficient mRNA included protein 4.9, which was also nearly undetectable, and aquaporin, reduced to less than 50% of the wild-type level (Table 4). Because AHSP was the most significantly altered gene in the differential subtraction, we selected it for further study.
TABLE 4.
Expression of differentially expressed nonglobin erythroid genes in EKLF-deficient fetal liver mRNA
| Genotype | Quantitative RT-PCRa
|
RNase protection (EKLF−/−)a | |
|---|---|---|---|
| EKLF+/− | EKLF−/− | ||
| AHSP | 86.0 ± 2.8 | 0.0001 ± 0.0 | 5.0 ± 2.6 |
| Protein 4.9 | 65.0 ± 1.4 | 1.0 ± 0.0 | 8.9 ± 4.3 |
| Rh30 | 86.0 ± 7.1 | 61.5 ± 4.9 | 77.4 ± 9.6 |
| Aquaporin | 45.0 ± 1.4 | 46.0 ± 2.8 | 54.1 ± 8.9 |
| Protein 4.2 | 86.5 ± 24.7 | 66.0 ± 4.2 | 87.6 ± 13.6 |
| P55 | 66.0 ± 1.4 | 76.5 ± 2.1 | 88.3 ± 12.3 |
| ERMAP | 112.5 ± 14.8 | 62.0 ± 4.2 | 91.0 ± 17.4 |
| Actin | 97.5 ± 7.8 | 116.5 ± 3.5 | |
Results are corrected with α-globin. Numbers shown are percentages of wild-type expression.
FIG. 1.
Target gene expression in wild-type and EKLF-deficient fetal livers. (A) RPA of wild-type and EKLF-deficient fetal liver RNA demonstrated a 95% ± 6.2% decrease in AHSP mRNA in EKLF-deficient cells. (B) Western blot analysis of fetal liver proteins with an anti-AHSP monoclonal antibody. Recombinant AHSP (rAHSP) was added as a positive control. Virtually no AHSP was found in EKLF-deficient cells.
Chromatin at the AHSP locus is altered in EKLF-deficient mice.
EKLF interacts with the proximal CACCC box of the β-globin gene promoter, establishing local chromatin structure and directing high-level β-globin transcription. In EKLF-deficient cells, there is loss of the DNase I hypersensitive site at the β-globin promoter and diminution of another HS site (HS3) over 50 kb away in the β-globin locus control region (26, 69). Thus, we hypothesized that chromatin across the AHSP locus would be perturbed in erythroid cells from EKLF-deficient mice. To interrogate the chromatin status of the AHSP gene, DNase I hypersensitive site mapping and ChIP were performed.
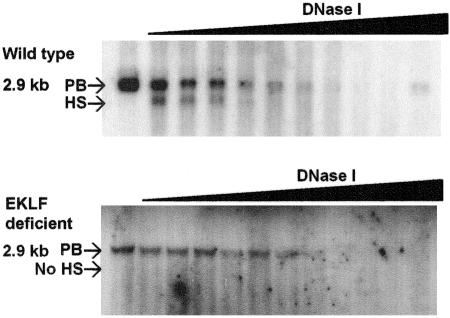
In chromatin from E13.5 WT fetal liver nuclei, a strong DNase I HS was demonstrated in the immediate 5′ flanking DNA in the core promoter region in a 162-bp HindIII/SmlI fragment (Fig. 2, upper panel) corresponding to coordinates 2187 to 2349 of a 6,472-bp murine AHSP fragment (GenBank accession no. AF485327) (59). Fine mapping of the HS localized it to a 43-bp HindIII/SmaI fragment corresponding to the core AHSP promoter containing a CACCC box (not shown). This HS site was absent in chromatin from EKLF-deficient fetal livers (Fig. 2, lower panel).
FIG. 2.
Hypersensitive site mapping across the AHSP locus. (Upper panel) In chromatin from E13.5 wild-type fetal liver nuclei digested with DNase I and BamHI (B), a strong DNase I HS was found in the 5′ flanking DNA of the AHSP gene generated from a 2.9-kb parent band (PB). (Lower panel) This HS site was absent in chromatin from EKLF-deficient fetal liver.
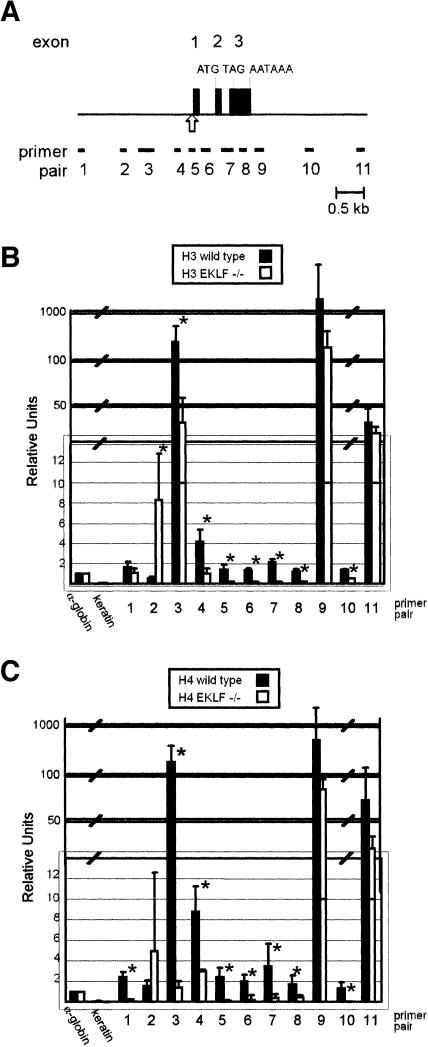
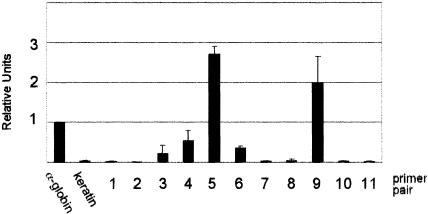
Histone modifications across 3.5 kb of the murine AHSP locus were examined using a ChIP assay with anti-diacetyl histone H3 and anti-tetraacetyl histone H4 antibodies and fetal liver chromatin from E13.5 WT and EKLF-deficient embryos. Eleven primer pairs spanning the murine AHSP locus, approximately 300 bp apart (Table 2 and Fig. 3A), were utilized for PCR. ChIP analysis of WT fetal liver chromatin demonstrated two regions where histones H3 and H4 were hyperacetylated relative to a control region from the mouse α-globin gene promoter (Fig. 3B and C; Table 5). The 5′ region corresponded to the 5′ flanking DNA and promoter region of the AHSP gene, and the second region mapped 3′ of the region of the AHSP cDNA in proximity to the polyadenylation signal (Fig. 3). Compared to the control, histones H3 and H4 were also acetylated in the interval between these peaks of hyperacetylation. In chromatin immunoprecipitated from EKLF-deficient fetal liver cells, there was hypoacetylation between primer pairs 4 through 8 (Fig. 3B and C; Table 5). This encompasses the core AHSP promoter and coding region, correlating with the severe reduction in AHSP gene expression. Significant H3 acetylation extends 5′ of the AHSP core promoter region, potentially indicating the interaction of EKLF with other regulatory elements in this region.
FIG. 3.
Histone acetylation across the murine AHSP locus in vivo. (A) Locations of primers used for quantitative PCR amplification after chromatin immunoprecipitation of wild-type and EKLF-deficient fetal liver chromatin. The DNase I hypersensitive site in the core promoter region is denoted by the arrow. (B) Pattern of acetylation of diacetylated histone H3. (C) Pattern of acetylation of tetraacetylated histone H4. In panels B and C, differences between wild-type and EKLF-deficient chromatin with P values of <0.15 are denoted by asterisks and the values are provided in Table 5.
TABLE 5.
Histone acetylation across the murine AHSP locus: dependence on EKLF
| Primera | H4
|
H3
|
||||
|---|---|---|---|---|---|---|
| WTb | EKLF−/−b | P value | WTb | EKLF−/−b | P value | |
| α-Globin | 1.0 | 1.0 | 1.0 | 1.0 | ||
| Keratin 14 | 0.065 ± 0.067 | 0.023 ± 0.01 | 0.34 | 0.05 ± 0.01 | 0.007 ± 0.0005 | 0.003 |
| 1 | 2.39 ± 0.53 | 0.25 ± 0.001 | 0.01 | 1.68 ± 0.48 | 1.07 ± 0.43 | 0.22 |
| 2 | 1.52 ± 0.61 | 4.91 ± 7.7 | 0.45 | 0.52 ± 0.18 | 8.27 ± 4.61 | 0.08 |
| 3 | 187.3 ± 54.5 | 1.41 ± 0.66 | 0.03 | 297.6 ± 9.6 | 38.3 ± 37.41 | 0.003 |
| 4 | 8.77 ± 2.5 | 2.97 ± 0.15 | 0.02 | 4.17 ± 1.19 | 0.99 ± 0.56 | 0.004 |
| 5 | 2.36 ± 0.94 | 0.11 ± 0.12 | 0.047 | 1.39 ± 0.47 | 0.077 ± 0.10 | 0.03 |
| 6 | 1.96 ± 0.67 | 0.23 ± 0.42 | 0.01 | 1.33 ± 0.21 | 0.103 ± 0.11 | 0.003 |
| 7 | 3.46 ± 2.18 | 0.43 ± 0.34 | 0.13 | 2.1 ± 0.38 | 0.09 ± 0.08 | 0.01 |
| 8 | 1.73 ± 0.83 | 0.49 ± 0.20 | 0.11 | 1.22 ± 0.26 | 0.17 ± 0.04 | 0.01 |
| 9 | 819.7 ± 250.6 | 78.4 ± 11.9 | 0.04 | 1,099.9 ± 444.9 | 144.2 ± 75.6 | 0.45 |
| 10 | 1.31 ± 0.59 | 0.06 ± 0.005 | 0.07 | 1.37 ± 0.11 | 0.55 ± 0.01 | 0.008 |
| 11 | 70.6 ± 42.3 | 19.0 ± 15.6 | 0.24 | 37.8 ± 15.6 | 35.13 ± 7.84 | 0.86 |
EKLF binds to a site in the 3′ region of the AHSP gene in vitro.
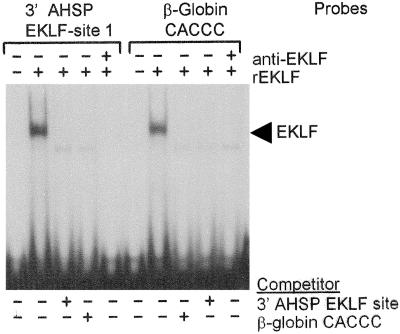
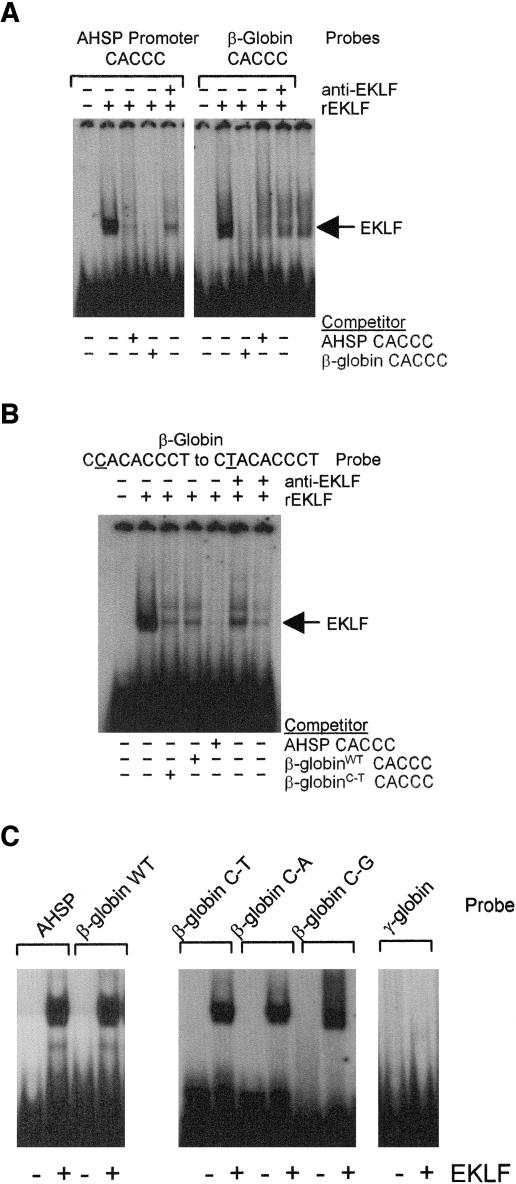
Analysis of the murine AHSP gene including 10 kb 5′ and 3′ of the coding region identified four canonical EKLF binding consensus sites, CCNCNCCCN, in the 3′ flanking region. To determine if EKLF could bind any of these EKLF sites in vitro, double-stranded oligonucleotide probes containing the AHSP 3′ flanking DNA EKLF sites or a β-globin promoter EKLF binding site probe as a positive control were prepared and used in EMSA with recombinant EKLF protein. The AHSP probe near the AHSP polyadenylation signal (AHSP 3′ flanking EKLF consensus site 1) (Table 3) yielded a single complex that migrated identically to a complex formed by the control β-globin promoter probe (Fig. 4). These complexes were effectively competed by both an excess of unlabeled AHSP probe, an excess of unlabeled control β-globin probe, and a monoclonal antibody against EKLF protein (not shown). The other three probes did not bind to recombinant EKLF (not shown). These data indicate that EKLF binds to a region of the 3′ flanking DNA region of the AHSP gene in vitro. This site is in the region of histone hyperacetylation identified by ChIP (primer pair 9) (Tables 2 and 5; Fig. 3).
FIG. 4.
Gel mobility shift assays of the EKLF consensus binding sites in the 3′ region of the AHSP gene. Gel mobility shift assays using oligonucleotide probes corresponding to the EKLF consensus binding sites in the 3′ flanking region of the murine AHSP gene were performed using recombinant EKLF protein (rEKLF). Results with site 1 are shown. A β-globin promoter-proximal CACCC box probe was used as a positive control. Excess, unlabeled probe or EKLF antibody was added where indicated. +, present; −, absent.
EKLF binds to the AHSP promoter CACCC box in vivo.
We wished to determine the region(s) where EKLF bound to the AHSP gene in vivo. However, we were unable to establish conditions for ChIP with anti-EKLF antibodies. Instead, ChIP across the AHSP locus was performed with HA-immunoprecipitated chromatin obtained from mice with an HA-EKLF-TAP-tagged knock-in allele (77). A peak of binding was seen in the 3′ region of the AHSP gene where a region of histone hyperacetylation was identified by ChIP and binding to a canonical EKLF consensus site was found (Fig. 5; Table 6).
FIG. 5.
EKLF occupancy across the murine AHSP locus in vivo. Quantitative chromatin immunoprecipitation across the AHSP locus was performed with fetal liver chromatin from mice with an HA-EKLF-TAP-tagged knock-in allele. Quantitative PCR amplification was performed with the primers shown in Fig. 3A.
TABLE 6.
EKLF occupancy across the murine AHSP locus determined by ChIP
Another region of EKLF binding extending from the 5′ region of the AHSP promoter HS to intron 1, peaking over the site of the AHSP promoter CACCC box, was identified (Fig. 5; Table 6). This peak is over the region of the DNase I hypersensitive site and histone hyperacetylation found in wild-type fetal liver cells.
A CACCC box in the AHSP gene promoter binds EKLF in vitro.
The AHSP promoter CACCC box, located in the region of the DNase I hypersensitive site, histone hyperacetylation, and EKLF binding, did not match the EKLF binding consensus, CCNCNCCCN, as it had a single mismatch at position 1, ACCCACCCT. To determine if EKLF could bind the imperfect EKLF site in the AHSP gene promoter in vitro, double-stranded oligonucleotide probes containing the AHSP 5′ flanking DNA imperfect EKLF site or a β-globin promoter EKLF binding site probe were prepared and used in EMSA with recombinant EKLF protein. The AHSP probe yielded a single complex that migrated identically to a complex formed by the control β-globin promoter probe (Fig. 6A). These complexes were effectively competed both by an excess of unlabeled AHSP probe, an excess of unlabeled control β-globin probe, and a monoclonal antibody against EKLF protein (Fig. 6A). These data indicate that EKLF binds to a region of the 5′ flanking DNA region of the AHSP gene in vitro.
FIG. 6.
Gel mobility shift assays of the AHSP promoter CACCC box. Gel mobility shift assays using oligonucleotide probes were performed using recombinant EKLF protein (rEKLF). Excess, unlabeled probe or EKLF antibody was added where indicated. (A) Probes corresponding to the AHSP promoter CACCC box and the β-globin promoter-proximal CACCC box. (B) A β-globin CACCC box probe with position 1 mutated from C to T to mimic the AHSP CACCC box. (C) Probes corresponding to the AHSP promoter CACCC box, the wild-type β-globin CACCC box, the β-globin promoter CACCC box with position 1 of the EKLF consensus sequence mutated to the other 3 possible nucleotides, and the γ-globin promoter CACCC box. +, present; −, absent.
To examine the influence of position 1 on EKLF-DNA binding, a β-globin oligonucleotide probe with a mutation of the EKLF consensus sequence to mimic the AHSP sequence (position 1, C to A) (Fig. 6B) or probes with mutation of position 1 to the other two possibilities (C to G and C to T) (Fig. 6C) were used in EMSA. All test probes yielded complexes similar to that of the wild-type β-globin probe. These complexes were effectively competed by an excess of unlabeled AHSP probe, an excess of unlabeled control β-globin probe, and a monoclonal antibody against EKLF protein (Fig. 6B and C). These data indicate that the C at position 1 of the EKLF consensus site is not critical for EKLF binding to the β-globin proximal CACCC box in vitro.
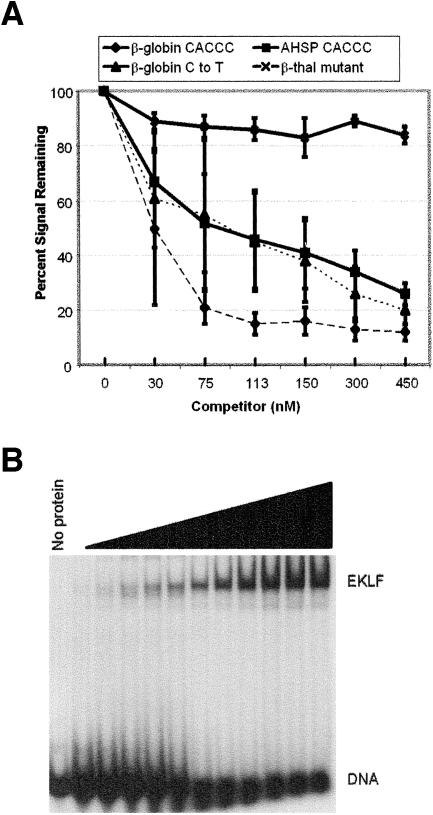
Competitive electrophoretic mobility shift assays were performed to compare the β-globin promoter CACCC site to the AHSP CACCC site. In this assay, the wild-type β-globin promoter CACCC site is used as a probe and different unlabeled probes are assayed for their ability to compete the EKLF-CACCC complex. Approximately 50 nM unlabeled wild-type β-globin CACCC probe was required to compete the EKLF-CACCC complex by 50% (Fig. 7A). Approximately 150 nM unlabeled wild-type AHSP probe or β-globin CACCC probe with position 2 C to T was required for similar competition (Fig. 7A). An unlabeled β-globin CACCC probe with a thalassemia mutation known to perturb EKLF binding (CACCC to CACGC) did not compete the EKLF-CACCC complex at the amounts added in these experiments. Quantitative EMSA were performed to further assess binding of the AHSP CACCC site. Recombinant EKLF-GST expressed in E. coli and purified on glutathione-Sepharose beads was used in DNA titration experiments to determine the concentration of protein capable of binding DNA (not shown). Active protein concentrations were then used in protein titrations to determine the KD for the various CACCC-containing oligonucleotide probes of interest (Fig. 7B). The highest-affinity site was the β-globin promoter CACCC box, with a KD of 3.98 × 10−4 ± 0.91 × 10−4 nM. The AHSP promoter CACCC box and the β-globin promoter CACCC boxes with position 2 mutated had affinities 30- to 40-fold less than the β-globin promoter CACCC box: AHSP promoter CACCC box, 1.1 × 10−3 ± 0.90 × 10−3 nM; β-globin C to T, 1.1 × 10−3 ± 0.18 × 10−3 nM; β-globin C to A, 1.3 × 10−3 ± 0.46 × 10−3 nM; and β-globin C to G, 1.2 × 10−3 ± 0.36 × 10−3 nM. The β-globin thalassemia mutant demonstrated a significantly lower binding affinity than the wild-type β-globin promoter CACCC box, 4.6 × 10−2 + 0.39 × 10−2. Together, these data indicate that the C at position 2 of the EKLF consensus site influences, but is not absolutely essential for, EKLF binding to the β-globin proximal CACCC box in vitro.
FIG. 7.
Quantitative electrophoretic mobility shift assays of the AHSP promoter CACCC box. (A) Competitive electrophoretic mobility shift assays were performed with the β-globin promoter CACCC box as a probe. Different unlabeled probes are assayed for their ability to compete the EKLF-CACCC complex. The amount of complex without competitor is defined as 100%. The points where curves cross the 50% line of the percent signal remaining was used as estimate the competitive ability of each oligonucleotide probe for binding to EKLF relative to the wild-type β-globin promoter CACCC box. β-thal, β-thalassemia. (B) EKLF protein titrations with β-globin and AHSP CACCC boxes. To determine the KD for the interaction between EKLF and each CACCC box, EKLF protein titrations were performed, gels scanned, and KD calculated with the rearranged mass action equation, PD/Dt = 1/(1 + KDP), using nonlinear least-square analyses. A sample gel with a wild-type β-globin promoter CACCC box probe is shown.
Transactivation of the AHSP gene promoter by EKLF in K562 cells.
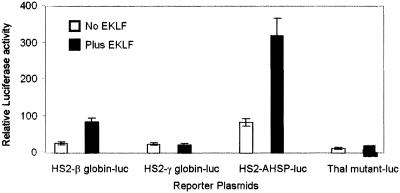
Luciferase reporter plasmids with an HS2-AHSP gene promoter fragment, (24) an HS2 human β-globin promoter fragment as a positive control, and an HS2 γ-globin promoter fragment as a negative control (4, 32) were transiently cotransfected into K562 cells with an EKLF cDNA expression plasmid. The AHSP promoter and the positive control β-globin promoter plasmids were transactivated by EKLF, whereas the negative control γ-globin promoter and thalassemia-mutant β-globin promoter plasmids were not (Fig. 8).
FIG. 8.
EKLF transactivates the AHSP promoter in K562 cells. HS2-AHSP promoter, HS2-γ-globin promoter, HS2-β-globin gene promoter, or mutant HS2-β-globin gene promoter/luciferase reporter plasmids were cotransfected into K562 cells with an EKLF cDNA expression plasmid. Luciferase activity was assayed 24 h after transfection and normalized to β-galactosidase to control for transfection efficiency. Thal, thalassemia.
DISCUSSION
Identification of AHSP as an EKLF target gene confirms the finding of differential expression identified by analysis of a noncommercial microarray (20) in a different genetically altered mouse line (57) and suggests that the hemolytic anemia in EKLF-deficient mice is due to both decreased transcription of the β-globin gene and decreased expression of other erythroid genes. AHSP deficiency could be part of the explanation why simply balancing globin chain synthesis does not rescue EKLF-deficient mice. Interestingly, in several studies of β-thalassemia patients with varying clinical severity, no mutations were identified in the AHSP gene (12, 23, 41, 68), even though in some reports discordant thalassemic patients were found to have decreased AHSP mRNA or protein expression in erythroid cells (23, 41). It is possible that the variation in clinical severity and AHSP expression are attributable to differences in EKLF, making EKLF another candidate modifier gene in the β-thalassemia syndromes. Deficiency of other erythroid genes such as protein 4.9 or other membrane proteins may also contribute to this anemia.
Like the β-globin locus, the AHSP locus demonstrates significant alterations in chromatin configuration in EKLF-deficient cells. Alterations in chromatin were not only found in the core AHSP promoter region at a site of EKLF-DNA binding but across the locus where there was global histone hypoacetylation. EKLF plays an important role in chromatin remodeling at the β-globin locus (1, 26, 36, 42, 47, 69, 75). EKLF is acetylated by CBP and p300, coactivator proteins that posses histone acetyltransferase activity (9, 75). It also interacts with other proteins, including those of the SWI/SNF chromatin remodeling complex (1, 10, 36, 42, 76). Recent evidence demonstrates that EKLF plays an important role in formation of an active chromatin hub in erythroid cells (21). Together, these data demonstrate support for the hypothesis that chromatin remodeling of the AHSP locus requires EKLF and demonstrate that EKLF may act as a transcription factor and a chromatin modulator for genes other than β-globin.
These data also demonstrate the value of an HA-EKLF-TAP-tagged knock-in allele mouse. In vivo histone acetylation data demonstrated two regions of hyperacetylation in wild-type mice that were absent in EKLF-deficient mice, one over the promoter and the other in the 3′ region of the gene. In vitro EMSA studies suggested that EKLF binding was occurring in the 3′ region. Generation of in vivo data from the HA-EKLF-TAP-tagged mouse permitted identification of EKLF binding to the core promoter CACCC box, which based on the published EKLF consensus sequence, was not initially considered.
The EKLF consensus sequence CCNCNCCCN was generated from data obtained from modeling of the crystal structure of DNA zinc finger contacts of Zif268 at 2.1 Å, as well as in vivo footprinting and methylation interference studies of the murine β-globin promoter (22, 54, 60). These studies demonstrated that β-thalassemia mutations associated with the β-globin promoter CACCC box disrupt specific contacts between guanine on the G-rich strand and arginine or histidine of the XYZ of fingers 2 or 3 (22). Identification of an EKLF binding site with a substitution in finger 1, where no thalassemia-associated mutations have been identified, suggests some relaxed freedom for DNA binding of finger 1 by EKLF. Finger 1 of EKLF is similar to another member of the C2H2-type zinc finger protein family, the ubiquitous transcription factor Sp1. Compared to fingers 2 and 3, finger 1 of Sp1 has more relaxed sequence and site specificity and contributes less to its DNA binding affinity (51, 67, 72).
Recent nuclear magnetic resonance structure of Sp1 revealed that, compared to fingers 2 and 3, which recognize four DNA base pairs by residues −1, 2, 3, and 6 of the recognition helix, finger 1 uses only residues −1 and 3 for DNA recognition (51). Differences in 3 amino acids of Sp1 finger 1 contribute to a broader recognition sequence than Zif268 and other C2H2-type zinc finger proteins. EKLF has a sequence identical to that of Sp1 at these 3 residues (see Fig. S1 in the supplemental material). Based on these data, it is possible that the EKLF consensus is more similar to the GGGCCG consensus of Sp1, e.g., NCNCCC, and that other factors, such as sequence context, composition of basal transcription machinery recruited to the promoter, and/or other EKLF-protein interactions, determine specificity and activity (4, 8). Several reports support this hypothesis, demonstrating that the function of the β-globin CACCC box is context dependent (4, 18, 29, 43, 64).
Supplementary Material
Acknowledgments
This work was supported in part by grants HL68965 (T.M.T.), HL65448, and DK/HL62039 (P.G.G.) from the National Institutes of Health and intramural funds from NHGRI.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Armstrong, J. A., J. J. Bieker, and B. M. Emerson. 1998. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell 95:93-104. [DOI] [PubMed] [Google Scholar]
- 2.Asano, H., X. S. Li, and G. Stamatoyannopoulos. 2000. FKLF-2: a novel Kruppel-like transcriptional factor that activates globin and other erythroid lineage genes. Blood 95:3578-3584. [PubMed] [Google Scholar]
- 3.Asano, H., X. S. Li, and G. Stamatoyannopoulos. 1999. FKLF, a novel Kruppel-like factor that activates human embryonic and fetal beta-like globin genes. Mol. Cell. Biol. 19:3571-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asano, H., and G. Stamatoyannopoulos. 1998. Activation of beta-globin promoter by erythroid Kruppel-like factor. Mol. Cell. Biol. 18:102-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basu, P., T. G. Sargent, L. C. Redmond, J. C. Aisenberg, E. P. Kransdorf, S. Z. Wang, G. D. Ginder, and J. A. Lloyd. 2004. Evolutionary conservation of KLF transcription factors and functional conservation of human gamma-globin gene regulation in chicken. Genomics 84:311-319. [DOI] [PubMed] [Google Scholar]
- 6.Basu, P., P. E. Morris, J. L. Haar, M. A. Wani, J. B. Lingrel, K. M. Gaensler, and J. A. Lloyd. 2005. KLF2 is essential for primitive erythropoiesis and regulates the human and murine embryonic beta-like globin genes in vivo. Blood 106:2566-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bieker, J. J. 2001. Kruppel-like factors: three fingers in many pies. J. Biol. Chem. 276:34355-34358. [DOI] [PubMed] [Google Scholar]
- 8.Bieker, J. J., and C. M. Southwood. 1995. The erythroid Kruppel-like factor transactivation domain is a critical component for cell-specific inducibility of a beta-globin promoter. Mol. Cell. Biol. 15:852-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blobel, G. A., T. Nakajima, R. Eckner, M. Montminy, and S. H. Orkin. 1998. CREB-binding protein cooperates with transcription factor GATA-1 and is required for erythroid differentiation. Proc. Natl. Acad. Sci. USA 95:2061-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown, R. C., S. Pattison, J. van Ree, E. Coghill, A. Perkins, S. M. Jane, and J. M. Cunningham. 2002. Distinct domains of erythroid Kruppel-like factor modulate chromatin remodeling and transactivation at the endogenous beta-globin gene promoter. Mol. Cell. Biol. 22:161-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cantor, A. B., and S. H. Orkin. 2002. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene 21:3368-3376. [DOI] [PubMed] [Google Scholar]
- 12.Cappellini, M. D., C. Refaldi, D. Bignamini, L. Zanaboni, and G. Fiorelli. 2004. Molecular analysis of alpha hemoglobin stabilizing protein (AHSP) in caucasian patients with different beta-thalassemia phenotypes. Blood 104:3770. [Google Scholar]
- 13.Chen, W. Y., and T. M. Townes. 2000. Molecular mechanism for silencing virally transduced genes involves histone deacetylation and chromatin condensation. Proc. Natl. Acad. Sci. USA 97:377-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, X., and J. J. Bieker. 2004. Stage-specific repression by the EKLF transcriptional activator. Mol. Cell. Biol. 24:10416-10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, X., and J. J. Bieker. 2001. Unanticipated repression function linked to erythroid Kruppel-like factor. Mol. Cell. Biol. 21:3118-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ciavatta, D. J., T. M. Ryan, S. C. Farmer, and T. M. Townes. 1995. Mouse model of human beta zero thalassemia: targeted deletion of the mouse beta maj- and beta min-globin genes in embryonic stem cells. Proc. Natl. Acad. Sci. USA 92:9259-9263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crossley, M., E. Whitelaw, A. Perkins, G. Williams, Y. Fujiwara, and S. H. Orkin. 1996. Isolation and characterization of the cDNA encoding BKLF/TEF-2, a major CACCC-box-binding protein in erythroid cells and selected other cells. Mol. Cell. Biol. 16:1695-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donze, D., T. M. Townes, and J. J. Bieker. 1995. Role of erythroid Kruppel-like factor in human gamma- to beta-globin gene switching. J. Biol. Chem. 270:1955-1959. [DOI] [PubMed] [Google Scholar]
- 19.dos Santos, C. O., and F. F. Costa. 2005. AHSP and beta-thalassemia: a possible genetic modifier. Hematology 10:157-161. [DOI] [PubMed] [Google Scholar]
- 20.Drissen, R., M. von Lindern, A. Kolbus, S. Driegen, P. Steinlein, H. Beug, F. Grosveld, and S. Philipsen. 2005. The erythroid phenotype of EKLF-null mice: defects in hemoglobin metabolism and membrane stability. Mol. Cell. Biol. 25:5205-5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drissen, R., R. J. Palstra, N. Gillemans, E. Splinter, F. Grosveld, S. Philipsen, and W. de Laat. 2004. The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes Dev. 18:2485-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng, W. C., C. M. Southwood, and J. J. Bieker. 1994. Analyses of beta-thalassemia mutant DNA interactions with erythroid Kruppel-like factor (EKLF), an erythroid cell-specific transcription factor. J. Biol. Chem. 269:1493-1500. [PubMed] [Google Scholar]
- 23.Galanello, R., L. Perseu, N. Giagu, G. Sole, and C. Perra. 2003. AHSP expression in beta-thalassemia carriers with thalassemia intermedia phenotype. Blood 104:516a. [Google Scholar]
- 24.Gallagher, P. G., E. Y. Wong, A. M. Pilon, M. J. Weiss, and D. M. Bodine. 2003. The human alpha hemoglobin stabilizing protein (AHSP) gene is regulated by GATA-1 and EKLF. Blood 102:267a. [Google Scholar]
- 25.Gell, D., Y. Kong, S. A. Eaton, M. J. Weiss, and J. P. Mackay. 2002. Biophysical characterization of the alpha-globin binding protein alpha-hemoglobin stabilizing protein. J. Biol. Chem. 277:40602-40609. [DOI] [PubMed] [Google Scholar]
- 26.Gillemans, N., R. Tewari, F. Lindeboom, R. Rottier, T. de Wit, M. Wijgerde, F. Grosveld, and S. Philipsen. 1998. Altered DNA-binding specificity mutants of EKLF and Sp1 show that EKLF is an activator of the beta-globin locus control region in vivo. Genes Dev. 12:2863-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goetz, T. L., T. L. Gu, N. A. Speck, and B. J. Graves. 2000. Auto-inhibition of Ets-1 is counteracted by DNA binding cooperativity with core-binding factor alpha2. Mol. Cell. Biol. 20:81-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gregory, R. C., K. A. Lord, L. B. Panek, P. Gaines, S. B. Dillon, and D. M. Wojchowski. 2000. Subtraction cloning and initial characterization of novel epo-immediate response genes. Cytokine 12:845-857. [DOI] [PubMed] [Google Scholar]
- 29.Guy, L. G., N. Delvoye, and L. Wall. 2000. Expression of a human beta-globin transgene in mice with the CACC motif and upstream sequences deleted from the promoter still depends on erythroid Kruppel-like factor. J. Biol. Chem. 275:3675-3680. [DOI] [PubMed] [Google Scholar]
- 30.Hanazawa, M., M. Mochii, N. Ueno, Y. Kohara, and Y. Iino. 2001. Use of cDNA subtraction and RNA interference screens in combination reveals genes required for germ-line development in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 98:8686-8691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harju, S., K. J. McQueen, and K. R. Peterson. 2002. Chromatin structure and control of beta-like globin gene switching. Exp. Biol. Med. (Maywood) 227:683-700. [DOI] [PubMed] [Google Scholar]
- 32.Jane, S. M., P. A. Ney, E. F. Vanin, D. L. Gumucio, and A. W. Nienhuis. 1992. Identification of a stage selector element in the human gamma-globin gene promoter that fosters preferential interaction with the 5′ HS2 enhancer when in competition with the beta-promoter. EMBO J. 11:2961-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson, K. D., H. M. Christensen, B. Zhao, and E. H. Bresnick. 2001. Distinct mechanisms control RNA polymerase II recruitment to a tissue-specific locus control region and a downstream promoter. Mol. Cell 8:465-471. [DOI] [PubMed] [Google Scholar]
- 34.Jonsen, M. D., J. M. Petersen, Q. P. Xu, and B. J. Graves. 1996. Characterization of the cooperative function of inhibitory sequences in Ets-1. Mol. Cell. Biol. 16:2065-2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaczynski, J., T. Cook, and R. Urrutia. 2003. Sp1- and Kruppel-like transcription factors. Genome Biol. 4:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kadam, S., G. S. McAlpine, M. L. Phelan, R. E. Kingston, K. A. Jones, and B. M. Emerson. 2000. Functional selectivity of recombinant mammalian SWI/SNF subunits. Genes Dev. 14:2441-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kihm, A. J., Y. Kong, W. Hong, J. E. Russell, S. Rouda, K. Adachi, M. C. Simon, G. A. Blobel, and M. J. Weiss. 2002. An abundant erythroid protein that stabilizes free alpha-haemoglobin. Nature 417:758-763. [DOI] [PubMed] [Google Scholar]
- 38.Kong, Y., S. Zhou, A. J. Kihm, A. M. Katein, X. Yu, D. A. Gell, J. P. Mackay, K. Adachi, L. Foster-Brown, C. S. Louden, A. J. Gow, and M. J. Weiss. 2004. Loss of alpha-hemoglobin-stabilizing protein impairs erythropoiesis and exacerbates beta-thalassemia. J. Clin. Investig. 114:1457-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koury, M. J., S. T. Sawyer, and S. J. Brandt. 2002. New insights into erythropoiesis. Curr. Opin. Hematol. 9:93-100. [DOI] [PubMed] [Google Scholar]
- 40.Kulozik, A. E., A. Bellan-Koch, S. Bail, E. Kohne, and E. Kleihauer. 1991. Thalassemia intermedia: moderate reduction of beta globin gene transcriptional activity by a novel mutation of the proximal CACCC promoter element. Blood 77:2054-2058. [PubMed] [Google Scholar]
- 41.Lai, M. I., S. Menzel, J. Jiang, M. J. Weiss, and S. L. Thein. 2005. Alpha haemoglobin stabilizing protein expression in thalassaemia intermedia. Blood Cells Mol. Dis. 34:99. [Google Scholar]
- 42.Lee, C. H., M. R. Murphy, J. S. Lee, and J. H. Chung. 1999. Targeting a SWI/SNF-related chromatin remodeling complex to the beta-globin promoter in erythroid cells. Proc. Natl. Acad. Sci. USA 96:12311-12315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee, J. S., H. Ngo, D. Kim, and J. H. Chung. 2000. Erythroid Kruppel-like factor is recruited to the CACCC box in the beta-globin promoter but not to the CACCC box in the gamma-globin promoter: the role of the neighboring promoter elements. Proc. Natl. Acad. Sci. USA 97:2468-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lim, S. K., J. J. Bieker, C. S. Lin, and F. Costantini. 1997. A shortened life span of EKLF-/- adult erythrocytes, due to a deficiency of beta-globin chains, is ameliorated by human gamma-globin chains. Blood 90:1291-1299. [PubMed] [Google Scholar]
- 45.Lowrey, C. H., D. M. Bodine, and A. W. Nienhuis. 1992. Mechanism of DNase I hypersensitive site formation within the human globin locus control region. Proc. Natl. Acad. Sci. USA 89:1143-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luzzatto, L., and R. Notaro. 2002. Haemoglobin's chaperone. Nature 417:703-705. [DOI] [PubMed] [Google Scholar]
- 47.McMorrow, T., A. van den Wijngaard, A. Wollenschlaeger, M. van de Corput, K. Monkhorst, T. Trimborn, P. Fraser, M. van Lohuizen, T. Jenuwein, M. Djabali, S. Philipsen, F. Grosveld, and E. Milot. 2000. Activation of the beta globin locus by transcription factors and chromatin modifiers. EMBO J. 19:4986-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller, I. J., and J. J. Bieker. 1993. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Kruppel family of nuclear proteins. Mol. Cell. Biol. 13:2776-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nilson, D. G., S. H. Orkin, P. G. Gallagher, and D. M. Bodine. 2001. The erythrocyte membrane protein genes ankyrin and band 3 (AE1) are non-globin erythroid krupple-like factor (EKLF) target genes. Blood 12:552a. [Google Scholar]
- 50.Nuez, B., D. Michalovich, A. Bygrave, R. Ploemacher, and F. Grosveld. 1995. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature 375:316-318. [DOI] [PubMed] [Google Scholar]
- 51.Oka, S., Y. Shiraishi, T. Yoshida, T. Ohkubo, Y. Sugiura, and Y. Kobayashi. 2004. NMR structure of transcription factor Sp1 DNA binding domain. Biochemistry 43:16027-16035. [DOI] [PubMed] [Google Scholar]
- 52.Orkin, S. H., S. E. Antonarakis, and H. H. Kazazian, Jr. 1984. Base substitution at position −88 in a beta-thalassemic globin gene. Further evidence for the role of distal promoter element ACACCC. J. Biol. Chem. 259:8679-8681. [PubMed] [Google Scholar]
- 53.Orkin, S. H., H. H. Kazazian, Jr., S. E. Antonarakis, S. C. Goff, C. D. Boehm, J. P. Sexton, P. G. Waber, and P. J. Giardina. 1982. Linkage of beta-thalassaemia mutations and beta-globin gene polymorphisms with DNA polymorphisms in human beta-globin gene cluster. Nature 296:627-631. [DOI] [PubMed] [Google Scholar]
- 54.Pavletich, N. P., and C. O. Pabo. 1991. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science 252:809-817. [DOI] [PubMed] [Google Scholar]
- 55.Perkins, A. 1999. Erythroid Kruppel like factor: from fishing expedition to gourmet meal. Int. J. Biochem. Cell. Biol. 31:1175-1192. [DOI] [PubMed] [Google Scholar]
- 56.Perkins, A. C., K. M. Gaensler, and S. H. Orkin. 1996. Silencing of human fetal globin expression is impaired in the absence of the adult beta-globin gene activator protein EKLF. Proc. Natl. Acad. Sci. USA 93:12267-12271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perkins, A. C., A. H. Sharpe, and S. H. Orkin. 1995. Lethal beta-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature 375:318-322. [DOI] [PubMed] [Google Scholar]
- 58.Perkins, A. C., K. R. Peterson, G. Stamatoyannopoulos, H. E. Witkowska, and S. H. Orkin. 2000. Fetal expression of a human agamma globin transgene rescues globin chain imbalance but not hemolysis in EKLF null mouse embryos. Blood 95:1827-1833. [PubMed] [Google Scholar]
- 59.Pilon, A. M., C. Wong, L. J. Garrett-Beal, M. Weis, P. G. Gallagher, and D. M. Bodine. 2004. Chromatin remodeling of the mouse ahsp gene requires EKLF. Blood 104:110a. [Google Scholar]
- 60.Reddy, P. M., and C. K. Shen. 1993. Erythroid differentiation of mouse erythroleukemia cells results in reorganization of protein-DNA complexes in the mouse beta maj globin promoter but not its distal enhancer. Mol. Cell. Biol. 13:1093-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rivella, S., C. May, A. Chadburn, I. Riviere, and M. Sadelain. 2003. A novel murine model of Cooley anemia and its rescue by lentiviral-mediated human beta-globin gene transfer. Blood 101:2932-2939. [DOI] [PubMed] [Google Scholar]
- 62.Shojaei, F., L. Gallacher, and M. Bhatia. 2004. Differential gene expression of human stem progenitor cells derived from early stages of in utero human hematopoiesis. Blood 103:2530-2540. [DOI] [PubMed] [Google Scholar]
- 63.Skow, L. C., B. A. Burkhart, F. M. Johnson, R. A. Popp, D. M. Popp, S. Z. Goldberg, W. F. Anderson, L. B. Barnett, and S. E. Lewis. 1983. A mouse model for beta-thalassemia. Cell 34:1043-1052. [DOI] [PubMed] [Google Scholar]
- 64.Tanimoto, K., Q. Liu, F. Grosveld, J. Bungert, and J. D. Engel. 2000. Context-dependent EKLF responsiveness defines the developmental specificity of the human epsilon-globin gene in erythroid cells of YAC transgenic mice. Genes Dev. 14:2778-2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tewari, R., N. Gillemans, M. Wijgerde, B. Nuez, M. von Lindern, F. Grosveld, and S. Philipsen. 1998. Erythroid Kruppel-like factor (EKLF) is active in primitive and definitive erythroid cells and is required for the function of 5′HS3 of the beta-globin locus control region. EMBO J. 17:2334-2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thein, S. L. 2004. Genetic insights into the clinical diversity of beta thalassaemia. Br. J. Haematol. 124:264-274. [DOI] [PubMed] [Google Scholar]
- 67.Thiesen, H. J., and C. Bach. 1990. Target detection assay (TDA): a versatile procedure to determine DNA binding sites as demonstrated on SP1 protein. Nucleic Acids Res. 18:3203-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Viprakasit, V., V. S. Tanphaichitr, W. Chinchang, P. Sangkla, M. J. Weiss, and D. R. Higgs. 2004. Evaluation of alpha hemoglobin stabilizing protein (AHSP) as a genetic modifier in patients with beta thalassemia. Blood 103:3296-3299. [DOI] [PubMed] [Google Scholar]
- 69.Wijgerde, M., J. Gribnau, T. Trimborn, B. Nuez, S. Philipsen, F. Grosveld, and P. Fraser. 1996. The role of EKLF in human beta-globin gene competition. Genes Dev. 10:2894-2902. [DOI] [PubMed] [Google Scholar]
- 70.Wong, E. Y., J. Lin, B. G. Forget, D. M. Bodine, and P. G. Gallagher. 2004. Sequences downstream of the erythroid promoter are required for high level expression of the human alpha-spectrin gene. J. Biol. Chem. 279:55024-55033. [DOI] [PubMed] [Google Scholar]
- 71.Yang, B., S. Kirby, J. Lewis, P. J. Detloff, N. Maeda, and O. Smithies. 1995. A mouse model for beta 0-thalassemia. Proc. Natl. Acad. Sci. USA 92:11608-11612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yokono, M., N. Saegusa, K. Matsushita, and Y. Sugiura. 1998. Unique DNA binding mode of the N-terminal zinc finger of transcription factor Sp1. Biochemistry 37:6824-6832. [DOI] [PubMed] [Google Scholar]
- 73.Zhang, L., J. Zhao, and H. J. Edenberg. 1999. A human Raf-responsive zinc-finger protein that binds to divergent sequences. Nucleic Acids Res. 27:2947-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang, P., P. Basu, L. C. Redmond, P. E. Morris, J. W. Rupon, G. D. Ginder, and J. A. Lloyd. 2005. A functional screen for Kruppel-like factors that regulate the human gamma-globin gene through the CACCC promoter element. Blood Cells Mol. Dis. 35:227-235. [DOI] [PubMed] [Google Scholar]
- 75.Zhang, W., and J. J. Bieker. 1998. Acetylation and modulation of erythroid Kruppel-like factor (EKLF) activity by interaction with histone acetyltransferases. Proc. Natl. Acad. Sci. USA 95:9855-9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang, W., S. Kadam, B. M. Emerson, and J. J. Bieker. 2001. Site-specific acetylation by p300 or CREB binding protein regulates erythroid Kruppel-like factor transcriptional activity via its interaction with the SWI-SNF complex. Mol. Cell. Biol. 21:2413-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhou, D., J. X. Ren, T. M. Ryan, N. P. Higgins, and T. M. Townes. 2004. Rapid tagging of endogenous mouse genes by recombineering and ES cell complementation of tetraploid blastocysts. Nucleic Acids Res. 32:e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhu, Y. Y., E. M. Machleder, A. Chenchik, R. Li, and P. D. Siebert. 2001. Reverse transcriptase template switching: a SMART approach for full-length cDNA library construction. BioTechniques 30:892-897. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.