Abstract
During leaf abscission in oilseed rape (Brassica napus), cell wall degradation is brought about by the action of several hydrolytic enzymes. One of these is thought to be polygalacturonase (PG). Degenerate primers were used to isolate a PG cDNA fragment by reverse transcriptase-polymerase chain reaction from RNA extracted from ethylene-promoted leaf abscission zones (AZs), and in turn a full-length clone (CAW471) from an oilseed rape AZ cDNA library. The highest homology of this cDNA (82%) was to an Arabidopsis sequence that was predicted to encode a PG protein. Analysis of expression revealed that CAW471 mRNA accumulated in the AZ of leaves and reached a peak 24 h after ethylene treatment. Ethylene-promoted leaf abscission in oilseed rape was not apparent until 42 h after exposure to the gas, reaching 50% at 48 h and 100% by 56 h. In floral organ abscission, expression of CAW471 correlated with cell separation. Genomic libraries from oilseed rape and Arabidopsis were screened with CAW471 and the respective genomic clones PGAZBRAN and PGAZAT isolated. Characterization of these PG genes revealed that they had substantial homology within both the coding regions and in the 5′-upstream sequences. Fusion of a 1,476-bp 5′-upstream sequence of PGAZAT to β-glucuronidase or green fluorescent protein and transformation of Arabidopsis revealed that this fragment was sufficient to drive expression of these reporter genes in the AZs at the base of the anther filaments, petals, and sepals.
Abscission is a critical process in the life cycle of a plant (Roberts et al., 2000). It culminates in the shedding of a range of organs including leaves, flowers, and fruits and is the consequence of a highly coordinated sequence of events. Although the precise sequence of biochemical and molecular changes that bring about cell separation have yet to be established, there is convincing evidence from histological studies that they are restricted to only a few layers of cells. The limited number of cells that undergo these changes has hampered the study of this important developmental event and the majority of our knowledge has come from studies of abscission in species such as Phaseolus vulgaris, Sambucus nigra, and tomato (Lycopersicon esculentum) (Gonzalez-Carranza et al., 1998).
Shedding takes place at predetermined sites called abscission zones (AZs) and during organ separation, the dissolution of the cell wall at the point of detachment is observed (Abeles, 1968; Sexton and Roberts, 1982; Osborne, 1989). Cell wall degradation is not restricted to AZs and has been observed to take place throughout development contributing to such processes as elongation growth (Cosgrove, 1998), fruit ripening (Fisher and Bennett, 1991), pod dehiscence (Meakin and Roberts, 1990; Petersen et al., 1996; Stolle-Smits et al., 1999), outgrowth of lateral roots (Peretto et al., 1992), pollen tube growth (Clarke and Gleeson, 1981), and seed germination (Sitrit et al., 1999).
It is well documented that during leaf, flower, and fruit abscission cell wall degradation is associated with an increase in the activity of several hydrolytic enzymes including β-1,4-glucanases (Bonghi et al., 1992, 1993; Lashbrook et al., 1994; Taylor et al., 1994; del Campillo and Bennett, 1996; del Campillo, 1999) and polygalacturonases (PGs; Taylor et al., 1993; Kalaitzis et al., 1995, 1997; Brown, 1997). Other abscission-related proteins recently have been identified such as expansins, which may either contribute to the cell separation process (Cho and Cosgrove, 2000; E.J. Belfield and J.A. Roberts, unpublished data) or play a role in protecting the exposed fracture surface from pathogen attack (Coupe et al., 1995, 1997).
The major components of the cell wall are pectins, cellulose, and hemicellulose (Carpita and Gibeaut, 1993; Chun and Huber, 1998; Hadfield et al., 1998; Torki et al., 1999). The structure of pectins may be modified by the ionic strength of the apoplast, or by exo- or endo-acting enzymes that degrade regions of the pectin (Hadfield and Bennett, 1998) by cleaving chain residues (De Veau et al., 1993). An increase in PG activity during abscission was first reported over 30 years ago and there is a now a considerable body of evidence that supports a key role for this enzyme in the cell separation process (Taylor et al., 1991, 1993; Hadfield and Bennett, 1998). Not only has the activity of PG been shown to increase specifically in AZ tissue during the shedding of organs such as leaves and flowers, but also the expression of PG genes has been found to increase prior to and during abscission (Bonghi et al., 1992; Coupe et al., 1995). The identification of seven different PG genes from tomato has been reported, TAPG1-TAPG6 and TPG7, from which TAPG1, TAPG2, TAPG4, and TAPG5 are expressed during ethylene-induced leaf and flower abscission (Kalaitzis et al., 1997; Hong and Tucker, 2000). An analysis of the spatial and temporal expression of two of these genes (TAPG1 and TAPG4) has been carried out recently by fusing their respective promoters to β-glucuronidase (GUS). A minimal promoter of 247 bp upstream of the start of transcription of TAPG1 was found to be sufficient to drive GUS expression in both the leaf and floral AZs (Hong et al., 2000).
Although Arabidopsis has been used successfully as a model system for the study of a range of developmental processes in plants, few studies on abscission have been undertaken on this species (Bleecker and Patterson, 1997; Patterson, 2001). This is primarily because this plant does not shed its leaves and the tissues that are lost (sepals, petals, and anthers) provide little AZ material for biochemical or molecular analysis. With this in mind, we undertook a study of the abscission process in oilseed rape (Brassica napus). This species undergoes both leaf and flower abscission and the size of the former zone readily allows the isolation and characterization of abscission-related genes. Moreover, our previous studies on dehiscence-related genes have shown that Arabidopsis and oilseed rape homologs have close sequence identity. In this paper, we describe the isolation and characterization of a gene encoding a PG that is specifically up-regulated during leaf and flower abscission in oilseed rape. The homologous PG gene has been isolated from Arabidopsis and fusion of the promoter to either GUS or green fluorescent protein (GFP) reveals that expression takes place at the site of floral organ separation.
RESULTS
Isolation of an Abscission-Related PG cDNA from Oilseed Rape
Reverse transcriptase (RT)-PCR using degenerate primers designed to anneal within highly conserved regions of PG genes was used to amplify a 150-bp product from RNA extracted from ethylene-treated oilseed rape leaf AZs. The DNA sequence of the putative PG PCR product was compared with other sequences in the EMBL database using the FASTA program (Pearson and Lipman, 1988) and found to exhibit 79.4% identity with the dehiscence zone PG SAC66.
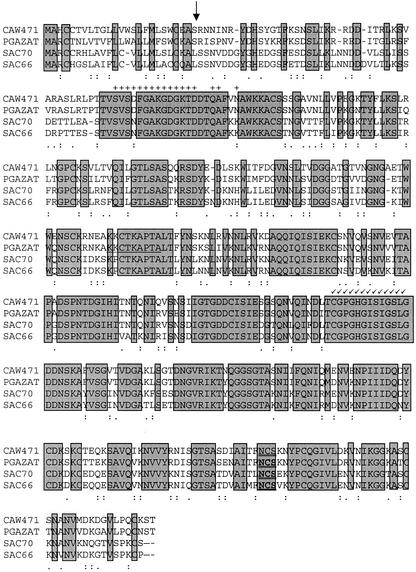
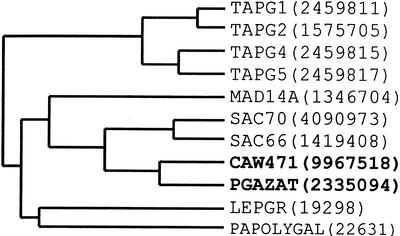
A cDNA library generated from mRNA extracted from ethylene-treated oilseed rape leaf AZ tissue was screened using the PCR fragment as a probe to isolate the equivalent full-length clone. Sequence analysis of one of the positive clones (CAW471, accession no. AJ250919) showed it to be a full-length cDNA and to include the 150-bp PCR fragment. The longest open reading frame in the 1,764-bp cDNA, CAW471, encodes a putative 434-amino acid protein with a predicted molecular mass of 46.6 kD. The amino terminal region had a hydrophobic region indicative of a signal peptide (the first 24 amino acids) and the presence of one potential glycosylation site (N-X-S) could be detected at amino acid residue 392. The 3′-untranslated region contains two putative polyadenylation signals at position 1,610 and 1,667. The predicted isoelectric point (pI) was 8.39 and the charge of the peptide at pH 7 was calculated to be 5.09. A comparison of the CAW471 nucleic acid sequence to the EMBL database revealed closest homology with a putative Arabidopsis PG (82%; GeneInfo Identifier [GI] no. 2335094; Lin et al., 1999) and similarity to PGs from the silique dehiscence zones of Arabidopsis SAC70 (66%; GI no. 4090973; Jenkins et al., 1999) and oilseed rape SAC66 (63%; GI no. 1419408; Jenkins et al., 1999; Fig. 1). Sequence identity with PGs from tomato leaf and flower AZs was much lower at 39%, 39%, 38%, and 36% with TAPG2, TAPG5, TAPG1, and TAPG4 (GI nos. 1575705, 2459817, 2459811, and 2459815) respectively (Kalaitzis et al., 1997). A phylogenetic tree of those PGs showing closest homology to CAW471 was obtained using Clustal method with PAM250 residue weight table (Fig. 2).
Figure 1.
Comparison of the predicted peptide sequence of CAW471 with that of PGAZAT, SAC70 (Jenkins et al., 1999), and SAC66 (Jenkins et al., 1999). Identical residues are in shaded boxes; a colon represents similar residues, whereas a period represents related residues. The predicted signal peptide cleavage site is indicated by an arrow (↓) and the conserved Asn glycosylation site (N-X-S/T) is double underlined. The exon previously omitted from the PGAZAT sequence is underlined and conserved residues in other endo- and exo-PGs are indicated by + and √, respectively. The alignment was generated using a Clustal method using the Pam 250 residue weight table on the DNAstar program. Dark shading indicates identical residues.
Figure 2.
Phylogenetic tree of PGs showing closest homology to CAW471. TAPG1, 2, 4, and 5 are expressed during leaf and flower abscission in tomato (Hong and Tucker, 1998). MAD14A, LEPGR, and PAPOLYGAP are fruit PGs from apple (Malus domestica), tomato, and avocado, respectively (Sheehy et al., 1987; Dopico et al., 1993; Atkinson, 1994). SAC70 and SAC66 are pod dehiscence-related PGs from Arabidopsis and oilseed rape, respectively (Jenkins et al., 1999). The numbers in parentheses are protein identification numbers and the tree was generated using the DNAstar software.
Time Course of Expression of CAW471 mRNA during Leaf and Flower Abscission in Oilseed Rape
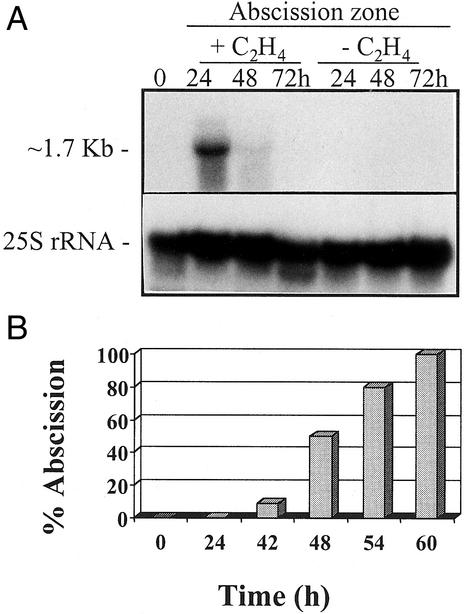
The time course of CAW471 expression during leaf abscission is shown in Figure 3A. An antisense strand-specific RNA probe of CAW471 hybridized to an mRNA of approximately 1.7 kb that accumulated specifically in the AZ tissue after exposure to ethylene (10 μL L−1). Expression was detected at 24 h decreasing by 48 h after ethylene treatment. No signal was detected in the absence of the gas (data not shown). Abscission of petioles commenced at 42 h after exposure to ethylene (10 μL L−1) reaching 50% at 48 h and 100% by 56 h (Fig. 3B).
Figure 3.
A, Northern blot showing expression of CAW471 in mRNA extracted from leaf AZs that had been incubated in the presence or absence of ethylene (10 μL L−1) for 0, 24, 48, and 72 h. The same membrane was probed with a 25S rRNA to show equal loading of the RNA. B, Time course of abscission in leaf explants exposed to ethylene (10 μL L−1) for 0, 24, 42, 48, 54, and 60 h.
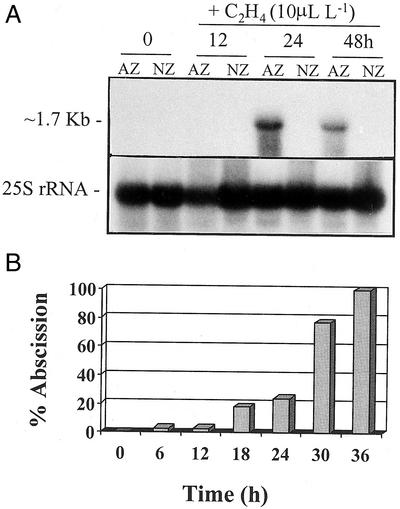
No expression of CAW471 mRNA could be detected using northern analysis in roots, laminar portion of leaves, pods (dehiscence zone or non-zone [NZ] material), or seeds (data not shown). However, accumulation of CAW471 could be detected in floral AZ tissues undergoing cell separation (Fig. 4A).
Figure 4.
A, Northern blot showing expression of CAW471 in mRNA extracted from flower AZ and NZ tissue that had been exposed to ethylene (10 μL L−1) for 0, 12, 24, and 48 h. The same membrane was probed with a 25S rRNA to show equal loading of the RNA. B, Time course of abscission in flower AZ explants exposed to ethylene (10 μL L−1) for 0, 6, 12, 18, 24, 30, and 30 h.
The time course of expression of CAW471 during the course of ethylene-stimulated flower abscission is shown in Figure 4A. As with leaf abscission, an antisense strand-specific RNA probe of CAW471 hybridized to an mRNA of approximately 1.7 kb that accumulated specifically in the AZ tissue during exposure to ethylene. Expression reached a peak at 24 h and then declined after 48 h of ethylene treatment. Abscission of floral parts commenced at 6 h, reaching 24% at 24 h, 72% by 28 h, and 100% at 34 h after exposure to ethylene (Fig. 4B).
Isolation and Characterization of the Gene Encoding CAW471 from Oilseed Rape and Its Homolog in Arabidopsis
To identify PG genes, genomic libraries generated from DNA extracted from oilseed rape and Arabidopsis were screened with the CAW471 cDNA as indicated in “Materials and Methods.” The oilseed rape gene corresponding to CAW471 (PGAZBRAN, accession no. AJ250918) consists of eight introns and nine exons. When a sequence of approximately 300 bp of the promoter of PGAZBRAN was used to search the database, a sequence from chromosome II of Arabidopsis (accession no. AC002339; Lin et al., 1999) exhibited high homology. Based on the assumption that the clones isolated from the Arabidopsis library might be homologous to this, primers were designed and used on positive clones, confirming them to be the same gene as that reported in the database. The predicted protein for this gene (PGAZAT) was compared with CAW471 and a small exon that had not been previously identified was detected in the sequence (KKCTKAPTA) from amino acids 183 through 192 (Fig. 1).
Spatial Expression Pattern of the PGAZAT Gene in Arabidopsis
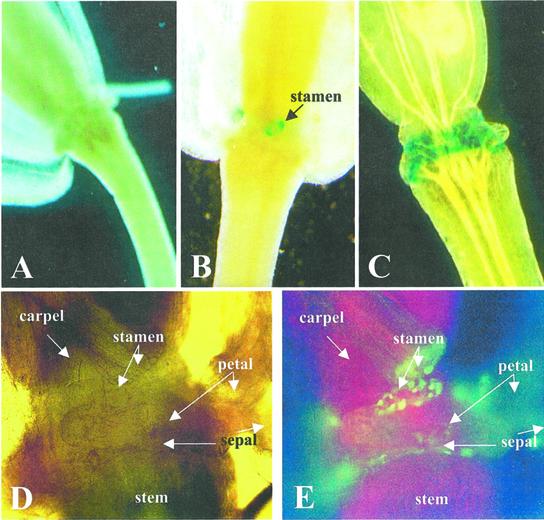
A 1,476-bp fragment of the promoter of the PGAZAT gene was fused to GUS or GFP separately and used to transform Arabidopsis plants via an infiltration technique. The expression of the reporter genes was examined in transgenic plants during the “natural” shedding of floral parts. No expression of either GUS or GFP was observed in young flowers (Fig. 5A); however, during the separation of floral organs, both reporter genes could be detected in a number of transgenic lines at the sites of shedding of the stamens, petals, and sepals. The accumulation of GUS could be detected prior to organ abscission (Fig. 5B) and expression was first observed at the base of the anther filament followed by the petal and finally the sepal (Fig. 5C). Expression at the base of the stamen and petal was restricted to a ring of cortical cells and this could be seen clearly using either GUS or GFP reporter genes (Fig. 5, B and E). After separation, one layer of cells expressing the PG was retained by the proximal tissues of the plant, while another remained attached to the distal separating organ (Fig. 5, D and E).
Figure 5.
PGAZAT-driven GUS (A–C) or GFP (D and E) expression in flowers of transgenic Arabidopsis. GUS activity is undetectable in flowers prior to “natural” abscission (A) but is evident firstly at the base of the anther filament (stamen; B) and subsequently at the site of shedding of petals and sepals (C). By comparing a bright field (D) and fluorescence image (E) of naturally abscising flowers, expression of GFP can be seen in cells that would be expressing PG, i.e. at the base of the anther filament (stamen), petals, and sepals.
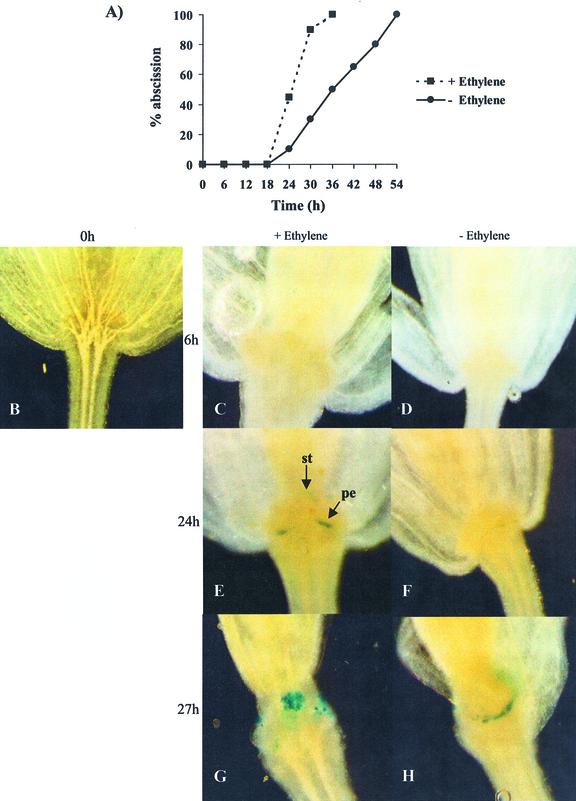
Transgenic Arabidopsis homozygous for the PGAZAT::GUS gene were incubated in either the presence or absence of ethylene (10 μL L−1) and the time course of floral organ abscission determined (Fig. 6A). After 24 h exposure to the gas, approximately 50% of the flowers showed signs of tissue abscission and reporter gene activity could be readily detected at the site of cell separation (Fig. 6). The onset of abscission was delayed by approximately 3 h in the absence of the gas with only 30% of flower parts undergoing shedding after 27 h. This delay correlated closely with the first signs of GUS activity in the flower tissues (Fig. 6).
Figure 6.
Floral organ abscission in transgenic Arabidopsis expressing PGAZAT::GUS in the presence (+) or absence (-) of ethylene (10 μL L−1). A, Time course of abscission; B, flower at 0 h; C, E, and G, flowers maintained in the presence of ethylene for 6, 24, or 27 h; D, F, and H, flowers maintained in the absence of ethylene for 6, 24, or 27 h.
A detailed analysis of transgenic PGAZAT::GUS and PGAZAT::GFP plants was carried out to determine reporter gene expression throughout the lifecycle of Arabidopsis plants from germination to silique senescence. A number of homozygous transgenic lines of each construct were studied and all showed reporter gene activity at the base of the stamens, petals, and sepals at the time of abscission of these tissues. Expression at other places including the root cap, site of lateral root emergence, and tips of cotyledons was also observed; however, these patterns were not observed consistently in every homozygous line examined (data not shown). None of the transgenic lines exhibited GUS or GFP accumulation at the base of the leaf petioles, in the dehiscence zone of anthers or pods, or at the junction between the seed and the funiculus.
DISCUSSION
It has been well documented that abscission is brought about by cell wall breakdown and that it is associated with an increase in the activity of hydrolytic enzymes such as PG and β-1,4-glucanase. However, the role of these enzymes and the mechanisms by which the genes encoding them are up-regulated has yet to be resolved. The wealth of information available about the Arabidopsis genome have yet to be applied to study the abscission process because organ shedding is restricted in this species to the floral parts (Patterson, 2001). The strategy that we have adopted is to study the process in the close relative oilseed rape because this material has a large AZ at the base of the leaf, enabling genes up-regulated in the cell separation process to be readily identified.
Using a degenerate primer strategy, a PCR product encoding a PG was amplified from RNA extracted from 48- to 72-h ethylene-treated AZ oilseed rape leaf tissue. Confirmation that the expression of the PG was abscission related was obtained by northern analysis and this indicated that the mRNA accumulated within 24 h of ethylene treatment. A full-length cDNA (CAW471) was isolated from an ethylene-induced leaf AZ cDNA library and this has 66% and 63% identity at the amino acid level with SAC70 (Jenkins et al., 1999) and SAC66 (Jenkins et al., 1996). These proteins are PGs expressed during pod dehiscence in Arabidopsis and oilseed rape, respectively. The cDNA CAW471 shares 82% identity at amino acid level with its homolog from Arabidopsis, which is described as a putative PG (GI no. 2335094; Lin et al., 1999; called PGAZAT here). When the analyses of the genomic sequences were performed, a missing exon in the putative polypeptide of Arabidopsis was detected (KKCTKAPYTA; Fig. 1).
The polypeptides predicted from oilseed rape cDNA (CAW471) and the PG from Arabidopsis (PGAZAT) show putative signal peptides of 24 amino acids and a site of potential glycosylation. It is interesting that a comparison with the SAC70 and SAC66 polypeptides reveals that their molecular weights and pI are similar (46.6 kD and pI of 8.39 in CAW471, 46.7 kD and pI of 9.039 in PGAZAT, 46.6 kD and pI of 8.2 in and SAC70, and 46.7 kD and pI of 8.34 in SAC66). The hydrophobicity index of these four peptides is also comparable and they all possess at least one glycosylation site (Fig. 1). These observations suggest that the PGs expressed at the sites of abscission and dehiscence might function in a biochemically comparable manner.
The phylogenetic tree (Fig. 2) shows that CAW471 is more related to its homologous gene in Arabidopsis (PGAZAT) and to the dehiscence PGs from oilseed rape and Arabidopsis than to other abscission-related PGs from leaf and flower AZ of tomato. This observation may indicate the close evolutionary relationship between these two species and the similarity between the processes of cell separation at these two sites. It is intriguing that PGAZAT is not expressed in the funiculus during seed abscission, suggesting that some of the events regulating flower and seed shedding may be different.
It is interesting to note that expression of CAW471 mRNA occurs well before the onset of leaf abscission. The accumulation of mRNA reaches a peak 24 h after ethylene treatment before declining over the following 48 h, whereas organ separation is not observed until 42 h after exposure to the gas (Fig. 3B). The pattern of PG expression preceding organ separation has also been reported in tomato, where TAPG4 mRNA accumulates prior to leaf abscission (Kalaitzis et al., 1997). In contrast, other abscission-related PGs that have been characterized have been reported to be up-regulated at the time of organ separation (Taylor et al., 1991, 1993; Bonghi et al., 1992; Hong and Tucker, 1998).
Although the expression of CAW471 in leaf AZ tissue takes place prior to organ shedding, the accumulation of the mRNA during flower abscission is more closely associated with the onset of organ separation. The significance of these differing temporal expression patterns remains to be established; however, it is possible that the accumulation of the CAW471 protein may be more closely related to the timing of the abscission process. In transgenic PGAZAT::GUS lines of Arabidopsis, activation of the PG promoter in floral parts coincides with the abscission process in the presence or absence of ethylene. This observation suggests that the expression of the abscission-related PGs in flowers of Arabidopsis and oilseed rape follows a similar time course.
Analysis of the sequences of the genomic clones obtained by screening of libraries from oilseed rape and Arabidopsis with CAW471 indicates that both genes (PGAZBRAN and PGAZAT) have eight introns and nine exons. In comparison, the structures of other AZ PGs from leaf and flower abscission of tomato have four introns and five exons (Kalaitzis et al., 1997; Hong and Tucker, 1998). The PGs from pod dehiscence zones of oilseed rape and Arabidopsis have the same number of introns and exons as PGAZBRAN and PGAZAT and this may be important at the evolutionary level.
Fusion of 1,476 bp of the promoter of PGAZAT is sufficient to drive expression of GUS or GFP specifically within the cells at the base of the anther filament, petal, or sepal at the time of “natural” and ethylene-promoted floral organ shedding. Expression in stamen and petals can be seen to occur in a ring of cortical cells located at the perimeter of the tissue and this suggests that not all individuals within the plate of cells where separation takes place may produce PG. Whether this is the case for other hydrolytic enzymes is not clear, but it may provide a mechanism by which cell wall hydrolysis does not occur in an uncontrolled fashion at the base of the silique. The spatial pattern of expression of AZ PGs may be different in Arabidopsis from that in tomato. For instance, it has been reported recently that during tomato flower or leaf abscission expression of TAPG4::GUS initially was observed in the vascular tissue, whereas expression of TAPG1::GUS took place throughout the zone of cell separation. There is no evidence that PGAZAT::GUS or PGAZAT::GFP expression is associated with the vascular trace in either ethylene-promoted or “natural” flower abscission. This may be a reflection of the fact that the tissues undergoing abscission are different or that other, as yet, unidentified PGs might contribute to the separation process in Arabidopsis.
Expression patterns of the promoter of the pod dehiscence zone-specific PG from oilseed rape (RDPG1 or SAC66 using our nomenclature) have been reported recently in transgenic Arabidopsis (Sander et al., 2001). These workers showed that reporter gene (GUS) expression could be detected in a range of tissues including the floral AZs in addition to the sites where pods and anthers dehisce. We have observed expression of PGAZAT at a number of sites where cell separation takes place, including the site of lateral root emergence and the margins of the root cap. However, GUS and GFP accumulation was only consistently observed at the site of floral organ abscission in every line analyzed and as a consequence, we are of the opinion that expression of PGAZAT is primarily restricted to these tissues. No expression was detected in the dehiscence zone of pods or anthers, and unlike RDPG1 there was no evidence that the AZ PG is up-regulated by wounding. These observations indicate that although PGAZAT shares close homology with PGDZAT, its spatial pattern of expression is different.
In this paper, we have described how the study of developmental processes in oilseed rape can be used to identify functional homologs in Arabidopsis. By comparing and contrasting the promoters of the equivalent genes in these two species, we intend to examine how abscission-related gene expression may be regulated and explore a molecular genetic strategy to determine the sequence of events that regulates the process of cell separation.
MATERIALS AND METHODS
Plant Material
Seedlings of oilseed rape (Brassica napus var. Rafal) were grown in a greenhouse with supplementary light to maintain a 16-h photoperiod and ensure a constant light regime throughout the year. Plants were raised for 3 weeks in Foremost John Innes compost no. 2 (The Scotts Company [UK] Ltd., Ipswich, Suffolk, UK) until the first pair of true leaves was fully expanded. Explants were prepared, supported on 1% (w/v) agar (Technological no. 3, Oxoid [UK] Basingstoke, Hampshire, UK), and incubated in the presence or absence of ethylene (10 μL L−1) at 24°C under a 16-h photoperiod. Solid potassium permanganate was used to absorb ethylene in experiments where explants were incubated in the absence of ethylene. Following exposure, the explants were dissected to collect NZ stem below the nodal junction and AZ for RNA extraction. To obtain flower tissue, plants were grown to the five-true-leaf stage in Foremost John Innes compost no. 2 and vermiculite (2:1, v/v), vernalized at 4°C for 6 weeks, then transferred to the greenhouse and maintained to maturity. Flower explants were prepared and exposed to ethylene as described earlier, then dissected to collect AZ and NZ for RNA extraction.
Arabidopsis plants (ecotype Wassilewskija) were grown in a greenhouse with supplementary light to maintain a 16-h photoperiod. Following transformation, plants were maintained to maturity and seeds collected. Transgenic seedlings were selected by their ability to grow on Murashige and Skoog media containing 40 mg L−1 kanamycin, then transferred to pots containing Foremost John Innes compost no. 2 and vermiculite (2:1, v/v). Plants were maintained at a 16-h photoperiod until maturity when flower tissue was collected for analysis.
RNA Extraction
Total RNA was extracted from leaf AZ and NZ tissue exposed to ethylene for 0, 24, 48, and 72 h and from flower AZ and NZ tissue exposed to ethylene for 0, 12, 24, and 48 h using procedures described previously (Logemann et al., 1987; Maliyakal, 1992).
Time Course of Leaf and Flower Abscission
Fifty leaf AZ and flower AZ explants of oilseed rape were supported on 1% (w/v) agar (Technological no. 3, Oxoid) and exposed to ethylene (10 μL L−1) for 0, 24, 42, 48, 54, and 60 h and 0, 6, 12, 18, 24, 30, and 36 h respectively. At each time point, the number of detached petioles (leaf AZ) or petals (flower AZ) was recorded. Arabidopsis flowers were excised at the base of the pedicel, which was then inserted into 1% (w/v) agar (Technological no. 3, Oxoid) supported by a 9-cm petri dish lid containing small holes through which the pedicels could penetrate. Abscission was monitored in the presence or absence of ethylene (10 μL L−1).
RT-PCR
First strand cDNA was prepared from total RNA extracted from leaf AZ and NZ tissue that had been incubated in the presence and absence of ethylene (10 μL L−1) for 48 to 72 h. The 20-μL reactions contained 1 μg of total RNA, 200 units of Moloney murine leukemia virus RT (Stratagene Europe, Amsterdam), 5 μm oligo-dT primer (GAGAGAGGA-TCCTCGAGTTTTTTTTTTTTTTTT; Genosys Biotechnologies [Europe] Ltd., Pampisford, Cambs, UK), 1× Moloney murine leukemia virus buffer, 0.5 mm dNTPs (Pharmacia Upjohn Ltd., Milton Keynes, UK), and 21 units of RNAse inhibitor (Pharmacia). The reaction conditions were as follows: 65°C for 5 min, 37°C for 90 min, and 95°C for 5 min (Crocodile III, Appligene Oncor, Durham, UK). RT was omitted from the control reactions. For PCR, 2 μL of cDNA was used as template in a 50-μL reaction containing 1× PCR buffer, 1.5 mm MgCl2, 0.2 mm dNTPs (Pharmacia), 2.5 units of Taq DNA polymerase (Invitrogen [UK] Ltd., Paisley, UK), 1.7 μm primer P1 (ggcgaatt-CCNAAYACNGAYGG), and 1.7 μm primer P3 (ggcggatcc-GGYAC-NGGNCCNGG). The thermal cycling conditions were as follows: 20 cycles of 94°C for 1 min, 45°C for 2 min, and 72°C for 2 min, followed by five cycles of 94°C for 1 min, 45°C for 2 min, 72°C for 2.5 min, and finally five cycles of 94°C for 1 min, 45°C for 2 min, and 72°C for 3 min (Crocodile III, Appligene). A PCR product of the expected size (approximately 150 bp) was eluted from the gel, cloned into pCRII (Invitrogen), and sequenced.
cDNA and Genomic Library Screening
A full-length cDNA clone of CAW471 was obtained by screening a oilseed rape ethylene-induced leaf AZ cDNA library. The library was plated at a density of 50,000 recombinant plaques per 13-cm plate, then transferred to Hybond N+ nylon membrane (Amersham, Buckinghamshire, UK). The duplicate filters were probed with 32P-labeled CAW471 PCR product. Hybridization and washes were carried out according to the manufacturers' instructions. Phagemid was excised in vivo from phage plaques that hybridized to the probe after a third round of screening; then, following reinfection of Escherichia coli, plasmid DNA was isolated (QIAGEN [UK] Ltd., Crawley, West Sussex, UK) and sequenced.
To obtain genes corresponding to the CAW471 cDNA, an oilseed rape genomic library (Dr. Tony Fawcett, University of Durham, Durham, UK) and an Arabidopsis genomic library (Dr. Rod Scott, University of Bath, Bath, UK) were plated as described earlier in this section and screened using a 32P-labeled 644-bp PCR product of CAW471. Southern analysis was carried out following the secondary and tertiary screens to confirm isolation of clones corresponding to CAW471. The resulting genomic clones were sequenced and designated PGAZBRAN (oilseed rape) and PGAZAT (Arabidopsis).
Northern-Blot Analysis
For northern-blot analysis, 10 μg of total RNA extracted from leaf AZ, flower AZ and the respective NZ tissue was fractionated on gels containing 1% (w/v) agarose, 3% (v/v) formaldehyde, and 20 mm sodium phosphate, pH 6.5. The RNA was transferred to nylon membranes (Genescreen, DuPont [UK] Ltd., Stevenage, UK) and hybridized to 32P-labeled antisense strand-specific RNA probe of CAW471. Hybridization and washing conditions were performed according to the manufacturers' instructions. Filters were stripped and hybridized with a 32P-labeled DNA probe of 25S rRNA to show equal loading of the RNA.
Construct Preparation and Transformation of Arabidopsis
A 1,476-bp fragment containing the 5′-upstream sequence was amplified from PGAZAT using primers ATPF (5′ CTA GAA GAA ACA CGA AAT GG 3′) and ATPR (5′ GTT GCT GGA AAA AGG AAT GCT 3′). The promoter fragment was cloned upstream of either the GUS (PGAZAT::GUS) or GFP (PGAZAT::GFP) reporter genes in the binary vector pMOG402. Plasmid DNA was isolated (QIAGEN) and used to transform Agrobacterium tumefaciens strain C58. Transformation of Arabidopsis (ecotype Wassilewskija stock no. N915) was performed as described by Clough and Bent (1998).
GUS and GFP Analysis
PGAZAT::GUS and PGAZAT::GFP analysis was carried out on plants harvested throughout their life cycle from germination until pod dehiscence. In particular, flowers were collected prior to and during “natural” and ethylene-promoted abscission and assayed for reporter gene activity. For GUS analysis, plant tissue was incubated in a solution containing 1 mm X-gluc (Medford Laboratories [UK] Ltd., Ipswich, UK); 200 mm sodium phosphate, pH 7.0; and 0.06% (v/v) Triton X-100 at 37°C for 12 h, then washed in 100% (v/v) ethanol. Tissue was viewed and photographed using a Wild M10 stereomicroscope (Leica, Solms, Germany) with Fujichrome 64T tungsten film (Fuji Photo Film, Tokyo). For GFP analysis, expression of the reporter gene was examined directly under the microscope using distilled water as mounting fluid. Plant material was viewed under bright-field or UV fluorescence filter system (blue) with Leitz PL-APO objectives using a Leica DMR microscope. The images were photographed using Fujichrome 64T tungsten film or 400 ASA Fujichrome color film (Fuji Photo Film).
Footnotes
This work was supported by Consejo Nacional de Ciencia y Tecnología (México; fellowship no. 94459/110150 to Z.H.G.-C.) and by the Biotechnology and Biological Science Research Council.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010610.
LITERATURE CITED
- Abeles FB. Role of RNA and protein synthesis in abscission. Plant Physiol. 1968;43:1577–1586. [PMC free article] [PubMed] [Google Scholar]
- Atkinson RG. A cDNA clone for endopolygalacturonase from apple. Plant Physiol. 1994;105:1437–1438. doi: 10.1104/pp.105.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleecker AB, Patterson S. Last exit: senescence, abscission, and meristem arrest in Arabidopsis. Plant Cell. 1997;9:1169–1179. doi: 10.1105/tpc.9.7.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonghi C, Casadoro G, Ramina A, Rascio N. Abscission in leaf and fruit explants of Prunus persica(L) Batsch. New Phytol. 1993;123:555–565. doi: 10.1111/j.1469-8137.1993.tb03768.x. [DOI] [PubMed] [Google Scholar]
- Bonghi C, Rascio N, Ramina A, Casadoro G. Cellulase and polygalacturonase involvement in the abscission of leaf and fruit explants of peach. Plant Mol Biol. 1992;20:839–848. doi: 10.1007/BF00027155. [DOI] [PubMed] [Google Scholar]
- Brown KM. Ethylene and abscission. Physiol Plant. 1997;100:567–576. [Google Scholar]
- Carpita NC, Gibeaut DM. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993;3:1–30. doi: 10.1111/j.1365-313x.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- Cho H-T, Cosgrove DJ. Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2000;97:9783–9788. doi: 10.1073/pnas.160276997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun J-P, Huber DJ. Polygalacturonase-mediated solubilization and depolymerization of pectic polymers in tomato fruit cell walls. Plant Physiol. 1998;117:1293–1299. doi: 10.1104/pp.117.4.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke AE, Gleeson PA. Molecular aspects of recognition and response in pollen-stigma interactions. Rec Adv Phytochem. 1981;15:161–211. [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–43. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Cell wall loosening by expansins. Plant Physiol. 1998;118:333–339. doi: 10.1104/pp.118.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coupe SA, Taylor JE, Roberts JA. Characterization of an mRNA encoding a metallothionein-like protein that accumulates during ethylene-promoted abscission of Sambucus nigraL. leaflets. Planta. 1995;197:442–447. doi: 10.1007/BF00196665. [DOI] [PubMed] [Google Scholar]
- Coupe SA, Taylor JE, Roberts JA. Temporal and spatial expression of mRNAs encoding pathogenesis-related proteins during ethylene-promoted leaflet abscission in Sambucus nigra. Plant Cell Environ. 1997;20:1517–1524. [Google Scholar]
- del Campillo E. Multiple endo-1,4-β-D-glucanase (cellulase) genes in Arabidopsis. Curr Top Dev Biol. 1999;46:39–61. doi: 10.1016/s0070-2153(08)60325-7. [DOI] [PubMed] [Google Scholar]
- del Campillo E, Bennett AB. Pedicel breakstrength and cellulase gene expression during tomato flower abscission. Plant Physiol. 1996;111:813–820. doi: 10.1104/pp.111.3.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veau EJI, Gross KC, Huber DJ, Warada AE. Degradation and solubilization of pectin of β-galacto-sidases purified from avocado mesocarp. Plant Physiol. 1993;87:279–285. [Google Scholar]
- Dopico B, Lowe AL, Wilson ID, Merodio C, Grierson D. Cloning and characterization of avocado fruit mRNAs and their expression during ripening and low-temperature storage. Plant Mol Biol. 1993;21:437–449. doi: 10.1007/BF00028802. [DOI] [PubMed] [Google Scholar]
- Fisher RL, Bennett AB. Role of cell wall hydrolases in fruit ripening. Ann Rev Plant Physiol. 1991;42:675–703. [Google Scholar]
- Gonzalez-Carranza ZH, Lozoya-Gloria E, Roberts JA. Recent developments in abscission: shedding light on the shedding process. Trends Plant Sci. 1998;3:10–13. [Google Scholar]
- Hadfield KA, Bennett AB. Polygalacturonases: many genes in search of a function. Plant Physiol. 1998;117:337–343. doi: 10.1104/pp.117.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield KA, Rose JCK, Yaver DS, Berka RM, Bennett AB. Polygalacturonase gene expression in ripe melon fruit supports a role for polygalacturonase in ripening-associated pectin disassembly. Plant Physiol. 1998;117:363–373. doi: 10.1104/pp.117.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S-B, Sexton R, Tucker ML. Analysis of gene promoters for two tomato polygalacturonases expressed in abscission zones and the stigma. Plant Physiol. 2000;123:869–880. doi: 10.1104/pp.123.3.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S-B, Tucker ML. Genomic organization of six tomato polygalacturonases and 5′ upstream sequence identity with TAP1 and win2genes. Mol Gen Genet. 1998;258:479–487. doi: 10.1007/s004380050758. [DOI] [PubMed] [Google Scholar]
- Hong S-B, Tucker ML. Molecular characterization of a tomato ploygalacturonase gene abundantly expressed in the upper third of pistils from opened and unopened flowers. Plant Cell Rep. 2000;19:680–683. doi: 10.1007/s002999900175. [DOI] [PubMed] [Google Scholar]
- Jenkins E, Paul W, Coupe SA, Bell SJ, Davies EC, Roberts JA. Characterization of an mRNA encoding a poly-galacturonase expressed during pod development in oilseed rape (Brassica napusL) J Exp Bot. 1996;47:111–115. [Google Scholar]
- Jenkins E, Paul W, Craze M, Whitelaw C, Weigand A, Roberts JA. Dehiscence-related expression of an Arabidopsis thaliana gene encoding a polygalacturonase in transgenic plants of Brassica napus. Plant Cell Environ. 1999;22:159–167. [Google Scholar]
- Kalaitzis P, Koehler SM, Tucker ML. Cloning of a tomato polygalacturonase expressed in abscission. Plant Mol Biol. 1995;28:647–656. doi: 10.1007/BF00021190. [DOI] [PubMed] [Google Scholar]
- Kalaitzis P, Solomos T, Tucker ML. Three different polygalacturonases are expressed in tomato leaf and flower abscission, each with a different temporal expression pattern. Plant Physiol. 1997;113:1303–1308. doi: 10.1104/pp.113.4.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashbrook CC, Gonzalez BC, Bennett AB. Two divergent endo-β-1,4-glucanase genes exhibit overlapping expression in ripening fruits and abscising flowers. Plant Cell. 1994;6:1485–1493. doi: 10.1105/tpc.6.10.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Kaul S, Rounsley S, Shea TP, Benito M-I, Town ChD, Fujii CY, Mason T, Bowman ChL, Bernstead M et al. Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature. 1999;402:761–767. doi: 10.1038/45471. [DOI] [PubMed] [Google Scholar]
- Logemann J, Schell J, Willmitzer L. Improved method for the isolation of RNA from plant tissues: analytical biochemistry. 1987;163:16–20. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- Maliyakal EJ. An efficient method for isolation of RNA and DNA from plants containing polyphenolics. Nucleic Acids Res. 1992;20:2381. doi: 10.1093/nar/20.9.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meakin PJ, Roberts JA. Dehiscence of fruit in oilseed rape (Brassica napusL.): I. Anatomy of pod dehiscence. J Exp Bot. 1990;41:995–1002. [Google Scholar]
- Osborne DJ. Abscission. Crit Rev Plant Sci. 1989;8:103–129. [Google Scholar]
- Patterson SE. Cutting loose: abscission and dehiscence in Arabidopsis. Plant Physiol. 2001;126:494–500. doi: 10.1104/pp.126.2.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ. Improved tools for sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peretto R, Favaron F, Bettini V, De Lonrenzo G, Marini S, Alghisi P, Cervone F, Bonfante P. Expression and localization of polygalacturonase during the outgrowth of lateral roots in Allium porrumL. Planta. 1992;188:164–172. doi: 10.1007/BF00216810. [DOI] [PubMed] [Google Scholar]
- Petersen M, Sander L, Child R, Van Onckelen H, Ulvskov P, Borkhardt B. Isolation and characterization of a pod dehiscence zone-specific polygalacturonase from Brassica napus. Plant Mol Biol. 1996;31:517–527. doi: 10.1007/BF00042225. [DOI] [PubMed] [Google Scholar]
- Roberts JA, Whitelaw CA, Gonzalez-Carranza ZH, McManus MT. Cell separation processes in plants: models, mechanisms and manipulation. Ann Bot. 2000;86:223–235. [Google Scholar]
- Sander L, Child R, Ulvskov P, Albrechtsen M, Borkhardt B. Analysis of a dehiscence zone endo-polygalacturonase in oilseed rape (Brassica napus) and Arabidopsis thaliana: evidence for roles in cell separation in dehiscence and abscission zones, and in stylar tissues during pollen tube growth. Plant Mol Biol. 2001;46:469–479. doi: 10.1023/a:1010619002833. [DOI] [PubMed] [Google Scholar]
- Sexton R, Roberts JA. Cell biology of abscission. Ann Rev Plant Physiol. 1982;33:133–162. [Google Scholar]
- Sheehy RE, Pearson J, Brady CJ, Hiatt WR. Molecular characterization of tomato fruit polygalacturonase. Mol Gen Genet. 1987;208:30–36. [Google Scholar]
- Sitrit Y, Hadfield KA, Bennett AB, Bradford KJ, Downie AB. Expression of a polygalacturonase associated with tomato seed germination. Plant Physiol. 1999;121:419–428. doi: 10.1104/pp.121.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolle-Smits T, Beekhuizen JG, Kok MTC, Pijnenburg M, Recourt K, Darkesen J, Voragen AGJ. Changes in cell wall polysaccharides of green bean pods during development. Plant Physiol. 1999;121:363–372. doi: 10.1104/pp.121.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JE, Coupe SA, Picton S, Roberts JA. Characterization and accumulation pattern of an mRNA encoding an abscission-related β-1,4-glucanase from leaflets of Sambucus nigra. Plant Mol Biol. 1994;24:961–964. doi: 10.1007/BF00014449. [DOI] [PubMed] [Google Scholar]
- Taylor JE, Tucker GA, Lasslett Y, Smith C, Arnold C, Watson C, Schuch W, Grierson D, Roberts JA. Polygalacturonase expression during leaf abscission of normal and transgenic tomato plants. Planta. 1991;183:133–138. doi: 10.1007/BF00197577. [DOI] [PubMed] [Google Scholar]
- Taylor JE, Webb TS, Coupe SA, Tucker GA, Roberts JA. Changes in polygalacturonase activity and solubility of polyuronides during ethylene-stimulated leaf abscission in Sambucus nigra. J Exp Bot. 1993;44:93–98. [Google Scholar]
- Torki M, Mandaron P, Thomas F, Quigley F, Mache R, Falconet D. Differential expression of a polygalacturonase family in Arabidopsis thaliana. Mol Gen Genet. 1999;261:948–952. doi: 10.1007/s004380051042. [DOI] [PubMed] [Google Scholar]