Abstract
The FAT10 gene encodes a diubiquitin-like protein containing two tandem head-to-tail ubiquitin-like domains. There is a high degree of similarity between murine and human FAT10 sequences at both the mRNA and protein levels. In various cell lines, FAT10 expression was shown to be induced by gamma interferon or by tumor necrosis factor alpha. In addition, FAT10 expression was found to be up-regulated in some Epstein-Barr virus-infected B-cell lines, in activated dendritic cells, and in several epithelial tumors. However, forced expression of FAT10 in cultured cells was also found to produce apoptotic cell death. Overall, these findings suggest that FAT10 may modulate cellular growth or cellular viability. Here we describe the steps to generate, by genetic targeting, a FAT10 gene knockout mouse model. The FAT10 knockout homozygous mice are viable and fertile. No gross lesions or obvious histological differences were found in these mutated mice. Examination of lymphocyte populations from spleen, thymus, and bone marrow did not reveal any abnormalities. However, flow cytometry analysis demonstrated that the lymphocytes of FAT10 knockout mice were, on average, more prone to spontaneous apoptotic death. Physiologically, these mice demonstrated a high level of sensitivity toward endotoxin challenge. These findings indicate that FAT10 may function as a survival factor.
The major histocompatibility complex (MHC) locus is composed of many genes that play various key roles in immune surveillance against cancer and infectious diseases. Among these, there are several non-class I genes that structurally belong to a variety of families (10, 11). One of these genes is the human HLA-F adjacent transcript 10 (FAT10) gene. This gene encodes a diubiquitin-like (UBD) protein containing two tandem head-to-tail ubiquitin-like domains that are substantially different from those of other members of the ubiquitin family (2, 7, 10). There are few reports on FAT10 in the literature, and we have a very limited understanding of its biological role.
Ubiquitin, a small protein consisting of 76 amino acids, has been found in all eukaryotic cells studied. It is one of the most conserved proteins known and is instrumental in regulating the degradation of variety of different proteins. This regulation is mediated by a covalent binding of ubiquitin molecules to a target protein. A glycine residue from the ubiquitin C terminus is conjugated to a lysine residue on the protein marked for degradation. Studies in recent years have shown that ubiquitin has increasingly important functions in eukaryotic cells, including cell differentiation, cell cycle regulation, embryogenesis, apoptosis, signal transduction, DNA repair, transmembrane and vesicular transport, stress responses, and immune responses (3, 15, 16, 19, 33, 35). Not all of these functions are mediated through protein degradation.
Following the discovery of ubiquitin, other genes that contain structural motifs that are related to ubiquitin were discovered. Ubiquitin-like (UBL) proteins have been found to play a role in various cell growth functions. Among them are the following: RAD23, which is involved in DNA repair following UV damage; elongin B, which regulates transcription elongation; SUMO, which is involved in regulating the cell cycle, nuclear transport, transcription, DNA repair, and apoptosis; ISG15, which is involved in pregnancy and innate immunity, including modulation of the response to alpha interferon (IFN-α); and NEDD8, which is involved in cell cycle control (4, 6, 14, 16-18, 21, 25, 26, 30, 31, 36).
The human FAT10 mRNA is expressed in mature B cells and in certain cell lines following stimulation with IFN-γ. At the tissue level, a high level of expression of human FAT10 was found in placenta, thymus, spleen, and embryos. FAT10 mRNA is expressed in organs where T and B cells develop into mature and active lymphocytes, suggesting that the FAT10 protein may have an important role in lymphocyte maturation. The expression of FAT10 protein may be transient and labile and is regulated at the transcriptional, translational and posttranslational levels by cytokines (e.g., IFN-γ and tumor necrosis factor alpha [TNF-α]) (2, 7, 23). In addition, induced overproduction of the human FAT10 was found to be lethal to the transfected cells that undergo apoptosis (27). However, the growth and apoptotic profiles of cells stably overexpressing FAT10 were reported to be similar to those of their parental cells (29). Interestingly, down-regulation of FAT10 expression was shown to reduce apoptosis in renal tubular epithelial cells infected by human immunodeficiency virus (32). Up-regulation of FAT10 expression was noticed in several tumors, suggesting a role for FAT10 in neoplastic processes (22). It was later suggested that FAT10 involvement in carcinogenesis is mediated by p53, which regulates FAT10 expression through its promoter and prevents it from reaching high levels in the cell (37). After translation, FAT10 protein was shown to rapidly degrade due to the interaction with NUB-1L, which is bound to the proteasome (14). However, subsequent study suggested that FAT10 provides a signal for proteasomal degradation of other proteins as an alternative route for protein degradation mediated by ubiquitin (13).
Our previous study revealed that human FAT10 bound specifically to the mitotic checkpoint protein MAD2, as well as to several other proteins (unpublished data). Subsequently, it was shown by others that FAT10 interaction with MAD2 takes place only during mitosis and that overexpression of FAT10 results in chromosomal instability (29).
Overall, the functional role of the FAT10 protein is poorly understood. To elucidate the overall biological function of the FAT10 gene in vivo, we generated FAT10 gene knockout mice by using gene-targeting technology (8, 9, 20, 34).
We obtained a full-length cDNA of the mouse FAT10 gene using rapid amplification of 5′ cDNA ends (5′-RACE), isolated one mouse FAT10 genomic clone from a 129/Ola genomic library, and generated a FAT10 gene-targeting vector to obtain several embryonic stem (ES) cell clones with targeted disruption of the endogenous gene. Two mouse lines were obtained, and both demonstrated similar characteristics. Mice homozygous for the disruption of the FAT10 gene were viable and fertile. We found no major pathological or histological differences. However, the FAT10 knockout mice were found to be susceptible to challenge with a low dose of endotoxin. In addition, lymphocytes from these mutated mice demonstrated a higher level of spontaneous apoptosis. Hence, we suggest that FAT10 may also function as a cellular survival factor.
MATERIALS AND METHODS
Mouse genomic library.
A mouse 129/Ola genomic library (2 × 106 PFU/ml) was kindly provided by Raju Kucherlapati, Department of Molecular Genetics, Albert Einstein College of Medicine. This library was constructed by a partial Sau3A digestion of murine 129/Ola genomic DNA, and the fragments had an average insert size of 9 to 15 kb. The genomic fragments were cloned into the BamHI site of the Charon 35 lambda phage.
Mouse FAT10 cDNA.
Mouse FAT10 cDNA clone (identification number 573650, AA119390) was purchased from Research Genetics, Inc. (Huntsville, AL). This cDNA clone was confirmed by sequencing and used as a probe for screening for mouse FAT10 genomic clones from the genomic library.
5′-RACE.
Total RNA was extracted with TRIzol (GIBCO BRL) from mouse (C57BL/6) spleen. The 3′ end of the first-strand cDNA was extended using terminal transferase (Roche) to form a poly(A) tail and then used as a PCR template, using a primer within exon2 of the mouse FAT10 cDNA (29). Purified PCR products were cloned into the TA vector (Invitrogen) and sequenced.
Isolation and characterization of the mouse FAT10 clone.
The 456-bp cDNA of the mouse FAT10 gene spanning the exon2-coding region was used as a probe to screen the 129/Ola mouse genomic library. One phage clone (termed B122) was isolated through four rounds of screening with the mouse FAT10 probe. A 6.4-kb EcoRV fragment encompassing the entire FAT10 cDNA region was subcloned into pBluescript II KS (Stratagene) and characterized by restriction digestion and Southern blot hybridization analysis using standard procedures (30). The 6.4-kb insert was sequenced by primer walking with serial primers at Keck Foundation Biotechnology Resource Laboratory, Yale University.
Construction of the mouse FAT10 gene-targeting vector.
The pGK-Neo neomycin phosphotransferase expression cassette was excised by EcoRI-HindIII from a pGEM7-KJ1 plasmid, and used to replace the entire second exon of FAT10 in the 6.4-kb insert within pBluescript II KS by digestion with AvrII-SwaI and blunting the ends of the DNA fragments. The pGK-HSV-TK herpes simplex virus thymidine kinase expression cassette was inserted upstream of the 6.4-kb fragment to generate the mouse FAT10 gene-targeting vector (termed pV2N4T6).
ES cell culture.
ES cells (TC1) from a 129S6 mouse were incubated in Dulbecco modified Eagle medium (GIBCO BRL) with 15% fetal bovine serum and 100 units of penicillin/streptomycin and supplemented with 0.5% of leukemia inhibitory factor (GIBCO BRL) to maintain undifferentiated cells.
Transfection of the gene-targeting vector into mouse ES cells.
The FAT10 gene-targeting vector pV2N4T6 was linearized by digestion with XhoI. Eight million ES cells were transfected by electroporation with 25 μg of pV2N4T6. Following 24 h of incubation, ES culture medium supplemented with 300 μg/ml of G418 (GIBCO BRL) was added to select for neomycin-resistant clones, and 2 μM of ganciclovir (GIBCO BRL) was added for negative selection. The selected ES clones were collected after 9 to 12 days in culture.
Screening of ES cell clones by Southern blotting.
The genomic DNA of ES cells was digested with several restriction enzymes (e.g., BamHI, HindIII, and NcoI) and fractionated on a 0.7% agarose gel. The nylon membrane (Hybond, Amersham Life Science) with transferred DNA was placed into Church-Gilbert buffer (0.7 M sodium phosphate buffer, pH 7.4, 7% sodium dodecyl sulfate, 1% bovine serum albumin, 1.0 mM EDTA) at 65°C overnight in a hybridization incubator. A genomic DNA probe amplified by PCR from the B122 genomic clone was labeled by using a random primed DNA labeling kit according to the manufacturer's recommendations (Boehringer Mannheim). Briefly, 60 ng of DNA was denatured at 100°C for 10 min and then placed on ice for 2 min. The DNA was mixed with 1 μl of dATP, dGTP, and dTTP, 2 μl of hexanucleotide mixture, 5 μl of [32P]dCTP (50 μCi; 3,000 Ci/mmol), 7 μl of sterile distilled H2O, and 1 μl of Klenow enzyme (2 units/μl) and placed at 37°C for 2 h. The labeled DNA probe was purified with Sephadex G-50 columns (Boehringer Mannheim), denatured at 100°C for 10 min, and hybridized overnight to the DNA membrane in hybridization solution at 65°C.
Karyotype of positive ES cells.
The targeted ES cells were treated with 50 μl of Colcemid (10 μg/ml; Sigma) for 2 to 3 h before they were washed with phosphate-buffered saline (PBS). The ES pellet was then treated with 10 ml of 0.075 M KCl for 10 min and cold methanol:acetic acid fixative (3:1) for 15 min. After two more fixation steps, the cell slides were stained with Wright-Giemsa stain (Sigma) for 15 min and observed under the microscope to determine the chromosome number.
Microinjection of ES cells into blastocysts.
ES cells positive for FAT10 disruption were injected into blastocysts by standard methods (31). Following an injection of 10 to 15 ES cells per blastocyst, embryos were incubated for 2 h in microdrops of SECM medium (GIBCO BRL) covered with paraffin oil at 37°C to allow regeneration. Ten blastocysts per uterus were transferred into 3-day pseudopregnant (B6 × C3H F1 mice; NCI) foster mothers to generate genetically chimeric mice.
Gene typing of mutant mice by PCR.
DNA from tail tips of mice was isolated by the method mentioned above for extraction of ES genomic DNA or by the Red-NAmp kit (Sigma). Mouse genotyping was carried out by PCR and analyzed on a 2% agarose gel. The following primer pairs were used. To detect the neomycin gene, NeoF primer (ATGGATTGCACGCAGGTTCTC) and NeoR primer (ACTCGTCAAGAAGGCGATAG) were used. To detect the second exon of the murine FAT10 gene, 419F primer (CTTTGAGACCACTGAGAAT) and 10R primer (CCAGTGCAGTGTGTTGTCAG were used. For a DNA template control, we amplified the intron 5-spanning sequence of the murine β-actin gene (MACTBF1 primer, GACCTCTATGCCAACACAGT; MACTNBR1 primer, TTGCTGATCCACATCTGCT).
Northern blot analysis and reverse transcription-PCR (RT-PCR).
Total RNA was extracted with TRIzol reagent from the spleen, thymus, liver, brain, lung, and kidney of a C57BL/6 mouse. A nylon membrane was blotted with 25 μg of total RNA and hybridized with the mouse FAT10 cDNA probe labeled with [α-32P]dCTP using the random DNA labeling kit (Roche) at 65°C overnight. Blots were washed three times under high-stringency conditions (0.2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate] and 0.1% sodium dodecyl sulfate) at 65°C. A sample of 6 μg total RNA was reverse transcribed into first-strand cDNA with Superscript II reverse transcription kit (GIBCO BRL), and 2 μl of the first-strand cDNA was used as templates for PCR of the mouse FAT10 cDNA using the following primers: Up424 (5′-CTCTTGATGTTGTAGTCACC-3′) from the second exon of the mouse FAT10 gene, and Lp165 primer (5′-TACAGACATGGCTTCTGTCCGCACCTGT-3′) within the first exon of the mouse FAT10 gene.
Pathological and histological evaluation.
Mice were euthanized by CO2 treatment, and soft tissues were fixed in neutral buffered 10% formalin (Fisher Scientific). Limbs and the skull with brain were fixed in Bouin's solution (Ricca Chemical Company, Arlington, TX). Tissues were processed, embedded in paraffin, and sectioned by routine methods (Yale Mouse Research Pathology, Section of Comparative Medicine, Yale University School of Medicine). Tissues were sectioned at 5 microns and stained with hematoxylin and eosin. Tissues were examined by routine light microscopy with an Axioskop microscope (Carl Zeiss Micro Imaging, Inc., Thornwood, NY). Digital light microscopic images were taken with a Zeiss Axioskop 2 Plus microscope, AxoCam HR camera, and AxioVision 5.05.10 imaging software (Carl Zeiss Micro Imaging, Inc., Thornwood, NY). Necropsy examination was performed for both genders and on mice that were 9 to 24 weeks old.
Flow cytometry analysis of lymphocyte populations.
The spleens, thymi, lymph nodes, and bone marrow were harvested from 8- to 14-week-old mice and digested with collagenase at 37°C for 25 min. The lymphocytes were collected through nylon mesh and plated in 96-well plates. Cells were blocked with anti-Fcγ for 15 min on ice and centrifuged at 1,500 rpm for 2 min to remove the blocking agent. After the cells were resuspended in the diluted antibodies and incubated on ice for 30 min, they were washed twice with fluorescence-activated cell sorting (FACS) buffer (1.5% bovine serum albumin in PBS) and resuspended in 300 μl of FACS buffer for analysis. FACS analysis was performed on a FACSCalibur flow cytometer (Becton Dickinson, San Jose, CA). FACS data were analyzed using CellQuest FACS software.
Dendritic cell cultures and FACS analysis.
Mouse bone marrow was isolated from the femurs and tibia of 8-week-old mice, depleted of erythrocytes with red blood cell lysis buffer and cultured in RPMI 1640 medium supplemented with 5% fetal calf serum, 10 mM HEPES, 10 mM sodium pyruvate, 50 μM β-mercaptoethanol, 100 U/ml of penicillin, 10 μg/ml of streptomycin, and 1% granulocyte-macrophage-colony-stimulating factor. The culture medium was replaced every 2 days. On day 5, dendritic cells were matured with 30 ng/ml of lipopolysaccharide (LPS) in PBS for about 24 h. The cells were harvested and incubated on ice with Cy-labeled anti-CD11c, phycoerythrin-labeled anti-CD86, and fluorescein isothiocyanate (FITC)-labeled anti-I-Ab antibodies. Stained cells were analyzed with a FACScabilur flow cytometer.
Cellular death analysis.
Cells were isolated from the spleen, thymus, and bone marrow of 8- to 14-week-old mice and cultured in RPMI 1640 medium supplemented with 10% fetal calf serum. Following 48 h of incubation at 37°C in 5% CO2, spontaneous apoptotic death was evaluated by FACS analysis double staining for annexin V-FITC and propidium iodide (PI) according to the manufacturer's recommendations (Molecular Probes).
Endotoxin challenge in vivo.
Eight-week-old mice (∼22 g) were subjected to several doses of endotoxin (LPS from Escherichia coli serotype O111:B4; Sigma, St. Louis, MO) to evaluate the 50% lethal dose for FAT10 knockout mice (see text and figures for details). All mice were injected intraperitoneally with 200 μl of endotoxin dissolved in PBS. Mortality was evaluated for 72 h after the endotoxin challenge.
RESULTS AND DISCUSSION
Characterization of the mouse FAT10 gene and generation of FAT10-deficient mice.
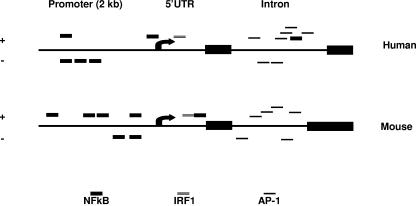
To isolate the full-length mouse FAT10 cDNA, we used 5′-RACE to reach the end of the first exon of the FAT10 gene. After designing the 5′ primer from the first exon and the 3′ primer from the second exon, we confirmed by PCR the presence of a 4-kb intron between these two exons. Using genomic walking, we sequenced the 5′ upstream genomic sequence of the mouse FAT10 gene. We then performed PCR with different 5′ primers and a specific 3′ primer within the second exon using mouse spleen cDNA (Clontech) as a template to check the 5′ sequence of the mouse FAT10 mRNA. Two of our 5′ primers produced positive PCR bands that showed the presence of a small open reading frame at the 5′ end of mouse FAT10 cDNA. Comparisons of the sequences of the mouse and human FAT10 genes revealed almost 77% identity in cDNA sequence and about 72% identity in amino acid sequence (Fig. 1). DNA sequence analysis for potential transcription factor binding sites that may be involved in the regulation of FAT10 expression revealed very similar patterns for mouse and human FAT10 exons, intron, and a 2-kb upstream region (includes the promoter area and the transcription initiation site). Regulation of FAT10 expression by TNF-α or IFN-γ can be mediated by NF-κB and AP-1 or interferon-regulating factor 1 (IRF-1), respectively. NF-κB binding sites were found to be located in the promoter area, while the IRF site was located in the 5′ untranslated region. Interestingly, the intron domain contains numerous AP-1 binding sites which are the target for binding by Jun (Fig. 2). The FAT10 gene encodes a protein containing tandem head-to-tail ubiquitin-like domains with the conservation of key functional residues (2). The 165-amino-acid polypeptide (mouse FAT10 consists of 162 amino acids) could be divided into two ubiquitin-like domains, separated by 5 amino acids. The N-terminal domain, which showed a lower degree of conservation (30%) with respect to ubiquitin, also had an initial extension of six amino acids. The C-terminal domain, which exhibited more conservation (41%), contained a carboxy-terminal glycine doublet. Unlike other ubiquitin molecules, for which specific hydrolases exist, the primary translation product of FAT10 mRNA did not contain a C-terminal extension beyond the diglycine sequence. Lysine residues shown by mutagenesis to be important in ubiquitin-protein binding were conserved in both domains (1, 5, 12, 24). The two domains were more closely related to ubiquitin than to each other (20%), suggesting an evolution towards domains with different functions.
FIG. 1.
Amino acid sequence comparison of human and mouse FAT10 proteins with human ubiquitin protein. The human and mouse FAT10 amino acid sequences are indicated as HFAT10 and MFAT10, respectively, to the left of the sequences. The shaded amino acids are identical. Gaps introduced to maximize alignment (∼) are indicated. *, stop condon.
FIG. 2.
DNA sequence analysis reveals high degree of similarity between human and mouse FAT10 for expression regulation. The human and mouse FAT10 sequences that were compared encompassed the two exons, the intermediate intron, and a 2-kb upstream sequence that includes the 5′ untranslated region (5′UTR) and the promoter regions. TRANSFAC analysis (http://www.gene-regulation.com/pub/databases.html) was performed for the regulatory binding sites. NF-κB, IRF, and AP-1 are shown schematically for both strands.
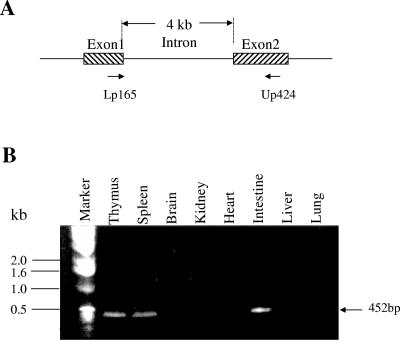
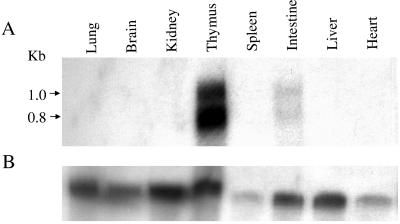
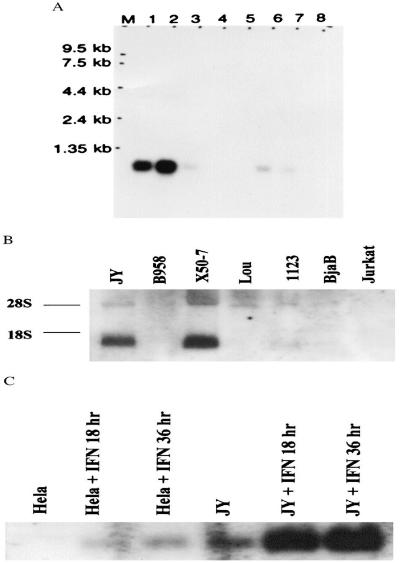
We evaluated the expression pattern of the FAT10 gene in various mouse tissues by isolating total RNA from of 6- to 8-week-old mice. RT-PCR results showed that FAT10 mRNA was expressed in mouse thymus, spleen, and small intestine but not in mouse heart, brain, lung, kidney and liver (Fig. 3). Northern blot analysis suggested that the mouse FAT10 mRNA was expressed more strongly in the thymus than in the small intestine and was expressed at a very low level in spleen. The low density of the FAT10 mRNA hybridization band in mouse spleen tissue could be due to an RNA loading issue according to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) internal control (Fig. 4). The mouse FAT10 mRNA showed two positive hybridization bands (about 1.0 kb and 0.8 kb), suggesting that the FAT10 gene might have different mRNA transcript forms in mouse tissues. In contrast, only a 1.0-kb size of the human FAT10 mRNA was strongly expressed in both human thymus and spleen (Fig. 5). The Northern blot data also showed that the expression of human FAT10 mRNA could be increased by IFN-γ treatment in HeLa and JY human cell lines (23).
FIG. 3.
RT-PCR analysis of FAT10 expression from total RNA from various mouse tissues. A. Schematic representation of the locations of RT-PCR primers on the mouse FAT10 gene. Intron-spanning primers were designed to eliminate the formation of genomic amplicons. B. Gel electrophoresis analysis of various mouse tissues for FAT10 gene expression. The origins of tissues are indicated above the gel, and the positions of the DNA markers are indicated to the left of the gel.
FIG. 4.
Northern blot analysis of FAT10 mRNA expression in different mouse tissues. A. The FAT10 mRNA expression pattern in different mouse tissues. The origins of tissues are indicated above the gel, and the positions of the kilobase markers are indicated to the left of the gel. B. GADPH probe hybridized to the same membrane as in panel A for RNA loading controls.
FIG. 5.
Northern blot analysis of human FAT10 mRNA. A. Northern blot of multiple tissues from healthy humans. Lane 1, spleen; 2, thymus; 3, prostate; 4, testis; 5, ovary; 6, small intestine; 7, colon mucosal lining; 8, peripheral blood leukocytes. Lane M contains molecular size markers. B. Analysis of total RNA blots from various cell lines. The origins of cell lines are indicated above the gel, and the two rRNA bands are indicated as 28S and 18S to the left of the gel. C. IFN-γ treatment for 0, 18, and 36 h induces progressive increases in FAT10 mRNA expression in HeLa and JY human cell lines.
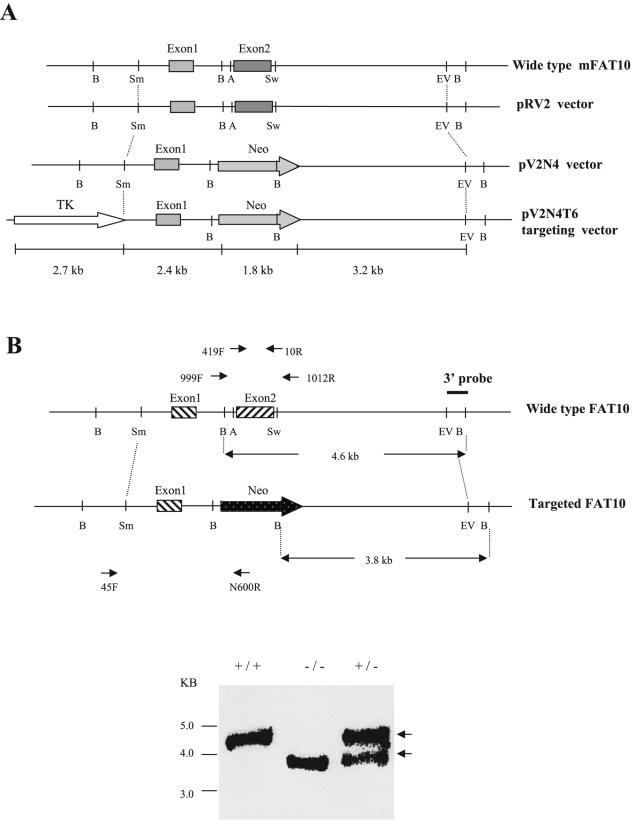
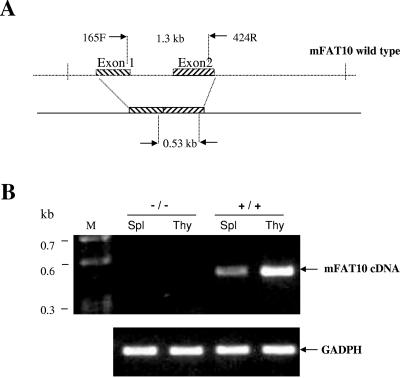
To isolate a mouse FAT10 genomic clone from the 129/Ola mouse genomic library, we used a mouse FAT10 cDNA clone (430 bp) as a probe. We screened the 129/Ola mouse genomic library and found one positive clone (termed B122) out of 1 × 105 PFU per plate. After the B122 clone was digested with different enzymes, we performed Southern blot analysis with a probe derived from the mouse FAT10 exon2. The results showed that a 6.4-kb EcoRV fragment contained the entire mouse FAT10 cDNA (including exon1 and exon2). The entire B122 genomic fragment was sequenced by primer walking and was confirmed to contain the entire cDNA sequence of the mouse FAT10 gene. The 6.4-kb genomic fragment that contained the entire mouse FAT10 gene was subcloned into pBluescript vector within an EcoRV site to generate the pRV2 vector (∼10.6 kb) (Fig. 6). The second exon (596 bp) was deleted by AvrII-SwaI digestion, which produced the 848-bp fragment which was replaced by the neomycin expression cassette as a positive selection marker to generate the pV2N4 vector (∼11.6 kb). Finally, the HSV-TK expression cassette for a negative selection marker was inserted into the SmaI site upstream of the mouse FAT10 genomic fragment. One clone (pV2N4T6, ∼14.0 kb) was obtained by PCR selection and confirmed by sequencing to contain the mutated FAT10 genomic fragment, the neomycin gene cassette. XhoI was used to linearize the pV2N4T6 vector. ES cells were transfected with the linear vector, and the neomycin-resistant cells were selected (using ganciclovir) and harvested for PCR and Southern blot analyses. We used the sense mFAT10-45F primer (derived from the 5′-flanking region of the targeting vector) and antisense N600R primer (derived from the neomycin gene) to perform PCR selection of targeted ES clones. Out of 150 ES clones screened, 4 were identified with a disruption in FAT10 (ES22, ES63, ES84, and ES86). There is one BamHI site within the neomycin gene cassette and one BamHI site just outside the 3′ end of the targeting vector. Thus, digestion of ES genomic DNA with BamHI will produce a 3.8-kb fragment for a disrupted FAT10 gene and a 4.6-kb fragment of a full-length FAT10 gene (Fig. 6). Using the 3′ external probe to perform Southern blot analysis, we found that targeted ES clones could produce the 4.6-kb and 3.8-kb hybridization bands, while the wild-type ES clones had only one 4.6-kb DNA band. This analysis was further confirmed by PCR analysis for the neomycin cassette and β-actin. The result of karyotype analysis verified that the two targeted ES clones (ES22 and ES86) had the normal number of chromosomes for mice. These two ES clones were microinjected into blastocysts of C57BL/6 mice, and the blastocysts were transferred into pregnant C57BL/6 foster mothers for generation of chimeras. A total of 12 chimeras were produced. Ten (83%) of them were male chimeras. Six male chimeras were bred with six female C57BL/6 mice to generate the FAT10 heterozygous mutant mice. We obtained about 70 (about 88%) agouti mice. The PCR analysis of genomic DNA for neomycin and Southern blot analysis following BamHI digestion showed 40% of these agouti mice to be heterozygous for FAT10 disruption. We inbred these heterozygous mice and genotyped their offspring by PCR analysis to find about 25% were homozygous mice, suggesting a Mendelian segregation. To further confirm that these mice were homozygotes, we performed Southern blot analysis of the genomic DNA to find only the 3.8-kb hybridization band (Fig. 6). In addition, we confirmed FAT10 disruption by performing RT-PCR for FAT10 from total RNA from these mice. The result showed that there was no FAT10 expression in the thymus or spleen of homozygous FAT10 knockout mice (Fig. 7). Hence, we have generated mutant mice that lack the FAT10 gene.
FIG. 6.
Disruption of murine FAT10 expression by gene targeting. A. Schematic cascade of the constructs generated in the process of generating the targeting vector pV2N4T6. Neomycin (Neo) expression cassette replaced the second exon of FAT10, and a thymidine kinase (TK) gene was placed upstream of the first exon of FAT10. Restriction enzyme sites: A, AvrII; B, BamHI; EV, EcoRV; Sm, SmaI; Sw, SwaI. B. Screening of ES cells for FAT10 disruption was carried out by Southern blotting and PCR analysis. Following BamHI digestion, the three external probes hybridized to a 4.6-kb fragment in the wild-type mice (+/+) and to a 3.8-kb fragment in the FAT10 knockout mice (−/−). The two fragments appeared in the heterozygous ES cell (+/−). PCR screenings for homozygous mice (−/−) and wild-type mice (+/+) were done using the oligonucleotide pair: 999F/1012R generated amplicons of 2.1 and 1.2 kb, respectively. Both amplicons were generated for the heterozygote. However, the use of the oligonucleotide pair 45F/N600R resulted in a 2.9-kb amplicon that was shared only by FAT10 knockout mice (−/−) and heterozygous mice.
FIG. 7.
>RT-PCR of mouse FAT10 cDNA from total RNA from FAT10 knockout mice. A. Schematic representation of mouse FAT10 (mFAT10) 165F/424R primers used for RT-PCR. B. RT-PCR of mouse spleen (Spl) and thymus (Thy). The expected 530-bp RT-PCR products were found in the wild-type mouse (+/+) thymus and spleen but not in the FAT10 knockout mouse (−/−) thymus and spleen. Lane M contains molecular size markers. The 500-bp band of GADPH was used as an internal control.
Histopathological analysis of FAT10-deficient mice.
We performed detailed histologic analyses of FAT10-deficient mice and wild-type littermate control mice, and there were no overt tissue phenotypic differences by genotype. The FAT10-deficient mice used were 36 and 12 weeks old and were from the 5th and 8th generations of a backcross to C57BL/6 mice respectively. FAT10-deficient mice are viable and do not demonstrate physical differences from wild-type mice in size or weight (data not shown). We bred FAT10 null mice, and they produced viable offspring.
FACS studies of various lymphocyte populations and dendritic cells reveal no differences from the lymphocytes and dendritic cells from wild-type mice in the distribution of cell types but did show an increase in apoptotic death in the absence of FAT10 expression.
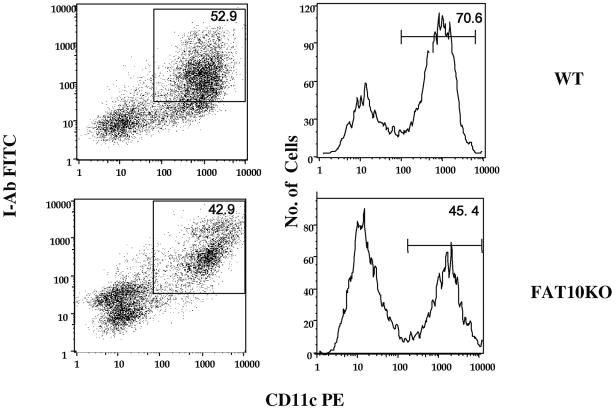
To investigate whether the FAT10 gene knockout has affected the hematological cellular profile, we performed blood differential counting analysis on FAT10 knockout mice and on heterozygous littermates and wild-type C57BL/6 mice. The analysis revealed no difference at all between the three groups of mice for the various hematological lineages (data not shown). In a further analysis to examine possible differences in lymphocyte subpopulations, we performed FACS analysis for a variety of cells and molecular markers. Cells were fractionated from mouse spleen, thymus, lymph node, and bone marrow. The following cell surface markers for T cells, B cells, NK cells, and dendritic cells were selected: CD4, CD8, immunoglobulin M, immunoglobulin D, CD25, CD44, CD69, CD86, B220, CD11b, and CD11c. However, our FACS analysis data, including the examination of T-lymphocyte subpopulations in the thymus (data not shown) did not show any significant difference between the FAT10-deficient mice and the wild-type mice. Harvested mouse bone marrow cells were cultured for 5 days in the presence of 1% of granulocyte-macrophage-colony-stimulating factor and stained with the dendritic cell marker CD11c-cry and dendritic cell maturation markers class II I-Ab labeled with FITC and CD86 labeled with phycoerythrin. FACS analysis data showed that although cell populations of CD11 and I-Ab from FAT10-deficient mice seemed lower than that from wild-type littermates, the difference was not statistically significant (Fig. 8). We also stimulated dendritic cells with LPS, IFN-γ, anti-CD40 antibody, and CpG but did not find any significant differences in the CD11c- and CD86- or I-Ab-positive cell populations of FAT10-deficient and wild-type mice (data not shown).
FIG. 8.
FACS analysis of FAT10 knockout mouse dendritic cell population in vitro. FACS analysis of dendritic cells cultured from the bone marrow of wild-type (WT) and FAT10 knockout (FAT10KO) mice. For details of the procedure, see text and Materials and Methods. CD11c PE, phycoerythrin-labeled CD11c.
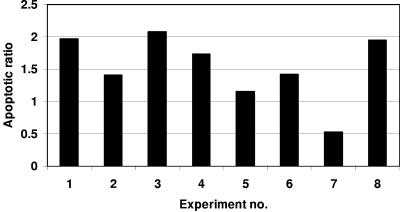
As mentioned earlier, recent studies suggested that expression of the human FAT10 gene was associated with cellular apoptosis or malignant growth (22, 28, 29, 32, 37). Therefore, it was of interest to check whether cells from the FAT10-deficient mice differed from cells from wild-type mice in their susceptibility to cellular death. Cells were obtained from spleens, thymuses, and bone marrow and were cultured prior to analyzing cellular death by FACS analysis of annexin V-FITC and PI staining. A significant difference in the apoptotic fraction (annexin V-FITC positive and PI negative) was found in the FAT10-deficient mice compared to the control mice. The FAT10-deficient mice demonstrate a higher level of spontaneous apoptotic death that is statistically significant (P < 0.05) (Fig. 9). In addition, we induced cell death with IFN-γ, TNF-α, and anti-Fas antibody with or without cycloheximide. However, the data from these studies were not conclusive (Table 1). Our results are consistent with the report of an increase in FAT10 expression in several tumors (22), suggesting that while FAT10 deficiency leads to susceptibility toward apoptosis, a high level of expression may contribute to neoplasms. However, it is not clear from that report whether FAT10 was mutated in those tumors or whether a high level of expression of FAT10 is the by-product of the neoplastic state or contributes to neoplastic growth. As for the seemingly contradicting report that tetracycline-induced FAT10 expression resulted in apoptosis, it is possible that the effects of FAT10 may be dependent on the cell type and physiologic state as well as the level of expression of FAT10. The relatively normal phenotype of FAT10-deficient mice is consistent with the expectation that the principal physiologic role for FAT10 is limited to effects on the immune system. It will be particularly important to understand whether the elevation of FAT10 levels reported in certain malignancies contributes to the malignant state or to the survival of malignant cells.
FIG. 9.
Leukocytes from FAT10-deficient mice demonstrate a higher level of apoptotic death. Bars represent the ratio of apoptotic leukocyte populations from FAT10-deficient mice to that of the FAT10-expressing control. Leukocytes from experiment 2 were obtained from bone marrow, while experiments 3 and 8 were performed on thymocytes. All other experiments were done on splenocytes. Analysis of variance analysis was performed on the raw data (two factors without replication, P = 0.042).
TABLE 1.
Summary of data from FACS analysis measurements performed in this studya<
| Expt. no. | Date (mo-day-yr) | Mouse and/or genotype | Organ | Culture time (h) | Treatmentb | Apoptotic fraction (%) | Viable fraction (%) | Necrotic fraction (%) | Note |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 8-15-03 | 129 +/+ | Spleen | 48 | Spontaneous | 11.33 | 4.4 | 84.26 | |
| 48 | TNF-α | 13.21 | 0.47 | 85.97 | |||||
| 48 | IFN-γ | 10.12 | 0.4 | 89.13 | |||||
| 48 | Anti-FAS | 9.96 | 0.41 | 89.3 | |||||
| FAT10ko −/− | Spleen | 48 | Spontaneous | 22.34 | 4.04 | 73.63 | |||
| 48 | TNF-α | 23.09 | 2.2 | 74.4 | |||||
| 48 | IFN-γ | 16.7 | 1.2 | 81.74 | |||||
| 48 | Anti-FAS | 18.1 | 1.28 | 80.44 | |||||
| 2 | 129 +/+ | Bone marrow | 48 | Spontaneous | 34.8 | 2.95 | 62.7 | ||
| 48 | TNF-α | 30.24 | 1.6 | 67.53 | |||||
| 48 | IFN-γ | 28.92 | 1.35 | 69.2 | |||||
| 48 | Anti-FAS | 36.42 | 0.65 | 62.6 | |||||
| FAT10ko −/− | Bone marrow | 48 | Spontaneous | 49.1 | 1.66 | 49.23 | |||
| 48 | TNF-α | 37.65 | 0.57 | 61.44 | |||||
| 48 | IFN-γ | 38.75 | 0.67 | 60.14 | |||||
| 48 | Anti-FAS | 39.29 | 0.42 | 59.94 | |||||
| 3 | 129 +/+ | Thymus | 48 | Spontaneous | 11.55 | 3.08 | 85.36 | ||
| 48 | TNF-α | 9.93 | 1.57 | 88.37 | |||||
| 48 | IFN-γ | 10.125 | 2.77 | 86.43 | |||||
| 48 | Anti-FAS | 8.6 | 1.45 | 89.64 | |||||
| FAT10ko −/− | Thymus | 48 | Spontaneous | 23.98 | 3.33 | 72.7 | |||
| 48 | TNF-α | 19.53 | 1.06 | 79.28 | |||||
| 48 | IFN-γ | 20.17 | 1.13 | 78.41 | |||||
| 48 | Anti-FAS | 17.14 | 1.3 | 81.15 | |||||
| 4 | 9-5-03 | 129 +/+ | Spleen | 18 | Spontaneous | 25.2 | 20.06 | 32.1 | Flow cyto problems |
| 18 | TNF-α | 5.37 | 48.8 | 41.66 | Flow cyto problems | ||||
| 18 | IFN-γ | 4.46 | 55.77 | 38.46 | Flow cyto problems | ||||
| 18 | Anti-FAS | 9.65 | 39.69 | 46.9 | Flow cyto problems | ||||
| FAT10ko −/− | Spleen | 18 | Spontaneous | 21.6 | 24.51 | 34.6 | Flow cyto problems | ||
| 18 | TNF-α | 9.33 | 55.56 | 26.6 | Flow cyto problems | ||||
| 18 | IFN-γ | 13.68 | 37.2 | 47.6 | Flow cyto problems | ||||
| 18 | Anti-FAS | 18.95 | 31.85 | 38.4 | Flow cyto problems | ||||
| 5 | 129 +/+ | Spleen | 18 | Cycloheximide | 23.25 | 16.33 | 46.2 | Flow cyto problems | |
| 18 | TNF-α + cycloheximide | 7.65 | 12.83 | 68.99 | Flow cyto problems | ||||
| 18 | IFN-γ + cycloheximide | 5.15 | 57.28 | 36.45 | Flow cyto problems | ||||
| 18 | Anti-FAS + cycloheximide | 12 | 14.13 | 71.52 | Flow cyto problems | ||||
| FAT10ko −/− | Spleen | 18 | Cycloheximide | 14.7 | 18.25 | 51 | Flow cyto problems | ||
| 18 | TNF-α + cycloheximide | 12.35 | 8.25 | 51.96 | Flow cyto problems | ||||
| 18 | IFN-γ + cycloheximide | 13.1 | 12.15 | 61.58 | Flow cyto problems | ||||
| 18 | Anti-FAS + cycloheximide | 7.54 | 18.76 | 65.75 | Flow cyto problems | ||||
| 6 | 9-10-03 | 129 +/+ | Spleen | 18 | Spontaneous | 5.37 | 48.79 | 45.84 | |
| FAT10ko −/− | Spleen | 18 | Spontaneous | 9.33 | 55.56 | 35 | |||
| 7 | 129 +/+ | Spleen | 18 | Cycloheximide | 7.65 | 12.83 | 79.5 | ||
| FAT10ko −/− | Spleen | 18 | Cycloheximide | 13.1 | 12.15 | 74.6 | |||
| 8 | 9-18-03 | 129 +/+ | Thymus | 48 | Spontaneous | 4.46 | 55.77 | 39.77 | |
| FAT10ko −/− | Thymus | 48 | Spontaneous | 5.15 | 57.28 | 37.67 | |||
| 9 | 129 +/+ | Spleen | 48 | Spontaneous | 9.65 | 39.69 | 50.66 | ||
| FAT10ko −/− | Spleen | 48 | Spontaneous | 13.68 | 37.19 | 49.1 | |||
| 10 | 129 +/+ | Spleen | 48 | Cycloheximide | 12.01 | 14.13 | 73.87 | ||
| FAT10ko −/− | Spleen | 48 | Cycloheximide | 17.39 | 13.34 | 69.27 | |||
| 11 | 2-13-04 | FAT10 +/+ | Spleen | 48 | Spontaneous | 11.79 | 13.66 | 74.55 | |
| FAT10ko −/− | Spleen | 48 | Spontaneous | 6.28 | 10.67 | 83 | |||
| 12 | 3-5-04 | C57BL/6 +/+ | Spleen | 45 | Spontaneous | 5.52 | 25.74 | 66.7 | |
| 129 +/+ | Spleen | 45 | Spontaneous | 5.27 | 27.02 | 65.7 | |||
| FAT10ko −/− | Spleen | 45 | Spontaneous | 10.78 | 26.31 | 62.9 |
Values shown in boldface type were used for the final statistical analysis. Spontaneous cultures were not stimulated during incubation.
The following concentrations were used: 100 ng/ml of either TNF-α or IFN-γ; 1 μg/ml of anti-Fas antibody; 1 μg/ml cycloheximide.
FAT10 knockout mice are sensitive to low doses of endotoxin.
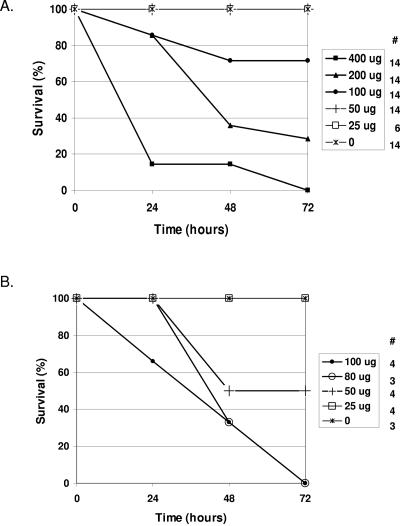
Following the studies of the acquired immune response, we set out to test the innate immune response in FAT10 knockout mice in vivo. Given the fact that TNF-α was reported to induce the expression of FAT10, we examined the mice in a septic model by challenging them with various levels of endotoxin. This manipulation has been well documented to induce the secretion of high levels of TNF-α into the circulation system, which later manifests in multiorgan failure and death. We set out to examine the susceptibility of the knockout mice (9th generation of a backcross to C57BL/6 mice) by evaluating their mortality in the 72 h after endotoxin administration. In comparison to the original C57BL/6 mice, which demonstrated a 50% lethal dose of ∼150 μg, this value for the FAT10 knockout mice was found to be ∼50 μg (Fig. 10).
FIG. 10.
FAT10 knockout mice are highly susceptible to endotoxin administration. Mice were subjected to various doses of endotoxin (from 0 to 400 μg). Mortality was evaluated daily, and the initial numbers of mice in each group are indicated under the # sign. Survival of C57BL/6 mice (A) and FAT10 knockout mice (B) is shown.
These data suggest that in vivo, FAT10 plays an important role in resisting sepsis from gram-negative bacteria as it is mediated by endotoxin. Further studies are needed to clarify the mechanism of action in which FAT10 provides resistance to gram-negative septicemia.
Acknowledgments
We thank Juli Unternaehrer and Aimin Jian from Ira Mellman's laboratory for their kind assistance in dendritic cell experiments.
REFERENCES
- 1.Amerik, A. Y., J. Nowak, S. Swaminathan, and M. Hochstrasser. 2000. The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways. Mol. Biol. Cell 11:3365-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bates, E. E., O. Ravel, M. C. Dieu, S. Ho, C. Guret, J. M. Bridon, S. Ait-Yahia, F. Briere, C. Caux, J. Banchereau, and S. Lebecque. 1997. Identification and analysis of a novel member of the ubiquitin family expressed in dendritic cells and mature B cells. Eur. J. Immunol. 27:2471-2477. [DOI] [PubMed] [Google Scholar]
- 3.Biggins, S., I. Ivanovska, and M. D. Rose. 1996. Yeast ubiquitin-like genes are involved in duplication of the microtubule organizing center. J. Cell Biol. 133:1331-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boddy, M. N., K. Howe, L. D. Etkin, E. Solomon, and P. S. Freemont. 1996. PIC 1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene 13:971-982. [PubMed] [Google Scholar]
- 5.Bryant, N. J., and T. H. Stevens. 1998. Vacuole biogenesis in Saccharomyces cerevisiae: protein transport pathways to the yeast vacuole. Microbiol. Mol. Biol. Rev. 62:230-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desterro, J. M., M. S. Rodriguez, and R. T. Hay. 1998. SUMO-1 modification of IκBα inhibits NF-κB activation. Mol. Cell 2:233-239. [DOI] [PubMed] [Google Scholar]
- 7.Fan, W., W. Cai, S. Parimoo, D. C. Schwarz, G. G. Lennon, and S. M. Weissman. 1996. Identification of seven new human MHC class I region genes around the HLA-F locus. Immunogenetics 44:97-103. [DOI] [PubMed] [Google Scholar]
- 8.Fassler, R., K. Martin, E. Forsberg, T. Litzenburger, and A. Iglesias. 1995. Knockout mice: how to make them and why. The immunological approach. Int. Arch. Allergy Immunol. 106:323-334. [DOI] [PubMed] [Google Scholar]
- 9.Galli-Taliadoros, L. A., J. D. Sedgwick, S. A. Wood, and H. Korner. 1995. Gene knock-out technology: a methodological overview for the interested novice. J. Immunol. Methods 181:1-15. [DOI] [PubMed] [Google Scholar]
- 10.Gruen, J. R., S. R. Nalabolu, T. W. Chu, C. Bowlus, W. F. Fan, V. L. Goei, H. Wei, R. Sivakamasundari, Y. Liu, H. X. Xu, S. Parimoo, G. Nallur, R. Ajioka, H. Shukla, P. Bray-Ward, J. Pan, and S. M. Weissman. 1996. A transcription map of the major histocompatibility complex (MHC) class I region. Genomics 36:70-85. [DOI] [PubMed] [Google Scholar]
- 11.Gruen, J. R., and S. M. Weissman. 1997. Evolving views of the major histocompatibility complex. Blood 90:4252-4265. [PubMed] [Google Scholar]
- 12.Hicke, L. 1997. Ubiquitin-dependent internalization and down-regulation of plasma membrane proteins. FASEB J. 11:1215-1226. [DOI] [PubMed] [Google Scholar]
- 13.Hipp, M. S., B. Kalveram, S. Raasi, M. Groettrup, and G. Schmidtke. 2005. FAT10, a ubiquitin-independent signal for proteasomal degradation. Mol. Cell. Biol. 25:3483-3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hipp, M. S., S. Raasi, M. Groettrup, and G. Schmidtke. 2004. NEDD8 ultimate buster-1L interacts with the ubiquitin-like protein FAT10 and accelerates its degradation. J. Biol. Chem. 279:16503-16510. [DOI] [PubMed] [Google Scholar]
- 15.Hochstrasser, M. 1998. There's the rub: a novel ubiquitin-like modification linked to cell cycle regulation. Genes Dev. 12:901-907. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, E. S., I. Schwienhorst, R. J. Dohmen, and G. Blobel. 1997. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 16:5509-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, K. I., N. V. Giannakopoulos, H. W. Virgin, and D. E. Zhang. 2004. Interferon-inducible ubiquitin E2, Ubc8, is a conjugating enzyme for protein ISGylation. Mol. Cell. Biol. 24:9592-9600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, K. I., and D. E. Zhang. 2003. ISG15, not just another ubiquitin-like protein. Biochem. Biophys. Res. Commun. 307:431-434. [DOI] [PubMed] [Google Scholar]
- 19.King, R. W., R. J. Deshaies, J. M. Peters, and M. W. Kirschner. 1996. How proteolysis drives the cell cycle. Science 274:1652-1659. [DOI] [PubMed] [Google Scholar]
- 20.Koller, B. H., and O. Smithies. 1992. Altering genes in animals by gene targeting. Annu. Rev. Immunol. 10:705-730. [DOI] [PubMed] [Google Scholar]
- 21.Lammer, D., N. Mathias, J. M. Laplaza, W. Jiang, Y. Liu, J. Callis, M. Goebl, and M. Estelle. 1998. Modification of yeast Cdc53p by the ubiquitin-related protein Rub1p affects function of the SCFCdc4 complex. Genes Dev. 12:914-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, C. G., J. Ren, I. S. Cheong, K. H. Ban, L. L. Ooi, S. Yong Tan, A. Kan, I. Nuchprayoon, R. Jin, K. H. Lee, M. Choti, and L. A. Lee. 2003. Expression of the FAT10 gene is highly upregulated in hepatocellular carcinoma and other gastrointestinal and gynecological cancers. Oncogene 22:2592-2603. [DOI] [PubMed] [Google Scholar]
- 23.Liu, Y. C., J. Pan, C. Zhang, W. Fan, M. Collinge, J. R. Bender, and S. M. Weissman. 1999. A MHC-encoded ubiquitin-like protein (FAT10) binds noncovalently to the spindle assembly checkpoint protein MAD2. Proc. Natl. Acad. Sci. USA 96:4313-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loayza, D., and S. Michaelis. 1998. Role for the ubiquitin-proteasome system in the vacuolar degradation of Ste6p, the a-factor transporter in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:779-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matunis, M. J., E. Coutavas, and G. Blobel. 1996. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 135:1457-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muller, S., M. J. Matunis, and A. Dejean. 1998. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 17:61-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raasi, S., G. Schmidtke, R. de Giuli, and M. Groettrup. 1999. A ubiquitin-like protein which is synergistically inducible by interferon-gamma and tumor necrosis factor-alpha. Eur. J. Immunol. 29:4030-4036. [DOI] [PubMed] [Google Scholar]
- 28.Raasi, S., G. Schmidtke, and M. Groettrup. 2001. The ubiquitin-like protein FAT10 forms covalent conjugates and induces apoptosis. J. Biol. Chem. 276:35334-35343. [DOI] [PubMed] [Google Scholar]
- 29.Ren, J., A. Kan, S. H. Leong, L. L. Ooi, K. T. Jeang, S. S. Chong, O. L. Kon, and C. G. Lee. 2006. FAT10 plays a role in the regulation of chromosomal stability. J. Biol. Chem. 281:11413-11421. [DOI] [PubMed] [Google Scholar]
- 30.Ritchie, K. J., C. S. Hahn, K. I. Kim, M. Yan, D. Rosario, L. Li, J. C. de la Torre, and D. E. Zhang. 2004. Role of ISG15 protease UBP43 (USP18) in innate immunity to viral infection. Nat. Med. 10:1374-1378. [DOI] [PubMed] [Google Scholar]
- 31.Rodriguez, M. S., J. M. Desterro, S. Lain, C. A. Midgley, D. P. Lane, and R. T. Hay. 1999. SUMO-1 modification activates the transcriptional response of p53. EMBO J. 18:6455-6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ross, M. J., M. S. Wosnitzer, M. D. Ross, B. Granelli, G. L. Gusella, M. Husain, L. Kaufman, M. Vasievich, V. D. D'Agati, P. D. Wilson, M. E. Klotman, and P. E. Klotman. 2006. Role of ubiquitin-like protein FAT10 in epithelial apoptosis in renal disease. J. Am. Soc. Nephrol. 17:996-1004. [DOI] [PubMed] [Google Scholar]
- 33.Saitoh, H., R. T. Pu, and M. Dasso. 1997. SUMO-1: wrestling with a new ubiquitin-related modifier. Trends Biochem. Sci. 22:374-376. [DOI] [PubMed] [Google Scholar]
- 34.Snouwaert, J. N., K. K. Brigman, A. M. Latour, N. N. Malouf, R. C. Boucher, O. Smithies, and B. H. Koller. 1992. An animal model for cystic fibrosis made by gene targeting. Science 257:1083-1088. [DOI] [PubMed] [Google Scholar]
- 35.Takayama, S., T. Sato, S. Krajewski, K. Kochel, S. Irie, J. A. Millan, and J. C. Reed. 1995. Cloning and functional analysis of BAG-1: a novel Bcl-2-binding protein with anti-cell death activity. Cell 80:279-284. [DOI] [PubMed] [Google Scholar]
- 36.Yeh, E. T., L. Gong, and T. Kamitani. 2000. Ubiquitin-like proteins: new wines in new bottles. Gene 248:1-14. [DOI] [PubMed] [Google Scholar]
- 37.Zhang, D. W., K. T. Jeang, and C. G. Lee. 2006. p53 negatively regulates the expression of FAT10, a gene upregulated in various cancers. Oncogene 25:2318-2327. [DOI] [PubMed] [Google Scholar]