Abstract
SIRK is a K+ channel identified in grapevine (Vitis vinifera), belonging to the so-called Shaker family. The highest sequence similarities it shares with the members of this family are found with channels of the KAT type, although SIRK displays a small ankyrin domain. This atypical feature provides a key to understand the evolution of the plant Shaker family. Expression in Xenopus laevis oocytes indicated that SIRK is an inwardly rectifying channel displaying functional properties very similar to those of KAT2. The activity of SIRK promoter region fused to the GUS reporter gene was analyzed in both grapevine and Arabidopsis. Like other KAT-like channels, SIRK is expressed in guard cells. In Arabidopsis, the construct is also expressed in xylem parenchyma. Semiquantitative reverse transcriptase-polymerase chain reaction experiments indicated that SIRK transcript was present at low levels in the berry, during the first stages of berry growth. After veraison, the period of berry development that corresponds to the inception of ripening and that is associated with large biochemical and structural modifications, such as evolution of stomata in nonfunctional lenticels and degeneration of xylem vasculature, the transcript was no longer detected. The whole set of data suggests that in the berries SIRK is expressed in guard cells and, possibly, in xylem tissues. The encoded channel polypeptide could therefore play a role in the regulation of transpiration and water fluxes in grapevine fruits.
The grapevine (Vitis vinifera) is a highly productive water stress-adapted plant and the most economically important fruit crop in the world (Coombe, 1989; Kanellis and Roubelakis-Angelakis, 1993). The grape berry provides, at the plant physiology level, a model for investigating non-climacteric fruit development, the present knowledge in this field being very poor. Also, with vacuolar pH values being very low (approximately 2.2–3.5; Hrazdina et al., 1994), the grape berry is of interest as a model for acid fruit physiology (Ros et al., 1995; Terrier et al., 2001).
The growth pattern of the grape berry follows a double sigmoidal curve, in which three successive phases can be distinguished: two growth periods, separated by a phase of slow growth (Kanellis and Roubelakis-Angelakis, 1993). Immediately after flowering, the first phase results from rapid cell division followed by marked cell enlargement, because of rapid accumulation of organic acids (tartaric and malic acids) in the vacuolar compartment. The second phase begins approximately 7 to 10 weeks after flowering. The berry displays slow or no growth. Further accumulation of organic acids makes the berry acidity reach a maximum at the end of this stage. The entry into the second growth period begins with the sudden onset of ripening, called veraison. This transition, which may occur within 24 h, results in and is characterized by berry softening (Coombe, 1992). Growth after veraison is rather due to flesh cell expansion than to cell division. A rapid accumulation of sugar (up to 1 m) and amino acids sets up, as well as a decrease in organic acid content, essentially due to malate degradation. Before veraison, the grape berry is photosynthetically active and its stomata are functional. The berry is connected to the rest of the plant mainly via the xylem vessels for water and solutes importation. After veraison, the berry becomes a strong sink and depends on phloem sap flux not only for sugars and other organic molecules but also for mineral ions and water, because xylem vessels are no longer functional. Discontinuities of xylem occur by the time of veraison in the vascular tissues that connect the berry to the stalk (Düring et al., 1987; Findlay et al., 1987). At the same time, berry stomata evolve into nonfunctional lenticels, the stomatal vestibule being progressively blocked by accumulation of polyphenolics, suberin, silicon, and calcium (Blanke et al., 1999).
The mechanisms of grape berry development, as those of other non-climacteric fruits, are still poorly understood at the molecular level. Most studies have focused on genes, the expression of which is modified at veraison (Robinson et al., 1997; Tattersall et al., 1997; Davies and Robinson, 2000). A particular attention has been given to sugar transporters and enzymes involved in Suc metabolism (Davies and Robinson, 1996; Fillion et al., 1999; Ageorges et al., 2000; Manning et al., 2001). The aim of the present study was to clone and characterize vine K+ transporters implicated in ionic homeostasis of the berry. K+ is the most abundant cation in the cell, being weakly chaotropic and compatible at high concentration, in the 0.1 m range, with water and protein structure (Clarkson and Hanson, 1980). It is involved in a number of basic functions linked together at the cellular or the whole plant level, e.g. control of cell turgor, and thereby control of cell enlargement or guard cell movements, and electrical neutralization of organic acids stored in the vacuolar compartment. At the agronomic level, and concerning wine quality, a large set of data has demonstrated that an increase in grape berry K+ content, due to e.g. increased K+ levels in the soil, or rootstock efficiency in taking up and transporting K+, leads to decreased vacuolar acidity and changes in relative concentrations of organic acids in the ripe berry, resulting in poor quality wine (Hale, 1977; Delas et al., 1989).
Here, we report the identification and characterization of a vine K+ channel, belonging to the so-called Shaker family. In Arabidopsis, channels of this family have been shown to play a role in K+ uptake from the soil solution, K+ secretion into the xylem sap, and probably K+ redistribution via the phloem sap, as well as stomatal movements (Zimmermann and Sentenac, 1999). When expressed in Xenopus laevis oocytes, the newly identified vine channel is gated by voltage and endowed with inwardly rectifying activity. A GUS reporter gene approach revealed that expression of this channel in vegetative organs is restricted to leaf and stem guard cells. Semiquantitative reverse transcriptase (RT)-PCR experiments indicated that its expression in the grape berry is strongly decreased after veraison. Finally, analysis of the vine channel sequence provided interesting clues regarding evolution of the Shaker K+ channel family in plants.
RESULTS
Cloning of a Gene Encoding a Shaker-Like K+ Channel from Vine
A vine genomic library was screened with a probe consisting in an equimolar mix of the Arabidopsis AKT1 (Sentenac et al., 1992) and KAT1 (Anderson et al., 1992) Shaker K+ channel cDNAs. Of 500,000 clones (equivalent to about 10 times the grapevine genome, i.e. 10 times 475 Mb, as estimated by Lodhi and Reisch [1995]), 10 positive phages were obtained after the first screening round, two of which were discarded during further purification rounds. Recombinant DNA from the purified phages was amplified and digested by several restriction enzymes (SacI, BamHI, EcoRI, NotI, PstI, and HindIII). The restriction patterns of five of the eight clones were identical, reducing to four the number of clones different from each other. Southern blots of these four clones displayed hybridization with the AKT1/KAT1 probe. For two of the clones, the hybridization signals corresponded to two SacI restriction fragments, exhibiting sizes of 2.8 and 3.4 kb. BamHI digestion of the third clone led to a 4-kb fragment also revealed by hybridization. Finally, a 12-kb fragment resulting from digestion of the fourth clone by NotI was also hybridized, but because of its size, it was not further characterized. PCR analyses and sequencing revealed overlaps of the SacI and BamHI fragments, allowing reconstruction of a single gene, with a coding sequence of about 5 kb and a promoter region of 3 kb. The predicted coding sequence showed similarities with K+ channels of the Shaker family. The gene was named SIRK, for Stomatal Inward Rectifying K+ channel (see below). SIRK shares the strongest similarities (see below) with members of the so-called KAT subfamily (Ache et al., 2000), which comprises KAT1 and KAT2 in Arabidopsis and KST1 in potato (Solanum tuberosum).
Cloning of SIRK cDNA
Assuming that SIRK was expressed in the same organs or cell types as the Arabidopsis KAT1 and KAT2 genes, i.e. mainly guard cells (Nakamura et al., 1995; Pilot et al., 2001), RT-PCR experiments were performed on leaf RNAs, with several primers designed from the predicted exons of the SIRK gene. Analyses of the sequence of the amplified fragments proved that SIRK was effectively expressed in grapevine leaves. Amplification of the complete cDNA in a single reaction failed, for unclear reasons. It was observed that SIRK mRNA accumulation level was low, and splicing intermediaries were often obtained when performing the above-mentioned RT-PCR experiments (not shown). The complete cDNA therefore was amplified as two fragments that were joined thanks to a unique NsiI restriction site in the coding sequence. Sequence comparison confirmed that the reconstructed cDNA corresponds to the SIRK gene, and not to an artifactual chimera obtained from transcripts of two different genes.
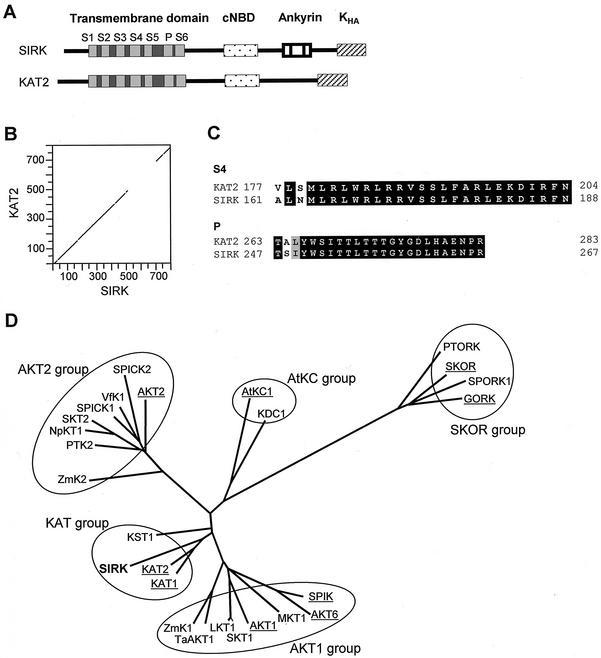
The resulting cDNA is 2.4 kb long, encoding a 791-amino acid protein, with a predicted molecular mass of 98.1 kD. The hydropathy profile of the polypeptide suggests the presence of a hydrophobic core composed of six membrane-spanning segments (not shown). Detailed analysis indicated that the protein exhibits the same structure as Shaker channels of the KAT type (Fig. 1A; see “Discussion”): (a) a short, cytoplasmic N-terminal region; (b) a hydrophobic core composed of six transmembrane segments and a pore-forming domain (P) between the last two transmembrane segments; and (c) a long C-terminal region containing a putative cyclic nucleotide binding domain (cNBD) and the so-called KHA domain, rich in hydrophobic and acidic amino acids (Fig. 1A). SIRK shares 70% identical amino acids with KAT2 and 56% with KAT1 (Table I). These similarities are mainly detected in the regions lying upstream of the putative cNBD (Fig. 1B). In particular, the P domain (pore) and the S4 segment (involved in voltage sensing) of SIRK differ from those of KAT2 by only two residues (Fig. 1C). However, SIRK displays a particular feature when compared with KAT2 and all the KAT-like channels cloned so far: It possesses a small ankyrin domain, located downstream from the putative cNBD, and reminiscent of the ankyrin domain found in plant Shaker channels of the AKT1, AKT2, and SKOR subfamilies. But whereas AKT- and SKOR-like polypeptides possess five to six ankyrin repeats (33 amino acids each; Lux et al., 1990) in tandem, SIRK displays only one complete repeat, surrounded by two one-half repeats. It is worthwhile to note that this ankyrin domain is responsible for a decrease in similarity between SIRK and KAT2: When this domain is not considered, the two proteins share 83% identity, a score higher than that observed between KAT1 and KAT2, the two members of the KAT subfamily in Arabidopsis. In other words, when the ankyrin domain is ignored, the grapevine channel SIRK is found more related to the Arabidopsis KAT2 channel than the twin Arabidopsis channels KAT1 and KAT2 to each other (72% identity; see Table I).
Figure 1.
The grapevine SIRK gene encodes a Shaker-like K+ channel related to the Arabidopsis KAT2 channel. A, Schematic representations of the predicted domains of SIRK and KAT2 channels. S1 through S6, Six membrane-spanning segments (light gray) and linkers (dark gray) forming the channel transmembrane domain. P, Conserved pore-forming domain. KHA, C-terminal domain supposed to be involved in channel subunit tetramerization and/or clustering in the membrane. B, Dot matrix comparison (DNA Strider program [Marck, 1990] polypeptide homology matrix; stringency = 7, window = 15 amino acids) of the deduced SIRK amino acid sequence (accession no. AF359521) with that of KAT2 (accession no. AJ288900). C, Alignment of SIRK and KAT2 voltage sensor (S4) and pore sequences. Identical amino acids are written in white, similar amino acids are written in black on a gray background. D, Phylogenetic tree of plant potassium channels deduced from full-length cDNA. This tree was generated by the Darwin program (Gonnet et al., 1992) with the complete protein sequences of the channels. Arabidopsis sequences are underlined. Protein accession numbers are AKT1, S23606; SKT1, T07651; ZmK1, T03939; SPIK, AJ309323; AKT6, CAA22577; LKT1, X96390; KAT1, S32816; KAT2, T04931; KST1, S55349; AKT2, AAA97865; NpKT1, BAA84085; SKT2, T07052; VfK1, T12177; SPICK1, AAD16278; SPICK2, AAD39492; ZmK2, CAB54856; AtKC1, G7488024; KDC1, AJ249962; SKOR, CAA11280; TaAKT1, AF207745; GORK, AJ279009; SPORK1, AJ299019; PTK2, AJ271447; PTORK, AJ271446; MKT1, AF267753; and SIRK, AF359521.
Table I.
Percentage of identities and similarities between protein sequences of AKT1, AKT2, SKOR, KAT1, KAT2, and SIRK
| Channel | AKT1 | AKT2 | SKOR | KAT1 | KAT2 | SIRK |
|---|---|---|---|---|---|---|
| AKT1 | – | 38 | 28 | 37 | 37 | 45 |
| AKT2 | 50 | – | 29 | 46 | 48 | 40 |
| SKOR | 42 | 43 | – | 37 | 37 | 33 |
| KAT1 | 45 | 36 | 24 | – | 72 | 56 |
| KAT2 | 45 | 37 | 24 | 76 | – | 70 |
| SIRK | 58 | 50 | 50 | 67 | 81 | – |
The close relationships between KAT1, KAT2, and SIRK were also assessed by a phylogenetic approach. These three channels are closely associated on the tree presented in Figure 1D, i.e. on the same branch, whereas KST1, which is a KAT-like channel expressed in potato guard cells, is located on a different branch. Thus, the SIRK small ankyrin domain did not influence the positioning of this channel in the phylogenetic tree, i.e. SIRK was not placed in a group of channels possessing ankyrin repeats. Therefore, the usual classification of Shaker channels in proteins with or without an ankyrin domain (Chérel et al., 1996) must be reconsidered.
Comparison of the cDNA and gene sequences allowed determination of intron positions, number and length. Like the KAT2 gene, SIRK contains 10 introns (KAT1 possesses only eight introns). In both KAT2 and SIRK genes, these introns are positioned exactly at the same places within the same codons and follow the AG/GT rule for splice junctions (Hanley and Schuler, 1988). It is therefore remarkable that the presence of an ankyrin domain in SIRK does not result in altered gene structure when compared with the KAT2 gene. The introns in SIRK are, in general, slightly longer than in KAT2, but remain in the range of plant intron mean lengths (Hawkins, 1988). Finally, the SIRK 5′-untranslated region was shown to be very long (data not shown). 5′-RACE experiments suggested that this region is about 1 kb long (the longest untranslated 5′ region we isolated was 850 bp long). When analyzing the corresponding genomic sequence, a putative transcription start site (ATATCA) was identified at 967 bp from the translation start codon. A putative TATA box (TTATTT) was localized at about 25 bp upstream from this transcription start site, which is in agreement with the consensus distances generally found between these two regulation sites.
Number of SIRK Copies in the Grapevine Genome
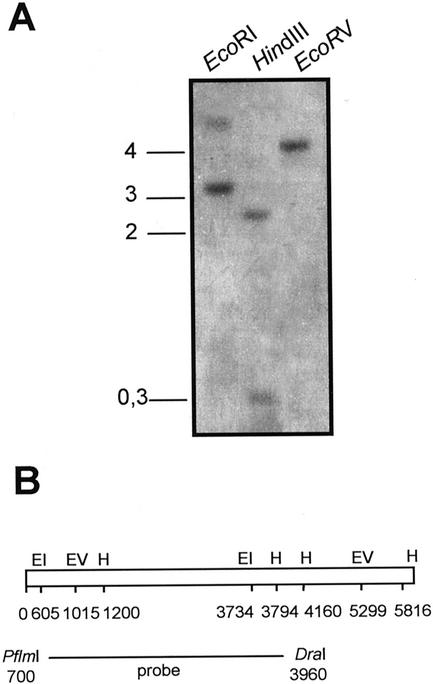
Southern blots of leaf genomic DNA (grapevine cv Pinot noir) were performed, using a fragment of SIRK cDNA (Fig. 2B) as a probe (Fig. 2). The banding pattern (Fig. 2A) for any of the three tested restriction enzymes is as expected from the restriction pattern of the SIRK gene (Fig. 2B). Furthermore, low-stringency Southern-blot analysis of grapevine genome with a KAT1/KAT2 probe resulted in the same banding pattern (not shown). These data indicate that SIRK is probably a single-copy gene. If there is more than one KAT-like gene in grapevine genome, the other gene(s) is (are) probably quite divergent from SIRK.
Figure 2.
Southern-blot analysis of SIRK gene copy number. A, Blot of total genomic DNA (10 μg per lane) digested with the indicated restriction enzymes. Hybridization was performed with the 3.2-kb PflmI/DraI fragment of SIRK cDNA. The blot was washed under high-stringency conditions. The positions of the size standards are indicated on the left in kilobases. B, Restriction map of SIRK gene. The localization of restriction enzyme sites (EI, EcoRI; EV, EcoRV; and H, HindIII) and the position of the probe are indicated in bases from the translational start codon of the gene.
Functional Characterization in X. laevis Oocytes
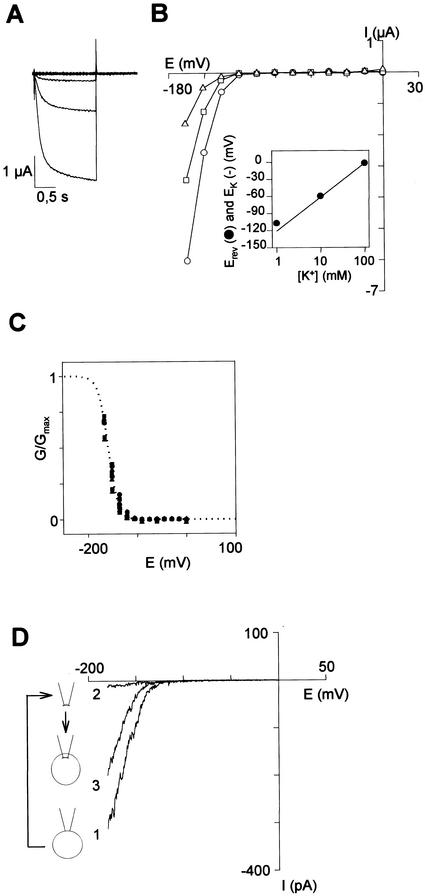
In X. laevis oocytes injected with SIRK cRNA, hyperpolarization of the membrane beyond −120 mV elicited an inward current (Fig. 3A) that was not recorded in control oocytes injected with water (data not shown). The exogenous macroscopic current displayed slow activation (Fig. 3A) and deactivation (data not shown). No inactivation could be seen even during hyperpolarizing pulses lasting 50 s (not shown). The steady-state current (Fig. 3B) and the G/Gmax-voltage plots (Fig. 3C) showed a strong inward rectification with a threshold potential of about −120 mV. The G/Gmax-voltage plot shows that the gating parameters (Ea50, the half-activation potential and za, the so-called gating charge) are insensitive to external K+ concentration (Fig. 3C). The reversal potential (Erev) of SIRK current was determined at different external K+ concentrations and was found to remain close to EK (Fig. 3B, inset), indicating that the inward current mediated by SIRK was mainly carried by K+ ions. Determination of reversal potential in pseudo bi-ionic conditions (Bruggemann et al., 1993) allowed determination of relative permeability ratios (not shown). SIRK displays the following permeability sequence: K+ > Rb+ ≫ Na+ (Table II).
Figure 3.
SIRK is an inward-rectifying voltage-gated channel, as assessed by macroscopic current analysis in X. laevis oocytes. A through C, Analysis by two-electrode voltage-clamp. The bath solution contained 1 mm CaCl2, 1.5 mm MgCl2, 5 mm HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid]-NaOH (pH 7.4), and 100 mm (K + Na) Cl (K+ concentration indicated below). A, SIRK currents elicited by 1.5-s voltage pulses from 0 to −165 mV in 12 steps (−15-mV increments) from a holding potential of −40 mV in a 100 mm K+ solution. B, The steady-state current at the end of the activation step was plotted against membrane potential for three different external K+ concentrations (in mm): 100 (circles), 10 (squares), and 2 (triangles). Inset, Plotting the reversal potential (Erev) for the SIRK current (black circles) versus the external concentration of K+ revealed that Erev shifted by 56.5 mV for a 10-fold increase in external K+ concentration, as expected for a highly selective K+ channel. Solid line in the inset, K+ equilibrium potential (EK) calculated with the Nernst equation assuming a 113 mm intracellular K+ concentration (i.e. Erev = EK in 100 mm external K+). C, SIRK activation level at steady state for three different K concentrations (in mm): 100 (diamonds), 10 (squares), and 2 (triangles); n = 3 oocytes. A two-state Boltzmann relation (dotted line; Lacombe and Thibaud, 1998) modeled the G/Gmax ratio. D, Analysis of macroscopic cur- rents assessed by patch-clamp (patch-excision and patch-cramming). Both bath and pipette solutions contained 100 mm KCl, 2 mm MgCl2, and 10 mm HEPES-NaOH (pH 7.4). In the cell-attached patch clamp configuration, the current-voltage curve (1) is reminiscent of those obtained by two-electrode voltage-clamp (see B). After patch excision (inside-out configuration), the current amplitude decreased very quickly (trace 2, obtained 10 s after the patch excision). Patch cramming into the oocyte produced an increase in current (trace 3). Data shown are representative of five independent patch-cramming experiments.
Table II.
Functional features of SIRK channel
| Selectivity | ||||
| PRb/PK | PNa/PK | |||
| 0.31 ± 0.01 (3) | <2.10−3 (3) | |||
| pH effect on gating (n = 4) | ||||
| pHe/pHi | 7.5/7.4 | 6.0/7.4 | 7.0/7.4 | 7.0/7.0 |
| Za | 2.79 ± 0.08 | 2.8 ± 0.07 | 2.8 ± 0.01 | 2.79 ± 0.02 |
| Ea50 | −158 ± 2 | −129 ± 2 | −151 ± 2 | −142 ± 2 |
| Caesium block (0.1 mm, inhibition in % of control, n = 3) | ||||
| −160 mV | −140 mV | −120 mV | ||
| 83 ± 2 | 92 ± 5 | 96 ± 2 | ||
Permeability ratios were determined from the reversal potential obtained in external solution containing 100 mm XCI (X+ being K+, Rb+, or Na+), 1 mm CaCl2, 1.5 mm MgCl2, and 5 mm HEPES-NaOH (pH 7.4), as previously described (Véry et al., 1995). Gating parameters were determined in solutions described in Fig. 3. The gating parameters za and Ea50 were obtained as previously described (Lacombe and Thibaud, 1998). Each value is mean ± se (no. of oocytes). Cs+ block was determined in 10 mm K+ solution.
SIRK gating parameters were determined and proved to be close to those of KAT1 and KAT2: the values of the gating charges for the three channels are 2.8 for SIRK, 2.5 for KAT2, and 1.6 for KAT1, and the half-activation potential values are −161 mV for SIRK, −152 mV for KAT2, and −130 mV for KAT1 (Table II; Pilot et al., 2001). Based on these values, the gating properties of SIRK are more related to those of KAT2 than to those of KAT1.
In our experimental conditions (expression level, size of patch), macroscopic SIRK currents mimicking whole-oocyte currents could be recorded in the cell-attached patch-clamp configuration (Fig. 3D, current trace labeled 1). Upon patch excision, however, the SIRK current decreased rapidly (inside-out configuration; Fig. 3D, current trace labeled 2). SIRK rundown could be overcome and the initial current partly restored by cramming the patch into the oocyte (Fig. 3D, current trace labeled 3). This suggested that SIRK opening required intracellular factors, available in the oocyte cytoplasm.
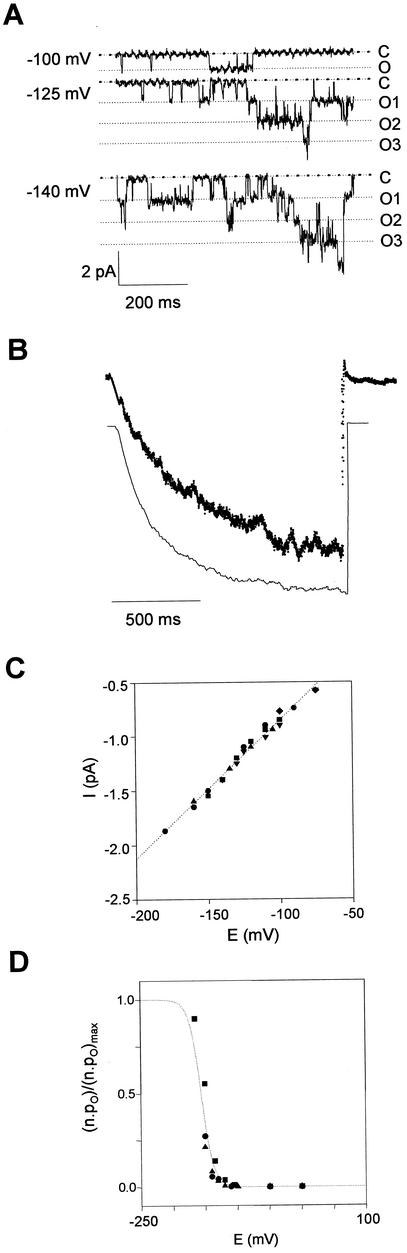
The patch-clamp technique was also used to investigate single-channel properties of SIRK expressed in X. laevis oocyte. Figure 4A shows an example of the most frequent single-channel activity recorded in membrane patches from SIRK-injected oocytes. At steady state, this activity resulted only in inward currents and was observed only at potentials negative to −80 mV. The sum of 200 current records obtained at −140 mV revealed that the single-channel current shown in Figure 4A was mediated by SIRK: The resulting curve was clearly reminiscent of the macroscopic current trace recorded by the two-electrode voltage-clamp technique (Fig. 4B). Analysis of the single-channel current-voltage relationship allowed determination of the single-channel slope conductance (Fig. 4C): 13 pS in 100 mm external potassium concentration. The voltage sensitivity of the single channel current (Fig. 4D) was also reminiscent of that of the macroscopic current (Fig. 3A).
Figure 4.
Single channel analysis of SIRK in X. laevis oocytes. Both bath and pipette solutions contained 100 mm KCl, 2 mm MgCl2, and 10 mm HEPES-NaOH (pH 7.4). A, SIRK single-channel currents in a X. laevis oocyte recorded at −100, −125, and −140 mV. C, Closed state; O1, O2, and O3, one, two, and three open channels, respec- tively. B, Upper trace, Sum of 200 pulses at −140 mV from a holding potential of 0 mV. Bottom trace, Current at −140 mV measured by two-electrode voltage-clamp (see Fig. 3A). Both traces were normalized to the steady-state value for easier comparison. C, Single-channel current-voltage relationship measured by patch-clamp experiments (n = 5). Single channel conductance: 13 pS in K+ 100 mm. D, Voltage-sensitivity of open probability of SIRK.
Block by external Cs+ is a classical feature of plant and animal K+ channels that is believed to involve binding of Cs+ to some site within the pore (Hille, 1992). Need of pore penetration by Cs+ for its blocking action is deduced from the voltage dependence of the K+ channel block as reported for the KAT1 channel (Véry et al., 1994; Becker et al., 1996). Addition of 0.1 mm Cs+ resulted in a strong block of SIRK inward currents (more than 80%; Table II). This effect was poorly voltage dependent, similar to the Cs+ block of the KAT2 channel (Pilot et al., 2001).
All the Arabidopsis Shaker-like K+ channels characterized so far at the functional level are sensitive to changes in external pH. The outwardly rectifying channels SKOR and GORK and the weakly inwardly rectifying channel AKT2 are inhibited by external acidification (Marten et al., 1999; Ache et al., 2000; Lacombe et al., 2000a, 2000b) while the inwardly rectifying channels KAT1 and KAT2 are activated (Pilot et al., 2001, and references therein). Decrease in external pH from 7.4 to 6.0 induced a positive shift of SIRK activation potential (Table II), leading to an increase in current amplitude at a given potential. Analyses of the corresponding G/Gmax versus potential curves (not shown) showed that, whereas the apparent gating charge (za) was not changed, the half-activation potential (Ea50) was shifted by approximately +30 mV when the pH was decreased from 7.5 to 6.0 (Table II). At external pH 7.0, a decrease in internal pH from 7.4 to 7.0 was obtained using acetate solution as described previously (Lacombe et al., 2000b). This cytosolic acidification also induced a positive shift of the half-activation potential (close to +10 mV) without affecting the gating charge (Table II).
Localization of SIRK Expression
Localization of SIRK expression was investigated by the reporter gene approach. The promoter sequence of SIRK (3 kb) was fused to GUS cDNA (encoding the Escherichia coli β-glucuronidase), and the resulting construct was used for Agrobacterium tumefaciens-mediated transformations of both Arabidopsis and grapevine. About 1 year is necessary after the transformation steps (performed on embryogenic calli) to regenerate transgenic vines.
In Arabidopsis, reporter gene activity was analyzed on F1 and F2 progeny of 10 independent transgenic plants. β-glucuronidase (GUS) activity was systematically detected in guard cells of leaves, stems, and petioles (Fig. 5, A–C). It was also detected in leaf xylem tissues, but with different patterns depending on the lines: The plants could present either a coloration in all the xylem tissues, or in the primary vessels, or only in the minor veins. Some leaves displayed no staining at all. GUS activity was never detected in roots.
Figure 5.
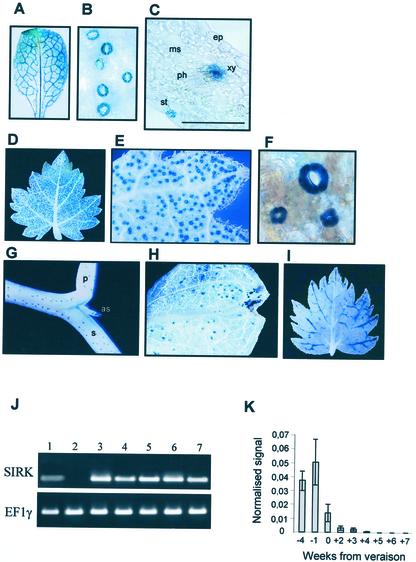
SIRK is expressed in guard cells and in grapevine berries until veraison. A through G, Localization of SIRK promoter activity in transgenic Arabidopsis (A–C) and vines (D–G) using GUS reporter gene. A, Mature leaf from 4-week-old Arabidopsis. B, Magnification of the leaf in A showing GUS-stained stomata. C, Thirty-micrometer- thick cross section of the leaf in A (bar = 30 μm). D, Eight-week-old vine plantlet leaf. E and F, Magnification of a part of the leaf with GUS-stained stomata. G, Stem and petiole with GUS-stained stomata. H, Cotyledon with GUS-stained stomata. I, Leaf of a 8-week-old control vine plantlet expressing a 35S:GUS construction. as, Axillary stem; ep, epidermis; ms, mesophyll; p, petiole; ph, phloem; s, stem; xy, xylem parenchyma. J, RT-PCR-detected expression of SIRK and EF1γ (control). 1, Stalks; 2, roots; 3, stems; 4, leaves; 5, berries 3 weeks before veraison; 6, berries at veraison; and 7, berries 3 weeks after veraison. K, Expression of SIRK in berry pericarp at different developmental stages analyzed by semiquantitative RT-PCR. Results (mean ± se, n = 3, P < 0.05) are expressed as a normalized signal (control, signal obtained for EF1γ transcripts; see “Materials and Methods”). Steps of berry development are expressed in weeks from veraison.
GUS activity was also detected in each of the 13 independent grapevine lines obtained. The staining was restricted to guard cells in cotyledons, leaves, stems, and petioles (Fig. 5, D–H). To our knowledge, this work is the first in which a reporter gene approach using a specific promoter has been developed in grapevine. Thus, in parallel experiments, we also transformed calli with a control construction, namely the fusion of a minimal 35S promoter to GUS cDNA. In the seven independent lines analyzed, GUS activity was systematically detected in the vascular tissues (particularly in the minor veins) of leaves (Fig. 5, I) and in root tips (not shown). This pattern is not as extended as previously observed with more strengthened versions of the 35S promoter (Baribault et al., 1990; Mauro et al., 1995; Scorza et al., 1996), but is in good agreement with results described by Jefferson et al. (1987). It can be therefore assumed that the data we obtained with the SIRK promoter construct do not result from an artifactual expression of GUS.
Attempts to investigate SIRK expression pattern by northern-blot analysis failed, suggesting that the amount of SIRK transcript was very low. Expression analyses were therefore performed using RT-PCR techniques, with RNAs extracted from leaves, young stalks, roots, and berries at three different stages of development (3 weeks before veraison, at veraison, and 3 weeks after veraison). SIRK transcripts were detected in all organs tested, except in roots (Fig. 5J). Concerning the vegetative parts, these results are in good agreement with the data obtained from the transgenic plants expressing the GUS reporter gene construct.
The time course of SIRK expression during berry maturation was studied using semiquantitative RT-PCR on nine samples collected along berry development (Fig. 5J). The resulting signals were expressed relatively to those obtained for the EF1γ transcript (control; Fig. 5J). The accumulation level of SIRK transcripts appeared to be very low in comparison to that of EF1γ transcripts. This could indeed explain failure of the previous northern-blot analyses. It is worth noting that the amount of SIRK transcript drastically decreased after veraison, and that the transcript was no longer detected 4 weeks later.
DISCUSSION
Very few transport systems have been molecularly characterized in vine. The published data actually concern one hexose transporter (Fillion et al., 1999) and three Suc transporters (Davies and Robinson, 1996; Ageorges et al., 2000; Manning et al., 2001). A few sequences corresponding to aquaporins and to a (H+) pyrophosphatase are accessible in databases. To our knowledge, SIRK is the first K+ transport system cloned in vine.
SIRK belongs to the Shaker-like family of plant K+ channels. The members of this family share sequence similarities and structural homologies with animal Shaker channels (Jan and Jan, 1997). Since the initial cloning of KAT1 (Anderson et al., 1992) and AKT1 (Sentenac et al., 1992) in Arabidopsis, the plant Shaker family has increased (Ache et al., 2000), with nine members identified in Arabidopsis (Zimmermann and Sentenac, 1999) and at least 16 in other plants (see Fig. 1D). As their animal counterparts, plant Shaker channels are thought to be tetrameric structures, formed by four α-subunits arranged around a central pore (MacKinnon, 1991; Daram et al., 1997; Urbach et al., 2000). The transmembrane hydrophobic core of the Shaker subunit harbors six membrane-spanning segments, named S1 to S6 and a pore-forming domain, P, between S5 and S6. S4 is characterized by the presence of regularly spaced basic amino acids rendering the channel sensitive to changes in transmembrane electrical potential (Zei and Aldrich, 1998; Bezanilla, 2000): Movements of S4 within the membrane in response to changes in membrane polarization result in conformational changes of the protein, leading to pore opening or closing. The channel is said to be voltage gated, and S4 is defined as the channel voltage sensor. The P domain is usually the most conserved domain in Shaker channels. It presents a hallmark motif, GYGD, typical of highly selective K+ channels. The C-terminal part of the polypeptide is cytoplasmic (Uozumi et al., 1998). In plant Shaker channels, three regions can be distinguished within it. The first one is a putative cyclic nucleotide binding site, followed by a variable domain, which may be composed of ankyrin motif repetitions (AKT1, AKT2, and SKOR subfamilies) or sequences sharing no similarity with other proteins (KAT and AtKC subfamilies). The third region corresponds to the so-called KHA domain, rich in hydrophobic and acidic residues. Two-hybrid (Daram et al., 1997) and green fluorescent protein fusion (Ehrhardt et al., 1997) experiments suggest that this domain is involved in Shaker polypeptide tetramerization and/or channel clustering in the membrane.
Some Arabidopsis Shaker channels have been shown to be involved in long-term, wholesale K+ transport. Integrated approaches combining electrophysiological analyses in planta and/or in heterologous systems, expression studies, and reverse genetics have led to the demonstration that AKT1 is involved in K+ nutrition (Lagarde et al., 1996; Hirsch et al., 1998; Spalding et al., 1999), SKOR in K+ secretion into the xylem sap (Gaymard et al., 1998) and SPIK in pollen tube growth (Mouline et al., 2002). Expression studies support the hypothesis that AKT2 plays a role in K+ transport in phloem tissues in both source and sink organs (Lacombe et al., 2000b). KAT1 (Nakamura et al., 1995), KAT2 (Pilot et al., 2001), and GORK (Ache et al., 2000) are expressed in guard cells. KAT1 and KAT2 are inwardly rectifying channels (Schachtman et al., 1992; Pilot et al., 2001) supposed to mediate K+ influx during stoma opening. On the other hand, the outwardly rectifying K+ channel GORK could mediate the K+ efflux leading to stoma closure (Ache et al., 2000). RT-PCR experiments suggest that other Shaker channel genes, AKT1, AKT2, and AtKC1, could be expressed in guard cells, and characterization of an Arabidopsis mutant knock-out for the KAT1 gene has recently revealed that the encoded polypeptide is not essential for stomatal opening (Szyroki et al., 2001).
SIRK shares the highest similarities with the members of the KAT subfamily within the plant Shaker family (Table I). It is most related to the Arabidopsis KAT2 channel, based on sequence analysis of the deduced polypeptides. Similarities are also found at the gene level, because SIRK and KAT2 have strictly the same number of introns, placed at the same positions, referring to the coding sequence. These data suggest that SIRK is the orthologous gene of KAT2 in vine.
As the Arabidopsis Shaker channels KAT1 and KAT2, SIRK expressed in X. laevis oocytes behaves as a voltage-gated, inwardly rectifying channel, highly selective for K+, in accordance with the presence of the tetrapeptide GYGD in its pore. Detailed comparison of the functional features of SIRK with those of KAT1 and KAT2 indicates that SIRK is more related to KAT2, with closer activation potentials, gating charges, and pH sensitivities. Furthermore, SIRK and KAT2 are both blocked by Cs+ in an essentially voltage-independent way, whereas the block of KAT1 by this cation is clearly voltage dependent (Véry et al., 1994). Therefore, sequence comparison between KAT1, KAT2, and SIRK could help in investigating the structure-function relationship of these channels.
In summary, both sequence analysis and functional characterization lead to the conclusion that SIRK is most related to KAT2 among the members of the Shaker family in Arabidopsis. KAT1 and KAT2 are both expressed in Arabidopsis guard cells (Nakamura et al., 1995; Pilot et al., 2001). SIRK promoter also directs expression of the GUS reporter gene in guard cells. In addition, KST1, a KAT-type Shaker channel cloned in potato, is expressed in guard cells (Müller-Röber et al., 1995). These data suggest that the expression pattern of KAT-like genes could be conserved among different plant species. Such a situation would be different from that described for the plasma membrane H+-ATPase gene family, because no relationship can be proposed between H+-ATPase gene expression patterns in Arabidopsis and in tobacco (Nicotiana tabacum; Morsomme and Boutry, 2000).
KAT1 and KAT2, and all other channels belonging to the KAT subfamily identified so far possess no ankyrin domain (KST1 in potato; sequence BAA96150 in rice [Oryza sativa]; ZmKT1 in maize [Zea mays]; Dr. Yan-Hua Su, personal communication). Despite its high similarities with KAT-type channels and its positioning on the same branch as KAT1 and KAT2 in the phylogenetic tree, SIRK harbors an ankyrin domain. When fewer plant Shaker channels had been identified, they were classified by taking into account the presence or absence of the ankyrin domain (Chérel et al., 1996), the channels being said of the AKT or KAT type, respectively. The cloning of SIRK demonstrates that KAT-like channels can have ankyrin domains. We propose that the presence of this ankyrin domain is an indication of the evolution of plant Shaker channels: The common ancestor of all plant Shaker channels may have possessed the ankyrin domain, and the ancestor of KAT-type genes would have lost this domain. Thus, SIRK could represent an intermediate step in the loss of this domain, with only a small remaining ankyrin domain (one complete motif, surrounded by two half-motifs, instead of six repeats as in AKT-type channels). Ankyrin domains are involved in protein-protein interactions (Hoffman, 1991). The ankyrin domain present in plant Shaker channels does not seem to be involved in interactions between the four polypeptides forming the functional channel (Daram et al., 1997). It is therefore likely that this domain interacts with other proteins, cytosolic or associated to plasma membrane. In the absence of any information regarding these targets and the role of the ankyrin domain in AKT type channels, the meaning of conservation or loss of this domain in plant Shaker channels cannot be assessed. It is worth noting that no ankyrin domain has been identified in any Shaker channel identified in animal cells up to now. The presence of such a domain seems to be typical of plant channels.
The expression pattern of the SIRK:GUS reporter gene construct was investigated here in both grapevine and Arabidopsis. In both the homologous and heterologous context, the SIRK promoter region was consistently active in guard cells. Thus, Arabidopsis could provide an interesting alternative for studying grapevine promoters and identifying regulatory boxes, considering the time necessary for regeneration of transgenic vines. In Arabidopsis, additional activity of the reporter gene was often detected in leaf xylem parenchyma. This expression could be artifactual, because of the heterologous genetic background. However, it cannot be excluded that SIRK is expressed in xylem tissues in grapevine, but in organs or at developmental stages that could not be tested, or in specific environmental conditions. Checking the latter hypothesis will be possible when the transgenic lines are older and grown in field conditions.
RT-PCR experiments showed that SIRK was expressed in the grape berries. This result will be verified by analysis of the vine transgenic lines expressing the SIRK:GUS construct, when the plants are old enough to produce berries (this requires about 2 years). Semiquantitative RT-PCR analysis indicates that SIRK transcript accumulation varies during berry development, the amount of SIRK transcript decreasing drastically by the time of veraison. As stated in the introduction, veraison is a period of deep physiological and histological modifications in the berry. Nutritional status of the berries switches from (photosynthetic) source to (no longer photosynthetic) sink, whereas functional stomata and xylem tissues are lost by these organs. Based on the available expression data (Fig. 5), we propose that the decrease in SIRK transcript accumulation in the berry after veraison is due to the fact that SIRK is expressed in guard cells and/or in xylem tissues of this organ (only before veraison). Therefore, SIRK could be involved in the regulation of berry water loss and/or K+ loading before veraison. The identification of the SIRK gene is therefore likely to provide the first molecular tool for investigating these aspects of berry development.
MATERIALS AND METHODS
Plant Material
Aerial tissue samples were collected from grapevine (Vitis vinifera cv Pinot noir), from Agro-M/Institut National de la Recherche Agronomique collections (Montpellier, France), in the 1999 season. Roots were collected from 1-year-old rooted canes planted in “Perlite” 10-L containers and maintained with Hoagland solution once a week. Embryogenic calli of grapevine cv Portan were maintained as described by Torregrosa (1998).
Library Screening
A genomic library prepared in λ-GEM-12 (Sarni-Manchado et al., 1997) was screened using standard plaque-lift method (Sambrook et al., 1989). The probe consisted of a mix of AKT1 (Sentenac et al., 1992) and KAT1 (Anderson et al., 1992) channel cDNAs radiolabeled with [32P]dATP and dCTP (random priming kit, Promega, Madison, WI). Filters were prehybridized at 65°C in Church buffer (Church and Gilbert, 1984) for 2 h and then hybridized in the same solution with added radiolabeled probe at 65°C for 48 h. Filters were washed twice in 2× SSC and 0.1% (w/v) SDS for 10 min each at room temperature and once in 0.1× SSC and 0.1% (w/v) SDS for 15 min at 65°C. The positive fragments were subcloned in pBS SK+, sequenced, and compared with the GenBank data library.
RNA Extraction
Samples were collected and immediately frozen in liquid nitrogen and stored at −80°C until use. As described by Fillion et al. (1999), two different procedures were performed to achieve RNA isolation, one for vegetative tissues and the other one for grapevine berry pericarp.
RT-PCR
Ten micrograms of RNA was denatured at 65°C for 5 min, and RT was performed for 30 min at 42°C in 30 μL in the presence of 200 units of Superscript II (Gibco-BRL, Cleveland) and 5 pmol of specific primer (SIRK reverse, 5′-CATTATAGTGTTTCAGTAACCATTAGG-3′). Then, 1 μL from this reaction was amplified in a final volume of 50 μL with 5 pmol of each primer (SIRK forward, 5′-GTTGCTGTCTGCAACGGGC-3′; SIRK reverse, see above) and 0.5 units of Thermus brokianus polymerase (Extrapol I, Eurobio, Les Ulis, France), following the manufacturer's protocol.
Semiquantitative RT-PCR
For each developmental stage of berries, three RT reactions were performed as described above, with three different batches of RNA. The products of these three reactions were pooled to alleviate the differences in reaction efficiency. One microliter of these pools was then used for PCR amplification. Three PCR reactions were repeated independently for each developmental stage and for both the SIRK gene and a control gene, EF1γ. The PCR reactions were performed as described above, with 24 cycles consisting of 45 s at 95°C, 45 s at 50°C, and 90 s at 72°C. The primers used for these amplifications were SIRK forward and SIRK reverse (see above), EF1γ forward (5′-AGCTTTTACCGCGGGCAAGAGATACC-3′) and EF1γ reverse (5′-TTTGGATAGGTAACGTATCACTTAAATAAC-3′). The amplified fragments were about 300 bp long. Every two cycles between cycle 10 and cycle 24, 2.5 μL was taken from the PCR reaction and blotted on a nylon membrane as dots. These dots were hybridized with 300-bp probes (SIRK or EF1γ) corresponding to the amplified fragments. Hybridization was performed at 45°C in 50% (v/v) formamide, 5× SSC, 10% (w/v) dextran sulfate, and 1% (w/v) N-lauroylsarcosine. Filters were washed twice in 2× SSC, 0.1% (w/v) SDS for 10 min each at room temperature and once in 0.1× SSC and 0.1% (w/v) SDS for 15 min at 50°C. Signals on the hybridization membranes were quantified by a PhosphorImager (STORM, Molecular Dynamics, Sunnyvale, CA) and analyzed with the software ImageQuant (Molecular Dynamics).
Statistical analyses indicated that the amplification was linear (as a semilogarithmic function) between cycles 14 and 22 for both cDNAs. We estimated the relative levels of the two cDNAs by calculating the ratio of the amount of amplified fragments for SIRK and EF1γ (SIRK/EF1γ) for each PCR reaction at each cycle. The mean value of these ratios was taken as an indicator of the relative abundance of SIRK compared with the abundance of EF1γ.
Southern-Blot Analysis
Genomic DNA was extracted as described by Steenkamp et al. (1994). DNA (10 μg) was digested with the specified enzymes according to the manufacturer's instruction (Promega), electrophoresed in a 0.8% (w/v) agarose gel, and blotted onto nylon membrane (ICN, Costa Mesa, CA). The blot was hybridized as described above with a probe corresponding to the 32P-radiolabeled SIRK cDNA.
Promoter-GUS Fusion Construction
SIRK promoter region was isolated from the genomic library and 3 kb were amplified with primers introducing a unique NcoI site just upstream from the ATG codon (5′-TTTTCCATGGTTTTGTATTTGAATTCCTCAAAGGC-3′) and a unique XhoI site at the 3′ extremity (5′-TTTTTCTCGAGAAGTGGGACTGGTTGGGGCTGC-3′). This fragment was introduced into pBI 320.X (Dr. Rick Derose, Aventis, Evry, France; this plasmid bears a unique NcoI site at the initiation codon of the promoterless GUS-3′ nopaline synthase gene), leading to a translational fusion between the SIRK promoter and GUS coding sequence. This construct was digested by KpnI and SacI and introduced into pBIB-Hygro binary vector (derivating from pBIN19).
Arabidopsis Transformation
The SIRK promoter:GUS construct was introduced into Agrobacterium tumefaciens MP90 (Höfgen and Willmitzer, 1988). Arabidopsis (ecotype Columbia) was transformed using the floral dip method (Clough and Bent, 1998). Selection of T1 seedlings was performed in vitro on Murashige and Skoog/2 medium (Murashige and Skoog, 1962) supplemented with 1% (w/v) Suc, 0.7% (w/v) agar, and 30 μg L−1 hygromycin under the following conditions: 21°C/18°C day/night temperature, 16-h photoperiod, and 150 μE m−2 s−1. For GUS assay, plants were cultivated either in vitro on the same medium and in the same conditions as described above or grown in greenhouse on attapulgite-peat compost (Gaymard et al., 1998).
Grapevine Transformation
The SIRK promoter:GUS fusion was introduced into A. tumefaciens EHA 105 (Li et al., 1992). Twenty milliliters of A. tumefaciens culture (A550 = 0.4 [≈1.106 colony forming units mL−1]) was added to 1 g of embryogenic callus, vigorously shaken and incubated for 10 min at room temperature. Calli were then separated from the liquid phase with a 100-μm nylon net (Millipore, Bedford, MA), and transferred onto GS1CA medium (Franks et al., 1998) for a 2-d coculture. Calli were then washed in half-strength Murashige and Skoog medium (Murashige and Skoog, 1962) with 1 mg mL−1 Augmentin (Duchefa, Haarlem, The Netherlands) and transferred to 55-mm petri dishes containing GS1CA plus 1 mg mL−1 Augmentin. After 3 weeks, calli were transferred onto 55-mm plates containing selective GS1CA plus 10 μg mL−1 hygromycin, cefotaxim, and Augmentin. Subsequent subcultures occurred every month under the same conditions except that hygromycin was progressively increased to 25 μg mL−1.
To stimulate germination of putative transformed embryos, the embryogenic calli were transferred on Murashige and Skoog/2 medium plus 25 μg mL−1 hygromycin and 10 μg mL−1 cefotaxim and Augmentin. Only developing embryo-like structures were further subcultured. To stimulate caulogenesis of germinating embryos, whole root meristem regions plus cotyledon upper parts were discarded and cotyledon basal parts containing the shoot meristem were scarified. This tissue was transferred on BFe2 medium (Torregrosa and Bouquet, 1995) and incubated under attenuated light (approximately 15 μE s−1 m−2). Axillary shoots emerged in a few weeks and elongated after two to three subcultures enabling the transfer to rooting medium (Murashige and Skoog/2 + 5 μm IAA) and plant regeneration.
GUS Assay
GUS histochemical staining was performed according to Lagarde et al. (1996). Cross sections of GUS-stained material were prepared on 4% (w/v) agarose embedded tissues with a Vibracut (Bio-Rad, Hercules, CA).
Expression in Xenopus laevis Oocytes and Electrophysiology
In vitro-transcribed SIRK cRNA were injected into X. laevis oocytes (purchased from Centre de Recherche de Biochimie Macromoléculaire, Centre National de la Recherche Scientifique, Montpellier, France) using a 10- to 15-μm tip-diameter micropipette and a pneumatic injector (20 nL of 1 μg μL−1 RNA solution per oocyte). Control oocytes were injected with 20 nL of deionized water.
Whole-cell currents were recorded as previously described (Lacombe and Thibaud, 1998) using the two-electrode voltage-clamp technique, 3 to 7 d after injection on oocytes continuously perfused with bath solution (see figure legends). Quantitative analyses of macroscopic current that yielded the gating parameters were performed as previously described (Lacombe and Thibaud, 1998). Intracellular pH was monitored using pH-sensitive microelectrodes prepared and used as previously described (Lacombe et al., 2000a).
Patch-clamp experiments were performed on devitellinized oocytes as previously described (Lacombe et al., 2000a). Voltage-pulse protocol application, data acquisition, and data analyses were performed using pClamp (Axon Instruments, Foster City, CA), Winascd (Dr. G. Droogmans, University of Leuven, Belgium) and Sigmaplot (Jandel Scientific, Erkrath, Germany) software.
ACKNOWLEDGMENTS
We are grateful to Drs. Guillaume Pilot and Nathalie Ollat for helpful scientific discussion and to Drs. Sabine Zimmermann and Isabel Lefèvre for critical reading of the manuscript.
Footnotes
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010529.
LITERATURE CITED
- Ache P, Becker D, Ivashikina I, Dietriech P, Roelfsema MRG, Hedrich R. GORK, a delayed outward rectifier expressed in guard cells of Arabidopsis thaliana, is a K+-selective, K+-sensing ion channel. FEBS Lett. 2000;486:93–98. doi: 10.1016/s0014-5793(00)02248-1. [DOI] [PubMed] [Google Scholar]
- Ageorges A, Issaly N, Picaud S, Delrot S, Romieu C. Identification and functional expression in yeast of a grape berry sucrose carrier. Plant Physiol Biochem. 2000;38:177–185. [Google Scholar]
- Anderson JA, Huprikar SS, Kochian LV, Lucas WJ, Gaber RF. Functional expression of a probable Arabidopsis thaliana potassium channel in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1992;89:3736–3740. doi: 10.1073/pnas.89.9.3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baribault TJ, Skene KGM, Cain PA, Steele SN. Transgenic grapevines: regeneration of shoots expressing β-glucuronidase. J Exp Bot. 1990;41:1045–1049. [Google Scholar]
- Becker D, Dreyer I, Hoth S, Reid JD, Busch H, Lehnen M, Palme K, Hedrich R. Changes in voltage activation, Cs+ sensitivity, and ion permeability in H5 mutants of the plant K+channel KAT1. Proc Natl Acad Sci USA. 1996;93:8123–8128. doi: 10.1073/pnas.93.15.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla F. The voltage sensor in voltage dependent ion channels. Physiol Rev. 2000;80:555–592. doi: 10.1152/physrev.2000.80.2.555. [DOI] [PubMed] [Google Scholar]
- Blanke MM, Pring RJ, Baker EA. Structure and elemental composition of grape berry stomata. J Plant Physiol. 1999;154:477–481. [Google Scholar]
- Bruggemann A, Pardo LA, Stuhmer W, Pongs O. Ether-a-go-go encodes a voltage-gated channel permeable to K+ and Ca2+and modulated by cAMP. Nature. 1993;365:445–448. doi: 10.1038/365445a0. [DOI] [PubMed] [Google Scholar]
- Chérel I, Daram P, Gaymard F, Horeau C, Thibaud JB, Sentenac H. Plant K+channels: structure, activity and function. Biochem Soc Trans. 1996;24:964–971. doi: 10.1042/bst0240964. [DOI] [PubMed] [Google Scholar]
- Church GW, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson DT, Hanson JB. The mineral nutrition of higher plants. Annu Rev Plant Physiol. 1980;31:239–298. [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Coombe BG. The grape berry as a sink. Acta Hortic. 1989;239:149–158. [Google Scholar]
- Coombe BG. Research on development and ripening of the grape berry. Am J Enol Vitic. 1992;43:101–110. [Google Scholar]
- Daram P, Urbach S, Gaymard F, Sentenac H, Chérel I. Tetramerization of the AKT1 plant potassium channel involves its C-terminal cytoplasmic domain. EMBO J. 1997;16:3455–3463. doi: 10.1093/emboj/16.12.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies C, Robinson SP. Sugar accumulation in grape berry: cloning of two putative vacuolar invertase cDNAs and their expression in grapevine tissues. Plant Physiol. 1996;111:275–283. doi: 10.1104/pp.111.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies C, Robinson SP. Differential screening indicates a dramatic change in mRNA profiles during grape berry ripening: cloning and characterization of cDNAs encoding putative cell wall and stress response proteins. Plant Physiol. 2000;122:803–812. doi: 10.1104/pp.122.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delas J, Molot C, Soyer JP. 4ème Symposium International d'Oenologie-Actualités Oenologiques, Bordeaux, France, Dunod, Paris. 1989. Qualité et constitution des raisins de cuve: fertilisation minérale de la vigne et teneurs en potassium des baies, des moûts et des vins; pp. 1–6. [Google Scholar]
- Düring H, Lang A, Oggionni F. Patterns of water flow in Riesling berries in relation to developmental changes in their xylem morphology. Vitis. 1987;26:123–131. [Google Scholar]
- Ehrhardt T, Zimmermann S, Müller-Röber B. Association of plant K+inchannels is mediated by conserved C-termini and does not affect subunit assembly. FEBS Lett. 1997;409:166–170. doi: 10.1016/s0014-5793(97)00502-4. [DOI] [PubMed] [Google Scholar]
- Fillion L, Ageorges A, Picaud S, Coutos-Thévenot P, Lemoine R, Romieu C, Delrot S. Cloning and expression of a hexose transporter gene expressed during the ripening of grape berry. Plant Physiol. 1999;120:1083–1093. doi: 10.1104/pp.120.4.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay N, Oliver KJ, Nii N, Coombe BG. Solute accumulation by grape pericarp cells: IV. Perfusion of pericarp apoplast via the pedicel and evidence for xylem malfunction in ripening berries. J Exp Bot. 1987;38:668–679. [Google Scholar]
- Franks T, He DG, Thomas M. Regeneration of transgenic Vitis viniferaL. Sultana plants: genotypic and phenotypic analysis. Mol Breeding. 1998;4:321–333. [Google Scholar]
- Gaymard F, Pilot G, Lacombe B, Bouchez D, Bruneau D, Boucherez J, Michaux-Ferriere N, Thibaud JB, Sentenac H. Identification and disruption of a plant Shaker-like outward channel involved in K+release into the xylem sap. Cell. 1998;94:647–655. doi: 10.1016/s0092-8674(00)81606-2. [DOI] [PubMed] [Google Scholar]
- Gonnet GM, Cohen MA, Benner SA. Exhaustive matching of the entire protein sequence database. Science. 1992;256:1443–1445. doi: 10.1126/science.1604319. [DOI] [PubMed] [Google Scholar]
- Hale CR. Relation between potassium and the malate and tartrate contents of grape berries. Vitis. 1977;16:9–19. [Google Scholar]
- Hanley BA, Schuler MA. Plant intron sequences: evidence for distinct groups of introns. Nucleic Acids Res. 1988;16:7159–7176. doi: 10.1093/nar/16.14.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins JD. A survey on intron and exon lengths. Nucleic Acids Res. 1988;16:9893–9908. doi: 10.1093/nar/16.21.9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille B. Ionic channels in excitable membranes. Ed 2. Sunderland, MA: Sinauer Associates; 1992. [Google Scholar]
- Hirsch RE, Lewis BR, Spalding EP, Sussman MR. A role for the AKT1 potassium channel in plant nutrition. Science. 1998;280:918–921. doi: 10.1126/science.280.5365.918. [DOI] [PubMed] [Google Scholar]
- Hoffman M. New role found for a common protein “motif.”. Science. 1991;253:742. doi: 10.1126/science.1831563. [DOI] [PubMed] [Google Scholar]
- Höfgen R, Willmitzer L. Storage of competent cells for Agrobacteriumtransformation. Nucleic Acids Res. 1988;20:9877. doi: 10.1093/nar/16.20.9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrazdina G, Parsons GF, Mattick LR. Physiological and biochemical events during development and maturation of grape berries. Am J Enol Vitic. 1994;35:220–227. [Google Scholar]
- Jan LY, Jan YN. Cloned potassium channels from eukaroytes and prokaryotes. Annu Rev Neurosci. 1997;20:91–123. doi: 10.1146/annurev.neuro.20.1.91. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kananagh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellis AK, Roubelakis-Angelakis KA. Grape. In: Seymour G, Taylor J, Tucker G, editors. Biochemistry of Fruit Ripening. London: Chapman & Hall; 1993. pp. 189–234. [Google Scholar]
- Lacombe B, Pilot G, Gaymard F, Sentenac H, Thibaud JB. pH control of the plant outwardly-rectifying potassium channel SKOR. FEBS Lett. 2000a;466:351–354. doi: 10.1016/s0014-5793(00)01093-0. [DOI] [PubMed] [Google Scholar]
- Lacombe B, Pilot G, Michard E, Gaymard F, Sentenac H, Thibaud JB. A Shaker-like K+ channel with weak rectification is expressed in both source and sink phloem tissues of Arabidopsis. Plant Cell. 2000b;12:837–851. doi: 10.1105/tpc.12.6.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe B, Thibaud JB. Evidence for a multi-ion pore behavior in the plant potassium channel KAT1. J Membr Biol. 1998;166:91–100. doi: 10.1007/s002329900451. [DOI] [PubMed] [Google Scholar]
- Lagarde D, Basset M, Lepetit M, Conejero G, Gaymard F, Astruc S, Grignon C. Tissue-specific expression of Arabidopsis AKT1 gene is consistent with a role in K+nutrition. Plant J. 1996;9:195–203. doi: 10.1046/j.1365-313x.1996.09020195.x. [DOI] [PubMed] [Google Scholar]
- Li XO, Liu CN, Ritchie SW, Peng JY, Gelvin SB, Hodges TK. Factors influencing Agrobacterium-mediated transient expression of gusAin rice. Plant Mol Biol. 1992;20:1037–1048. doi: 10.1007/BF00028891. [DOI] [PubMed] [Google Scholar]
- Lodhi MA, Reisch BI. Nuclear DNA content of Vitisspecies, cultivars and other genera of the Vitaceae. Theor Appl Genet. 1995;90:11–16. doi: 10.1007/BF00220990. [DOI] [PubMed] [Google Scholar]
- Lux SE, John KM, Benett V. Analysis of cDNA for human erythrocyte ankyrin indicates a repeated structure with homology to tissue-differentiation and cell-cycle control proteins. Nature. 1990;344:36–42. doi: 10.1038/344036a0. [DOI] [PubMed] [Google Scholar]
- MacKinnon R. Determination of the subunit stoichiometry of a voltage-activated potassium channel. Nature. 1991;350:232–235. doi: 10.1038/350232a0. [DOI] [PubMed] [Google Scholar]
- Manning K, Davies C, Bowen HC, White PJ. Functional characterization of two ripening-related sucrose transporters from grape berries. Ann Bot. 2001;87:125–129. [Google Scholar]
- Marck C. DNA Strider: a “C” program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 1990;16:1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marten I, Hoth S, Deeken R, Ache P, Ketchum KA, Hoshi T, Hedrich R. AKT3, a phloem-localized K+channel, is blocked by protons. Proc Natl Acad Sci USA. 1999;96:7581–7586. doi: 10.1073/pnas.96.13.7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro MC, Toutain S, Waletr B, Pinck L, Otten L, Coutos-Thevenot P, Deloire A, Barbier P. High efficiency regeneration of grapevine plants transformed with the GFLV coat protein gene. Plant Sci. 1995;112:97–106. [Google Scholar]
- Morsomme P, Boutry M. The plant plasma membrane H+-ATPase: structure, function and regulation. Biochim Biophys Acta. 2000;1465:1–16. doi: 10.1016/s0005-2736(00)00128-0. [DOI] [PubMed] [Google Scholar]
- Mouline K, Véry AA, Gaymard F, Boucherez J, Pilot G, Devic M, Bouchez D, Thibaud JB, Sentenac H (2002) Pollen tube development and competitive ability are impaired by disruption of a Shaker K+ channel in Arabidopsis. Genes Dev (in press) [DOI] [PMC free article] [PubMed]
- Murashige T, Skoog F. A revised medium for rapid growth and bio-assays with tobacco tissues culture. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Müller-Röber B, Ellenberg J, Provart N, Willmitzer L, Busch H, Becker D, Dietrich P, Hoth S, Hedrich R. Cloning and electrophysiological analysis of KST1, an inward rectifying K+channel expressed in potato guard cells. EMBO J. 1995;14:2409–2416. doi: 10.1002/j.1460-2075.1995.tb07238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura RL, Mc Kendree WL, Hirsch RE, Sedbrook JC, Gaber RF, Sussman MR. Expression of an Arabidopsispotassium channel gene in guard cells. Plant Physiol. 1995;109:371–374. doi: 10.1104/pp.109.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilot G, Lacombe B, Gaymard F, Chérel I, Boucherez J, Thibaud JB, Sentenac H. Guard cell inward K+ channel activity in Arabidopsisinvolves expression of the twin channel subunits KAT1 and KAT2. J Biol Chem. 2001;276:3215–3221. doi: 10.1074/jbc.M007303200. [DOI] [PubMed] [Google Scholar]
- Robinson SP, Jacobs AK, Dry IB. A class IV chitinase is highly expressed in grape berries during ripening. Plant Physiol. 1997;114:771–778. doi: 10.1104/pp.114.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ros R, Romieu C, Gibrat R, Grignon C. The plant inorganic pyrophosphatase does not transport K+ in vacuole membrane vesicles multilabeled with fluorescent probes for H+, K+, and membrane potential. J Biol Chem. 1995;270:4368–4374. doi: 10.1074/jbc.270.9.4368. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sarni-Manchado P, Verries C, Tesnière C. Molecular characterization and structural analysis of one alcohol dehydrogenase gene (GV-Adh1) expressed during ripening of grapevine (Vitis viniferaL.) Plant Sci. 1997;125:177–187. [Google Scholar]
- Schachtman DP, Schroeder JI, Lucas WJ, Anderson JA, Gaber RF. Expression of an inward-rectifying potassium channel by the Arabidopsis KAT1cDNA. Science. 1992;258:1654–1658. doi: 10.1126/science.8966547. [DOI] [PubMed] [Google Scholar]
- Scorza R, Cordts JM, Gray DJ, Gonsalves D, Emershad RL, Ramming DW. Producing transgenic “Thompson Seedless” grape (Vitis viniferaL.) plants. J Am Soc Hortic Sci. 1996;121:616–619. [Google Scholar]
- Sentenac H, Bonneaud N, Minet M, Lacroute F, Salmon JM, Gaymard F, Grignon C. Cloning and expression in yeast of a plant potassium ion transport system. Science. 1992;256:663–665. doi: 10.1126/science.1585180. [DOI] [PubMed] [Google Scholar]
- Spalding EP, Hirsch RE, Lewis DE, Qi Z, Sussman MR. Potassium uptake supporting plant growth in the absence of AKT1 channel activity. J Gen Physiol. 1999;113:909–918. doi: 10.1085/jgp.113.6.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenkamp J, Wiid I, Lourens A, Vanhelden P. Improved method for DNA extraction from Vitis vinifera. Am J Enol Vitic. 1994;45:102–106. [Google Scholar]
- Szyroki A, Ivashikina N, Dietrich P, Roelfsema MRG, Ache P, Reintanz B, Deeken R, Godde M, Felle H, Steinmeyer R et al. KAT1 is not essential for stomatal opening. Proc Natl Acad Sci USA. 2001;98:2917–2921. doi: 10.1073/pnas.051616698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattersall DB, van Heeswijck R, Hoj PB. Identification and characterization of fruit-specific, thaumatin-like protein that accumulates at very high levels in conjunction with the onset of sugar accumulation and berry softening in grapes. Plant Physiol. 1997;114:759–769. doi: 10.1104/pp.114.3.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrier N, Sauvage FX, Ageorges A, Romieu C. Changes in acidity and in proton transport at the tonoplast of grape berries during development. Planta. 2001;213:20–28. doi: 10.1007/s004250000472. [DOI] [PubMed] [Google Scholar]
- Torregrosa L. A simple and efficient method to obtain stable embryogenic cultures from anthers of Vitis viniferaL. Vitis. 1998;37:91–92. [Google Scholar]
- Torregrosa L, Bouquet A. In vitro propagation of Vitis × Muscadiniahybrids by microcuttings or axillary budding. Vitis. 1995;34:237–238. [Google Scholar]
- Uozumi N, Nakamura T, Schroeder JI, Muto S. Determination of transmembrane topology of an inward-rectifying potassium channel from Arabidopsis thaliana based on functional expression in Escherichia coli. Proc Natl Acad Sci USA. 1998;95:9773–9778. doi: 10.1073/pnas.95.17.9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbach S, Chérel I, Sentenac H, Gaymard F. Biochemical characterization of the Arabidopsis K+channels KAT1 and AKT1 expressed or co-expressed in insect cells. Plant J. 2000;23:527–538. doi: 10.1046/j.1365-313x.2000.00828.x. [DOI] [PubMed] [Google Scholar]
- Véry AA, Bosseux C, Gaymard F, Sentenac H, Thibaud JB. Level of expression in Xenopus oocytes affects some characteristics of a plant inward-rectifying voltage-gated K+channel. Pflügers Arch. 1994;428:422–424. doi: 10.1007/BF00724528. [DOI] [PubMed] [Google Scholar]
- Véry AA, Gaymard F, Bosseux C, Sentenac H, Thibaud JB. Expression of cloned plant K+ channel in Xenopusoocytes: analysis of macroscopic currents. Plant J. 1995;7:321–332. doi: 10.1046/j.1365-313x.1995.7020321.x. [DOI] [PubMed] [Google Scholar]
- Zei PC, Aldrich RW. Voltage-dependent gating of single wild-type and S4 mutant KAT1 inward rectifier potassium channels. J Gen Physiol. 1998;112:679–713. doi: 10.1085/jgp.112.6.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann S, Sentenac H. Plant ion channels: from molecular structures to physiological functions. Curr Opin Plant Biol. 1999;2:477–482. doi: 10.1016/s1369-5266(99)00020-5. [DOI] [PubMed] [Google Scholar]