Abstract
A single, recessive mutation in soybean (Glycine max L. Merr.), which confers a seed phenotype of increased inorganic phosphate, decreased phytic acid, and a decrease in total raffinosaccharides, has been previously disclosed (S.A. Sebastian, P.S. Kerr, R.W. Pearlstein, W.D. Hitz [2000] Soy in Animal Nutrition, pp 56–74). The genetic lesion causing the multiple changes in seed phenotype is a single base change in the third base of the codon for what is amino acid residue 396 of the mature peptide encoding a seed-expressed myo-inositol 1-phospate synthase gene. The base change causes residue 396 to change from lysine to asparagine. That amino acid change decreases the specific activity of the seed-expressed myo-inositol 1-phosphate synthase by about 90%. Radio tracer experiments indicate that the supply of myo-inositol to the reaction, which converts UDP-galactose and myo-inositol to galactinol is a controlling factor in the conversion of total carbohydrate into the raffinosaccharides in both wild-type and mutant lines. That same decrease in myo-inositol 1-phosphate synthetic capacity leads to a decreased capacity for the synthesis of myo-inositol hexaphosphate (phytic acid) and a concomitant increase in inorganic phosphate.
Phytic acid is a nearly ubiquitous component of plant seeds and is usually the most abundant form of phosphate in those seeds (Raboy et al., 2000). Similarly, the α-galactoside series of sugars that includes raffinose and stachyose are found in most terrestrial plant seeds (Kuo et al., 1988). The wide distribution of these compounds among seeds of many plant species along with their abundance within those seeds suggest that they may be important for some aspect of seed or seedling physiology.
Phytic acid is presumed to act as a phosphate storage compound, and phytic acid can account for more than 60% of the total seed phosphate in many cases (Rayboy et al., 1984). At the same time, however, Raboy and Dickinson (1987) used phosphate starvation during seed development to produce soybean (Glycine max) seeds with greatly reduced phytic acid and convincingly showed that phytic acid is not a requirement for seed viability or germination. Mutant lines with greatly reduced levels of phytic acid have been described in maize (Zea mays; Raboy et al., 2000), barley (Hordeum vulgare; Larson et al., 1998), rice (Oryza sativa; Larson et al., 2000), and most recently soybean (Sebastian et al., 2000; Wilcox et al., 2000). The fact that these mutants were recovered and can be seed propagated suggests that a high level of stored phosphate is not a requirement for seed germination or seedling growth.
The raffinose series of sugars as well as cyclitols based on myo-inositol have been suggested to be involved in protection of seeds from damage during desiccation (for review, see Obendorf, 1997). The physical reasoning behind this suggestion stems from the ability of sugars to stabilize membrane structures during desiccation (Crowe et al., 1987). Correlation between the onset of seed viability after premature dry down and the appearance of raffinose and stachyose have also been cited as evidence of the involvement of these sugars in desiccation tolerance (Koster and Leopold, 1988). Bentsink et al. (2000), however, found no correlation between seed storability and raffinosaccharide content in Arabidopsis recombinant inbred lines. Sebastian et al. (2000) also described the recovery of soybean mutant lines with greatly decreased levels of raffinose and stachyose.
Whereas the wide occurrences of these two classes of compounds across many plant families make it seem likely that they play some role in seed viability or seedling nutrition, at least partial decreases do not result in seed or seedling lethality. Both classes of compounds are poorly digested by monogastric animals and are considered to be antinutritional factors in animals fed rations that contain high levels of grains that contain them so the reduction or elimination of these compounds in seeds are attractive breeding targets.
Mutant lines with much lower levels of both raffinosaccharides and phytic acid have been described (Sebastian et al., 2000). In the most highly characterized of these lines, raffinose and stachyose are decreased to about 10 and 4 μmol per gram of seed dry weight as compared with 20 and 75 μmol per gram in normal soybean seeds. The phytic acid content is reduced to about 50% of that in normal soybean seeds. The lines have been shown to have increased available phosphate and metabolizable energy when fed as a component of poultry diets (Cromwell et al., 2000a, 2000b; Spencer et al., 2000a). Dietary available phosphate was also increased when the low phytic acid soybeans were included in swine diets, especially when fed in combination with low phytic acid corn (Spencer et al., 2000b).
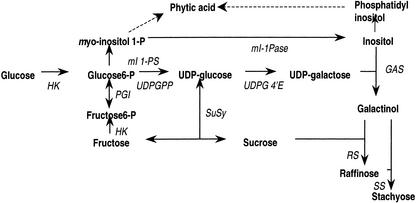
A schematic of the biosynthetic pathways to phytic acid, raffinose, and stachyose is shown in Figure 1. Whereas details of the conversion of Glc into raffinose and stachyose are known and involve myo-inositol as the recycled carrier of activated Gal (Lehele and Tanner 1973), the exact pathways to phytic acid remain less precisely defined and are shown as the dotted lines in Figure 1. The exact path to phytic acid almost certainly consists of multiple phosphorylation steps (Brearley and Hanke, 1996a, 1996b) and could either start with myo-inositol-1-phosphate and proceed by way of soluble intermediates or could go by way of free myo-inositol through phosphatidyl-inositol with the first phosphorylations occurring at the membrane-bound intermediate level. The two sources of myo-inositol-1-phosphate are its synthesis from Glc 6-phosphate and from phosphorylation of free myo-inositol (Loewus and Murthy, 2000).
Figure 1.
A schematic diagram of the interactions involved inconversion of carbon from Glc into either phytic acid or Suc, raffinose, and stachyose. HK, Hexokinase; PGI, phospho-Glc iosmerase; UDPGP, uridine diphospho-Glc phyrophosphorylase; UDPG 4′E, uridine diphospho-Glc 4′ epimerase; SuSy, Suc synthase; GAS, galactinol synthase; RS, raffinose synthase; SS, stachyose synthase; mI-1pase, myo-inositol 1-phosphate phosphatase; mI 1PS, myo-inositol 1-phosphate synthase. The product of the reaction catalyzed by myo-inositol 1-phosphate synthase (EC 5.5.1.4) is l-myo-inositol 1-phosphate in the l-convention nomenclature or d-myo-inositol 3-phosphate in the d-convention nomenclature. The naming of genes that catalyze this reaction has followed the l-convention nomenclature. Because the soybean genes described in this paper were cloned by homology to already named genes, the l-convention nomenclature is maintained here and extends to the reaction products.
Because phytic acid and the raffinosaccharides share at least myo-inositol-1-phosphate and possibly free myo-inositol as a common intermediate, we characterized the production of these metabolites in wild type and in the mutant soybean line.
RESULTS
Characterization of Soluble Carbohydrate Mutants
Screening of bulk seed from about 8,000 single plant, M3 generation plants and of the soybean Plant Introduction collection held by the U.S. Department of Agriculture as described (Sebastian et al., 2000) produced two types of modified carbohydrate profile soybean seeds. Both classes of mutants were backcrossed to commercial soybean cultivars, and the mutant-altered soluble carbohydrate profiles were followed in the segregating populations derived from the crosses. Typical analysis of bulked seeds from these two types and a wild-type soybean cultivar is shown in Table I. Several mutants of the type represented by LR28 were found, but in the carbohydrate screen, only one of the type represented by LR33 was found.
Table I.
Soluble carbohydrate content of bulk seeds from several soybean lines
| Line | Sucrose | Galactinol | Raffinose | Stachyose |
|---|---|---|---|---|
| μmol sugar g−1 dry seed | ||||
| Commercial ave. | 165.2 ± 7 | 0 | 24.2 ± 6 | 70.5 ± 6 |
| LR28 | 212.6 ± 10 | 59.5 ± 8 | 3.5 ± 0.7 | 17.5 ± 3 |
| LR33 derived ave. | 244 ± 16 | 0 | 10.6 ± 1.5 | 3.8 ± 0.8 |
| LR33xLR28 | 269.0 ± 10.5 | 0 | 4.0 ± 1 | 0.7 ± 0.6 |
Wild-type soybeans are the average from four commercial lines: A2872, A2242, A2704, and A1923. The average values for the LR33 derived lines are bulk seed from five lines each of which were crossed one or more times to the four commercial lines that make up the control averages. The LR28 values are the average individual plot bulks for the non-backcrossed mutant line. The values for the LR33xLR28 lines are from three plots confirmed as homozygotes for both mutations and backcrossed once to one of the four commercial lines in the study. All plants were grown in the field near Newark, Delaware.
Three characteristics of the dry seed phenotype are suggestive of the points in the metabolic pathway to the raffinosaccarides that might be effected by the mutations. In the LR28 mutant, galactinol is increased from levels that were not detected in wild-type soybeans to amounts that exceed that of raffinose in wild-type seeds on a weight basis. Suc is increased in the LR28 mutation as it is in the LR33 mutation and the cross of the two mutations. The LR33 mutation leads to a greater decrease in total raffinosaccharide synthesis without an increase in galactinol accumulation than does LR28 and in combination with the LR28 mutation eliminates the galactinol accumulated in that line. Further, the LR33 mutation causes a greater decrease in the amount of stachyose produced than it does raffinose, whereas the LR28 mutation has the opposite effect. The increase in galactinol and Suc might be anticipated if blockage of the pathway were at the raffinose synthase or stachyose synthase steps in the LR28 mutant because an accumulation of the immediate precursors might occur (Fig. 1.). Raffinose synthase is the more probable candidate step because both raffinose content and stachyose content are decreased. The LR33 mutation accomplishes a greater reduction in total raffinosaccharide, galactinol accumulation is not increased, and the amount of stachyose accumulated is less than the amount of raffinose accumulated. All of these observations are consistent with a mutation that affects a step required for galactinol accumulation in the case of LR33.
Enzyme Activities in the Raffiosaccharide Synthesis Pathway
Seeds from a wild-type line (A1923), LR28, and seeds homozygous for both the LR28 and the LR33 mutations were harvested during the late pod filling stage at which raffinose and stachyose accumulate. Enzyme activities for the three enzymes in the pathway that are essential for raffinose and stachoyse synthesis only were measured in vitro on freshly harvested seeds.
Of these three enzymes, only raffinose synthase shows a clear decrease in activity in the LR28 mutant. That same difference is born out in the double mutant line but neither of the other two enzymes measured is decreased in activity in the double mutant (Table II).
Table II.
Activities of galactinol synthase, raffinose synthase, and stachyose synthase in yellowing seeds of three soybean lines
| Line | Galactinol Synthase | Raffinose Synthase | Stachyose Synthase |
|---|---|---|---|
| A1923 | 7.5 ± 4.1 | 0.10 ± 0.05 | 0.65 ± 0.18 |
| LR28 | 16.5 ± 5.2 | 0.004 ± 0.007 | 0.81 ± 0.24 |
| LR33xLR28 | 22.6 ± 8.8 | 0.006 ± 0.008 | 0.87 ± 0.21 |
The wild-type control is line A1923. Enzyme activities are expressed as μmol of product produced per gram fresh wt per hour under the assay conditions. Values are mean ± sd for five replicates.
In separate experiments that measured the rate of raffinose and stachyose appearance, the rate of total raffinosaccharide accumulation in wild-type seeds was estimated at just less than 0.1 μmol g−1 fresh weight h−1 (data not shown). Whereas the in vitro measured activity of galactinol synthase is much greater than the estimated flux through that step in the pathway, both raffinose synthase activity and stachyose synthase activity are similar to flux in the wild type. Raffinose synthase activity in the LR28 mutant is considerably below the estimated flux required to maintain normal accumulation. Both the decrease in measurable raffinose synthase activity and the accumulation of both substrates for that reaction are consistent with the LR28 mutation effecting raffinose synthase activity. The only conclusion regarding the LR33 mutation that can be drawn from the data is that it does not affect galactinol synthase activity.
Myo-Inositol Content of Soybeans during Raffinosaccharide Accumulation
The LR33 mutation could control raffinosaccharide synthesis indirectly by controlling the synthesis of one of the substrates for the first committed step in the pathway, galactinol synthesis (Fig. 1). Myo-inositol content was estimated in the wild-type line A2872 and in LR33 during the period of raffinosaccharide synthesis. Because the period of active raffinosaccharide synthesis is short and not easily predicted from time after pollination, single seeds taken at the stage of early pod yellowing were analyzed to check for variation as shown in Table III.
Table III.
Water content, dry weight, and myo-inositol content of 7 individual seeds from A2872 and LR33
| Seed No. | Genotype | H2O | Dry Wt | Inositol |
|---|---|---|---|---|
| % | mg | μmole g−1 dry wt | ||
| 1 | Wt | 60.2 | 196 | 4.2 |
| 2 | Wt | 58.6 | 208 | 2.2 |
| 3 | Wt | 58.8 | 212 | 2.5 |
| 4 | Wt | 55.9 | 201 | 1.3 |
| 5 | Wt | 55.3 | 169 | 1.6 |
| 6 | Wt | 57.6 | 151 | 2.0 |
| 7 | Wt | 54.2 | 182 | 1.4 |
| 8 | LR33 | 57.2 | 165 | 1.0 |
| 9 | LR33 | 48.2 | 174 | 0.5 |
| 10 | LR33 | 60.4 | 161 | 0.5 |
| 11 | LR33 | 57.7 | 172 | 0.6 |
| 12 | LR33 | 59.2 | 157 | 0.6 |
| 13 | LR33 | 55.2 | 154 | 1.4 |
| 14 | LR33 | 58.0 | 152 | 0.8 |
Wt, Wild type.
The myo-inositol content on a single seed basis was quite variable although each of the seeds was of a similar developmental stage as judged by a water content of near 60% and a dry weight near the maximum expected for the mature seed. Although the content is variable, the mean myo-inositol content for LR33 seeds is only 36% of that of wild type (wild type, 2.2 ± 0.1 μmol g−1 dry weight; LR33, 0.8 ± 0.3 μmol g−1 dry weight). In a separate experiment done with bulk seeds, the LR33 myo-inositol content was again 20% of the bulk wild-type seeds in the experiment (data not shown). There is clearly a substantial pool of myo-inositol during the period of raffinose synthesis, but it does appear to be reduced in the LR33 line.
Myo-Inositol Perfusion and in Vivo Labeling of Raffinose Saccharides
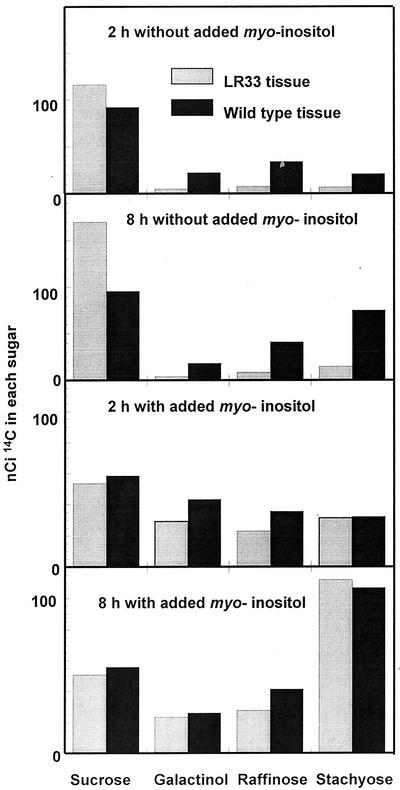
To ascertain whether or not the observed decrease in free myo-inositol content of the LR33-derived seeds might be the cause of the decreased raffinosaccharide content at maturity, tissue slice feeding studies were done using 1-mm tissue slices taken from developing soybean embryos during the period of raffinosaccharide synthesis. In preliminary experiments, it was observed that trace [14C]Glc is effectively converted first into Suc and then into the products and intermediates in the stachyose pathway in this tissue. Under the conditions of incubation, the cotyledon tissue slices continued to metabolize added [14C]Glc to oligosaccharides for at least 24 h after addition of the label. Results of one of two experiments done to compare rate of conversion of Glc to oligosaccharide products in wild-type and LR33 seeds are shown in Figure 2.
Figure 2.
The conversion of trace amounts of [14C]Glc into Suc, galactinol, raffinose, and stachyose by maturing seeds of wild-type or LR33 mutant soybeans. The lower two panels are the conversions from tissue slices of both genotypes that were done concurrently but in which 50 mm inositol was included with the Glc tracer.
Both wild-type and LR33-derived tissue slices converted label from supplied [14C]Glc to Suc and then on into the raffinosaccharide sugars. There was little difference between the two lines in terms of amount of 14C converted to oligosaccharides. Without the addition of exogenous myo-inositol, the line containing the LR33 mutation converts very little label into the raffinossaccarides (14.7% after 2 h) in comparison with the wild-type line (45.9% after 2 h). In the next 6 h, both lines converted added 14C to oligosaccharides at a reduced rate, converted Suc into higher oligosaccharides, and decreased the amount of label in galactinol. Both genotypes respond to exogenous myo-inositol by converting more of the supplied label to raffinosaccharide sugars. With the addition of myo-inositol, the two genotypes become essentially equal in their ability to convert the supplied label to Gal-containing sugars. The data may be interpreted to indicate that the supply of myo-inositol to the galactinol synthase reaction imposes a fairly strong control over flux to the raffinosaccharides in soybean seed. The decreased myo-inositol content conferred by the LR33 mutation further decreases the raffinosaccharide synthetic capacity in the mutant line, presumably enough to produce the low raffinosaccharide phenotype in the dry seed.
The Influence of the LR33 Mutation on Seed Phytic Acid and Inorganic Phosphate Levels
Soybean seeds contain phytic acid at levels of 20 to 30 μmol g−1, corresponding to 120 to 180 μmol of phytic acid contained phosphate per gram. This constitutes from 50% to 70% of the total seed phosphate (Raboy et al., 1984). It seemed likely, therefore, that mutations effecting myo-inositol synthesis might also affect seed phytic acid and possibly inorganic phosphate levels. The levels of phytic acid and inorganic phosphate in three wild-type soybean lines and three lines that contain the LR33 mutation were measured. The experiment was repeated three times and the results are given in Table IV.
Table IV.
Seed phosphate in inorganic phosphate and in phytic acid
| Seed Line | Genotype | Replicates | Inorganic Phosphate | Phytic Acid Phosphate |
|---|---|---|---|---|
| μmol phosphate g−1 | ||||
| A2872 | Wild type | 7 | 2.7 ± 3 | 125 ± 8.4 |
| A1923 | Wild type | 1 | 2.0 | 126 |
| A1929 | Wild type | 2 | 1.5 | 155 |
| 5ST-1309 | LR33 | 4 | 76.0 ± 5.7 | 62.5 ± 12.4 |
| GxE-117 | LR33 | 7 | 36.7 ± 6.4 | 55.8 ± 13.9 |
| GxE-76 | LR33 | 6 | 32.2 ± 3.5 | 72.8 ± 14 |
Values are mean ± sd where more than two replicates were obtained or the average value when two replicates were obtained.
The LR33 mutation also causes a decrease in seed phytic acid level of about 2-fold with a concomitant increase in the seed inorganic phosphate content of about 15- to 35-fold depending upon the genetic background in which the mutation is contained.
Molecular Identification of the LR33 Mutation
The above evidence is consistent with the LR33 mutation decreasing free myo-inositol and compounds derived from it. The decrease in enzymic capacity might be at the myo-inositol 1-phosphate forming step or it could be in the conversion of the cyclitol phosphate to free myo-inositol (Fig. 1). Attempts at measuring the total myo-inositol 1-phosphate synthase activity in crude extracts from developing soybeans were not successful and initial measurements of myo-inositol 1-phosphate phosphatase activity indicated no difference between wild-type and LR33 seeds (data not shown). cDNA clones of the seed-expressed myo-inositol 1-phosphate synthase were obtained by homology to the Arabidopsis enzyme from wild type and from LR33 seeds. Their sequences are on average 85% identical to other plant sequences in the GenBank database, and the wild-type sequence is deposited in GenBank with the designation Gm mI 1-PS-1A (GenBank accession no. AY 038802). The cDNA clone of the mI 1-PS message recovered from line LR33 is identical at the nucleic acid level to the clone from the soybean line chosen to represent wild type with the exception of a single base change from G to T at position 1,188 from the start codon. The change in the third position of the Lys-396 codon in the wild-type peptide places a Gln residue in the LR33 peptide. As shown in Figure 3, K396 and the peptide sequence immediately surrounding it is highly conserved in mI 1-PS sequences even across kingdoms.
Figure 3.
The comparison of the amino acids sequences from Arabidopsis, maize, fruitfly (Drosophila melanogaster), and yeast (Saccharomyces cerevisiae) myo-inositol 1-phosphate synthase in the region corresponding to the conserved Lys residue at position 396 of the soybean peptide. All sequences were deduced from the GenBank nucleic acid sequences.
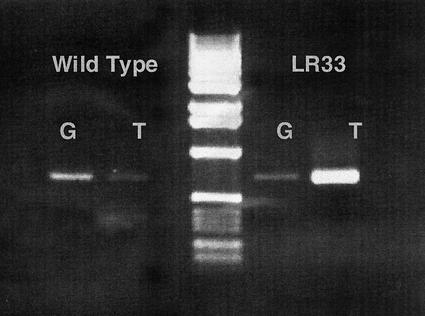
A 17-base PCR primer set ending with base 1,188 at the 3′ end with either the wild-type (G) or LR33 base (T) and a common reverse primer was capable of identifying segregating plants from crosses of wild type by LR33 plants. The PCR product of about 600 bp was obtained in much higher yield if the final base of the primer matched that of the template (Fig. 4.). PCR reactions done using DNA from known heterozygotes as primer produced products in both reactions (data not shown).
Figure 4.
The PCR reaction products obtained using a 17-bp primer corresponding to bases 1,171 to 1,188 of the open reading frame of mI 1-PS-1b and a second, reverse strand primer where the 1,171 to 1,188 primer ends either in G (wild-type sequence) or T (LR33 sequence). The reaction was run with an annealing temperature of 62°C. Both primers give roughly equal reaction products in lines that are heterozygous for the LR33 mutation.
The effect of the K396 to N396 change on the activity of mI 1-PS was measured using the wild-type protein, the LR33 protein, and the protein from the wild-type sequence in which K396 was changed to N396 by site-directed mutagenesis. All three proteins expressed in Escherichia coli using the T7 promoter system to give an approximately 55-kD peptide that remained in the soluble fraction. The wild-type mI 1-PS was somewhat toxic to E. coli growth. Growth at decreased temperature and on simple media during the cell mass accumulation phase of fermentation was required to achieve acceptable cell growth. E. coli cultures containing the LR33 protein or the created mutant protein consistently achieved slightly higher levels of recombinant protein expression than did cultures containing the wild-type soybean gene.
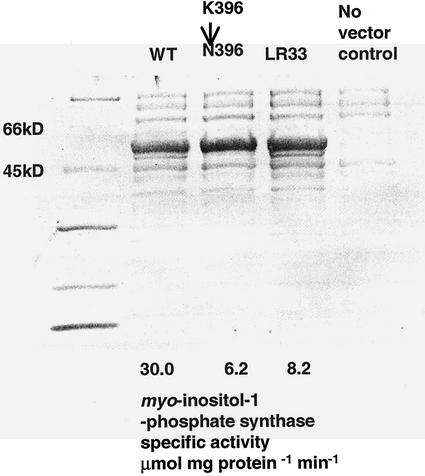
A Coomassie Blue-stained SDS-PAGE gel of typical E. coli extracts after an ammonium sulfate precipitation step is shown in Figure 5 along with the specific activity of myo-inositol 1-phosphate synthase from the same fraction. Direct conversion of K396 to N396 decreased the activity of the protein to a level very similar to the protein produced by the LR33 mutant clone. Whereas the protein expressed from the LR33 clone does retain activity, that activity is between about 10% and 30% of that of the wild-type protein as judged by the measured specific activity and the relative purity of the three fractions.
Figure 5.
A photograph of the Coomassie Blue-stained SDS-PAGE protein gel of soluble proteins from E. coli after cell disruption and ammonium sulfate fractionation. Lane 1, Mr markers; lane 2, extract from cells expressing wild-type GM-mI-PS-1a under control of the T7 promoter; lane 3, extract from wild-type GM-mI-PS-1a in which residue K396 was changed to N396 by site directed mutation; lane 4, GM-mI-PS-1a from the LR33 mutant; lane 5, an extract from control E. coli DE3 cells. The specific activity of myo-inositol 1-phosphate synthase in lanes 2 through 4 is listed below the lane.
Detailed kinetics to determine whether the decreased reaction rate observed under the standard reaction conditions is due to Km or Vmax changes were not done. The activity of both proteins responded similarly to altered NAD concentration in the reaction mix, indicating that the mutation does not alter binding of the required but unchanged cofactor in the reaction (Loewus and Loewus, 1971). The decreased catalytic capacity and the cosegregation of the G to T change with the high Pi and low raffinosaccharide phenotype indicate that this single base change is responsible for the altered seed phenotype in the LR33 mutant.
Characteristics of the mI 1-PS Gene Family in Soybean
Observations made while cloning the first soybean Gm mI 1-PS cDNA, subsequent analysis using the DuPont soybean expressed sequence tag (EST) collection, and the above-mentioned PCR analysis of the segregation pattern reveal some details of the mI 1-PS gene in soybean. First, during the analysis of the G to T change in populations that were segregating for the LR33 phenotype, it was observed that the PCR product obtained was larger than that predicted by the placement of the two PCR primers in the cDNA sequence. Second, the size of the PCR product obtained from the wild-type primer set was the same size as the PCR product derived using DNA from the LR33 mutant as template in some lines but in others it was smaller. The two different sized PCR products from wild-types lines were sequenced and their sequence, when aligned with the cDNA sequences indicates that there are two introns within the Gm mI 1-PS gene in this region. The first intron follows base 1,352 of the open reading frame of either sequence. Both sequences have identical 97-bp introns followed by 62-bp exons. The next intron is identical between the two products for 39 bases but is extended to a total of 87 bases in the longer of the two PCR products. The exon/intron arrangement through this region of the soybean mI 1-PS gene appears to be the same as one of the mI 1-PS genes from maize. The 62-bp exon in the described PCR product corresponds to the ninth exon of the corn gene (GenBank accession no. AF323175).
In PCR analysis of eight commercial, wild-type lines using primers designed to amplify the above described region, the longer intron product was observed six times and the short product two times. There were no lines that contained both. In segregating populations of LR33 into a short product background, both products were observed. A cDNA sequence was obtained by RT-PCR using message from immature seeds from a short PCR product soybean line. Over the 1,533 bases of the open reading frame, there are 40 base changes between the two sequences that lead to seven amino acid changes. Based on the PCR fragment data and on searches within cDNAs libraries made from single soybean cultivars, it appears that the short intron form is an allelic variant at a single locus. We tentatively call the two forms Gm mI 1-PS-1a and Gm mI 1-PS-1b.
Blast searches of the DuPont soybean EST database indicate that there is at least one other class of mI 1-PS genes expressed in soybean, the Gm mI 1-PS-1a/b class and a second (Gm mI 1-PS-2) that is about 90% identical in the coding region of the cDNA. Gm mI 1-PS-2 may consist of a two- or perhaps three-member gene family; the exact number cannot be determined from the tagged sequence data.
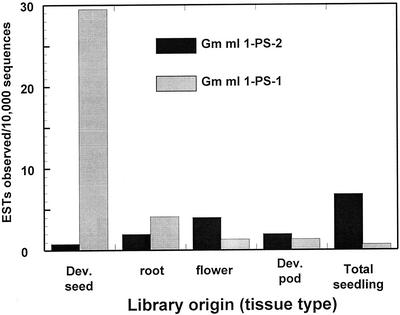
The frequency of observation of ESTs corresponding to the Gm mI 1-PS-1a/b class and of Gm mI 1-PS-2 is shown in Figure 6. Sixty-two libraries and about 150,000 sequences were queried in the blast search. Blast analysis was done at stringency sufficient to largely distinguish the class 1 from class 2, and individual sequence inspection was used to confirm the class. Data from 15 libraries where the number of mI 1-PS sequence observations was high enough to give statistically useful data were combined by tissue type. Gm mI 1-PS-1a is the seed preferred gene, although it is expressed in other tissue types. The Gm mI 1-PS-2 class is essentially not expressed in developing seeds but is expressed in an amount that is at least equal to Gm mI 1-PS-1a in many other tissues. The level of expression of mI 1-PS in developing seeds is also high relative to the expression level in other tissues.
Figure 6.
Relative expression level of the mI 1-PS-1 sequence and the mI 1-PS-2 sequence family in cDNA libraries prepared from different soybean tissues. Five seed libraries are combined for the developing seed data; four root libraries are combined; and two leaf, one flower, two whole seedling, and one developing pod library comprise the whole data set.
DISCUSSION
Myo-inositol synthesis is fairly central in metabolism both at the structural level and as a precursor to the myo-inositol-polyphosphates involved in second message signaling at the regulatory level. In that sense, it is interesting that mutations that decrease its synthesis are tolerated. The specific mutation described here is unique in fitting into a pattern that allows a decrease but at the same time retains viability. First, there is a single seed preferred gene so effective, single gene mutations could be recovered using a phenotype screen. Second, the mutation does not result in complete loss of activity, only a significant decrease within the developing seed and the Gm mI 1-PS-2 is expressed in the remainder of the plant. Finally, for both phytic acid synthesis and raffinosaccharide synthesis, the availability of either myo-inositol itself or of myo-inositol 1-phosphate appears to be one of the controlling factors in the synthesis of the end products. It is this last factor that allows even partial inactivation of the gene to be effective in producing measurable and useful phenotypes.
The existence of this mutant still does not resolve questions about the role of raffinosaccharides or cyclitol derivatives in seed viability. The originally isolated mutant did have significant problems with seedling vigor though not seed viability per se. Some of those problems have been overcome during crossing and selection to derive commercial lines, so it is not yet clear that the mutation results in seed or seedling changes that cannot be compensated within the plant. Also, with this mutant there are two sets of changes occurring. On a mass basis, the increase in inorganic phosphate is the most significant change followed by the raffinosaccharide decrease. Given that, conclusions should not be drawn until the two end product changes can be assessed independently for their effects. There are possible genetic and physiological approaches to those experiments.
Whatever the details of the involvement of phytic acid and the Gal-containing sugars in seed and seedling growth, their conservation across species does merit consideration as to their playing a role. The questions that may present themselves as we attempt to use crop species that are altered in these compounds may run to the practical one of: Are the roles of these compounds unconditional or just highly advantageous in specific environments? And: Are they required in the modern agronomic environment?
MATERIALS AND METHODS
Plant Materials
The soybean (Glycine max) line containing the mutation conferring the low raffinosaccharide and low phytic acid phenotype was produced by a chemical mutagenesis and screening protocol detailed in Sebastian et al. (2000). In brief, soybean seeds were soaked in 2.5 mmN-nitroso-N-methylurea, rinsed, and field planted. M2 seeds were harvested from the surviving plants from the mutagenized seed, planted again, and grown. The seed from the M3 plants (M3:4) was single-plant harvested and bulked, and a subsample of each plant was analyzed for stachyose content using a high-throughput, semiquantitative test. Putative low stachyose lines were confirmed using HPLC. Subsequent generations were either field grown or grown in growth chambers with a 30°C/22°C 16-h-light cycle. Soybean lines Williams 82, Wye, A1923, A2872, A2242, and A2704 were used in various studies as indicated in the discussion or detail of the methods.
Chromatographic Analysis of Soluble Sugars
Soluble sugars were quantified either using differential refractive index detection on a chromatographic system consisting of a Zorbax Amino column (DuPont, Wilmington, DE) eluted at 1 mL min−1 with 70% (v/v) acetonitrile or a pulsed ampheroteric system (Dionex Corp. Sunnyvale, CA). The Dionex system used a Dionex PA1 column eluted at 1.3 mL min−1 with 150 mm NaOH and the pulsed ampheroteric detector set as recommended by the manufacturer for sugar detection. Trehalose was used as an internal standard in both systems.
In Vitro Assay of Activity of Enzymes in the Raffinose Saccharide Pathway
Seeds from pods that had just begun to yellow and that contained seeds that were also loosing green color or just beginning to yellow were removed from the pod, weighed to obtain fresh weight, and then ground in a mortar and pestle in 10 volumes of 50 mm HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid]-NaOH, pH 7, buffer, which was also 5 mm in 2-mercaptoethanol. The ground samples were centrifuged at 10,000g for 10 min, and the supernatant was desalted by passage through Sephadex G-25, which had been equilibrated in the grinding buffer.
For the assay of galactinol synthase, 10 μL of the desalted extract was added to 90 μL of the pH 7 HEPES buffer that was also 20 mm in myo-inositol, 10 mm in dithiothreitol, 1 mm in MnCl2, and 1 mm in UDP-[14C]Gal (1.25 μCi μmol−1). The assay mixture was incubated for 10 min at 25°C then stopped by the addition of 400 μL of ethanol. To the stopped reactions was added 200 μL of Dowex AG-1x8 anion-exchange resin (Bio-Rad, Hercules, CA), and the mixture was shaken for 25 min. The resin was removed by centrifugation, and the supernatant was taken for scintillation counting. Nonanionic, myo-inositol-dependent radioactivity was taken as a measure of galactinol synthesis.
For the assay of raffinose synthase and stachyose synthase, 50 μL of the desalted extract was added to 50 μL of 25 mm HEPES buffer at pH 7 that was also 10 mm in dithiothreitol, 10 mm in galactinol, and 40 mm in Suc for the assay of raffinose synthase or 40 mm in raffinose for the assay of stachyose synthase. After a 1-h incubation at 25°C, the reactions were stopped by the addition of 40 μL of ethanol and then placed in a boiling water bath for 1 min to precipitate proteins. The reaction mixes were centrifuged to clear, and the supernatant was passed through a 0.22-micron filter and then reduced to dryness under vacuum. The residue was redissolved in 0.5 mL of water and 20 μL was separated on the Dionex HPLC system as described above for the quantitation of raffinose and stachyose.
Assay of the Free myo-Inositol Content of Maturing Soybean Seeds
Seeds of the maturity described above were chosen for assay. Individual seeds were weighed, crushed, and lyophilized. The dry residue was extracted in 1 mL of 80% (v/v) methanol at 60°C for 1 h. The extraction solution contained 1 μmol of trehalose, which was added to act as a myo-inositol retention marker on the Zorbax amino column and as an internal standard. After heating, the extract was reground with a small pestle in the microfuge tube and centrifuged to pellet the insoluble material. The supernatant was transferred to a second microfuge tube containing about 100 μL of mixed bed resin, the mixture was briefly shaken and 20 μL of the solution above the resin was removed and taken to dryness under vacuum. The residue was redissolved in 110 μL total volume and 55 μL was injected onto the Zorbax Amino column run as described above. The trehalose peak was trapped, and 20 μL of the column fraction was reinjected on the Dionex system. The myo-inositol peak was quantitated by comparison with the trehalose internal standard.
myo-Inositol Perfusion and in Vivo Labeling of Raffinose Saccharides
Four seeds each from wild-type line A2242 and LR33-derived line 5ST-1434 were harvested by the maturity criterion described above. The seed coats were removed and the cotyledons rinsed in 5 mm potassium phosphate buffer at pH 5.5, then sliced into approximately 1-mm slices and again rinsed with buffer. The tissue slices were divided into two groups. One group was immersed in the potassium phosphate buffer and the second group was immersed in the potassium phosphate buffer containing 50 mm myo-inositol. The tissue slices were vacuum infiltrated and incubated for 30 min at room temperature. After the pre-incubation period, 5 μCi of [14C]Glc (58 μCi μmol−1) was added to each grouping and the tissue was again vacuum infiltrated. Ten tissue slices from each group were taken at 2 and 8 h after addition of the labeled Glc. The tissue slices were placed in tarred, 1.5-mL microfuge tubes to obtain fresh weight and ground in 300 μL of 80% (v/v) methanol. The tubes were centrifuged, the supernatant removed to a second tube, the extraction was repeated twice more, and the supernatants were combined. The combined supernatants were reduced to dryness under vacuum, redissolved in 50% (v/v) acetonitrile, and separated on the Zorbax amino HPLC system. Fractions were collected for scintillation counting at 15-s intervals through the region of the chromatogram containing Glc, Suc, raffinose, and galactinol and at 1-min intervals through the region containing stachyose. Radioactive fractions corresponding to the sugar standard peaks were grouped for analysis. The results are expressed as total radiolable in the four product sugars.
Seed Phytic Acid and Inorganic Phosphate Assays
Phytic acid and inorganic were assayed in dry seeds by a modification of the method described by Raboy et al. (1984). Approximately 20 seeds from each line were ground in a small impact mill and 100 mg of the resulting powder was weighed into a 15-mL screw cap tube. Five milliliters of 0.4 n HCl with 0.7 m Na2SO4 was added to the powder, and the mixture was shaken overnight on a rocker platform. The tubes were centrifuged at 3,900g for 15 min and 2 mL of the supernatant was removed to a second, glass tube. Two milliliters of water and 1 mL of 15 mm FeCl2 were added, and the tubes were heated for 20 min in a boiling water bath. The tubes were centrifuged as above to precipitate the iron-phytic acid complex and the supernatant was discarded. The pellets were resuspended in 2 mL of 0.2 n HCl and heated for 10 min in the boiling water bath. The precipitate was again removed by centrifugation and resuspended in 2 mL of 5 mm EDTA. Fifteen microliters of the EDTA suspension was taken for wet ashing to obtain phytic acid phosphate. To each tube containing the 15-μL aliquot was added 10 μL of a 10% (w/v) solution of calcium nitrate. The water was allowed to evaporate, and the residue was heated in a flame until a white ash remained in the tube. The ash was dissolved in 0.3 mL of 0.5 n HCL and heated at 90°C for 20 to 30 min. Acid molybdate reagent (0.7 mL of 0.36% [w/v] ammonium molybdate and 1.42% [w/v] ascorbic acid in 0.86 n sulfuric acid) was added and the color was allowed to develop for 1 h at 37°C before being read at 820 nm.
Inorganic phosphate was determined by adding 0.3 mL of 0.5 n HCl and 0.7 mL of the phosphate color reagent to either 20- or 40-μL aliquots of the initial 5-mL extract. Potassium phosphate standards were developed at the same time.
cDNA Cloning of the Wild-Type Soybean myo-Inositol 1-Phosphate Synthase
A cDNA library prepared from developing soybean seeds (Hitz et al., 1994) was used to infect Escherichia coli BB4 cells. Duplicate plaque lifts were made onto nitrocellulose filters (Schleicher & Schuell, Keene, NH) and the filters were prehybridized in 25 mL of hybridization buffer consisting of 6× SSPE, 5× Denhardt's solution, 0.5% (w/v) SDS, 5% (w/v) dextran sulfate, and 0.1 mg mL−1 denatured salmon sperm DNA (Sigma, St. Louis) at 60°C for 2 h.
The blocked filters were then hybridized to a radiolabeled probe made from a cDNA from Arabidopsis that had been identified as a myo-inositol-1-phosphate synthase by homology to yeast myo-inositol-1-phosphate synthase (Johnson and Sussex, 1995). The Arabidopsis clone was obtained from the Arabidopsis Biological Resource Center, DNA Stock Center (Columbus, OH), clone number 181C18T7113E9T7. The 1.2-kB cDNA insert was removed from the vector DNA by digestion with SalI and Not and radiolabeled with [32P]dCTP with a random primer labeling kit (Bethesda Research Laboratory, Gaithersburg, MD). The filters were allowed to hybridize overnight under the same conditions as described for prehybridization and finally washed in 0.2× SSC and 0.1% (w/v) SDS at 60°C. Approximately 200 positive signals were observed, and of these, six were purified by excision of the area around the signal, replating the phage, and rescreening as above. Two clones were excised to phagmids and used to infect E. coli to obtain plasmid clones using the protocols described by the manufacturer (Stratagene, La Jolla, CA). Of the two clones, one designated p5bmi-1-ps was sequenced using Applied Biological Instruments (PerkinElmer, Foster City, CA) methodology and equipment.
cDNA Cloning of myo-Inositol 1-Phosphate Synthase from Immature Seeds of LR33 Soybeans
Seeds from the LR33 plants were harvested at about 50% through the seed filling period, removed from the pod, and stored frozen at −80°C. Total RNA was isolated by the cetyltrimethylammonium bromide method (Sambrook et al., 1989), and mRNA was purified by oligo(dT) binding using an mRNA purification kit (Pharmacia, Piscataway, NJ).
Thirteen nanograms of the polyadenylated mRNA was used as template for amplification from oligo(dT) using a GeneAmp RNA-PCR kit (part no. N808–0017, PerkinElmer Cetus, Boston). The reverse transcriptase reaction was run for 30 min at 42°C. For the PCR amplification, Vent DNA polymerase (New England Biolabs, Beverly, MA) was substituted for the DNA polymerase supplied by the kit manufacturer and an additional 2 μL of 100 mm magnesium sulfate was added to each 100-μL reaction. The 5′ primer had the sequence 5′-GGGAATTCCATATGTTCATCGA-GAATTTTAAGGTT-3′, wherein the underlined ATG is the translation start of soybean m-I-1PS and the additional 5′ bases are added to encode an NdeI site. The 3′ primer had the sequence 5′-AAGGAAAAAAGCGGCCGCTCACTTGTACTCGAGAATCAT-3′, which is the reverse complement of the 3′ end of the soybean mI-1PS sequence plus added bases to provide a NotI site and additional bases to enhance restriction digestion. The PCR reaction was run for 35 cycles at a 52°C annealing temperature and 1.5-min extension time. A product of about 1,550 bp was obtained and purified by passage through a 50 microfuge filter (Amicon, Beverly, MA) followed by extraction with an equal volume of 1:1 (v/v) phenol:chloroform, extraction of the upper layer of the phenol-chloroform separation with 1 volume of chloroform, and precipitation with ethanol. Five micrograms of the resulting PCR product was digested overnight at 37°C with both NdeI and NotI. The restriction enzyme digest was deproteinized and ligated into 2 μg of pET24a T7 expression vector (Novagen, Madison, WI) that had also been digested with NdeI and NotI and treated with calf intestine alkaline phosphatase. The ligation mixture was used to transform electrocompetant DH 10B E. coli cells and plasmid was purified from confirmed transformants. Plasmid preparations from six transformants were used to transform electrocompetant DE 3 E. coli cells for protein expression.
Functional Expression of the myo-Inositol-1-Phosphate Synthase from Wild-Type and LR33 Soybeans in E. coli
Myo-inositol-1-phosphate synthase from p5bmi-1-ps was placed into the pET24a T7 expression vector by PCR amplification of p5bmi-1-ps using the PCR primers described for RT-PCR amplification and used to transform DE E. coli cells. Wild-type and LR33-derived clones were screened for mI-1-PS activity using an assay consisting of conversion of 33P-labeled Glc-6-phosphate to 33P-myo-inositol-1-phosphate. Active clones from both the wild-type and LR33 groups were chosen on the basis of this screen for further analysis. Residue K396 of the wild-type Gm mI-1PS sequence was converted to N396 by site-directed mutagenesis using a PCR primer set with the forward primer corresponding to bases 1,165 to 1,201of Gm mI-1PS in which base 1,188 was changed from G to T and the reverse primer was the 3′-end primer with the added NotI site described above. The reaction produced a 360-bp product with an existing SnaBI site 16 bases 5′ to the introduced mutation. The PCR product and pET24a plasmid containing wild-type Gm mI1-PS sequence were digested with SnaBI and NotI followed by agarose gel purification of the digested PCR product and the pET24a vector with the 360-bp SnaBI to NotI fragment removed. The PCR product was ligated back into the pET24a Gm mI1-PS fragment to give the sequence with the single G to T change at 1,188.
Fifty-milliliter cultures in M9 (Sambrook et al., 1989) media supplemented with 0.2% (w/v) Glc, 2 mg mL−1 casamino acids (DIFCO Laboratories, Detroit), and 30 mg L−1 kanamycin were started from single colonies and grown for 10 h at 28°C. Cultures were made to 1 mm in isopropyl thiogalactoside at between 1.5 and 1. 8 OD at 600 nm and transferred to 37°C for 3 h. Cells were harvested by centrifugation and resuspended in 0.6 mL of 50 mm Tris-HCl at pH 8.0 containing 1 mm dithiothreitol, 0.5 mm EDTA, and 0.2 mm phenyl methylsulfonyl fluoride. The cells were disrupted by sonication and centrifuged at 12,000g for 15 min to remove insoluble debris. The supernatant was made to 30% saturation with solid ammonium sulfate, the resulting precipitate was removed by centrifugation, and the resulting supernatant was adjusted to 50% saturation in ammonium sulfate. The precipitate was removed by centrifugation and redissolved in 0.4 mL of 5 mm Tris-HCl at pH 8.0 with 1 mm 2-mercaptoethanol.
Myo-inositol 1-phosphate synthase activity was measured as the disappearance of Glc 6-phosphate in a 100-μL assay, which was 0.32 mm in Glc 6-phosphate, 0.4 mm in NAD, and 15 mm in ammonium acetate, all in the starting elution buffer. Twenty micrograms of protein from each of the three expression mixtures and a DE3 cell-only culture were incubated for 20 min in the reaction mix before 1 mL of a Glc 6-phosphate assay mix consisting of 2 units of Glc 6-phosphate dehydrogenase (G7877, Sigma), and 1 mm NADP was added. A340 was measured after 5 min at room temperature. The reaction rate was determined after subtraction of the small blank reaction from the DE3 cell extract control. Ten micrograms of protein of protein from each ammonium sulfate fractionated culture was separated by SDS-PAGE and Coomassie Blue stained to estimate relative purity of the 55-kD Gm mI 1-PS protein.
Construction of cDNA Libraries and EST Analysis
cDNA libraries were constructed using standard methods (Sambrook et al., 1989) typically using the Lambda Zap II kit (Stratagene). mRNA representing a variety of tissue types was isolated from soybean grown under various conditions. Libraries were converted into plasmid libraries according to the protocol provided by Stratagene. cDNA inserts from randomly picked bacterial colonies containing recombinant pBluescript plasmids were amplified via PCR using primers specific for vector sequences flanking the inserted cDNA sequences or plasmid DNA was purified from randomly selected colonies using R.E.A.L Prep 96 System (QIAGEN, Valencia, CA). Amplified insert DNAs or plasmid DNAs were sequenced in either dye-primer sequencing or dye terminator reactions to generate partial cDNA sequences (ESTs; Adams et al., 1991). The resulting ESTs were analyzed using a fluorescent sequencer (model 377, PerkinElmer, Norwalk, CT). Three hundred to 10,000 clones were sequenced per library. Sixty-two soybean cDNA libraries were sampled. Typically, libraries continued to be sampled at least until the percentage of novel genes was less then 30%. Over 150,000 ESTs of soybean were created and queried, but only ESTs from libraries that had not been normalized before sequencing were included in the analysis. All sequences were used to query the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov/Entrez/) using the BLAST program. Approximately 85% of the sequences were analyzed using both BLASTX and BLASTN (Altshul et al., 1990), and 15% of the sequences were analyzed using Gapped BLASTX and Gapped BLASTN (Altshul et al., 1997).
Footnotes
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010585.
LITERATURE CITED
- Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RF et al. Complementary DNA sequencing: expressed sequence tags and human genome project. Science. 1991;252:1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- Altshul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altshul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L, Alonson-Blanco C, Vreugdenhil D, Tesnier K, Groot S, Koorneef M. Genetic analysis of seed-soluble oligosaccharides in relation to seed storability of Arabidopsis. Plant Physiol. 2000;124:1595–1604. doi: 10.1104/pp.124.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brearley CA, Hanke DE. Metabolic evidence for the order of addition of individual phosphate esters to the myo-inositol moiety of inositol hexakisphosphate in the duckweed Spirodela polyrhizaL. Biochem J. 1996a;314:227–233. doi: 10.1042/bj3140227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brearley CA, Hanke DE. Inositol phosphates in barley (Hordeum vulgare L.) aleurone tissue are stereochemical similar to the products of breakdown of Ins P6 in vitroby wheat bran phytase. Biochem J. 1996b;318:279–286. doi: 10.1042/bj3180279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromwell G, Traylor S, Lindermann M, Stilborn H, Sauber T. Bioavailability of phosphorus in low oligosaccharide, low-phytate soybean meal for chicks. Poultry Sci Suppl. 2000a;79:127. [Google Scholar]
- Cromwell G, Xavier E, Souza L, Lindemann M, Sauber T. Effects of low-phytate corn and low-oligosaccharide, low-phytate soybean meal on performance bone traits and phosphorus excretion by growing chicks. Poultry Sci Suppl. 2000b;79:22. [Google Scholar]
- Crowe JH, Crowe LM, Carpenter JF, Wistrom CA. Stabilization of dry phospholipid bilayers and proteins by sugars. Biochem J. 1987;242:1–10. doi: 10.1042/bj2420001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitz WD, Carlson TJ, Booth JR, Kinney AJ, Stecca KL, Yadav NS. Cloning of a higher-plant plastid ω-6 fatty acid desaturase cDNA and its expression in a cyanobacterium. Plant Physiol. 1994;105:635–641. doi: 10.1104/pp.105.2.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MD, Sussex IM. 1l-myo-Inositol 1-phosphate synthase from Arabidopsis thaliana. Plant Physiol. 1995;107:613–619. doi: 10.1104/pp.107.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster KL, Leopold AC. Sugars and desiccation tolerance in seeds. Plant Physiol. 1988;88:829–832. doi: 10.1104/pp.88.3.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo TM, VanMiddlesworth JF, Wolf WJ. Content of raffinose oligosaccharides and sucrose in various plant seeds. J Agric Food Chem. 1988;36:32–36. [Google Scholar]
- Larson S, Rutger J, Young K, Raboy V. Isolations and genetic mapping of an non-lethal rice low phytic acid Imutation. Crop Sci. 2000;40:1397–1405. [Google Scholar]
- Larson S, Young K, Cook A, Blake T, Raboy V. Linkage mapping of two mutations that reduce phytic acid content of barley grain. Theor Appl Genet. 1998;97:141–146. [Google Scholar]
- Lehele L, Tanner W. The function of myo-inositol in the biosynthesis of raffinose. Eur J Biochem. 1973;38:103–110. doi: 10.1111/j.1432-1033.1973.tb03039.x. [DOI] [PubMed] [Google Scholar]
- Loewus FA, Murthy PPN. myo-Inositol metabolism in plants. Plant Sci. 2000;150:1–19. [Google Scholar]
- Loewus MW, Loewus F. The isolation and characterization ofd-glucose 6-phosphate cycloaldolase (NAD-dependent) from Acer pseudoplatanusL. cell cultures. Plant Physiol. 1971;48:255–260. doi: 10.1104/pp.48.3.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obendorf RL. Oligosaccharides and galactosyl cyclitols in seed desiccation tolerance. Seed Sci Res. 1997;7:63–74. [Google Scholar]
- Raboy V, Dickinson D. The timing and rate of phytic acid accumulation in developing soybean seeds. Plant Physiol. 1987;85:841–844. doi: 10.1104/pp.85.3.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raboy V, Dickinson D, Below F. Variation in seed total P: phytic acid, zinc, calcium, magnesium, and protein among lines of Glycine max and G. sojae. Crop Sci. 1984;24:431–434. [Google Scholar]
- Raboy V, Gerbasi P, Young K, Stoneberg S, Pickett S, Bauman A, Murthy P, Sheridan W, Ertl D. Origin and Seed Phenotype of Maize low phytic acid 1-1 and low phytic acid 2-1. Plant Physiol. 2000;124:355–368. doi: 10.1104/pp.124.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sebastian SA, Kerr PS, Pearlstein RW, Hitz WD. Soybean germplasm with novel genes for improved digestibility. In: Drackley JK, editor. Soy in Animal Nutrition. Savoy, IL: Federation of Animal Science Societies; 2000. pp. 56–74. [Google Scholar]
- Spencer J, Allee G, Frank J, Sauber T. Use of low-phytate corn and low-phytate/low-oligosaccharide soybean meal in broiler diets reduces phosphorus excretion. Poultry Sci Suppl. 2000a;79:11–12. [Google Scholar]
- Spencer J, Allee G, Frank J, Sauber T. Nutrient retention and growth performance of pigs fed diets formulated with low-phytate corn and/or low-phytate/low oligosaccharides soybean meal. J Anim Sci Suppl. 2000b;78:73. [Google Scholar]
- Wilcox J, Premachandra G, Yound K, Raboy V. Isolation of high seed inorganic P, low-phytate soybean mutants. Crop Sci. 2000;40:1601–1605. [Google Scholar]