Abstract
Purification of abscisic acid (ABA)-binding proteins is considered to constitute a major step toward isolating ABA receptors. We report here that an ABA-binding protein was for the first time, to our knowledge, purified from the epidermis of broad bean (Vicia faba) leaves via affinity chromatography. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis, isoelectric focusing electrophoresis, and isoelectric focusing/sodium dodecyl sulfate-polyacrylamide gel electrophoresis two-dimensional electrophoresis of the purified ABA-binding protein all identified a single protein band with a molecular mass of 42 kD and an isoelectric point 4.86. The Scatchard plot for the purified protein showed a linear function with a maximum binding activity of 0.87 mol mol−1 protein and an equilibrium dissociation constant of 21 nm, indicating that the purified protein may be a monomeric one, possessing one binding site. The ABA-binding protein was enriched more than 300-fold with a yield of 14%. (−)ABA and trans-ABA were substantially incapable of displacing 3H-(±)ABA bound to the ABA-binding protein, and (±)ABA was less effective than (+)ABA in the competition. These findings allow establishment of the stereospecificity of the 42-kD protein and suggest its ABA receptor nature. Pretreatment of the guard cell protoplasts of broad bean leaves with the monoclonal antibody raised against the 42-kD protein significantly decreased the ABA specific-induced phospholipase D activity in a dose-dependent manner. This physiological significance provides more clear evidence for the potential ABA-receptor nature of the 42-kD protein.
Abscisic acid (ABA) plays a major role in various aspects of plant growth and development, including seed maturation and germination, adaptation to environmental stresses, and fruit development (for reviews, see Coome, 1976, 1992; Zeevaart and Creelman, 1988; McCarty, 1995; Rock and Quatrano, 1995; Leung and Giraudat, 1998). ABA signal transduction has been extensively studied in the past years (Giraudat et al., 1992, 1994; Leung et al., 1994, 1997; Meyer et al., 1994; Bertauche et al., 1996; Cutler et al., 1996; Ingram and Bartels, 1996; Merlot and Giraudat, 1997; Finkelstein et al., 1998; Leung and Giraudat, 1998; Leube et al., 1998; Rodriguez et al., 1998a, 1998b; Sheen, 1998; Gosti et al., 1999; Li et al., 2000). Investigations on ABA-induced stomatal movement have led to considerable progress in understanding the ABA signaling pathway, revealing the involvement of second messengers such as Ca2+, IP3, and reversible protein phosphorylation (Gehring et al., 1990; Gilroy et al., 1990; Meyer et al., 1994; Allen et al., 1995; Allen and Sanders, 1995; Lee et al., 1996; Li and Assmann, 1996; Leung and Giraudat, 1998; Li et al., 2000). Explorations on more downstream elements of ABA signaling in stress responses, especially drought and cold tolerance, identified numerous ABA responsive cis-acting elements and trans-acting factors (Ingram and Bartels, 1996; Merlot and Giraudat, 1997; Leung and Giraudat, 1998). However, ABA signal perception, a key step in the signal transduction, remains unknown.
Studies on ABA perception so far have mainly focused on the biochemical analysis of ABA-binding proteins or ABA-binding sites that are considered putative ABA receptors. As early as the 1970s, ABA-binding sites was reported to be present in subcellular fractions of broad bean (Vicia faba) leaves (Hocking et al., 1978). Hornberg and Weiler (1984) detected high-affinity, specific ABA-binding sites on plasmalemma of guard cell protoplasts of broad bean. More recent studies reported that ABA receptors might also exist inside guard cells (Allan et al., 1994; Anderson et al., 1994; MacRobbie, 1995). Using some conventional methods such as the labeled-ligand technique, some studies were carried out on biochemical analysis of ABA-binding proteins with various plant tissues (Pedron et al., 1998; Zhang et al., 1999, 2001a). These studies provided some useful information for characterization of ABA-binding proteins. Nevertheless, the ABA-binding proteins have not been purified to date, and the genes of ABA receptors have not been cloned. Purification of ABA-binding proteins will constitute a major step toward isolating ABA receptors and elucidating the mechanism of ABA signal perception. We report here that using an specially designed ABA-linked AEH-Sepharose 4B as the affinity chromatography medium, a selected plant tissue, and a series of correspondingly modified procedures, an ABA specific-binding protein of 42 kD has been purified and identified from broad bean leaves for the first time, to our knowledge.
RESULTS
Purification
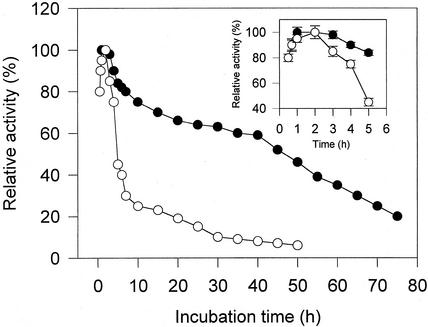
A preliminary experiment showed that the lower epidermis of broad bean leaves, more enriched with ABA-binding proteins, was a suitable tissue for purification of ABA-binding proteins probably because of its high content of ABA-responsive guard cells (all data not shown; see Fig. 6 and Table II). For extraction of the ABA-binding proteins, three methods were compared. Results shown in Table I demonstrated that the ABA-binding activity was much higher in the crude extract solubilized only with the detergent 0.5% (v/v) Triton X-100 (method 1) than that in the crude extract concentrated with the cold acetone (method 2) or than that in the crude extract salted out with ammonium sulfate (method 3). As shown in Figure 1, the results indicated that ABA was bound rapidly to the ABA-binding proteins solubilized only by 0.5% (v/v) Triton X-100, with 80% of the maximum being attained within 30 min, a subsequent 3-h duration of the maximum binding (Fig. 1), and a period of relatively higher (>60%) and stable binding activity that was maintained from the 10th to 40th h (Fig. 1). But the duration of the maximum activity of ABA-binding protein extracted with the cold acetone was short (only 30 min approximately), and the subsequent binding decrease was rapid (Fig. 1). The ABA-binding proteins salted out with ammonium sulfate showed a similar time course of the binding activity (data not shown) to the cold acetone extracted ABA-binding proteins. So the solubilization with 0.5% (v/v) Triton X-100 only (method 1) was adopted for the ABA-binding protein extraction from the epidermis of the leaves.
Figure 6.
Scatchard plots of ABA-binding activities to the crude (a) and purified (b) proteins. B, 3H-ABA bound; F, 3H-ABA free. For the crude proteins (a), all points except for the first two were fitted with a linear relationship (r2 = 0.98), and according to the Scatchard plot, the maximum binding activity (Bmax) and the equilibrium dissociation constant (Kd) were calculated: Bmax = 68 nmol g−1 protein and Kd = 19 nm. The fitted relationship of the purified protein (b) was also linear (r2 = 0.99) with Bmax = 0.87 mol mol −1 protein and Kd = 21 nm. Points represent the means ± sd of five determinations.
Table II.
Summary of the purification of the crude ABA-binding proteins from the lower epidermis of bean leaves
| Purification Step | Protein | Bmax | Kd | ABA-Binding Activity | Yield | Purification |
|---|---|---|---|---|---|---|
| mg | nmol g−1 protein | nm | nmol | % | -fold | |
| Crude extra | 2,531.2 | 68 | 19 | 172 | 100 | 1 |
| Affinity | 1.17 | 20,700 | 21 | 24 | 14 | 304 |
| Chromatography |
Bmax, Maximum ABA-binding activity; Kd, equilibrium dissociation constant.
Table I.
Effects of different extraction methods on the 3H-ABA binding to the crude extract proteins
| Extraction Methods | Specific Binding Activity | Relative Binding Activity |
|---|---|---|
| nmol g−1 protein | % | |
| 0.5% (w/v) Triton X-100 | 0.487 ± 0.025 | 100 |
| Cold acetone | 0.325 ± 0.028 | 66 |
| 80% (w/v) (NH4)2SO4 | 0.223 ± 0.032 | 45 |
Values are means ± sd of five determinations.
Figure 1.
Time course of 3H-ABA binding to the crude ABA-binding proteins extracted with two different methods. Extraction with the buffer containing 0.5% (w/v) Triton X-100 only as the solubilizing medium (●). Extraction first with 0.5% (w/v) Triton X-100 and then with the cold acetone (○). The crude extracts (equivalent to 10 μg of protein) were incubated at 4°C in the buffer containing 70 nm 3H-(±)ABA with the different incubation time. The 3H-ABA binding was counted after each incubation. The time course of 3H-ABA binding during the first 5 h is shown more clearly in the inset. Points in the figure represent the means of three determinations, and those in the inset represent means ± sd of three determinations.
The pH of both the extraction medium and the elution medium was critical. We used a medium pH 6.5 because the maximum ABA-binding activity was attained at this pH value (data not shown for the crude extract).
An appropriate affinity-chromatography column should be of special importance. We chose EAH-Sepharose 4B as the affinity medium. The EAH-Sepharose 4B contains a 10-atom spacer arm to which a free amino group was conjugated. Therefore, ABA molecules can be coupled through their carboxyl groups at C1 to the free amino groups located in the spacer arm of EAH-Sepharose 4B by the coupling reagent 1-ethyle-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride. It should be particularly noted that a high coupling efficiency of ABA to EAH-Sepharose 4B was a key step for the successful purification of ABA-binding proteins. The coupling efficiency we obtained reached approximately 80%, being about 6 to 8 μmol linked ABA mL−1 drained EAH-Sepharose 4B containing 7 to 11 μmol of active amino groups.
NaCl was used to remove the nonspecific-bound proteins. The effects of NaCl (0–500 mm) on ABA-binding protein activity was assayed to determine a suitable concentration of NaCl in the elution buffer. The results shown in Figure 2 indicated that there was no significant inhibiting effect of NaCl from 0 to 100 mm on ABA-binding protein activity and that the effect on ABA-binding protein activity by NaCl concentrations between 100 and 300 mm was small, with a relative activity at 300 mm NaCl being still 65% of the control. However, the decrease in the activity above 300 mm NaCl was sharp. Therefore, we adopted a concentration gradient of NaCl from 100 to 300 mm for the purification.
Figure 2.
Effects of NaCl on ABA-binding activity in the crude extracts. A range of NaCl concentrations from 0 to 500 mm was tested. ABA-binding activity of the control (without NaCl) was taken as 100%. Points are means ± sd of three determinations.
The elution curve of proteins is shown in Figure 3. The affinity chromatography column was first thoroughly washed with NaCl at concentrations 100 mm (400 mL), 150 mm (200 mL), 250 mm (100 mL), and 300 mm (300 mL), respectively, until the A280 of the effluent became almost zero. In this step the nonspecific-bound proteins were efficiently removed, but the ABA-binding activity was practically not taken off from the column, which demonstrated that our affinity chromatography column was effective in removing the nonspecific-binding proteins. The subsequent elution with 1 mm ABA in the presence of 150 mm NaCl eluted a small amount of protein with high ABA-binding activity (Fig. 3), suggesting that the ABA-binding proteins present in a low quantity were efficiently eluted. The purification procedures can be summarized as follows: crude extraction with 0.5% (v/v) Triton X-100 → wash of the unspecific-binding proteins with NaCl → affinity elution with ABA → Sephadex G-25 chromatography to remove free ABA → ultrafiltration for concentrating the purified proteins.
Figure 3.
Affinity chromatography on the ABA-linked EAH-Sepharose 4B column of the crude ABA-binding proteins. The crude ABA-binding proteins for affinity chromatography were solubilized only with Triton X-100 contained in the buffer described in “Materials and Methods” (method 1). After equilibration, the column was first eluted with a total volume of 1,000 mL MES/NaCl (pH 6.5) buffer containing NaCl in a step concentration gradient from 100 to 300 mm, and then the ABA-binding protein was eluted with 300 mL of the same buffer containing 1 mm (±)ABA and 150 mm NaCl. NaCl concentrations (dashed line). Elution profile expressed on the optical density at 280 nm (solid line). Relative 3H-ABA-binding activity of the eluted protein (○), taking the maximum binding activity obtained at the point of about 1,150 mL of the elution volume, as 100%. Points for the relative 3H-ABA-binding activity are means ± sd of three determinations.
Purity, Molecular Mass, and pI
The result of SDS-PAGE of the crude extract and purified ABA-binding protein is shown in Figure 4A. The purified ABA-binding protein migrated as a single protein band on SDS-PAGE (Fig. 4A, lane b). It had an estimated molecular mass of 42 kD and possessed a high degree of purity (Fig. 4A). The most shaded band with a molecular mass of 68 kD on the lane c of the gel (Fig. 4A) is that of bovine serum albumin (BSA) added to the extraction medium. This band disappeared from the purified ABA-binding protein lane (Fig. 4A, lane b), which is an additional measure of the purity and specificity of the ABA-binding proteins from the affinity column. As shown in Figure 4B, the native isoelectric focusing (IEF) electrophoresis of the purified ABA-binding protein also detected a single protein band with a native pI of 4.86. In Figure 4C, IEF/SDS-PAGE two-dimensional gel electrophoresis, which is of higher resolution and more accurate than either alone, showed further a substantial single protein band for the ABA-binding protein. It is particularly noteworthy that the multiblot of the ABA-binding protein on the gel (Fig. 4C) was probably due to the presence of urea in IEF sample buffer. In the preparation of the first dimension sample, urea was occasionally exposed to relatively high temperature or high pH that could induce a decomposition of the urea into isocyanate, then the isocyanate could result in a carbamylation of amino acid residues possessing positive charges that could be displaced by carbamyls from the peptide. As a result, the positive charges of the peptide were decreased by heterogeneous carbamylation, which led to the “carbamylation train” of the same peptide during IEF in the first dimension (Carbamylation calibration kit for 2D electrophoresis, Pharmacia AB, Uppsala; Smith et al., 1991). So it is true that the ABA-binding protein migrated as a substantial single band of the IEF in spite of the appearance of the multiblot. The measured molecular mass and pI were, respectively, 42 kD and 4.86 (Fig. 4C), identical with that obtained respectively in SDS-PAGE and native IEF.
Figure 4.
Commassie blue-stained SDS-PAGE (A), silver-stained native IEF (B), and silver-stained IEF/SDS-PAGE (C) of the purified ABA-binding protein. In A: a, molecular mass standards; b, purified ABA-binding proteins (3 μg), of which the calculated molecular mass was 42 kD; c, proteins in the crude extract (15 μg). In B: a, the purified ABA-binding protein, of which the measured pI was 4.86; b, the protein standards. C, The purified ABA-binding protein (2 μg) was resolved by IEF in the first dimension followed by SDS-PAGE; a, molecular mass markers; b, the purified ABA-binding protein. The measured molecular mass and pI were, respectively, 42 kD and 4.86.
Kinetic Properties
In Figure 5, the 3H-ABA binding to the crude proteins (Fig. 5a) and the purified protein (Fig. 5b) were shown to be both saturable with increasing 3H-ABA concentrations. The nonspecific binding was lower than 10% and was linear (Fig. 5, a and b). In Figure 6a, the Scatchard plot for the crude proteins seems to show two different linear functions, suggesting that more than one type of ABA-binding protein may exist in the crude extracts. Anyway all points except for the first two can be well fitted with a linear relationship (r2 = 0.97), from which were calculated a maximum ABA-binding activity (Bmax) of 68 nmol g−1 protein and an equilibrium dissociation constant (Kd) of 19 nm (Fig. 6a). However, the Scatchard plot for the purified protein illustrated a linear function, with a Bmax of 0.87 mol mol −1 protein and a Kd of 21 nm (Fig. 6b). The linear Scatchard plot of the purified protein suggests that the purified protein contains only one type of ABA-binding protein. The Kd of the purified ABA-binding protein (21 nm) and that of the ABA-binding protein in the crude extract (19 nm) were comparable, suggesting that the two proteins may be the same. In Table II, a comparison of the Bmax of the purified ABA-binding protein (0.87 mol mol −1 protein = 20,700 nmol g−1 protein) with that of the crude protein (68 nmol g−1 protein) indicates an enrichment of more than 300-fold of the ABA-binding protein after the purification, with a yield of purification being 14%.
Figure 5.
Saturation curves (●) of the ABA-binding activities to the crude (a) and purified ABA-binding proteins (b). 3H-(±)ABA at a gradient of concentrations was added to the incubation medium. The free 3H-ABA concentrations were adjusted to 3H-(+)ABA concentrations (one-half of the 3H-[±]ABA concentrations). As indicated in both of the figures above, the nonspecific binding (○) was determined with 1,000-fold molar excess of cold ABA and was lower than 10%. Points are means ± sd of five determinations.
pH Dependence and Bivalent Cation Requirement
As shown in Figure 7A, the ABA binding to the purified ABA-binding protein was highly sensitive to the pH of incubation medium. The optimum pH for the ABA binding was 6.5. Whether below or above this optimum pH, the ABA-binding activity was sharply reduced (Fig. 7A). The ABA binding to the prepurified ABA-binding proteins showed the same pH dependence (data not shown). Thus, as mentioned previously, a rigorous pH at 6.5 is needed for the purification of ABA-binding proteins.
Figure 7.
Effects of pH (A) and Ca2+ and Mg2+ (B) on ABA-binding activity to the purified ABA-binding protein. A, The pH of incubation medium was modulated linearly from 4 to 8 for binding assays. The ABA-binding activity attained maximum at pH 6.5, and this maximum binding was taken as 100%. Assays were performed at 4°C for 1 h. Points are means ± sd of five assays. B, CaCl2 or MgCl2 at a linear concentration gradient from 0 to 10 mm were added in the incubation medium containing 250 mm mannitol, 10 mm Tris/MES (pH 6.5), 70 nm 3H-(±)ABA and 30 ng of purified protein. ABA-binding activity at 2 mm Ca2+ or Mg2+ was taken as 100%. Points are means ± sd of three determinations.
Ca2+ and Mg2+ are two well-known bivalent cations that play important roles in the regulation of many enzyme activities (Fersht, 1985). In Figure 7B, the results of the effects of Ca2+ and Mg2+ on ABA binding to the purified ABA-binding protein showed that the ABA-binding activity can be stimulated by both Ca2+ and Mg2+ at relatively lower concentrations (below 2 mm), but higher concentrations than 2 mm of Ca2+ or Mg2+ did not show any correspondingly stronger effects.
Stereospecificity
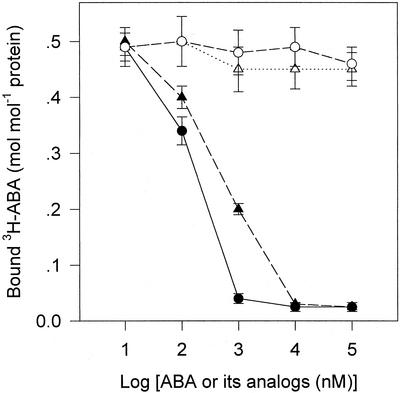
In Figure 8, (−)ABA and trans-ABA were shown to have scarcely competing effects on 3H-(±)ABA binding to the purified ABA-binding protein. In the incubation buffer containing only 70 nm 3H-(±)ABA, the binding was not significantly reduced even by 104 to 105 nm (−)ABA or trans-ABA (more than 140–1,400 times as much as 3H-[±]ABA), whereas the 3H-(±)ABA binding was significantly reduced by only 100 nm unlabeled (+)ABA or (±)ABA, and nearly completely displaced by 103 nm (+)ABA or 104 nm (±)ABA. (±)ABA was shown to be less effective in the competition than (+)ABA (Fig. 8), which was probably because of lower levels of (+)ABA in the only (±)ABA-contained binding medium.
Figure 8.
Influence of two ABA analogs on ABA-binding activity to the purified ABA-binding protein. (−)ABA (○), trans-ABA (▵), taking (±)ABA (▴), and (+)ABA (●) as the controls. Points are means ± sd of five determinations.
Effect of ABA-Binding Protein Antibody on Phospholipase D (PLD) Activity of Guard Cell Protoplasts Treated by ABA
In Figure 9, the level of (7-nitro-2–1,3-benz-oxadiazol-4-yl)amino-phosphatidylbutanol (NBD-PtdBut) production as a measure of PLD activity, by the guard cell protoplasts not treated with (±)ABA (nor with the antibody against the purified ABA-binding protein) and using NBD-phosphatidyl-choline (PtdCho) and 1-butanol (buOH) as the substrates, was shown to be low, but 10 μm (±)ABA treatment enhanced more than five times the NBD-PtdBut production by the protoplasts in the case that the protoplasts had not been pretreated with the antibody. However, the pretreatment of the protoplasts with the antibody against the purified ABA-binding protein led a substantial linear reduction of this enhanced-NBD-PtdBut level with increasing amounts of the antibody added in the protoplasts reaction mixture, and the pretreatment with 40 to 50 μg of antibody reduced the NBD-PtdBut production up to approximately the same level as that by the protoplasts not treated with (±)ABA nor with antibody (Fig. 9). In contrast, the pretreatment of the protoplasts with either preimmune mouse IgG or BSA had no significant effect on the NBD-PtdBut production enhanced by the treatment with (±)ABA (Fig. 9).
Figure 9.
The effect of the monoclonal antibody raised against the purified ABA-binding protein on in vivo levels of PtdBut production induced by ABA in guard cell protoplasts of broad bean leaves. The protoplasts were pretreated with the antibody, incubated in 0.5 mg mL−1 NBD-PtdCho, treated with 0.1% (v/v) 1-buOH, and then incubated in 10 μm (±)ABA for 20 min. The levels of NBD-labeled PtdBut were determined according to Ritchie and Gilroy (1998), from protoplasts pretreated with different amounts of the ABA-binding protein antibody (●) with those of either preimmune mouse IgG (○) or BSA (▵). The point ▴ represents NBD-PtdBut level from the protoplasts treated with neither(±)ABA nor the antibody (and treated with neither preimmune mouse IgG nor BSA). Points are means ± sd of five determinations.
DISCUSSION
Whereas auxin- and cytokinin-binding proteins were purified many years ago (Selivankina et al., 1982; Romanov et al., 1986; Shimomura et al., 1986; Mitsui and Sugiura, 1993), there is no report on successful purification of ABA-binding proteins to date (see Leung and Giraudat, 1998). The lower abundance and higher sensitivity to various factors of ABA-binding proteins may constitute the major obstacles. Because of this, an appropriate plant material and a suitable affinity chromatography medium should be of primary importance for an efficient purification. The lower epidermis should be an ideal material because it is one of the most important target tissues of ABA and therefore may have more ABA-binding sites. As a matter of fact, the Bmax detected in this research in the crude extract of lower epidermis of broad bean leaves (68 nmol g−1 protein, see Table II) is much higher than that in the membranous preparations of whole leaf of broad bean (3.5 × 10−3 nmol g−1 protein; Hocking et al., 1978). As the affinity medium, EAH-Sepharose 4B was adopted in the present study instead of the conventionally used cyanogen bromide-activated Sepharose 4B. EAH-Sepharose 4B possess a 10-atom spacer arm that may be able to provide a better spacing effect for the efficient reaction of ABA-binding proteins with the coupled ABA to this arm. Such an ABA-linked EAH-Sepharose 4B and a high coupling efficiency of ABA to this gel (about 80%) may greatly improve the purification efficiency. Furthermore, the lower epidermis tissue with concentrated ABA-binding proteins and the efficient affinity column allowed also a reduction in the number of steps for the protein purification, which could avoid the possible denaturation or loss of binding activity of these fragile ABA-binding proteins.
The successful purification of ABA-binding protein in the present study showed that an efficient extraction, an optimum pH in the extraction and purification, and an appropriate concentration of NaCl used in elution were also important for the purification. The cold acetone washing, ammonium sulfate precipitation, and Triton X-100 washing were often used in the extraction of plant hormone-binding proteins (Shimomura et al., 1986; Sugaya and Sakai, 1996; Brault et al., 1997, 1999), among which we adopted Triton X-100 as an unique washing medium because both the cold acetone washing and ammonium sulfate precipitation were shown to reduce or even to abolish the ABA-binding activity. This may be because the ABA-binding proteins are generally membrane-bound proteins (Hocking et al., 1978; Hornberg and Weiler, 1984; Anderson et al., 1994; MacRobbie, 1995; Pedron et al., 1998; Zhang et al., 1999), and the membrane lipids may be important to maintain their functional conformation. It is likely that Triton X-100, as a mild nonionic detergent, can partly take the place of the membrane lipids (Hjelmeland, 1984) to supply the ABA-binding proteins with a suitable lipid environment. The suitable concentration of Triton X-100 was determined as 0.5% (w/v) for both a higher ABA-binding activity and a higher protein yield.
Regarding the NaCl washing of the affinity column, a higher concentration than 300 mm of NaCl was shown to be harmful to ABA-binding activity, therefore the NaCl concentration during elution should be below this value. The purification of the ABA-binding proteins should also meet the needs of the marked pH dependence of the proteins, of which the optimum is at 6.5. It is finally noteworthy that the ABA concentration in the elution buffer is also of importance. In fact, the eluted quantity of the purified ABA-binding protein, estimated by protein concentration assay or SDS-PAGE, increased from 1 to 5 mm ABA contained in the elution buffer, but the ABA-binding activity of the purified ABA-binding protein was shown to be unstable when the protein was eluted with the buffer containing more than 1 mm ABA (data not shown). The detailed reason for this is currently unknown; nevertheless, we adopted 1 mm as the ABA concentration in the elution buffer.
Here we describe an ABA specific-binding protein that was for the first time, to our knowledge, purified to homogeneity from the lower epidermis of broad bean leaves under the selected conditions. The purified ABA-binding protein, possessing a Kd of 21 nm with a linear function of the Scatchard plot, should be one of the two classes of proteins existing in the crude extract, namely the protein with a Kd of 19 nm. But another class of ABA-binding protein having a higher affinity to bind ABA (lower Kd value, see Fig. 6a) in the crude extract was lost during the purification probably because its content was too low or it was too sensitive to in vitro environmental conditions. The homogeneity of the purified ABA-binding protein deduced from its Scatchard plot was confirmed by SDS-PAGE, IEF electrophoresis, and IEF/SDS-PAGE two-dimensional electrophoresis (Fig. 4). Both the Scatchard plot and electrophoresis indicated that the purified ABA-binding protein may be a monomeric one. There was found to be about one binding site per monomer according to its maximum binding activity (0.87 mol mol−1 protein).
The purified ABA-binding protein was shown to have the properties of saturability, reversibility, and high affinity. It is particularly noteworthy that the competition assay revealed a stereospecificity of the purified ABA-binding protein to bind the physiologically active (+)ABA: two ABA analogs, (−)ABA and trans-ABA, had substantially no competing capacity with (+)ABA for binding to the ABA-binding protein, and (±)ABA was also shown to be less effective in the competition (Fig. 8). These results strongly suggest the nature of the ABA-binding protein to perceive physiological ABA signal. All these properties closely meet the expected primary criteria of a hormone receptor (Venis, 1985).
The most important criterion of a hormone receptor is undoubtedly its cell physiological significance (Venis, 1985). To determine preliminarily the potential role of the purified ABA-binding protein in ABA signal cascades, PLD activity induced by ABA in guard cell protoplasts of broad bean leaves in relation to the ABA-binding protein was measured. In fact, PLD, hydrolyzing phospholipids, producing phosphatidic acid and the head group, plays a pivotal role in regulating many critical cellular functions (Wang, 1999). PLD activity recently was implicated in the initial steps of ABA signal transduction in barley aleurone cells (Ritchie and Gilroy, 1998, 2000) and in broad bean guard cells (Jacob et al., 1999). It has been shown that ABA can activate the enzyme PLD, triggering subsequent ABA response of the guard cells via the PLD-catalyzing product phosphatidic acid (Jacob et al., 1999). In this experiment, guard cell protoplasts carefully prepared according to Ling and Assmann (1992) and Kruse et al. (1989) were shown to be of good quality (data not shown). The monoclonal antibody raised against the purified ABA-binding protein was shown to be able to block 3H-ABA-binding activity of this protein (data not shown), indicating the specificity of the antibody. The in vivo PLD activity in guard cell protoplasts, assessed by NBD-PtdBut production using NBD-PtdCho and 1-buOH as the substrates, was shown to be activated by ABA treatment of the protoplasts (Fig. 9), indicating that this system was effective. The pretreatment of the protoplasts with the antibody significantly decreased this ABA-induced PtdBut production from PLD activity in a dose-dependent manner and even completely blocked this ABA-induced effect on the cells when the amount of the antibody was sufficient (Fig. 9). The inefficacy of the pretreatments of the protoplasts with either preimmune mouse IgG or BSA in decreasing ABA-induced PLD activity assessed by PtdBut production (Fig. 9), indicated the specificity of the ABA-binding protein antibody. These results suggest that ABA receptor perceiving ABA signal may be localized at outside of the guard cell plasma membrane because the antibodies are macromolecules impossible to traverse the plasma membrane. This is somewhat consistent with previous reports providing evidence for plasmalemma surface perception of ABA (Anderson et al., 1994; MacRobbie, 1995; Schultz and Quatrano, 1997; Jeannette et al., 1999; Ritchie and Gilroy, 2000). The binding of the antibodies to ABA receptors may block the ABA-binding sites on ABA receptors, therefore reducing or even blocking the ABA-mediating physiological response that, in this case, was the induction of PLD activity. This finding provides more clear evidence for the potential receptor nature of the purified ABA-binding protein. It is certain that for clarification of the receptor nature of the ABA-binding protein and elucidation of its mechanism in ABA signal transduction further evidence will be necessary, especially the information about the ABA-binding protein-encoding gene (s). Further studies on the 42-kD ABA-binding protein-encoding gene and its cell physiological functions should shed new light on first events of ABA signal transduction.
MATERIALS AND METHODS
Chemicals
Sephadex G-25, EAH-Sepharose 4B, Dextran T70-coated charcoal (DCC), glycerol, polyacrylamide gel, acrylamide, ampholytes, β-mercaptoethanol, 3H-(±)ABA (2.37×1012 Bq mmol−1, purity 98.4%) were purchased from Amersham Pharmacia Biotech Ltd (Little Chalfont, Buckinghamshire, UK). Cis,trans-(+)ABA (abbreviated to [+]ABA, purity 98%), cis,trans-(−)ABA ([−]ABA, purity 98%), cis,trans-(±)ABA ([±]ABA, purity 99%), 2-trans,4-trans-ABA (trans-ABA, purity 99%), and all other chemicals were purchased from Sigma (St. Louis), unless otherwise indicated.
Plant Material
Broad bean (Vicia faba) plants were grown in pots filled with soil that was composed of loam, peat, and coarse sand in 7:3:2 volume ratio and was supplemented with nitrogen-phospho-potash complete chemical fertilizer, in an illuminated green house under a 14-h light (approximately 300 μmol m−2 s−1)/10-h dark cycle at 25/18°C(day/night temperature). The plants were watered once a day and used for experiments when they were 4 weeks old.
Preparation of Leaf Epidermis
The leaves were harvested from 4-week-old plants. The lower epidermis was peeled off from the leaves and immersed for 3 to 4 h in cold buffer (4°C) containing 10 mm MES/NaOH (pH 6.0), 0.02% (w/v) BSA, 0.25 m mannitol, 0.1 mm CaCl2, and 1 mm EGTA. The epidermis samples were either kept at 0°C for immediate use or frozen in liquid nitrogen and stored at −80°C until use.
Crude Extraction
For extraction of the ABA-binding proteins, we compared three methods as follows. Unless otherwise indicated, all of the procedures described below were done at 4°C.
Method 1: Extraction with Triton X-100
The epidermis sample was ground with a pestle and mortar in MES/NaOH buffer with a sample weight:buffer volume ratio of 1:3. The buffer medium was composed of 10 mm MES/NaOH (pH 6.5), 100 mm NaCl, 1 mm MgCl2, 2 mm EGTA, 1 mm phenylmethylsulfonyl fluoride, 2 mm dithiothreitol, and 0.5% (v/v) Triton X-100. After grinding the sample was centrifuged for 15 min at 15,000g and the supernatant was centrifuged again at 100,000g for 30 min, then the supernatant was concentrated to 3 to 4 mg protein mL−1 by ultrafiltration.
Method 2: Extraction with Cold Acetone
To the same supernatant of 15,000g prepared as described above, a volume of five times (based on the supernatant) of cold acetone (−40°C) was added with vigorous stirring in an ice-bath for 5 min, and the mixture was centrifuged at 15,000g for 5 min. The precipitate was dried under vacuum to remove the acetone, ground in the same MES/NaOH buffer containing 0.2% (v/v) Triton X-100 as described above for 30 min, and finally centrifuged at 100,000g for 30 min to obtain the supernatant for use.
Method 3: Salting Out with Ammonium Sulfate
To the same supernatant of 100,000g prepared as described above in method 1, the powder of ammonium sulfate was slowly added with stirring to a final concentration of 80% saturation, and then the gentle stirring continued for 10 min. The mixture was centrifuged at 15,000g for 10 min to obtain the precipitate. The precipitate was then dissolved in the same MES/NaOH buffer containing 0.2% (v/v) Triton X-100 as described above. The solution was applied to a Sephadex G-25 column to remove the ammonium sulfate and then was concentrated to 3 to 4 mg protein mL−1 by ultrafiltration.
Preparation of ABA-Linked EAH-Sepharose 4B
EAH-Sepharose 4B (containing 7–11 μmol conjugated amino groups in 1 mL of drained gel) was adopted as the affinity medium to couple ABA. ABA-linked EAH-Sephrose 4B was prepared according to the method of preparing NAA-linked AH-Sephrose 4B for purification of auxin-binding protein by Shimomura et al. (1986) with the following modifications. The coupling reaction of ABA to EAH-Sepharose 4B was performed as follows: (±)ABA (1 g) dissolved in 60 mL of 50% (w/v) dimethylformamide solution was mixed with 50 mL of drained EAH-Sepharose 4B. 1-Ethyle-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride (4 g) was added to the ABA-EAH-Sepharose 4B solution, of which the pH was adjusted to 8.0 with 1 n NaOH. The ABA-EAH-Sepharose 4B solution was shaken for 20 h at 4°C in the dark. After the coupling reaction had finished, the ABA-EAH-Sepharose 4B gel was washed with 50% (w/v) dimethylformamide and then again with both 0.5 m NaCl in 0.1 m Tris/HCl buffer (pH 8.3) and 0.5 m NaCl in 0.1 m sodium acetate-acetic acid buffer (pH 4.0). Finally, the gel was extensively washed with double distilled water.
The coupling amount of ABA to EAH-Sepharose 4B was determined essentially according to Nilsson and Mosbach (1984): 40 mg ABA-EAH-Sepharose 4B was dissolved in 80% (w/v) glycerol, and then the UV A252 of the solution was measured with an UV-Photometer (UV-240, Shimandz, Tokyo) using 80% (w/v) glycerol as the control. The amount of the coupled ABA was calculated according to the standard UV absorbance per millimolar ABA at 252 nm. The tested coupling efficiency, which is referred to the ratio of the coupling amount of ABA to total amount of the amino groups conjugated to the gel, was approximately 80%.
Purification
Purification was performed at 4°C. The ABA-EAH-Sepharose 4B gel was packed into a column of 1.6 × 50 cm for affinity chromatography. This affinity column was equilibrated with the buffer A solution containing 10 mm MES/NaOH (pH 6.5), 150 mm NaCl, 2 mm MgCl2, 2 mm CaCl2, and 5 mm KCl, and then the crude extract obtained with Triton X-100 only as the extraction medium (method 1) was loaded onto the affinity column. The column was first eluted with a step gradient of NaCl in buffer A (1,000 mL), from 100 to 300 mm, to remove the nonspecific-bound proteins. ABA-binding proteins were then eluted with the same buffer A containing 1 mm (±)ABA (300 mL). The eluting solutions were assayed for ABA-binding activity, and all the eluting fractions containing ABA-binding activity were pooled. The free ABA in the eluting solution was removed by passing through a Sephadex G-25 column, and the eluting solution was finally concentrated by ultrafiltration.
ABA-Binding Assay
The activity of ABA-binding proteins was assayed according to Zhang et al. (1999) with modifications. The incubation medium for binding assays contained 250 mm mannitol, 5 mm MgCl2, and 1 mm CaCl2 (except when determining the effects of Mg2+ and Ca2+ on ABA binding), 10 mm Tris/MES (pH 6.5, except when analyzing ABA binding at different pH), 70 nm 3H-(±)ABA (except when analyzing ABA-binding kinetics where a step gradient of concentrations of 3H-[±]ABA was used), and 30 ng of purified protein or the crude extract equivalent to 10 μg of protein. The total incubation volume of each assay was 200 μL. The mixtures were incubated at 4°C for 1 h (an incubation of 1–2 h gave almost the same results, see below in Fig. 1) and then quickly placed in ice. After the addition of 50 μL of 0.5% (w/v) DCC to remove the free 3H-ABA by adsorption, the mixtures were maintained in ice for 10 min and then centrifuged to remove DCC. The radioactivity of the supernatant was counted using a liquid scintillometer (LS-5801, Beckman Instruments, Fullerton, CA). The specific binding was determined by the difference between the radioactivity bound to the purified proteins or crude extract incubated only with 3H-ABA (total binding) and the radioactivity bound in the presence of 1,000-fold molar excess of unlabeled (±)ABA (Sigma; unspecific binding). The unlabeled (±)ABA was added into the incubation medium at the same time with 3H-ABA. The activity of crude ABA-binding protein was expressed as the number of nanomoles of 3H-ABA specifically bound per gram of protein and that of the purified protein as the number of moles of 3H-ABA per mole of protein (specific binding activity). A preliminary experiment for validating the methods of ABA-binding assay showed that the Dextran-charcoal absorption method mentioned above gave substantially the same results as those by the equilibrium dialysis technique (Venis, 1985), but the latter was not used mainly because of its long duration to attain binding equilibrium (about 3–4 h).
The stereospecificity of the purified ABA-binding protein was assayed using two ABA analogs competing possibly for the same binding sites of the protein: (−)ABA and trans-ABA. These two related compounds are structurally similar to (+)ABA but are shown to be functionally inactive by many experiments (Walton, 1983; Balsevich et al., 1994; Hill et al., 1995; Walker-Simmons et al., 1997). The two ABA analogs and (+)ABA and (±)ABA (as controls) were assayed in the concentrations ranging linearly from 10 to 105 nm. The conditions of incubation were the same as those described above (the incubation buffer containing 70 nm 3H-[±]ABA). Protein concentration was determined according to Markwell et al. (1978) using bovine serum albumin as the standard.
SDS-PAGE
The SDS-PAGE of the purified ABA-binding proteins was carried out essentially according to Laemmli (1970) with the Mini-PROTEAN II System (Bio-Rad, Richmond, CA). The concentration of running slab gel was 12% and that of stacking gel (Amersham Pharmacia, Uppsala) was 2.5%. Protein was stained with Coomassie Brilliant Blue R-250 (Amersham Pharmacia). Molecular mass standards (Molecular Weight Calibration Kit, Amersham Pharmacia) were: myosin (H-chain), 200 kD; phosphorylase B, 97.4 kD; BSA, 68 kD; ovalbumin, 43 kD; and carbonic anhydrase, 29 kD.
IEF Electrophoresis
The pI of the purified ABA-binding protein was determined with IEF using the model III Mini-IEF System (Bio-Rad) on polyacrylamide gels containing ampholytes (pH 3.0–10.0). The protein standards (Broad pI Kit, Amersham Pharmacia) were: amylglucosidase, pI 3.5; trypsin inhibitor, pI 4.55; β-lactoglobulin A, pI 5.20; carbonic anhydrase B (bovine), pI 5.85; carbonic anhydrase B (human), pI 6.55; myoglobin acidic-band, pI 6.85; and basic-band, pI 7.35. The pIs of the standards were assumed to be as designated by the supplier (Amersham Pharmacia).
IEF/SDS-PAGE Two-Dimensional Electrophoresis
Two-dimensional electrophoresis IEF/SDS-PAGE was performed essentially according to O'Farrell (1975) with the Mini-PROTEAN II 2-D System (Bio-Rad). Samples were run from cathodic reservoir (100 mm NaOH) to anodic reservoir (10 mm H3PO4). IEF tube gel was composed of 9.2 m urea, 4% (w/v) acrylamide, 2% (v/v) Triton X-100, 1.6% (v/v) ampholyte, pH 5.0 to 7.0, and 0.4% (v/v) ampholyte, pH 3.0 to 10.0. After pre-electrophoresis at 200 V for 10 min, 300 V for 15 min, and 400 V for 15 min, the ABA-binding protein was dissolved in the sample buffer (IEF buffer) containing 9.5 m urea, 2% (v/v) Triton X-100, 5% (v/v) β-mercaptoethanol, 1.6% (v/v) ampholyte, pH 5.0 to 7.0, and 0.4% (v/v) ampholyte, pH 3.0 to 10.0, and then the sample dissolved was loaded into the IEF tube gel. The IEF was conducted at 500 V for 10 min and at 750 V for 3.5 h. After the electrophoresis the tube gel was equilibrated with SDS-sample buffer consisting of 62.5 mm Tris-HCl (pH 6.8), 2.3% (w/v) SDS, 5.0% (v/v) β-mercaptoethanol, and 10% (w/v) glycerol and then applied to 12% (v/v) SDS-PAGE for the electrophoresis of second dimension. Proteins in the gel were detected by silver-staining using the Silver-Staining Kit of Protein (Amersham Pharmacia). Molecular mass markers used (Amersham Pharmacia) were: phosphorylase B, 94.0 kD; albumin, 67.0 kD; ovalbumin, 43.0 kD; carbonic anhydrase, 30.0 kD; and trypsin inhibitor, 20.1 kD.
Preparation of Monoclonal Antibody against the Purified ABA-Binding Protein
BALB/c mice were immunized by intraperitoneal injection of 20 μg of the purified ABA-binding protein emulsified with complete Freund's adjuvant. Four weeks later, the purified ABA-binding protein (20 μg) emulsified with incomplete adjuvant was injected intraperitoneally. After the second injection, one intraperitoneal booster injection of 20 μg of the protein without adjuvant was given 4 d before fusion. This final injection could be administered up to 4 months after the initial injection. The spleen was removed after the whole blood was collected. The serum could be used as polyclonal antibody. Spleen cells were fused to a myeloma cell line, X63Ag8.653 (Kearney et al., 1979), according to a standard method described by Higashihara et al. (1989). The fused cells were resuspended in RPMI 1640 (Flow, Irvine, Scotland) medium with 15% (v/v) fetal bovine serum and then were distributed into 96-well flat-bottom polystyrene tissue plates (Iwaki, Chiba, Japan) at a density of 5 × 105 cells/well. Selection with hypoxanthine-aminopterin-thymidine medium was begun 24 h after fusion. Throughout the cloning procedures, the cells were cultured in RPMI medium supplement with 15% (v/v) fetal bovine serum, 0.225% (w/v) NaHCO3, 2 mm l-Gln, 60 mg L−1 kanamycin sulfate, and 50 mg L−1 gentamicin sulfate. Screening of antibody-producing cells was carried out using an ELISA as described by Higashihara et al. (1989), and then the hybridoma cells were recloned twice by limiting dilution using BALB/c mouse splenocytes as a feeder layer. An established hybridoma clone was cultured in RPMI medium with 15% (v/v) fetal bovine serum or injected intraperitoneally into pristane-primed BALB/c mice. The cultured supernatant and the ascitic fluid were used as the monoclonal antibody source. IgG class antibody was purified by a protein G-Sepharose column (Pharmacia-LKB, Uppsala) from the monoclonal antibody source according to the manufacture's protocol. The isotype of monoclonal antibody was determined by using a mouse antibody isotyping kit (Gibco-BRL, Cleveland). The antibody used in this study was found to contain IgG1 heavy chain and k light chain (data not shown).
Preparation of Guard Cell Protoplasts
Young broad bean leaves were excised from 3- to 4-week-old plants. Guard cell protoplasts were prepared according to Ling and Assmann (1992) based on the blender method of Kruse et al. (1989). This blender method has been used in our laboratory (Zhang et al., 2001b). Guard cell protoplasts released by two-stage pectolytic and cellulytic digestion were suspended into 0.45% (w/v) mannitol, 1 mm CaCl2, and 0.5 mm ascorbic acid and then purified by centrifugation at 200g for 15 min onto a 100% Histopaque 1077 cushion. Healthy protoplasts were collected at the interface between the mannitol buffer and Histopaque 1077. These protoplasts were rewashed in 0.6 m mannitol and 1 mm CaCl2 buffer, resuspended in 0.6 m mannitol and 1 mm CaCl2, examined, and measured by light microscopy, and quantitated with a hemocytometer. Contaminating protoplasts in preparations were clearly discernible by morphology. Enriched protoplasts were concentrated by centrifugation at 200g. The purity of guard cell protoplasts was 99.8% based on counting a sample of about 9,000 cells. The protoplasts were either immediately used or frozen at −80°C.
Assay of PLD Activity of Guard Cell Protoplasts Treated with Anti-ABA-Binding Protein Antibody
NBD-PtdCho (Avanti Polar Lipids, Birmingham, AL) was stored at −80°C in chloroform. Before use it was dried under a stream of N2 and emulsified by sonication in H2O. In vivo measurement of PtdBut production was conducted for assessing PLD activity according to Jacob et al. (1999) and Ritchie and Gilroy (1998). Protoplasts (100 μL, approximately 2.5 × 105 protoplasts) were pretreated with 5 to 50 μg of soluble ABA-binding protein antibody expressed as protein content for 10 min at 4°C. Pretreatments of protoplasts with either preimmune mouse IgG or BSA (at an equal protein content to ABA-binding protein antibody in both cases) instead of the ABA-binding protein antibody were taken as the controls. Afterward, the protoplasts were incubated in 0.5 mg mL−1 NBD-PtdCho for 80 min on ice, and then they were transferred to 22°C for 10 min. 1-buOH (0.1%, v/v) also was added at the start of the 22°C incubation. (±)ABA (10 μm) was then added into the mixture from a stock of 50 mm in 95% (v/v) ethanol (final [ethanol], 0.02% [v/v]). After 20 min incubation in (±)ABA, the samples were processed and NBD-labeled PtdBut was quantified according to Ritchie and Gilroy (1998).
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 39730340, 39870487, and 30070532) and a grant from the China National Key Basic Research Program (grant no. G1999011700).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010531.
LITERATURE CITED
- Allan AC, Fricker MD, Ward JL, Beale MH, Trewavas AJ. Two transduction pathways mediate rapid effects of abscisic acid in Commelinaguard cells. Plant Cell. 1994;6:1319–1328. doi: 10.1105/tpc.6.9.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen GJ, Muir SR, Sanders D. Release of Ca2+from individual plant vacuoles by both IP3 and cyclic ADP-ribose. Science. 1995;268:735–737. doi: 10.1126/science.7732384. [DOI] [PubMed] [Google Scholar]
- Allen GJ, Sanders D. Calcineurin, a type 2B protein phosphatase, modulates the Ca2+-permeable slow vacuolar ion channel of stomatal guard cells. Plant Cell. 1995;7:1473–1483. doi: 10.1105/tpc.7.9.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson BE, Ward JM, Schroeder JI. Evidence for an extra-cellular reception site for abscisic acid in Commelinaguard cells. Plant Physiol. 1994;104:1177–1183. doi: 10.1104/pp.104.4.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsevich JL, Cutler AJ, Lamb N, Friesen LJ, Kurz EU, Perras MR, Abrams SR. Response of cultured maize cells to (+)-abscisic acid, (−)-abscisic acid, and their metabolites. Plant Physiol. 1994;106:135–142. doi: 10.1104/pp.106.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertauche N, Leung J, Giraudat J. Protein phosphatase activity of abscisic acid insensitive 1 (ABI1) protein from Arabidopsis thaliana. Eur J Biochem. 1996;24:193–200. doi: 10.1111/j.1432-1033.1996.0193t.x. [DOI] [PubMed] [Google Scholar]
- Brault M, Caiveau O, Pedron J, Maldiney R, Sotta B, Miginiac E. Detection of membrane-bound cytokinin-binding proteins in Arabidopsis thalianacells. Eur J Biochem. 1999;260:512–519. doi: 10.1046/j.1432-1327.1999.00190.x. [DOI] [PubMed] [Google Scholar]
- Brault M, Maldiney R, Miginiac E. Cytokinin-binding proteins. Physiol Plant. 1997;100:520–527. [Google Scholar]
- Coome BG. The development of flesh fruits. Annu Rev Plant Physiol. 1976;27:507–528. [Google Scholar]
- Coome BG. Research on development and ripening of the grape berry. Am J Enol Vitic. 1992;43:101–110. [Google Scholar]
- Cutler S, Ghassemian M, Bonetta D, Cooney S, McCourt P. A protein farnesyl transferase involved in abscisic acid signal transduction in Arabidopsis. Science. 1996;273:1239–1241. doi: 10.1126/science.273.5279.1239. [DOI] [PubMed] [Google Scholar]
- Fersht A. Enzyme Structure and Mechanism. Ed 2. New York: WH Freeman; 1985. pp. 47–97. [Google Scholar]
- Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman HM. The Arabidopsisabscisic acid response locus ABI4 encodes an APETALA2 domain protein. Plant Cell. 1998;10:1043–1054. doi: 10.1105/tpc.10.6.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring CA, Irving HR, Parish RW. Effects of auxin and abscisic acid on cytosolic calcium and pH in plant cells. Proc Natl Acad Sci USA. 1990;87:9645–9649. doi: 10.1073/pnas.87.24.9645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy S, Read ND, Trewavas AJ. Elevation of cytoplasmic calcium by caged calcium or caged inositol trisphosphate initiates stomatal closure. Nature. 1990;343:769–771. doi: 10.1038/346769a0. [DOI] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM. Isolation of the ArabidopsisABI3 gene by positional cloning. Plant Cell. 1992;4:1251–1261. doi: 10.1105/tpc.4.10.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat J, Parcy F, Bertauche N, Gosti F, Leung J. Current advances in abscisic acid action and signaling. Plant Mol Biol. 1994;26:1557–1577. doi: 10.1007/BF00016490. [DOI] [PubMed] [Google Scholar]
- Gosti F, Beaudoin N, Serizet C, Webb AAR, Vartanian N, Giraudat J. ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell. 1999;11:1897–1910. doi: 10.1105/tpc.11.10.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashihara M, Frado LLY, Craig R, Ikebe M. Inhibition of conformational change in smooth muscle myosin by a monoclonal antibody against the 17 kD light chain. J Biol Chem. 1989;264:5218–5225. [PubMed] [Google Scholar]
- Hill RD, Liu JH, Durnin D, Lamb N, Shaw A, Abrams SR. Abscisic acid structure-activity relationships in barley aleurone layers and protoplasts. Plant Physiol. 1995;108:573–579. doi: 10.1104/pp.108.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjelmeland LM. Solubilization of functional membrane proteins. Methods Enzymol. 1984;104:305–318. doi: 10.1016/s0076-6879(84)04097-0. [DOI] [PubMed] [Google Scholar]
- Hocking J, Clapham J, Cattell KJ. Abscisic acid-binding to sub-cellular fractions from leaves of Vicia faba. Planta. 1978;138:303–304. doi: 10.1007/BF00386826. [DOI] [PubMed] [Google Scholar]
- Hornberg C, Weiler EW. High affinity binding sites for abscisic acid at the plasmalemma of Vicia fabaguard cells. Nature. 1984;310:321–324. [Google Scholar]
- Ingram J, Bartels D. The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:377–403. doi: 10.1146/annurev.arplant.47.1.377. [DOI] [PubMed] [Google Scholar]
- Jacob T, Ritchie S, Assmann SM, Gilroy S. Abscisic acid signal transduction in guard cells is mediated by phospholipase D activity. Proc Natl Acad Sci USA. 1999;96:12192–12197. doi: 10.1073/pnas.96.21.12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeannette E, Rona JP, Bardat F, Cornel D, Sotta B, Miginiac E. Induction of RAB18 gene expression and activation of K+ outward rectifying channels depend on an extracellular perception of ABA in Arabidopsis thalianasuspension cells. Plant J. 1999;18:13–22. doi: 10.1046/j.1365-313x.1999.00423.x. [DOI] [PubMed] [Google Scholar]
- Kearney JF, Radbruch A, Liesegang B, Rajewsky K. A new mouse myeloma cell line that has lost immunoglobulin expression but permits the construction of antibody-secreting hybrid cell lines. J Immunol. 1979;123:1548–1550. [PubMed] [Google Scholar]
- Kruse T, Tallman G, Zeiger E. Isolation of guard cell protoplasts from mechanically prepared epidermis of Vicia fabaleaves. Plant Physiol. 1989;90:1382–1386. doi: 10.1104/pp.90.4.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Active-staining cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–682. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee YS, Choi YB, Suh S, Lee J, Assmann SM. Abscisic acid-induced phosphoinostide turnover in guard cell protoplast of Vicia faba. Plant Physiol. 1996;110:987–996. doi: 10.1104/pp.110.3.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Bouvier-Durand M, Morris PC, Guerrier D, Chefdor F, Giraudat J. ArabidopsisABA-response gene ABI1: features of a calcium-modulated protein phosphatase. Science. 1994;264:1448–1452. doi: 10.1126/science.7910981. [DOI] [PubMed] [Google Scholar]
- Leung J, Giraudat J. Abscisic acid signal transduction. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:199–222. doi: 10.1146/annurev.arplant.49.1.199. [DOI] [PubMed] [Google Scholar]
- Leung J, Merlot S, Giraudat J. The ArabidopsisABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode redundant protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell. 1997;9:759–771. doi: 10.1105/tpc.9.5.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leube MP, Grill E, Amrhein N. ABI1 of Arabidopsisis a protein serine/threonine phosphatase highly regulated by the proton and magnesium ion concentration. FEBS Lett. 1998;424:100–104. doi: 10.1016/s0014-5793(98)00149-5. [DOI] [PubMed] [Google Scholar]
- Li J, Assmann SM. An abscisic acid-activated and calcium-independent protein kinase from guard cells of fava bean. Plant Cell. 1996;8:2359–2368. doi: 10.1105/tpc.8.12.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang XQ, Watson MB, Assmann SM. Regulation of abscisic acid-induced stomatal closure and anion channels by guard cell AAPK kinase. Science. 2000;287:300–303. doi: 10.1126/science.287.5451.300. [DOI] [PubMed] [Google Scholar]
- Ling V, Assmann SM. Cellular distribution of calmodulin and calmodulin-binding proteins in Vicia fabaL. Plant Physiol. 1992;100:970–978. doi: 10.1104/pp.100.2.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRobbie EAC. ABA-induced ion efflux in stomatal guard cells: multiple actions of ABA inside and outside the cell. Plant J. 1995;7:565–576. [Google Scholar]
- Markwell AK, Hass SM, Bieber LL, Tolbert NE. A modification of Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem. 1978;87:206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- McCarty DR. Genetic control and integration of maturation and germination pathways in seed development. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:71–93. [Google Scholar]
- Merlot S, Giraudat J. Gene analysis of abscisic acid signal transduction. Plant Physiol. 1997;114:751–757. doi: 10.1104/pp.114.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K, Leube MP, Grill E. A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science. 1994;264:1452–1455. doi: 10.1126/science.8197457. [DOI] [PubMed] [Google Scholar]
- Mitsui S, Sugiura M. Purification and properties of cytokinin-binding proteins from tobacco leaves. Plant Cell Physiol. 1993;34:543–547. [Google Scholar]
- Nilsson K, Mosbach K. Immobilization of ligands with organic sulfonyl chlorides. Methods Enzymol. 1984;104:56–59. doi: 10.1016/s0076-6879(84)04083-0. [DOI] [PubMed] [Google Scholar]
- O'Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Pedron J, Brault M, Nake C, Miginiac E. Detection of abscisic acid-binding proteins in the microsomal protein fraction of Arabidopsis thalianawith abscisic acid-protein conjugates used as affinity probes. Eur J Biochem. 1998;252:385–390. doi: 10.1046/j.1432-1327.1998.2520385.x. [DOI] [PubMed] [Google Scholar]
- Ritchie S, Gilroy S. Abscisic acid signal transduction in the barley aleurone is mediated by phospholipase D activity. Proc Natl Acad Sci USA. 1998;95:2697–2702. doi: 10.1073/pnas.95.5.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie S, Gilroy S. Abscisic acid stimulation of phospholipase D in the barley aleurone is G-protein-mediated and localized to the plasma membrane. Plant Physiol. 2000;124:639–702. doi: 10.1104/pp.124.2.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock CD, Quatrano RS. The role of hormones during seed development. In: Davies PJ, editor. Plant Hormones. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 671–697. [Google Scholar]
- Rodriguez PL, Benning G, Grill E. ABI2, a second protein phosphatase 2C involved in abscisic acid signal transduction in Arabidopsis. FEBS Lett. 1998a;421:185–190. doi: 10.1016/s0014-5793(97)01558-5. [DOI] [PubMed] [Google Scholar]
- Rodriguez PL, Benning G, Grill E. Molecular cloning in Arabidopsis thalianaof a new protein phosphatase 2C (PP2C) with homology to ABI1 and ABI2. Plant Mol Biol. 1998b;38:879–883. doi: 10.1023/a:1006012218704. [DOI] [PubMed] [Google Scholar]
- Romanov GA, Tara V, Chvojika L, Kulaeva ON. Specific binding of zeatin by a protein fraction of barley leaves and purification of cytokinin-binding proteins. Soviet Plant Physiol. 1986;33:75–85. [Google Scholar]
- Schultz TF, Quatrano RS. Evidence for surface perception of abscisic acid by rice suspension cells as assayed by Em gene expression. Plant Sci. 1997;130:63–71. [Google Scholar]
- Selivankina SY, Romanko EG, Ovcharov AM, Khachenko VI. Participation of cytokinin-binding protein from barley leaves in cytokinin activation of chromatin-bound RNA-polymerase. Soviet Plant Physiol. 1982;29:208–214. [Google Scholar]
- Sheen J. Mutational analysis of protein phosphatase 2C involved in abscisic acid signal transduction in higher plants. Proc Natl Acad Sci USA. 1998;95:975–980. doi: 10.1073/pnas.95.3.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura S, Sotobayashi T, Futai M, Fukui T. Purification and properties of an auxin-binding protein from maize shoot membranes. J Biochem. 1986;99:1513–1524. doi: 10.1093/oxfordjournals.jbchem.a135621. [DOI] [PubMed] [Google Scholar]
- Smith JB, Thevenon G, Smith DV, Green B. Elucidation of the primary structure of proteins by mass spectrometry. Anal Biochem. 1991;192:118–124. doi: 10.1016/0003-2697(91)90050-4. [DOI] [PubMed] [Google Scholar]
- Sugaya S, Sakai S. A soluble auxin-binding protein from mung bean hypocotyls has indole-3-acetadehade reductase activity. Physiol Plant. 1996;97:433–439. [Google Scholar]
- Venis M. Methods in receptor research. In: Venis M, editor. Hormone Binding Sites in Plants. New York: Longman; 1985. pp. 24–40. [Google Scholar]
- Walker-Simmons MK, Holappa LD, Abrams GD, Abrams SR. ABA metabolites induce group 3 LEAmRNA and inhibit germination in wheat. Physiol Plant. 1997;100:474–480. [Google Scholar]
- Walton DC. Structure-activity relationships of abscisic acid analogs and metabolites. In: Addicott FT, editor. Abscisic Acid. New York: Praeger Scientific; 1983. pp. 113–146. [Google Scholar]
- Wang X. The role of phospholipase D in signal cascades. Plant Physiol. 1999;120:645–651. doi: 10.1104/pp.120.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart JAD, Creelman RA. Metabolism and physiology of abscisic acid. Annu Rev Plant Physiol Plant Mol Biol. 1988;39:439–473. [Google Scholar]
- Zhang DP, Chen SW, Peng YB, Shen YY. Abscisic acid-specific binding sites in the flesh of developing apple fruits. J Exp Bot. 2001a;52:2097–2103. doi: 10.1093/jexbot/52.364.2097. [DOI] [PubMed] [Google Scholar]
- Zhang DP, He FL, Jia WS. Cell biological mechanism for triggering of ABA accumulation under water stress in Vicia fabaleaves. Sci China (ser C) 2001b;44:421–428. doi: 10.1007/BF02879609. [DOI] [PubMed] [Google Scholar]
- Zhang DP, Zhang ZL, Chen J, Jia WS. Specific abscisic acid-binding sites in mesocarp of grape berry: properties and sub-cellular localization. J Plant Physiol. 1999;155:324–331. [Google Scholar]