Abstract
Methanogenic granules from an anaerobic bioreactor that treated wastewater of a beer brewery consisted of different morphological types of granules. In this study, the microbial compositions of the different granules were analyzed by molecular microbiological techniques: cloning, denaturing gradient gel electrophoresis and fluorescent in situ hybridization (FISH), and scanning and transmission electron microscopy. We propose here that the different types of granules reflect the different stages in the life cycle of granules. Young granules were small, black, and compact and harbored active cells. Gray granules were the most abundant granules. These granules have a multilayer structure with channels and void areas. The core was composed of dead or starving cells with low activity. The brown granules, which were the largest granules, showed a loose and amorphous structure with big channels that resulted in fractured zones and corresponded to the older granules. Firmicutes (as determined by FISH) and Nitrospira and Deferribacteres (as determined by cloning and sequencing) were the predominant Bacteria. Remarkably, Firmicutes could not be detected in the brown granules. The methanogenic Archaea identified were Methanosaeta concilii (70 to 90% by FISH and cloning), Methanosarcina mazei, and Methanospirillum spp. The phenotypic appearance of the granules reflected the physiological condition of the granules. This may be valuable to easily select appropriate seed sludges to start up other reactors.
High-rate anaerobic bioreactors are commonly applied for the purification of industrial wastewaters. The upflow anaerobic sludge bed (UASB) is one of the most common types of bioreactors (17). In these reactors, methanogenic microbial communities are present as granules, typically 0.2 to 5 mm. The granules are characterized by a high density and high methanogenic activity (23). The different physiological types of microorganisms are in close vicinity of each other, enhancing interspecies electron transfer and thus facilitating high-rate methane formation. The microbial composition of methanogenic granules has been studied by several groups (32, 39, 41, 47, 56), and practical information on how to achieve granule formation in anaerobic bioreactors was obtained (2, 30, 45).
Methanogenic granules are complex microbial communities. Molecular biological techniques are very appropriate to gain insight into the phylogenetic position of microorganisms that are nutritionally dependent on each other in a complex food chain. Some of these microorganisms are difficult to cultivate due to their special growth requirements, such as a low hydrogen partial pressure, which is maintained by methanogenic archaea (44). Some molecular techniques, like cloning and sequencing of extracted DNA and RNA, give insights into which microorganisms are present and which are most active (15, 24, 39, 47, 54). Denaturing gradient gel electrophoresis (DGGE) and fluorescent in situ hybridization (FISH) are becoming routinely applied to microbial ecological studies. They are very appropriate to detect changes in the microbial composition with time or to compare the microbial composition of different bioreactors. DGGE has been used to analyze the biodiversity of granular sludge (12, 39, 56), to compare the start-up and evolution of mesophilic and thermophilic UASB reactors (27, 29), to investigate changes in the microbial population in continuously stirred tank reactors (37, 52), to determine the diversity of sulfate-reducing populations in anaerobic biofilms (42), and to analyze the microbial population dynamics in anaerobic reactors treating landfill leachate or trichloroethane (10, 51). The granule structure (19, 46), the microbial distribution of sulfate reducers and methanogens (19, 38, 43, 46, 49), and the topography and biodiversity (11, 13, 38, 41) have been analyzed by FISH.
It has frequently been observed that methanogenic granules from one single full-scale or laboratory-scale bioreactor contain granules with different sizes (from a hundred micrometers to a few millimeters) and different colors varying from light brown to black. The aim of this research was to gain insight into the microbial composition and structure of the different types of granules in a UASB reactor from an industrial brewery. We combined different molecular techniques (FISH, DGGE, and cloning) and electron microscopy to gain insight into the structure, function, and physical appearance of methanogenic granules. The combination of complementary techniques was important to elucidate the structure-function relationships of the different granules.
MATERIALS AND METHODS
Sludge source.
Anaerobic granular sludge from full-scale UASB reactors treating brewery wastewaters (MAHOU, Guadalajara, Spain) was used. The reactor was operated at an organic loading rate of 11 kg chemical oxygen demand (COD)/m3 · day and a hydraulic residence time of 8 h. The upflow velocity applied was 0.75 m/h. The production of biogas was 1.28 nm3/m3, with a CH4:CO2 content of 75:20. The efficiencies were 80% for total COD removal and 85% for biological oxygen demand removal.
The average composition of the industrial wastewater was 4,000 mg/liter COD, 2,400 mg/liter biological oxygen demand, 1.3 g/liter total suspended solids, 100 mg/liter N-Kjeldahl, 15 mg/liter NH4+, and15 mg/liter P, pH 7.2. The specific methanogenic activity of the sludge with a mixture of fatty acids was 0.15 g COD/g volatile suspended solids · day.
Samples of granular sludge (3 g of wet weight), which contained about 500 to 600 granules, were taken and separated into three different-colored fractions: gray, brown, and black. The percentage of each type of granule was determined. The size, shape, and morphology of each fraction were determined. Sedimentation velocity was calculated using the volumes of several weights of granules (taken at random), and the Stokes equation was applied. Different granules were chosen for different analyses. A mixture of granules, taken completely at random from the reactor, was used for cloning and sequencing. After classifying granules by color and size, representative granules of each class were selected for FISH, DGGE, and transmission electron microscopy. Transmission electron microscopy was performed only with the gray granules.
FISH.
Two different hybridization experiments were performed, one with crushed granules in order to quantify different microbial groups and one with thin sections of granules to study the spatial distribution of the different microbial groups. FISH was applied according to protocols described previously by Amann et al. (4, 5, 6). Granular sludge was fixed with formaldehyde (4% in phosphate-buffered saline [PBS]) for 6 h. The formamide concentration used for hybridization (2 h at 46°C) and the NaCl concentrations used for washing (15 min at 48°C) are listed in Table 1. The probes, labeled at the 5′ end with fluorescein or Cy3, were purchased from Genotek (Barcelona, Spain). After hybridization, the specimens were stained with 4′,6′-diamidino-2-phenylindole (DAPI) (1 mg/ml). Acidiphilium cryptum DSM2389 (α-proteobacteria), Alcaligenes xylosoxidans (β-Proteobacteria), Pseudomonas aeruginosa PAO1 (γ-Proteobacteria), Bacillus cereus (Firmicutes), Micrococcus luteus (Actinobacteria), and Methanobacterium formicicum (DSM 1535) (Archaea) were used as positive controls.
TABLE 1.
rRNA gene oligonucleotide probes used for FISH and PCR amplificationa
| Probe | Target organism(s) | Probe sequence (5′-3′) | Formamide (%)/NaCl (mM) | Reference |
|---|---|---|---|---|
| EUB338 | Bacteria | GCTGGCTCCCGTAGGAGT | 35/70 | 4 |
| ARC915 | Archaea | GTGCTCCCCCGCCAATTCCT | 20/215 | 48 |
| NON338 | Negative control | ACTCCTACGGGAGGCAGC | 35/70 | 4 |
| ALF968 | α-Proteobacteria | GGTAAGGTTCTGCGCGTT | 20/215 | 31 |
| BET42a | β-Proteobacteria | GCCTTCCCACTTCGTTT | 35/70 | 31 |
| GAM42a | γ-Proteobacteria | GCCTTCCCACATCGTTT | 35/70 | 31 |
| SRB385 | Sulfate-reducing bacteria | CGGCGTCGCTGCGTCAGG | 35/70 | 5 |
| DSS658 | Desulfosarcina, Desulfococcus | TCCACTTCCCTCTCCCAT | 60/102 | 31 |
| DSV698 | Desulfovibrio spp. | GTTCCTCCAGATATCTACGG | 35/70 | 31 |
| BAC1080 | Bacteroides | GCACTTAAGCCGACACCT | 20/215 | 14 |
| SYN835 | Syntrophobacter | GCGGGTACTCATTCCTG | 35/70 | 22 |
| LGC354A | Gram-positive bacteria with low G+C content | TGGAAGATTCCCTACTGC | 20/215 | 34 |
| HGC69A | Gram-positive bacteria with high G+C content | TATAGTTACCACCGCCGT | 25/149 | 40 |
| MEB859 | Methanobacteriales (except Methanothermaceae) | GGACTTAACAGCTTCCCT | 25/149 | 9 |
| MC1109 | Methanococcales | GCAACATAGGGCACGGGTCT | 35/70 | 36 |
| MG1200 | Methanomicrobiales | CGGATAATTCGGGGCATGCTG | 5/630 | 36 |
| MSSH859 | Methanosarcinales | CTCACCCATACCTCACTCGGG | 35/70 | 9 |
| MS1414 | Methanosarcinales (except Methanosaeta) | CTCACCCATACCTCACTCGGG | 35/70 | 36 |
| MX825 | Methanosaeta | TCGCACCGTGGCCGACACCTAGC | 20/215 | 36 |
| 341F(GC)b | Bacteria | CCTACGGGAGGCAGCAG | 12 | |
| 907R | Bacteria | CCGTCAATTCMTTTGAGTTTc | 12 | |
| 622F(GC)b | Archaea | TGAAATCYYRTAATCCCc | 12 | |
| 1492R | Universal | TACGGYTACCTTGTTACGACTTc | 12 | |
| 27F | Bacteria | AGAGTTTGATCMTGGCTCAGc | 26 | |
| 25F | Archaea | CYGGTTGATCCTGCCRGc | 26 |
Specificity and hybridization/washing conditions are shown. PCR details can be found in Materials and Methods.
GC clamp at the 5′ end: 5′-CGC CCG CCG CGC CCC GCG CCC GTC CCG CCC CCG CCC-3′.
M, C/A; Y, C/T; R, A/G.
Cryo coupes.
The fixed granules were embedded in the cryogel OCT resin (11% polyvinyl alcohol, 5% carbowax; Sakura Tissue-Tek) by keeping it in dry ice at −70°C during 72 h. The block was cut into 50-μm slices (Cryotomo 2800 Frigocut N; Reichert-Jung) at −23°C. The OCT resin of the slices was removed by washing for 30 min in PBS. The slices were stored at −20°C.
DGGE.
Three different types of granules (Table 2) were separated and independently examined. Granular sludge was resuspended in PBS, and cells were disrupted using a Fastprep FP120 bead beater (BIO101-Savant) (six times for 40 s, each at 5.5 cycles/s). The DNA was extracted using the FastDNA kit for soil (Q-BIOgene). A fragment of the 16S rRNA gene was amplified by PCR with primer pairs 341F(GC)-907R (annealing temperature [T*] = 52°C) for Bacteria and 622F(GC)-1492R (T* = 42°C) for Archaea (12). The amplification reaction was performed according to the Taq DNA polymerase protocol (Promega, Madison, Wis.). The PCR conditions were as follows: 94°C for 10 min; 30 cycles at 94°C for 1 min, T* for 1 min, and 72°C for 3 min; and 72°C for 10 min. The PCR products (566 bp for bacteria and 870 bp for archaea) were analyzed by DGGE using a D-Code Universal system (Bio-Rad). Six percent polyacrylamide (37.5:1 acrylamide:bisacrylamide) gels with a 30 to 60% urea-formamide denaturant gradient were used. The electrophoresis was performed at 100 V for 12 h at 60°C. Bands detected by fluorescence using a UV transilluminator were excised and reamplified for sequencing.
TABLE 2.
Physical characteristics of the three types of granules
| Granule | General characteristic(s) | Diam (mm) | Sedimentation velocity | Abundance (%) |
|---|---|---|---|---|
| Black | Smooth and shiny surface, spherical shape | 0.1-1 | High | 4 |
| Gray | Spherical and elliptical shape | Elliptical, 1 × 2, 2 × 3; spherical, 0.1-3 | Medium | 81 |
| Brown | Variable shape | 0.1-2.5 | Low | 15 |
Cloning.
DNA was extracted from a representative granule sample. The 16S rRNA gene was amplified using the oligonucleotide primer pairs 27F and 1492R (annealing T, 56°C) for the Bacteria domain (26) and 25F and 1492R (annealing T, 52°C) for the Archaea domain (26). The amplified fragments (length, 1,465 to 1,467 bp) were cloned using a TOPO cloning kit (Invitrogen Corporation, San Diego, Calif.) and then transformed using competent Escherichia coli cells. Plasmid inserts were grouped according to their restriction patterns with EcoRI and Sau3AI enzymes (Biolabs Inc.) and subsequently amplified by PCR using the M13 primer set (Invitrogen).
Sequencing and sequence analysis.
The sequences were automatically analyzed on an ABI model 377 sequencer (Applied Biosystems) and were thereafter corrected manually. Sequences were compared with the NCBI databases using the Basic Local Alignment Search Tool (BLAST) to identify the closest sequence (http://www.ncbi.nlm.nih.gov/BLAST/).
Microscopy and cell counting.
A representative sample of granules (0.03 g of sludge in 1 ml of PBS:ethanol, 1:1) was disrupted by sonication (30 to 60 s at a power of 37 W). The cell suspension was diluted 1/50 for DAPI counting (total cell number) and 1/5 for hybridized cell counting. About 10 at-random fields were analyzed (500 to 1,000 DAPI-stained cells) using a Zeiss Axioskop fluorescence microscope to determine the average number of cells in the samples. For spatial reconstruction of the granules and topological (geographical) distribution of the microbial groups, slides were examined with a Radiance2000 confocal laser scanning microscope equipped with He-Ne lasers and a reversed microscope (Zeiss Axiovert S100).
Transmission electron microscopy was done according to a protocol described previously (20) by using a MET JEM1010 microscope on semithin (1-μm) and ultrathin (70-nm) sections. Scanning electron microscopy (SEM) was performed with a Philips XL30 microscope as described elsewhere previously (3).
Nucleotide sequence accession numbers.
Sequences of cloning and DGGE bands have been deposited in the NCBI database under accession numbers DQ478735 to DQ478750 for clones and DQ478751 to DQ478760 for DGGE.
RESULTS
Morphological characterization.
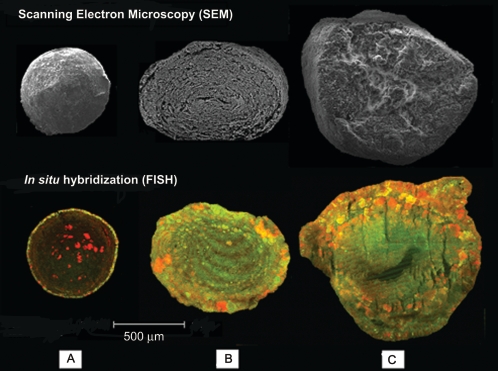
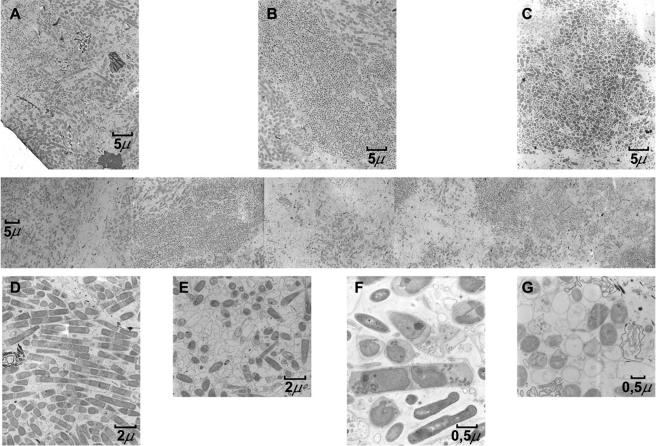
The characterization of a brewery wastewater treatment UASB reactor showed the existence of different types of granules. Besides color, the granules differed in sedimentation rate and size. The granules were black, gray, or brown. Some of the general properties are listed in Table 2 (the Gaussian distribution of the three color classes of granules by size is presented in Fig. SM1 in the supplemental material). Figure 1 shows representative SEM and confocal laser scanning microscope images of the three types of granules. The black granules were small and compact. The brown granules were less compact and porous. The gray granules were composed of defined concentric layers of microorganisms. The gray granules were most abundant in the sludge. Figure 2 shows some transmission electron micrographs of thin sections of gray granules from the edge (left side) to the core (right side). Dense and sparse areas were visible in the granules. The presence of microcolonies with both low (Fig. 2B) and high (Fig. 2C) morphological diversity as well as areas with a high heterogeneity (Fig. 2A, D, and F) can be seen. Empty, presumably dead, cells were present as well (Fig. 2E and G).
FIG. 1.
Structures of different types of methanogenic granules analyzed by scanning electron microscopy (upper part) and confocal laser microscopy of fluorescent in situ-hybridized granules (lower part). (A) Black granule. (B) Gray granule. (C) Brown granule. FISH Eub338-fluorescein, green, universal probe for Bacteria; Arch915-Cy3, red, universal probe for Archaea.
FIG. 2.
Transmission electron microscopy of a gray granule. Reconstruction from the external layer (left) to the core (right) is shown in the middle row. Heterogeneous colonies (A, C, and F) and a colony composed of microorganisms with similar morphologies (C) are shown in the upper part of the figure. Details of several shapes and cell morphologies are displayed at the bottom of the figure. Filaments and sheathed rods with sharp ends, typical for the class Methanomicrobia (D), and coccoid cells (G) are shown. Dead or void cells can be observed (E and G). Magnifications: A, B, and C, ×1,500; D and E, ×4,000; F and G, ×10,000.
In situ hybridization.
Using DAPI staining, high numbers of microorganisms were counted in all three types of granules (Table 3). The cell number in the black granules was somewhat higher than those in the gray and brown granules, confirming its more compact structure and density. However, only a low percentage of the cells that were stained with DAPI hybridized clearly with the bacterial and archaeal probes. This indicates that the number of ribosomes in the cells is low, which might reflect the low metabolic activity of the cells.
TABLE 3.
Quantification of the total (DAPI-stained) and hybridized (active) cells with universal bacterial (EUB338) and archaeal (ARCH915) probes in the granules
| Granule type | No. of DAPI cells/g of sludge | No. of EUB338 cells/g of sludge | No. of ARCH915 cells/g of sludge | Total active cells (%) | Active bacteria (%) | Active archaea (%) |
|---|---|---|---|---|---|---|
| Black | 3.5 × 1011 | 1.3 × 1010 | 1.4 × 1010 | 7.7 | 49 | 51 |
| Grey | 2.3 × 1011 | 8.8 × 109 | 8.8 × 109 | 7.5 | 50 | 50 |
| Brown | 2.5 × 1011 | 5.3 × 109 | 1.9 × 1010 | 9.6 | 22 | 78 |
The localization of bacteria and archaea in the granules was done using hybridization probes with their corresponding specificities (Fig. 1). In the black granules, bacteria appeared in the outer shell of the granule, while archaea formed dense bright clusters inside the granules. FISH as well as SEM revealed a multilayer structure of the gray granules. The core of these granules showed archaea with low activity, as indicated by the low fluorescence with the archaeal probes. By contrast, brown granules seemed to have higher numbers of active methanogenic archaea.
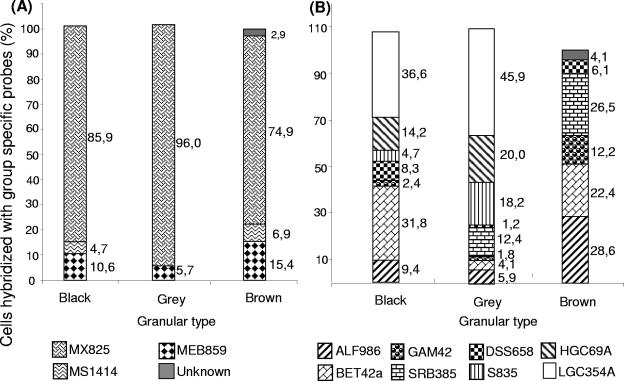
Group-specific oligonucleotide probes were used to identify the relative abundance of different groups of archaea and bacteria in the granules (Fig. 3). Gram-positive bacteria were the most abundant type of bacteria in the black and gray granules. Remarkably, gram-positive bacteria were almost absent in the brown granules. These granules contained mainly proteobacteria. With respect to the archaeal domain, specific probes for the four orders of methanogenic archaea, Methanobacteriales, Methanococcales, Methanomicrobiales, and Methanosarcinales, were tested. In all three types of granules, Methanosaeta was the dominant genus (between 75 and 95% of total archaeal cells). Methanosarcina was detected only in black and brown granules.
FIG. 3.
Microbial compositions of the three types of granules determined by using group-specific oligonucleotide probes for Archaea (A) and Bacteria (B). The solid gray bars represent the difference between the number of cells hybridized with the universal probes (ARC915 for panel A and EUB338 for panel B) and the sum of the cells hybridized with the specific probes assayed.
Microbial biodiversity.
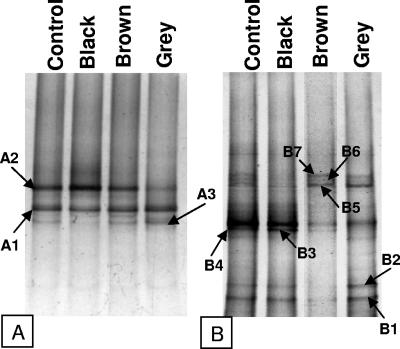
Figure 4 shows the band patterns obtained with DGGE after DNA amplification using specific primers for the domains Archaea [primers 622F(GC) and 1492R] and Bacteria [primers 341F(GC) and 907R]. The three types of granules had a similar bacterial diversity, although quantitative differences occurred; e.g., band A2 in the gray granule had a lower intensity than in the other granules. This band was identified as Methanosarcina, which was not detected by FISH in the gray granules. Similarly, it has previously been shown that the biodiversity of archaea changed little in response to changes in the feed composition of an anaerobic reactor, but the relative proportion of organisms identified in the DGGE profiles did change (1). The bands were excised, and the DNA was eluted, PCR amplified, and sequenced. Bands of the DGGE gels that had a weak fluorescence under a UV lamp were not analyzed. The sequences were compared with those found in the NCBI database by using BLAST. The taxonomic affiliation of the partial 16S rRNA sequences is shown in Table 4. Remarkably, the only proteobacteria identified were δ-Proteobacteria. Among the gram-positive clostridia, Clostridium butyricum (Fig. 4, band B2, and Table 4) and Clostridium difficile (Fig. 4, band B3, and Table 4) were the most dominant species. It should be mentioned these two bands (B2 and B3) were very weak in the brown granules. This is in accordance with the FISH results. Gram-positive bacteria were not detected by FISH in these granules. The methanogens detected by this technique, Methanosaeta, Methanosarcina, and Methanospirillum, were the same as those detected by FISH and SEM.
FIG. 4.
DGGE fingerprints of the different types of granules (black, gray, and brown) for Archaea (A) and Bacteria (B). A mixture of the granular sludge was used as a control. Arrows indicate the bands excised and sequenced.
TABLE 4.
16S rRNA gene sequence-based phylogenetic affiliations of DGGE bands using a BLAST search of the NCBI database
| Band | Phylum, class, or order | Closest species or taxon (NCBI accession no.) | Similarity (%) |
|---|---|---|---|
| A1 | Methanosarcinales | Methanosaeta concilii (M59146) | 97 |
| A2 | Methanosarcinales | Methanosarcina mazei (AF028691) | 93 |
| A3 | Methanomicrobiales | Methanospirillum hungatei (M60880) | 94 |
| B1 | Nitrospira | Uncultured bacterium (L22045) | 97 |
| Nitrospira marina (L35501) | 97 | ||
| B2 | Firmicutes | Clostridium butyricum (M59085) | 97 |
| B3 | Firmicutes | Clostridium difficile (X73450) | 96 |
| B4 | δ-Proteobacteria | Syntrophobacter wolinii (X70905) | 93 |
| B5 | δ-Proteobacteria | Syntrophobacter wolinii (X70905) | 93 |
| B6 | Deferribacteres | Uncultured bacterium (AF280863) | 95 |
| Synergistes jonesii (L08066) | 91 | ||
| B7 | Deferribacteres | Uncultured bacterium (U81735) | 97 |
The phylogeny of the almost-complete sequences of the cloned 16S rRNA gene is shown in Table 5. The 96 clones obtained for the Bacteria domain were grouped into 13 different restriction patterns, which were sequenced and analyzed. Interestingly, 85% of the total bacterial clones obtained (nine of the restriction patters found) belonged to the poorly known phyla Nitrospira and Deferribacteres. The methanogenic archaea detected were the same by either technique; Methanosaeta concilii (70% of the archaeal clones) was most predominant.
TABLE 5.
16S rRNA gene sequence-based phylogenetic affiliations of clones using a BLAST search of the NCBI database
| Clone | Abundance (%) | Phylum, class, or order | Closest species or taxon (NCBI accession no.) | Similarity (%) |
|---|---|---|---|---|
| G35 | 5 | Methanomicrobiales | Uncultured archaeon (AF229775) | 96 |
| Methanospirillum hungatei (M60880) | 90 | |||
| G46 | 70 | Methanosarcinales | Methanosaeta concilii (X51423)a | 99 |
| G74 | 25 | Methanosarcinales | Methanosarcina mazei (AF028691) | 98 |
| G33 | 5 | α-Proteobacteria | Sphingopyxis witflariensis (AJ416410) | 99 |
| G9 | 2.5 | Firmicutes | Uncultured bacterium (AY134903) | 93 |
| G19 | 5 | Firmicutes | Acetobacterium paludosum (X96958) | 91 |
| G3 | 2.5 | Choroflexi | Uncultured bacterium (AY08260) | 92 |
| G6, 49, G59, and G98 | 51 | Nitrospira | Uncultured bacterium (AF351225) | 97 |
| Magnetobacterium bavaricum (X71838) | 93 | |||
| G30, G36, and G39 | 18 | Deferribacteres | Uncultured bacterium (AF280863) | 98 |
| Synergistes jonesii (L08066) | 93 | |||
| G50 | 8 | Deferribacteres | Uncultured bacterium (AF229792) | 96 |
| G96 | 8 | Deferribacteres | Uncultured bacterium (AF229792) | 98 |
Cited with the former name of Methanothrix soehngenii.
DISCUSSION
Methanogenic granular sludge is composed of a variety of granules with different physical properties (8, 12, 16, 50). In this work, we paid special attention to the structural features, microbial diversity, and evolution of methanogenic granules from a UASB reactor.
Microbial diversity.
The microbial diversity present in the diverse granules has been studied using complementary molecular ecology tools: DGGE, FISH, and cloning. The methanogenic Archaea identified by DGGE were Methanosaeta concilii, Methanosarcina mazei, and Methanospirillum hungatei. Sequences obtained by cloning DNA and RNA extracted from the reactor granules confirmed these results. The clones obtained from DNA showed a 98 to 99% sequence homology with Methanosaeta concilii and Methanosarcina mazei. Furthermore, the analysis of the clones obtained from RNA showed the presence of only two species, Methanosaeta concilii and Methanosarcina mazei, indicating that these two species correspond to active cells and suggesting that they play an important role in the functional granule. In general, acetoclastic methanogens are more abundant than hydrogenotrophic methanogens. Quantification using fluorescence in situ hybridization showed that Methanosaeta was predominant (75 to 96% of the total active Archaea) in all types of granules. This suggests that the acetate concentration inside the granules is limiting. Methanosarcina has a higher maximum growth rate but a lower affinity for acetate (μmax, 0.21 day−1; Ks, 4 mM) than Methanosaeta (μmax, 0.11 day−1; Ks, 0.44 mM) (53, 55).
The analysis of the DGGE bacterial sequences showed that members of the phyla Firmicutes, Nitrospira, and Deferribacter and the class δ-Proteobacteria were present in the granules. Fermenting bacteria like Clostridium and H2-producing fatty acid-oxidizing bacteria, such as Syntrophobacter, are commonly present in methanogenic environments. The retrieved Nitrospira and Deferribacteres sequences were closely related to those of uncultured bacteria found in anaerobic digesters (18, 27, 43). These results agree with previous DGGE data (12). Two clones belonging to the class Clostridia and one clone belonging to the class α-Proteobacteria were obtained from DNA extracted from different granules. The latter had 99% similarity with Sphingopyxis witflariensis, an anaerobic decomposer of carbohydrates isolated from urban wastewaters (25). The majority (87%) of the bacteria detected by cloning were related to the phyla Deferribacteres, Nitrospira, and Chloroflexi. Moreover, all the clones obtained from rRNA genes belonged to these phyla. Some clones were related to the genera Synergistes, Aminobacterium, and Aminovorans (Deferribacteres), which are anaerobic amino acid-degrading bacteria (7, 27, 33). Other clones showed similarity with green nonsulfur bacteria, which have also been found in methanogenic granular sludge (39, 47) and which may contribute to the degradation of carbohydrates and other cellular components (54). The identification and quantification using FISH complemented our previous observations (13) and showed the presence of gram-positive bacteria with low and high G+C content, δ-proteobacteria (sulfate reducers and syntrophic bacteria), β-proteobacteria, and γ-proteobacteria. The relatively high value (40%) obtained for gram-positive bacteria with low G+C content was observed previously (28). Remarkably, gram-positive bacteria were present only in low numbers in brown granules.
Evolution of methanogenic granules.
There are indications that the different granule types reflect different steps in the development of granular sludge. A model to explain the growth of methanogenic granules has recently been described (35). Those authors stated that granule formation follows a phase of nucleation and homogeneous growth followed by a fully active granule and finally a granular stage in which only the outer part is active. After 4 weeks, the center of the granules was mostly empty due to biomass decay. In the case reported here, black granules correspond to “young” granules. Bacteria associated with the first steps of granule development are gram negative (mainly α-proteobacteria). The first methanogenic Archaea that appear to be associated with the granule structure are Methanospirillum and Methanosarcina (N. Fernández, unpublished results). All the microorganisms present in these granules are active, and consequently, black granules are compact and dense.
As a consequence of diffusion problems and the corresponding lack of nutrients, growth occurs mainly at the periphery of the granule, giving rise to multilayered structures. Active metabolic bacteria colonize the granule edges, where compounds from the media are actively degraded, and the methanogenic Archaea develop in the interior part of the granule, forming large colonies. It is probable that the syntrophic relationships between proton-reducing acetogenic bacteria and hydrogen-consuming methanogens caused multispecies microcolonies, as observed previously (21, 22). The predominant Bacteria are gram positive bacteria, and the Archaea belong mainly to the genus Methanosaeta. These granules are gray, have an ellipsoidal or spherical shape, and correspond to the majority of the granules in the sludge.
The brown granules seem to correspond to “old” granules. They are relatively large and have lost the spherical-ellipsoidal shape, and the structure has become soft and fluffy. Probably in response to the diffusion problems, the well-defined layered structure breaks, resulting in empty areas and channels that penetrate toward the interior of the granule. Methanosaeta might play an important role in these phenomena, since large tufts of Methanosaeta-like filaments in these fractured areas can be observed using SEM (see Fig. SM2 in the supplemental material). No gram-positive bacteria have been detected in these brown granules. At present, we do not have any explanation for the radical bacterial population change between gray and brown granules.
Hybridization with universal probes for bacterial and archaeal domains showed that the inner areas of the brown and gray granules exhibit little or no microbial activity, probably as a consequence of the limitation or lack of nutrients in these internal areas of the granules. Hybridizations were restricted mainly to the periphery of the granules, the zone where cell growth takes place. This observation correlates with the presence of high numbers of empty cells detected by SEM, probably corresponding to inactive or dead cells (35, 41). Only the black granules appeared to harbor active microorganisms in the interior of the granule. As a concluding remark, it can be stated that the combination of different complementary techniques (cloning, DGGE, FISH, transmission electron microscopy, and SEM) allowed us to monitor the structural and microbial diversity and functional evolution of methanogenic granular sludge from a UASB reactor.
Supplementary Material
Acknowledgments
The work was partially supported by Spanish Ministry for Science and Technology grant REN2001-2980-C02-02/HID. The Agencia Española de Cooperación Internacional (AECI) provided a predoctoral fellowship to E. E. Díaz. An institutional grant to the CBM from the Fundación Areces is also appreciated.
We thank J. Ciriza and Maohu S.A. for the sludge supplied and information on the wastewater treatment plant.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Akarsubasi, A. T., O. Ince, B. Kirdar, N. A. Oz, D. Orhon, T. P. Curtis, I. M. Head, and B. K. Ince. 2005. Effect of wastewater composition on archaeal population diversity. Water Res. 39:1576-1584. [DOI] [PubMed] [Google Scholar]
- 2.Alphenaar, P. A. 1994. Anaerobic granular sludge: characterization, and factors affecting its functioning. Ph.D. thesis. Wageningen Agricultural University. Wageningen, The Netherlands.
- 3.Alphenaar, P. A., N. Groeneveld, and A. C. van Aelst. 1994. Scanning electron microscopical method for internal structure analysis of granular sludge. Micron 25:129-133. [Google Scholar]
- 4.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial population. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amann, R. I., I. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amann, R. I. 1995. In situ identification of microorganisms by whole cell hybridization with rRNA-targeted nucleic acid probes, p. 1-15. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbiology ecology manual, 3.3.6. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 7.Baena, S., M. L. Fardeau, B. Ollivier, M. Labat, P. Thomas, J. L. Garcia, and B. K. Patel. 1999. Aminomonas paucivorans gen. nov., sp. nov., a mesophilic, anaerobic, amino-acid-utilizing bacterium. Int. J. Syst. Bacteriol. 49:975-982. [DOI] [PubMed] [Google Scholar]
- 8.Batstone, D. J., J. Keller, and L. L. Blackall. 2004. The influence of substrate kinetics in the microbial community structure on granular anaerobic biomass. Water Res. 38:1390-2404. [DOI] [PubMed] [Google Scholar]
- 9.Boetius, A., K. Ravenschlag, C. Schubert, D. Rickert, F. Widdel, A. Gieseke, R. Amann, B. Jørgensen, U. Witte, and O. Pfannkuche. 2000. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407:623-626. [DOI] [PubMed] [Google Scholar]
- 10.Calli, B., B. Mertoglu, N. Tas, B. Inanc, O. Yenigun, and I. Oztyrk. 2003. Investigation of variations in microbial diversity in anaerobic reactors treating landfill leachate. Water Sci. Technol. 48(4):105-112. [PubMed] [Google Scholar]
- 11.Calli, B., B. Mertoglu, K. Roest, and B. Inanc. 2006. Comparison of long-term performances and final microbial compositions of anaerobic reactors treating landfill leachate. Bioresour. Technol. 97:641-647. [DOI] [PubMed] [Google Scholar]
- 12.Chan, O. C., W. T. Liu, and H. H. P. Fang. 2001. Study of microbial community of brewery-treating granular sludge by denaturing gradient gel electrophoresis of 16S rRNA gene. Water Sci. Technol. 43(1):77-82. [PubMed] [Google Scholar]
- 13.Díaz, E., R. Amils, and J. L. Sanz. 2003. Molecular ecology of anaerobic granular sludge grown at different conditions. Water Sci. Technol. 48(6):57-64. [PubMed] [Google Scholar]
- 14.Doré, J., A. Sghir, G. Hannequart-Gramet, G. Corthier, and P. Pochart. 1998. Design and evaluation of a 16S rRNA-targeted oligonucleotide probe for specific detection and quantization of human faecal Bacteroides populations. Syst. Appl. Microbiol. 21:65-71. [DOI] [PubMed] [Google Scholar]
- 15.Ferrera, I., R. Massana, E. O. Casamayor, V. Balague, O. Sánchez, C. Pedrós-Alió, and J. Más. 2004. High-diversity biofilm for the oxidation of sulfide-containing effluents. Appl. Microbiol. Biotechnol. 64:726-734. [DOI] [PubMed] [Google Scholar]
- 16.Foster, C. F., and J. Quarmby. 1995. The physical characteristics of anaerobic granular sludges in relation to their internal architecture. Antonie Leeuwenhoek 67:103-110. [DOI] [PubMed] [Google Scholar]
- 17.Frankin, R. J. 2001. Full-scale experiences with anaerobic treatment of industrial wastewater. Water Sci. Technol. 44(8):1-6. [PubMed] [Google Scholar]
- 18.Godon, J. J., E. Zumstein, P. Dabert, F. Habouzit, and R. Moletta. 1997. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit sequence analysis. Appl. Environ. Microbiol. 63:2802-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.González-Gil, G., P. N. L. Lens, A. van Aelst, H. van As, A. I. Versprille, and G. Lettinga. 2001. Cluster structure of anaerobic aggregates of an expanded granular sludge bed reactor. Appl. Environ. Microbiol. 67:3683-3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grotenhuis, J. T. C., M. Smit, A. A. M. van Lammeren, A. J. M. Stams, and A. J. B. Zehnder. 1991. Localization and quantification of extracellular polymers in methanogenic granular sludge. Appl. Microbiol. Biotechnol. 36:115-119. [Google Scholar]
- 21.Grotenhuis, J. T. C., M. Smit, C. M. Plugge, Y. Xu, A. A. M. van Lammeren, A. J. M. Stams, and A. J. B. Zehnder. 1991. Bacteriological composition and structure of granular sludge adapted to different substrates. Appl. Environ. Microbiol. 57:1942-1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harmsen, H. J. M., H. M. P. Kengen, A. D. L. Akkermans, A. J. M. Stams, and W. M. de Vos. 1996. Detection and localization of syntrophic propionate-oxidizing bacteria in granular sludge by in situ hybridization using 16S rRNA-based oligonucleotide probes. Appl. Environ. Microbiol. 62:1656-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hulshoff Pol, L. W., S. I. de Castro Lopes, G. Lettinga, and P. N. L. Lens. 2004. Anaerobic sludge granulation. Water Res. 38:1376-1389. [DOI] [PubMed] [Google Scholar]
- 24.Ito, T., S. Okabe, H. Satoh, and Y. Watanabe. 2002. Successional development of sulfate-reducing bacterial populations and their activities in a wastewater biofilm growing under microaerophilic conditions. Appl. Environ. Microbiol. 68:1392-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kämpfer, P., R. Witzenberger, E. B. M. Denner, H. J. Busse, and A. Neef. 2002. Sphingopyxis witflariensis sp. nov., isolated from wastewater. Int. J. Syst. Evol. Microbiol. 52:2029-2043. [DOI] [PubMed] [Google Scholar]
- 26.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics, John Wiley & Sons, New York, N.Y.
- 27.LaPara, T. M., C. H. Nakatsu, L. Pantea, and J. E. Alleman. 2000. Phylogenetic analysis of bacterial communities in mesophilic and thermophilic bioreactors treating pharmaceutical wastewater. Appl. Environ. Microbiol. 66:3951-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, W. T., O. C. Chan, and H. H. P. Fang. 2002. Characterization of microbial community in granular sludge treating brewery wastewater. Water Res. 36:1767-1775. [DOI] [PubMed] [Google Scholar]
- 29.Liu, W. T., O. C. Chan, and H. H. P. Fang. 2002. Microbial community dynamics during start-up of acidogenic anaerobic reactors. Water Res. 36:3203-3210. [DOI] [PubMed] [Google Scholar]
- 30.Liu, Y., H. L. Xu, S. F. Yang, and J. H. Tay. 2003. Mechanisms and models for anaerobic granulation in upflow anaerobic sludge blanket reactor. Water Res. 37:661-673. [DOI] [PubMed] [Google Scholar]
- 31.Manz, W., M. Eisenbrecher, T. R. Neu, and U. Szewzyk. 1998. Abundance and spatial organization of gram-negative sulphate-reducing bacteria in activated sludge investigated by in situ probing with specific 16S rRNA targeted oligonucleotides. FEMS Microbiol. Ecol. 25:43-61. [Google Scholar]
- 32.McMahon, K. D., D. Zheng, A. J. Stams, R. I. Mackie, and L. Raskin. 2004. Microbial population dynamics during start-up and overload conditions of anaerobic digesters treating municipal solid waste and sewage sludge. Biotechnol. Bioeng. 87:823-834. [DOI] [PubMed] [Google Scholar]
- 33.McSweeny, C. S., M. J. Allison, and R. I. Mackei. 1993. Amino acid utilization by the rumen bacterium Synergiste jonessi strain 78-1. Arch. Microbiol. 159:131-135. [DOI] [PubMed] [Google Scholar]
- 34.Meier, H., R. Amann, W. Ludwig, and K. H. Schleifer. 1999. Specific oligonucleotide probes for in situ detection of a major group of gram-positive bacteria with low DNA G+C content. Syst. Appl. Microbiol. 22:186-196. [DOI] [PubMed] [Google Scholar]
- 35.Picioreanu, C., D. J. Batstone, and M. C. M. van Loosdrecht. 2005. Multidimensional modelling of anaerobic granules. Water Sci. Technol. 52(1-2):501-507. [PubMed] [Google Scholar]
- 36.Raskin, L., J. M. Stromley, B. E. Rittmann, and D. A. Stahl. 1994. Group-specific 16S RNA hybridization probes to describe natural communities of methanogens. Appl. Environ. Microbiol. 60:1232-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rincon, B., F. Raposo, R. Borja, J. M. Gonzalez, M. C. Portillo, and C. Saiz-Jiménez. 2006. Performance and microbial communities of a continuous stirred tank anaerobic reactor treating two-phases olive mill solid wastes at low organic loading rates. J. Biotechnol. 121:534-543. [DOI] [PubMed] [Google Scholar]
- 38.Rocheleau, S., C. W. Greer, J. R. Lawrence, C. Cantin, L. Laramee, and S. R. Guiot. 1999. Differentiation of Methanosaeta concilii and Methanosarcina barkeri in anaerobic mesophilic granular sludge by fluorescent in situ hybridization and confocal scanning laser microscopy. Appl. Environ. Microbiol. 65:2222-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roest, K., H. G. Heilig, H. Smidt, W. M. de Vos, A. J. M. Stams, and A. D. L. Akkermans. 2005. Community analysis of a full-scale anaerobic bioreactor treating paper mill wastewater. Syst. Appl. Microbiol. 28:175-185. [DOI] [PubMed] [Google Scholar]
- 40.Roller, C., M. Wagner, R. Amann, W. Ludwing, and K. H. Schleifer. 1994. In situ probing of gram bacteria with high DNA G+C content using 23S rRNA targeted oligonucleotides. Microbiology 140:2849-2858. (Erratum, 141:1267, 1995.) [DOI] [PubMed] [Google Scholar]
- 41.Saiki, Y., C. Iwabuchi, A. Katami, and Y. Kitagawa. 2002. Microbial analyses by fluorescence in situ hybridisation of well-settled granular sludge in brewery wastewater treatment plants. J. Biosci. Bioeng. 93:601-606. [DOI] [PubMed] [Google Scholar]
- 42.Santegoeds, C. M., T. G. Ferdelman, G. Muyzer, and D. de Beer. 1998. Structural and functional dynamics of sulfate-reducing populations in bacterial biofilms. Appl. Environ. Microbiol. 64:3731-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Santegoeds, C. M., L. R. Damgaard, G. Hesselink, J. Zopfi, P. Lens, G. Muyzer, and D. de Beer. 1999. Distribution of sulfate-reducing and methanogenic bacteria in anaerobic aggregates determined by microsensor and molecular analyses. Appl. Environ. Microbiol. 65:4618-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schink, B., and A. J. M. Stams. 2002. Syntrophism among prokaryotes. In M. Dworkin, K. H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, 3rd ed. Springer Verlag, New York, N.Y. [Online.] http://141.150.157.117:8080/prokPUB/index.htm.
- 45.Schmidt, J. E. and B. K. Ahring. 1996. Granular sludge formation in upflow anaerobic sludge blanket (UASB) reactors. Biotechnol. Bioeng. 49:229-246. [DOI] [PubMed] [Google Scholar]
- 46.Sekiguchi, Y., Y. Kamagata, K. Nakamura, A. Ohashi, and H. Harada. 1999. Fluorescence in situ hybridization using 16S rRNA-targeted oligonucleotides reveals localization of methanogens and selected uncultured bacteria in mesophilic and thermophilic sludge granules. Appl. Environ. Microbiol. 65:1280-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sekiguchi, Y., H. Takahashi, Y. Kamagata, A. Ohashi, and H. Harada. 2001. In situ detection, isolation, and physiological properties of a thin filamentous microorganism abundant in methanogenic granular sludge: a novel isolate affiliated with a clone cluster, the green non-sulfur bacteria, subdivision I. Appl. Environ. Microbiol. 67:5740-5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stahl, D. A., and R. Amann. 1991. Development and application of nucleic acid probes, p. 207-248. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics, vol. 8. John Wiley & Sons, London, England. [Google Scholar]
- 49.Tagawa, T., K. Syutsubo, Y. Sekiguchi, A. Ohashi, and H. Harada. 2000. Quantification of methanogen cell density in anaerobic granular sludge consortia by fluorescence in-situ hybridization. Water Sci. Technol. 42(3-4):77-82.
- 50.Ten Brummeler, E., L. W. Hulshoff Pol, J. Dolfing, G. Lettinga, and A. J. B. Zehnder. 1985. Methanogenesis in an upflow anaerobic sludge blanket reactor at pH 6 on an acetate-propionate mixture. Appl. Environ. Microbiol. 49:1472-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tresse, O., F. Mounien, M. J. Levesque, and S. Guiot. 2005. Comparison of the microbial population dynamics and phylogenetic characterization of a CANOXIS reactor and a UASB reactor degrading trichloroethane. J. Appl. Microbiol. 98:440-449. [DOI] [PubMed] [Google Scholar]
- 52.Ueno, Y., S. Haruta, M. Ishii, and Y. Igarashi. 2001. Changes in product formation and bacterial community by dilution rate on carbohydrate fermentation by methanogenic microflora in continuous flow stirred tank reactor. Appl. Microbiol. Biotechnol. 57:65-73. [DOI] [PubMed] [Google Scholar]
- 53.Wandrey, C., and A. Aivasidis. 1983. Continuous anaerobic digestion with Methanosarcina barkeri. Ann. N. Y. Acad. Sci. 413:489-500. [Google Scholar]
- 54.Yamada T., Y. Sekiguchi, H. Imachi, Y. Kamagata, A. Ohashi, and H. Harada. 2005. Diversity, localization, and physiological properties of filamentous microbes belonging to Chloroflexi subphylum I in mesophilic and thermophilic methanogenic sludge granules. Appl. Environ. Microbiol. 71:7493-7503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zehnder, A. J. B., B. A. Huser, T. D. Brock, and K. Wuhrmann. 1980. Characterization of an acetate-decarboxylating, non-hydrogen-oxidizing methane bacterium. Arch. Microbiol. 124:1-11. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, Y., S. Aiyuk, H. Xu, G. Chen, and W. Verstraete. 2005. Study of microbial community structures in UASB sludge treating municipal wastewaters by denaturing gradient gel electrophoresis of 16S rRNA. Sci. China C Life Sci. 48:128-135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.