Abstract
The most notable characteristic of Bacillus thuringiensis is its ability to produce insecticidal proteins. More than 300 different proteins have been described with specific activity against insect species. We report the molecular and insecticidal characterization of a novel cry gene encoding a protein of the Cry1I group with toxic activity towards insects of the families Noctuidae, Tortricidae, Plutellidae, and Chrysomelidae. PCR analysis detected a DNA sequence with an open reading frame of 2.2 kb which encodes a protein with a molecular mass of 80.9 kDa. Trypsin digestion of this protein resulted in a fragment of ca. 60 kDa, typical of activated Cry1 proteins. The deduced sequence of the protein has homologies of 96.1% with Cry1Ia1, 92.8% with Cry1Ib1, and 89.6% with Cry1Ic1. According to the Cry protein classification criteria, this protein was named Cry1Ia7. The expression of the gene in Escherichia coli resulted in a protein that was water soluble and toxic to several insect species. The 50% lethal concentrations for larvae of Earias insulana, Lobesia botrana, Plutella xylostella, and Leptinotarsa decemlineata were 21.1, 8.6, 12.3, and 10.0 μg/ml, respectively. Binding assays with biotinylated toxins to E. insulana and L. botrana midgut membrane vesicles revealed that Cry1Ia7 does not share binding sites with Cry1Ab or Cry1Ac proteins, which are commonly present in B. thuringiensis-treated crops and commercial B. thuringiensis-based bioinsecticides. We discuss the potential of Cry1Ia7 as an active ingredient which can be used in combination with Cry1Ab or Cry1Ac in pest control and the management of resistance to B. thuringiensis toxins.
Bacillus thuringiensis is a spore-forming bacterium that has been isolated from many different natural habitats (32). The main interesting characteristic of this bacterium is that during sporulation, it produces one or sometimes more crystalline protein inclusions that exhibit high insecticidal activity upon ingestion. B. thuringiensis-susceptible species range across a wide variety of insects belonging to the orders Lepidoptera, Diptera, Coleoptera, and Hymenoptera (19) and include other types of invertebrates, such as nematodes and mites (31). The protoxins that form the crystal are dissolved in the alkaline insect midgut and are then proteolytically activated to yield a toxic fragment (18, 45). The activated toxin binds to specific receptors on the brush border membrane of gut epithelial cells and is partially inserted into the membrane, generating pores. This results in colloid osmotic lysis of gut epithelial cells followed by the death of the insect (17, 36).
The crystal is composed of Cry and Cyt proteins, in different combinations and proportions. To date, these proteins have been classified in 49 Cry groups and 2 Cyt groups and in different subgroups depending on their amino acid sequence homologies (http://www.biols.susx.ac.uk/home/Neil_Crickmore/Bt/toxins2.html). Over the past 20 years, more than 300 different Cry and Cyt proteins have been identified, and several of them have been successfully employed in biological insecticides in integrated pest management programs (36). Generally, crystals are composed of protoxins of 130 to 140 kDa (corresponding to the expression of cry1 genes), 65 to 70 kDa (from cry2 genes), and 70 or 130 kDa (from cry3 genes). However, not all insecticidal proteins produced by B. thuringiensis clump together in the crystal. Some B. thuringiensis strains also secrete insecticidal proteins during the vegetative growth phase; these are called VIP proteins (10). In addition, some cry genes, named cry1I genes (formerly cryV genes), encode proteins of around 70 to 81 kDa that do not accumulate in the crystal (5, 12, 23, 34, 35, 37, 39, 41, 43, 46); these have been classified as Cry1I proteins due to their similarity with those in the Cry1 group (6). Their lack of involvement in the crystal structure has prevented these proteins from being included as active ingredients of B. thuringiensis-based insecticides. However, the effectiveness of Cry1I in protecting transformed plants from insect attack has been demonstrated (25, 29, 38). cry1I genes are usually located approximately 500 bp downstream of other cry1 genes, but cry1I genes sometimes may not be expressed due to the lack of an upstream promoter-like sequence (23). In fact, cry1I genes are usually either silent or expressed in the vegetative phase and secreted into the growth suspension (23, 37, 41, 46). Cry1I proteins have a broader host range than most other Cry1 proteins, and the hosts include important species of lepidopteran and coleopteran pests (43). Strains containing novel cry1I genes have been evaluated as a source of new proteins with a broad host range (39, 41).
The intensive use of B. thuringiensis-based insecticides has already given rise to resistance in field populations of the diamondback moth, Plutella xylostella (11). Since B. thuringiensis-transgenic crops (Bt crops) generally express one or two types of Cry proteins, the development of pest resistance is viewed as a major threat for this technology. The search for novel B. thuringiensis strains harboring new cry genes has received considerable attention during the last 2 decades, not only to find toxins with novel activity spectra but also to increase the arsenal of toxins that can be used for resistance management (11, 12). The most common mechanism of resistance is the reduction of binding of the toxin to its specific midgut receptor(s). This may also confer cross-resistance to other toxins that share the same receptor (11). It is therefore important to study toxin binding properties to determine the utility of novel toxins and the possibility of their being used appropriately in combination with other commercially used B. thuringiensis toxins. In the present paper, we characterize the expression, toxicity spectrum, and binding site characteristics of a novel Cry1I protein from a Spanish strain, B. thuringiensis HU4-2, originally described by Martínez et al. (33). This strain was selected for study because it contains a wide variety of cry genes and has a broad spectrum of activity against lepidopteran pest species (33).
MATERIALS AND METHODS
Bacillus thuringiensis strains.
Strain HU4-2 was isolated from a dust sample originating from a maize grain silo in the Spanish province of Huesca as part of a countrywide screening program involving the isolation and characterization of B. thuringiensis strains suitable for use in biological control (21). Strain HU4-2 was classified as B. thuringiensis serovar aizawai (33) and deposited in the Spanish collection of type cultures (accession number CECT5950). Recombinant B. thuringiensis strains EG7077, EG11070, EG11916, and EG1081 (from Ecogen Inc., Langhorne, Pa.) expressing single proteins (Cry1Ab, Cry1Ac, Cry1Ba, and Cry1Ca, respectively) were used for comparative bioassays and binding experiments.
Preparation of parasporal crystals from strain HU4-2.
Single colonies from Luria-Bertani (LB) plates were inoculated in 500 ml of CCY sporulation medium (42) and grown for 3 days until lysis was complete. A 1/5 volume of 5 M NaCl was added to the culture medium, which was then mixed. Spores and crystals were harvested by centrifugation at 15,000 × g for 20 min. The pellet was washed twice with sterile bi-distilled water, resuspended in sterile Milli-Q water, and finally stored at −20°C until used. Crystals were purified by ultracentrifugation in a sucrose discontinuous gradient as previously described (44). Briefly, the spore-crystal mixture was sonicated for 20 s in a Soniprep 150 MSE apparatus (Curtin Matheson Scientific) and immediately loaded onto centrifuge tubes containing two layers of sucrose solutions at 67% and 79% (wt/vol). After centrifugation at 70,000 × g for 16 h, the interphase containing the crystals was recovered with a Pasteur pipette, mixed with bi-distilled sterile water to a final volume of 200 ml, and centrifuged again (15,000 × g, 15 min). This step was repeated twice, and the crystal pellet was finally resuspended in sterile bi-distilled water. Crystal purity was checked by phase-contrast microscopy at a magnification of ×400, and the crystal samples were stored at −20°C until required.
Production, purification, and analysis of crystal proteins.
Single-protein-expressing strains were grown for 48 h at 29°C in CCY medium (42) supplemented with the appropriate antibiotic (3 μg/ml of chloramphenicol for EG11070 and EG1081, 10 μg/ml of tetracycline for EG7077, and 25 μg/ml of erythromycin for EG11916). After centrifugation to concentrate the spores and crystals, crystal protein solubilization and trypsin activation were carried out as described previously (9). For binding analyses, Cry1Ab and Cry1Ac were further purified by anion-exchange chromatography with a MonoQ HR5/5 column by fast protein liquid chromatography (Pharmacia, Uppsala, Sweden).
For the production of the Cry1I-type protein, fresh Escherichia coli BL21(DE3) cells were transformed with pPC-ire1 plasmid (see “Identification, cloning, and sequencing of the novel cry gene” below). Cells were grown overnight in LB with kanamycin (50 μg/ml) at 37°C and used to inoculate 750 ml of tryptone-yeast extract (2× TY) culture medium (26). The culture was grown at 37°C until the optical density at 600 nm was 0.5 to 0.6 and then incubated at 25°C for 45 min. Isopropyl-β-d-thiogalactopyranoside (IPTG) (1 mM) was added and the incubation continued for 2 h at 25°C. Cold phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4, pH 7.5) (0.5 volume) was added to the culture, and cells were recovered by centrifugation (16,000 × g, 15 min). The pellet was resuspended in cold PBS (1/10 volume), centrifuged, and stored at −80°C. Cells were thawed on ice with 1/33 volume of cold binding buffer (40 mM imidazole, 4 M NaCl, 160 mM Tris-HCl, pH 7.9; Novagen, Darmstadt, Germany) and sonicated for 60 s in 15-s pulses. Protein purification was performed using a His Bind purification kit (Novagen) according to the manufacturer's instructions. Finally, the buffer was changed to carbonate (50 mM NaCO3, 100 mM NaCl, pH 11.3) with a Sephadex G-25 prepacked column (Amersham Biosciences, Uppsala, Sweden) and the protein solution stored at 4°C until used. This protein was trypsin activated for binding assays but used as protoxin for insect bioassays.
Protein quantification was performed with the Bradford assay (3) using bovine serum albumin (BSA) as a standard. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10% polyacrylamide gel, 100:1 acrylamide/bis-acrylamide ratio) was run at 50 mA for 1 h in a mini-Protean III apparatus (Bio-Rad, Hercules, CA) as previously described (24). Gels were stained with a solution containing 50% (vol/vol) ethanol, 10% (vol/vol) acetic acid, and 0.1% (wt/vol) Coomassie brilliant blue R250 for 40 min and destained with a solution containing 6.75% (vol/vol) glacial acetic acid and 9.45% (vol/vol) ethanol. Protein sizes were determined by comparison with a broad-range protein marker (Bio-Rad).
To investigate the presence of insecticidal proteins in the growth medium, samples were taken every 12 h from a culture of the HU4-2 strain grown in CCY medium at 28°C over a total period of 72 h. Samples were purified and concentrated by centrifugation in tubes with a polyethersulfone membrane with a pore size corresponding to a cutoff of 10 kDa (Vivascience, Hannover, Germany). Noninoculated medium was used as the negative control. Proteins in the supernatant were analyzed by 10% SDS-PAGE and bioassayed as described below.
Identification, cloning, and sequencing of the novel cry gene.
The presence of a cry1I-type gene in the B. thuringiensis HU4-2 strain was detected by PCR. A general primer pair [1I(98)Fw and 1I(98)Rv] recognizing both cry1Ia and cry1Ib genes was used in combination with oligonucleotides specifically recognizing cry1Ia [primers 1Ia(10)Fw and 1Ia(11)Rv)] or cry1Ib [primers 1Ib(8)Fw and 1Ib(9)Rv)] (Table 1). These primers were selected because no other toxin sequences from the Cry1I group had been published at the moment that the amplifications were performed. Template DNA was obtained directly from a loopful of cells from an overnight LB plate, suspended in 100 μl of sterile water, and boiled for 10 min. Five microliters of this suspension was added to 20 μl of the PCR mix containing 0.25 mM deoxynucleoside triphosphates, 1 mM MgCl2, 0.6 to 1 mM of each primer, and 1 U of Taq DNA polymerase (Amersham Biosciences). Amplification was performed using an Eppendorf Mastercycler thermal cycler with the following program: a 3-min denaturation step at 95°C; 30 amplification cycles of 1 min at 95°C, 1 min at 45 to 50°C, and 1 min at 72°C; and a final extension step of 10 min at 72°C.
TABLE 1.
PCR oligonucleotides designed and used for cloning the cry1Ia7 gene
| Primer | Sequence (5′-3′) | Positions | Sourcee |
|---|---|---|---|
| 1I(98)Fw | CACTAAAAAATGAAACAGATATAGA | 74-98a | Van Rie, pers. comm. |
| 1I(98)Rv | CCACATATTCATATACTGAGTGRTT | 1057-1080a | Van Rie, pers. comm. |
| 1Ia(10)Fw | TGTCTGAGTATGAAAATGTAGA | 134-155c | Van Rie, pers. comm. |
| 1Ia(11)Rv | GTTTTAATTGGATACATTTG | 841-860c | Van Rie, pers. comm. |
| 1Ib(8)Fw | TCTGAGCATGAGAGTATTGA | 136-155b | Van Rie, pers. comm. |
| 1Ib(9)Rv | GTGGTTTTAATAGGATATACAA | 842-862b | Van Rie, pers. comm. |
| 1I(A)Fw | GTATGAATAAAATTATATCTG | 300-320c | This study |
| 1I(B)Rv | GCAACAAATGTAAATTTGCAGC | 928-949c | This study |
| 1I(C)Fw | TTCTCTACCATAGAGTCTGC | 1318-1337c | This study |
| 1I(D)Rv | TTATAGTCTAAGTCCTCTCC | 2134-2153c | This study |
| 1I(E)Rv | CTACATGTTACGCTCAATATGGA | 2138-2160a | This study |
| 1I(F)Rv | GCAGACTCTATGGTAGAG | 1320-1337c | This study |
| 1I(G)Fw | GGTCTAAATAACTTGAGGGG | 1078-1097c | This study |
| 1I(H)Rv | GAAGAGAAGTTCCAAGCACC | 2221-2240c | This study |
| MUTFw | GGGCTAGCATGAAACTAAAGAATCCd | 1-17a | This study |
The positions given correspond to the cry1Ia7 gene.
The positions given correspond to the cry1Ib1 gene.
The positions given correspond to the cry1Ia1 gene.
The underlined nucleotides represent the NheI site.
Van Rie, pers. comm., personal communication with J. Van Rie (Bayer, Belgium).
PCR products were sequenced and, according to the sequence information obtained, several primers were designed to specifically amplify the whole gene (Table 1). PCR with all possible primer combinations was performed under the conditions described above, and for positive reactions, primers were modified for NheI site inclusion. Three independent PCRs were carried out with primers MUTFw and 1I(E)Rv, and the amplified products were cloned into pGEM-T Easy vector (Promega, Madison, WI). The resulting plasmids were named pPC-ire1, pPC-ire2, and pPC-ire3. These three cloned amplicons were sequenced.
Nucleotide and amino acid sequence analysis.
Sequence homology was determined using the NCBI nucleotide-nucleotide BLAST and protein-protein BLAST online services at http://www.ncbi.nlm.nih.gov/BLAST. Protein alignments were performed by use of ClustalW from the European Bioinformatics Institute at http://www.ebi.ac.uk/services/.
Bioassays.
The activity of the Cry1I-type protein obtained from the recombinant E. coli strain was tested against 10 lepidopteran species, including 1 of the family Bombycidae (Bombyx mori), 6 of Noctuidae (Earias insulana, Helicoverpa armigera, Spodoptera exigua, Spodoptera frugiperda, Spodoptera littoralis, and Trichoplusia ni), 1 of Sphingidae (Manduca sexta), 1 of Tortricidae (Lobesia botrana), and 1 of Plutellidae (P. xylostella). This protein was also tested against two species from other insect orders, namely, Leptinotarsa decemlineata (Coleoptera: Chrysomelidae) and Tipula oleracea (Diptera: Tipulidae). The Cry1I-type protein was not trypsin activated because we were most interested in determining the toxicity spectrum of the Cry1Ia7 protein in its nonprocessed form.
Cry1I-type protein concentrations for bioassays were adjusted by diluting the protein stock with 50 mM NaCO3, 100 mM NaCl, pH 11.3. A preliminary test was performed to determine the specificity of this protein at a relatively high protein concentration (100 μg/ml). For the resulting susceptible species, five different protein concentrations were prepared to determine the mortality responses. Different bioassay methods were chosen depending on the insect tested. For E. insulana, H. armigera, L. botrana, S. exigua, S. frugiperda, S. littoralis, and T. ni, bioassays were performed with neonate larvae by incorporating the protein into an artificial diet (30). A diet surface contamination assay and neonate larvae were used for M. sexta. Bioassays with B. mori, L. decemlineata, P. xylostella, and T. oleracea were carried out by dipping leaf disks prepared from white mulberry, potato, cabbage, or lettuce, respectively, into the protein solution (21). B. mori and P. xylostella were tested with larvae in the second and third instars, respectively, whereas for L. decemlineata and T. oleracea, neonate larvae were used. Positive controls for the bioassays were included in the study by use of the same conditions as described above but with the following toxins: Cry1Ab for B. mori, L. botrana, M. sexta, and P. xylostella; Cry1Ac for E. insulana and H. armigera; Cry1Ba for L. decemlineata; Cry1Ca for S. exigua, S. frugiperda, and S. littoralis; and a crystal suspension of B. thuringiensis serovar israelensis for T. oleracea. Cry1A, Cry1Ba, and Cry1Ca proteins were trypsin activated because, in this state, they represent the toxins with the highest known insecticidal activities against the tested insect species. Negative controls for all the insects tested were included using the same conditions but without any toxin. For each insect species, at least 25 larvae were tested per concentration, and the bioassays were repeated at least three times. Bioassays were conducted at 25°C, 60 to 70% rH, and a 16:8 (light/dark [h]) photoperiod. For all insects bioassayed, mortality was evaluated after 5 days, except for P. xylostella, which was scored after 48 h. Concentration-mortality data for the four most susceptible insect species were analyzed by Probit analysis (28). Additionally, to test for toxins present in the growth medium, purified protein fractions recovered from growth medium supernatant that had been subjected to SDS-PAGE were bioassayed at concentrations of 8 to 11 μg/ml against L. botrana, as described above.
Midgut isolation and preparation of BBMV.
Midguts of E. insulana and L. botrana were dissected from final instar larvae, washed with MET buffer (250 mM mannitol, 17 mM Tris-HCl, 5 mM EDTA, pH 7.5), frozen in liquid nitrogen, and kept at −80°C until used. Brush border membrane vesicles (BBMV) were prepared by the MgCl2 precipitation method (49).
Toxin labeling and binding assays with E. insulana and L. botrana BBMV.
Trypsin-activated Cry1Ia7 and trypsin-activated and fast protein liquid chromatography-purified Cry1Ab and Cry1Ac were labeled with a protein biotinylation kit (Amersham Biosciences) according to the manufacturer's instructions. Binding assays were carried out in a final volume of 0.1 ml binding buffer (PBS [1 mM KH2PO4, 10 mM Na2HPO4, 137 mM NaCl, 2.7 mM KCl, pH 7.4], 0.1% BSA) by incubating the biotinylated protein with the appropriate amount of BBMV (25 μg for both insect species) for 1 h at room temperature. The amounts of biotinylated protein were 20 ng for Cry1Ab, 50 ng for Cry1Ac, and 140 ng for Cry1Ia7 when E. insulana BBMV were used and 30 ng for Cry1Ab and 140 ng for Cry1Ia when L. botrana BBMV were used. The same binding conditions were used for competition assays, but an excess of at least 400-fold of unlabeled protein was added to the reaction mixture. After BBMV incubation, toxin bound to BBMV was recovered by centrifugation at 11,000 × g for 10 min at 4°C followed by two washes with 0.5 ml of cold binding buffer, as described elsewhere (16). The final pellet was solubilized in 10 μl sample buffer (24) and boiled for 10 min, and the proteins were separated by 10% SDS-PAGE. Proteins were electrotransferred onto a nitrocellulose membrane (Hybond ECL; Amersham Biosciences). The membrane was blocked overnight with a solution containing 3% ECL blocking agent (Amersham Biosciences), 0.1% BSA, and 0.1% Tween 20 in PBS; incubated for 1 h with a streptavidin-AP conjugate (Roche Diagnosis, Indiana); and washed three times with 0.1% BSA, 0.1% Tween 20 in PBS. Toxin bound to the BBMV was revealed by incubating the membrane with a Nitro Blue Tetrazolium-BCIP (5-bromo-4-chloro-3-indolylphosphate) solution (Roche Diagnostics).
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper have been filed in the GenBank database under accession number AF278797.
RESULTS
Analysis of the proteins produced by the HU4-2 strain.
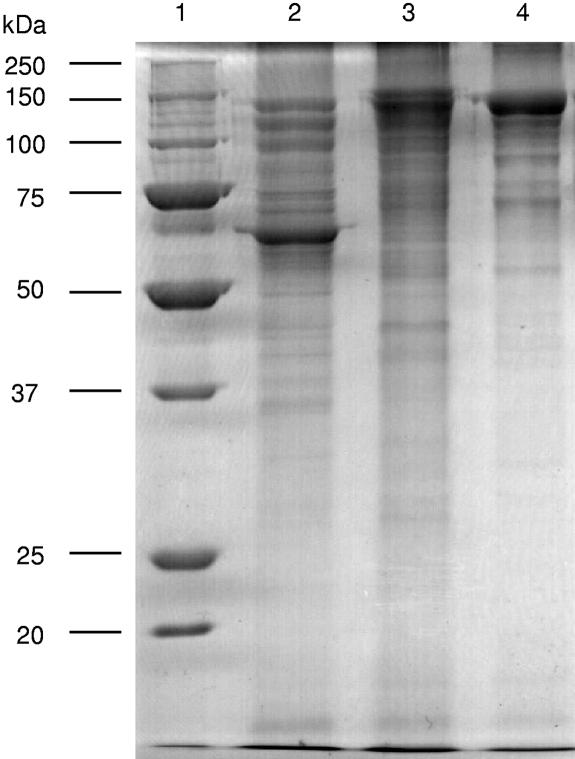
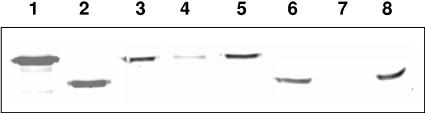
The purified crystals from HU4-2 produced a three-band pattern corresponding to peptides with sizes ranging between 130 and 145 kDa. A band corresponding to the expected molecular mass of a Cry1I-type protein was not detected as a component of the crystal (Fig. 1). However, a protein of ∼75 kDa was observed following SDS-PAGE of purified samples of the growth medium of the HU4-2 strain (data not shown) and was subjected to insect bioassay as described below.
FIG. 1.
SDS-PAGE of parasporal crystals obtained from the commercial biopesticides Dipel (lane 2) and Xentari (lane 3) and from the strain HU4-2 (lane 4) and purified by ultracentrifugation in a sucrose gradient as described in Materials and Methods. Molecular masses of the protein markers (lane 1) are given on the left.
Cloning and nucleotide sequence of the cry1I gene.
The cry1I general primer pair, 1I (98)Fw and 1I(98)Rv, produced an amplification fragment of 983 bp (data not shown), whereas cry1Ia- and cry1Ib-specific primers (Table 1) did not show any amplification, suggesting the presence of a new cry1I gene. Three independent PCRs performed with the primers MUTFw and 1I(E)Rv resulted in the same 2,200-bp fragment being amplified in all of them. The amplified fragments were independently cloned, and plasmids pPC-ire1, pPC-ire2 and pPC-ire3 were obtained. All of these fragments showed identical sequences. The cloned fragments contained an open reading frame in the nucleotide sequence, and these data were deposited into GenBank. Comparison with other available sequences indicated 97.2% identity to cry1Ia1, 93.6% to cry1Ib1, 92.5% to cry1Ic1, 90% to cry1Id1, and 93.7% to cry1Ie.
Deduced amino acid sequence of Cry1Ia7.
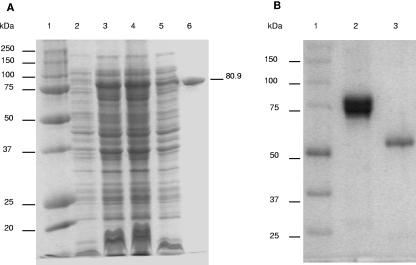
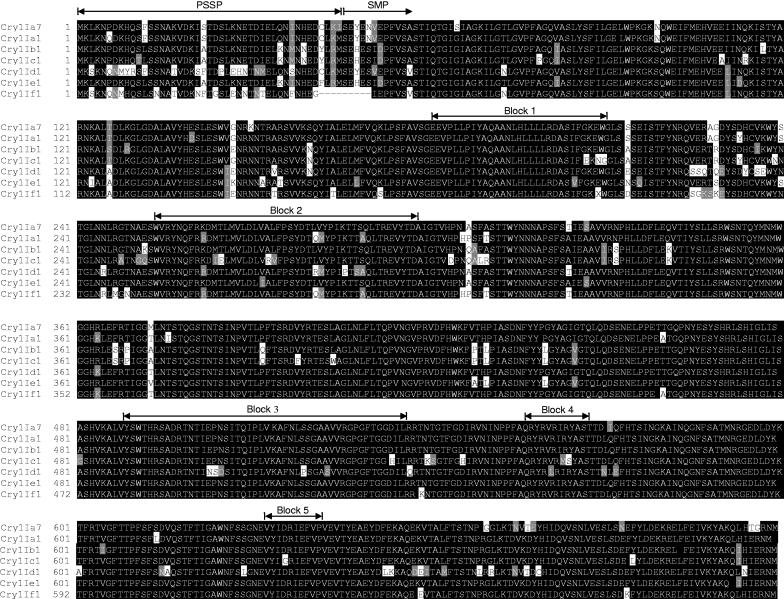
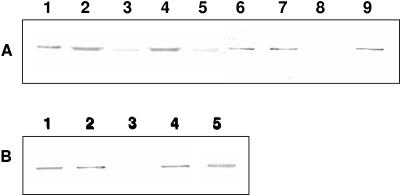
A fragment from pPC-ire1 encoding amino acids 1 to 719 of the Cry1I-type protein was cloned into pET28b(+) vector (Novagen) to construct pPC-ire1I plasmid for T7 polymerase-driven overexpression. The Cry1I-type polypeptide has a predicted molecular mass of 80.9 kDa, which is in agreement with the 80-kDa band observed by SDS-PAGE analysis of the expression and purification procedure (Fig. 2A). Trypsin digestion of this protein resulted in a fragment of ca. 60 kDa, which is typical of activated Cry1 proteins (Fig. 2B). Alignment of the deduced amino acid sequence with those of other known Cry1I proteins showed that the Cry1I-type protein had 96.1% sequence identity to Cry1Ia1, 92.8% to Cry1Ib1, 89.6% to Cry1Ic1, 89% to Cry1Id1, and 93% to Cry1Ie1. According to current classification criteria based on amino acid sequence similarity (6), the name Cry1Ia7 was assigned to the novel polypeptide. Cry1Ia7 had from 28 to 47 amino acid differences from other Cry1I proteins, and these appeared randomly distributed throughout the peptide sequence (Fig. 3). Some amino acid substitutions were found in the five conserved blocks identified. Block 2 was the most variable one, with one to six amino acid substitutions, and block 5 was the most conserved. The N-terminal domain contains features such as a positively charged stretch of amino acids (from M-1 to Q-10) followed by relatively hydrophobic residues (from S-11 to A-16) and a more-polar region (from K-17 to K-44). This region is highly conserved in Cry1I proteins, as confirmed in the amino acid sequence analysis (Fig. 3), and may function as a secretion signal peptide in Bacillus species (40). Thus, in B. thuringiensis, the mature putative protein would start at amino acid 46 in the predicted amino acid sequence and may be secreted as a protein with a theoretical molecular mass of ∼75 kDa.
FIG. 2.
SDS-PAGE showing the expression and purification of Cry1Ia7. (A) Lanes: 1, molecular mass markers; 2, E. coli BL21(DE3) native strain; 3, E. coli transformed with plasmid pPC-ire1 after IPTG-induced expression of the cloned cry1Ia7 gene; 4, pellet of the culture that expressed Cry1Ia7 after sonication; 5, supernatant of this culture after sonication; 6, Cry1Ia7 protein after nickel affinity column purification and Sephadex column buffer exchange. (B) Cry1Ia7 protein after purification (lane 2) and after trypsin digestion (lane 3). Molecular sizes of the markers (lanes 1) are given in kDa.
FIG. 3.
Comparison of the deduced amino acid sequences of Cry1I proteins. Conserved amino acid blocks for Cry1I proteins, predicted secretion signal peptides (PSSP), and starts of mature proteins (SMP) are shown. Letters highlighted in black are conserved amino acids, and those in gray are semiconserved.
Cry1Ia7 insecticidal activity.
The Cry1Ia7 protoxin was active against the lepidopterans E. insulana, L. botrana, and P. xylostella and the coleopteran L. decemlineata (Table 2). However, Cry1Ia7 was not toxic at the highest doses assayed (100 μg/ml) to the lepidopterans B. mori, H. armigera, M. sexta, S. exigua, S. frugiperda, S. littoralis, and T. ni or to the dipteran T. oleracea. Cry1Ia7 50% lethal concentration (LC50) values were calculated for the four susceptible species and then compared to the LC50 values of activated Cry proteins known to be active against these species (Table 2). With an LC50 of 10.0 μg/ml, Cry1Ia7 was 1.8-fold more active than Cry1Ba against L. decemlineata but less active than the control Cry1A proteins chosen for being among the most toxic ones for these species. Against L. botrana, Cry1Ia7 (LC50 of 8.6 μg/ml) was sixfold less active than activated Cry1Ab. Against E. insulana, Cry1Ia7 (LC50 of 21.1 μg/ml) was 19-fold less active than activated Cry1Ac. Against P. xylostella, Cry1Ia7 (LC50 of 12.2 μg/ml) was around 150-fold less active than activated Cry1Ab.
TABLE 2.
Toxicity of Cry1Ia7 against first-instar larvae of L. botrana, E. insulana, P. xylostella, and L. decemlineataa
| Treatment for indicated species | Regression line value
|
LC50 (μg/ml) | Distribution value
|
Relative potency | Confidence limit (95%)
|
|||
|---|---|---|---|---|---|---|---|---|
| Slope ± SE | a↓ ± SEb | χ2 | df | Lower | Upper | |||
| L. botrana | ||||||||
| Cry1Ab | 1.87 ± 0.20 | 4.71 ± 0.12 | 1.4 | 0.30 | 3 | 1 | ||
| Cry1Ia7 | 2.93 ± 0.19 | 2.65 ± 0.20 | 8.6 | 0.75 | 3 | 0.18 | 0.13 | 0.23 |
| E. insulana | ||||||||
| Cry1Ac | 1.87 ± 0.17 | 4.92 ± 0.06 | 1.1 | 0.09 | 3 | 1 | ||
| Cry1Ia7 | 3.25 ± 0.35 | 0.71 ± 0.47 | 21.1 | 1.27 | 3 | 0.05 | 0.04 | 0.06 |
| P. xylostella | ||||||||
| Cry1Abc | 1.90 ± 0.20 | ND | 0.08 | ND | ND | ND | ND | ND |
| Cry1Ia7 | 2.74 ± 0.25 | 2.01 ± 0.26 | 12.3 | 1.62 | 3 | ND | ND | ND |
| L. decemlineata | ||||||||
| Cry1Ba | 1.32 ± 0.09 | 3.35 ± 0.14 | 17.9 | 1.33 | 3 | 1 | ||
| Cry1Ia7 | 1.32 ± 0.09 | 3.68 ± 0.12 | 10.0 | 1.22 | 3 | 1.79 | 1.2 | 2.4 |
Relative potencies were calculated with respect to positive control Cry1 proteins that have been trypsin activated. ND, not determined.
a↓, intercept of the regression line.
Data are from reference 14.
Bioassay of the ∼75-kDa protein component purified from the HU4-2 growth medium resulted in 47 to 53% mortality of L. botrana larvae that had consumed 8 to 11 μg protein/ml, which is comparable with the insecticidal activity of Cry1Ia7 expressed in E. coli.
Binding experiments with E. insulana and L. botrana BBMV.
Biotinylated Cry1Ia7 bound to BBMV from the two insect species, and this binding was specific, since it was completely displaced by the addition of an excess of the same unlabeled toxin (Fig. 4, lanes 6 and 7, for L. botrana and Fig. 5B, lanes 2 and 3, for E. insulana). Heterologous competition experiments were also performed with biotinylated Cry1Ia7 and an excess of unlabeled Cry1A toxins. In these experiments, neither Cry1Ab nor Cry1Ac displaced Cry1Ia7 binding to BBMV (Fig. 4, lane 8, for L. botrana, and Fig. 5B, lanes 4 and 5, for E. insulana). Similarly, experiments with biotinylated Cry1Ab and biotinylated Cry1Ac (the former only with E. insulana BBMV) were carried out, and the binding was also shown to be specific (Fig. 4, lanes 3 and 4, and Fig. 5A, lanes 2, 3, 7, and 8). Cry1Ia7 could not displace Cry1Ab binding to L. botrana BBMV (Fig. 4, lane 5), nor could it displace Cry1Ab or Cry1Ac binding to E. insulana BBMV (Fig. 5A, lanes 4 and 9, respectively). Cry1Ac could displace Cry1Ab binding in E. insulana (Fig. 5A, lane 5), in agreement with previous data (20). Taken together, these results indicate that Cry1Ia7 binds to sites different from those of Cry1Ab in both species. In the case of E. insulana, where Cry1Ac was also included in the analysis, this toxin shared binding sites with Cry1Ab but not with Cry1Ia7.
FIG. 4.
Binding and competition experiments with biotinylated Cry1Ab and Cry1Ia7 with L. botrana BBMV. Lanes: 1, biotinylated Cry1Ab (control without BBMV); 2, biotinylated Cry1Ia7 (control without BBMV); 3 to 5, binding of biotinylated Cry1Ab to BBMV (lane 3) and in the presence of an excess of unlabeled Cry1Ab (lane 4) or Cry1Ia7 (lane 5); 6 to 8, binding of biotinylated Cry1Ia7 to BBMV (lane 6) and in the presence of an excess of unlabeled Cry1Ia7 (lane 7) or Cry1Ab (lane 8).
FIG. 5.
Binding and competition experiments with biotinylated Cry1Ab, Cry1Ac, and Cry1Ia7 with E. insulana BBMV. (A) Lanes: 1, biotinylated Cry1Ab (control without BBMV); 2 to 5, binding of biotinylated Cry1Ab to BBMV either without further addition (lane 2) or in the presence of an excess of unlabeled Cry1Ab (lane 3), Cry1Ia7 (lane 4), or Cry1Ac (lane 5); 6, biotinylated Cry1Ac (control without BBMV); 7 to 9, binding of biotinylated Cry1Ac to BBMV either without further addition (lane 7) or and in the presence of an excess of unlabeled Cry1Ac (lane 8) or Cry1Ia7 (lane 9). (B) Lanes: 1, biotinylated Cry1Ia7 (control without BBMV); 2 to 5, binding of biotinylated Cry1Ia7 to BBMV either without further addition (lane 2) or and in the presence of an excess of unlabeled Cry1Ia7 (lane 3), Cry1Ab (lane 4), or Cry1Ac (lane 5).
DISCUSSION
A novel gene encoding a protein of the Cry1I group has been cloned and sequenced. The new protein has interesting insecticidal properties, particularly with regard to its wide host range, a feature of some proteins in this group. Moreover, its binding sites are different from those of the Cry1A toxins commonly found in Bt crops and B. thuringiensis-based insecticides.
Our studies did not detect the protein (predicted molecular mass of 81 kDa) by SDS-PAGE analysis after the crystal was formed in HU4-2 cells. However, the cry1Ia7 gene was identified in this strain, which contains an important level of diversity of additional cry genes. Sometimes the cry gene complex of a strain is not reflected in the protein content of the crystal (33), because not all cry genes are expressed or because not all Cry proteins take part in crystal formation, and some may be secreted into the growth medium (10, 23). In fact, our results indicated a significant contribution of 130- to 145-kDa proteins in the parasporal crystal that may correspond to the expression of other cry1 genes, but not cry1I, which code for proteins of approximately 80 kDa (6). This could be due to the lack of significant expression of this gene or to the fact that the protein is secreted into the medium (23, 37, 41, 46).
The complete open reading frame of the new cry1Ia7 gene was cloned and expressed in E. coli cells. SDS-PAGE analysis of the purified protein generated in E. coli revealed a major peptide of ca. 81 kDa which is similar to other proteins of the Cry1I group (5, 12, 41, 43). Cry1I proteins produce inclusion bodies when they are expressed in E. coli. However, these proteins do not participate in crystal formation, since they are secreted into the medium during the vegetative growth phase of the bacterium (5, 23, 37). Cry1Ia7 also produced inclusion bodies in E. coli after being induced by IPTG at 37°C, but at a lower temperature (25°C), the protein produced is soluble. Lower growth temperatures, a different culture medium (2× TY instead of LB), and a lack of overexpression could be some of the reasons for this difference. Amino acid sequence analysis of Cry1Ia7 showed characteristics similar to those of other secreted proteins from the same group. However, Cry1Ia7 was different at several positions from the reported Cry1I proteins (46). Moreover, minor changes in amino acid sequences can produce important variations in the host range or insecticidal properties of these toxins. Cry1Ia1 and Cry1Ia2 differ by a single amino acid in domain II and exhibit differing insecticidal activity spectra (12). Cry1Ia7 differs from Cry1Ia1 by four amino acids which are scattered throughout block II, so we may have expected modifications in the toxicity and specificity of the encoded Cry protein. However, these changes in the sequence did not appear to modify the activity spectrum. The regions corresponding to the three domains of Cry1Ia7 were highly similar to those of Cry1Ia1. Loop regions in domain II have been reported to be involved in receptor binding and toxicity (36), and the three potential loop regions in domain II of Cry1Ia7 matched those of Cry1Ia1. Similar results were obtained by Choi et al. (5), who observed no amino acid differences in loop regions of Cry1Id1 compared with Cry1Ia3, although the latter protein exhibited a significantly higher activity against B. mori. Amino acid substitutions in regions adjacent to the loops of domain II might also affect the protein conformation, and thus the toxicity could also be altered. In addition, domain III has been reported to be involved in receptor binding (7), and some of the amino acid substitutions in the corresponding region of Cry1Ia7 could therefore correlate with differences in receptor binding and toxicity.
Cry1I toxins are particularly interesting from an agricultural perspective because of their wide host range. Other proteins, such as Cry1B, Cry1C, and Cry2A, also exhibit host ranges spanning more than one insect order (1, 4, 48, 50). Cry1I, Cry1B, Cry1C, and Cry2A protein groups include the only native proteins which, independent of their differential proteolytic processing, are toxic for insects of different orders. Cry1I proteins were initially characterized by their dual activity towards Lepidoptera and Coleoptera (43). The first protein reported by Tailor et al. (43) had dual activity against L. decemlineata and Ostrinia nubilalis, but the activity of the remaining Cry1I-type proteins that have been characterized subsequently has been restricted to lepidopteran species (5, 12, 23). Cry1Ic2 is the only protein for which insecticidal activity against a beetle, Diabrotica virgifera, has been reported (34). Cry1Ia7 protein has been found not to be toxic against H. armigera, T. ni, and Spodoptera spp. However, Cry1Ia7 was toxic against P. xylostella, an insect which is usually susceptible to Cry1I proteins (5, 23, 37, 39, 41). Nevertheless, Cry1Ia7 showed activity against L. decemlineata, supporting the dual activity against Lepidoptera and Coleoptera described by Tailor et al. (43). Moreover, Cry1Ia7 was toxic to L. botrana larvae, whose susceptibility had not been previously reported in the Cry1I group.
Despite the fact that binding site competition studies involving Cry1A toxins are numerous in the literature (8, 9, 11, 15, 16, 47), this study is the first involving a Cry1I toxin. Cry1A proteins compete for common binding sites in all species studied, and this forms the basis of cross-resistance or multiple resistance among these toxins (2, 9, 13, 27). To determine the compatibility, in terms of resistance management, of Cry1Ia7 with other Cry1A proteins widely used in Bt crops, we performed heterologous binding assays with BBMV from L. botrana and E. insulana. The results showed that Cry1Ia7 does not compete for Cry1Ab or Cry1Ac binding sites. It is therefore unlikely that development of resistance to Cry1A toxins would confer cross-resistance to Cry1Ia7 or vice versa (11). In view of the similarities found in binding site models among insect species, the results obtained with L. botrana and E. insulana seem likely to apply to other species.
The fact that Cry1I proteins are secreted and not crystallized impedes their use in biopesticide spray applications. Nevertheless, their interesting insecticidal characteristics can be successfully exploited if Cry1I proteins are expressed in transgenic plants (25, 29, 38). Alternatively, cry1I genes could be cloned in Pseudomonas spp., thus expressing the Cry1I proteins and microencapsulating them in the bacterial cell wall (22). Microencapsulated Cry1I could be used in spray applications alone or in rotations with B. thuringiensis-based insecticides containing Cry1A toxins.
Acknowledgments
We thank Eloi Erro, Ainara Nepote, and Noelia Gorría for technical assistance. We are grateful to Trevor Williams for critically reading the manuscript.
This study was funded by MCyT projects (AGL2000-0840-C03 and AGL2003-09282-CO3) and the Generalitat Valenciana (GRUPOS2004-21). I.R.D.E. received a Government of Navarra fellowship. A.E. was supported by a fellowship (FP2000-5497) and B.E. and J.A.O. received support from the Spanish Ministry of Education and Culture Ramón y Cajal program.
REFERENCES
- 1.Abdul-Rauf, M., and D. J. Ellar. 1999. Toxicity and receptor binding properties of a Bacillus thuringiensis CryIC toxin active against both Lepidoptera and Diptera. J. Invertebr. Pathol. 73:52-58. [DOI] [PubMed] [Google Scholar]
- 2.Ballester, V., F. Granero, B. E. Tabashnik, T. Malvar, and J. Ferré. 1999. Integrative model for binding of Bacillus thuringiensis toxins in susceptible and resistant larvae of the diamondback moth (Plutella xylostella). Appl. Environ. Microbiol. 65:1413-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Bradley, D., M. A. Harkey, M. K. Kim, K. D. Biever, and L. S. Bauer. 1995. The insecticidal CrylB crystal protein of Bacillus thuringiensis ssp thuringiensis has dual specificity to Coleopteran and Lepidopteran larvae. J. Invertebr. Pathol. 65:162-173. [DOI] [PubMed] [Google Scholar]
- 5.Choi, S. K., B. S. Shin, E. M. Kong, H. M. Rho, and S. H. Park. 2000. Cloning of a new Bacillus thuringiensis Cry1I-type crystal protein. Curr. Microbiol. 41:65-69. [DOI] [PubMed] [Google Scholar]
- 6.Crickmore, N., D. R. Zeigler, J. Feitelson, E. Schnepf, J. Van Rie, D. Lereclus, J. Baum, and D. H. Dean. 1998. Revision of the nomenclature for the Bacillus thuringiensis pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Maagd, R. A., H. van der Klei, P. L. Bakker, W. J. Stiekema, and D. Bosch. 1996. Different domains of Bacillus thuringiensis δ-endotoxins can bind to insect midgut membrane proteins on ligand blots. Appl. Environ. Microbiol. 62:2753-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Escriche, B., J. Ferré, and F. J. Silva. 1997. Occurrence of a common binding site in Mamestra brassicae, Phthorimaea operculella, and Spodoptera exigua for the insecticidal crystal proteins CryIA from Bacillus thuringiensis. Insect Biochem. Mol. Biol. 27:651-656. [DOI] [PubMed] [Google Scholar]
- 9.Estela, A., B. Escriche, and J. Ferré. 2004. Interaction of Bacillus thuringiensis toxins with larval midgut binding sites of Helicoverpa armigera (Lepidoptera: Noctuidae). Appl. Environ. Microbiol. 70:1378-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Estruch, J. J., G. W. Warren, M. A. Mullins, G. J. Nye, J. A. Craig, and M. G. Koziel. 1996. Vip3A, a novel Bacillus thuringiensis vegetative insecticidal protein with a wide spectrum of activities against lepidopteran insects. Proc. Natl. Acad. Sci. USA 93:5389-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferré, J., and J. Van Rie. 2002. Biochemistry and genetics of insect resistance to Bacillus thuringiensis. Annu. Rev. Entomol. 47:501-533. [DOI] [PubMed] [Google Scholar]
- 12.Gleave, A. P., R. Williams, and R. J. Hedges. 1993. Screening by polymerase chain reaction of Bacillus thuringiensis serotypes for the presence of cryV-like insecticidal protein genes and characterization of a cryV gene cloned from Bacillus thuringiensis subsp. kurstaki. Appl. Environ. Microbiol. 59:1683-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.González-Cabrera, J., B. Escriche, B. E. Tabashnik, and J. Ferré. 2003. Binding of Bacillus thuringiensis toxins in resistant and susceptible strains of pink bollworm (Pectinophora gossypiella). Insect Biochem. Mol. Biol. 33:929-935. [DOI] [PubMed] [Google Scholar]
- 14.González-Cabrera, J., S. Herrero, A. H. Sayyed, B. Escriche, Y. B. Liu, S. K. Meyer, D. J. Wright, B. E. Tabashnik, and J. Ferré. 2001. Variation in susceptibility to Bacillus thuringiensis toxins among unselected strains of Plutella xylostella. Appl. Environ. Microbiol. 67:4610-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Granero, F., V. Ballester, and J. Ferré. 1996. Bacillus thuringiensis crystal proteins Cry1Ab and Cry1Fa share a high affinity binding site in Plutella xylostella (L.). Biochem. Biophys. Res. Commun. 224:779-783. [DOI] [PubMed] [Google Scholar]
- 16.Herrero, S., J. González-Cabrera, B. E. Tabashnik, and J. Ferré. 2001. Shared binding sites in Lepidoptera for Bacillus thuringiensis Cry1Ja and Cry1A toxins. Appl. Environ. Microbiol. 67:5729-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofmann, C., and P. Lüthy. 1986. Binding and activity of Bacillus thuringiensis delta-endotoxin to invertebrate cells. Arch. Microbiol. 146:7-11. [DOI] [PubMed] [Google Scholar]
- 18.Hofmann, C., P. Lüthy, R. Hütter, and V. Pliska. 1988. Binding of the d-endotoxin from Bacillus thuringiensis to brush-border membrane vesicles of the cabbage butterfly (Pieris brassicae). Eur. J. Biochem. 173:85-91. [DOI] [PubMed] [Google Scholar]
- 19.Höfte, H., and H. R. Whiteley. 1989. Insecticidal crystal proteins of Bacillus thuringiensis. Microbiol. Rev. 53:242-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ibargutxi, M. A., A. Estela, J. Ferré, and P. Caballero. 2006. Bacillus thuringiensis toxins for the control of the cotton pest Earias insulana (Boisd.) (Lepidoptera: Noctuidae). Appl. Environ. Microbiol. 72:437-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iriarte, J., Y. Bel, M. D. Ferrandis, R. Andrew, J. Murillo, J. Ferré, and P. Caballero. 1998. Environmental distribution and diversity of Bacillus thuringiensis in Spain. Syst. Appl. Microbiol. 21:97-106. [DOI] [PubMed] [Google Scholar]
- 22.Kaur, S. 2000. Molecular approaches towards development of novel Bacillus thuringiensis biopesticides. World J. Microb. Biotechnol. 16:781-793. [Google Scholar]
- 23.Kostichka, K., G. W. Warren, M. Mullins, A. D. Mullins, J. A. Craig, M. G. Koziel, and J. J. Estruch. 1996. Cloning of a cryV-type insecticidal protein gene from Bacillus thuringiensis: the cryV-encoded protein is expressed early in stationary phase. J. Bacteriol. 178:2141-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laemmli, U. K. 1970. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 25.Lagnaoui, A., V. Cañedo, and D. S. Douches. 2001. Evaluation of Bt-cry1Ia1 (cryV) transgenic potatoes on two species of potato tuber moth, Phthorimaea operculella and Symmetrischema tangolias (Lepidoptera: Gelechiidae) in Peru. CIP Program Rep. 1999-2000:117-121.
- 26.Lech, K., and R. Brent. 1992. Escherichia coli, plasmids and bacteriophages, p. 1-51. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Short protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 27.Lee, M. K., F. Rajamohan, F. Gould, and D. H. Dean. 1995. Resistance to Bacillus thuringiensis CryIA δ-endotoxins in a laboratory-selected Heliothis virescens strain is related to receptor alteration. Appl. Environ. Microbiol. 61:3836-3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.LeOra Software. 1987. POLO-PC: a user's guide to Probit or Logit analysis. LeOra Software, Berkeley, Calif.
- 29.Liu, Y. J., F. P. Song, K. L. He, Y. Yuan, X. X. Zhang, P. Gao, J. H. Wang, and G. Y. Wang. 2004. Expression of a modified cry1Ie gene in E. coli and in transgenic tobacco confers resistance to corn borer. Acta Biochim. Biophys. Sin. 36:309-313. [DOI] [PubMed] [Google Scholar]
- 30.MacIntosh, S. C., T. B. Stone, S. R. Sims, P. L. Hunst, J. T. Greenplate, P. G. Marrone, F. J. Perlak, D. A. Fischhoff, and R. L. Fuchs. 1990. Specificity and efficacy of purified Bacillus thuringiensis proteins against agronomically important insects. J. Invertebr. Pathol. 56:258-266. [DOI] [PubMed] [Google Scholar]
- 31.Marroquin, L. D., D. Elyassnia, J. S. Griffitts, J. S. Feitelson, and R. V. Aroian. 2000. Bacillus thuringiensis (Bt) toxin susceptibility and isolation of resistance mutants in the nematode Caenorhabditis elegans. Genetics 155:1693-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin, P. A. W., and R. S. Travers. 1989. Worldwide abundance and distribution of Bacillus thuringiensis isolates. Appl. Environ. Microbiol. 55:2437-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martínez, C., M. Porcar, A. López, I. Ruiz de Escudero, F. J. Pérez-Llarena, and P. Caballero. 2004. Characterization of a Bacillus thuringiensis strain with a broad spectrum of activity against lepidopteran insects. Entomol. Exp. Appl. 111:71-77. [Google Scholar]
- 34.Osman, Y. A., M. A. Madkour, J. Bulla, and A. Lee. May 2001. Bacillus thuringiensis isolates with broad spectrum. U.S. patent 6,232,439.
- 35.Payne, J., D. A. Cummings, R. Cannon, J. C. Raymond, K. E. Narva, and S. Stelman. March 1998. Bacillus thuringiensis genes encoding lepidopteran-active toxins. U.S. patent 5,723,758.
- 36.Schnepf, E., N. Crickmore, J. Van Rie, D. Lereclus, J. Baum, J. Feitelson, D. R. Zeigler, and D. H. Dean. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:775-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Selvapandiyan, A., N. Arora, R. Rajagopal, S. K. Jalali, T. Venkatesan, S. P. Singh, and R. K. Bhatnagar. 2001. Toxicity analysis of N- and C-terminus-deleted vegetative insecticidal protein from Bacillus thuringiensis. Appl. Environ. Microbiol. 67:5855-5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Selvapandiyan, A., V. S. Reddy, P. A. Kumar, K. K. Tewari, and R. K. Bhatnagar. 1998. Transformation of Nicotiana tabacum with a native cry1Ia5 gene confers complete protection against Heliothis armigera. Mol. Breed. 4:473-478. [Google Scholar]
- 39.Shin, B. S., S. H. Park, S. K. Choi, B. T. Koo, S. T. Lee, and J. I. Kim. 1995. Distribution of cryV-type insecticidal protein genes in Bacillus thuringiensis and cloning of cryV-type genes from Bacillus thuringiensis subsp. kurstaki and Bacillus thuringiensis subsp. entomocidus. Appl. Environ. Microbiol. 61:2402-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simonen, M., and I. Palva. 1993. Protein secretion in Bacillus species. Microbiol. Mol. Biol. Rev. 57:109-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song, F. P., J. Zhang, A. X. Gu, Y. Wu, L. L. Han, K. L. He, Z. Y. Chen, J. Yao, Y. Q. Hu, G. X. Li, and D. F. Huang. 2003. Identification of cry1I-type genes from Bacillus thuringiensis strains and characterization of a novel cry1I-type gene. Appl. Environ. Microbiol. 69:5207-5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stewart, G. S. A., K. Johnstone, E. Hagelberg, and D. J. Ellar. 1981. Commitment of bacterial spores to germinate. Biochem. J. 198:101-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tailor, R., J. Tippett, G. Gibb, S. Pells, D. Pike, L. Jordan, and S. Ely. 1992. Identification and characterization of a novel Bacillus thuringiensis d-endotoxin entomocidal to coleopteran and lepidopteran larvae. Mol. Microbiol. 6:1211-1217. [DOI] [PubMed] [Google Scholar]
- 44.Thomas, W. E., and D. J. Ellar. 1983. Mechanism of action of Bacillus thuringiensis var israelensis insecticidal δ-endotoxin. FEBS Lett. 154:362-368. [DOI] [PubMed] [Google Scholar]
- 45.Tojo, A., and K. Aizawa. 1983. Dissolution and degradation of Bacillus thuringiensis δ-endotoxin by gut juice protease of the silkworm Bombyx mori. Appl. Environ. Microbiol. 45:576-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tounsi, S., N. Zouari, and S. Jaoua. 2003. Cloning and study of the expression of a novel cry1Ia-type gene from Bacillus thuringiensis subsp kurstaki. J. Appl. Microbiol. 95:23-28. [DOI] [PubMed] [Google Scholar]
- 47.Van Rie, J., S. Jansens, H. Höfte, D. Degheele, and H. Van Mellaert. 1989. Specificity of Bacillus thuringiensis delta-endotoxins. Importance of specific receptors on brush border membrane of the mid-gut of target insects. Eur. J. Biochem. 186:239-247. [DOI] [PubMed] [Google Scholar]
- 48.Widner, W. R., and H. R. Whiteley. 1990. Location of the dipteran specificity region in a lepidopteran-dipteran crystal protein from Bacillus thuringiensis. J. Bacteriol. 172:2826-2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolfersberger, M., P. Luethy, A. Maurer, P. Parenti, F. V. Sacchi, B. Giordana, and G. M. Hanozet. 1987. Preparation and partial characterization of amino-acid transporting brush border membrane vesicles from the larval midgut of the cabbage butterfly (Pieris brassicae). Comp. Biochem. Physiol. 86:301-308. [Google Scholar]
- 50.Zhong, C. H., D. J. Ellar, A. Bishop, C. Johnson, S. S. Lin, and E. R. Hart. 2000. Characterization of a Bacillus thuringiensis d-endotoxin which is toxic to insects in three orders. J. Invertebr. Pathol. 76:131-139. [DOI] [PubMed] [Google Scholar]