Abstract
The titan mutants of Arabidopsis exhibit striking defects in seed development. The defining feature is the presence of abnormal endosperm with giant polyploid nuclei. Several TTN genes encode structural maintenance of chromosome proteins (condensins and cohesins) required for chromosome function at mitosis. Another TTN gene product (TTN5) is related to the ARL2 class of GTP-binding proteins. Here, we identify four additional TTN genes and present a general model for the titan phenotype. TTN1 was cloned after two tagged alleles were identified through a large-scale screen of T-DNA insertion lines. The predicted gene product is related to tubulin-folding cofactor D, which interacts with ARL2 in fission yeast (Schizosaccharomyces pombe) and humans to regulate tubulin dynamics. We propose that TTN5 and TTN1 function in a similar manner to regulate microtubule function in seed development. The titan phenotype can therefore result from disruption of chromosome dynamics (ttn3, ttn7, and ttn8) or microtubule function (ttn1 and ttn5). Three other genes have been identified that affect endosperm nuclear morphology. TTN4 and TTN9 appear to encode plant-specific proteins of unknown function. TTN6 is related to the isopeptidase T class of deubiquitinating enzymes that recycle polyubiquitin chains following protein degradation. Disruption of this gene may reduce the stability of the structural maintenance of chromosome complex. Further analysis of the TITAN network should help to elucidate the regulation of microtubule function and chromosome dynamics in seed development.
Seed development in Arabidopsis requires coordinated differentiation of the embryo proper, suspensor, endosperm tissue, and seed coat. Interactions between these components have been explored in part through the analysis of embryo-defective mutants (Meinke, 1995). Some of these mutants have provided insights into the maintenance of cellular identity during seed development. Suspensor cell identity has been examined in twin mutants (Vernon and Meinke, 1994; Zhang and Somerville, 1997), meristem identity explored in stm mutants (Long et al., 1996), and cotyledon identity analyzed in lec mutants (Meinke, 1992; Lotan et al., 1998). Embryo-defective mutants have also been used to identify large numbers of genes with essential functions during seed development (McElver et al., 2001). Gene products identified to date include a variety of metabolic enzymes (Patton et al., 1998; Jang et al., 2000; Schrick et al., 2000; Boisson et al., 2001; Lukowitz et al., 2001), transcription factors (Long et al., 1996; Hardtke and Berleth, 1998; Li and Thomas, 1998; Lotan et al., 1998), chloroplast and mitochondrial proteins (Tsugeki et al., 1996; Uwer et al., 1998; Albert et al., 1999; Apuya et al., 2001), and proteins required for vesicle trafficking (Lauber et al., 1997; Assaad et al., 2001; Rojo et al., 2001). These essential genes represent an important subset of the minimal gene set needed to make a functional plant.
Early endosperm development in Arabidopsis is characterized by specialized patterns of nuclear division, nuclear migration, and delayed cellularization (Brown et al., 1999; Otegui and Staehelin, 2000; Boisnard-Lorig et al., 2001; Olsen, 2001). Endosperm identity is therefore modulated to some extent by factors that regulate mitosis and cell division. The TITAN genes described in this report play an important role in this process of endosperm differentiation. Genetic analysis of endosperm development in Arabidopsis has focused in recent years on mutants with defects in gene imprinting and inappropriate endosperm development in the absence of fertilization (Grossniklaus et al., 1998; Luo et al., 1999; Ohad et al., 1999; Yadegari et al., 2000; Sorensen et al., 2001). These studies have underscored the importance of polycomb proteins and associated factors in regulating gene expression and nuclear division during early stages of endosperm development.
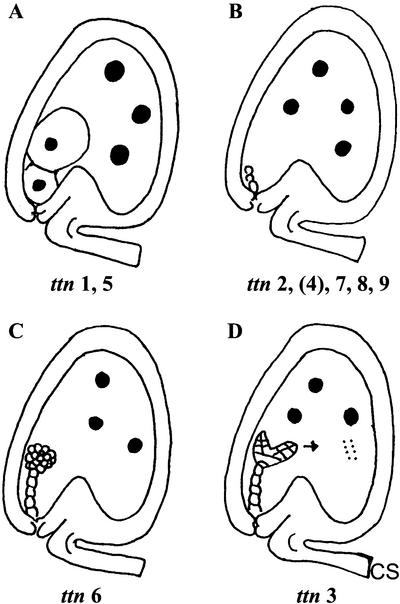
Three titan mutants with striking defects in embryo and endosperm development were originally described by Liu and Meinke (1998). These mutants are characterized by dramatic enlargement of endosperm nuclei (Fig. 1). Embryo phenotypes depend on the locus involved: giant cells with enlarged nuclei (ttn1), small cells arrested early in development (ttn2), or viable cells that survive seed desiccation (ttn3). The tagged ttn3 mutant is disrupted in a gene that encodes a chromosome scaffold protein (SMC2) related to structural maintenance of chromosome (SMC) proteins in Saccharomyces cerevisiae, which are required for normal chromosome function at mitosis (Liu et al., 2002). The weak embryo phenotype appears to result from expression of a duplicate gene with overlapping functions. Another tagged mutant (ttn5) with a phenotype similar to ttn1 was identified in a separate collection of insertion lines (McElver et al., 2001). This gene encodes a small GTP-binding protein (ARL2) related to ADP ribosylation factors (McElver et al., 2000). Related mutants (pilz) with large embryo cells and defects in microtubule organization have also been described by Mayer et al. (1999). Comparison of map locations suggests that hal corresponds to ttn5 and that cho is ttn1.
Figure 1.
Phenotypic classes of titan mutants identified. Large black dots represent enlarged nucleoli. Small dots in ttn3 endosperm correspond to condensed mitotic chromosomes. Arrow indicates continued embryo development in ttn3 seeds. An intermediate embryo phenotype is observed in ttn4 seeds late in development.
To establish a more complete picture of TITAN functions in seed development, we performed a forward genetic screen for additional knockouts within a large collection of insertion lines generated at Syngenta (McElver et al., 2001). This screen expanded the total number of titans to include at least 17 mutants defective in nine different genes. Two of these genes encode SMC1 and SMC3 cohesins, which are known to interact with condensins in other eukaryotes to regulate chromosome dynamics (Liu et al., 2002). We therefore have established a strong connection between loss of SMC function during seed development and the appearance of a titan endosperm phenotype.
We describe in this report the identification of four additional TITAN genes represented by tagged mutant alleles. One of these (TTN1) encodes a regulatory protein known as tubulin-folding cofactor D, which interacts with ARL2 in fission yeast (Schizosaccharomyces pombe; Radcliffe et al., 2000a, 2000b) and humans (Homo sapiens; Bhamidipati et al., 2000) to modulate microtubule dynamics. This discovery makes it possible to explain much of the ttn1 phenotype (Liu and Meinke, 1998), clarify the role of TTN5 in seed development (McElver et al., 2000), and explain the loss of microtubules noted in pilz mutants (Mayer et al., 1999). A second gene (TTN6) encodes a deubiquitinating enzyme related to human isopeptidase T (Wilkinson, 1997). Knockouts in this gene (AtUBP14) have recently been noted to result in embryonic lethality (Doelling et al., 2001), but the titan phenotype was not identified. The ttn6 mutant establishes a connection between TITAN protein networks and the ubiquitin pathway. TTN4 corresponds to a senescence-associated gene (SAG18) that encodes a novel protein (Weaver et al., 1998; Miller et al., 1999) with an unknown function in seed development. Another gene (TTN9) with a titan endosperm phenotype also appears to encode a novel protein. These results are consistent with a model in which titan abnormalities result from disruption of either microtubule function or chromosome dynamics during seed development. The novel proteins may influence these central pathways indirectly through mechanisms unique to plants. Elucidation of additional TITAN functions should provide further insights into the regulation of mitosis and cytokinesis during endosperm development and the complex network of proteins required to differentiate endosperm from other parts of the seed.
RESULTS
Isolation of Tagged titan Mutants
A forward genetic screen of T-DNA insertion lines produced at Syngenta was performed to identify tagged titan mutants amenable to gene isolation. This project was part of a large-scale effort to isolate tagged embryo-defective mutants and to identify essential genes in Arabidopsis (McElver et al., 2001). Two strategies were used to find titan mutants within this collection: screening immature siliques from heterozygous plants for glassy seeds indicative of a titan phenotype; and examining cleared seeds with Nomarski optics for the presence of enlarged endosperm nucleoli (Liu and Meinke, 1998). The second approach was generally reserved for tagged mutants with flanking sequence information. Tagging status was resolved by identifying mutant lines with a low ratio of resistant-to-sensitive seedlings, transplanting resistant seedlings to soil, and looking for linkage between the resistance gene and mutant phenotype (McElver et al., 2001). Results of this insertional mutagenesis project are summarized in Table I. Mutations in at least nine different genes have been identified that result in a strong titan phenotype. These mutants can be placed into four phenotypic classes as illustrated in Figure 1. Additional mutants with variable and intermediate titan phenotypes have also been recovered.
Table I.
Summary of titan mutants
| Mutant | Source | Classa | Linkageb | Gene | Putative Protein Function |
|---|---|---|---|---|---|
| ttn1-1 | Feldmann | A | NA | At3g60740 | Tubulin-folding cofactor D |
| ttn1-2 | Syngenta | A | 103 /103 | At3g60740 | Tubulin-folding cofactor D |
| ttn1-3 | Syngenta | A | 105 /105 | At3g60740 | Tubulin-folding cofactor D |
| ttn2 | Feldmann | B | NA | Unknown | Gene identity unknown |
| ttn3 | Feldmann | D | 196 /196 | At5g62410 | SMC2 condensin |
| ttn4 | Syngenta | B | 123 /123 | At1g71190 | SAG18; unknown function |
| ttn5-1 | Syngenta | A | 206 /206 | At2g18390 | ARL2 GTPase |
| ttn5-2 | Lukowitz | A | NA | At2g18390 | ARL2 GTPase |
| ttn6-1 | Syngenta | C | 126 /126 | At3g20625 | Deubiquitinating enzyme |
| ttn6-2 | Syngenta | C | NA | At3g20625 | Deubiquitinating enzyme |
| ttn6-3 | Syngenta | C | 33 /33 | At3g20625 | Deubiquitinating enzyme |
| ttn6-4 | Syngenta | C | 102 /102 | At3g20625 | Deubiquitinating enzyme |
| ttn7-1 | Syngenta | B | 178 /178 | At2g27170 | SMC3 cohesin |
| ttn7-2 | Syngenta | B | 177 /177 | At2g27170 | SMC3 cohesin |
| ttn8-1 | Syngenta | B | 170 /170 | At3g54670 | SMC1 cohesin |
| ttn8-2 | Syngenta | B | 127 /127 | At3g54670 | SMC1 cohesin |
| ttn9 | Syngenta | B | 103 /103 | At3g20070 | Unknown function |
Phenotype class as defined in Figure 1.
For tagged mutants, evidence of genetic linkage between T-DNA insert carrying resistance gene and mutant locus. Nos. represent plants heterozygous for mutation/total resistant plants screened. NA, Not applicable because the mutant is not tagged.
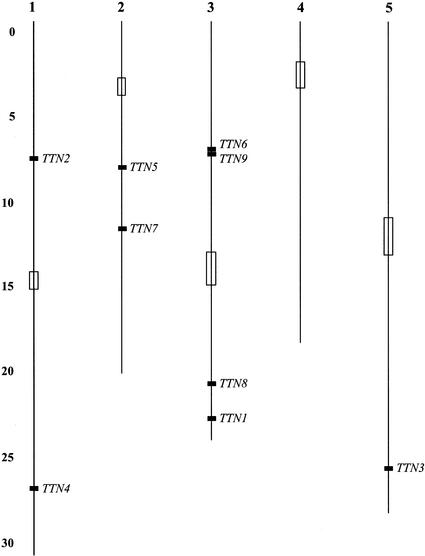
Duplicate mutant alleles were expected to be found given the large number of insertion lines screened. Allelism was demonstrated through a combination of genetic complementation tests and flanking sequence information. The ethyl methanesulfonate-induced ttn5-2 allele obtained from Wolfgang Lukowitz (Carnegie Institution of Washington, Stanford, CA) was confirmed by direct sequencing of amplified DNA from heterozygotes (McElver et al., 2000). Allelism between the untagged ttn6-2 allele and the complex ttn6-3 insertion allele was established by crossing heterozygotes. Approximately 22% of the 228 seeds produced from ttn6-1 × ttn6-2 crosses and 26% of the 235 seeds produced from ttn6-1 × ttn6-3 crosses appeared mutant. Similar crosses revealed allelism between ttn1-1 and ttn1-2. In contrast, ttn2 and ttn4 complemented when crossed, and the two genes mapped to different chromosomal regions. Analysis of F2 plants produced from crosses with visible markers placed ttn4 near the bottom of chromosome 1. A pilz mutant (pfi) with related phenotype has also been mapped to this region (Mayer et al., 1999). The genetic map position of ttn4, 12 cM below clv2 (180 F2 plants scored) and 15 cM above clv1 (190 F2 plants), is consistent with the physical location based on sequence analysis. A composite genetic and physical map of TTN genes is shown in Figure 2.
Figure 2.
Localization of TTN genes on a sequence-based chromosome map of Arabidopsis. Open rectangles correspond to centromeric regions as defined by genetic analysis (Arabidopsis Genome Initiative, 2000). Numbers indicate the estimated length of each chromosome in Megabases. The position of TTN2 was estimated from genetic linkage data.
Phenotypic Characterization of titan Mutants
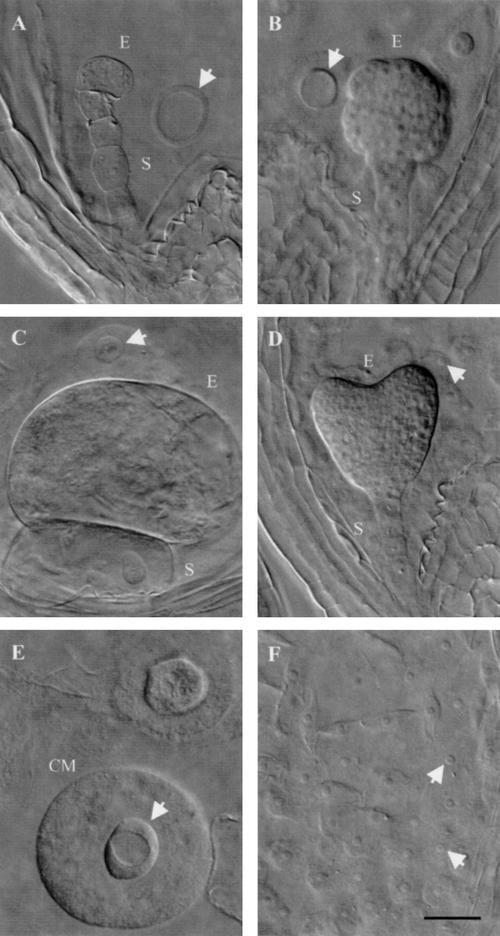
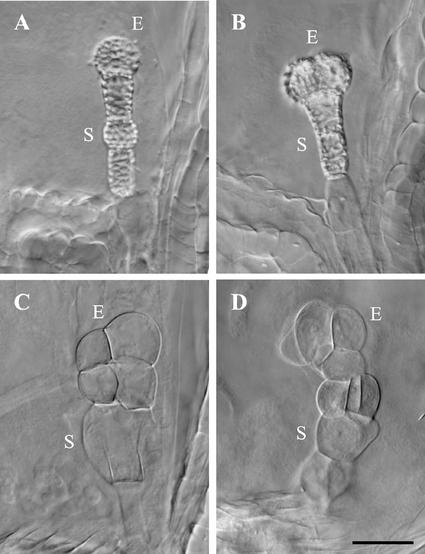
Three phenotypic classes of titan mutants were recognized by Liu and Meinke (1998). Differences were found in embryo morphology, seed viability, chromosome condensation, nucleolar appearance, and endosperm nuclear migration. Screening of the Syngenta collection yielded additional examples of the ttn1 (Fig. 1A) and ttn2 (Fig. 1B) classes. Another class (ttn6) characterized by a globular arrested embryo (Fig. 1C) was also identified. Our failure to recover mutants with a ttn3 pattern (Fig. 1D) was not surprising given the subtle embryo phenotype. Nomarski images of ttn1, ttn4, and ttn6 seeds at the heart-to-cotyledon stage of normal development are shown in Figure 3.
Figure 3.
Phenotypes of mutant seeds examined with Nomarski optics. Embryo (E) and suspensor (S) cells, enlarged endosperm nucleoli (arrows), and endosperm cytoplasmic masses (CM) are visible in cleared mutant seeds from heterozygous siliques at the heart-to-cotyledon stages of normal development. A, ttn4 embryo; B, ttn6-1 embryo; C, ttn1-2 embryo; D, wild-type embryo; E, ttn1-2 endosperm; F, wild-type endosperm. Scale bar = 30 μm.
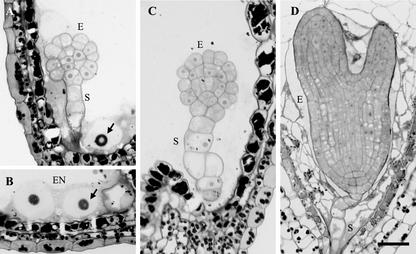
The ttn6 phenotype was examined in most detail because it defined a new titan class. Embryo cells often appeared rounded and disorganized. Endosperm cellularization was also disrupted. These abnormalities were confirmed in sectioned material, as shown in Figure 4. Table II documents developmental changes observed in cleared mutant seeds from tagged (ttn6-1) and untagged (ttn6-2) alleles. Defects visible at the heart stage of normal development included: increased size and reduced number of endosperm nuclei and nucleoli; and developmental arrest of the embryo proper. Endosperm nuclear enlargement was similar to that observed with other titans (Liu and Meinke, 1998; McElver et al., 2000). The average size of the embryo proper and largest endosperm nucleolus increased somewhat following the heart stage. A number of small nucleoli with a diameter of 5 to 6 μm were also found in the mutant endosperm, and their size remained constant between the heart and cotyledon stages. This variability in nuclear size within a single seed is a common feature of titan mutants. Most ttn6 seeds at the heart stage contained between 20 and 50 endosperm nuclei. This number did not increase later in development and remained far below the number found in wild-type seeds. Therefore, endosperm nuclear division is completed at about the same time in mutant and wild-type seeds.
Figure 4.
Light microscopy of ttn6-2 seeds. A through C, Stained sections of mutant seeds at the cotyledon stage of normal development. Abnormal cells of the embryo proper (E) and suspensor (S) are visible. Enlarged endosperm nuclei (EN) and nucleoli (arrows) are present. The image in B was rotated 90o counterclockwise. The vacuolated cell (right) is part of the suspensor. D, Wild-type embryo and cellularized endosperm from a seed at the equivalent time in development. Scale bar = 30 μm.
Table II.
Analysis of ttn6 seeds at different stages of development
| Allele | Stagea | Diameter
of Embryo Proper
|
Diameter of Largest Endosperm Nucleolus
|
||
|---|---|---|---|---|---|
| Average | Range | Average | Range | ||
| μm | |||||
| ttn6-1 | Heart | 44 | 23–58 | 18 | 14–35 |
| Linear cotyledon | 52 | 32–64 | 21 | 12–41 | |
| Curled cotyledon | 58 | 37–104 | 24 | 14–46 | |
| ttn6-2 | Heart | 40 | 30–51 | 14 | 9–20 |
| Linear cotyledon | 48 | 35–74 | 16 | 12–23 | |
| Curled cotyledon | 69 | 69–108 | 23 | 14–32 | |
Developmental stage of normal seeds obtained from the same silique. No. of seeds analyzed: ttn6-1: heart (24), linear cotyledon (47), and curled cotyledon (40); ttn6-2: heart (31), linear cotyledon (38), and curled cotyledon (38). The diameter of a normal embryo cell is about 8 μm, and that of an endosperm nucleolus is 4 μm.
The most common titan embryo phenotype in the Syngenta collection was early lethality without dramatic cell enlargement (Fig. 1B). This pattern was characteristic of knockouts in five different TTN genes (Table I). Several of these mutants escaped our initial screen for glassy seeds and were identified as titans only after examination with Nomarski optics. Mutant embryos contained a few small cells and were often difficult to find in cleared seeds. Enlargement of endosperm nucleoli was pronounced but somewhat variable. Cellularization of the endosperm was also blocked. The ttn9 embryo, which contained at most four small cells, was typical of this class and resembled the cohesin (ttn7 and ttn8) knockouts described by Liu et al. (2002). The ttn4 embryo was larger and more vacuolated late in development and therefore represented an intermediate class. In addition, embryo cells often accumulated wall thickenings that resulted in birefringence when viewed under Nomarski optics. These features are highlighted in Figure 5. Variations in titan seed phenotypes observed within each mutant are summarized in Table III. Typically, 10% to 20% of mutant seeds with an arrested embryo failed to exhibit a titan endosperm phenotype. The cellular basis for this variation remains to be explained. Globular embryos were found only in ttn6 seeds. The tagged ttn1-2 allele (Fig. 3C) exhibited a seed phenotype identical to ttn1-1 (Liu and Meinke, 1998). Arrested embryos were found in 84% of 100 cleared ttn1-2 seeds examined (Table III). Fifty-six percent of these embryos were composed of two cells (Fig. 3C). The remainder contained a single large cell. Embryo cell enlargement in ttn1-3 was similar. Over 90% of these embryos contained two cells.
Figure 5.
Late phenotypes of ttn4 mutant embryos. A and B, Cell wall thickenings appear as bright regions on the surface of the embryo proper (E) and suspensor (S) in cleared seeds viewed with Nomarski optics. C and D, Embryo cells become enlarged and distorted in shape prior to seed desiccation. Scale bar = 30 μm.
Table III.
Phenotypic variation observed in mutant seeds
| Mutant | No. Seeds Examined | Percentage of Seeds
Observed with Specified Mutant Phenotype
|
|||||
|---|---|---|---|---|---|---|---|
| Endosperm
titan
phenotypea
|
Arrested embryo
phenotype
|
||||||
| Strong | Moderate | Weak | Preglobular | Globular | NDb | ||
| ttn1-2 | 100 | 78 | 0 | 22 | 84 | 0 | 16 |
| ttn1-3 | 100 | 67 | 9 | 24 | 96 | 0 | 4 |
| ttn4 | 96 | 72 | 12 | 16 | 73 | 0 | 27 |
| ttn6-1 | 100 | 81 | 14 | 5 | 28 | 70 | 2 |
| ttn6-4 | 93 | 68 | 22 | 10 | 13 | 87 | 0 |
| ttn9 | 100 | 73 | 14 | 13 | 50 | 0 | 50 |
Seeds with giant endosperm nucleoli were classified as strong, those with nucleoli of intermediate sizes were called moderate, and those with smaller nucleoli were considered weak.
ND, Not detected because the embryo was too small.
Molecular Identification of TTN1
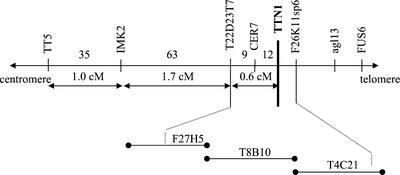
We first attempted to isolate the TTN1 gene through map-based cloning because the original ttn1-1 allele from the Feldmann collection was not tagged. Mapping with visible markers placed ttn1 below tt5, close to cer7 on chromosome 3 (Franzmann et al., 1995; Liu and Meinke, 1998). Rare recombinants obtained from crosses between ttn1-1 heterozygotes (Wassilewskija [WS] ecotype) and tt5 or cer7 homozygotes (Ler ecotype) were analyzed with a series of linked molecular markers. From 1,852 F2 plants examined, 119 crossovers between tt5 and ttn1 were obtained. The combined results, as summarized in Figure 6, enabled us to localize ttn1 below cer7 and likely on bacterial artificial chromosome (BAC) T4C21 within a region spanned by markers T22D23T7 and F26K11sp6. One gene in this region (T4C21.150) encodes a protein that resembles tubulin-folding cofactor D. This gene became a TTN1 candidate when we learned that ARL2 (TTN5) interacts with cofactor D to regulate microtubule assembly in yeast and humans (Bhamidipati et al., 2000; Radcliffe et al., 2000b). Two knockouts were later found in the Syngenta collection of embryo defectives. Allelism between these tagged mutants and ttn1-1 was demonstrated through genetic complementation tests. Approximately 22% of the 510 seeds produced from reciprocal crosses between heterozygotes were mutant. These results confirmed that TTN1 had been identified.
Figure 6.
Map-based localization of TTN1. The TTN1 gene was localized to BAC T4C21 on chromosome 3 by analyzing recombinants produced from crosses with visible markers for the presence of linked molecular markers as described in the text.
TTN1 Resembles Tubulin-Folding Cofactor D
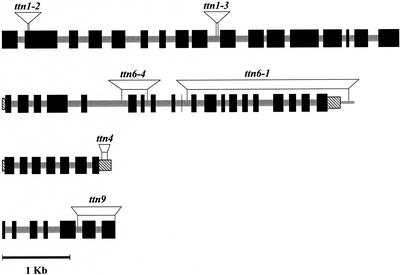
The predicted structure for TTN1 (At3g60740) is shown in Figure 7. The gene is approximately 5.9 kb in length, contains 16 introns based on AGI gene models, and encodes a predicted protein of 1,249 amino acids. The ttn1-2 allele contains a T-DNA insertion in exon 2 and lacks 12 bp around the insertion site. The ttn1-3 allele has an insertion in intron 9 and lacks 18 bp adjacent to the insertion site. The existence of two mutants with similar phenotypes and defined insertions in the same gene provides confirmation of gene identity. The location of the mutation in ttn1-1 has not been determined but the strong phenotype is consistent with a null allele.
Figure 7.
Gene structures and T-DNA insertion sites for TTN1, TTN6, TTN4, and TTN9. Large black boxes designate exons, stippled boxes are introns, striped rectangles correspond to untranslated regions, and thin rectangles represent adjacent genomic DNA. Insertion sites and associated deletions are shown above the predicted gene structures. Gene structures for TTN6 and TTN4 have been confirmed by cDNA sequence analysis. Models for TTN1 and TTN9 intron and exon boundaries are based on the Arabidopsis Genome Initiative (2000).
From BLAST sequence analysis, TTN1 appears to be a single copy gene in Arabidopsis. Expression has been confirmed through identification of expressed sequence tags (ESTs) from vegetative structures (Asamizu et al., 2000), seedling hypocotyl (Newman et al., 1994), roots (Asamizu et al., 2000), and seedlings exposed to salt stress (Gong et al., 2001). BLASTP searches against all GenBank sequences revealed a high level of sequence identity to cofactor D from human (35% identity; e = 0.0), bovine (35% identity; 0.0), fruit fly (Drosophila melanogaster; 31% identity; −148), Caenorhabditis elegans (26% identity; −81), and S. pombe (27% identity; −48). A number of conserved protein domains were found when these sequences were compared. Results of this analysis are presented in Figure 8. The high degree of sequence conservation observed in these domains is consistent with a critical cellular function for this protein in eukaryotes.
Figure 8.
Conserved protein domains identified in tubulin-folding cofactor D. Each segment corresponds to a conserved domain identified by BLOCKS (Henikoff et al., 1995). Bold letters represent amino acids conserved in at least five of the six sequences. Numbers mark the amino acid location within the protein. Species and GenBank accessions: Arabidopsis (CAB82678), human (NP005984), Bos taurus (AAB17537), fruit fly (AAF51300), C. elegans (T21018), and S. pombe (Q10197).
TTN6 (AtUBP14) Resembles Isopeptidase T
The predicted structure for TTN6 (At3g20625) is shown in Figure 7. This gene model was compiled from AGI sequence of two adjacent BACs (K10D20 and F3H11). The predicted gene is approximately 5.0 kb in length, contains 19 introns, and encodes a predicted protein of 797 amino acids. The protein sequence is based on the availability of a full-length cDNA (AF302664). The ttn6-1 allele contains a large deletion (approximately 2.7 kb) at the insertion site that removes 10 exons coding for the C-terminal half of the protein. The ttn6-4 allele has a smaller deletion (approximately 0.4 kb) that eliminates exons 6 and 7. The ttn6-3 allele appears to be tagged from genetic evidence, but it contains a complex T-DNA insert that remains to be resolved.
Twenty-seven deubiquitinating enzymes (DUBs) of the ubiquitin-specific protease (UBP) class have been identified in Arabidopsis (Yan et al., 2000). TTN6 (AtUBP14) is most similar in protein sequence to the isopeptidase T class of enzymes (Wilkinson et al., 1995) from human (49% identity; e = 0.0), mouse (47% identity; 0.0), fruit fly (44% identity; 0.0), Dictyostelium discoideum (UbpA; 40% identity; −180), C. elegans (34% identity; −119), and S. cerevisiae (UBP14; 31% identity; −69). The most closely related protein is derived from genomic sequencing of rice (Oryza sativa; 65% identity; 0.0). Figure 9 illustrates conserved protein domains identified by Pfam analysis (Bateman et al., 2000): a zinc finger UBP domain, ubiquitin carboxyl-terminal hydrolases (UCH)-1 domain with conserved “Cys” box, UBA domains, and C-terminal UCH-2 domain with conserved “His” box. The absence of an N-terminal extension in the rice protein may reflect an incorrect gene model. Sequence comparisons of these conserved motifs have recently been published by Doelling et al. (2001).
Figure 9.
Conserved Pfam domains identified in TTN6 (UBP14)-related proteins from different organisms. TTN6 contains all of the protein domains expected for DUBs of the isopeptidase T class. Pfam analysis (Bateman et al., 2000) revealed the presence of a zinc finger UBP domain (ellipse), UCH-1 domain (diamond) with conserved “Cys” box, two UBA domains (hexagons), and a C-terminal UCH-2 domain with conserved “His” box in the expected locations. Organisms and GenBank accession numbers: A, Arabidopsis, TTN6, AAG42755; B, rice, BAB17073; C, human, XP_006971; D, Mus musculus, NP_038728; E, S. cerevisiae, UBP14, NP_009614; F, D. discoideum, P54201; G, fruit fly, AAF47720.
TTN4 and TTN9 Appear to Be Plant-Specific Proteins
Although the identities of TTN4 and TTN9 are each based on analysis of a single mutant allele, the genetic data summarized in Table I are consistent with tagging, and both sides of each insert were recovered and found to match a single locus. The isolation of single mutant alleles in contrast to duplicate alleles for other titans is also consistent with the small size of these genes. T-DNA insertion sites and predicted gene structures are presented in Figure 7. The TTN4 gene model predicted from the sequencing project (Arabidopsis Genome Initiative, 2000) was confirmed through isolation of a full-length cDNA. Two amino acid differences identified were attributed to errors in sequencing of the cDNA. The SAG18 partial cDNA sequence (AF053063) described by Weaver et al. (1998) in their screen for senescence-associated genes corresponds to the 3′ end of the full-length transcript. The T-DNA insert in ttn4 is located in the 3′-untranslated region. The predicted protein product contains 281 amino acids and lacks defined domains and sequence similarity to known proteins. BLASTP analysis revealed a related Arabidopsis gene (F14F18.40) with 47% identity (e = −60) and a corresponding EST. A similar gene has also been identified in rice (BAB56093; 47% identity; e = −23). No significant matches were found with any proteins identified from other organisms. TTN9 appears to be a single copy gene that is expressed in siliques based on EST data. The predicted protein is 282 amino acids in length and lacks known motifs. One BLASTP match was identified in GenBank: an EST (AF325722; 32% identity; e = −10) from pistils of an apomictic grass (Pennisetum ciliare). These results are consistent with the conclusion that TTN9 and TTN4 are plant-specific proteins of unknown function.
DISCUSSION
TITAN Proteins Have Diverse but Overlapping Functions in Seed Development
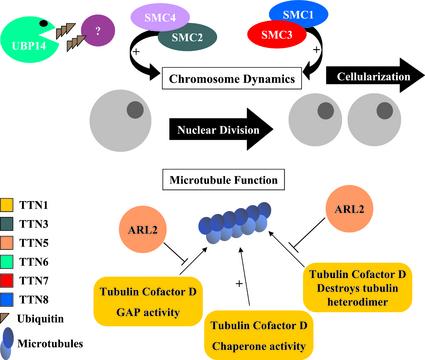
We have identified two networks of TITAN proteins in Arabidopsis that regulate endosperm nuclear division and cellularization. A model illustrating the functions of these proteins is presented in Figure 10. One network involves chromosomal scaffold proteins known as condensins (SMC2 and SMC4) and cohesins (SMC1 and SMC3). These myosin-like ATPases play a central role in chromosome condensation, sister chromatid cohesion, dosage compensation, and recombination repair (Hirano, 2000). The importance of SMC proteins in endosperm development became apparent when TTN3 was identified as an SMC2 condensin and was later confirmed when additional titans were found to be disrupted in SMC cohesins (Liu et al., 2002). A second network of TITAN proteins involves the regulation of microtubule assembly. To our knowledge, the importance of this network in plants is described for the first time in this report. The titan phenotype therefore can result from disruption of either chromosomal proteins or cytoskeletal organization. This conclusion is consistent with the contrasting models of gene functions presented when titan mutants were first identified (Liu and Meinke, 1998).
Figure 10.
Model of TITAN gene functions in Arabidopsis. Nuclear division in the developing endosperm requires at least two networks of TITAN proteins. One modulates chromosome integrity through scaffold proteins known as cohesins (SMC1 and SMC3) and condensins (SMC2). Another regulates microtubule assembly through interactions between ARL2 and tubulin-folding cofactor D. Knockouts in AtSMC4, protein targets of TTN6 activity, and cellular functions of TTN4 and TTN9 remain to be identified.
Two defining features of early endosperm development in angiosperms are nuclear migration and the suppression of phragmoplast formation following nuclear division (Olsen, 2001). These processes require appropriate coordination between cytoskeletal organization and cell-cycle progression. The complex networks of TITAN proteins described here perform an essential role in maintaining chromosome structure and function throughout the cell cycle and in regulating the establishment of the microtubule arrays required for chromosome movement and phragmoplast formation. TITAN proteins therefore can be viewed as central mediators in processes that help to distinguish endosperm tissue from adjacent parts of the seed.
TTN1 and TTN5 Encode Proteins That Interact in Yeast and Humans
Many proteins have been identified that regulate microtubule dynamics in eukaryotes (Nogales, 2000). The formation of α/β-tubulin heterodimers begins with the appearance of chaperonin complexes and proceeds through interactions with specialized folding cofactors (Tian et al., 1996; Radcliffe et al., 2000a). Cofactor D associates with β-tubulin subunits and is encoded by Alp1 in S. pombe (Hirata et al., 1998) and CIN1 (for chromosome instability) in S. cerevisiae (Fleming et al., 2000). Loss of Alp1 activity is lethal and results in abnormal mitoses, destruction of microtubule structures, and defects in cell division (Hirata et al., 1998). In contrast, CIN1 mutations are not lethal (Stearns et al., 1990; Fleming et al., 2000). In addition to modulating assembly of tubulin heterodimers, cofactor D functions as a GTP-activating protein for hydrolysis of GTP by β-tubulin and subsequent release of free heterodimers (Nogales, 2000). Cofactor D can also interact with native tubulin, alter the ratio of free subunits by sequestering β-tubulin from GTP-bound heterodimers, and stimulate destruction of heterodimers (Bhamidipati et al., 2000; Martin et al., 2000).
ARL2 interacts directly with human cofactor D in culture, prevents degradation of tubulin heterodimers, and reduces the GTP-activating protein activity of cofactor D in vitro (Bhamidipati et al., 2000). Deletion of the ARL2 homolog in S. pombe (Alp41) results in defects in cell division similar to those found in cofactor mutants (Radcliffe et al., 2000a, 2000b). Therefore, ARL2 (Alp41) and cofactor D (Alp1) are essential proteins in fission yeast. The subtle phenotype of CIN4 (ARL2) knockouts is consistent with the nonessential role of tubulin cofactors in budding yeast (Stearns et al., 1990; Fleming et al., 2000).
Nuclear and cytoskeletal defects observed in ttn1 and ttn5 seeds are consistent with known roles of ARL2 and cofactor D in regulating microtubule dynamics in fission yeast and humans. Enlargement of endosperm nuclei and nucleoli appears to result from microtubule-associated defects in chromosome mechanics and cell plate formation coupled with continued progression through the cell cycle. Defects in microtubule organization have been documented with fluorescence microscopy in the corresponding pilz mutants (Mayer et al., 1999). The dramatic changes in embryo cell morphology described here are consistent with known functions of microtubules in plants. These functions have been difficult to address from a genetic perspective in Arabidopsis because of redundancy in the tubulin gene family (Kopczak et al., 1992; Snustad et al., 1992). Several mutants defective in microtubule organization have nevertheless been identified, including ton2/fass (Torres-Ruiz and Jurgens, 1994; Traas et al., 1995), mor1 (Whittington et al., 2001), bot1 (Bichet et al., 2001), zwi (Oppenheimer et al., 1997), and fra2/AtKTN1 (Burk et al., 2001). Changes in cell morphology have also been noted following exposure of roots to microtubule inhibitors (Baskin et al., 1994). We describe here a genetic system for studying the consequences of a dramatic loss of microtubule function, demonstrate the importance of ARL2 and cofactor D in seed development, and clarify the connection between ARL2 function and microtubule dynamics in plants.
The Ubiquitin Pathway Is Linked to TITAN Functions
The ubiquitin pathway plays a key role in selective degradation of proteins in eukaryotic cells (Hershko and Ciechanover, 1998). Targeted proteins are modified through the formation of an isopeptide bond between the C terminus of ubiquitin and the ε-amino group of Lys on the target protein (Naviglio et al., 1998). DUBs are hydrolyzing proteases that process primary ubiquitin gene products, edit the ubiquitination state of cellular proteins, and recycle ubiquitin released following hydrolysis of proteins targeted for destruction via the proteasome. Isopeptidase DUBs have specificity for substrates containing ε-amide bonds to a side chain Lys (Wilkinson, 1997). Some isopeptidases can also disassemble specific ubiquitin-protein conjugates before proteolysis by the proteasome. This process is thought to have either a regulatory function for essential proteins or a salvaging function for incorrectly ubiquitinated proteins (Hershko and Ciechanover, 1998). Two general classes of DUBs that differ in sequence and substrate specificity have been identified: small UCH proteins and UBP proteins with conserved Cys and His boxes (Wilkinson, 1997). These DUBs have the ability to cleave ubiquitin linked to target proteins by either peptide or isopeptide bonds. TTN6 (AtUBP14) is a large protein with unknown substrate specificity but characteristic UBP domains.
A number of DUB genes have already been identified by mutation. These include fruit fly fat facets, which is required for reproductive development and eye differentiation (Fischer-Vize et al., 1992) and is thought to act by preventing degradation of its target regulatory protein (Huang et al., 1995); S. cerevisiae DOA4 and UBP3, which are required for a variety of cellular processes including control of DNA replication (see Wilkinson, 1997) and regulation of gene silencing (Moazed and Johnson, 1996); and D. discoideum UbpA, which is required for normal development but not for continued growth (Lindsey et al., 1998; Chung and Baek, 1999). The UBP family of Arabidopsis consists of at least 27 genes with the conserved protein domains expected for catalytic activity (Chandler et al., 1997; Rao-Naik et al., 2000; Yan et al., 2000). Knockouts in two of these genes (AtUBP1 and AtUBP2) exhibit increased sensitivity to the amino acid analog canavanine, which can increase the concentration of abnormal proteins produced during translation (Yan et al., 2000). Therefore, these family members appear to function in the removal of abnormal proteins from the cell. Although substrate specificities and cellular localizations of several Arabidopsis UPBs have been described (Chandler et al., 1997; Rao-Naik et al., 2000), much remains to be learned about the precise functions of specific UBP proteins in Arabidopsis. Doelling et al. (2001) recently described two allelic UBP14 (TTN6) knockouts that resulted in embryonic lethality at the globular stage, demonstrated that mutant seeds accumulated multi-ubiquitin chains, consistent with a defect in ubiquitin cycling, and found that Arabidopsis UBP14 complements the corresponding yeast mutant. We demonstrate here the connection between UBP14 function and a titan seed phenotype.
We propose two models to explain the relationship between ubiquitin pathways and a titan phenotype. These models are based on two observations: the absence of dramatic cell enlargement in ttn6 embryos, which suggests that a disruption of microtubule function is not involved; and the connection between chromosome stability and protein degradation recently established for the SSC1 cohesin of yeast (Rao et al., 2001). According to the first model, accumulation of free multiubiquitin chains enhances the stability of a target protein that under normal circumstances modulates SMC function. An alternative model is that TTN6 removes ubiquitin directly from a target protein that influences chromosome dynamics in wild-type seeds and the resulting accumulation of this regulatory protein in mutant seeds is responsible for the mutant phenotype. This model could involve the same target protein as described for the first model but a different mechanism for altering the stability of this protein.
Embryo Phenotypes Reflect Differences in TITAN Functions
The titan endosperm phenotype is consistent with known functions of microtubules and SMC proteins in eukaryotes. Even the atypical ttn3 endosperm phenotype can be explained by the presence of a related gene with overlapping functions. Differences observed between titan embryo phenotypes, however, are more problematic. Two questions remain to be addressed: Why are giant cells found only in ttn1 and ttn5 seeds; and why do nuclei in many titan embryos fail to enlarge? With respect to the second question, we propose that different cell-cycle checkpoints are involved in the embryo and endosperm. Disruption of the SMC complex in the embryo interferes with essential cell functions and consequently results in cell abortion. DNA replication and nuclear enlargement continue in the endosperm because cellularization is not required. Disruption of the SMC complex may also be the cause of abnormalities seen in ttn2 and ttn9 seeds, which have similar embryo phenotypes. The intermediate ttn4 embryo phenotype is intriguing because the wall thickenings seen late in development are reminiscent of changes associated with programmed cell death and differentiation of tracheary elements (Fukuda, 2000; Roberts and McCann, 2000). The most dramatic embryo phenotype observed to date is the striking cell enlargement found in ttn1 and ttn5 seeds. The continuation of DNA replication in these embryos indicates that the SMC-related checkpoint is bypassed. The progressive cell enlargement demonstrates that elimination of ARL2-cofactor D-mediated regulation of microtubule assembly is not immediately lethal. Whether a similar mechanism is used in the formation of giant feeding cells exposed to root-knot nematodes (Niebel et al., 1996) remains to be explored.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The ttn1-1, ttn2, and ttn3 mutants were generated through Agrobacterium tumefaciens-mediated seed transformation of the WS ecotype (Feldmann, 1991) and were identified and maintained as described (Liu and Meinke, 1998). The ttn5-2 mutant was isolated by Wolfgang Lukowitz (Carnegie Institution of Washington) in the Landsberg erecta ecotype following seed mutagenesis with ethyl methanesulfonate (McElver et al., 2000). The remaining titan mutants described in this report were produced at Syngenta through A. tumefaciens-mediated plant transformation of the Columbia ecotype using the vacuum infiltration (Bechtold and Pelletier, 1998) and floral dip (Clough and Bent, 1998) methods. Seeds can be obtained through the Arabidopsis Biological Resource Center. Details of plant transformation, vector design, and screening of insertion lines for seed defects are presented in McElver et al. (2001). Additional information on mutants defective in SMC genes (ttn3, ttn7, and ttn8) can be found in Liu et al. (2002). Plants were grown in pots containing a mixture of vermiculite, soil, and sand, placed in a growth room at 24° ± 2°C under fluorescent lights on 16-h light/8-h dark cycles, and watered daily with a fertilizer solution (Heath et al., 1986). Heterozygotes were identified by screening immature siliques for the presence of 25% defective seeds following self pollination (Meinke, 1994).
Genetic and Phenotypic Characterization
T-DNA vectors used for transformation experiments conferred resistance to kanamycin (ttn3), hygromycin (ttn4), or Basta (ttn1-2, ttn1-3, ttn6-1, ttn6-3, ttn6-4, and ttn9). Linkage between the T-DNA insert and mutant phenotype was demonstrated by transplanting resistant seedlings from selection plates to soil and scoring the resulting plants for the presence of the seed mutation (McElver et al., 2001). Mapping of ttn4 with visible markers was performed as described by Franzmann et al. (1995). Complementation tests were performed by crossing heterozygotes and scoring the resulting siliques for 25% defective seeds with the expected phenotype. Mutant seeds cleared for observations were treated with Hoyer's solution (Meinke, 1994) and examined with a compound microscope (model E600; Nikon, Tokyo) equipped with Nomarski optics. Images were captured with a DXM1200 digital imaging system (Nikon). Sections of embedded mutant seeds were prepared as noted by Liu and Meinke (1998).
Map-Based Localization of TTN1
Crosses were made between ttn1 heterozygotes (WS ecotype) and either dis1, clv2, er, tt5 homozygotes or er, gl1, cer7 homozygotes (Landsberg) to identify crossovers in the vicinity of TTN1. Known RFLP markers (CD2-12 and pCIT1210), cleaved-amplified polymorphic sequence markers (IMK2 and IMK3), and SSLP markers (nga6) were used to estimate the position of TTN1 on the physical map. Eight cleaved-amplified polymorphic sequence markers (TT5, T22D23T7, F26K11sp6, agl13, FUS6, 2A19E, ACS1, and 2A19B) based on the BAC contig and genomic sequences in this region were then used to initiate a walk toward the TTN1 gene. Sequence details, PCR primer sequences, cycling conditions, and information on restriction enzymes used can be obtained upon request from the authors.
TTN Gene Identification and Sequence Analysis
Plant sequences flanking T-DNA insertion sites in tagged mutants were obtained through plasmid rescue or thermal asymmetric interlaced-PCR and confirmed by direct PCR sequencing using a combination of genome-specific and T-DNA primers as described in detail by McElver et al. (2001). The TTN4 full-length cDNA was isolated and sequenced according to standard methods (McElver et al., 2000). Sequence comparisons were performed using the BLAST 2.0 algorithm (Altschul et al., 1997) with default settings and the low complexity filter removed. Conserved protein motifs were identified with Pfam (Bateman et al., 2000) and were subjected to CLUSTALW (Thompson et al., 1994) and BLOCKS (Henikoff et al., 1995) analyses through the Baylor College of Medicine (Houston; Smith et al., 1996; http://searchlauncher.bcm.tmc.edu).
ACKNOWLEDGMENTS
We thank the many members of the Patton laboratory at Syngenta, in particular George Aux, for assistance with the production of insertion lines, initial screening for seed mutations, and isolation of flanking plant sequences; Mike Rumbaugh and Mary Ann Cushman for assistance with map-based localization of TTN1; and Steven Hutchens, Cathy Sonleitner, Becky Rogers, and Shkelzen Shabani for assistance with phenotypic characterization of tagged mutants.
Footnotes
This research was supported in part by grants from the National Science Foundation, Developmental Mechanisms Program, and by the Plant Biology Division of the S.R. Noble Foundation.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010911.
LITERATURE CITED
- Albert S, Despres B, Guilleminot J, Bechtold N, Pelletier G, Delseny M, Devic M. The EMB506 gene encodes a novel ankyrin repeat containing protein that is essential for the normal development of Arabidopsisembryos. Plant J. 1999;17:169–179. doi: 10.1046/j.1365-313x.1999.00361.x. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apuya NR, Yadegari R, Fischer RL, Harada JJ, Zimmerman JL, Goldberg RB. The Arabidopsis embryo mutant schlepperless has a defect in the chaperonin-60αgene. Plant Physiol. 2001;126:717–730. doi: 10.1104/pp.126.2.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Asamizu E, Nakamura Y, Sato S, Tabata S. A large scale analysis of cDNA in Arabidopsis thaliana: generation of 12,028 non-redundant expressed sequence tags from normalized and size-selected cDNA libraries. DNA Res. 2000;7:175–180. doi: 10.1093/dnares/7.3.175. [DOI] [PubMed] [Google Scholar]
- Assaad FF, Huet Y, Mayer U, Jurgens G. The cytokinesis gene KEULEencodes a Sec1 protein that binds the syntaxin KNOLLE. J Cell Biol. 2001;152:531–543. doi: 10.1083/jcb.152.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin TI, Wilson JE, Cork A, Williamson RE. Morphology and microtubule organization in Arabidopsisroots exposed to oryzalin or taxol. Plant Cell Physiol. 1994;35:935–942. [PubMed] [Google Scholar]
- Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thalianaplants by vacuum infiltration. Methods Mol Biol. 1998;82:259–266. doi: 10.1385/0-89603-391-0:259. [DOI] [PubMed] [Google Scholar]
- Bhamidipati A, Lewis SA, Cowan NJ. ADP ribosylation factor-like protein 2 (Arl2) regulates the interaction of tubulin-folding cofactor D with native tubulin. J Cell Biol. 2000;149:1087–1096. doi: 10.1083/jcb.149.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bichet A, Desnos T, Turner S, Grandjean O, Hofte H. BOTERO1 is required for normal orientation of cortical microtubules and anisotropic cell expansion in Arabidopsis. Plant J. 2001;25:137–148. doi: 10.1046/j.1365-313x.2001.00946.x. [DOI] [PubMed] [Google Scholar]
- Boisnard-Lorig C, Colon-Carmona A, Bauch M, Hodge S, Doerner P, Bancharel E, Dumas C, Haseloff J, Berger F. Dynamic analyses of the expression of the HISTONE::YFP fusion protein in Arabidopsisshow that syncytial endosperm is divided in mitotic domains. Plant Cell. 2001;13:495–509. doi: 10.1105/tpc.13.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson M, Gomord V, Audran C, Berger N, Dubreucq B, Granier F, Lerouge P, Faye L, Caboche M, Lepiniec L. Arabidopsis glucosidase 1mutants reveal a critical role of N-glycan trimming in seed development. EMBO J. 2001;20:1010–1019. doi: 10.1093/emboj/20.5.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RC, Lemmon BE, Nguyen H, Olsen O-A. Development of endosperm in Arabidopsis thaliana. Sex Plant Reprod. 1999;12:32–42. [Google Scholar]
- Burk DH, Liu B, Zhong R, Morrison WH, Ye ZH. A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell. 2001;13:807–828. [PMC free article] [PubMed] [Google Scholar]
- Chandler JS, McArdle B, Callis J. AtUBP3 and AtUBP4 are two closely related Arabidopsis thalianaubiquitin-specific proteases present in the nucleus. Mol Gen Genet. 1997;255:302–310. doi: 10.1007/s004380050501. [DOI] [PubMed] [Google Scholar]
- Chung CH, Baek SH. Deubiquitinating enzymes: their diversity and emerging roles. Biochem Biophys Res Commun. 1999;266:633–640. doi: 10.1006/bbrc.1999.1880. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Doelling JH, Yan N, Kurepa J, Walker J, Vierstra RD. The ubiquitin-specific protease UBP14 is essential for early embryo development in Arabidopsis thaliana. Plant J. 2001;27:393–405. doi: 10.1046/j.1365-313x.2001.01106.x. [DOI] [PubMed] [Google Scholar]
- Feldmann KA. T-DNA insertion mutagenesis in Arabidopsis: mutational spectrum. Plant J. 1991;1:71–82. [Google Scholar]
- Fischer-Vize JA, Rubin GM, Lehmann R. The fat facets gene is required for Drosophilaeye and embryo development. Development. 1992;116:985–1000. doi: 10.1242/dev.116.4.985. [DOI] [PubMed] [Google Scholar]
- Fleming JA, Vega LR, Solomon F. Function of tubulin binding proteins in vivo. Genetics. 2000;156:69–80. doi: 10.1093/genetics/156.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzmann LH, Yoon ES, Meinke DW. Saturating the genetic map of Arabidopsis thalianawith embryonic mutations. Plant J. 1995;7:341–350. [Google Scholar]
- Fukuda H. Programmed cell death of tracheary elements as a paradigm in plants. Plant Mol Biol. 2000;44:245–253. doi: 10.1023/a:1026532223173. [DOI] [PubMed] [Google Scholar]
- Gong Z, Koiwa H, Cushman MA, Ray A, Bufford D, Kore-eda S, Matsumoto TK, Zhu J, Cushman JC, Bressan RA. Genes that are uniquely stress regulated in salt overly sensitive (sos) mutants. Plant Physiol. 2001;126:363–375. doi: 10.1104/pp.126.1.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus U, Vielle-Calzada JP, Hoeppner MA, Gagliano WB. Maternal control of embryogenesis by MEDEA, a polycomb-group gene in Arabidopsis. Science. 1998;280:446–450. doi: 10.1126/science.280.5362.446. [DOI] [PubMed] [Google Scholar]
- Hardtke CS, Berleth T. The Arabidopsis gene MONOPTEROSencodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 1998;17:1405–1411. doi: 10.1093/emboj/17.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath JD, Weldon R, Monnot C, Meinke DW. Analysis of storage proteins in normal and aborted seeds from embryo-lethal mutants of Arabidopsis thaliana. Planta. 1986;169:304–312. doi: 10.1007/BF00392124. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG, Alford WJ, Pietrokovski S. Automated construction and graphical presentation of protein blocks from unaligned sequences. Gene. 1995;163:GC17–GC26. doi: 10.1016/0378-1119(95)00486-p. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hirano T. Chromosome cohesion, condensation, and separation. Annu Rev Biochem. 2000;69:115–144. doi: 10.1146/annurev.biochem.69.1.115. [DOI] [PubMed] [Google Scholar]
- Hirata D, Masuda H, Eddison M, Toda T. Essential role of tubulin-folding cofactor D in microtubule assembly and its association with microtubules in fission yeast. EMBO J. 1998;17:658–666. doi: 10.1093/emboj/17.3.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Baker RT, Fischer-Vize JA. Control of cell fate by a deubiquitinating enzyme encoded by the fat facetsgene. Science. 1995;270:1828–1831. doi: 10.1126/science.270.5243.1828. [DOI] [PubMed] [Google Scholar]
- Jang JC, Fujioka S, Tasaka M, Seto H, Takatsuto S, Ishii A, Aida M, Yoshida S, Sheen J. A critical role of sterols in embryonic patterning and meristem programming revealed by the fackel mutants of Arabidopsis thaliana. Genes Dev. 2000;14:1485–1497. [PMC free article] [PubMed] [Google Scholar]
- Kopczak SD, Haas NA, Hussey PJ, Silflow CD, Snustad DP. The small genome of Arabidopsiscontains at least six expressed α-tubulin genes. Plant Cell. 1992;4:539–547. doi: 10.1105/tpc.4.5.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber MH, Waizenegger I, Steinmann T, Schwarz H, Mayer U, Hwang I, Lukowitz W, Jurgens G. The ArabidopsisKNOLLE protein is a cytokinesis-specific syntaxin. J Cell Biol. 1997;139:1485–1493. doi: 10.1083/jcb.139.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Thomas TL. PEI1, an embryo-specific zinc finger protein gene required for heart-stage embryo formation in Arabidopsis. Plant Cell. 1998;10:383–398. doi: 10.1105/tpc.10.3.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey DF, Amerik A, Deery WJ, Bishop JD, Hochstrasser M, Gomer RH. A deubiquitinating enzyme that disassembles free polyubiquitin chains is required for development but not growth in Dictyostelium. J Biol Chem. 1998;273:29178–29187. doi: 10.1074/jbc.273.44.29178. [DOI] [PubMed] [Google Scholar]
- Liu CM, McElver J, Tzafrir I, Joosen R, Wittich P, Patton D, Van Lammeren AAM, Meinke DW (2002) Condensin and cohesin knockouts in Arabidopsis exhibit a titan seed phenotype. Plant J (in press) [DOI] [PubMed]
- Liu CM, Meinke DW. The titan mutants of Arabidopsisare disrupted in mitosis and cell cycle control during seed development. Plant J. 1998;16:21–31. doi: 10.1046/j.1365-313x.1998.00268.x. [DOI] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature. 1996;379:66–69. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]
- Lotan T, Ohto M, Matsudaira Yee K, West MAL, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB. ArabidopsisLEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell. 1998;93:1195–1205. doi: 10.1016/s0092-8674(00)81463-4. [DOI] [PubMed] [Google Scholar]
- Lukowitz W, Nickle TC, Meinke DW, Last RL, Conklin PL, Somerville CR. Arabidopsis cyt1mutants are deficient in a mannose-1-phosphate guanylyltransferase and point to a requirement of N-linked glycosylation for cellulose biosynthesis. Proc Natl Acad Sci USA. 2001;98:2262–2267. doi: 10.1073/pnas.051625798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Bilodeau P, Koltunow A, Dennis ES, Peacock WJ, Chaudhury AM. Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1999;96:296–301. doi: 10.1073/pnas.96.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin L, Fanarraga ML, Aloria K, Zabala JC. Tubulin folding cofactor D is a microtubule destabilizing protein. FEBS Lett. 2000;470:93–95. doi: 10.1016/s0014-5793(00)01293-x. [DOI] [PubMed] [Google Scholar]
- Mayer U, Herzog U, Berger F, Inzé D, Jurgens G. Mutations in the PILZ group genes disrupt the microtubule cytoskeleton and uncouple cell cycle progression from cell division in Arabidopsisembryo and endosperm. Eur J Cell Biol. 1999;78:100–108. doi: 10.1016/S0171-9335(99)80011-9. [DOI] [PubMed] [Google Scholar]
- McElver J, Patton D, Rumbaugh M, Liu CM, Yang LJ, Meinke D. The TITAN5 gene of Arabidopsisencodes a protein related to the ADP ribosylation factor family of GTP binding proteins. Plant Cell. 2000;12:1379–1392. doi: 10.1105/tpc.12.8.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElver J, Tzafrir I, Aux G, Rogers R, Ashby C, Smith K, Thomas C, Schetter A, Zhou Q, Cushman MA et al. (2001) Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics (in press) [DOI] [PMC free article] [PubMed]
- Meinke DW. A homoeotic mutant of Arabidopsis thalianawith leafy cotyledons. Science. 1992;258:1647–1650. doi: 10.1126/science.258.5088.1647. [DOI] [PubMed] [Google Scholar]
- Meinke DW. Seed development in Arabidopsis. In: Meyerowitz EM, Somerville CR, editors. Arabidopsis. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. pp. 253–295. [Google Scholar]
- Meinke DW. Molecular genetics of plant embryogenesis. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:369–394. [Google Scholar]
- Miller JD, Arteca RN, Pell EJ. Senescence-associated gene expression during ozone-induced leaf senescence in Arabidopsis. Plant Physiol. 1999;120:1015–1024. doi: 10.1104/pp.120.4.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D, Johnson D. A deubiquitinating enzyme interacts with SIR4 and regulates silencing in S. cerevisiae. Cell. 1996;86:667–677. doi: 10.1016/s0092-8674(00)80139-7. [DOI] [PubMed] [Google Scholar]
- Naviglio S, Mattecucci C, Matoskova B, Nagase T, Nomura N, Di Fiore PP, Draetta GF. UBPY: a growth-regulated human ubiquitin isopeptidase. EMBO J. 1998;17:3241–3250. doi: 10.1093/emboj/17.12.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman T, de Bruijn FJ, Green P, Keegstra K, Kende H, McIntosh L, Ohlrogge J, Raikhel N, Somerville S, Thomashow M. Genes galore: a summary of methods for accessing results from large-scale partial sequencing of anonymous Arabidopsis cDNA clones. Plant Physiol. 1994;106:1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niebel A, de Almeida Engler J, Hemerly A, Ferreira P, Inze D, Van Montagu M, Gheysen G. Induction of cdc2a and cyc1At expression in Arabidopsis thalianaduring early phases of nematode-induced feeding cell formation. Plant J. 1996;10:1037–1043. doi: 10.1046/j.1365-313x.1996.10061037.x. [DOI] [PubMed] [Google Scholar]
- Nogales E. Structural insights into microtubule function. Annu Rev Biochem. 2000;69:277–302. doi: 10.1146/annurev.biochem.69.1.277. [DOI] [PubMed] [Google Scholar]
- Ohad N, Yadegari R, Margossian L, Hannon M, Michaeli D, Harada JJ, Goldberg RB, Fischer RL. Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell. 1999;11:407–416. doi: 10.1105/tpc.11.3.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen O-A. Endosperm development: cellularization and cell fate specification. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:233–267. doi: 10.1146/annurev.arplant.52.1.233. [DOI] [PubMed] [Google Scholar]
- Oppenheimer DG, Pollock MA, Vacik J, Szymanski DB, Ericson B, Feldmann K, Marks MD. Essential role of a kinesin-like protein in Arabidopsistrichome morphogenesis. Proc Natl Acad Sci USA. 1997;94:6261–6266. doi: 10.1073/pnas.94.12.6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui M, Staehelin LA. Syncytial-type cell plates: a novel kind of cell plate involved in endosperm cellularization of Arabidopsis. Plant Cell. 2000;12:933–947. doi: 10.1105/tpc.12.6.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton DA, Schetter AL, Franzmann LH, Nelson K, Ward ER, Meinke DW. An embryo-defective mutant of Arabidopsis disrupted in the final step of biotin synthesis. Plant Physiol. 1998;116:935–946. doi: 10.1104/pp.116.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radcliffe PA, Garcia MA, Toda T. The cofactor-dependent pathways for α- and β-tubulins in microtubule biogenesis are functionally different in fission yeast. Genetics. 2000a;156:93–103. doi: 10.1093/genetics/156.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radcliffe PA, Vardy L, Toda T. A conserved small GTP-binding protein Alp41 is essential for the cofactor-dependent biogenesis of microtubules in fission yeast. FEBS Lett. 2000b;468:84–88. doi: 10.1016/s0014-5793(00)01202-3. [DOI] [PubMed] [Google Scholar]
- Rao H, Uhlmann F, Nasmyth K, Varshavsky A. Degradation of a cohesin subunit by the N-end rule pathway is essential for chromosome stability. Nature. 2001;410:955–959. doi: 10.1038/35073627. [DOI] [PubMed] [Google Scholar]
- Rao-Naik C, Chandler JS, McArdle B, Callis J. Ubiquitin-specific proteases from Arabidopsis thaliana: cloning of AtUBP5 and analysis of substrate specificity of AtUBP3, AtUBP4, and AtUBP5 using Escherichia coliin vivo and in vitro assays. Arch Biochem Biophys. 2000;379:198–208. doi: 10.1006/abbi.2000.1874. [DOI] [PubMed] [Google Scholar]
- Roberts K, McCann MC. Xylogenesis: the birth of a corpse. Curr Opin Plant Biol. 2000;3:517–522. doi: 10.1016/s1369-5266(00)00122-9. [DOI] [PubMed] [Google Scholar]
- Rojo E, Gillmor CS, Kovaleva V, Somerville CR, Raikhel NV. VACUOLELESS1 is an essential gene required for vacuole formation and morphogenesis in Arabidopsis. Dev Cell. 2001;1:303–310. doi: 10.1016/s1534-5807(01)00024-7. [DOI] [PubMed] [Google Scholar]
- Schrick K, Mayer U, Horrichs A, Kuhnt C, Bellini C, Dangl J, Schmidt J, Jurgens G. FACKEL is a sterol C-14 reductase required for organized cell division and expansion in Arabidopsisembryogenesis. Genes Dev. 2000;14:1471–1484. [PMC free article] [PubMed] [Google Scholar]
- Smith RF, Wiese BA, Wojzynski MK, Davison DB, Worley KC. BCM Search Launcher: an integrated interface to molecular biology data base search and analysis services available on the World Wide Web. Genome Res. 1996;6:454–462. doi: 10.1101/gr.6.5.454. [DOI] [PubMed] [Google Scholar]
- Snustad DP, Haas NA, Kopczak SD, Silflow CD. The small genome of Arabidopsiscontains at least nine expressed β-tubulin genes. Plant Cell. 1992;4:549–556. doi: 10.1105/tpc.4.5.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen MB, Chaudhury AM, Robert H, Bancharel E, Berger F. Polycomb group genes control pattern formation in plant seed. Curr Biol. 2001;11:277–281. doi: 10.1016/s0960-9822(01)00072-0. [DOI] [PubMed] [Google Scholar]
- Stearns T, Hoyt MA, Botstein D. Yeast mutants sensitive to antimicrotubule drugs define three genes that affect microtubule function. Genetics. 1990;124:251–262. doi: 10.1093/genetics/124.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G, Huang Y, Rommelaere H, Vandekerckhove J, Ampe C, Cowan NJ. Pathway leading to correctly folded β-tubulin. Cell. 1996;86:287–296. doi: 10.1016/s0092-8674(00)80100-2. [DOI] [PubMed] [Google Scholar]
- Torres-Ruiz RA, Jurgens G. Mutations in the FASS gene uncouple pattern formation and morphogenesis in Arabidopsisdevelopment. Development. 1994;120:2967–2978. doi: 10.1242/dev.120.10.2967. [DOI] [PubMed] [Google Scholar]
- Traas J, Bellini C, Nacry P, Kronenberger J, Bouchez D, Caboche M. Normal differentiation patterns in plants lacking microtubular preprophase bands. Nature. 1995;375:676–677. [Google Scholar]
- Tsugeki R, Kochieva EZ, Fedoroff NV. A transposon insertion in the Arabidopsis SSR16gene causes an embryo-defective lethal mutation. Plant J. 1996;10:479–489. doi: 10.1046/j.1365-313x.1996.10030479.x. [DOI] [PubMed] [Google Scholar]
- Uwer U, Willmitzer L, Altmann T. Inactivation of a glycyl-tRNA synthetase leads to an arrest in plant embryo development. Plant Cell. 1998;10:1277–1294. doi: 10.1105/tpc.10.8.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernon DM, Meinke DW. Embryogenic transformation of the suspensor in twin, a polyembryonic mutant of Arabidopsis. Dev Biol. 1994;165:566–573. doi: 10.1006/dbio.1994.1276. [DOI] [PubMed] [Google Scholar]
- Weaver LM, Gan S, Quirino B, Amasino RM. A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol Biol. 1998;37:455–469. doi: 10.1023/a:1005934428906. [DOI] [PubMed] [Google Scholar]
- Whittington AT, Vugrek O, Wei KJ, Hasenbein NG, Sugimoto K, Rashbrooke MC, Wasteneys GO. MOR1 is essential for organizing cortical microtubules in plants. Nature. 2001;411:610–613. doi: 10.1038/35079128. [DOI] [PubMed] [Google Scholar]
- Wilkinson KD. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J. 1997;11:1245–1256. doi: 10.1096/fasebj.11.14.9409543. [DOI] [PubMed] [Google Scholar]
- Wilkinson KD, Tashayev VL, O'Connor LB, Larsen CN, Kasperek E, Pickart CM. Metabolism of the polyubiquitin degradation signal: structure, mechanism, and role of isopeptidase T. Biochemistry. 1995;34:14535–14546. doi: 10.1021/bi00044a032. [DOI] [PubMed] [Google Scholar]
- Yadegari R, Kinoshita T, Lotan O, Cohen G, Katz A, Choi Y, Katz A, Nakashima K, Harada JJ, Goldberg RB. Mutations in the FIE and MEAgenes that encode interacting polycomb proteins cause parent-of-origin effects on seed development by distinct mechanisms. Plant Cell. 2000;12:2367–2382. doi: 10.1105/tpc.12.12.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan N, Doelling JH, Falbel TG, Durski AM, Vierstra RD. The ubiquitin-specific protease family from Arabidopsis: AtUBP1 and 2 are required for the resistance to the amino acid analog canavanine. Plant Physiol. 2000;124:1828–1843. doi: 10.1104/pp.124.4.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JZ, Somerville CR. Suspensor-derived polyembryony caused by altered expression of valyl-tRNA synthetase in the twn2 mutant of Arabidopsis. Proc Natl Acad Sci USA. 1997;94:7349–7355. doi: 10.1073/pnas.94.14.7349. [DOI] [PMC free article] [PubMed] [Google Scholar]