Abstract
An extracellular matrix composed of a layered meshwork of β-glucans, chitin, and mannoproteins encapsulates cells of the yeast Saccharomyces cerevisiae. This organelle determines cellular morphology and plays a critical role in maintaining cell integrity during cell growth and division, under stress conditions, upon cell fusion in mating, and in the durable ascospore cell wall. Here we assess recent progress in understanding the molecular biology and biochemistry of cell wall synthesis and its remodeling in S. cerevisiae. We then review the regulatory dynamics of cell wall assembly, an area where functional genomics offers new insights into the integration of cell wall growth and morphogenesis with a polarized secretory system that is under cell cycle and cell type program controls.
INTRODUCTION
Cell structure seldom stops at the plasma membrane boundary but usually extends beyond it in an extracellular network of proteins and polysaccharides. These extracellular matrices have compositions that are highly variable between taxonomic groups. For example, the distinct structures of the cell walls of pathogenic bacteria have been the focus of much study as targets for antibiotics. Although the bacterial cell wall field is mature, its distinctness has limited its contribution to eukaryotic extracellular matrix work. Even among the eukaryotes, extracellular matrix composition is sufficiently different between the metazoan, fungal, and plant kingdoms that a research area has been independently developed for each, with little common ground. Fungi and plants have extracellular matrices containing polysaccharide-protein complexes termed cell walls. Cell walls allow fungi and plants to build structures based on the use of cells as hydrostatic bricks, a process that is particularly well elaborated in woody plants (265). Despite a common body plan, the actual polymers used by fungi and plants to construct cell walls are often different. Such structural diversity has forced cell wall research to develop in a fragmented way that lacks the benefits of the conserved cell biology that links much eukaryotic function. However, all eukaryotic extracellular matrices are shaped by a common underlying cytoskeleton. This positions a highly conserved secretory system that moves the relevant construction machinery, which in turn lays down the extracellular matrix. Hence, much of the evolutionary diversity seemingly comes only at the level of the extracellular matrices themselves. It remains to be seen how similar are the underlying cytoskeletal and secretory pathway strategies for constructing such matrices and whether they will offer a more unifying view of their biology.
This review primarily covers work on the cell wall of the yeast Saccharomyces cerevisiae but, when appropriate, draws on studies of other fungi for comparison. A powerful attraction of S. cerevisiae is that the whole organism is under intensive study, providing information on all aspects of its biology as they relate to the cell wall. While an in-depth study of the structure and function of the cell wall of one species is critical, fungal cell walls are highly diverse, so yeast studies will capture only a fraction of the fungal cell wall repertoire.
COMPONENTS AND SYNTHESIS
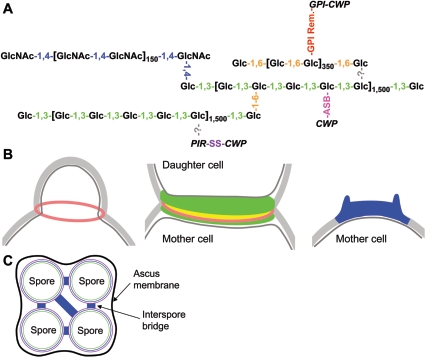
The S. cerevisiae cell wall represents ∼30% of the dry weight of the cell and is composed largely of polysaccharides (∼85%) and proteins (∼15%) (206). Extensive biochemical analyses reveal that glucose, N-acetylglucosamine (GlcNAc), and mannose residues represent 80 to 90%, 1 to 2%, and 10 to 20% of the total polysaccharide, respectively. Glucose residues are linked to other glucose molecules through β-1,3 and β-1,6 linkages and to GlcNAc via β-1,4 bonds. Mannoproteins can be linked to β-1,6-glucose chains through a processed glycosylphosphatidylinositol (GPI) anchor or to β-1,3-glucan through an alkali-labile bond (see references 73, 130, 136-138, and 194 and references therein). Based on these analyses, a structure for the mature cell wall has been proposed (36, 73, 136, 256) and is illustrated in Fig. 1A. Chains of β-1,3-linked glucose residues are branched by β-1,6 linkages, forming a fibrillar β-1,3-glucan which serves as a backbone to which are linked chitin, β-1,6-glucan, and some mannoproteins. In addition, the β-1,6-glucan and GPI mannoproteins are linked together via a remnant of their GPI anchors. The vegetative cell wall has a layered ultrastructure as observed by electron microscopy (212), with an inner layer of glucans and chitin and an outer layer of mannoproteins.
FIG. 1.
Schematic representation of cell wall components and their linkages. (A) Architecture of the lateral cell wall. The β-1,3-, β-1,4-, and β-1,6-glucosidic bonds are represented as green, blue, and orange, respectively. Cell wall mannoproteins (CWP) can be linked to the β-1,3-glucan via alkali-sensitive bonds (ASB) or to PIR proteins (PIR) via a disulfide link (SS). GPI cell wall proteins (GPI-CWP) are attached to the β-1,6-glucan through a remnant GPI anchor (GPI Rem.). The links between β-1,3-glucan and β-1,6-glucan or PIR proteins are still uncharacterized (?). Note that the figure qualitatively illustrates the cell wall linkages but does not reflect their stoichiometry. (B) Assembly of cell wall constituents at the septum during bud emergence and at the mother/daughter interface (left and middle panels, respectively) and in the bud scar (right panel). Color representation: lateral cell wall, light gray; plasma membrane, dark gray; chitin ring, pink; chitinous primary septum, yellow; glucan- and mannoprotein-based secondary septum, green; chitinous bud scar, blue. (C) Ascospore wall organization. The glucan-mannoprotein, chitosan, and dityrosine layers are shown in green, blue, and violet, respectively. Chitosan-rich interspore bridges connecting the spores are shown in blue. For clarity the ascus cell wall has been omitted.
Components of the lateral cell wall are also found in the septum, a specialized structure critical for cell separation that is located at the bud neck and is synthesized in several steps (36, 51). A ring of chitin is initially deposited at the time and site of bud emergence (Fig. 1B, left panel). A primary septum, a disk of chitin which physically separates the mother cell from the daughter cell, is then laid down after mitosis is completed. Following primary septum synthesis, a secondary septum of β-glucans and mannoproteins is assembled (Fig. 1B, middle panel). After cytokinesis, the primary septum is degraded by chitinase, leading to cell separation. As relics of this process, a chitinous bud scar is left on the mother cell wall (Fig. 1B, right panel) and a less prominent birth scar remains on the daughter cell.
During sporulation of diploid cells, meiotic ascospores are encapsulated in the ascus by a specialized cell wall, the ascospore wall (51, 264) (Fig. 1C). The ascospores are connected by chitosan-containing structures, the interspore bridges (54). The ascospore wall, which is more robust than the vegetative cell wall, has a multilayered structure. β-Glucans and mannoproteins form the innermost layer, which is surrounded by fibrils of chitosan, a deacetylated chitin polymer. The outermost layer is composed of dityrosine, an insoluble arrangement of cross-linked d- and l-tyrosine residues.
β-1,3-Glucan
The β-1,3-glucan chains, with a degree of polymerization of ∼1,500 glucose units/chain, have a coiled spring-like structure that confers elasticity and tensile strength to the cell wall (136, 137). In cell wall extracts, β-1,3-glucan is found as a branched polymer with β-1,6 interchain links. β-1,3-Glucan is covalently linked to the other wall components (Fig. 1A): its nonreducing ends are cross-linked to the reducing ends of chitin chains through a β-1,4 link; β-1,6- and β-1,3-glucan chains are attached by a still-uncharacterized link. Some O-mannosylated cell wall proteins (CWPs) are attached to β-1,3-glucan via an alkali-sensitive bond (73, 136-138, 194).
Glucan synthase.
β-1,3-Glucan synthase (β-1,3-GS) (Table 1) subunits have been identified using an in vitro assay for β-1,3-GS activity. Basically, membrane extracts are incubated with radiolabeled UDP-glucose in the presence of GTP, and the accumulation of acid-insoluble radioactive β-1,3-glucan follows (35). By using such an assay, β-1,3-GS was separated into membrane-bound and cytosolic fractions, containing catalytic and regulatory GTP-binding subunits, respectively (127). Cytoplasmic UDP-glucose is likely the in vivo sugar donor, since mutants with defects in UDP-glucose synthesis show cell wall defects (210).
TABLE 1.
Enzymatic activities and genes required for cell wall polymer synthesis and remodeling in S. cerevisiae
| Process | Activitya | Geneb | Protein function/comment |
|---|---|---|---|
| β-1,3-Glucan synthesis | β-1,3-GS | FKS1 | Catalytic subunit of β-1,3-glucan synthase |
| GSC2 | Catalytic subunit of β-1,3-glucan synthase | ||
| RHO1 | GTP-binding protein, regulatory subunit of β-1,3-glucan synthase | ||
| FKS3 | Protein with similarity to Fks1p and Gsc2p | ||
| β-1,6-Glucan synthesis | β-1,6 GS | RHO1 | Probable regulatory subunit of β-1,6-synthase |
| Unknown | BIG1 | ER membrane protein required for cell wall β-1,6-glucan synthesis | |
| KRE1 | Cell wall glycoprotein involved in β-glucan assembly | ||
| KRE5 | UGGT-related protein required for β-1,6-glucan biosynthesis | ||
| KRE6 | Protein required for β-1,6-glucan biosynthesis | ||
| KRE9 | Glycoprotein involved in cell wall β-glucan assembly | ||
| KNH1 | Kre9p homologue | ||
| ROT1 | Protein required for cell wall function | ||
| SKN1 | Type II membrane protein with similarity to Kre6p | ||
| Chitin synthesis | CSI | CHS1 | Required for repairing the chitin septum after separation |
| CSII | CHS2 | Makes chitin in the primary septum during cytokinesis | |
| CSIII | CHS3 | Makes the majority of cell wall chitin, the chitin ring during bud emergence, and spore wall chitosan | |
| SKT5 | Activator of Chs3p, recruits Chs3p to the bud neck via an interaction with Bni4p | ||
| SHC1 | Sporulation-specific activator of Chs3p with similarity to Skt5p | ||
| Chitosan synthesis | Chitin deacetylase | CDA1 | Involved in biosynthesis of ascospore wall chitosan |
| CDA2 | Involved in biosynthesis of ascospore wall chitosan | ||
| Protein mannosylation | α-1,6-MT | OCH1 | N-glycan outer chain initiation |
| α-1,2/6-MT | MNN9 | ManPol I and II subunits involved in N glycosylation | |
| α-1,6-MT | VAN1 | ManPol I subunit involved in N glycosylation | |
| ANP1 | ManPol II subunit involved in N glycosylation | ||
| HOC1 | ManPol II subunit involved in N glycosylation | ||
| MNN10 | ManPol II subunit involved in N glycosylation | ||
| MNN11 | ManPol II subunit involved in N glycosylation | ||
| α-1,3-MT | MNN1 | Involved in N glycosylation termination and O mannosylation | |
| α-1,2-MT | MNN2 | Branching of outer chain of N glycosylation | |
| MNN5 | N-glycan outer chain extension | ||
| MNT2 | α-1,3-Mannosyltransferase involved in O mannosylation | ||
| MNT3 | α-1,3-Mannosyltransferase involved in O mannosylation | ||
| α-1,2 MT | KRE2 | KTR family member involved in N and O glycosylation | |
| KTR1 | KTR family member involved in N and O glycosylation | ||
| MT | KTR2 | KTR family member involved in N and O glycosylation | |
| KTR3 | KTR family member involved in N and O glycosylation | ||
| YUR1 | KTR family member involved in N and O glycosylation | ||
| KTR4 | Putative mannosyltransferase | ||
| KTR5 | Putative mannosyltransferase | ||
| KTR7 | Putative mannosyltransferase | ||
| α-1,2-MpT | KTR6 | α-1,2-Mannosylphosphate transferase involved in N glycosylation | |
| Glucan processing/remodeling | β-1,3-Glucan elongase | GAS1 | β-1,3-Glucanosyltransferase |
| Exo-β-1,3/1,6-glucanase | EXG1 | Major exo-β-1,3-glucanase of the cell wall | |
| EXG2 | GPI-anchored plasma membrane exo-β-1,3-glucanase | ||
| SPR1 | Sporulation-specific exo-β-1,3-glucanase | ||
| Endo-β-1,3-glucanase | BGL2 | Can link in vitro β-1-3-glucan chains through a β-1,6-linkage | |
| DSE4 | Involved in septum degradation | ||
| Glucanase like | DSE2 | Glucanase-like secreted protein involved in septum degradation | |
| EGT2 | GPI-anchored cell wall endoglucanase required for proper cell separation after cytokinesis | ||
| GAS2 | GAS family member | ||
| GAS3 | GAS family member | ||
| GAS4 | GAS family member | ||
| GAS5 | GAS family member | ||
| SCW3 | Glucanase-like “soluble” cell wall protein | ||
| SCW4 | Glucanase-like “soluble” cell wall protein | ||
| SCW10 | Glucanase-like “soluble” cell wall protein | ||
| SCW11 | Glucanase-like protein | ||
| Chitin processing | Chitinase | CTS1 | Endochitinase, required for cell separation after mitosis |
| CTS2 | Sporulation-specific endochitinase-like protein | ||
| Glucan/chitin cross-linking | Transglycosidase | CRH1 | Putative glycosidase of the cell wall |
| UTR2 | Putative GPI-anchored glycosidase localized to the bud neck | ||
| CRR1 | Protein with similarity to Crh1p |
MT, mannosyltransferase; MpT, mannosylphosphate transferase.
Genes whose products have no known or established enzymatic activity are in boldface.
(i) Catalytic subunit: the FKS family.
A search for components of the membrane-bound fraction revealed that two highly homologous genes, FKS1 and GSC2/FKS2, likely encode catalytic subunits of the β-1,3-GS. Despite a large body of genetic, molecular, cell biological, biochemical, and comparative genomic evidence, the case for the Fks proteins being the catalytic subunits of β-1,3-GS, while strong, remains circumstantial (summarized in reference 73). Unlike chitin synthase (see below), β-1,3-GS has never been purified to homogeneity, and thus its activity has not been reconstituted in vitro from purified components. This is the situation for all β-glucan synthases, including plant cellulose synthase (265), and resolving this problem will require improvements in the technology of purifying enzymatically active membrane-bound complexes.
FKS1 (for FK506 sensitive) was originally cloned by complementation of a mutant that was hypersensitive to the calcineurin inhibitor FK506 (216). The link between FKS1 and the cell wall was established by the finding that ETG1 and PBR1 (whose mutants are resistant to β-1,3-GS inhibitors), CWH53 (required for resistance to calcofluor white), and CND1 (with mutant alleles requiring a functional calcineurin pathway) are all identical to FKS1 (44, 76, 80, 91, 231). FKS1 mutants show decreased β-1,3-GS activity, while Fks1 cofractionates with GS activity, implicating Fks1 as a β-1,3-GS catalytic subunit (76, 119). FKS1 encodes a 1,876-amino-acid (aa) protein which localizes at the plasma membrane and is predicted to have 16 transmembrane domains (TMDs) (76, 119, 228). An Fks1-green fluorescent protein (GFP) fusion protein expressed from the FKS1 promoter localizes to sites of polarized growth and colocalizes dynamically with actin patches (71, 286). Fks1 shows a four-domain structure: it includes an ∼400-aa N-terminal cytosolic tail thought to be involved in activation of the GS activity, an ∼300-aa region containing six TMDs and required for Fks1 localization to sites of polarized growth, an ∼600-aa cytosolic putative catalytic domain, and a C-terminal ∼600-aa domain containing 10 TMDs and involved in localization to the cell surface (71).
GSC2 encodes a 1,895-residue protein that is 88% identical to Fks1 and has a similar topology and domain organization. As with Fks1, a GFP-Gsc2 fusion protein is localized to the sites of polarized growth (71). Gsc2 and Fks1 form a redundant essential pair, with a double fks1Δ gsc2Δ mutant being unviable (119. 178). This synthetic lethality and the finding that purified membranes from an fks1Δ mutant, which express only Gsc2, show β-1,3-GS activity which can be immunodepleted on treatment with anti-Gsc2 antibodies (119, 178) indicate that Gsc2 is involved in β-1,3-glucan synthesis.
FKS1 and GSC2 show differential patterns of expression. The FKS1 gene is expressed during mitotic growth, consistent with its proposed function as the major β-1,3-GS subunit. Upon α-factor release of a synchronized population, levels of the FKS1 transcript peak at the late G1/S phase of the cell cycle, in a way similar to that for the G1/S cyclin CLN2, thymidylate synthase CDC21, and histone H2A HTA2 transcripts (178, 231). This expression pattern is dependent on Swi4, the DNA-binding component of the Swi4-Swi6 cell cycle box binding factor (SBF) transcription factor (115). A genome-wide survey of SBF binding sites by chromatin immunoprecipitation followed by microarray hybridization revealed that the FKS1 promoter is bound in vivo by SBF (121). These data indicate that during the cell cycle, FKS1 expression is regulated by the SBF transcription factor. In addition, a weak Rlm1-dependent increase of FKS1 mRNA levels upon hyperactivation of the mitogen-activated protein (MAP) kinase Slt2 has been reported (123), suggesting that the protein kinase C (PKC) cell integrity pathway through its principal transcription factor Rlm1 also regulates FKS1 expression.
In contrast, GSC2 mRNA, which is undetectable during mitotic growth on glucose, accumulates upon glucose exhaustion, during growth on low levels of glucose, or during growth on galactose, glycerol, or acetate. These expression changes coincide with decreased levels of the FKS1 transcript (178, 313). Thus, GSC2 expression appears to be under catabolite repression. Consistent with this, deletion of the catabolic-repressor-encoding gene MIG1 results in a high basal transcription and a weak low-glucose induction of a GSC2-driven reporter. Further, the induction of GSC2 at low glucose concentrations is regulated by the Snf1 protein kinase, mediating derepression of glucose-repressed genes (313). The regulators of GSC2 induction upon entry into stationary phase and on nonglucose substrates have not been identified. Whether GSC2 transcript levels are cell cycle regulated during growth on a nonglucose carbon source is unknown.
GSC2 can also be induced by a number of environmental stimuli. The addition of extracellular calcium or α-factor leads to strong GSC2 induction that is dependent on the calcineurin signaling pathway (178) and is mediated by Crz1, a zinc finger transcription factor that binds to a calcineurin-dependent response element in the GSC2 promoter (176, 268).
Shift of a wild-type strain from 23°C to 39°C also induces GSC2 mRNA accumulation (313). A block in the calcineurin pathway by addition of the drug FK506 delays this induction, indicating that the calcineurin pathway is responsible for the rapid GSC2 response upon heat shock. In contrast, an slt2 mutant, which is defective in PKC cell integrity pathway signaling, exhibits a rapid but transient increase of GSC2 mRNA levels upon temperature shift, suggesting that the PKC cell integrity pathway is required for the maintenance of GSC2 expression during growth at high temperature (313). Further, exposure to cell wall- and cell surface-damaging agents, such as caspofungin and amphotericin B, or deletion of genes required for cell wall assembly, such as fks1, gas1, kre6, mnn9, or smi1, also results in high levels of GSC2 mRNA (5, 145, 178, 232). These effects, too, are thought to be mediated by the PKC cell integrity and/or the calcineurin pathway (see below). For example, exposure of wild-type cells to calcofluor white (a toxic chitin-binding agent) or to Zymolyase (a β-1,3-glucanase enzymatic preparation) increases expression of a GSC2-lacZ reporter in an Slt2-dependent manner (68). A promoter dissection study revealed that both Rlm1, the target of the PKC cell integrity pathway, and Crz1 mediate calcofluor induction of a GSC2-lacZ reporter (145). Finally, treatment with the reducing agent dithiothreitol induced high levels of the GSC2 transcript (282). Possible explanations for this include cell wall stress caused by perturbing the function of disulfide-bridged cell wall proteins, Fks1 misfolding, or a disruption of intracellular calcium homeostasis caused by malfunction of the endoplasmic reticulum (ER).
Consistent with Fks1 function in β-1,3-glucan synthesis, an fks1Δ mutant shows a reduced β-1,3-GS activity and an altered cell wall composition, with decreased β-1,3- and β-1,6-glucan levels and increased chitin and mannan levels (30, 59, 71, 76, 151, 213, 232). In contrast, during vegetative growth on glucose, a gsc2 mutant has no cell wall defect or growth phenotype. However, a homozygous gsc2/gsc2 null diploid fails to sporulate. The effects of the deletion of FKS1 or GSC2 can be partially suppressed by overexpression of the other member of the pair (71, 178), indicating their functional similarity. However, the strength of the phenotypes observed in an fks1 mutant compared to a gsc2 mutant shows that Fks1 is the major player during vegetative growth on rich medium, with Gsc2 being the protein functioning under more stressful conditions. As mentioned above, fks1 mutants show hypersensitivity to FK506 or calcofluor white (91, 231), two phenotypes explained by the calcineurin-dependent GSC2 induction and by the increased cell wall chitin found in the fks1 mutant. Interestingly, an fks1Δ mutant is more sensitive to caspofungin, echinocandins, or aerothricin 3 than is a gsc2Δ mutant (139, 150, 178). The differential sensitivity of the two proteins to these compounds and their expression balance likely explain these phenotypes.
The S. cerevisiae FKS family has a third member, FKS3, whose product is 55% identical to both Fks1 and Gsc2. A search for Fks3 function in glucan or cell wall synthesis under vegetative growth conditions has been unsuccessful: no phenotype was associated with an FKS3 deletion, and no activity could be attributed to Fks3 (71). However, in a large-scale search for sporulation mutants, a homozygous fks3/fks3 null diploid was found to be sporulation deficient, indicating that Fks3 is required for optimal sporulation (70). FKS3 expression is regulated by Ste12 upon pheromone exposure (311), implying that Fks3 may also have some role in mating.
(ii) Rho1, a regulatory subunit.
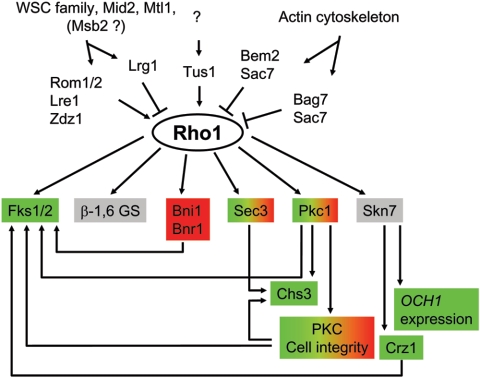
Considerable biochemical and genetic evidence identifies the Rho1 GTPase as a regulator of β-1,3-GS (78, 177, 228): (i) Rho1 colocalizes and copurifies with Fks1 and cofractionates with β-1,3-GS activity, (ii) a thermosensitive rho1 mutation leads to thermolabile β-1,3-GS activity, and (iii) a hyperactive rho1 allele shows a GTP-independent β-1,3-GS activity. The essential 24-kDa Rho1 is the prototype of small G proteins which in their active GTP-bound state bind and activate their effectors (for reviews, see references 34 and 274). Although Fks1 and Gsc2 are well established as Rho1 effectors, the interaction domains between the catalytic and the regulatory subunits of the β-1,3-GS and the basis for Rho1 activation on β-1,3-GS catalytic subunits remain unknown. In addition to its role as a subunit of the β-1,3-GS, Rho1 plays a critical role in the coordination of cell wall organization and the polarization of the actin cytoskeleton by regulating signaling through the cell integrity pathway (see “The PKC cell integrity pathway” below and Fig. 4). These Rho1 functions are separable, and a phenotypic and complementation analysis of 12 rho1 temperature-sensitive alleles identified 4 rho1 alleles with direct effects on β-1,3-GS activity (245). The Cabib group reported seven additional temperature-sensitive rho1 alleles specifically affecting β-1,3-GS regulation (239). These showed bud lysis and decreased incorporation of cell wall component phenotypes at the restrictive temperature.
FIG. 4.
The central role of Rho1 in integrating multiple signals. Rho1 transmits signals from the cell wall sensors and the actin cytoskeleton through GAPs and GEFs to its effectors. Rho1 effectors are colored according to signal input provenance as follows: green, cell wall; red, actin; gray, unknown. Two colors indicate a dual regulation.
Like other small G proteins, Rho1 cycles between its active GTP-bound state and its inactive GDP-bound state under modulation by GDP-GTP exchange factors (GEFs) and GTPase-activating protein (GAPs). Specific GAPs and GEFs are thought to regulate G-protein-effector pairs, and to date, four Rho1 GAPs and three Rho1 GEFs have been identified (152, 243). Of those, Lrg1 and Rom2 are the respective GAP and GEF specialized for the regulation of β-1,3-glucan synthesis. Indeed, using a thermosensitive fks1-1154 gsc2Δ double mutant, which shows a thermolabile β-1,3-GS activity (253), Oyha's group reported that mutation in LRG1 or overexpression of ROM2 restored the impaired β-1,3-glucan synthesis of this mutant (253, 301). Two additional multicopy suppressors of the fks1-1154 gsc2Δ phenotype, LRE1 and ZDS1, activate the GS activity through Rho1 (253). However, no GEF activity has been described for the products of these two genes, and how they act to stimulate β-1,3-glucan synthesis is still unclear.
Finally, Rho1 is membrane anchored via a C-terminal prenylyl tail, which is required for Rho1 membrane association and for its function in regulating β-1,3-GS activity (118).
(iii) Fungal glucan synthases.
β-1,3-Glucan is widely used by fungi, and a BLAST search for Fks1 homologues in the NCBI nonredundant database of fungal protein sequences on 1 February 2006 returned over 90 matches with E values of <0.001 in 40 organisms (http://www.ncbi.nlm.nih.gov/BLAST/). Yarrowia lipolytica, Cryptococcus neoformans, Coccidiodes posadasii, and Pneumocystis carinii all possess a single essential FKS gene (132, 149, 218, 278). A single FKS orthologue is also found in the completed genomes of Aspergillus nidulans, Aspergillus fumigatus, Neurospora crassa, and Rhizopus oryzae, indicating that these organisms also possess a single FKS gene that is likely essential (17, 114, 133, 249). The FKS protein of N. crassa bound the UDP-glucose analog 5-azido-UDP-glucose in a cross-linking study using a partially purified FKS preparation (249). However, work is needed to confirm the specificity and functional relevance of this substrate binding. The genome of Candida albicans contains three putative FKS homologues. Of these, the GSC1 gene is essential, and mutant alleles show an altered sensitivity to echinocandins; in contrast, GSL1 encodes a truncated protein, and GSL2 expression, if any, is very low (183). Thus, it is likely that a single and essential catalytic GS subunit, encoded by GSC1, functions in this organism. Finally, some species, such as Schizosaccharomyces pombe, Ashbya gossypii, and members of the genus Saccharomyces, have an FKS gene family. For example, Schizosaccharomyces pombe has four FKS homologues, three of which (bsg+, bsg2+, and bsg3+) are expressed during vegetative growth and sporulation and one of which (bsg4+) is sporulation specific. The differential localization of these proteins indicates that each has a specialized function in polarized cell wall growth, septum synthesis and cytokinesis, and ascospore wall assembly (56, 57, 158, 173, 174). This presence of an FKS family in some organisms probably reflects the selective retention of duplicated genes through increased fitness conferred by their specialized application of GS function. This complexity is also found in plants, the cell walls of which can contain callose, a β-1,3-glucan induced in response to wounding. Plant cells possess multiple FKS genes, with many of the 12 callose synthase genes present in the genome of Arabidopsis thaliana being tissue and development specific (for a review, see reference 292).
The absence of β-1,3-glucan in mammals and its widespread distribution and essentiality in the fungal kingdom has made GS a favored antifungal target. Echinocandins, such as caspofungin, have been developed as drugs targeting GS. Caspofungin is active against many Candida and Aspergillus species and is approved for the treatment of fungal infections in patients whose infections are refractory to other therapies (for a review, see reference 69). Despite the β-1,3-GS activity from Cryptococcus neoformans membranes being effectively inhibited in vitro, Cryptococcus neoformans is resistant to caspofungin (169). However, in Cryptococcus neoformans, caspofungin has been found to be synergistic in combination with membrane-interfering drugs such as amphotericin B or azoles (88).
β-1,6-Glucan
In addition to having β-1,3-glucan, many fungi, including S. cerevisiae, have a second β-linked glucan, β-1,6-glucan. In contrast to the microfibrillar structure of β-1,3-glucan, in S. cerevisiae this β-1,6-polymer is shorter than β-1,3-glucan, is amorphous in structure, and acts as a flexible glue by forming covalent cross-links to β-1,3-glucan and chitin and to cell wall mannoproteins (138) (Fig. 1A). Studies on S. cerevisiae β-1,6-glucan have been reviewed (210, 256), and earlier work will only be summarized here. The polymer has an average chain length of ∼350 glucose residues (138), with the β-1,6 backbone branched with β-1,6 side chains via 3,6-substituted glucose residues on ∼15% of the residues (167, 170). In vegetative cells, the β-1,6-polymer comprises ∼12% of the cell wall polysaccharide (167).
Data from C. albicans show that its β-1,6-glucan polymer has a more linear structure than that found in S. cerevisiae, with branch points on ∼7% of the residues, and is more abundant, comprising ∼21% of the total cell wall carbohydrate (108, 184). Recent structural studies on the cell wall of Schizosaccharomyces pombe clarify the nature and structure of its β-1,6 polymer (168, 271), now called diglucan by Magnelli et al. (168). Diglucan has a β-1,6-linked backbone, but 75% of these residues are also β-1,3 linked so that the polymer is highly branched, far more so than in S. cerevisiae and C. albicans. Schizosaccharomyces pombe diglucan is moderately abundant, comprising 15% of the cell wall polysaccharide.
These comparative studies indicate the variability of β-1,6-glucan structure among yeasts, with the more highly branched Schizosaccharomyces pombe diglucan likely a variant of the β-1,6-glucan found in S. cerevisiae and C. albicans. A simplifying general view is that these polymers share a common path of synthesis of the β-1,6 backbone, with much fungus-specific variation in frequency of branching and cross-linking. If this common-pathway view is true, then the immunodetection of a β-1,6-polymer in the Golgi and vesicles of Schizosaccharomyces pombe (113) shows that synthesis of the β-1,6 backbone occurs at least partially in the secretory pathway. However, Magnelli et al. (168) point out that the diglucan structure also raises the possibility of a synthesis from preformed building blocks, for example, from repetitive core units that are linked to each other from a lipid-linked oligosaccharide or by a transglycosidase.
The β-1,6 polymer has been reported to occur widely among the fungi, including the Hemi-, Archae-, and Euascomycetes and the Basidiomycetes (see discussion in reference 297). An analysis of the alkali-insoluble fraction of the cell wall of the filamentous euascomycete A. fumigatus found no β-1,6-glucan (87). However, there are reports of wall proteins covalently linked to β-1,6-glucan in the related species Aspergillus niger and Fusarium oxysporum (26, 252). Thus, a comparative structural analysis of the cell walls of filamentous ascomycetes for the presence of β-1,6-glucan is needed among this large and divergent group of fungi. More generally, the widespread occurrence of the polymer across fungal lineages shows that it was present early in fungal evolution prior to the Basidiomycetes-Ascomycetes divergence and that it may have subsequently been lost from some groups.
Genetic analysis.
Extracellular polysaccharides are made either in secretory pathway compartments (e.g., N- and O-glycans) or at the cell surface (e.g., β-1,3-glucan and chitin). While it remains unclear which synthetic pathway is used to make β-1,6-glucan, components both in the secretory pathway and at the cell surface are required.
In S. cerevisiae, genes affecting β-1,6-glucan levels are found throughout the secretory pathway to the cell wall (213, 256), and it has been proposed that these define a biosynthetic pathway (21, 210). However, most of these genes are not directly involved in β-1,6-glucan synthesis, and this has prevented identification of the gene(s) encoding the β-1,6-glucan synthase. On the more positive side, β-1,6-glucan genetics have uncovered much interactional complexity of cell surface assembly in which the synthesis and regulation of this polymer are embedded (Table 1).
For example, a cluster of genes involved in protein glucosylation-dependent ER quality control in other organisms (KRE5, CNE1, CWH41, and ROT2, encoding UDP-glucose:glycoprotein glucosyltransferase [UGGT]- and calnexin-related proteins and glucosidases I and II, respectively) are required for normal levels of β-1,6-glucan synthesis in S. cerevisiae (2, 256, 257). Why this is so is unknown, but they likely act indirectly to maintain wild-type levels of some unidentified components of the β-1,6-glucan synthetic machinery. Such a situation resembles the finding for Arabidopsis thaliana that glucosidases I and II, the plant equivalents of S. cerevisiae Cwh41 and Rot2, are required for cellulose synthesis (32, 96). Further, in S. cerevisiae, a cluster of genes involved in core N-glycan synthesis also affects β-glucan levels (45, 213, 257). Possible explanations include the synthetic components being N-glycoproteins or the need for a glucosylated acceptor structure for β-1,6-glucan chain initiation.
(i) KRE5.
One gene, KRE5, which has quality control functions in other organisms but a strong β-1,6-glucan phenotype in S. cerevisiae, has been particularly intensively studied. Kre5 is a large soluble ER-resident protein (153, 180) that shows weak but significant sequence similarity to eukaryotic UGGTs and is a highly diverged member of this family (10). Classic UGGTs monoglucosylate misfolded N-glycoproteins in the ER as part of a protein-refolding pathway (reviewed in reference 217). This pathway has not been found to operate in S. cerevisiae; rather, mutations in KRE5 are lethal or cause very severe growth defects and greatly reduced levels of β-1,6-glucan (11, 153, 180).
Comparative studies strongly suggest that the S. cerevisiae Kre5 is a diverged UGGT and that the role of KRE5 in β-1,6-glucan synthesis is indirect. The C. albicans Kre5 sequence far more closely resembles the well-characterized UGGTs (108) and, although this has not been shown directly, is likely a UGGT. C. albicans homozygous kre5/kre5 null mutants are viable and show reduced β-1,6-glucan levels, though the residual 20% of the polymer in the mutant appears identical in structure to that of the wild type. This glucan phenotype argues that Kre5 is not the C. albicans β-1,6-glucan synthase. The C. albicans KRE5 gene complements an S. cerevisiae kre5 null mutant (108, 153) and allows β-1,6-glucan levels to be restored (108). Thus, as the C. albicans protein can substitute for the S. cerevisiae one, the S. cerevisiae Kre5p is also likely to be a UGGT. Both Levinson et al. (153) and Herrero et al. (108) speculate that the diverged S. cerevisiae protein is a more specialized UGGT in these yeasts, perhaps serving as an ER protein glucosyltransferase/chaperone dedicated to proteins involved in β-1,6-glucan synthesis.
(ii) BIG1 and ROT1.
BIG1 and ROT1 are fungus-specific essential genes whose null mutants can be osmotically rescued by adding 0.6 M sorbitol to the growth medium to give very slowly growing cells that, with patience, are amenable to experimentation. Both genes were initially identified as suppressors of target of rapamycin-related phenotypes by the Hall group (18). Consistent with their invoking a cell wall stress response, both big1 and rot1 mutants have increased β-1,3-glucan and chitin levels (18, 165, 213). Mutants with mutations in BIG1 and ROT1 have large reductions in β-1,6-glucan compared to a wild-type strain, though, importantly, there is a small amount of residual polymer, suggesting that, like KRE5, neither gene encodes the β-1,6-glucan synthase. Both Big1 and Rot1 are small integral membrane proteins. Big1 is an ER resident, while the location of Rot1 is unknown. A big1 mutant synthetically interacts with a kre5 mutant but not with a rot1 mutant, implying that Big1 and Rot1 act in a related pathway, perhaps in protein folding or quality control, that is at least partially distinct from that of Kre5 in the ER.
(iii) KRE6 and SKN1.
The duplicated pair of type 2 Golgi proteins encoded by KRE6 and SKN1 is required for normal levels of β-1,6-glucan. Both proteins have a luminal domain with sequence and structural similarity to family 16 glycoside hydrolases, which are likely glucosyl hydrolases or transglucosylases (186). Such an activity might indicate some glucose-processing step, rather than a glucan synthase. Possibilities include processing of some precursor or acceptor for β-1,6-glucan polymer synthesis or the processing of a β-1,6-glucan synthase-associated glycoprotein.
(iv) KRE1.
KRE1 is linked biosynthetically with β-1,6-glucan. The kre1 mutant has both a reduced level and an altered structure of the residual polymer, with the mutant polymer being shorter than the wild type (21). This suggests that the plasma membrane-associated, GPI-anchored Kre1 acts to extend the chain by adding linear β-1,6-glucan onto a highly branched acceptor glucan.
(v) KRE9 and KNH1.
The duplicated pair of fungus-specific genes KRE9 and KNH1 is synthetically lethal and shows a strong β-1,6-glucan reduction, with a kre9 null mutant having shorter β-1,6-glucan chains as shown by gel filtration (25, 256). They likely encode secreted or cell surface O-glycoproteins, but this has been shown only by their overexpression (72). Possible roles for these proteins include anchoring or cross-linking the newly synthesized β-1,6-glucan in the wall or as extracellular components of a β-1,6-glucan synthase.
(vi) Overview.
Why has genetic analysis been so difficult here? Potential problems hindering gene identification are that the β-1,6-glucan synthase may have an essential gene for which hypomorphic alleles are difficult to obtain or that these synthases form a gene family where mutants with mutations in single genes have weak phenotypes. Further, the area lacks any complementary biochemistry. To begin biochemical approaches, a sensitive dot blot immunoassay for the in vitro synthesis of β-1,6-glucan has been developed using an anti-β-1,6-glucan specific antibody (297). This assay requires a crude cell-free membrane preparation, UDP-glucose, and GTP. The nature of the in vitro reaction has not been investigated, but in defective mutants the rate of de novo synthesis correlates with their reduced levels of endogenous β-1,6-glucan. This suggests that β-1,6 chain elongation is being measured, with the rate of the in vitro reaction being proportional to the number of β-1,6-glucan chains present at the start of the reaction in the membrane extracts of the various mutants. The GTP dependence of the reaction implies involvement of a GTP-binding protein in regulating β-1,6-glucan synthesis. The reaction was independent of RHO2 to -5 but was stimulated by overexpression of RHO1, suggesting that like β-1,3-glucan synthase, β-1,6-glucan synthase is Rho1 dependent. It is hoped that this assay will be the spur for some much-needed biochemical investigation in this area.
Chitin and Chitosan
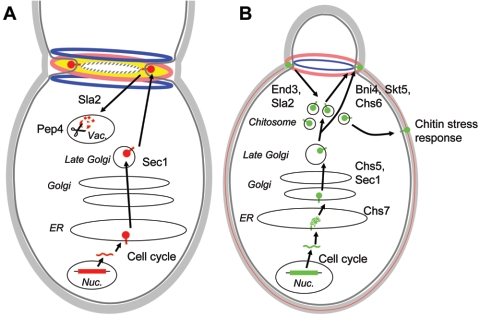
Chitin is a linear polymer of β-1,4-linked GlcNAc. It forms microfibrils which are stabilized by hydrogen bonds. In the crystalline form, microfibrils can orient in a parallel, an antiparallel, or an alternating parallel and antiparallel manner (for a review, see reference 142). Chitin is a minor constituent of the S. cerevisiae lateral wall (1 to 2% cell wall dry weight) and is concentrated at the bud neck and at the septum. The lengths of chitin chains in the cell wall and in bud scars are estimated to be ∼100 and 190 GlcNAc residues, respectively (128, 136). In the cell wall, about 40 to 50% of the chitin chains are linked to the nonreducing end of β-1,3-glucan via a β-1,4 bond engaging the reducing end of the chitin polymer (137) (Fig. 1A). Its crystalline structure confers stretching resistance to the cell wall. Chitin deposition within the cell is precisely controlled both spatially and temporally (for a review, see reference 29). At late G1, it is deposited as a ring at the site of bud emergence, then as a disk (the primary septum), and finally in the lateral cell wall of the mother cell after septation (Fig. 1B and 2).
FIG. 2.
Trafficking and biosynthetic pathways for chitin synthases 2 (A) and 3 (B). The CHS2 and CHS3 genes and their products are represented in red (A) and green (B), respectively. The chitin made by Chs2 is shown in yellow, and that made by Chs3 is shown in pink. The cell wall, the plasma membrane, and septin rings are shown in light and dark gray and as a blue ring, respectively (A and B), and the actin-myosin ring is represented as a hatched gray ring (A). Nucleus, Nuc.; vacuole, Vac.
Chitin synthase.
The reaction catalyzed by chitin synthase uses UDP-GlcNAc as the sugar donor. The synthesis of UDP-GlcNAc occurs in the cytosol (210), whereas chitin is found extracellularly. Analysis of chitin synthesis in spheroplasts or by the enrichment of chitin synthase in membranes indicates that, in addition to catalyzing chitin polymerization per se, chitin synthase has the capacity to extrude the polymer extracellularly through the plasma membrane (210). Three chitin synthase activities have been identified in S. cerevisiae membrane extracts and distinguished by their in vitro biochemical properties (Table 1): chitin synthase I (CSI) has a cryptic activity that can be detected by trypsin activation and incubation in the presence of Mg2+ at an optimum pH of 6.5, CSII activity is detected after proteolytic activation in the presence of Co2+ at an optimum pH of 8, and CSIII is active at pH 8 in the presence of Co2+ without proteolytic activation (for a review, see reference 29). Although CSI and CSII activities can be detected in vitro upon proteolytic treatment, it is unclear whether these proteins are synthesized as precursors and whether and how they are converted from zymogens to active forms in vivo. The genes encoding the catalytic subunits of CSI, CSII, and CSIII are CHS1, CHS2, and CHS3, respectively, and encode 1,131-, 963-, and 1,165-residue integral membrane proteins with six or seven putative transmembrane domains (28, 260, 289).
A C-terminally tagged Chs2 can be detected throughout the secretory pathway in cells that are unbudded or have a small bud, while it is found at the bud neck in large-budded cells (Fig. 2A), indicating that Chs2 localization is regulated during the cell cycle. Analyses of α-factor-synchronized cells show that the levels of CHS2 mRNA and Chs2 fluctuate with the cell cycle, peaking during M phase and at the end of mitosis, respectively, while pulse-chase analysis indicates that Chs2 is synthesized only at the end of mitosis (50, 215, 267). Chs2 levels are stabilized by sec1-1, sla2/end4-1, and pep4Δ mutations, affecting Golgi-plasma membrane transport, endocytosis, and protein degradation in the vacuole, respectively. Collectively, these results indicate that Chs2 is transported through the secretory pathway to the plasma membrane at the bud neck, internalized by endocytosis, and then targeted to the vacuole for degradation (50) (Fig. 2A). CHS2 is essential in many strain backgrounds, and chs2 mutants show major septum disorganization, indicating that Chs2 makes the chitin in the primary septum (258, 261). In addition to Chs2, the process of septation requires an actin-myosin ring, which provides the contractile force required for cytokinesis and whose function is intimately linked to that of Chs2 (251). The localization of both Chs2 and the actin-myosin ring requires the assembly of 10-nm septin filaments consisting of five conserved GTP-binding domain-containing proteins (the CDC3, -10, -11, and -12 and SHS1/SEP7 gene products) (see references 77, 160, and 294 for reviews on septin function and assembly). Septin filaments organize as a ring that forms prior to bud formation at the incipient bud site and acts as a scaffold recruiting proteins to the bud neck, including actin-myosin ring components, such as Myo1 and filamentous actin (157; see references 77, 160, and 294 for reviews). At the start of cytokinesis, the septin ring splits into two rings which act to restrict Chs2 localization, thus guiding septum synthesis between the split rings (74, 240). This process is illustrated in Fig. 2A. In septin mutants, localization of Chs2 and other components of the septation apparatus is perturbed, leading to ectopic septum synthesis (240).
Exit of Chs3 from the ER and its further transport to the Golgi requires Chs7, which prevents Chs3 aggregation (140, 284). Examination of Chs3 localization by indirect immunofluorescence reveals intracellular and punctuate labeling in most cells, with a ring structure at the bud neck of small-budded cells (50) (see Fig. 2B for a cartoon of Chs3 trafficking dynamics). Though the levels of CHS3 mRNA fluctuate during the cell cycle, with a peak in G1 (215), the steady-state levels of Chs3 are stable, with a half-life of 90 min, and are unaffected by deletion of PEP4, indicating that Chs3 is not degraded in the vacuole (50). Rather, a large fraction of the protein accumulates in intracellular vesicles, loosely termed chitosomes, with a minor fraction found at the plasma membrane (50, 315). The intracellular transit of Chs3 has been elucidated using mutants with secretory defects. A sec1-1 mutant accumulates Chs3 in Golgi-derived vesicles (50), whereas a chs5Δ mutant accumulates Chs3 in an inactive form in Golgi-derived vesicles (47, 247). In a chs6 null mutant, Chs3 accumulates in chitosomes and does not reach the plasma membrane (316). A mutation in END3 or SLA2 decreases the amount of Chs3 in purified chitosomes (315, 316). The interaction between Chs3 and Skt5 at the plasma membrane is required for CSIII activity (65, 209, 283). Finally, the appearance and activity of Chs3 at the bud neck depend on the association of the Chs3 activator Skt5 with Bni4, a protein recruited by the Cdc10 septin (65). This data set is consistent with Chs3 being transported through the secretory pathway, stored in chitosomes, and then mobilized to the plasma membrane in a Chs6- and septin-dependent process where it is fully activated upon interaction with Skt5 (Fig. 2B).
A chs3 mutant lacks chitin at the incipient bud site and in the lateral wall, with chitin detectable only at the septum (258). This indicates that Chs3 is responsible both for the synthesis of the chitin ring at the bud base and for lateral wall chitin. The chitin ring is thought to provide stretching resistance at the bud neck, thereby ensuring cell integrity during bud expansion (Fig. 2A and B [note the presence of the ring]). A chs2 chs3 double mutant is inviable, indicating that these two chitin synthases have partially redundant function. Indeed, in a chs2 mutant, Chs3 synthesizes a “remedial septum,” which is disorganized and much thicker than the wild-type primary septum (258). Under conditions of cell wall stress, such as defective glucan synthesis or exposure to cell surface-perturbing agents, the levels of chitin in the cell wall increase dramatically in a Chs3-dependent manner (211, 221). Thus, Chs3 is a component of the normal cell wall synthesis machinery that can be stress activated (Fig. 2B). In addition, CHS3 mRNA is induced during sporulation, and the spore walls of homozygous chs3/chs3 null diploids lack chitosan (46, 215). Thus, the role of Chs3 during sporulation is to make the chitin which is deacetylated to chitosan during ascospore maturation (see below). Furthermore, CSIII activity can be stimulated by the sporulation-specific Skt5 homologue Shc1p, indicating a dedicated Chs3 activation pathway in ascospore synthesis (248).
The third CHS gene, CHS1, shows an expression pattern similar to that of CHS3, peaking in early G1, while the Chs1 level remains constant during the cell cycle (215, 315). Chs1 shows the same dual subcellular localization as Chs3, with most of the protein found in chitosomes and a minor fraction found at the plasma membrane (147, 315). Since Chs1 and Chs3 both populate chitosome vesicles, they are thought to follow the same route. However, the localization of Chs1 is independent of Chs6 (316), hinting that Chs1 may require specific trafficking factors.
A chs1 null mutant shows a cell wall defect at the birth scar, which can lead to daughter cell lysis (37). This phenotype is dependent on the chitinase-encoding gene CTS1 and is almost completely suppressed in a chs1 cts1 double mutant (38). These data led to a model in which Chs1 repairs the chitinase-mediated septum degradation that occurs during cell separation. CHS1 mRNA levels are induced by extracellular calcium in a Crz1-dependent process (309) and by mating pheromones (9, 46, 236). The CHS1 promoter contains pheromone-responsive elements. CSI is the only chitin synthase activity detectable in vitro from extracts of cells in stationary phase (29), but the biological significance of this finding remains unclear.
In addition to fungal cell walls, chitin is found in nematode eggshells and in insect and crustacean exoskeletons, and to date, the SwissProt (http://au.expasy.org/sprot/) and BRENDA (http://www.brenda.uni-koeln.de/) databases contain more than 50 sequences of chitin synthase genes. Since chitin is critical for development and morphogenesis, chitin synthesis inhibitors and chitinolytic enzymes have been developed as fungicides and insecticides.
Using Chs2 as a model, Nagahashi and colleagues mapped the catalytic domain of this chitin synthase, which contains three motifs (QXXEY, EDRXL, and QXRRW) that are found in all chitin synthases and believed to be necessary for the transfer of GlcNAc to the growing chitin chain (198). An alignment of amino acid sequences of 48 fungal chitin synthases extends the consensus sequence (QXXEYXnLPG/A, LA/GEDRXL, QR/GRRWL/IN) and reveals that this signature domain is included in a ∼250-amino-acid region with conserved predicted secondary structure (244). A second region located C terminal to a putative transmembrane domain is thought to play a role in chitin extrusion across the plasma membrane (306). Two families occur, based on the position of the chitin synthase signature domain, with family 1 and 2 proteins having their motifs in their middle or in their C-terminal half, respectively (244). S. cerevisiae Chs1 and Chs2 belong to family 1 and Chs3 to family 2. Chs1 shares 58% and 28% identity with Chs2 and Chs3, respectively, and Chs2 shares 32% identity with Chs3 over the signature domain. In addition to the position of the chitin synthase signature domain, the two families are distinguished by the presence of specific motifs. These chitin synthase families can be further subdivided in five classes (23, 181, 266), with families 1 and 2 containing three and two distinct classes, respectively.
Classes I and II, originating from family 1, are found in yeasts and filamentous fungi. Chs1 and Chs2 are found in classes I and II, respectively. The third family 1 class, class III, is found only in filamentous fungi. Family 2 enzymes of class IV are found in yeasts and filamentous fungi, whereas class V Chs proteins are found in filamentous fungi only. Class IV contains Chs3 and Chs3-like proteins. Interestingly, a number of class IV members, including Chs3, bear a C-terminal region whose deletion leads to a dramatic decrease in chitin synthase activity (58). This hydrophilic region, which is absent from Chs1- or Chs2-like proteins, may constitute a regulatory domain. Chs3 can self-interact in a two-hybrid test, showing that it can dimerize (65). Whether the protein functions as a dimer and whether other Chs proteins also dimerize remain to be demonstrated. Class V members are characterized by the presence of a large myosin motor-like domain which has a presumed role in the actin-based localization of these proteins to sites of active chitin synthesis during the cell wall growth of hyphal filaments.
Chitin deacetylases.
Two sporulation-specific S. cerevisiae genes, CDA1 and CDA2, confer chitin deacetylase activity when ectopically expressed during vegetative growth and together are required for chitosan formation during sporulation (Table 1) (48, 185). An homozygous cda1/cda1 cda2/cda2 double mutant shows increased spore lysis upon treatment with the lytic enzyme preparation Zymolyase or Glusulase, indicating the protective role of the chitosan layer (48, 185).
Mannoproteins
Linked to the cell wall polysaccharides are a varied set of mannoproteins that collectively form yeast mannan, the electron-dense, fibrillar outer layer of the wall (212). Carbohydrate chemistry and genetic studies pioneered by Ballou have elucidated the structure of the polysaccharides on S. cerevisiae mannoproteins (reviewed in reference 14), a structure that can vary considerably among yeast species (93). Below we briefly describe protein mannosylation, followed by a review of studies on the groups of cell wall mannoproteins.
Protein glycosylation and mannan synthesis.
Two types of oligosaccharidic protein modifications have been described for S. cerevisiae. N-glycosylated proteins receive an oligosaccharide through an N-glycosidic bond between a GlcNAc and an asparagine residue, while O-mannoslyated proteins receive short mannose chains onto the hydroxyl side chains of serine or threonine residues through an α-mannosyl bond (255). These processes have been extensively reviewed (60, 66, 270) and are covered in outline here.
(i) Protein N glycosylation.
All N-modified glycoproteins acquire the same initial oligosaccharide in the ER, and subsequently most intracellular glycoproteins are additionally modified, with the final chains, containing 9 to 13 mannose residues, being referred to as “core”-type oligosaccharides (60). In contrast, cell wall and secreted mannoproteins are extensively mannosylated in the Golgi, with a final structure of α-1,6-linked mannose chain of up to 50 mannose residues extending from the N-glycan core and to which are attached shorter chains of α-1,2 residues terminating in α-1,3-linked mannose residues, forming a highly branched structure containing as many as 200 mannose residues (14; see reference 60 for a review).
As a first step, the oligosaccharyl transferase complex transfers a Glc3Man9GlcNAc2 structure from a dolichyl-phosphate-linked precursor (see references 31 and 210 for reviews on precursor synthesis) onto an asparagine residue of N-X-S/T sequons, where X represents any amino acid residue except proline (reviewed in reference 66). Oligosaccharides are then trimmed in the ER by the action of glucosidases I and II and mannosidase I, encoded by CWH41, ROT2, and MNS1, respectively (see reference 109 for a review), leading to a Man8GlcNAc2 structure. This processing is part of a protein-folding quality control process that expedites protein export to the Golgi apparatus (see reference 79 for a review). Trimmed oligosaccharides are in some cases extended by the action of Golgi mannosyltransferases (Table 1).
Upon entry into the cis-Golgi of N-glycosylated proteins that are destined to be further elaborated, the Och1 α-1,6-mannosyltransferase catalyzes the transfer of a mannose residue to the Man8GlcNAc2 structure (199, 200). Glycoproteins produced by a null och1 mutant lack α-1,6-polymannose, indicating that the addition of this initial mannose residue is critical for other reactions to take place and/or that Och1 is involved in the catalysis of subsequent α-1,6 linkages (199). OCH1 is important for cell wall function, as an och1 null mutant shows a slow- and temperature-sensitive-growth phenotype that can be rescued by sorbitol addition (148, 200).
Subsequently, an α-1,6-mannose backbone of up to 50 residues is synthesized by the action of two cis-Golgi mannose polymerase (ManPol) complexes (124, 125). A striking feature of these complexes is that all known proteins resemble mannosyltransferases, with ManPol I containing Van1 and Mnn9 and ManPol II containing Mnn10, Mnn11, Anp1, Hoc1, and Mnn9. Assays of these complexes indicate that both have α-1,6-mannosyltransferase activity that can be compromised when some individual genes are deleted (124, 125). Components of the ManPol I complex exhibit differential in vitro activities, with Van1 being an α-1,6-mannosyltransferase and Mnn9 showing dual α-1,2- and α-1,6-mannosyltransferase activity (269). The α-1,2-mannosyltransferase activity conferred by Mnn9 may also play a role in the “core”-type modification of intracellular glycoproteins (269).
The α-1,6-mannose backbone shows α-1,2 branched chains, added by the α-1,2-mannosyltransferase Mnn2. Branches are then extended by α-1,2-linked mannose residues by Mnn5, a close homologue of Mnn2 (234). Five additional related proteins also participate in outer-chain extension (107, 110): Kre2/Mnt1, an α-1,2-mannosyltransferase whose structure has recently been determined and mechanism of catalytic action proposed (159); Ktr1, a demonstrated α-1,2-mannosyltransferases (241); and Ktr2, Ktr3, and Yur1, which have been implicated as mannosyltransferases in vitro and in vivo (163). The outer chain can be further branched by the action of the mannosylphosphate transferase Ktr6/Mnn6, another Kre2-related protein (300). Together with the uncharacterized Ktr4, Ktr5, and Ktr7, the Kre2-related proteins form the KTR family (162). Whereas Ktr2, Ktr6, and Yur1 affect protein N glycosyl-ation, Kre2, Ktr1, and Ktr3 are involved in both protein N and O glycosylation (see below).
Finally, outer chains are terminated in the late Golgi by addition of an α-1,3-linked mannose by Mnn1. MNN1 with five additional genes forms a family consisting of two subfamilies (162). The first of these contains the α-1,2-mannosyltransferases Mnn2 and Mnn5, which are involved in protein N glycosylation and mentioned above. The second subfamily contains Mnn1, which are involved in both N and O glycosylation (see below), and Mnt2, Mnt3 and Mnt4, which are involved in protein O mannosylation (see below).
(ii) Protein O mannosylation.
O-linked oligosaccharides consist of short chains of up to five mannose units, with the first two residues being α-1,2 linked and subsequent ones α-1,3 linked. Despite the small size of the O-linked chains, many cell wall proteins have serine/threonine-rich domains, and so the number of O chains per protein can be high and the amount of O-linked mannose in the cell wall significant (reviewed in reference 270).
The initial step of O mannosylation occurs in the ER, where a single mannose residue is transferred from a dolichyl-phosphate-mannose precursor (see references 31 and 210 for reviews of precursor synthesis) onto serine/threonine residues by a family of seven protein-O-mannosyltransferases, the PMT family (see reference 270 for a review). Subsequent steps occur in the Golgi and are catalyzed by mannosyltransferases using GDP-mannose as the sugar donor (Table 1). Three KTR family members, Kre2/Mnt1, Ktr1, and Ktr3, have been shown to be involved in catalyzing the two α-1,2 links (see reference 162 for a review). The two terminal α-1,3 links are catalyzed by the MNN1 family members Mnn1, Mnt2, and Mnt3 (100, 242, 308).
As discussed below, mannoproteins can be linked in numerous ways with other cell wall polymers: in some cases, the polymannose structure is thought to form a direct link with β-1,3-glucan chains, while some proteins are linked through a remnant of their GPI anchor and others appear to be directly linked to β-1,3-glucan through their protein moiety.
GPI-cell wall proteins.
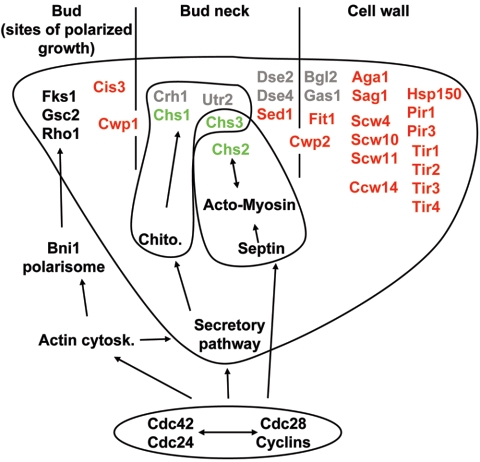
Cell wall digestion with laminarinase releases proteins that bind to anti-β-1,6-glucan antibodies (130). Nuclear magnetic resonance analysis revealed that the protein-β-1,6-glucan bond involves an ethanolamine-PO4-oligomannoside composed of four or five mannose residues, three of which are decorated with ethanolamine phosphate (83, 86, 117, 138). This structure is a GPI anchor remnant, indicating that GPI-modified proteins form a class of cell wall proteins (Fig. 1A). Acquisition of the GPI anchor occurs in the ER, where a preassembled anchor is transferred to the C terminus of target proteins (see reference 210 for a review). The GPI anchor is usually attached to an N/S/G/A/D/C residue found in the 40 C-terminal amino acids of the protein and in the context N/S/G/A/D/C-G/A/S/V/I/E/T/K/D/L/F-G/A/S/V-X4-19-φ10 (where φ represents a hydrophobic amino acid). The attachment reaction involves a proteolytic cleavage C terminal of the attachment residue and a transamidation between the generated amino acid and the ethanolamine phosphate of the GPI anchor (62, 210). A GPI anchorage signal can be found in ∼70 secretory proteins in the yeast genome (40, 62). Of these, about a third either are demonstrated CWPs or contain a signal sufficient to direct a fusion protein to the cell wall (105), a third are suspected to be CWPs based on sequence homology, and a third are known or putative plasma membrane proteins. So far, no cell wall-targeting signal has been described for GPI-CWPs, whereas an additional signal, usually a dibasic motif, seems to be required for the correct GPI anchoring in the plasma membrane, suggesting that the default destination for GPI-modified proteins is the cell wall (40, 62). In the ER, GPI-modified proteins are packaged in specialized COPII-coated vesicles for transport to the Golgi, where the N-and O-linked carbohydrate chains and the GPI anchor are extended and modified (272). A number of known GPI-proteins, including GPI-CWPs such as Ccw12, Ccw14, and Cwp2 have been shown to be extensively O mannosylated (187, 192, 290). Other GPI-CWPs contain serine/threonine-rich domains, which are sites of mannose attachment, suggesting that O mannosylation is a common feature of GPI-CWPs.
Based on amino acid sequence similarity, some GPI-CWPs can be grouped into families, including the flocculins, the agglutinins, and the CRH1-UTR2, the TIR, and the SED1-SPI1 families (40, 62). The flocculins, or Flo proteins, are lectin-like adhesion proteins with a role in flocculence and invasive growth (for a review, see reference 295). The critical role of the mating-type-specific agglutinins Aga1 and Sag1 in cell-cell adhesion prior to mating is well established (for a review, see reference 156). Crh1 and Utr2 are candidate cross-linking enzymes that are involved in cell wall remodeling during vegetative growth (see below). The TIR family contains 10 members which are differentially expressed depending upon growth conditions. Two TIR genes, CWP1 and CWP2, are expressed during vegetative growth. Singly, their deletion, as well as that of the TIR gene TIP1, confers hypersensitivity to calcofluor white and Congo red (290). When viewed by electron microscopy, a cwp2 null mutant has a thinner cell wall than a wild-type strain (290). CWP1 and CWP2 are repressed under anaerobiosis, whereas other TIR genes (DAN1, DAN4, TIP1, TIR1, TIR2, TIR3, and TIR4) and two additional genes encoding non-GPI but Tir-related proteins (DAN2 and DAN3) are induced and thus show differential responses to aerobic and anaerobic conditions (3). Supporting this hypothesis, tir1, tir3, and tir4 single null mutants all show growth defects when cultivated anaerobically (3). Transcription profiling and studies of sterol uptake suggest that Dan1, and maybe Dan3, Dan4, and Tir4, could play a role in sterol influx under anaerobic conditions by binding sterols in the cell wall (304). In a similar way, the Fit set of GPI-anchored cell wall mannoproteins assist in iron uptake (223). These proteins are encoded by the FIT1, FIT2, and FIT3 genes, whose expression is induced in an Aft1-dependent way upon iron deprivation. Though not directly involved in iron transport through the plasma membrane, these Fit proteins bind iron in the cell wall, and in their absence, there is reduced uptake of iron bound to the siderophores ferrioxamine B and ferrichrome. The iron bound to these cell wall proteins is thought to increase the concentration of iron at the cell surface and thus facilitate iron uptake into the cell. The binding of ergosterol and iron by cell wall proteins raises the possibility that other small-molecule nutrients may also be sequestered in this way. The fifth family includes SED1 and SPI1, two stress-responsive genes induced by glucose limitation (227, 259). These gene products have a protective role under starvation and stress conditions, with mutants being hypersensitive to oxidative stress or Zymolyase (82, 259, 262). A pair of GPI-CWPs which do not fall in these families are Ccw12 and Ccw14. Compared to a wild-type strain, ccw12 and ccw14 null mutants show hypersensitivity to calcofluor white and Congo red, and the ccw12Δ mutant has a reduced mating efficiency (192).
Collectively, the GPI-CWPs appear to have a general function in cell wall stability. The hypersensitivity to calcofluor white of a ccw12Δ mutant is partially suppressed in a ccw12 ccw14 mutant and completely rescued in a ccw12 ccw14 dan1 triple null mutant. Interestingly, the additional deletion of TIP1 or CWP1 in the ccw12 ccw14 dan1 triple mutant had no additional effect on the calcofluor white sensitivity. This suggests that a compensation process is triggered when a defect threshold is reached. The CCW12 deletion alone may not be sufficient to trigger this compensatory process, and it probably involves the two stress GPI-CWPs Sed1 and Spi1, since ccw12 ccw14 dan1 sed1 and ccw12 ccw14 dan1 spi1 quadruple null mutants both show calcofluor white hypersensitivity (104). In another study, a ccw12 ccw14 cwp1 dan1 tip1 quintuple mutant showed an elevated mortality in stationary phase (276).
The PIR family and mild-alkali-extractable cell wall proteins. (i) The PIR family.
Treatment of biotinylated cell wall extracts with mild alkali results in the release of six proteins (194), with the four most abundant being Pir1/Ccw6, Hsp150/Pir2/Ccw7, Pir3/Ccw8, and Cis3/Pir4/Ccw5. These Pir proteins (for proteins with internal repeats) are structurally related, are serine/threonine rich, and are thought to be extensively O mannosylated (94, 194). They consist of a putative N-terminal signal peptide, followed by a Kex2 endoprotease cleavage site, an internal repeat region, and a C-terminal cysteine-based motif (194, 279). In a kex2 mutant, a shift in electrophoretic mobility of the Pir proteins was observed. However, their relative abundance was unchanged compared to that in the wild type. Thus, the Kex2 cleavage site is used in vivo but is not required for cell wall localization, leaving a role for this processing event open (194).
Pir1, Hsp150, and Pir3 can be detected with an anti-Pir3 antibody and visualized in the lateral wall of wild-type cells by immunoelectron microscopy (310). In contrast, a specific anti-Cis3 antibody localized Cis3 exclusively in the growing buds of vegetative cells (188). The nature of the links between the Pir proteins and the cell wall is unclear (Fig. 1A). The presence of Pir proteins linked to β-1,3-glucan that are releasable by mild alkali treatment suggests that a glycosidic bond links the β-1,3-glucan and a mannose residue of the O-linked chains of the Pir proteins (136). The internal repeats are probably involved in connecting Pir proteins to β-1,3-glucan, as suggested by a CIS3 mutational analysis (43). To date, neither the identity of nor the enzyme catalyzing the Pir protein-β-1,3-glucan link are known. Hsp150 and Cis3 were also found among the proteins released upon treatment of the cell wall with β-mercaptoethanol or dithiothreitol, showing that they are linked to other CWPs through disulfide bonds (Fig. 1A), probably involving the cysteine-rich C terminus (39, 43, 189).
The PIR genes show cell-cycle-dependent expression, with levels of the CIS3 transcript peaking in G2 and those of the PIR1, HSP150, and PIR3 transcripts peaking in early G1 (267). The levels of the four PIR mRNAs are elevated in a constitutively active mkk1 mutant in an Rlm1-dependent manner, indicating that their expression is under PKC cell integrity pathway control (123). This is consistent with the finding that these genes belong to a group of cell wall perturbation-responsive genes, with their expression being stimulated by exposure to calcofluor white or Zymolyase (22). Furthermore, PIR1, HSP150, and PIR3 are induced by high temperature (310). Apart from a hypersensitivity phenotype to aluminum-mediated oxidative stress seen in an hsp150Δ mutant (82), no effects on growth rate, cell morphology, or viability were found in mutants singly deleted for any of the PIR genes (195, 279). However, a pir1 hsp150 double null mutant shows reduced growth on solid medium and hypersensitivity to heat shock (279). A cis3 pir1 hsp150 triple null mutant shows hypersensitivity to calcofluor white and Congo red (195), and a pir1 hsp150 pir3 triple null mutant is hypersensitive to tobacco osmotin and shows an elevated mortality phenotype during stationary phase (204, 276, 310). Finally, a quadruple pir mutant shows a sorbitol-suppressible increased mortality both during exponential growth and in stationary phase (276). These data suggest a protective role for the Pir proteins, for example, as barrier proteins influencing cell wall permeability.
(ii) Other cell wall proteins with alkali-sensitive links.
The proteins released from isolated cell walls treated with sodium dodecyl sulfate and then HF or NaOH were identified using mass spectrometry (307). The set of proteins released by HF treatment, which cleaves phosphodiester bonds, contains known GPI-CWPs such as Ccw14 and Cwp1 as well as Pry3, a protein whose GPI anchor signal can target a fusion protein to the cell wall (105). In addition, other GPI-modified proteins such as GAS family members, the glucosidases Crh1 and Utr2 (see below), and the phospholipase Plb2 were also found. In addition to the Pir proteins, the set of NaOH-released proteins includes Cwp1, the putative glucanases Scw4 and Scw10, and the agglutinin-related Tos1. This work suggests that a GPI-CWP can be linked to the cell wall through either its GPI remnant or its oligosaccharide chains. The finding of several glucanases covalently linked to the cell wall implies that they function in remodeling the glucan matrix.
Roles of cell wall mannoproteins.
O-linked mannose chains are short and rigid, and by cross-linking proteins to the β-1,3-glucan, they may play a role in cell wall strengthening. In addition, mannoproteins usually show extensive N-linked glycosylation. The roles of fungal covalently linked cell wall proteins in processes such as water retention, adhesion, and virulence have been reviewed (63). Many of these functions require proper glycosylation of the respective proteins. Mutations of the synthetic glycosylation and mannosylation pathways collectively affect all mannoproteins and cause multiple cell wall phenotypes, such as compound or stress hypersensitivity and altered cell wall composition (63, 210). However, some steps in protein glycosylation are catalyzed by gene families whose members' substrate specificity can directly affect cell wall assembly. For example, the PMT family members Pmt4p and the Pmt1-Pmt2 pair act in vitro on Gas1 and Pir2, respectively (270). In addition, high redundancy occurs within families of Golgi mannosyltransferases, with most single mutants showing weak or no phenotypes. However, the impact of these gene families on cell wall synthesis is seen by the severe cell wall phenotypes found in triple or quadruple mutants (8, 162). The proliferation of genes and gene products with overlapping function in Golgi N and/or O mannosylation suggests that such gene duplications have conferred fitness and have been selected for during fungal evolution.
Cell Wall-Remodeling Enzymes
Glucan-remodeling enzymes.
The product synthesized in vitro by the minimal β-1,3-GS is linear (73). However, the branched structure of mature β-1,3-glucan necessitates enzymes responsible for branching and/or processing of these linear chains (Table 1). Subsequently, cell wall assembly requires the linkage of β-1,3-glucan to other cell wall polymers (Table 1).
(i) GAS1 and the GAS gene family.
The following observations implicate GAS1 in cell wall synthesis: (i) a mutation in GAS1/CWH52 confers hypersensitivity to calcofluor white and to caspofungin (150, 233), (ii) a gas1 mutant shows decreased β-1,3-glucan levels and increased mannan and chitin contents in its cell wall (231), and (iii) a gas1 mutant secretes high levels of β-1,3-glucan (232). The last observation implies that the β-1,3-glucan chains made in a gas1 mutant are abnormally assembled. An in vitro assay for transglucosidase activity using recombinant enzyme shows that Gas1 can catalyze the formation of a β-1,3-glycosidic bond between the reducing end of a donor β-1,3-glucan chain and the nonreducing end of a acceptor β-1,3-glucan molecule (42, 190). These data point toward Gas1 acting in vivo as a β-1,3-glucan elongase.
Gas1 is synthesized as a 559-residue precursor, with a cleavable N-terminal signal peptide. During its transit through the ER, Gas1 is N and O glycosylated and receives a C-terminal GPI anchor (92, 207, 208, 287). The ER-modified Gas1, likely along with other GPI-modified proteins, is packaged into specialized COPII secretory vesicles and transported from the ER to the Golgi, where the N- and O-linked glycans are further elaborated (196, 272). Mature Gas1 is a 115-kDa protein, anchored in the outer leaflet of the plasma membrane (for a review, see reference 222). This localization is consistent with the presumed role of Gas1 in cell wall remodeling. The Gas1 GPI anchor is required for function, since expression of a tailless Gas1 fails to complement a gas1Δ mutant (222). Gas1 was recently identified in cell walls after sodium dodecyl sulfate treatment, suggesting that a fraction of Gas1 is covalently attached to the cell wall (307). Transcription of GAS1 during the cell cycle is SBF dependent, peaks at G1/S as does FKS1 (115), and is repressed during sporulation (222).
The S. cerevisiae genome contains four additional GAS1-related genes, GAS2, -3, -4, and -5, whose products are 44, 33, 37, and 32% identical to Gas1, respectively. The GAS gene products belong to glycoside hydrolase family 72, which includes 70 plant and fungal enzymes with β-1,3-glucanosyltransglycosylase activity (http://afmb.cnrs-mrs.fr/CAZY/). Roles for these other GAS family members in S. cerevisiae cell wall synthesis remain unknown.
(ii) Glucanases and glucanase-related proteins.
Remodeling of the glucan network presumably requires both the shortening of the polymer by nicks and the introduction of polymer branch points. Such reactions are catalyzed by exo- and endoglucanase activities (4).
Three gene products, Exg1, Exg2, and Spr1, have exo-β-1,3-glucanase activity in vitro (for a review, see reference 146). The abundant extracellular N-glycoprotein Exg1 is released with other soluble cell wall proteins upon treatment of intact cells with dithiothreitol (39). EXG1 mRNA levels are elevated in cell wall mutants, such as those with fks1, kre6, or mnn9 null alleles, implicating Exg1 in a compensatory process triggered by cell wall damage (145). Exg2 is highly N glycosylated and membrane bound. The truncation of a 69-aa C-terminal domain which encompasses a putative site for GPI modification (40, 62) results in the secretion of Exg2 to the culture medium (146). Exg1 and Exg2 also act in vitro on pustulan, a β-1,6-glucan polymer, indicating that they may play a role in β-1,3- and/or β-1,6-glucan remodeling and explaining the finding that deletion and overexpression of EXG1 lead to increased and reduced in vivo β-1,6-glucan levels, respectively (122). SPR1/SSG1 mRNA has been detected only in sporulating diploids, and a homozygous spr1/spr1 null diploid shows retarded sporulation, indicating that Spr1 functions specifically in spore wall assembly (197).
Analysis of proteins released by intact cells treated with dithiothreitol identified seven “soluble” cell wall proteins (39), including Exg1, Bgl2, and a set of three proteins (Scw3, Scw4, and Scw10) with sequence similarity to glucanases. BGL2 encodes an abundant extracellular protein with an in vitro endo-β-1,3-glucanase activity (135, 193). Interestingly, at high substrate concentrations Bgl2 can also catalyze the formation of a β-1,6 bond linking two β-1,3 chains together (98). Since in the cell wall, the local polymer concentration is very high, these results imply that Bgl2 acts in vivo as a branching enzyme. In agreement with this hypothesis, a bgl2Δ mutant, while having wild-type glucan levels, shows increased chitin levels (126). Such a stress response may result from cell wall disorganization caused by inefficient glucan branching. A phenotypic analysis revealed that a scw4Δ scw10Δ double mutant exhibits hypersensitivity to calcofluor white (39). The yeast genome contains an additional glucanase-related gene, SCW11, with homology to SCW4 and SCW10 and whose deletion leads to cell separation defects (39). So far, no enzymatic activity has been associated with the Scw proteins.
Among other genes with sequence similarity to β-glucanase genes, DSE2 (encoding a putative GPI-anchored cell surface protein) (62) and DSE4/ENG1 (encoding a secreted and glycosylated endo-β-1,3-glucanase) (13) are believed to be involved in septum degradation. This is supported by (i) the cell separation defects observed in dse2 and dse4 mutants, (ii) the localization of Dse2 and Dse4 at the mother-daughter interface, and (iii) the levels of DSE2 and DSE4 mRNA peaking at the M/G1 transition (13, 52, 75). EGT2 was identified as an early G1 gene required for cell separation, and based on cell separation defects observed in an egt2 null mutant, it was suggested that EGT2 either encodes or regulates a glucanase function (141). However, a search for Egt2 glucanase activity was unsuccessful (13), leaving its role in cell separation open.
(iii) Fungal remodeling enzymes.
As mentioned above, remodeling enzymes form large families of glycoside hydrolases. In some cases, such as the A. fumigatus and Candida orthologues of Gas1, enzymatic characteristics indicate a conservation of catalytic activity (42, 191). In addition to this, mutants with mutations in GAS-related genes, such as C. albicans PHR1 or PHR2, Candida glabrata GAS genes, Candida maltosa EPD genes, or A. fumigatus GEL genes, show an abnormal cell shape and morphogenesis and defects in hyphal growth (190, 201, 202, 302). A number of related glucanases are found in other fungi (for a review, see reference 4). A recent proteomics study showed that a number of putative C. albicans glucanases are covalently linked to the cell wall, suggesting that they function in wall remodeling (61).
Chitinases.
A screen for elevated glycosidase activity uncovered CTS1, a gene encoding a secreted endochitinase (143, 144). The CTS1 gene product has a signal sequence, a region with similarity to plant chitinases that is the likely catalytic domain; a serine/threonine-rich domain, which is a site for O mannosylation; and a C-terminal chitin-binding domain. Cts1 is processed in the secretory pathway, and the mature protein is secreted and binds to chitin in the cell walls of growing cells from where chitinase activity can be recovered (55, 144). In exponentially growing cells, transcription of CTS1 peaks in early G1 and is dependent on Ace2, a transcription factor which controls the onset of a daughter-specific expression program. Consistent with this, a yellow fluorescent protein-tagged version of Cts1 is detected exclusively at the daughter side of the bud neck (52). Phenotypic analysis indicates that Cts1 has a role in cell separation, since a cts1Δ mutant grows as multicellular aggregates in which cells are held together by their septa. This growth pattern is also observed when wild-type cells are treated with the chitinase inhibitor dimethylallosamidin (144). As mentioned above, deletion of CTS1 suppresses the lysis phenotype of a chs1 mutant (38). Together, these data implicate Cts1 in septum degradation and suggest the existence of a balance between Cts1 and Chs1 activities. However, the chitinase activity of a chs1Δ mutant and the CSI activity of a cts1Δ mutant are close to wild-type levels, indicating that chitinase and CSI activities are not reciprocally regulated (254).
The product of a second yeast gene, CTS2, has sequence similarity to chitinases, and the gene product may be sporulation specific (http://www.yeastgenome.org).
Yeasts and filamentous fungi possess families of chitinase genes, an area recently reviewed (4). Fungal chitinases have been proposed to fall in two categories. Enzymes from the first group share structural similarities with plant chitinases and are putatively cell wall associated, suggesting a role as wall-remodeling enzymes. A second group of chitinases are more related to the bacterial chitinases involved in the degradation of exogenous chitin.
Glucan/chitin-cross-linking enzymes.
The products of the related genes CRH1 and UTR2/CRH2 bear a motif found in glycoside hydrolase family 16 (http://afmb.cnrs-mrs.fr/CAZY/). This family includes a number of prokaryotic, plant, and fungal enzymes, some of which are endo-β-1,3- or endo-β-1,3-1,4-glucanases and endo-xyloglucan transferases. Though no enzymatic activity has been assigned to Crh1 or Utr2, several lines of evidence support their candidacy as transglycosidases which cross-link β-glucan and chitin.
First, the cellular localization of these proteins is compatible with their having a role as cell wall-remodeling enzymes. Crh1 and Utr2 C termini contain a motif sufficient for the phospholipase C-sensitive localization of α-galactosidase to the cell wall, suggesting that Crh1 and Utr2 are GPI-anchored cell wall proteins (105). When fused to GFP, both proteins localized to the sites of polarized growth in exponentially growing cells (237). The localization pattern of Crh1 and Utr2 resembles that of chitin synthase III (Chs3), and the localization of Utr2 requires Chs5 and Bni4, two proteins that are also involved in the bud neck localization of Chs3 (238). This raises the interesting possibility that Utr2 is transported through chitosomes, a set of specialized secretory vesicles involved in chitin synthesis (see below).
Second, crh1 and utr2 mutants are hypersensitive to calcofluor white and Congo red. Furthermore, the deletion of CRH1, alone or in combination with UTR2, does not change cell wall composition but rather alters the proportion of alkali-soluble versus -insoluble β-glucan. These data show that a lack of Crh1 and/or Utr2 leads to deficient cross-linking of cell wall components which results in cell wall weakening (237). As the alkali insolubility of β-glucan is due to its cross-links to chitin, those authors postulate that Crh1 and Utr2 act as glucan/chitin-cross-linking enzymes. During vegetative growth, CRH1 shows cell cycle-regulated expression peaking at early G1 and at the M/G1 transition, whereas UTR2 is constitutively expressed, implying that Crh1 and Utr2 have specialized functions during cell growth (237).
An additional glycoside hydrolase family 16 member, CRR1, is related to CRH1 and UTR2, with Crr1 detected in the ascospore wall (99). Examination of the crr1Δ spore wall by transmission electron microscopy reveals that the layers of glucan, chitosan, and dityrosine are irregularly deposited, suggesting that Crr1 functions in spore wall construction to cross-link glucan to chitosan (99). CRR1 is strongly induced during sporulation and is expressed at very low levels in exponentially growing cells, with a peak at the G2/M transition (237). Further, the levels of UTR2 mRNA show a twofold increase in exponentially growing crr1Δ cells relative to the wild type, hinting at an interplay between these two genes and a role for CRR1 during vegetative growth.
CELL WALL BIOLOGY AS AN INTEGRATED PROCESS
During vegetative growth, cell shape and volume changes are accommodated by differential cell wall deposition in the mother and bud. S. cerevisiae also has a repertoire of specialized cell types, such as mating projections, pseudohyphae, and meiotic spores, which develop under specific conditions. The function of these specialized cells is intimately linked with their morphology. Mutants incapable of forming mating projections are, for example, unable to fuse with their partners and fail to mate. Though filamentation ability has been lost in many laboratory strains, in the wild, pseudohyphae are critical to deal with a nutrient-depleted or harmful environment. The morphological characteristics of these vegetative and specialized cells depend upon orderly cell wall deposition. Thus, regulation of cell wall synthesis is part of cellular growth programs and involves timing through coordination with the cell cycle and through spatial cues that involve coordination with the cell polarity machinery and the secretory pathway. In addition, the cell wall transduces external status information to the cell and thus controls intracellular processes through a feedback loop. An overview of regulatory pathways involving components of the cell wall machinery is described below.
Regulation Occurs at Multiple Levels
Cell cycle.
The cyclin-dependent protein kinase (CDK) Cdc28/Cdk1 is the key regulator of the vegetative S. cerevisiae cell cycle (for a review, see reference 182). CDK activity requires physical association of Cdc28 with cyclins. Sets of cyclins can bind to Cdc28. Three G1 cyclins (the Cln1-Cln2 pair and Cln3) regulate Cdc28 activity during the G1 phase, with Cln3 regulating the expression and activity of the other G1 cyclins and Cln1-Cln2 playing a role at the G1-S transition. Subsequently, three pairs of B-type cyclins (Clb1-Clb2, Clb3-Clb4, and Clb5-Clb6) regulate G2-M events, with Clb5-Clb6 initiating the S phase, Clb3-Clb4 playing a role in mitotic spindle assembly, and Clb1-Clb2 promoting entry into mitosis. Cyclin-activated Cdc28 selectively phosphorylates various substrates, including other cyclins and transcription factors, resulting in cell cycle-dependent cyclin activity and gene expression.
The transcription of many cell wall genes fluctuates during the cell cycle (Fig. 3A), with a subset of transcripts peaking in G1 when isotropic cell wall synthesis allows daughter cell expansion (267). The SBF transcription factor, composed of the DNA-binding protein Swi4 and the regulatory subunit Swi6, is responsible for G1 expression of many of these genes, including CRH1, FKS1, and GAS1 (115, 121). Specific induction of SBF-mediated transcription in G1 follows from its release from inhibition. During the cell cycle, SBF is maintained in an inactive form by the action of Whi5. In G1, Whi5p is inactivated through phosphorylation by the Cln3-bound Cdc28 (for a review, see reference 305), and in addition, Cln-activated Cdc28 may directly regulate SBF (305). A second level of regulation is through the action of Slt2, the MAP kinase of the PKC cell integrity pathway (see below). Slt2-dependent phosphorylation of Swi6, resulting in Swi6 nuclear exclusion, may provide an additional downregulation mechanism for SBF-dependent genes outside G1 (152, 166).
FIG. 3.
Regulation of cell wall gene expression during the cell cycle (A) and in response to external stimuli (B). Boxes contain genes whose expression is controlled by the same pathway or transcription factor. Genes are color coded: black, glucan synthesis; green, chitin synthesis; red, mannoprotein biosynthesis; gray, cell wall remodeling.
A set of genes transcribed in early G1 (CTS1, DSE2, DSE4, EGT2, and SCW11) (Fig. 3A) is expressed only in daughter cells (52). This transcription pattern is consistent with the proposed role of these gene products in cytokinesis and cell separation. Their expression depends on Ace2, a transcription factor whose activity and localization are restricted to the daughter nucleus by a signaling network named RAM (for regulation of Ace2 and cell morphogenesis) (see below), which centers on the serine/threonine protein kinase Cbk1 (52, 205, 303) and which is involved in cell polarized growth, bud site selection, and mating projection morphology (19, 229).
Construction of the primary septum is also tightly cell cycle regulated, with the specialized chitin synthase Chs2 being expressed, translated, and active only at the end of mitosis and subsequently rapidly degraded. CHS2 is part of the CLB2 cluster, a gene set which is transcribed from the end of S phase until nuclear division (267). However, unlike the case for many other CLB2 cluster members, oscillation of the CHS2 mRNA level does not depend on the forkhead transcription factors Fkh1 and Fkh2 (314), leaving the basis for its cell cycle expression unclear. However, the observation that Chs2 is a substrate for Clb2-activated Cdc28 suggests a possible means of regulation of Chs2 level and/or activity during the cell cycle (285).
Cell polarity and isotropic/apical growth.
During the mitotic cell cycle, cell growth alternates from a polarized (apical) to an unpolarized (isotropic) state within the bud. The former directs bud emergence, while the latter allows bud and subsequent daughter cell expansion. Monitoring of cell polarization during the cell cycle occurs through a morphogenesis checkpoint, which delays bud formation when the actin cytoskeleton, which determines cell polarity, is perturbed (for a review, see reference 154). Organelles attach to the actin cytoskeleton, and secretory vesicles aided by motor proteins are transported along actin cables. Changes in cell polarity induce relocalization of the secretory apparatus and affect the transport of cell wall synthetic components (for reviews, see references 225 and 226).
Actin cables anchored at the bud neck and directed toward the cell interior polarize the transport of secretory vesicles. During bud expansion, additional actin cables are found anchored at the bud tip and directed toward the neck. Disruption of the actin cytoskeleton leads to isotropic growth and cell wall synthesis. One protein involved in polarization of the secretory apparatus is the Rho1 GTPase, which activates the formin Bni1 (116) and the exocyst component Sec3 (102). Bni1 is required for actin nucleation to cables and is part of the polarisome, a complex located at the bud tip and determining cell polarity (for reviews, see references 225 and 226), while Sec3 defines sites of active exocytosis (85). In addition, the mobilization of Chs3 from the chitosomes to the plasma membrane requires Rho1 and its effector Pkc1 (288). Two Rho1 GAPs, Sac7 and Bag7, have been proposed to regulate actin cytoskeleton assembly (250). Thus, Rho1, in addition to its role as a regulatory subunit of the β-1,3-GS, also functions in the regulation of anterograde secretion and thus effects the transport of cell wall synthesis components (Fig. 4).
Actin can also assemble into cortical patches, which are highly dynamic structures that localize close to the cell cortex, converge to the bud tip during periods of apical growth, and are associated with sites of active endocytosis (for a review, see reference 225). Evidence for their relationship with regulated cell wall synthesis comes from mutants having abnormally thickened cell walls in mother cells and thin cell walls in daughters. For example, las17Δ, vrp1Δ, or sla1Δ mutants show a new layer of cell wall being deposited in mother cells at each cell division, indicating that the laying down of a new cell wall is no longer confined to daughter cells (155), a phenotype also observed in pan1 and end3 mutants (275). Some actin patch mutants show an altered cell wall composition, a phenotype compatible with a defect in recycling polymer synthases from the plasma membrane (30, 151, 155, 213). Further, mutants defective in cortical actin patch or endocytosis function have elevated levels of cell wall chitin and require CHS3 for survival, suggesting that these mutations generate cell wall stress due to defects in recycling cell wall synthetic components (151).
The coupling of endocytosis with exocytosis allows the targeting and the dynamic relocalization of cell wall-synthetic and -remodeling systems. The organization of the actin cytoskeleton, governed mainly by the Rho-type GTPase Cdc42, is coordinated with the cell cycle by cyclin-bound Cdc28 (for a review, see reference 224). Perturbation of actin polymerization with rapamycin or latrunculin B induces activation of the PKC cell integrity pathway (106, 281). This may result in the activation of cell wall genes, providing a regulatory link between the actin cytoskeleton and cell wall assembly.
A number of actin assembly factors (Act1 and Sac6), endocytosis regulators (Rvs167 and Hua2), chitin synthases (Chs1, Chs2, and Chs3), and glucan synthesis-related proteins (Bgl2, Fks1, Gas1, Gsc2, Kre6, and Skn1) were found in a large-scale search for ubiquitinylated membrane-associated proteins (111). This posttranslational modification may participate in regulating the turnover of these proteins and/or their internalization.
Septins are key players in the coordination of cell polarity with the cell cycle (16, 97). They assemble as filaments appearing at the incipient bud site prior to bud formation as a ring, which enlarges to a collar during S, G2, and M phases, when the bud expands (see references 77, 160, and 294 for reviews). At the start of cytokinesis, the septin collar splits into two rings which disassemble after cell separation and then reassemble at the next bud site. Septin filament assembly dynamics are regulated by septin phosphorylation, by a number of protein kinases (including the Cdc42 effector Cla4 and Cln-activated Cdc28), and by septin dephosphorylation via the type 2A protein phosphatase (Pph21/22) bound to its regulatory subunit Rts1. In addition to having a role in the function of Chs3 and the septation apparatus, which includes Chs2 and the contractile actin-myosin ring, the septin rings are involved in the maintenance of cell polarity by recruiting many factors, such as bud site selection proteins and sensors for the spindle position and cell morphogenesis checkpoints (77, 160, 294).
Coordination of the regulatory pathways affecting the transport and localization of the cell wall assembly machinery is illustrated in Fig. 5.
FIG. 5.
Regulation of transport and localization of proteins involved in cell wall assembly. Proteins are grouped according to their final localization (bud, bud neck, or in the cell wall; note that Cwp1 and Cwp2 show dual localizations) and colored according to their function in cell wall assembly as in Fig. 3. Actin cytosk. actin cytoskeleton; Chito., chitosomes.
Mating, pseudohyphal growth, and sporulation.
During the development of specialized cell types, the shape of the cell wall changes radically, altering cell morphology. Upon exposure to mating pheromone, haploid cells arrest in G1, polarize, and grow a projection, the “shmoo,” toward the pheromone source. Synthesis and remodeling of cell wall material during this pro-cess are critical to ensure shmoo elongation and integrity. As a result, a number of cell wall assembly mutants, such as chs3, chs5, chs7, crh2, kre9, and skt5 mutants, show a reduced mating efficiency (25, 237, 246, 283, 284), and a mid2 (for mating-induced death) mutant, defective in signaling to the PKC cell integrity pathway, exhibits pheromone-induced cell lysis (134). Shmoo formation is initiated by activation of the pheromone signaling pathway involving the G-protein-coupled receptors for α- and a-factors (Ste2 and Ste3, respectively), a trimeric G-protein (with Gpa1, Ste4, and Ste18 being the α, β, and γ subunits, respectively), and a MAP kinase cascade (consisting of the MEKK Ste11, the MEK Ste7, and the MAP kinase Fus3) (for a review, see reference 103). Fus3 activates, among others, the Cdc28 inhibitor Far1, resulting in G1 arrest. During this arrest period, Gβγ subunits also recruit Cdc42 through interactions with Far1 and the Cdc42 GEF Cdc24, allowing polarization of the actin cytoskeleton toward the shmoo tip (33). The finding that Slt2 is activated upon pheromone exposure in a polarisome-dependent way implies a sequential activation of the mating pathway, the polarity pathway, and then the PKC cell integrity pathway (27). Gβγ has been also found to recruit Rho1 and Pkc1 to the shmoo tip, providing a mechanism for concerted regulation of the pheromone response with the cell polarity and PKC cell integrity pathways (15). Chs3-dependent chitin synthesis occurs at the shmoo neck to preserve shmoo neck integrity and relies on septin filament assembly in bundles parallel to the shmoo axis (see reference 77 for a review).
Upon starvation, haploid and diploid cells can develop specialized cells that exhibit invasive and pseudohyphal growth, respectively. As seen with mating projections, cell integrity during filamentous growth requires cell wall synthesis and remodeling to be coordinated with changes in cell polarity. Sensing of nutrient depletion activates Ras2 signaling to the cyclic AMP-dependent protein kinase (Tpk1-3) and to a Cdc42-activated MAP kinase pathway involving Kss1. The Kss1 pathway regulates cell morphogenesis through the polarisome complex. In addition, inhibition of the Ace2 transcription factor leads to a downregulation of CTS1 and EGT2 and the subsequent lack of septum degradation typical of pseudohyphae (Fig. 3A). Kss1 inhibits Clb1/2 and maintains high levels of Cln1/2, thereby prolonging the G2 phase and maintaining Cdc42 activity (for a review, see reference 214). This results in the extension of the apical phase of growth and cell elongation. During sporulation, spore wall synthesis machinery is triggered, with genes involved in spore wall formation usually induced in the middle-late phase of sporulation (49). Some of these, including CDA1, CDA2, CWP1, DIT1, DIT2, SHC1, and SPR1, are candidates for regulation by the Ndt80 transcription factor, as they are induced by its ectopic expression ((49) (Fig. 3A). Deposition of the spore wall is dependent upon the MAP kinase Smk1 and the CDK-activating kinase Cak1, with smk1 and cak1 mutants showing defects in spore wall synthesis (298, 299). In addition, the sporulation-specific serine/threonine protein kinase Sps1 is involved in transport of Gsc2, Chs3, and the Chs3 activator, Shc1 (120). The spatial regulation of cell wall deposition is thought to rely on the assembly of septin filaments, including the two sporulation-specific septins Spr3 and Spr28, which act as a scaffold in the recruiting of spore wall synthesis proteins (77, 84). In genetic studies focused on the spore cell wall, the formation of the dityrosine layer was examined in ∼600 mutants (24), and a collection of ∼300 mutants with mutations in sporulation-specific genes was screened for sensitivity to ether vapor (53), which identified ∼60 deletion mutants with spore wall defects (24, 53). Detailed phenotypic analysis of spore wall mutants has led to the formulation of a developmental pathway for spore wall assembly (53).
Adaptation to external conditions.
The cell wall composition changes with carbon source, nutrient, or oxygen availability; with external pH; and upon stress such as heat shock or exposure to cell wall-perturbing agents, low osmolarity, and mutation in cell wall genes (6, 136, 263, 264). For example, depending on growth conditions, different sets of mannoproteins are used, and they likely have specific patterns of function. In other cases, stress-induced synthetic machinery is switched on under stress while the constitutive machinery is turned down, as exemplified by the differential expression of FKS1 and GSC2 discussed above. These glucan synthases are related but distinct proteins and show different Rho1 binding levels and caspofungin sensitivities (76, 139, 178), and Fks1 and Gsc2 have likely been selected for improved function under constitutive or stress conditions, respectively. In addition to changing the cell wall synthetic machinery (Fig. 3B), stressed cells also change the relative amount of their cell wall polymers. In many cases, the cell wall is reinforced by an increase in chitin, in a chitin stress response (30, 211, 221), but the levels of mannoproteins and β-1,3-glucan can also change (3, 129).
Modifications of the nature and the amount of cross-linking between cell wall polymers occur, indicating regulation of the relative activities of synthases and remodeling enzymes. For example, the susceptibility of cells to Zymolyase under various growth conditions correlates with the β-1,6-glucan level, suggesting that the amount of β-1,6 cross-linking between polymers, rather than the absolute levels of the polymers per se, is critical for resistance to lytic enzymes (6).
Some compensation occurs among related synthases, as exemplified by the synthetic lethal interaction between FKS1 and GSC2 and between CHS2 and CHS3. Moreover, a defect in the synthesis of one cell wall polymer can be compensated for by another. Such cross-polymer compensation is shown by the synthetic lethal interactions between components of the β-1,3-glucan and the chitin synthesis pathways (41, 151, 211, 221).
(i) The PKC cell integrity pathway.
The major signaling pathway regulating cell wall assembly is the PKC cell integrity pathway (for a review, see reference 152). This pathway contains a sensing module which activates Rho1, which in turn triggers a MAP kinase cascade consisting of protein kinase C Pkc1, the MEKK Bck1, the redundant MEKs Mkk1 and Mkk2, and the MAP kinase Slt2. Activated Slt2 phosphorylates downstream transcription factors, the MADS box family member Rlm1, and the SBF subunits Swi4 and Swi6.
Sensing is achieved by two groups of transmembrane proteins, the WSC family (Slg1/Wsc1, Wsc2, and Wsc3) and the Mid2-Mtl1 pair. These families of sensors appear to respond to distinct stimulations, as suggested by the differential sensitivity to external stress exhibited by mutants with mutations in these genes (235, 293, 296). A search using the expression connection tool available from the Saccharomyces genome database (http://www.yeastgenome.org/) reveals that MID2, MTL1, and the WSC genes show distinct expression patterns, indicating that these sensors mediate signaling under different conditions. Further, these genes engage in a web of genetic interactions, such as the synthetic lethal interaction of a mid2 slg1 double mutant; the phenotypic exacerbation of slg1 wsc2, slg1 wsc3, and mid2 mtl1 double mutants; and the dosage suppression of slg1 or wsc2 by MID2 (134, 230, 293). Together, these data indicate both distinct and partially redundant functions among these genes. These sensor proteins are highly O mannosylated on extracellular serine/threonine-rich domains. This modification is believed to confer rigidity to the proteins, leading to the idea that they act as mechanosensors (219). However, the nature of the signal(s) sensed by these proteins and how the signal is subsequently transduced are still unknown.
These sensors activate Rho1 by recruiting the Rho1-GEF Rom2 (219). Rho1, with at least four effectors (Pkc1, Fks1, Bni1, and Sec3) and many GAP-GEF pairs, is broadly involved in processes such as cell polarization, cell wall synthesis, and secretion. Rho1 is thus at the hub of many signaling pathways and acts as a signal integrator (Fig. 4). For example, suppression analysis indicates that Mid2 and Slg1 signaling through Rho1 lead to different outputs, with Mid2-activated Rho1 signaling to Pkc1 and Slg1-activated Rho1 stimulating Fks1 and Pkc1 (101, 253).
Pkc1 acts as a regulator of cell wall assembly in at least three ways. First, Pkc1 transmits Rho1 signaling to the MAP kinase pathway. Second, Pkc1, with Rho1, is required for heat-induced mobilization of Chs3 from chitosome vesicles to the plasma membrane. Third, Chs3 is phosphorylated in vivo in a Pkc1-dependent manner. Though the physiological relevance of this modification is unclear, since it does not alter Chs3 localization, it may affect Chs3 activity (288). In addition, Pkc1, but not the downstream components of the PKC cell integrity pathway, is involved in the regulation of a range of cellular processes, such as cell cycle progression, secretion, and recombination. For example, a block in secretion triggers a growth arrest mediated by accumulation of Slg1 and Wsc2 in the secretory pathway and signaling through Pkc1 (203).
Slt2 has a clear role in signaling upon cell wall stress, as reflected by the lysis phenotype and the hypersensitivity to cell wall stress of slt2 mutants. A genome-wide search for genes upregulated by a constitutively active allele of slt2 identified 25 genes involved in cell wall assembly whose expression depends on Rlm1, indicating that this transcription factor regulates much of the transcriptional change upon Slt2 activation (123). In addition to Rlm1-dependent gene expression, Slt2 regulates other cellular processes. First, the SBF subunits Swi4 and Swi6 are substrates for Slt2, implicating Slt2 in cell cycle progression (166). Second, Slt2 associates with Swi4, suggesting that Slt2 may regulate Swi4 activity independently of Swi6 (12, 152). Third, Slt2 has a role in calcium homeostasis through the phosphorylation of Cch1-Mid1, a calcium channel whose activation triggers the calcineurin pathway (20). Fourth, detection of a pool of Slt2 at sites of polarized growth together with a defect in actin polarization exhibited by an slt2 mutant involve Slt2 in the regulation of assembly of the actin cytoskeleton (179, 291). Last, an slt2 null mutant is defective in the G2 arrest induced by perturbation of the actin cytoskeleton, indicating that Slt2 plays a role in triggering the morphogenesis checkpoint (106). A candidate integrator of Slt2 function is Smi1/Knr4. A smi1 mutant has reduced β-1,3-glucan and increased chitin levels (112, 172), implicating Smi1 as a positive regulator of β-1,3-glucan synthesis. In addition, a smi1 null mutant shows enhanced activation of both Slt2 and SBF-mediated transcription, suggesting that Smi1 has a negative role in cell cycle regulation (175).
Since its components are connected to many cellular functions, the PKC cell integrity pathway effectively coordinates cell wall assembly with the cell cycle, cell polarity, and secretion. Compartmentalization of signaling and regulatory components resulting in several pools of Rho1, Pkc1, and Slt2 can explain how these proteins harmoniously play their multiple roles. For example, Rho1 and Rom2 have independent trafficking pathways, allowing Rho1 to be kept in an inactive form until it reaches a Rom2-containing compartment (1). By using a series of Pkc1-GFP fusion proteins, the domains required for Pkc1 localization to the sites of polarized growth, to the nucleus, and to mitotic spindles have been mapped (67). During the cell cycle, an Slt2-GFP fusion protein shuttles between the nucleus and cytosol and can be found at sites of polarized growth in a polarisome-dependent way (291). A Smi1p-GFP also localizes to sites of polarized growth (172).
(ii) Other regulatory pathways.
Additional signaling pathways also regulate cell wall assembly. The transcription profiles of mutants deleted for FKS1, GAS1, KRE6, MNN9, or SMI1 (145, 277) reveal the upregulation of a “cell wall-compensatory” cluster of 79 genes, many of which are also induced under stress conditions, such as the presence of caspofungin, calcofluor white, Congo red, or Zymolyase (5, 22, 89, 235). Promoter analysis identifies a number of regulatory elements, such as Rlm1-binding sites and calcineurin-dependent, stress, and heat shock response elements, and points to a network of transcriptional controls, including the PKC cell integrity pathway, the general stress response pathway, and the calcineurin pathway (22, 145). Genetic evidence also illustrates a multiplicity of signaling pathways regulating cell wall assembly. First, the overexpression of LRE1 (which is known to play a role in Rho1 activation) suppressed the hypersensitivity to high osmolarity of a ste11 ssk2 ssk22 triple null mutant that is defective in the activation of the high-osmolarity glycerol response (HOG) pathway, which is required for glycerol synthesis upon osmotic stress (7). Second, mutants with mutations in genes of the HOG pathway show resistance to calcofluor white while having wild-type chitin levels, indicating that a functional HOG pathway modulates calcofluor toxicity (90). Third, cell growth at low pH leads to a Hog1-dependent acquisition of Zymolyase resistance and the induction of a number of cell wall proteins (129). Together, the data suggest cross talk between the PKC and the HOG pathways. The two-component transcription factor Skn7 is a likely Rho1 effector that is also activated by Sln1 and Ypd1 kinases of the HOG signaling pathway and is thus a candidate for an integrator of the PKC and HOG signaling pathways (152).
The calcineurin pathway, which is involved in GSC2 expression, is also involved in cell wall gene regulation (309). This pathway is activated by pheromone exposure or heat shock and following activation of the PKC cell integrity pathway. Indeed, when phosphorylated by activated Slt2, the Cch1-Mid1 channel triggers activation of the calcineurin pathway, resulting in the induction of Crz1-responsive genes, including GSC2. In addition, Skn7 is implicated in the regulation of Crz1-regulated genes, such as the mannosyltransferase OCH1 (152).
Finally, a temperature-sensitive fks1-1154 gsc2Δ double mutant shows cell cycle arrest in G2 under restrictive conditions, suggesting the existence of a “cell wall integrity” checkpoint (273). Though this checkpoint remains to be elaborated, cell cycle arrest is reportedly due to inhibition of Clb2 induction and requires components of the dynactin complex (273).
Towards an Integrative Analysis
Functional genomic technologies enable the experimenter to assemble large genome-wide data sets on cellular components and genetic phenotypes and to see how they vary under changing conditions. The challenge for systems biology is to understand how these many cellular components functionally interact to give the observed phenotypic outputs. A beginning has been made with attempts to integrate large-scale data sets. For example, the Gerstein group examined changes in interactions between transcription factors and target genes under conditions that include the cell cycle, sporulation, the diauxic shift, and DNA damage and stress responses (161). In other work, the timing of protein complex formation during the cell cycle was analyzed using a network of physical interactions among cell cycle-regulated gene products (64).
To get such an overview of processes regulating cell wall biology, large-scale phenotypic analyses have been conducted. These include genome-wide surveys of genes required for growth in the presence of caspofungin (150, 171), Congo red (89), K1 toxin (213), and calcofluor white (164). The Saccharomyces deletion collection has also enabled large-scale double mutant construction and screens for interacting double mutant combinations (280), with networks of genetic interactions of FKS1, CHS1, CHS3, GAS1, GSC2, and SMI1 having been analyzed (150, 151). These studies reveal many of the interlocking cellular functions required to cope with cell wall stress or mutation. A large number of the genes found in these studies are cell wall genes, reflecting the underlying compensatory processes between wall polymers. A second group of genes include those encoding the cell wall stress sensors and components of the PKC cell integrity pathway, genes required to trigger the onset of the wall compensatory repertoire. Other categories found contain genes required for cell polarization or with roles in vesicular transport or endocytosis. Wider analysis of this genetic network shows that many of these processes are themselves intimately interconnected, with, for example, genes involved in endocytosis being required for the survival of mutants with mutations in genes involved in cell polarity (280).
Initial efforts at data integration compare phenotypes with transcription profiles (89, 150, 171). The overlapping gene sets capture the essence of cell wall assembly, with CHS3, CRH1, CRZ1, FKS1, GSC2, PIR3, and SLT2 being both induced by and required for resistance to a number of cell wall-perturbing conditions. Not all genes induced under a given condition are required for survival under it, likely reflecting the existence of a limited number of stereotyped responses leading to the induction of many genes, only a fraction of which are required to cope with a specific stress. Importantly, not all genes required for survival are induced, indicating that other regulatory events, such as posttranscriptional modifications, occur. For example, the mobilization of Chs3 from chitosome vesicles to the plasma membrane involves activation of a constitutively present trafficking pathway, whose key players (CHS5, CHS6, CHS7, and SKT5) are all required for the survival of fks1 and gas1 mutants or in the presence of caspofungin (150). A similar weak correlation between phenotype and modulated transcription has also been observed under other conditions, such as growth in the presence of high concentrations of NaCl or sorbitol, at high pH, on galactose, or during sporulation (24, 81, 95).
Two studies integrate more than two types of data. Kelley and Ideker (131) combined genetic interaction data, protein-protein and protein-DNA interaction data, and shared-reaction metabolic relationships in a large network. The Roth group created a network of genetic, protein-protein, and protein-DNA interactions, pooled with sequence homology (312). Analysis of these networks reveals the presence of protein complexes linked to other protein complexes by genetic interactions, demonstrating how compensatory capacity is built into cellular function through such protein modules.
A global view of cell wall biology can be drawn (Fig. 6) by connecting the cellular processes affecting cell wall assembly. The complexity of these interactions is already evident, but the trend toward a global analysis of S. cerevisiae function will likely serve to increase it.
FIG. 6.
Integrated view of regulatory pathways involved in cell wall biology. Five levels of regulation are represented (cell wall synthetic machinery, brown; surface signaling, blue; cell cycle, red; cell polarity, purple; secretion/endocytosis, black). Interconnections among processes are indicated by arrows, which are numbered from their starting points and colored according to the type of interconnection (regulation of activity, green; expression regulation, gray; spatial regulation, yellow; vesicular transport, blue; undefined regulatory event, pink). Protein transport and glycosylation through the secretory pathway and protein recycling through the endocytic pathway affect the synthesis of glucan, chitin, and mannoprotein (1). The PKC cell integrity pathway is activated by glucan and chitin defects (2). β-1,3-GS and mannoprotein expression is regulated by glucose, nutrients, and oxygen (3). Nutrient availability affects filamentation through Kss1 and cyclin regulation (4). Sporulation regulates GSC2 and CHS3 expression through Ndt80 (5) and affects polarisome function through Kss1 (6). Slt2 acts through Rlm1 on glucan, chitin, and mannoprotein synthesis (7) and on the actin cytoskeleton (8). Pkc1 plays a role in the cell cycle (9). Slt2 acts on the SBF transcription factor (10). Slt2 affects calcineurin signaling through Cch1-Mid1 (11). Pkc1 phosphorylates Chs3 (12). Rho1 is the regulatory subunit of the β-1,3-GS (13). The chitosome is mobilized by the action of Rho1 and Pkc1 on the exocyst and on the secretory pathway, respectively (14). Bni1 is a Rho1 effector (15). The Rho1 effector Skn7 has a role in OCH1 expression (16). GSC2 expression is controlled by the calcineurin-dependent transcription factor Crz1 (17). Pkc1 and Rho1 are recruited at the shmoo tip (18). Pheromone exposure induces CHS1 (19), leads to recruitment of the cell polarity machinery at the shmoo tip (20), and induces a cell cycle arrest (21). The PIR genes are cell cycle regulated (22). Many cell wall genes are expressed in G1 (23). Cdc28 regulates the function of Cdc42 under vegetative growth (24), during mating (25), and during filamentation (26), respectively. The morphogenesis and the cell wall checkpoints affect the cell cycle through the septins and dynactin, respectively (27). Slt2 is regulated by the polarisome (28). Bud neck assembly regulates Chs2 function (29), and cell polarity directs secretion (30). CNR, calcineurin pathway; FIL, filamentous growth program; Fus3, mating pheromone pathway; Glc-Nut, glucose and nutrient sensing; MAT, mating program; PKC, PKC cell integrity pathway; Pkc1-Rho1, signaling through Pkc1 or Rho1 and not through the cell integrity pathway; Spo, sporulation cascade; VG, vegetative growth.
CONCLUSION
Genomics and systems biology and their genome-wide application in bacterial, fungal, and plant model organisms augur well for the future of cell wall studies, since representative walls of all these groups will be included in the increasingly global descriptions of function. This is true even for S. cerevisiae, which, unlike bacteria and plants, is studied primarily as a model eukaryotic cell. S. cerevisiae is of course a fungus, but to tap its full potential as a “universal cell” necessitates a global analysis, which means integrating the cell wall and other intrinsically fungal features into wider functional studies. A pertinent example is the detailed study of the fungal use of the PKC pathway for regulation of cell wall integrity. The broad outlines of yeast wall construction and regulation are apparent, but as seen here, at every level much remains to be clarified. Yeast cell wall studies will also inform more generally about fungal cell walls. A recent example comes from the genomic comparison of S. cerevisiae and A. gossypii, which despite a highly conserved and syntenic gene set are, respectively, a budding yeast and a mycelial fungus (220). Thus, a common gene set can be molded through regulated expression, cellular localization, and timing to fashion either a yeast or a hyphal cell wall.
A traditional and still compelling reason to study the fungal cell wall is because it is a specific target for antifungal drugs. However, genomic studies show that there are relatively few global essential gene targets in this fungus-specific organelle. Rather, cell wall integrity is buffered at many levels by redundancy and by compensatory pathways. Fortunately, our growing understanding of the molecular basis of cell wall robustness will reveal systems weaknesses that allow the possibility of more-sophisticated interventions that cut away such buffering, for example, through the use of combination therapies that synergistically target both glucan and chitin synthesis.
Acknowledgments
We thank F. Klis, C. Specht, and R. Rodriguez-Suarez for comments on the manuscript and for helpful discussions.
H.B. thanks the Natural Sciences and Engineering Research Council of Canada for long-term support of cell wall studies.
REFERENCES
- 1.Abe, M., H. Qadota, A. Hirata, and Y. Ohya. 2003. Lack of GTP-bound Rho1p in secretory vesicles of Saccharomyces cerevisiae. J. Cell Biol. 162:85-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abeijon, C., and L. Y. Chen. 1998. The role of glucosidase I (Cwh41p) in the biosynthesis of cell wall β-1,6-glucan is indirect. Mol. Biol. Cell 9:2729-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abramova, N., O. Sertil, S. Mehta, and C. V. Lowry. 2001. Reciprocal regulation of anaerobic and aerobic cell wall mannoprotein gene expression in Saccharomyces cerevisiae. J. Bacteriol. 183:2881-2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adams, D. J. 2004. Fungal cell wall chitinases and glucanases. Microbiology 150:2029-2035. [DOI] [PubMed] [Google Scholar]
- 5.Agarwal, A. K., P. D. Rogers, S. R. Baerson, M. R. Jacob, K. A. T. H. Barker, J. D. Cleary, L. A. Walker, D. G. Nagle, and A. M. Clark. 2003. Genome-wide expression profiling of the response to polyene, pyrimidine, azole, and echinocandin antifungal agents in Saccharomyces cerevisiae. J. Biol. Chem. 278:34998-35015. [DOI] [PubMed] [Google Scholar]
- 6.Aguilar-Uscanga, B., and J. M. Francois. 2003. A study of the yeast cell wall composition and structure in response to growth conditions and mode of cultivation. Lett. Appl. Microbiol. 37:268-274. [DOI] [PubMed] [Google Scholar]
- 7.Alonso-Monge, R., E. Real, I. Wojda, J. P. Bebelman, W. H. Mager, and M. Siderius. 2001. Hyperosmotic stress response and regulation of cell wall integrity in Saccharomyces cerevisiae share common functional aspects. Mol. Microbiol. 41:717-730. [DOI] [PubMed] [Google Scholar]
- 8.Angioni, C. F. 2003. Glycosylation in Saccharomyces cerevisiae: characterization of the KTR family of mannosyltransferases. Ph.D. thesis. University of Saarland, Saarbrücken, Germany.
- 9.Appeltauer, U., and T. Achstetter. 1989. Hormone-induced expression of the CHS1 gene from Saccharomyces cerevisiae. Eur. J. Biochem. 181:243-247. [DOI] [PubMed] [Google Scholar]
- 10.Aravind, L., H. Watanabe, D. J. Lipman, and E. V. Koonin. 2000. Lineage-specific loss and divergence of functionally linked genes in eukaryotes. Proc. Natl. Acad. Sci. USA 97:11319-11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Azuma, M., J. N. Levinson, N. Page, and H. Bussey. 2002. Saccharomyces cerevisiae Big1p, a putative endoplasmic reticulum membrane protein required for normal levels of cell wall β-1,6-glucan. Yeast 19:783-793. [DOI] [PubMed] [Google Scholar]
- 12.Baetz, K., J. Moffat, J. Haynes, M. Chang, and B. Andrews. 2001. Transcriptional coregulation by the cell integrity mitogen-activated protein kinase Slt2 and the cell cycle regulator Swi4. Mol. Cell. Biol. 21:6515-6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baladron, V., S. Ufano, E. Duenas, A. B. Martin-Cuadrado, F. del Rey, and C. R. Vazquez De Aldana. 2002. Eng1p, an endo-1,3-β-glucanase localized at the daughter side of the septum, is involved in cell separation in Saccharomyces cerevisiae. Eukaryot. Cell 1:774-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ballou, C. E. 1990. Isolation, characterization, and properties of Saccharomyces cerevisiae mnn mutants with nonconditional protein glycosylation defects. Methods Enzymol. 185:440-470. [DOI] [PubMed] [Google Scholar]
- 15.Bar, E. E., A. T. Ellicott, and D. E. Stone. 2003. Gβγ recruits Rho1 to the site of polarized growth during mating in budding yeast. J. Biol. Chem. 278:21798-21804. [DOI] [PubMed] [Google Scholar]
- 16.Barral, Y., V. Mermall, M. S. Mooseker, and M. Snyder. 2000. Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol. Cell 5:841-851. [DOI] [PubMed] [Google Scholar]
- 17.Beauvais, A., J. M. Bruneau, P. C. Mol, M. J. Buitrago, R. Legrand, and J. P. Latge. 2001. Glucan synthase complex of Aspergillus fumigatus. J. Bacteriol. 183:2273-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bickle, M., P. A. Delley, A. Schmidt, and M. N. Hall. 1998. Cell wall integrity modulates RHO1 activity via the exchange factor ROM2. EMBO J. 17:2235-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bidlingmaier, S., E. L. Weiss, C. Seidel, D. G. Drubin, and M. Snyder. 2001. The Cbk1p pathway is important for polarized cell growth and cell separation in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:2449-2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonilla, M., and K. W. Cunningham. 2003. Mitogen-activated protein kinase stimulation of Ca(2+) signaling is required for survival of endoplasmic reticulum stress in yeast. Mol. Biol. Cell 14:4296-4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boone, C., S. S. Sommer, A. Hensel, and H. Bussey. 1990. Yeast KRE genes provide evidence for a pathway of cell wall β-glucan assembly. J. Cell Biol. 110:1833-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boorsma, A., H. H. Nobel, B. B. Riet, B. Bargmann, S. Brul, K. J. Hellingwerf, and F. M. Klis. 2004. Characterization of the transcriptional response to cell wall stress in Saccharomyces cerevisiae. Yeast 21:413-427. [DOI] [PubMed] [Google Scholar]
- 23.Bowen, A. R., J. L. Chen-Wu, M. Momany, R. Young, P. J. Szaniszlo, and P. W. Robbins. 1992. Classification of fungal chitin synthases. Proc. Natl. Acad. Sci. USA 89:519-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Briza, P., E. Bogengruber, A. Thur, M. Rutzler, M. Munsterkotter, I. W. Dawes, and M. Breitenbach. 2002. Systematic analysis of sporulation phenotypes in 624 non-lethal homozygous deletion strains of Saccharomyces cerevisiae. Yeast 19:403-422. [DOI] [PubMed] [Google Scholar]
- 25.Brown, J. L., and H. Bussey. 1993. The yeast KRE9 gene encodes an O glycoprotein involved in cell surface β-glucan assembly. Mol. Cell. Biol. 13:6346-6356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brul, S., A. King, J. M. van der Vaart, J. Chapman, F. Klis, and C. T. Verrips. 1997. The incorporation of mannoproteins in the cell wall of S. cerevisiae and filamentous Ascomycetes. Antonie Leeuwenhoek 72:229-237. [DOI] [PubMed] [Google Scholar]
- 27.Buehrer, B. M., and B. Errede. 1997. Coordination of the mating and cell integrity mitogen-activated protein kinase pathways in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:6517-6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bulawa, C. E. 1992. CSD2, CSD3, and CSD4, genes required for chitin synthesis in Saccharomyces cerevisiae: the CSD2 gene product is related to chitin synthases and to developmentally regulated proteins in Rhizobium species and Xenopus laevis. Mol. Cell. Biol. 12:1764-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bulawa, C. E. 1993. Genetics and molecular biology of chitin synthesis in fungi. Annu. Rev. Microbiol. 47:505-534. [DOI] [PubMed] [Google Scholar]
- 30.Bulik, D. A., M. Olczak, H. A. Lucero, B. C. Osmond, P. W. Robbins, and C. A. Specht. 2003. Chitin synthesis in Saccharomyces cerevisiae in response to supplementation of growth medium with glucosamine and cell wall stress. Eukaryot. Cell 2:886-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burda, P., and M. Aebi. 1999. The dolichol pathway of N-linked glycosylation. Biochim. Biophys. Acta 1426:239-257. [DOI] [PubMed] [Google Scholar]
- 32.Burn, J. E., U. A. Hurley, R. J. Birch, T. Arioli, A. Cork, and R. E. Williamson. 2002. The cellulose-deficient Arabidopsis mutant rsw3 is defective in a gene encoding a putative glucosidase II, an enzyme processing N-glycans during ER quality control. Plant J. 32:949-960. [DOI] [PubMed] [Google Scholar]
- 33.Butty, A. C., P. M. Pryciak, L. S. Huang, I. Herskowitz, and M. Peter. 1998. The role of Far1p in linking the heterotrimeric G protein to polarity establishment proteins during yeast mating. Science 282:1511-1516. [DOI] [PubMed] [Google Scholar]
- 34.Cabib, E., J. Drgonova, and T. Drgon. 1998. Role of small G proteins in yeast cell polarization and wall biosynthesis. Annu. Rev. Biochem. 67:307-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cabib, E., and M. S. Kang. 1987. Fungal 1,3-β-glucan synthase. Methods Enzymol. 138:637-642. [DOI] [PubMed] [Google Scholar]
- 36.Cabib, E., D. H. Roh, M. Schmidt, L. B. Crotti, and A. Varma. 2001. The yeast cell wall and septum as paradigms of cell growth and morphogenesis. J. Biol. Chem. 276:19679-19682. [DOI] [PubMed] [Google Scholar]
- 37.Cabib, E., A. Sburlati, B. Bowers, and S. J. Silverman. 1989. Chitin synthase 1, an auxiliary enzyme for chitin synthesis in Saccharomyces cerevisiae. J. Cell Biol. 108:1665-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cabib, E., S. J. Silverman, and J. A. Shaw. 1992. Chitinase and chitin synthase 1: counterbalancing activities in cell separation of Saccharomyces cerevisiae. J. Gen. Microbiol. 138:97-102. [DOI] [PubMed] [Google Scholar]
- 39.Cappellaro, C., V. Mrsa, and W. Tanner. 1998. New potential cell wall glucanases of Saccharomyces cerevisiae and their involvement in mating. J. Bacteriol. 180:5030-5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caro, L. H., H. Tettelin, J. H. Vossen, A. F. Ram, H. van den Ende, and F. M. Klis. 1997. In silicio identification of glycosyl-phosphatidylinositol-anchored plasma-membrane and cell wall proteins of Saccharomyces cerevisiae. Yeast 13:1477-1489. [DOI] [PubMed] [Google Scholar]
- 41.Carotti, C., L. Ferrario, C. Roncero, M. H. Valdivieso, A. Duran, and L. Popolo. 2002. Maintenance of cell integrity in the gas1 mutant of Saccharomyces cerevisiae requires the Chs3p-targeting and activation pathway and involves an unusual Chs3p localization. Yeast 19:1113-1124. [DOI] [PubMed] [Google Scholar]
- 42.Carotti, C., E. Ragni, O. Palomares, T. Fontaine, G. Tedeschi, R. Rodriguez, J. P. Latge, M. Vai, and L. Popolo. 2004. Characterization of recombinant forms of the yeast Gas1 protein and identification of residues essential for glucanosyltransferase activity and folding. Eur. J. Biochem. 271:3635-3645. [DOI] [PubMed] [Google Scholar]
- 43.Castillo, L., A. I. Martinez, A. Garcera, M. V. Elorza, E. Valentin, and R. Sentandreu. 2003. Functional analysis of the cysteine residues and the repetitive sequence of Saccharomyces cerevisiae Pir4/Cis3: the repetitive sequence is needed for binding to the cell wall β-1,3-glucan. Yeast 20:973-983. [DOI] [PubMed] [Google Scholar]
- 44.Castro, C., J. C. Ribas, M. H. Valdivieso, R. Varona, F. del Rey, and A. Duran. 1995. Papulacandin B resistance in budding and fission yeasts: isolation and characterization of a gene involved in (1,3)β-d-glucan synthesis in Saccharomyces cerevisiae. J. Bacteriol. 177:5732-5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chavan, M., T. Suzuki, M. Rekowicz, and W. Lennarz. 2003. Genetic, biochemical, and morphological evidence for the involvement of N-glycosylation in biosynthesis of the cell wall β1,6-glucan of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 100:15381-15386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi, W. J., B. Santos, A. Duran, and E. Cabib. 1994. Are yeast chitin synthases regulated at the transcriptional or the posttranslational level? Mol. Cell. Biol. 14:7685-7694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choi, W. J., A. Sburlati, and E. Cabib. 1994. Chitin synthase 3 from yeast has zymogenic properties that depend on both the CAL1 and the CAL3 genes. Proc. Natl. Acad. Sci. USA 91:4727-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Christodoulidou, A., V. Bouriotis, and G. Thireos. 1996. Two sporulation-specific chitin deacetylase-encoding genes are required for the ascospore wall rigidity of Saccharomyces cerevisiae. J. Biol. Chem. 271:31420-31425. [DOI] [PubMed] [Google Scholar]
- 49.Chu, S., J. DeRisi, M. Eisen, J. Mulholland, D. Botstein, P. O. Brown, and I. Herskowitz. 1998. The transcriptional program of sporulation in budding yeast. Science 282:699-705. [DOI] [PubMed] [Google Scholar]
- 50.Chuang, J. S., and R. W. Schekman. 1996. Differential trafficking and timed localization of two chitin synthase proteins, Chs2p and Chs3p. J. Cell Biol. 135:597-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cid, V. J., A. Duran, F. del Rey, M. P. Snyder, C. Nombela, and M. Sanchez. 1995. Molecular basis of cell integrity and morphogenesis in Saccharomyces cerevisiae. Microbiol. Rev. 59:345-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Colman-Lerner, A., T. E. Chin, and R. Brent. 2001. Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell 107:739-750. [DOI] [PubMed] [Google Scholar]
- 53.Coluccio, A., E. Bogengruber, M. N. Conrad, M. E. Dresser, P. Briza, and A. M. Neiman. 2004. Morphogenetic pathway of spore wall assembly in Saccharomyces cerevisiae. Eukaryot. Cell 3:1464-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Coluccio, A., and A. M. Neiman. 2004. Interspore bridges: a new feature of the Saccharomyces cerevisiae spore wall. Microbiology 150:3189-3196. [DOI] [PubMed] [Google Scholar]
- 55.Correa, J. U., N. Elango, I. Polacheck, and E. Cabib. 1982. Endochitinase, a mannan-associated enzyme from Saccharomyces cerevisiae. J. Biol. Chem. 257:1392-1397. [PubMed] [Google Scholar]
- 56.Cortes, J. C., E. Carnero, J. Ishiguro, Y. Sanchez, A. Duran, and J. C. Ribas. 2005. The novel fission yeast (1,3)β-d-glucan synthase catalytic subunit Bgs4p is essential during both cytokinesis and polarized growth. J. Cell Sci. 118:157-174. [DOI] [PubMed] [Google Scholar]
- 57.Cortes, J. C., J. Ishiguro, A. Duran, and J. C. Ribas. 2002. Localization of the (1,3)β-d-glucan synthase catalytic subunit homologue Bgs1p/Cps1p from fission yeast suggests that it is involved in septation, polarized growth, mating, spore wall formation and spore germination. J. Cell Sci. 115:4081-4096. [DOI] [PubMed] [Google Scholar]
- 58.Cos, T., R. A. Ford, J. A. Trilla, A. Duran, E. Cabib, and C. Roncero. 1998. Molecular analysis of Chs3p participation in chitin synthase III activity. Eur. J. Biochem. 256:419-426. [DOI] [PubMed] [Google Scholar]
- 59.Dallies, N., J. Francois, and V. Paquet. 1998. A new method for quantitative determination of polysaccharides in the yeast cell wall. Application to the cell wall defective mutants of Saccharomyces cerevisiae. Yeast 14:1297-1306. [DOI] [PubMed] [Google Scholar]
- 60.Dean, N. 1999. Asparagine-linked glycosylation in the yeast Golgi. Biochim. Biophys. Acta 1426:309-322. [DOI] [PubMed] [Google Scholar]
- 61.De Groot, P. W., A. D. de Boer, J. Cunningham, H. L. Dekker, L. de Jong, K. J. Hellingwerf, C. de Koster, and F. M. Klis. 2004. Proteomic analysis of Candida albicans cell walls reveals covalently bound carbohydrate-active enzymes and adhesins. Eukaryot. Cell 3:955-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.De Groot, P. W., K. J. Hellingwerf, and F. M. Klis. 2003. Genome-wide identification of fungal GPI proteins. Yeast 20:781-796. [DOI] [PubMed] [Google Scholar]
- 63.De Groot, P. W., A. F. Ram, and F. M. Klis. 2005. Features and functions of covalently linked proteins in fungal cell walls. Fungal Genet. Biol. 42:657-675. [DOI] [PubMed] [Google Scholar]
- 64.de Lichtenberg, U., L. J. Jensen, S. Brunak, and P. Bork. 2005. Dynamic complex formation during the yeast cell cycle. Science 307:724-727. [DOI] [PubMed] [Google Scholar]
- 65.DeMarini, D. J., A. E. Adams, H. Fares, C. De Virgilio, G. Valle, J. S. Chuang, and J. R. Pringle. 1997. A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J. Cell Biol. 139:75-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dempski, R. E., Jr., and B. Imperiali. 2002. Oligosaccharyl transferase: gatekeeper to the secretory pathway. Curr. Opin. Chem. Biol. 6:844-850. [DOI] [PubMed] [Google Scholar]
- 67.Denis, V., and M. S. Cyert. 2005. Molecular analysis reveals localization of Saccharomyces cerevisiae protein kinase C to sites of polarized growth and Pkc1p targeting to the nucleus and mitotic spindle. Eukaryot. Cell 4:36-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.de Nobel, H., C. Ruiz, H. Martin, W. Morris, S. Brul, M. Molina, and F. M. Klis. 2000. Cell wall perturbation in yeast results in dual phosphorylation of the Slt2/Mpk1 MAP kinase and in an Slt2-mediated increase in FKS2-lacZ expression, glucanase resistance and thermotolerance. Microbiology 146:2121-2132. [DOI] [PubMed] [Google Scholar]
- 69.Deresinski, S. C., and D. A. Stevens. 2003, Caspofungin. Clin. Infect. Dis. 36:1445-1457. [DOI] [PubMed] [Google Scholar]
- 70.Deutschbauer, A. M., R. M. Williams, A. M. Chu, and R. W. Davis. 2002. Parallel phenotypic analysis of sporulation and postgermination growth in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 99:15530-15535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dijkgraaf, G. J., M. Abe, Y. Ohya, and H. Bussey. 2002. Mutations in Fks1p affect the cell wall content of β-1,3- and β-1,6-glucan in Saccharomyces cerevisiae. Yeast 19:671-690. [DOI] [PubMed] [Google Scholar]
- 72.Dijkgraaf, G. J., J. L. Brown, and H. Bussey. 1996. The KNH1 gene of Saccharomyces cerevisiae is a functional homolog of KRE9. Yeast 12:683-692. [DOI] [PubMed] [Google Scholar]
- 73.Dijkgraaf, G. J., H. Li, and H. Bussey. 2002. Cell-wall β-glucans of Saccharomyces cerevisiae, p. 179-213. In A. Steinbuchel (ed.), Biopolymers. Wiley-VCH, Weinheim, Germany.
- 74.Dobbelaere, J., and Y. Barral. 2004. Spatial coordination of cytokinetic events by compartmentalization of the cell cortex. Science 305:393-396. [DOI] [PubMed] [Google Scholar]
- 75.Doolin, M. T., A. L. Johnson, L. H. Johnston, and G. Butler. 2001. Overlapping and distinct roles of the duplicated yeast transcription factors Ace2p and Swi5p. Mol. Microbiol. 40:422-432. [DOI] [PubMed] [Google Scholar]
- 76.Douglas, C. M., F. Foor, J. A. Marrinan, N. Morin, J. B. Nielsen, A. M. Dahl, P. Mazur, W. Baginsky, W. Li, M. el Sherbeini, J. A. Clemas, S. M. Mandala, B. R. Frommer, and M. B. Kurtz. 1994. The Saccharomyces cerevisiae FKS1 (ETG1) gene encodes an integral membrane protein which is a subunit of 1,3-β-d-glucan synthase. Proc. Natl. Acad. Sci. USA 91:12907-12911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Douglas, L. M., F. J. Alvarez, C. McCreary, and J. B. Konopka. 2005. Septin function in yeast model systems and pathogenic fungi. Eukaryot. Cell 4:1503-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Drgonova, J., T. Drgon, K. Tanaka, R. Kollar, G. C. Chen, R. A. Ford, C. S. Chan, Y. Takai, and E. Cabib. 1996. Rho1p, a yeast protein at the interface between cell polarization and morphogenesis. Science 272:277-279. [DOI] [PubMed] [Google Scholar]
- 79.Ellgaard, L., and A. Helenius. 2003. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 4:181-191. [DOI] [PubMed] [Google Scholar]
- 80.el Sherbeini, M., and J. A. Clemas. 1995. Nikkomycin Z supersensitivity of an echinocandin-resistant mutant of Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 39:200-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Enyenihi, A. H., and W. S. Saunders. 2003. Large-scale functional genomic analysis of sporulation and meiosis in Saccharomyces cerevisiae. Genetics 163:47-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ezaki, B., R. C. Gardner, Y. Ezaki, H. Kondo, and H. Matsumoto. 1998. Protective roles of two aluminum (Al)-induced genes, HSP150 and SED1 of Saccharomyces cerevisiae, in Al and oxidative stresses. FEMS Microbiol. Lett. 159:99-105. [DOI] [PubMed] [Google Scholar]
- 83.Fankhauser, C., S. W. Homans, J. E. Thomas-Oates, M. J. McConville, C. Desponds, A. Conzelmann, and M. A. Ferguson. 1993. Structures of glycosylphosphatidylinositol membrane anchors from Saccharomyces cerevisiae. J. Biol. Chem. 268:26365-26374. [PubMed] [Google Scholar]
- 84.Fares, H., L. Goetsch, and J. R. Pringle. 1996. Identification of a developmentally regulated septin and involvement of the septins in spore formation in Saccharomyces cerevisiae. J. Cell Biol. 132:399-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Finger, F. P., T. E. Hughes, and P. Novick. 1998. Sec3p is a spatial landmark for polarized secretion in budding yeast. Cell 92:559-571. [DOI] [PubMed] [Google Scholar]
- 86.Fontaine, T., T. Magnin, A. Melhert, D. Lamont, J. P. Latge, and M. A. Ferguson. 2003. Structures of the glycosylphosphatidylinositol membrane anchors from Aspergillus fumigatus membrane proteins. Glycobiology 13:169-177. [DOI] [PubMed] [Google Scholar]
- 87.Fontaine, T., C. Simenel, G. Dubreucq, O. Adam, M. Delepierre, J. Lemoine, C. E. Vorgias, M. Diaquin, and J. P. Latge. 2000. Molecular organization of the alkali-insoluble fraction of Aspergillus fumigatus cell wall. J. Biol. Chem. 275:27594-27607. [DOI] [PubMed] [Google Scholar]
- 88.Franzot, S. P., and A. Casadevall. 1997. Pneumocandin L-743,872 enhances the activities of amphotericin B and fluconazole against Cryptococcus neoformans in vitro. Antimicrob. Agents Chemother. 41:331-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Garcia, R., C. Bermejo, C. Grau, R. Perez, J. Manuel Rodriguez-Pena, J. Francois, C. Nombela, and J. Arroyo. 2004. The global transcriptional response to transient cell wall damage in Saccharomyces cerevisiae and its regulation by the cell integrity signaling pathway. J. Biol. Chem. 279:15183-15195. [DOI] [PubMed] [Google Scholar]
- 90.Garcia-Rodriguez, L. J., A. Duran, and C. Roncero. 2000. Calcofluor antifungal action depends on chitin and a functional high-osmolarity glycerol response (HOG) pathway: evidence for a physiological role of the Saccharomyces cerevisiae HOG pathway under noninducing conditions. J. Bacteriol. 182:2428-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Garrett-Engele, P., B. Moilanen, and M. S. Cyert. 1995. Calcineurin, the Ca2+/calmodulin-dependent protein phosphatase, is essential in yeast mutants with cell integrity defects and in mutants that lack a functional vacuolar H+-ATPase. Mol. Cell. Biol. 15:4103-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gatti, E., L. Popolo, M. Vai, N. Rota, and L. Alberghina. 1994. O-linked oligosaccharides in yeast glycosyl phosphatidylinositol-anchored protein gp115 are clustered in a serine-rich region not essential for its function. J. Biol. Chem. 269:19695-19700. [PubMed] [Google Scholar]
- 93.Gemmill, T. R., and R. B. Trimble. 1999. Overview of N- and O-linked oligosaccharide structures found in various yeast species. Biochim. Biophys. Acta 1426:227-237. [DOI] [PubMed] [Google Scholar]
- 94.Gentzsch, M., and W. Tanner. 1997. Protein-O-glycosylation in yeast: protein-specific mannosyltransferases. Glycobiology 7:481-486. [DOI] [PubMed] [Google Scholar]
- 95.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 96.Gillmor, C. S., P. Poindexter, J. Lorieau, M. M. Palcic, and C. Somerville. 2002. α-Glucosidase I is required for cellulose biosynthesis and morphogenesis in Arabidopsis. J. Cell Biol. 156:1003-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gladfelter, A. S., L. Kozubowski, T. R. Zyla, and D. J. Lew. 2005. Interplay between septin organization, cell cycle and cell shape in yeast. J. Cell Sci. 118:1617-1628. [DOI] [PubMed] [Google Scholar]
- 98.Goldman, R. C., P. A. Sullivan, D. Zakula, and J. O. Capobianco. 1995. Kinetics of β-1,3 glucan interaction at the donor and acceptor sites of the fungal glucosyltransferase encoded by the BGL2 gene. Eur. J. Biochem. 227:372-378. [DOI] [PubMed] [Google Scholar]
- 99.Gomez-Esquer, F., J. M. Rodriguez-Pena, G. Diaz, E. Rodriguez, P. Briza, C. Nombela, and J. Arroyo. 2004. CRR1, a gene encoding a putative transglycosidase, is required for proper spore wall assembly in Saccharomyces cerevisiae. Microbiology 150:3269-3280. [DOI] [PubMed] [Google Scholar]
- 100.Graham, T. R., M. Seeger, G. S. Payne, V. L. MacKay, and S. D. Emr. 1994. Clathrin-dependent localization of α 1,3 mannosyltransferase to the Golgi complex of Saccharomyces cerevisiae. J. Cell Biol. 127:667-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Green, R., G. Lesage, A. M. Sdicu, P. Menard, and H. Bussey. 2003. A synthetic analysis of the Saccharomyces cerevisiae stress sensor Mid2p, and identification of a Mid2p interacting protein, Zeo1p, that modulates the PKC1-MPK1 cell integrity pathway. Microbiology 149:2487-2499. [DOI] [PubMed] [Google Scholar]
- 102.Guo, W., F. Tamanoi, and P. Novick. 2001. Spatial regulation of the exocyst complex by Rho1 GTPase. Nat. Cell Biol. 3:353-360. [DOI] [PubMed] [Google Scholar]
- 103.Gustin, M. C., J. Albertyn, M. Alexander, and K. Davenport. 1998. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62:1264-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hagen, I., M. Ecker, A. Lagorce, J. M. Francois, S. Sestak, R. Rachel, G. Grossmann, N. C. Hauser, J. D. Hoheisel, W. Tanner, and S. Strahl. 2004. Sed1p and Srl1p are required to compensate for cell wall instability in Saccharomyces cerevisiae mutants defective in multiple GPI-anchored mannoproteins. Mol. Microbiol. 52:1413-1425. [DOI] [PubMed] [Google Scholar]
- 105.Hamada, K., S. Fukuchi, M. Arisawa, M. Baba, and K. Kitada. 1998. Screening for glycosylphosphatidylinositol (GPI)-dependent cell wall proteins in Saccharomyces cerevisiae. Mol. Gen. Genet. 258:53-59. [DOI] [PubMed] [Google Scholar]
- 106.Harrison, J. C., E. S. Bardes, Y. Ohya, and D. J. Lew. 2001. A role for the Pkc1p/Mpk1p kinase cascade in the morphogenesis checkpoint. Nat. Cell Biol. 3:417-420. [DOI] [PubMed] [Google Scholar]
- 107.Hausler, A., and P. W. Robbins. 1992. Glycosylation in Saccharomyces cerevisiae: cloning and characterization of an α-1,2-mannosyltransferase structural gene. Glycobiology 2:77-84. [DOI] [PubMed] [Google Scholar]
- 108.Herrero, A. B., P. Magnelli, M. K. Mansour, S. M. Levitz, H. Bussey, and C. Abeijon. 2004. KRE5 gene null mutant strains of Candida albicans are avirulent and have altered cell wall composition and hypha formation properties. Eukaryot. Cell 3:1423-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Herscovics, A. 1999. Processing glycosidases of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1426:275-285. [DOI] [PubMed] [Google Scholar]
- 110.Hill, K., C. Boone, M. Goebl, R. Puccia, A. M. Sdicu, and H. Bussey. 1992. Yeast KRE2 defines a new gene family encoding probable secretory proteins, and is required for the correct N-glycosylation of proteins. Genetics 130:273-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hitchcock, A. L., K. Auld, S. P. Gygi, and P. A. Silver. 2003. A subset of membrane-associated proteins is ubiquitinated in response to mutations in the endoplasmic reticulum degradation machinery. Proc. Natl. Acad. Sci. USA 100:12735-12740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hong, Z., P. Mann, N. H. Brown, L. E. Tran, K. J. Shaw, R. S. Hare, and B. DiDomenico. 1994. Cloning and characterization of KNR4, a yeast gene involved in (1,3)-β-glucan synthesis. Mol. Cell. Biol. 14:1017-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Humbel, B. M., M. Konomi, T. Takagi, N. Kamasawa, S. A. Ishijima, and M. Osumi. 2001. In situ localization of β-glucans in the cell wall of Schizosaccharomyces pombe. Yeast 18:433-444. [DOI] [PubMed] [Google Scholar]
- 114.Ibrahim, A. S., J. C. Bowman, V. Avanessian, K. Brown, B. Spellberg, J. E. Edwards, Jr., and C. M. Douglas. 2005. Caspofungin inhibits Rhizopus oryzae 1,3-β-d-glucan synthase, lowers burden in brain measured by quantitative PCR, and improves survival at a low but not a high dose during murine disseminated zygomycosis. Antimicrob. Agents Chemother. 49:721-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Igual, J. C., A. L. Johnson, and L. H. Johnston. 1996. Coordinated regulation of gene expression by the cell cycle transcription factor Swi4 and the protein kinase C MAP kinase pathway for yeast cell integrity. EMBO J. 15:5001-5013. [PMC free article] [PubMed] [Google Scholar]
- 116.Imamura, H., K. Tanaka, T. Hihara, M. Umikawa, T. Kamei, K. Takahashi, T. Sasaki, and Y. Takai. 1997. Bni1p and Bnr1p: downstream targets of the Rho family small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae. EMBO J. 16:2745-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Imhof, I., I. Flury, C. Vionnet, C. Roubaty, D. Egger, and A. Conzelmann. 2004. Glycosylphosphatidylinositol (GPI) proteins of Saccharomyces cerevisiae contain ethanolamine phosphate groups on the α1,4-linked mannose of the GPI anchor. J. Biol. Chem. 279:19614-19627. [DOI] [PubMed] [Google Scholar]
- 118.Inoue, S. B., H. Qadota, M. Arisawa, T. Watanabe, and Y. Ohya. 1999. Prenylation of Rho1p is required for activation of yeast 1,3-β-glucan synthase. J. Biol. Chem. 274:38119-38124. [DOI] [PubMed] [Google Scholar]
- 119.Inoue, S. B., N. Takewaki, T. Takasuka, T. Mio, M. Adachi, Y. Fujii, C. Miyamoto, M. Arisawa, Y. Furuichi, and T. Watanabe. 1995. Characterization and gene cloning of 1,3-β-d-glucan synthase from Saccharomyces cerevisiae. Eur. J. Biochem. 231:845-854. [DOI] [PubMed] [Google Scholar]
- 120.Iwamoto, M. A., S. R. Fairclough, S. A. Rudge, and J. Engebrecht. 2005. Saccharomyces cerevisiae Sps1p regulates trafficking of enzymes required for spore wall synthesis. Eukaryot. Cell. 4:536-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Iyer, V. R., C. E. Horak, C. S. Scafe, D. Botstein, M. Snyder, and P. O. Brown. 2001. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409:533-538. [DOI] [PubMed] [Google Scholar]
- 122.Jiang, B., A. F. Ram, J. Sheraton, F. M. Klis, and H. Bussey. 1995. Regulation of cell wall beta-glucan assembly: PTC1 negatively affects PBS2 action in a pathway that includes modulation of EXG1 transcription. Mol. Gen. Genet. 248:260-269. [DOI] [PubMed] [Google Scholar]
- 123.Jung, U. S., and D. E. Levin. 1999. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signalling pathway. Mol. Microbiol. 34:1049-1057. [DOI] [PubMed] [Google Scholar]
- 124.Jungmann, J., and S. Munro. 1998. Multi-protein complexes in the cis Golgi of Saccharomyces cerevisiae with α-1,6-mannosyltransferase activity. EMBO J. 17:423-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jungmann, J., J. C. Rayner, and S. Munro. 1999. The Saccharomyces cerevisiae protein Mnn10p/Bed1p is a subunit of a Golgi mannosyltransferase complex. J. Biol. Chem. 274:6579-6585. [DOI] [PubMed] [Google Scholar]
- 126.Kalebina, T. S., V. Farkas, D. K. Laurinavichiute, P. M. Gorlovoy, G. V. Fominov, P. Bartek, and I. S. Kulaev. 2003. Deletion of BGL2 results in an increased chitin level in the cell wall of Saccharomyces cerevisiae. Antonie Leeuwenhoek 84:179-184. [DOI] [PubMed] [Google Scholar]
- 127.Kang, M. S., and E. Cabib. 1986. Regulation of fungal cell wall growth: a guanine nucleotide-binding, proteinaceous component required for activity of (1-3)-β-d-glucan synthase. Proc. Natl. Acad. Sci. USA 83:5808-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kang, M. S., N. Elango, E. Mattia, J. Au-Young, P. W. Robbins, and E. Cabib. 1984. Isolation of chitin synthetase from Saccharomyces cerevisiae. Purification of an enzyme by entrapment in the reaction product. J. Biol. Chem. 259:14966-14972. [PubMed] [Google Scholar]
- 129.Kapteyn, J. C., B. ter Riet, E. Vink, S. Blad, H. de Nobel, H. van den Ende, and F. M. Klis. 2001. Low external pH induces HOG1-dependent changes in the organization of the Saccharomyces cerevisiae cell wall. Mol. Microbiol. 39:469-479. [DOI] [PubMed] [Google Scholar]
- 130.Kapteyn, J. C., H. van den Ende, and F. M. Klis. 1999. The contribution of cell wall proteins to the organization of the yeast cell wall. Biochim. Biophys. Acta 1426:373-383. [DOI] [PubMed] [Google Scholar]
- 131.Kelley, R., and T. Ideker. 2005. Systematic interpretation of genetic interactions using protein networks. Nat. Biotechnol. 23:561-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kellner, E. M., K. I. Orsborn, E. M. Siegel, M. A. Mandel, M. J. Orbach, and J. N. Galgiani. 2005. Coccidioides posadasii contains a single 1,3-β-glucan synthase gene that appears to be essential for growth. Eukaryot. Cell 4:111-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kelly, R., E. Register, M. J. Hsu, M. Kurtz, and J. Nielsen. 1996. Isolation of a gene involved in 1,3-β-glucan synthesis in Aspergillus nidulans and purification of the corresponding protein. J. Bacteriol. 178:4381-4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ketela, T., R. Green, and H. Bussey. 1999. Saccharomyces cerevisiae Mid2p is a potential cell wall stress sensor and upstream activator of the PKC1-MPK1 cell integrity pathway. J. Bacteriol. 181:3330-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Klebl, F., and W. Tanner. 1989. Molecular cloning of a cell wall exo-β-1,3-glucanase from Saccharomyces cerevisiae. J. Bacteriol. 171:6259-6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Klis, F. M., P. Mol, K. Hellingwerf, and S. Brul. 2002. Dynamics of cell wall structure in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 26:239-256. [DOI] [PubMed] [Google Scholar]
- 137.Kollar, R., E. Petrakova, G. Ashwell, P. W. Robbins, and E. Cabib. 1995. Architecture of the yeast cell wall. The linkage between chitin and β(1-3)-glucan. J. Biol. Chem. 270:1170-1178. [DOI] [PubMed] [Google Scholar]
- 138.Kollar, R., B. B. Reinhold, E. Petrakova, H. J. Yeh, G. Ashwell, J. Drgonova, J. C. Kapteyn, F. M. Klis, and E. Cabib. 1997. Architecture of the yeast cell wall. β(1-6)-Glucan interconnects mannoprotein, β(1-3)-glucan, and chitin. J. Biol. Chem. 272:17762-17775. [DOI] [PubMed] [Google Scholar]
- 139.Kondoh, O., T. Takasuka, M. Arisawa, Y. Aoki, and T. Watanabe. 2002. Differential sensitivity between Fks1p and Fks2p against a novel β-1,3-glucan synthase inhibitor, aerothricin3. J. Biol. Chem. 277:41744-41749. [DOI] [PubMed] [Google Scholar]
- 140.Kota, J., and P. O. Ljungdahl. 2005. Specialized membrane-localized chaperones prevent aggregation of polytopic proteins in the ER. J. Cell Biol. 168:79-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kovacech, B., K. Nasmyth, and T. Schuster. 1996. EGT2 gene transcription is induced predominantly by Swi5 in early G1. Mol. Cell. Biol. 16:3264-3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kramer, K. J., and D. Koga. 1986. Insect chitin: Physical state, synthesis, degradation and metabolic regulation. Insect Biochem. 16:851-877. [Google Scholar]
- 143.Kuranda, M. J., and P. W. Robbins. 1987. Cloning and heterologous expression of glycosidase genes from Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 84:2585-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kuranda, M. J., and P. W. Robbins. 1991. Chitinase is required for cell separation during growth of Saccharomyces cerevisiae. J. Biol. Chem. 266:19758-19767. [PubMed] [Google Scholar]
- 145.Lagorce, A., N. C. Hauser, D. Labourdette, C. Rodriguez, H. Martin-Yken, J. Arroyo, J. D. Hoheisel, and J. Francois. 2003. Genome-wide analysis of the response to cell wall mutations in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 278:20345-20357. [DOI] [PubMed] [Google Scholar]
- 146.Larriba, G., E. Andaluz, R. Cueva, and R. D. Basco. 1995. Molecular biology of yeast exoglucanases. FEMS Microbiol. Lett. 125:121-126. [DOI] [PubMed] [Google Scholar]
- 147.Leal-Morales, C. A., C. E. Bracker, and S. Bartnicki-Garcia. 1994. Subcellular localization, abundance and stability of chitin synthetases 1 and 2 from Saccharomyces cerevisiae. Microbiology 140:2207-2216. [DOI] [PubMed] [Google Scholar]
- 148.Lee, B. N., and E. A. Elion. 1999. The MAPKKK Ste11 regulates vegetative growth through a kinase cascade of shared signaling components. Proc. Natl. Acad. Sci. USA 96:12679-12684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Leon, M., R. Sentandreu, and J. Zueco. 2002. A single FKS homologue in Yarrowia lipolytica is essential for viability. Yeast 19:1003-1014. [DOI] [PubMed] [Google Scholar]
- 150.Lesage, G., A. M. Sdicu, P. Menard, J. Shapiro, S. Hussein, and H. Bussey. 2004. Analysis of β-1,3 glucan assembly in Saccharomyces cerevisiae using a synthetic interaction network and altered sensitivity to caspofungin. Genetics 167:35-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lesage, G., J. Shapiro, C. A. Specht, A. M. Sdicu, P. Menard, S. Hussein, A. H. Tong, C. Boone, and H. Bussey. 2005. An interactional network of genes involved in chitin synthesis in Saccharomyces cerevisiae. BMC Genet. 6:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Levin, D. E. 2005. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69:262-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Levinson, J. N., S. Shahinian, A. M. Sdicu, D. C. Tessier, and H. Bussey. 2002. Functional, comparative and cell biological analysis of Saccharomyces cerevisiae Kre5p. Yeast 19:1243-1259. [DOI] [PubMed] [Google Scholar]
- 154.Lew, D. J. 2003. The morphogenesis checkpoint: how yeast cells watch their figures. Curr. Opin. Cell Biol. 15:648-653. [DOI] [PubMed] [Google Scholar]
- 155.Li, H., N. Page, and H. Bussey. 2002. Actin patch assembly proteins Las17p and Sla1p restrict cell wall growth to daughter cells and interact with cis-Golgi protein Kre6p. Yeast 19:1097-1112. [DOI] [PubMed] [Google Scholar]
- 156.Lipke, P. N., and J. Kurjan. 1992. Sexual agglutination in budding yeasts: structure, function, and regulation of adhesion glycoproteins. Microbiol. Rev. 56:180-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lippincott, J., and R. Li. 1998. Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis. J. Cell Biol. 140:355-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Liu, J., X. Tang, H. Wang, and M. Balasubramanian. 2000. Bgs2p, a 1,3-β-glucan synthase subunit, is essential for maturation of ascospore wall in Schizosaccharomyces pombe. FEBS Lett. 478:105-108. [DOI] [PubMed] [Google Scholar]
- 159.Lobsanov, Y. D., P. A. Romero, B. Sleno, B. Yu, P. Yip, A. Herscovics, and P. L. Howell. 2004. Structure of Kre2p/Mnt1p: a yeast α 1,2-mannosyltransferase involved in mannoprotein biosynthesis. J. Biol. Chem. 279:17921-17931. [DOI] [PubMed] [Google Scholar]
- 160.Longtine, M. S., and E. Bi. 2003. Regulation of septin organization and function in yeast. Trends Cell Biol. 13:403-409. [DOI] [PubMed] [Google Scholar]
- 161.Luscombe, N. M., M. M. Babu, H. Yu, M. Snyder, S. A. Teichmann, and M. Gerstein. 2004. Genomic analysis of regulatory network dynamics reveals large topological changes. Nature 431:308-312. [DOI] [PubMed] [Google Scholar]
- 162.Lussier, M., A. M. Sdicu, and H. Bussey. 1999. The KTR and MNN1 mannosyltransferase families of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1426:323-334. [DOI] [PubMed] [Google Scholar]
- 163.Lussier, M., A. M. Sdicu, A. Camirand, and H. Bussey. 1996. Functional characterization of the YUR1, KTR1, and KTR2 genes as members of the yeast KRE2/MNT1 mannosyltransferase gene family. J. Biol. Chem. 271:11001-11008. [DOI] [PubMed] [Google Scholar]
- 164.Lussier, M., A. M. White, J. Sheraton, T. di Paolo, J. Treadwell, S. B. Southard, C. I. Horenstein, J. Chen-Weiner, A. F. Ram, J. C. Kapteyn, T. W. Roemer, D. H. Vo, D. C. Bondoc, J. Hall, W. W. Zhong, A. M. Sdicu, J. Davies, F. M. Klis, P. W. Robbins, and H. Bussey. 1997. Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics 147:435-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Machi, K., M. Azuma, K. Igarashi, T. Matsumoto, H. Fukuda, A. Kondo, and H. Ooshima. 2004. Rot1p of Saccharomyces cerevisiae is a putative membrane protein required for normal levels of the cell wall 1,6-β-glucan. Microbiology 150:3163-3173. [DOI] [PubMed] [Google Scholar]
- 166.Madden, K., Y. J. Sheu, K. Baetz, B. Andrews, and M. Snyder. 1997. SBF cell cycle regulator as a target of the yeast PKC-MAP kinase pathway. Science 275:1781-1784. [DOI] [PubMed] [Google Scholar]
- 167.Magnelli, P., J. F. Cipollo, and C. Abeijon. 2002. A refined method for the determination of Saccharomyces cerevisiae cell wall composition and β-1,6-glucan fine structure. Anal. Biochem. 301:136-150. [DOI] [PubMed] [Google Scholar]
- 168.Magnelli, P. E., J. F. Cipollo, and P. W. Robbins. 2005. A glucanase-driven fractionation allows redefinition of Schizosaccharomyces pombe cell wall composition and structure: assignment of diglucan. Anal. Biochem. 336:202-212. [DOI] [PubMed] [Google Scholar]
- 169.Maligie, M. A., and C. P. Selitrennikoff. 2005. Cryptococcus neoformans resistance to echinocandins: (1,3)β-glucan synthase activity is sensitive to echinocandins. Antimicrob. Agents Chemother. 49:2851-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Manners, D. J., A. J. Masson, J. C. Patterson, H. Bjorndal, and B. Lindberg. 1973. The structure of a β-(1-6)-d-glucan from yeast cell walls. Biochem. J. 135:31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Markovich, S., A. Yekutiel, I. Shalit, Y. Shadkchan, and N. Osherov. 2004. Genomic approach to identification of mutations affecting caspofungin susceptibility in Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 48:3871-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Martin, H., A. Dagkessamanskaia, G. Satchanska, N. Dallies, and J. Francois. 1999. KNR4, a suppressor of Saccharomyces cerevisiae cwh mutants, is involved in the transcriptional control of chitin synthase genes. Microbiology 145:249-258. [DOI] [PubMed] [Google Scholar]
- 173.Martin, V., B. Garcia, E. Carnero, A. Duran, and Y. Sanchez. 2003. Bgs3p, a putative 1,3-β-glucan synthase subunit, is required for cell wall assembly in Schizosaccharomyces pombe. Eukaryot. Cell 2:159-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Martin, V., J. C. Ribas, E. Carnero, A. Duran, and Y. Sanchez. 2000. bgs2+, a sporulation-specific glucan synthase homologue is required for proper ascospore wall maturation in fission yeast. Mol. Microbiol. 38:308-321. [DOI] [PubMed] [Google Scholar]
- 175.Martin-Yken, H., A. Dagkessamanskaia, F. Basmaji, A. Lagorce, and J. Francois. 2003. The interaction of Slt2 MAP kinase with Knr4 is necessary for signalling through the cell wall integrity pathway in Saccharomyces cerevisiae. Mol. Microbiol. 49:23-35. [DOI] [PubMed] [Google Scholar]
- 176.Matheos, D. P., T. J. Kingsbury, U. S. Ahsan, and K. W. Cunningham. 1997. Tcn1p/Crz1p, a calcineurin-dependent transcription factor that differentially regulates gene expression in Saccharomyces cerevisiae. Genes Dev. 11:3445-3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Mazur, P., and W. Baginsky. 1996. In vitro activity of 1,3-β-d-glucan synthase requires the GTP-binding protein Rho1. J. Biol. Chem. 271:14604-14609. [DOI] [PubMed] [Google Scholar]
- 178.Mazur, P., N. Morin, W. Baginsky, M. el Sherbeini, J. A. Clemas, J. B. Nielsen, and F. Foor. 1995. Differential expression and function of two homologous subunits of yeast 1,3-β-d-glucan synthase. Mol. Cell. Biol. 15:5671-5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Mazzoni, C., P. Zarov, A. Rambourg, and C. Mann. 1993. The SLT2 (MPK1) MAP kinase homolog is involved in polarized cell growth in Saccharomyces cerevisiae. J. Cell Biol. 123:1821-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Meaden, P., K. Hill, J. Wagner, D. Slipetz, S. S. Sommer, and H. Bussey. 1990. The yeast KRE5 gene encodes a probable endoplasmic reticulum protein required for (1-6)-β-d-glucan synthesis and normal cell growth. Mol. Cell. Biol. 10:3013-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Mellado, E., A. Aufauvre-Brown, C. A. Specht, P. W. Robbins, and D. W. Holden. 1995. A multigene family related to chitin synthase genes of yeast in the opportunistic pathogen Aspergillus fumigatus. Mol. Gen. Genet. 246:353-359. [DOI] [PubMed] [Google Scholar]
- 182.Mendenhall, M. D., and A. E. Hodge. 1998. Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62:1191-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Mio, T., M. Adachi-Shimizu, Y. Tachibana, H. Tabuchi, S. B. Inoue, T. Yabe, T. Yamada-Okabe, M. Arisawa, T. Watanabe, and H. Yamada-Okabe. 1997. Cloning of the Candida albicans homolog of Saccharomyces cerevisiae GSC1/FKS1 and its involvement in β-1,3-glucan synthesis. J. Bacteriol. 179:4096-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Mio, T., T. Yamada-Okabe, T. Yabe, T. Nakajima, M. Arisawa, and H. Yamada-Okabe. 1997. Isolation of the Candida albicans homologs of Saccharomyces cerevisiae KRE6 and SKN1: expression and physiological function. J. Bacteriol. 179:2363-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Mishra, C., C. E. Semino, K. J. McCreath, H. de la Vega, B. J. Jones, C. A. Specht, and P. W. Robbins. 1997. Cloning and expression of two chitin deacetylase genes of Saccharomyces cerevisiae. Yeast 13:327-336. [DOI] [PubMed] [Google Scholar]
- 186.Montijn, R. C., E. Vink, W. H. Muller, A. J. Verkleij, H. van den Ende, B. Henrissat, and F. M. Klis. 1999. Localization of synthesis of β1,6-glucan in Saccharomyces cerevisiae. J. Bacteriol. 181:7414-7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Moukadiri, I., J. Armero, A. Abad, R. Sentandreu, and J. Zueco. 1997. Identification of a mannoprotein present in the inner layer of the cell wall of Saccharomyces cerevisiae. J. Bacteriol. 179:2154-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Moukadiri, I., L. Jaafar, and J. Zueco. 1999. Identification of two mannoproteins released from cell walls of a Saccharomyces cerevisiae mnn1 mnn9 double mutant by reducing agents. J. Bacteriol. 181:4741-4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Moukadiri, I., and J. Zueco. 2001. Evidence for the attachment of Hsp150/Pir2 to the cell wall of Saccharomyces cerevisiae through disulfide bridges. FEMS Yeast Res. 1:241-245. [DOI] [PubMed] [Google Scholar]
- 190.Mouyna, I., T. Fontaine, M. Vai, M. Monod, W. A. Fonzi, M. Diaquin, L. Popolo, R. P. Hartland, and J. P. Latge. 2000. Glycosylphosphatidyl-inositol-anchored glucanosyltransferases play an active role in the biosynthesis of the fungal cell wall. J. Biol. Chem. 275:14882-14889. [DOI] [PubMed] [Google Scholar]
- 191.Mouyna, I., M. Monod, T. Fontaine, B. Henrissat, B. Lechenne, and J. P. Latge. 2000. Identification of the catalytic residues of the first family of β(1-3)glucanosyltransferases identified in fungi. Biochem. J. 347:741-747. [PMC free article] [PubMed] [Google Scholar]
- 192.Mrsa, V., M. Ecker, S. Strahl-Bolsinger, M. Nimtz, L. Lehle, and W. Tanner. 1999. Deletion of new covalently linked cell wall glycoproteins alters the electrophoretic mobility of phosphorylated wall components of Saccharomyces cerevisiae. J. Bacteriol. 181:3076-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Mrsa, V., F. Klebl, and W. Tanner. 1993. Purification and characterization of the Saccharomyces cerevisiae BGL2 gene product, a cell wall endo-β-1,3-glucanase. J. Bacteriol. 175:2102-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Mrsa, V., T. Seidl, M. Gentzsch, and W. Tanner. 1997. Specific labelling of cell wall proteins by biotinylation. Identification of four covalently linked O-mannosylated proteins of Saccharomyces cerevisiae. Yeast 13:1145-1154. [DOI] [PubMed] [Google Scholar]
- 195.Mrsa, V., and W. Tanner. 1999. Role of NaOH-extractable cell wall proteins Ccw5p, Ccw6p, Ccw7p and Ccw8p (members of the Pir protein family) in stability of the Saccharomyces cerevisiae cell wall. Yeast 15:813-820. [DOI] [PubMed] [Google Scholar]
- 196.Muniz, M., C. Nuoffer, H. P. Hauri, and H. Riezman. 2000. The Emp24 complex recruits a specific cargo molecule into endoplasmic reticulum-derived vesicles. J. Cell Biol. 148:925-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Muthukumar, G., S. H. Suhng, P. T. Magee, R. D. Jewell, and D. A. Primerano. 1993. The Saccharomyces cerevisiae SPR1 gene encodes a sporulation-specific exo-1,3-β-glucanase which contributes to ascospore thermoresistance. J. Bacteriol. 175:386-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Nagahashi, S., M. Sudoh, N. Ono, R. Sawada, E. Yamaguchi, Y. Uchida, T. Mio, M. Takagi, M. Arisawa, and H. Yamada-Okabe. 1995. Characterization of chitin synthase 2 of Saccharomyces cerevisiae. Implication of two highly conserved domains as possible catalytic sites. J. Biol. Chem. 270:13961-13967. [DOI] [PubMed] [Google Scholar]
- 199.Nakanishi-Shindo, Y., K. Nakayama, A. Tanaka, Y. Toda, and Y. Jigami. 1993. Structure of the N-linked oligosaccharides that show the complete loss of α-1,6-polymannose outer chain from och1, och1 mnn1, and och1 mnn1 alg3 mutants of Saccharomyces cerevisiae. J. Biol. Chem. 268:26338-26345. [PubMed] [Google Scholar]
- 200.Nakayama, K., T. Nagasu, Y. Shimma, J. Kuromitsu, and Y. Jigami. 1992. OCH1 encodes a novel membrane bound mannosyltransferase: outer chain elongation of asparagine-linked oligosaccharides. EMBO J. 11:2511-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Nakazawa, T., H. Horiuchi, A. Ohta, and M. Takagi. 1998. Isolation and characterization of EPD1, an essential gene for pseudohyphal growth of a dimorphic yeast, Candida maltosa. J. Bacteriol. 180:2079-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Nakazawa, T., M. Takahashi, H. Horiuchi, A. Ohta, and M. Takagi. 2000. Cloning and characterization of EPD2, a gene required for efficient pseudohyphal formation of a dimorphic yeast, Candida maltosa. Biosci. Biotechnol. Biochem. 64:369-377. [DOI] [PubMed] [Google Scholar]
- 203.Nanduri, J., and A. M. Tartakoff. 2001. The arrest of secretion response in yeast: signaling from the secretory path to the nucleus via Wsc proteins and Pkc1p. Mol. Cell 8:281-289. [DOI] [PubMed] [Google Scholar]
- 204.Narasimhan, M. L., B. Damsz, M. A. Coca, J. I. Ibeas, D. J. Yun, J. M. Pardo, P. M. Hasegawa, and R. A. Bressan. 2001. A plant defense response effector induces microbial apoptosis. Mol. Cell 8:921-930. [DOI] [PubMed] [Google Scholar]
- 205.Nelson, B., C. Kurischko, J. Horecka, M. Mody, P. Nair, L. Pratt, A. Zougman, L. D. McBroom, T. R. Hughes, C. Boone, and F. C. Luca. 2003. RAM: a conserved signaling network that regulates Ace2p transcriptional activity and polarized morphogenesis. Mol. Biol. Cell 14:3782-3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Nguyen, T. H., G. H. Fleet, and P. L. Rogers. 1998. Composition of the cell walls of several yeast species. Appl. Microbiol. Biotechnol. 50:206-212. [DOI] [PubMed] [Google Scholar]
- 207.Nuoffer, C., A. Horvath, and H. Riezman. 1993. Analysis of the sequence requirements for glycosylphosphatidylinositol anchoring of Saccharomyces cerevisiae Gas1 protein. J. Biol. Chem. 268:10558-10563. [PubMed] [Google Scholar]
- 208.Nuoffer, C., P. Jeno, A. Conzelmann, and H. Riezman. 1991. Determinants for glycophospholipid anchoring of the Saccharomyces cerevisiae GAS1 protein to the plasma membrane. Mol. Cell. Biol. 11:27-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Ono, N., T. Yabe, M. Sudoh, T. Nakajima, T. Yamada-Okabe, M. Arisawa, and H. Yamada-Okabe. 2000. The yeast Chs4 protein stimulates the trypsin-sensitive activity of chitin synthase 3 through an apparent protein-protein interaction. Microbiology 146:385-391. [DOI] [PubMed] [Google Scholar]
- 210.Orlean, P. 1997. Biogenesis of yeast cell wall and surface components, p. 229-362. In J. R. Pringle, J. R. Broach, and E. W. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces. Cell cycle and cell biology. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 211.Osmond, B. C., C. A. Specht, and P. W. Robbins. 1999. Chitin synthase III: synthetic lethal mutants and “stress related” chitin synthesis that bypasses the CSD3/CHS6 localization pathway. Proc. Natl. Acad. Sci. USA 96:11206-11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Osumi, M. 1998. The ultrastructure of yeast: cell wall structure and formation. Micron 29:207-233. [DOI] [PubMed] [Google Scholar]
- 213.Page, N., M. Gerard-Vincent, P. Menard, M. Beaulieu, M. Azuma, G. J. Dijkgraaf, H. Li, J. Marcoux, T. Nguyen, T. Dowse, A. M. Sdicu, and H. Bussey. 2003. A Saccharomyces cerevisiae genome-wide mutant screen for altered sensitivity to K1 killer toxin. Genetics 163:875-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Palecek, S. P., A. S. Parikh, and S. J. Kron. 2002. Sensing, signalling and integrating physical processes during Saccharomyces cerevisiae invasive and filamentous growth. Microbiology 148:893-907. [DOI] [PubMed] [Google Scholar]
- 215.Pammer, M., P. Briza, A. Ellinger, T. Schuster, R. Stucka, H. Feldmann, and M. Breitenbach. 1992. DIT101 (CSD2, CAL1), a cell cycle-regulated yeast gene required for synthesis of chitin in cell walls and chitosan in spore walls. Yeast 8:1089-1099. [DOI] [PubMed] [Google Scholar]
- 216.Parent, S. A., J. B. Nielsen, N. Morin, G. Chrebet, N. Ramadan, A. M. Dahl, M. J. Hsu, K. A. Bostian, and F. Foor. 1993. Calcineurin-dependent growth of an FK506- and CsA-hypersensitive mutant of Saccharomyces cerevisiae. J. Gen. Microbiol. 139:2973-2984. [DOI] [PubMed] [Google Scholar]
- 217.Parodi, A. J. 2000. Protein glucosylation and its role in protein folding. Annu. Rev. Biochem. 69:69-93. [DOI] [PubMed] [Google Scholar]
- 218.Pereira, M., M. S. Felipe, M. M. Brigido, C. M. Soares, and M. O. Azevedo. 2000. Molecular cloning and characterization of a glucan synthase gene from the human pathogenic fungus Paracoccidioides brasiliensis. Yeast 16:451-462. [DOI] [PubMed] [Google Scholar]
- 219.Philip, B., and D. E. Levin. 2001. Wsc1 and Mid2 are cell surface sensors for cell wall integrity signaling that act through Rom2, a guanine nucleotide exchange factor for Rho1. Mol. Cell. Biol. 21:271-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Philippsen, P., A. Kaufmann, and H. P. Schmitz. 2005. Homologues of yeast polarity genes control the development of multinucleated hyphae in Ashbya gossypii. Curr. Opin. Microbiol. 8:370-377. [DOI] [PubMed] [Google Scholar]
- 221.Popolo, L., D. Gilardelli, P. Bonfante, and M. Vai. 1997. Increase in chitin as an essential response to defects in assembly of cell wall polymers in the ggp1Δ mutant of Saccharomyces cerevisiae. J. Bacteriol. 179:463-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Popolo, L., and M. Vai. 1999. The Gas1 glycoprotein, a putative wall polymer cross-linker. Biochim. Biophys. Acta 1426:385-400. [DOI] [PubMed] [Google Scholar]
- 223.Protchenko, O., T. Ferea, J. Rashford, J. Tiedeman, P. O. Brown, D. Botstein, and C. C. Philpott. 2001. Three cell wall mannoproteins facilitate the uptake of iron in Saccharomyces cerevisiae. J. Biol. Chem. 276:49244-49250. [DOI] [PubMed] [Google Scholar]
- 224.Pruyne, D., and A. Bretscher. 2000. Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J. Cell Sci. 113:365-375. [DOI] [PubMed] [Google Scholar]
- 225.Pruyne, D., and A. Bretscher. 2000. Polarization of cell growth in yeast. II. The role of the cortical actin cytoskeleton. J. Cell Sci. 113:571-585. [DOI] [PubMed] [Google Scholar]
- 226.Pruyne, D., A. Legesse-Miller, L. Gao, Y. Dong, and A. Bretscher. 2004. Mechanisms of polarized growth and organelle segregation in yeast. Annu. Rev. Cell Dev. Biol. 20:559-591. [DOI] [PubMed] [Google Scholar]
- 227.Puig, S., and J. E. Perez-Ortin. 2000. Stress response and expression patterns in wine fermentations of yeast genes induced at the diauxic shift. Yeast 16:139-148. [DOI] [PubMed] [Google Scholar]
- 228.Qadota, H., C. P. Python, S. B. Inoue, M. Arisawa, Y. Anraku, Y. Zheng, T. Watanabe, D. E. Levin, and Y. Ohya. 1996. Identification of yeast Rho1p GTPase as a regulatory subunit of 1,3-β-glucan synthase. Science 272:279-281. [DOI] [PubMed] [Google Scholar]
- 229.Racki, W. J., A. M. Becam, F. Nasr, and C. J. Herbert. 2000. Cbk1p, a protein similar to the human myotonic dystrophy kinase, is essential for normal morphogenesis in Saccharomyces cerevisiae. EMBO J. 19:4524-4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Rajavel, M., B. Philip, B. M. Buehrer, B. Errede, and D. E. Levin. 1999. Mid2 is a putative sensor for cell integrity signaling in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:3969-3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Ram, A. F., S. S. Brekelmans, L. J. Oehlen, and F. M. Klis. 1995. Identification of two cell cycle regulated genes affecting the β 1,3-glucan content of cell walls in Saccharomyces cerevisiae. FEBS Lett. 358:165-170. [DOI] [PubMed] [Google Scholar]
- 232.Ram, A. F., J. C. Kapteyn, R. C. Montijn, L. H. Caro, J. E. Douwes, W. Baginsky, P. Mazur, H. van den Ende, and F. M. Klis. 1998. Loss of the plasma membrane-bound protein Gas1p in Saccharomyces cerevisiae results in the release of β 1,3-glucan into the medium and induces a compensation mechanism to ensure cell wall integrity. J. Bacteriol. 180:1418-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Ram, A. F., A. Wolters, R. Ten Hoopen, and F. M. Klis. 1994. A new approach for isolating cell wall mutants in Saccharomyces cerevisiae by screening for hypersensitivity to calcofluor white. Yeast 10:1019-1030. [DOI] [PubMed] [Google Scholar]
- 234.Rayner, J. C., and S. Munro. 1998. Identification of the MNN2 and MNN5 mannosyltransferases required for forming and extending the mannose branches of the outer chain mannans of Saccharomyces cerevisiae. J. Biol. Chem. 273:26836-26843. [DOI] [PubMed] [Google Scholar]
- 235.Reinoso-Martin, C., C. Schuller, M. Schuetzer-Muehlbauer, and K. Kuchler. 2003. The yeast protein kinase C cell integrity pathway mediates tolerance to the antifungal drug caspofungin through activation of Slt2p mitogen-activated protein kinase signaling. Eukaryot. Cell 2:1200-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Roberts, C. J., B. Nelson, M. J. Marton, R. Stoughton, M. R. Meyer, H. A. Bennett, Y. D. He, H. Dai, W. L. Walker, T. R. Hughes, M. Tyers, C. Boone, and S. H. Friend. 2000. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science 287:873-880. [DOI] [PubMed] [Google Scholar]
- 237.Rodriguez-Pena, J. M., V. J. Cid, J. Arroyo, and C. Nombela. 2000. A novel family of cell wall-related proteins regulated differently during the yeast life cycle. Mol. Cell. Biol. 20:3245-3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Rodriguez-Pena, J. M., C. Rodriguez, A. Alvarez, C. Nombela, and J. Arroyo. 2002. Mechanisms for targeting of the Saccharomyces cerevisiae GPI-anchored cell wall protein Crh2p to polarised growth sites. J. Cell Sci. 115:2549-2558. [DOI] [PubMed] [Google Scholar]
- 239.Roh, D. H., B. Bowers, H. Riezman, and E. Cabib. 2002. Rho1p mutations specific for regulation of β(1-3)glucan synthesis and the order of assembly of the yeast cell wall. Mol. Microbiol. 44:1167-1183. [DOI] [PubMed] [Google Scholar]
- 240.Roh, D. H., B. Bowers, M. Schmidt, and E. Cabib. 2002. The septation apparatus, an autonomous system in budding yeast. Mol. Biol. Cell 13:2747-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Romero, P. A., M. Lussier, A. M. Sdicu, H. Bussey, and A. Herscovics. 1997. Ktr1p is an α-1,2-mannosyltransferase of Saccharomyces cerevisiae. Comparison of the enzymic properties of soluble recombinant Ktr1p and Kre2p/Mnt1p produced in Pichia pastoris. Biochem. J. 321:289-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Romero, P. A., M. Lussier, S. Veronneau, A. M. Sdicu, A. Herscovics, and H. Bussey. 1999. Mnt2p and Mnt3p of Saccharomyces cerevisiae are members of the Mnn1p family of α-1,3-mannosyltransferases responsible for adding the terminal mannose residues of O-linked oligosaccharides. Glycobiology 9:1045-1051. [DOI] [PubMed] [Google Scholar]
- 243.Roumanie, O., C. Weinachter, I. Larrieu, M. Crouzet, and F. Doignon. 2001. Functional characterization of the Bag7, Lrg1 and Rgd2 RhoGAP proteins from Saccharomyces cerevisiae. FEBS Lett. 506:149-156. [DOI] [PubMed] [Google Scholar]
- 244.Ruiz-Herrera, J., J. M. Gonzalez-Prieto, and R. Ruiz-Medrano. 2002. Evolution and phylogenetic relationships of chitin synthases from yeasts and fungi. FEMS Yeast Res. 1:247-256. [DOI] [PubMed] [Google Scholar]
- 245.Saka, A., M. Abe, H. Okano, M. Minemura, H. Qadota, T. Utsugi, A. Mino, K. Tanaka, Y. Takai, and Y. Ohya. 2001. Complementing yeast rho1 mutation groups with distinct functional defects. J. Biol. Chem. 276:46165-46171. [DOI] [PubMed] [Google Scholar]
- 246.Santos, B., A. Duran, and M. H. Valdivieso. 1997. CHS5, a gene involved in chitin synthesis and mating in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:2485-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Santos, B., and M. Snyder. 1997. Targeting of chitin synthase 3 to polarized growth sites in yeast requires Chs5p and Myo2p. J. Cell Biol. 136:95-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Sanz, M., J. A. Trilla, A. Duran, and C. Roncero. 2002. Control of chitin synthesis through Shc1p, a functional homologue of Chs4p specifically induced during sporulation. Mol. Microbiol. 43:1183-1195. [DOI] [PubMed] [Google Scholar]
- 249.Schimoler-O'Rourke, R., S. Renault, W. Mo, and C. P. Selitrennikoff. 2003. Neurospora crassa FKS protein binds to the (1,3)β-glucan synthase substrate, UDP-glucose. Curr. Microbiol. 46:408-412. [DOI] [PubMed] [Google Scholar]
- 250.Schmidt, A., T. Schmelzle, and M. N. Hall. 2002. The RHO1-GAPs SAC7, BEM2 and BAG7 control distinct RHO1 functions in Saccharomyces cerevisiae. Mol. Microbiol. 45:1433-1441. [DOI] [PubMed] [Google Scholar]
- 251.Schmidt, M., B. Bowers, A. Varma, D. H. Roh, and E. Cabib. 2002. In budding yeast, contraction of the actomyosin ring and formation of the primary septum at cytokinesis depend on each other. J. Cell Sci. 115:293-302. [DOI] [PubMed] [Google Scholar]
- 252.Schoffelmeer, E. A., J. C. Kapteyn, R. C. Montijn, B. J. Cornelissen, and F. M. Klis. 1996. Glucosylation of fungal proteins as potential target for novel antifungal agents, p. 157-162. In H. Lyr, P. E. Russel, and H. D. Sisler (ed.), Modern fungicides and antifungal compounds. Intercept, Andover, United Kingdom.
- 253.Sekiya-Kawasaki, M., M. Abe, A. Saka, D. Watanabe, K. Kono, M. Minemura-Asakawa, S. Ishihara, T. Watanabe, and Y. Ohya. 2002. Dissection of upstream regulatory components of the Rho1p effector, 1,3-β-glucan synthase, in Saccharomyces cerevisiae. Genetics 162:663-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Selvaggini, S., C. A. Munro, S. Paschoud, D. Sanglard, and N. A. Gow. 2004. Independent regulation of chitin synthase and chitinase activity in Candida albicans and Saccharomyces cerevisiae. Microbiology 150:921-928. [DOI] [PubMed] [Google Scholar]
- 255.Sentandreu, R., and D. H. Northcote. 1968. The structure of a glycopeptide isolated from the yeast cell wall. Biochem. J. 109:419-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Shahinian, S., and H. Bussey. 2000. β-1,6-Glucan synthesis in Saccharomyces cerevisiae. Mol. Microbiol. 35:477-489. [DOI] [PubMed] [Google Scholar]
- 257.Shahinian, S., G. J. Dijkgraaf, A. M. Sdicu, D. Y. Thomas, C. A. Jakob, M. Aebi, and H. Bussey. 1998. Involvement of protein N-glycosyl chain glucosylation and processing in the biosynthesis of cell wall β-1,6-glucan of Saccharomyces cerevisiae. Genetics 149:843-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Shaw, J. A., P. C. Mol, B. Bowers, S. J. Silverman, M. H. Valdivieso, A. Duran, and E. Cabib. 1991. The function of chitin synthases 2 and 3 in the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 114:111-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Shimoi, H., H. Kitagaki, H. Ohmori, Y. Iimura, and K. Ito. 1998. Sed1p is a major cell wall protein of Saccharomyces cerevisiae in the stationary phase and is involved in lytic enzyme resistance. J. Bacteriol. 180:3381-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Silverman, S. J. 1989. Similar and different domains of chitin synthases 1 and 2 of S. cerevisiae: two isozymes with distinct functions. Yeast 5:459-467. [DOI] [PubMed] [Google Scholar]
- 261.Silverman, S. J., A. Sburlati, M. L. Slater, and E. Cabib. 1988. Chitin synthase 2 is essential for septum formation and cell division in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 85:4735-4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Simoes, T., M. C. Teixeira, A. R. Fernandes, and I. Sa-Correia. 2003. Adaptation of Saccharomyces cerevisiae to the herbicide 2,4-dichlorophenoxyacetic acid, mediated by Msn2p- and Msn4p-regulated genes: important role of SPI1. Appl. Environ. Microbiol. 69:4019-4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Smits, G. J., J. C. Kapteyn, H. van den Ende, and F. M. Klis. 1999. Cell wall dynamics in yeast. Curr. Opin. Microbiol. 2:348-352. [DOI] [PubMed] [Google Scholar]
- 264.Smits, G. J., H. van den Ende, and F. M. Klis. 2001. Differential regulation of cell wall biogenesis during growth and development in yeast. Microbiology 147:781-794. [DOI] [PubMed] [Google Scholar]
- 265.Somerville, C., S. Bauer, G. Brininstool, M. Facette, T. Hamann, J. Milne, E. Osborne, A. Paredez, S. Persson, T. Raab, S. Vorwerk, and H. Youngs. 2004. Toward a systems approach to understanding plant cell walls. Science 306:2206-2211. [DOI] [PubMed] [Google Scholar]
- 266.Specht, C. A., Y. Liu, P. W. Robbins, C. E. Bulawa, N. Iartchouk, K. R. Winter, P. J. Riggle, J. C. Rhodes, C. L. Dodge, D. W. Culp, and P. T. Borgia. 1996. The chsD and chsE genes of Aspergillus nidulans and their roles in chitin synthesis. Fungal Genet. Biol. 20:153-167. [DOI] [PubMed] [Google Scholar]
- 267.Spellman, P. T., G. Sherlock, M. Q. Zhang, V. R. Iyer, K. Anders, M. B. Eisen, P. O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9:3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Stathopoulos, A. M., and M. S. Cyert. 1997. Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes Dev. 11:3432-3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Stolz, J., and S. Munro. 2002. The components of the Saccharomyces cerevisiae mannosyltransferase complex M-Pol I have distinct functions in mannan synthesis. J. Biol. Chem. 277:44801-44808. [DOI] [PubMed] [Google Scholar]
- 270.Strahl-Bolsinger, S., M. Gentzsch, and W. Tanner. 1999. Protein O-mannosylation. Biochim. Biophys. Acta 1426:297-307. [DOI] [PubMed] [Google Scholar]
- 271.Sugawara, T., S. Takahashi, M. Osumi, and N. Ohno. 2004. Refinement of the structures of cell-wall glucans of Schizosaccharomyces pombe by chemical modification and NMR spectroscopy. Carbohydr. Res. 339:2255-2265. [DOI] [PubMed] [Google Scholar]
- 272.Sutterlin, C., T. L. Doering, F. Schimmoller, S. Schroder, and H. Riezman. 1997. Specific requirements for the ER to Golgi transport of GPI-anchored proteins in yeast. J. Cell Sci. 110:2703-2714. [DOI] [PubMed] [Google Scholar]
- 273.Suzuki, M., R. Igarashi, M. Sekiya, T. Utsugi, S. Morishita, M. Yukawa, and Y. Ohya. 2004. Dynactin is involved in a checkpoint to monitor cell wall synthesis in Saccharomyces cerevisiae. Nat. Cell Biol. 6:861-871. [DOI] [PubMed] [Google Scholar]
- 274.Takai, Y., T. Sasaki, and T. Matozaki. 2001. Small GTP-binding proteins. Physiol. Rev. 81:153-208. [DOI] [PubMed] [Google Scholar]
- 275.Tang, H. Y., J. Xu, and M. Cai. 2000. Pan1p, End3p, and S1a1p, three yeast proteins required for normal cortical actin cytoskeleton organization, associate with each other and play essential roles in cell wall morphogenesis. Mol. Cell. Biol. 20:12-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Teparic, R., I. Stuparevic, and V. Mrsa. 2004. Increased mortality of Saccharomyces cerevisiae cell wall protein mutants. Microbiology 150:3145-3150. [DOI] [PubMed] [Google Scholar]
- 277.Terashima, H., N. Yabuki, M. Arisawa, K. Hamada, and K. Kitada. 2000. Up-regulation of genes encoding GPI-attached proteins in response to cell wall damage caused by disruption of FKS1 in Saccharomyces cerevisiae. Mol. Gen. Genet. 264:64-74. [DOI] [PubMed] [Google Scholar]
- 278.Thompson, J. R., C. M. Douglas, W. Li, C. K. Jue, B. Pramanik, X. Yuan, T. H. Rude, D. L. Toffaletti, J. R. Perfect, and M. Kurtz. 1999. A glucan synthase FKS1 homolog in Cryptococcus neoformans is single copy and encodes an essential function. J. Bacteriol. 181:444-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Toh-e, A., S. Yasunaga, H. Nisogi, K. Tanaka, T. Oguchi, and Y. Matsui. 1993. Three yeast genes, PIR1, PIR2 and PIR3, containing internal tandem repeats, are related to each other, and PIR1 and PIR2 are required for tolerance to heat shock. Yeast. 9:481-494. [DOI] [PubMed] [Google Scholar]
- 280.Tong, A. H., G. Lesage, G. D. Bader, H. Ding, H. Xu, X. Xin, J. Young, G. F. Berriz, R. L. Brost, M. Chang, Y. Chen, X. Cheng, G. Chua, H. Friesen, D. S. Goldberg, J. Haynes, C. Humphries, G. He, S. Hussein, L. Ke, N. Krogan, Z. Li, J. N. Levinson, H. Lu, P. Menard, C. Munyana, A. B. Parsons, O. Ryan, R. Tonikian, T. Roberts, A. M. Sdicu, J. Shapiro, B. Sheikh, B. Suter, S. L. Wong, L. V. Zhang, H. Zhu, C. G. Burd, S. Munro, C. Sander, J. Rine, J. Greenblatt, M. Peter, A. Bretscher, G. Bell, F. P. Roth, G. W. Brown, B. Andrews, H. Bussey, and C. Boone. 2004. Global mapping of the yeast genetic interaction network. Science 303:808-813. [DOI] [PubMed] [Google Scholar]
- 281.Torres, J., C. J. Di Como, E. Herrero, and M. A. De La Torre-Ruiz. 2002. Regulation of the cell integrity pathway by rapamycin-sensitive TOR function in budding yeast. J. Biol. Chem. 277:43495-43504. [DOI] [PubMed] [Google Scholar]
- 282.Travers, K. J., C. K. Patil, L. Wodicka, D. J. Lockhart, J. S. Weissman, and P. Walter. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101:249-258. [DOI] [PubMed] [Google Scholar]
- 283.Trilla, J. A., T. Cos, A. Duran, and C. Roncero. 1997. Characterization of CHS4 (CAL2), a gene of Saccharomyces cerevisiae involved in chitin biosynthesis and allelic to SKT5 and CSD4. Yeast 13:795-807. [DOI] [PubMed] [Google Scholar]
- 284.Trilla, J. A., A. Duran, and C. Roncero. 1999. Chs7p, a new protein involved in the control of protein export from the endoplasmic reticulum that is specifically engaged in the regulation of chitin synthesis in Saccharomyces cerevisiae. J. Cell Biol. 145:1153-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Ubersax, J. A., E. L. Woodbury, P. N. Quang, M. Paraz, J. D. Blethrow, K. Shah, K. M. Shokat, and D. O. Morgan. 2003. Targets of the cyclin-dependent kinase Cdk1. Nature 425:859-864. [DOI] [PubMed] [Google Scholar]
- 286.Utsugi, T., M. Minemura, A. Hirata, M. Abe, D. Watanabe, and Y. Ohya. 2002. Movement of yeast 1,3-β-glucan synthase is essential for uniform cell wall synthesis. Genes Cells 7:1-9. [DOI] [PubMed] [Google Scholar]
- 287.Vai, M., E. Gatti, E. Lacana, L. Popolo, and L. Alberghina. 1991. Isolation and deduced amino acid sequence of the gene encoding gp115, a yeast glycophospholipid-anchored protein containing a serine-rich region. J. Biol. Chem. 266:12242-12248. [PubMed] [Google Scholar]
- 288.Valdivia, R. H., and R. Schekman. 2003. The yeasts Rho1p and Pkc1p regulate the transport of chitin synthase III (Chs3p) from internal stores to the plasma membrane. Proc. Natl. Acad. Sci. USA 100:10287-10292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Valdivieso, M. H., P. C. Mol, J. A. Shaw, E. Cabib, and A. Duran. 1991. CAL1, a gene required for activity of chitin synthase 3 in Saccharomyces cerevisiae. J. Cell Biol. 114:101-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.van der Vaart, J. M., L. H. Caro, J. W. Chapman, F. M. Klis, and C. T. Verrips. 1995. Identification of three mannoproteins in the cell wall of Saccharomyces cerevisiae. J. Bacteriol. 177:3104-3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.van Drogen, F., and M. Peter. 2002. Spa2p functions as a scaffold-like protein to recruit the Mpk1p MAP kinase module to sites of polarized growth. Curr. Biol. 12:1698-1703. [DOI] [PubMed] [Google Scholar]
- 292.Verma, D. P., and Z. Hong. 2001. Plant callose synthase complexes. Plant Mol. Biol. 47:693-701. [DOI] [PubMed] [Google Scholar]
- 293.Verna, J., A. Lodder, K. Lee, A. Vagts, and R. Ballester. 1997. A family of genes required for maintenance of cell wall integrity and for the stress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94:13804-13809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Versele, M., and J. Thorner. 2005. Some assembly required: yeast septins provide the instruction manual. Trends Cell Biol. 15:414-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Verstrepen, K. J., G. Derdelinckx, H. Verachtert, and F. R. Delvaux. 2003. Yeast flocculation: what brewers should know. Appl. Microbiol. Biotechnol. 61:197-205. [DOI] [PubMed] [Google Scholar]
- 296.Vilella, F., E. Herrero, J. Torres, and M. A. De La Torre-Ruiz. 2005. Pkc1 and the upstream elements of the cell integrity pathway in Saccharomyces cerevisiae, Rom2 and Mtl1, are required for cellular responses to oxidative stress. J. Biol. Chem. 280:9149-9159. [DOI] [PubMed] [Google Scholar]
- 297.Vink, E., R. J. Rodriguez-Suarez, M. Gerard-Vincent, J. C. Ribas, H. de Nobel, H. van den Ende, A. Duran, F. M. Klis, and H. Bussey. 2004. An in vitro assay for (1-6)-β-d-glucan synthesis in Saccharomyces cerevisiae. Yeast 21:1121-1131. [DOI] [PubMed] [Google Scholar]
- 298.Wagner, M., P. Briza, M. Pierce, and E. Winter. 1999. Distinct steps in yeast spore morphogenesis require distinct SMK1 MAP kinase thresholds. Genetics 151:1327-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Wagner, M., M. Pierce, and E. Winter. 1997. The CDK-activating kinase CAK1 can dosage suppress sporulation defects of smk1 MAP kinase mutants and is required for spore wall morphogenesis in Saccharomyces cerevisiae. EMBO J. 16:1305-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Wang, X. H., K. Nakayama, Y. Shimma, A. Tanaka, and Y. Jigami. 1997. MNN6, a member of the KRE2/MNT1 family, is the gene for mannosylphosphate transfer in Saccharomyces cerevisiae. J. Biol. Chem. 272:18117-18124. [DOI] [PubMed] [Google Scholar]
- 301.Watanabe, D., M. Abe, and Y. Ohya. 2001. Yeast Lrg1p acts as a specialized RhoGAP regulating 1,3-β-glucan synthesis. Yeast 18:943-951. [DOI] [PubMed] [Google Scholar]
- 302.Weig, M., K. Haynes, T. R. Rogers, O. Kurzai, M. Frosch, and F. A. Muhlschlegel. 2001. A GAS-like gene family in the pathogenic fungus Candida glabrata. Microbiology 147:2007-2019. [DOI] [PubMed] [Google Scholar]
- 303.Weiss, E. L., C. Kurischko, C. Zhang, K. Shokat, D. G. Drubin, and F. C. Luca. 2002. The Saccharomyces cerevisiae Mob2p-Cbk1p kinase complex promotes polarized growth and acts with the mitotic exit network to facilitate daughter cell-specific localization of Ace2p transcription factor. J. Cell Biol. 158:885-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Wilcox, L. J., D. A. Balderes, B. Wharton, A. H. Tinkelenberg, G. Rao, and S. L. Sturley. 2002. Transcriptional profiling identifies two members of the ATP-binding cassette transporter superfamily required for sterol uptake in yeast. J. Biol. Chem. 277:32466-32472. [DOI] [PubMed] [Google Scholar]
- 305.Wittenberg, C., and S. I. Reed. 2005. Cell cycle-dependent transcription in yeast: promoters, transcription factors, and transcriptomes. Oncogene 24:2746-2755. [DOI] [PubMed] [Google Scholar]
- 306.Yabe, T., T. Yamada-Okabe, T. Nakajima, M. Sudoh, M. Arisawa, and H. Yamada-Okabe. 1998. Mutational analysis of chitin synthase 2 of Saccharomyces cerevisiae. Identification of additional amino acid residues involved in its catalytic activity. Eur. J. Biochem. 258:941-947. [DOI] [PubMed] [Google Scholar]
- 307.Yin, Q. Y., P. W. De Groot, H. L. Dekker, L. de Jong, F. M. Klis, and C. G. de Koster. 2005. Comprehensive proteomic analysis of Saccharomyces cerevisiae cell walls: identification of proteins covalently attached via GPI remnants or mild-alkali sensitive linkages. J. Biol. Chem. 280:20894-20901. [DOI] [PubMed] [Google Scholar]
- 308.Yip, C. L., S. K. Welch, F. Klebl, T. Gilbert, P. Seidel, F. J. Grant, P. J. O'Hara, and V. L. MacKay. 1994. Cloning and analysis of the Saccharomyces cerevisiae MNN9 and MNN1 genes required for complex glycosylation of secreted proteins. Proc. Natl. Acad. Sci. USA 91:2723-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Yoshimoto, H., K. Saltsman, A. P. Gasch, H. X. Li, N. Ogawa, D. Botstein, P. O. Brown, and M. S. Cyert. 2002. Genome-wide analysis of gene expression regulated by the calcineurin/Crz1p signaling pathway in Saccharomyces cerevisiae. J. Biol. Chem. 277:31079-31088. [DOI] [PubMed] [Google Scholar]
- 310.Yun, D. J., Y. Zhao, J. M. Pardo, M. L. Narasimhan, B. Damsz, H. Lee, L. R. Abad, M. P. D'Urzo, P. M. Hasegawa, and R. A. Bressan. 1997. Stress proteins on the yeast cell surface determine resistance to osmotin, a plant antifungal protein. Proc. Natl. Acad. Sci. USA 94:7082-7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Zeitlinger, J., I. Simon, C. T. Harbison, N. M. Hannett, T. L. Volkert, G. R. Fink, and R. A. Young. 2003. Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell 113:395-404. [DOI] [PubMed] [Google Scholar]
- 312.Zhang, L., O. D. King, S. L. Wong, D. S. Goldberg, A. H. Tong, G. Lesage, B. Andrews, H. Bussey, C. Boone, and F. P. Roth. 2005. Motifs, themes and thematic maps of an integrated S. cerevisiae interaction network. J. Biol. 4:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Zhao, C., U. S. Jung, P. Garrett-Engele, T. Roe, M. S. Cyert, and D. E. Levin. 1998. Temperature-induced expression of yeast FKS2 is under the dual control of protein kinase C and calcineurin. Mol. Cell. Biol. 18:1013-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Zhu, G., P. T. Spellman, T. Volpe, P. O. Brown, D. Botstein, T. N. Davis, and B. Futcher. 2000. Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature 406:90-94. [DOI] [PubMed] [Google Scholar]
- 315.Ziman, M., J. S. Chuang, and R. W. Schekman. 1996. Chs1p and Chs3p, two proteins involved in chitin synthesis, populate a compartment of the Saccharomyces cerevisiae endocytic pathway. Mol. Biol. Cell 7:1909-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Ziman, M., J. S. Chuang, M. Tsung, S. Hamamoto, and R. Schekman. 1998. Chs6p-dependent anterograde transport of Chs3p from the chitosome to the plasma membrane in Saccharomyces cerevisiae. Mol. Biol. Cell 9:1565-1576. [DOI] [PMC free article] [PubMed]