Abstract
The prevalence of Escherichia coli O157 associated with feedlot cattle in Saskatchewan was determined in a 10-month longitudinal study (3 feedlots) and a point prevalence study (20 feedlots). The prevalence of E. coli O157 at the three different sites in the horizontal study varied from 2.5 to 45%. The point prevalence of E. coli O157 among Saskatchewan cattle from 20 different feedlots ranged from 0% to a high of 57%. A statistically significant (P = 0.003) positive correlation was determined to exist between the density of cattle and the E. coli O157 prevalence rate. A significant correlation (P = 0.006) was also found between the E. coli O157 percent prevalence and the number of cattle housed/capacity ratio. All 194 E. coli O157 isolates obtained were highly virulent, and random amplified polymorphic DNA PCR analysis revealed that the isolates grouped into 39 different E. coli O157 subtypes, most of which were indigenous to specific feedlots. Two of the most predominant subtypes were detected in 11 different feedlots and formed distinct clusters in two geographic regions in the province. Antimicrobial susceptibility testing of the E. coli O157 isolates revealed that 10 were multidrug resistant and that 73 and 5 were resistant to sulfisoxazole and tetracycline, respectively.
In 1982, two outbreaks of bloody diarrhea in Oregon and Michigan (41) and another in Ottawa (47) led to the recognition of a new pathogenic Escherichia coli serotype, E. coli O157:H7. Since its discovery as an etiologic agent of hemorrhagic colitis in 1982, the clinical importance of E. coli O157:H7 has escalated rapidly. In little more than a decade, recognition of E. coli O157:H7 underwent a major transformation, from that of a newly described agent of diarrheal disease (1982) to that of the leading bacterial cause of bloody diarrhea in the United States in 1992 (18).
The pathogenicity of E. coli O157:H7 is associated with a powerful combination of different virulence factors. Several inducible mechanisms of acid resistance (24, 25) potentially contribute to the low infectious dose of this organism (1 to 100 CFU) (16, 36) and to its survival in the harsh environment of the stomach. In addition, E. coli O157:H7 efficiently adheres to intestinal epithelial cells, causing attaching and effacing lesions (46). The host cells undergo dramatic changes, losing microvilli on the cell surface, followed by the tight attachment of cells to the epithelial surface and deformation of the cell cytoskeletal structure, resulting in the formation of pedestals (30). A cluster of genes involved in the generation of attaching and effacing lesions are chromosomally carried on a “pathogenicity island” named the locus of enterocyte effacement (29). This pathogenicity island also carries the eae gene, which is responsible for the generation of an outer membrane protein called intimin that is necessary for attachment to enterocytes. Undoubtedly, the most potent virulence feature of E. coli O157:H7 is the ability to produce Shiga toxins (Stx1 and/or Stx2 and variants), a family of unique, heterodimeric protein toxins (33) that cause a wide spectrum of clinical symptoms, including hemorrhagic colitis (bloody diarrhea) and life-threatening complications such as hemolytic-uremic syndrome and thrombotic thrombocytopenic purpura (37, 48).
Ruminants, especially cattle, have been implicated as the primary reservoir of E. coli O157:H7 isolates that infect humans. The fecal excretion of these organisms by cattle appears to be seasonal, with excretion rates highest in spring and late summer (17, 45). Human infection with E. coli O157:H7 has been associated with various transmission routes, including direct exposure to infected animals (40), person-to-person passage (4), and, most commonly, consumption of contaminated foods and waters (5, 10, 21). From an epidemiological point of view, it is interesting that Shiga toxin-producing Escherichia coli (STEC) infections in humans seem to be different worldwide. Escherichia coli O157:H7 is the predominant serotype of STEC in the United States, Canada, the United Kingdom, and Japan, but in continental Europe, Australia, and Latin America, non-O157:H7 serotypes are much more common than E. coli O157:H7 (20). The incidence of E. coli O157:H7 disease has risen in Canada. According to the Public Health Agency of Canada (2005), the majority (95%) of pathogenic E. coli isolates from human cases are serovar O157.
As a consequence of the fast growth of the cattle industry in western Canada, the health risk from E. coli O157:H7 has increased, especially among residents of rural communities. There is virtually no information on the occurrence of E. coli O157 in Saskatchewan cattle herds and its presence in adjacent environments. This study was conducted to determine the frequency of occurrence of E. coli O157 in Saskatchewan cattle and environmental samples (pristine soil, soil treated with cattle manure, and water from dugouts) by a combination of longitudinal (time course) and point prevalence (multiple feedlots analyzed at approximately the same time) studies over a 2-year period. This study also sought to characterize the E. coli O157 isolates obtained in terms of their virulence profiles, antibiotic susceptibilities, cytotoxicity potentials, and random amplified polymorphic DNA (RAPD) genomic profiles. RAPD DNA banding patterns, in conjunction with virulence and antibiotic resistance patterns, may assist in the epidemiologic tracing of E. coli O157 isolates of medical concern.
MATERIALS AND METHODS
Longitudinal prevalence study.
A longitudinal study was designed to examine the prevalence of E. coli O157 over a 10-month period at two feedlots (randomly given the identifiers L and V) and a cow-calf operation (WB). At each sampling time, 20 cattle fecal samples were randomly collected from the pen floor from each facility (for a total of 400 fecal samples). An additional 15 environmental samples, consisting of soil samples from fields treated with manure as well as water from dugouts and natural runoff, were collected per sampling period (for a total of 165 environmental samples).
Point prevalence study.
The point prevalence study targeted a number of feedlots, ranging in size from 80 to 5,800 head of cattle. Twenty Saskatchewan feedlots were sampled once during the summer of 2004 (from late June 2004 to the end of September 2004). Forty fecal samples were collected from each feedlot. The percent prevalence of E. coli O157 in the point and longitudinal (described above) studies was calculated as follows: % prevalence = (number of samples positive for E. coli O157/total number of samples tested in a feedlot) × 100.
Collection of samples.
Over a 2-year period, 1,200 bovine fecal and 165 environmental samples were collected from 23 different cattle facilities as part of both the longitudinal and point prevalence studies. All bovine fecal samples were obtained from freshly defecated feces. Approximately 40 g of feces was aseptically collected per sample by using a sterile tongue depressor (Baxter Diagnostics Inc.) and was transferred to a sterile 80-ml plastic container (Starplex, Canada). Typically, two to six randomly chosen samples were obtained per pen so that each pen in multipen feedlots was screened for E. coli O157. Samples from lagoons, dugouts, and soil were sampled directly, using sterile 80-ml plastic containers. All samples were stored in a cooler with frozen cold packs, transported to the laboratory, and tested within 2 to 6 h of collection. All environmental and bovine fecal samples were processed in parallel by cultural methods, with and without immunomagnetic separation (IMS).
IMS.
Samples (1 g or 1 ml) were aseptically added to 10 ml of buffered peptone water (Becton Dickinson) supplemented with cefixime (0.5 mg liter−1; Rhone-Poulenc Rorer, Canada), vancomycin (8 mg liter−1; Sigma-Aldrich Chemie, Germany), and cefsuludin (10 mg liter−1; Sigma-Aldrich Chemie) (7). After homogenization of samples, the suspensions were incubated at 37°C for 6 to 8 h without agitation. IMS was performed according to the manufacturer's instructions, using a BeadRetriever (Dynal Biotech ASA, Oslo, Norway). One hundred microliters of the concentrated bead-bacterium complex was plated onto sorbitol-MacConkey (Becton Dickinson) agar plates supplemented with cefixime (0.5 mg liter−1) and potassium tellurite (2.5 mg liter−1; Oxoid Ltd., Hampshire, United Kingdom). After 24 h of incubation at 37°C, discrete, gray, non-sorbitol-fermenting colonies were selected and tested by seroagglutination using O157 latex reagent (Oxoid Ltd.). All seropositive isolates were saved in 10% glycerol at −70°C for further analysis.
Cultural technique.
Samples (1 g or 1 ml) were aseptically added to 10 ml of modified EC broth (Becton Dickinson) containing novobiocin (20 mg liter−1; Sigma Chemical Co.), vortexed, and then incubated at 37°C for 18 to 24 h. After incubation, a 1-ml aliquot of each enrichment culture was serially diluted to 10−5 in sterile 0.85% saline to facilitate the recovery of isolates. One-hundred-microliter aliquots of the 10−3, 10−4, and 10−5 dilutions were spread plated in duplicate onto sorbitol-MacConkey agar and incubated at 37°C overnight. Each non-sorbitol-fermenting colony with the characteristic morphology of E. coli O157 was serologically tested for the somatic O157 antigen, and seropositive isolates were saved as described above.
Multiplex PCR.
All presumptive E. coli O157-positive isolates were tested for the presence of four virulence determinants, including stx1, stx2, eaeA, and hlyA. Template DNAs for multiplex PCR and RAPD fingerprinting were prepared from isolates that were grown at 37°C for 24 h in lauryl tryptose broth (Becton Dickinson), with agitation. After incubation, the optical density at 600 nm of each culture was standardized by dilution with broth. Aliquots (400 μl) of stationary-phase cultures with an optical density value of 0.6 were centrifuged, and total DNAs were extracted according to the manufacturer's instructions, using a DNeasy tissue kit (QIAGEN Inc., Mississauga, Canada). For the longitudinal study, multiplex PCR was performed as described by Fagan et al. (12). Escherichia coli O157:H7 ATCC 43894 (which possesses the above virulence determinants) and Pseudomonas fluorescens strain cc848406E were used as positive and negative controls, respectively. In a recent study (11), it was shown that the Fagan PCR assay was unreliable for the detection of stx2d-positive STEC strains. Thus, a multiplex PCR protocol by Paton and Paton (35) which can identify all known subtypes of Shiga toxins was used during the analysis of isolates obtained from the point prevalence study.
Genotyping of E. coli O157 isolates.
In preliminary experiments, the following four decamer primers were tested against a panel of 15 different E. coli O157 isolates for the random amplification of polymorphic DNA: GEN 15001 (5′-GTGCAATGAG-3′) (39), 1283 (5′-GCGATCCCCA-3′) (15), 1290 (5′-GTGGATGCGA-3′) (34), and 1254 (5′-CCGCAGCCAA-3′) (1, 2, 27). Primer 1283 provided the greatest discriminatory ability among the E. coli O157 test isolates and was therefore used in this study. To further validate the reproducibility of this primer and procedure, three colonies of the same E. coli O157 isolate were amplified by primer 1283 in three independent PCRs. The band patterns of the three colonies were consistent both among the colonies and within the triplicate PCRs. The preparation of DNA templates for RADP PCR was described above. PCRs were carried out in 50-μl reaction volumes containing 5 μl of DNA template, 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 3 mM MgCl2, a 250 μM concentration of each deoxynucleoside triphosphate, 0.8 μM of primer, and 2 units of Taq polymerase. PCR amplification was performed in a Techne TC-412 thermal cycler under the following conditions: 45 cycles of 94°C for 1 min, 36°C for 1 min, and 72°C for 2 min, followed by an elongation step of 72°C for 5 min. Amplified products were then electrophoresed at 90 V for 2 h in 1.5% (wt/vol) agarose gels containing 0.125 μg ml−1 ethidium bromide. The 1 Kb Plus DNA ladder was used as a molecular weight marker.
Vero cell cytotoxicity assay.
The cytotoxicities of the E. coli O157 isolates were determined by the method previously described by Roberts et al. (42). Cytopathic effects of the E. coli O157 isolates were examined using a Zeiss inverted microscope (×200).
Antibiotic susceptibility testing.
Escherichia coli O157 isolates obtained from the point prevalence study (n = 131) were examined for antibiotic resistance by using Sensititre CMV1AGNF plates (TREK Diagnostic Systems, Cleveland, OH) containing 17 antimicrobial agents dosed in 96 wells at appropriate dilutions, as specified by the National Antimicrobial Resistance Monitoring System of the CDC. Each well of the Sensititre microtiter plate was inoculated according to the instructions of the manufacturer, followed by incubation at 37°C for 24 h. The MIC was manually determined for each isolate as the lowest concentration of each antibiotic that inhibited visible growth. The MIC breakpoints were determined according to the CLSI (formerly NCCLS) M100 (31) and M31 (32) standards.
Statistical analysis.
Data were analyzed with SAS (SAS Institute Inc., Cary, NC) by using multiple regression to determine the dependence of the prevalence of E. coli O157 on the size of feedlots, density of cattle in pens, and ratio between the number of housed cattle and the feedlot capacity. Each predictor (size of feedlots, density of cattle in pens, and ratio between housed cattle and feedlot capacity) was also evaluated using stepwise regression. The analysis of DNA patterns obtained by RAPD PCR was performed by using the Numerical Taxonomy and Multivariate Analysis system (Exeter Software, Setauket, N.Y.). Similarities between RAPD patterns were determined based on the Dice similarity coefficient. The resulting similarities in the matrix were further processed by employing the unweighted-pair group method using average linkages to create a dendrogram that graphed the genetic relatedness between E. coli O157 isolates.
RESULTS
Prevalence of E. coli O157. (i) Longitudinal study.
A total of 565 fecal bovine and environmental samples were collected over a 10-month period. Of 400 fecal samples, E. coli O157 was found in 57 samples (14.2%). At feedlots L and V, E. coli O157 isolates were detected during every visit, and the frequencies of detection fluctuated from 10 to 40% and 30 to 65%, respectively. The overall prevalence rates of E. coli O157 for feedlots L and V over the 10-month period were 22.5 and 45%, respectively. Escherichia coli O157 was found at the cow-calf operation WB only in August, resulting in an overall prevalence of 1.4%. The occurrences of E. coli O157 at all three cattle facilities over the 10 collection periods are summarized in Table 1. Of 165 environmental samples, none were found to be positive for E. coli O157.
TABLE 1.
Characteristics of feedlots examined and prevalence (%) of E. coli O157 detected at each facility
| Feedlota | Diet | Capacity | No. of cattle | Ratio of no. of housed cattle to lot capacityb | Cattle density (no./100 m2)b | Prevalence (%) |
|---|---|---|---|---|---|---|
| Z | Concentrate | 2,000 | 1,000 | 0.50 | 4.9 | 22.5 |
| T | Concentrate | 6,000 | 5,000 | 0.83 | 4.8 | 10 |
| P | Concentrate | 3,500 | 1,600 | 0.45 | 6.5 | 42.5 |
| B | Concentrate | 8,000 | 3,000 | 0.37 | 6.7 | 30 |
| G | Concentrate | 3,000 | 200 | 0.06 | 3.6 | 0 |
| S | Concentrate | 3,500 | 3,500 | 1 | 7.4 | 57.5 |
| M | Concentrate | 4,000 | 4,000 | 1 | 5.8 | 55 |
| W | Concentrate | 3,500 | 1,500 | 0.42 | 4.3 | 5 |
| HM | Concentrate | 7,500 | 5,800 | 0.77 | 5.4 | 40 |
| PP | Pasture | 1,600 | 80 | ND | ND | 0 |
| MT | Concentrate | 4,000 | 2,500 | 0.62 | 5.9 | 32.5 |
| MO | Pasture | 800 | 70 | ND | ND | 0 |
| K | Concentrate | 3,400 | 1,300 | 0.38 | 5 | 15 |
| ML | Pasture | 1,000 | 120 | ND | ND | 0 |
| SC | Concentrate | 2,500 | 420 | 0.16 | 4.7 | 0 |
| H | Concentrate | 700 | 100 | 0.14 | 3 | 0 |
| WE | Concentrate | 3,600 | 1,300 | 0.36 | 6.5 | 17.5 |
| TK | Concentrate | 2,200 | 1,000 | 0.45 | 4.3 | 0 |
| SB | Pasture | 3,000 | 80 | ND | ND | 0 |
| I | Pasture | 1,500 | 500 | ND | ND | 2.5 |
| L* | Concentrate | 28,000 | 15,000 | 0.53 | 5.6 | 22.5 |
| V* | Concentrate | 17,000 | 12,000 | 0.70 | 5.8 | 45 |
| WB | Pasture | 1,200 | 300 | ND | ND | 1.4 |
Asterisks indicate feedlots examined during the longitudinal study. WB refers to a cattle research station examined during the longitudinal study. All remaining letter designations refer to different feedlots examined during the point study.
ND, not determined.
(ii) Point study.
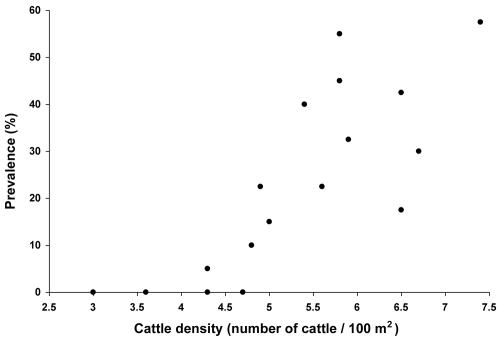
The prevalence rates of E. coli O157 in feedlots located throughout Saskatchewan are summarized in Table 1 and Fig. 1. A total of 800 samples were collected from 20 feedlots during this study. Of 20 feedlots, 12 were found to be positive for E. coli O157, and the prevalence rates of E. coli O157 ranged from 2.5 to 57.5%. A significant positive correlation (P = 0.003) was observed between the prevalence rate of E. coli O157 and the density of cattle in pens (Fig. 2, constructed using data obtained from both point prevalence and longitudinal studies). There was another statistically significant correlation (P = 0.006) found between the prevalence rate and the number of pens occupied by cattle within a feedlot (expressed as the ratio between the number of housed cattle and the feedlot capacity) (Table 1).
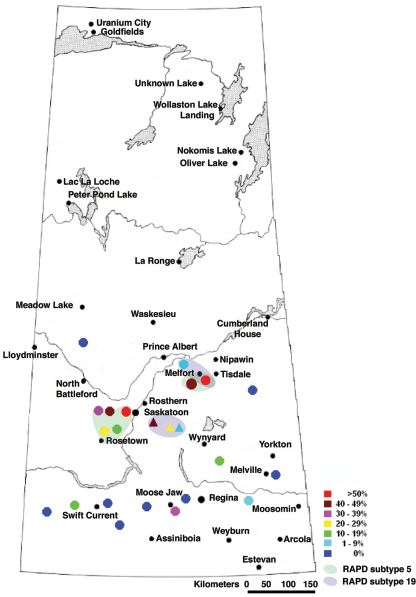
FIG. 1.
Map of Saskatchewan showing the prevalence rates of E. coli O157 detected (see color-coded legend) at feedlots/farms examined over a 2-year period. Triangles correspond to feedlots/farms examined during the longitudinal study, whereas circles represent feedlots examined during the point prevalence study. Note also the colored shaded regions showing the geographic locations of feedlots where E. coli O157 RAPD subtypes 5 and 19 were detected.
FIG. 2.
Relationship between the prevalence of E. coli O157 and the density of cattle (number of cattle/100 m2) housed at various feedlots (statistically significant [P = 0.003]).
Virulence profile of E. coli O157 isolates.
Multiplex PCR confirmed that all 194 serologically positive isolates (from both longitudinal and point studies) were indeed E. coli O157. These 194 E. coli O157 isolates exhibited five different virulence profiles. The majority of the isolates (161, or 82.9%) possessed all four virulence-associated genes (stx1, stx2, eaeA, and hlyA). The second most predominant virulence profile was a combination of the stx2, eaeA, and hlyA genes, with 22, or 11.4%, of the isolates having these genes. Six isolates (3.1%) possessed the stx1, eaeA, and hlyA genes, and four isolates (2.1%) were determined to have the stx1 and hlyA virulence markers. A single isolate (0.5%) had a unique virulence profile, with only the hlyA gene being present. Of the 194 E. coli O157 isolates obtained from across Saskatchewan over the 2-year study, 193 (99.4%) possessed at least one Shiga toxin gene and 187 (96.4%) had a combination of eaeA and stx1 and/or stx2, indicating a very high virulence potential of these isolates for humans.
Cytotoxicity.
The cytotoxicity effects of all 194 E. coli O157 isolates on Vero cells were observed, scaled from a value of 0 (no effect) to 10 (greatest effect), and compared to the reference strain E. coli O157 ATCC 43894, known to possess all four virulence determinants. All isolates from this study expressed cytotoxicities that ranged from 5.1 to 10. The majority of isolates (n = 107 [55%]) expressed very high toxicity values of 9.1 to 10; two other groups (n = 39 [20%] and n = 36 [19%]) had slightly lower cytotoxicities, ranging from 8.1 to 9.0 and 7.1 to 8.0, respectively. Interestingly, the lowest toxicities (5.1 to 6.0) were observed for isolate L-9 (longitudinal study), an isolate that possessed both Shiga toxins (data not shown). In contrast, isolate L-12, which possessed only the hlyA gene, had a somewhat greater toxic effect (7.1 to 8.0) (data not shown).
RAPD-PCR.
The panel of E. coli O157 isolates from the longitudinal and point studies was genetically characterized using the RAPD-PCR technique. All isolates were grouped into three major clusters that encompassed 39 different subtypes or RAPD-PCR patterns. Amplification reactions using primer 1283 produced a set of 18 DNA fragments ranging from 450 to 4,000 bp. Generally, all isolates displayed a basic total DNA banding pattern, with two doublets, migrating at 520 to 560 bp and 650 to 700 bp, one triplet at 1,400, 1,500, and 1,600 bp, and a final band at 2,900 bp. Genetic similarity between the E. coli O157 isolates ranged from 0.76 to 1.00. Fourteen isolates from nine feedlots (B, HM, M, P, MT, WE, K, V, and L) exhibited 14 distinct genetic patterns, whereas 11 other subtypes (3, 10, 12, 14, 18, 25, 29, 32, 33, 34, and 35) consisted of groups of isolates that were endemic to their feedlots. The two most predominant subtypes, 5 and 19, were detected at 6 feedlots each (12 total), among which 11 feedlots formed four distinct geographic clusters (each of <50 km in diameter) in central and north central Saskatchewan (Fig. 1).
Antibiotic susceptibility.
The antibiotic resistance patterns of the 131 isolates from the point prevalence study are shown in Table 2. Most of the 85 isolates (65%) were resistant to at least one antibiotic, whereas 46 isolates (35%) were sensitive to all 17 antibiotics tested. No resistance was observed for amikacin, ampicillin, amoxicillin-clavulanic acid, ceftriaxone, ciprofloxacin, cefoxitin, gentamicin, kanamycin, nalidixic acid, trimethoprim-sulfamethoxazole, and ceftiofur. The antibiotic for which resistance was most frequently observed was sulfisoxazole (61%), followed by tetracycline (12%). The prevalence rate of resistance to chloramphenicol and streptomycin among the E. coli O157 isolates was 2.3%. Two patterns of multidrug resistance, involving (i) chloramphenicol, sulfisoxazole, streptomycin, and tetracycline and (ii) sulfisoxazole and tetracycline, were observed in three (2.3%) and eight (6%) isolates, respectively (Table 2).
TABLE 2.
Characteristics of E. coli O157 isolates obtained from the point prevalence study
| Feedlot | Isolate | Isolation method
|
Virulence profile (presence of gene)
|
RAPD pattern | Antibiotic resistance | Cytotoxicity value (range) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| IMS | Culture | stx1 | stx2 | hlyA | eaeA | |||||
| Z | Z-6 | + | + | + | + | + | 1 | Sulfisoxazole | 9.1-10 | |
| Z-8 | + | + | + | + | + | 3 | Sulfisoxazole | 8.1-9.0 | ||
| Z-11 | + | + | + | + | + | + | 1 | Sulfisoxazole | 9.1-10 | |
| Z-14 | + | + | + | + | + | 1 | Sulfisoxazole | 8.1-9.0 | ||
| Z-19 | + | + | + | + | + | 3 | Sulfisoxazole | 8.1-9.0 | ||
| Z-27 | + | + | + | + | + | + | 5 | Sulfisoxazole | 9.1-10 | |
| Z-30 | + | + | + | + | + | + | 3 | Sulfisoxazole | 7.1-8.0 | |
| Z-37 | + | + | + | + | + | 3 | Sulfisoxazole | 7.1-8.0 | ||
| Z-38 | + | + | + | + | + | 5 | Sulfisoxazole | 8.1-9.0 | ||
| T | T-16 | + | + | + | + | + | 1 | Susceptible | 9.1-10 | |
| T-20a | + | |||||||||
| T-22 | + | + | + | + | + | 1 | Tetracycline | 8.1-9.0 | ||
| T-33 | + | + | + | + | + | 5 | Sulfisoxazole | 8.1-9.0 | ||
| T-36 | + | + | + | + | + | + | 5 | Sulfisoxazole | 9.1-10 | |
| B | B-1 | + | + | + | + | + | 5 | Susceptible | 7.1-8.0 | |
| B-3 | + | + | + | + | + | + | 5 | Tetracycline | 8.1-9.0 | |
| B-4 | + | + | + | + | + | 5 | Tetracycline | 9.1-10 | ||
| B-16 | + | + | + | + | + | 4 | Tetracycline | 9.1-10 | ||
| B-19 | + | + | + | + | + | + | 2 | Tetracycline | 9.1-10 | |
| B-22 | + | + | + | + | + | 6 | Susceptible | 8.1-9.0 | ||
| B-23 | + | + | + | + | + | 6 | Susceptible | 7.1-8.0 | ||
| B-27 | + | + | + | + | + | 6 | Susceptible | 9.1-10 | ||
| B-30 | + | + | + | + | 6 | Sulfisoxazole | 9.1-10 | |||
| B-33 | + | + | + | + | + | + | 6 | Sulfisoxazole | 9.1-10 | |
| B-36 | + | + | + | + | + | + | 6 | Sulfisoxazole, tetracycline | 9.1-10 | |
| B-38 | + | + | + | + | + | 6 | Sulfisoxazole, tetracycline | 7.1-8.0 | ||
| P | P-2 | + | + | + | + | 6 | Susceptible | 7.1-8.0 | ||
| P-3 | + | + | + | + | + | 6 | Sulfisoxazole | 9.1-10 | ||
| P-4 | + | + | + | + | + | 16 | Susceptible | 9.1-10 | ||
| P-5 | + | + | + | + | + | 38 | Sulfisoxazole | 9.1-10 | ||
| P-7 | + | + | + | + | + | 6 | Sulfisoxazole | 9.1-10 | ||
| P-9 | + | + | + | + | + | 18 | Sulfisoxazole | 8.1-9.0 | ||
| P-11 | + | + | + | + | + | 17 | Sulfisoxazole | 9.1-10 | ||
| P-13 | + | + | + | + | + | 18 | Sulfisoxazole | 9.1-10 | ||
| P-16 | + | + | + | + | 17 | Susceptible | 6.1-7.0 | |||
| P-21 | + | + | + | + | + | + | 18 | Sulfisoxazole | 8.1-9.0 | |
| P-26 | + | + | + | + | 17 | Susceptible | 9.1-10 | |||
| P-27 | + | + | + | + | 17 | Sulfisoxazole | 9.1-10 | |||
| P-28 | + | + | + | + | + | 17 | Sulfisoxazole | 9.1-10 | ||
| P-29 | + | + | + | + | + | 18 | Sulfisoxazole | 9.1-10 | ||
| P-36 | + | + | + | + | + | 18 | Sulfisoxazole | 9.1-10 | ||
| P-37 | + | + | + | + | + | 18 | Sulfisoxazole | 9.1-10 | ||
| P-40 | + | + | + | + | 17 | Sulfisoxazole | 9.1-10 | |||
| S | S-1a | + | + | |||||||
| S-4 | + | + | + | + | + | 5 | Susceptible | 9.1-10 | ||
| S-5 | + | + | + | + | + | + | 5 | Susceptible | 9.1-10 | |
| S-6 | + | + | + | + | + | 13 | Chloramphenicol, tetracycline, streptomycin, sulfisoxazole | 7.1-8.0 | ||
| S-7 | + | + | + | + | 5 | Susceptible | 7.1-8.0 | |||
| S-8 | + | + | + | + | + | + | 5 | Susceptible | 6.1-7.0 | |
| S-10 | + | + | + | + | + | + | 5 | Susceptible | 6.1-7.0 | |
| S-11 | + | + | + | + | + | 5 | Susceptible | 7.1-8.0 | ||
| S-14 | + | + | + | + | + | + | 5 | Susceptible | 6.1-7.0 | |
| S-16 | + | + | + | + | + | + | 5 | Susceptible | 6.1-7.0 | |
| S-17 | + | + | + | + | + | + | 5 | Susceptible | 8.1-9.0 | |
| S-19 | + | + | + | + | + | 5 | Susceptible | 7.1-8.0 | ||
| S-26 | + | + | + | + | + | + | 7 | Susceptible | 8.1-9.0 | |
| S-27 | + | + | + | + | + | 5 | Susceptible | 9.1-10 | ||
| S-28 | + | + | + | + | 5 | Susceptible | 7.1-8.0 | |||
| S-30 | + | + | + | + | + | 5 | Sulfisoxazole | 7.1-8.0 | ||
| S-32 | + | + | + | + | + | + | 5 | Sulfisoxazole | 8.1-9.0 | |
| S-33 | + | + | + | + | + | + | 5 | Susceptible | 7.1-8.0 | |
| S-34 | + | + | + | + | + | 5 | Susceptible | 9.1-10 | ||
| S-37 | + | + | + | + | + | + | 7 | Susceptible | 8.1-9.0 | |
| S-38 | + | + | + | + | 13 | Chloramphenicol, tetracycline, streptomycin, sulfisoxazole | 8.1-9.0 | |||
| S-39 | + | + | + | + | 5 | Sulfisoxazole | 8.1-9.0 | |||
| S-40 | + | + | + | + | + | 5 | Sulfisoxazole | 7.1-8.0 | ||
| M | M-2 | + | + | + | + | + | 7 | Susceptible | 9.1-10 | |
| M-4 | + | + | + | + | + | 14 | Susceptible | 9.1-10 | ||
| M-5 | + | + | + | + | 14 | Susceptible | 8.1-9.0 | |||
| M-6 | + | + | + | + | + | 5 | Sulfisoxazole | 9.1-10 | ||
| M-8 | + | + | + | + | 5 | Chloramphenicol, tetracycline, streptomycin, sulfisoxazole | 9.1-10 | |||
| M-9 | + | + | + | + | 5 | Susceptible | 9.1-10 | |||
| M-13 | + | + | + | + | 7 | Susceptible | 9.1-10 | |||
| M-14 | + | + | + | + | 5 | Sulfisoxazole | 7.1-8.0 | |||
| M-15 | + | + | + | + | 10 | Susceptible | 9.1-10 | |||
| M-16 | + | + | + | + | 7 | Susceptible | 6.1-7.0 | |||
| M-17 | + | + | + | + | + | 10 | Sulfisoxazole | 9.1-10 | ||
| M-19 | + | + | + | + | + | 11 | Sulfisoxazole | 9.1-10 | ||
| M-21 | + | + | + | + | + | 9 | Sulfisoxazole | 9.1-10 | ||
| M-22 | + | + | + | + | + | + | 19 | Susceptible | 9.1-10 | |
| M-23 | + | + | + | + | + | 39 | Susceptible | 8.1-9.0 | ||
| M-26 | + | + | + | + | + | 19 | Sulfisoxazole | 7.1-8.0 | ||
| M-27 | + | + | + | + | + | 14 | Sulfisoxazole | 8.1-9.0 | ||
| M-29 | + | + | + | + | 19 | Sulfisoxazole | 8.1-9.0 | |||
| M-31 | + | + | + | + | + | 10 | Sulfisoxazole | 9.1-10 | ||
| M-33 | + | + | + | + | 11 | Susceptible | 9.1-10 | |||
| M-34 | + | + | + | + | + | 15 | Susceptible | 9.1-10 | ||
| M-40 | + | + | + | + | + | 11 | Sulfisoxazole | 7.1-8.0 | ||
| W | W-12 | + | + | + | + | + | + | 19 | Sulfisoxazole | 7.1-8.0 |
| W-37 | + | + | + | + | + | + | 19 | Sulfisoxazole | 9.1-10 | |
| HM | HM-3 | + | + | + | + | + | + | 12 | Sulfisoxazole | 7.1-8.0 |
| HM-6 | + | + | + | + | + | 11 | Susceptible | 6.1-7.0 | ||
| HM-8 | + | + | + | + | + | 11 | Susceptible | 8.1-9.0 | ||
| HM-11 | + | + | + | + | + | 11 | Sulfisoxazole | 8.1-9.0 | ||
| HM-13 | + | + | + | + | + | 11 | Sulfisoxazole | 9.1-10 | ||
| HM-14 | + | + | + | + | + | + | 11 | Sulfisoxazole | 8.1-9.0 | |
| HM-19 | + | + | + | + | + | + | 11 | Sulfisoxazole | 9.1-10 | |
| HM-20 | + | + | + | + | + | + | 11 | Sulfisoxazole | 7.1-8.0 | |
| HM-22 | + | + | + | + | + | + | 11 | Sulfisoxazole | 8.1-9.0 | |
| HM-23 | + | + | + | + | + | 11 | Sulfisoxazole | 9.1-10 | ||
| HM-25 | + | + | + | + | + | + | 5 | Sulfisoxazole | 9.1-10 | |
| HM-26 | + | + | + | + | + | 12 | Sulfisoxazole | 9.1-10 | ||
| HM-27 | + | + | + | + | + | + | 8 | Sulfisoxazole | 8.1-9.0 | |
| HM-28 | + | + | + | + | + | 12 | Sulfisoxazole | 8.1-9.0 | ||
| HM-34 | + | + | + | + | + | 12 | Sulfisoxazole | 7.1-8.0 | ||
| HM-35 | + | + | + | + | + | + | 12 | Sulfisoxazole | 9.1-10 | |
| MT | MT-2 | + | + | + | + | + | + | 23 | Sulfisoxazole | 9.1-10 |
| MT-3 | + | + | + | + | + | + | 25 | Sulfisoxazole | 9.1-10 | |
| MT-4 | + | + | + | + | + | 25 | Sulfisoxazole | 9.1-10 | ||
| MT-6 | + | + | + | + | + | 25 | Sulfisoxazole | 9.1-10 | ||
| MT-7 | + | + | + | + | + | 25 | Susceptible | 9.1-10 | ||
| MT-9 | + | + | + | + | + | 26 | Susceptible | 9.1-10 | ||
| MT-17 | + | + | + | + | + | + | 26 | Susceptible | 9.1-10 | |
| MT-18 | + | + | + | + | + | + | 25 | Sulfisoxazole | 9.1-10 | |
| MT-21 | + | + | + | + | + | + | 17 | Susceptible | 9.1-10 | |
| MT-25 | + | + | + | + | + | 2 | Susceptible | 9.1-10 | ||
| MT-30 | + | + | + | + | + | + | 25 | Susceptible | 9.1-10 | |
| MT-34 | + | + | + | + | + | 25 | Sulfisoxazole | 9.1-10 | ||
| MT-38 | + | + | + | + | + | + | 25 | Susceptible | 9.1-10 | |
| K | K-4 | + | + | + | + | + | 28 | Tetracycline, sulfisoxazole | 9.1-10 | |
| K-5 | + | + | + | + | + | 29 | Tetracycline, sulfisoxazole | 9.1-10 | ||
| K-7 | + | + | + | + | + | 29 | Tetracycline, sulfisoxazole | 9.1-10 | ||
| K-11 | + | + | + | + | + | + | 29 | Tetracycline, sulfisoxazole | 9.1-10 | |
| K-12 | + | + | + | + | + | 29 | Tetracycline, sulfisoxazole | 9.1-10 | ||
| K-28 | + | + | + | + | + | 29 | Tetracycline, sulfisoxazole | 8.1-9.0 | ||
| WE | WE-4 | + | + | + | + | + | 26 | Sulfisoxazole | 8.1-9.0 | |
| WE-14 | + | + | + | + | + | + | 26 | Sulfisoxazole | 9.1-10 | |
| WE-18 | + | + | + | + | + | 26 | Sulfisoxazole | 9.1-10 | ||
| WE-23 | + | + | + | + | + | + | 19 | Sulfisoxazole | 9.1-10 | |
| WE-27 | + | + | + | + | + | 26 | Sulfisoxazole | 9.1-10 | ||
| WE-28 | + | + | + | + | + | 26 | Sulfisoxazole | 9.1-10 | ||
| WE-29 | + | + | + | + | + | 24 | Sulfisoxazole | 9.1-10 | ||
| I | I-31 | + | + | + | + | + | + | 17 | Susceptible | 9.1-10 |
Could not be subcultured after initial recovery.
DISCUSSION
The combined prevalence rate of E. coli O157 (15.6%) for the longitudinal and point prevalence studies was slightly higher than that in a recent study conducted in Canada (23). However, it is generally difficult, and sometimes impossible, to extrapolate results obtained from different studies due to the large variety of screening procedures for E. coli O157 surveys. As reported by Sanderson et al. (43), using large-volume (10-g) fecal samples increased the detection sensitivity by 5.4% over that for cotton-tipped swabbing of fecal samples. To increase the survey sensitivity, we tested two small fecal samples per sampling unit in parallel by using two different enrichment steps, with and without IMS. Our combined approach (which involved using both culture and culture-IMS methods) increased the sensitivities of E. coli O157 detection in the longitudinal and point studies by 19.3% and 25.2%, respectively, over the use of culture-IMS alone. The results from this study confirmed the recent findings of Pearce et al. (38), who noted a marked improvement in the sensitivity of detection via testing multiple small fecal samples per fecal sampling unit.
The frequencies of occurrence of E. coli O157 among surveyed feedlots showed a high degree of variation, ranging from 0 to 57.5%, without any geographic distribution pattern (Fig. 1). Two determinants were found to have an important impact on the prevalence of E. coli O157 in Saskatchewan feedlots. Statistically, the most significant correlation (P = 0.003) was observed between the density of cattle in pens (Fig. 2) and the prevalence rate of E. coli O157. The second statistically significant correlation (P = 0.006) was found between the ratio of housed cattle to feedlot capacity and the prevalence of the pathogen (Table 1). The effects of these two densities on the prevalence of E. coli O157 among feedlot cattle may be illustrated by an example. Feedlots S and M, which operated at full capacity, with 13.6 and 17.3 m2/head, respectively, resulted in the highest prevalence rates (57.5 and 55%, respectively). In contrast, feedlots H and G, which operated at 14 and 6% of capacity (33.2 and 27.8 m2/head), respectively, resulted in no detectable E. coli O157 samples. Performing stepwise regression statistical analysis, we found that the percentage of variance (R2) in the prevalence explained by the effect of cattle density in pens alone was 62.6%, whereas the percentage of variance in the prevalence explained by both densities was 78.7%. These results suggest that both densities have an additive effect on pathogen spread among feedlot cattle. High pen densities increase contact between infected and noninfected animals as well as pose a stressful condition for cattle, resulting in a higher shedding rate of the pathogen. Increases in the other density-related factor (the ratio of number of housed cattle to feedlot capacity) most likely accelerate the dissemination of E. coli O157 between pens via some other mechanism (air dust, runoff, etc.). To avoid a diet effect on the analyses of the prevalence distribution rates, all results obtained from pasture-fed cattle were not statistically processed.
Results from multiplex PCR analyses indicated that the majority of E. coli O157 isolates obtained from this study likely represent a significant public health risk. Several authors have previously pointed out that enterohemorrhagic E. coli (EHEC) isolates able to produce functional stx2-encoded toxin and intimin are strongly associated with human morbidity (6, 14, 36). Of 194 E. coli O157 isolates recovered from Saskatchewan cattle, 94% (n = 183) harbored both the eaeA and stx2 genes, two of the most potent EHEC virulence determinants. Two recent studies carried out in Australia (13) and at three large Midwestern beef processing plants in the United States (3) reported prevalence rates of 56% and 64%, respectively, for E. coli O157 isolates with both stx1 and stx2, whereas in the present study, 83% of the isolates carried the same combination of toxins. These results suggest that the virulence potential of isolates from Saskatchewan cattle herds is relatively high.
A Vero cell assay confirmed this assumption, as all E. coli O157 isolates expressed a strong cytopathic effect against Vero cells. We further observed high cytotoxicities, ranging from 7.1 to 8.0, for one stx1/stx2-negative isolate (data not shown). This paradoxical situation has been observed elsewhere (19, 28) and suggests that the pathogenic mechanisms of E. coli O157 are not completely understood.
Based on the genetic diversity that arose from the E. coli O157 isolates, we classified Saskatchewan's feedlots into three categories. The first category represented feedlots having an indigenous population of E. coli O157, where no genetically divergent isolates were observed to be present. The RAPD-PCR patterns of E. coli O157 isolates recovered from the K feedlot displayed only two unique genetic subtypes (subtypes 28 and 29) (Table 2), indicating their endemic character. The majority of feedlots (Z, HM, P, S, L, and V) were classified into a second category, where genetically more or less unrelated isolates were introduced and combined with a group of endemic isolates. It is possible that genetically more related isolates evolved from an existing endemic population. The third observed category of feedlots included those with large numbers of genetically unrelated E. coli O157 subtypes, implying that genetically diverse isolates originated from different ancestors (22) and were introduced into the feedlot, most probably by incoming cattle. Feedlot M, which included nine different genetic subtypes (5, 7, 9, 10, 11, 14, 15, 19, and 39) that were found in all three clusters, provides an example of this.
The use of antibiotics in food-producing animals for growth promotion, chemotherapy, and prophylaxis is linked to the emergence and dissemination of antibiotic resistance genes (49). Although Shiga toxin-producing E. coli infections do not involve the use of antimicrobial therapy (8, 50), E. coli O157 may easily acquire drug resistance genes in its natural habitat and transfer them to other organisms of clinical relevance. In this study, we provide insight into the antibiotic profiles of E. coli O157 isolates collected over a considerable geographic area that comprised a cattle population originating from across Canada and the United States. The most prevalent antibiotic determinant was sulfisoxazole resistance. Our RAPD-PCR results suggested that resistance to this drug is widely distributed among 22 different subtypes of E. coli O157 found in all three genetic clusters. The ubiquitous dissemination of sulfisoxazole resistance among genetically diverse E. coli O157 isolates clearly suggests the convergent acquisition of drug resistance genes by these different isolates rather than the clonal spread of a sulfisoxazole-resistant isolate across this geographic region. The high rate of resistance to sulfisoxazole may be attributed to the long history of sulfa drug use (since the mid-1930s) in the treatment of infectious diseases. Furthermore, the sul gene that encodes dihydropteroate (sulfonamide inhibitor) has been found within the conserved segments of several types of integrons (9, 26), implying the effective flux of sul in the microbial population. The prevalence rate of tetracycline resistance (12%) in E. coli O157 isolates from this study was similar to the findings of another study carried out in the United States (44). Based on an examination of 361 E. coli O157 isolates recovered from humans, cattle, swine, and food, Schroeder et al. (44) observed 9% resistance to tetracycline in STEC O157 organisms. Three isolates obtained during this study (S-6, S-38, and M-8) from two separate feedlots were found to have multiantibiotic resistance to chloramphenicol, tetracycline, streptomycin, and sulfisoxazole. The same virulence profiles and multidrug resistance traits of these organisms could indicate epidemic spread of a multidrug-resistant organism through herds in this region. Based on genetic typing, isolates S-6 and S-38 comprised a distinct genetic subtype (subtype 13), confirming the above-mentioned indication. In contrast, isolate M-8 was genetically different from isolates S-6 and S-38. Interestingly, isolate M-8 had the same RAPD-PCR pattern (subtype 5) as most isolates from feedlot S, indicating a possible ancestral source of multidrug resistance determinants for all three isolates. Most of the surveyed feedlots used the ionophore antibiotic monensin (Rumensin) (personal communication). Monensin inhibits gram-positive microfloras that tend to proliferate under concentrated (high starch) diets, and because of its general mode of action (at the level of the cell membrane), acquired mechanisms of resistance are doubtful. The use of monensin is not likely to have contributed to any of the antibiotic resistance profiles observed in this study.
In conclusion, this study clearly demonstrates a high impact of cattle density and the ratio between the number of housed cattle and feedlot capacity on the prevalence rate of E. coli O157 and thus may serve as a baseline for risk assessment modeling and reducing the prevalence rate of E. coli O157 among cattle in feedlots. Furthermore, in addition to providing current insight into the antibiotic susceptibilities of E. coli O157 isolates originating from across Canada and the United States, this work also illustrates how combining antibiotic resistance and genetic profiles provides a more comprehensive basis for comparing the relatedness and dissemination of pathogenic (resistant) E. coli strains.
Acknowledgments
The Beef Cattle Research Council, NSERC, and NSERC-CRD are acknowledged for financial support.
We also thank Gord Crockford, Andrew Potter, and Philip Willson (VIDO) for technical and laboratory support.
REFERENCES
- 1.Akopyanz, N., N. O. Bukanov, T. U. Westblom, S. Kresovich, and D. E. Berg. 1992. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based fingerprinting. Nucleic Acids Res. 20:5137-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aslam, M., F. Nattress, G. Greer, C. Yost, C. Gill, and M. McMullen. 2003. Origin of contamination and genetic diversity of Escherichia coli in beef cattle. Appl. Environ. Microbiol. 69:2794-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barkocy-Gallagher, G. A., T. M. Arthur, M. Rivera-Betancourt, X. Nou, S. D. Shackelford, T. L. Wheeler, and M. Koohmaraie. 2004. Characterization of O157:H7 and other Escherichia coli isolates recovered from cattle hides, feces, and carcasses. J. Food Prot. 67:993-998. [DOI] [PubMed] [Google Scholar]
- 4.Belongia, E. A., M. T. Osterholm, J. T. Soler, D. A. Ammend, J. E. Braun, and K. L. MacDonald. 1993. Transmission of Escherichia coli O157:H7 infection in Minnesota child day-care facilities. JAMA 269:883-888. [PubMed] [Google Scholar]
- 5.Besser, R. E., M. S. Lett, J. T. Weber, M. P. Doyle, T. J. Barrett, J. G. Wells, and P. M. Griffin. 1993. An outbreak of diarrhea and hemolytic uremic syndrome from Escherichia coli O157:H7 in fresh pressed apple cider. JAMA 269:2217-2220. [PubMed] [Google Scholar]
- 6.Boerlin, P., S. A. Mcewen, F. Boerlin-Petzold, J. B. Wilson, R. P. Johnson, and C. L. Gyles. 1999. Association between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J. Clin. Microbiol. 37:497-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapman, P. A., D. J. Wright, and C. A. Siddons. 1994. A comparison of immunomagnetic separation and direct culture for the isolation of verocytotoxin-producing Escherichia coli O157 from bovine faeces. J. Med. Microbiol. 40:424-427. [DOI] [PubMed] [Google Scholar]
- 8.Cimolai, N., J. E. Carter, B. J. Morrison, and J. D. Anderson. 1990. Risk factors for the progression of Escherichia coli O157:H7 enteritis to hemolytic-uremic syndrome. J. Pediatr. 116:1496-1500. [DOI] [PubMed] [Google Scholar]
- 9.Davies, J. 1994. Inactivation of antibiotics and the dissemination of resistance genes. Science 264:375-382. [DOI] [PubMed] [Google Scholar]
- 10.Dev, V. J., M. Main, and I. Gould. 1991. Waterborne outbreak of Escherichia coli O157. Lancet 337:1412. [DOI] [PubMed] [Google Scholar]
- 11.Djordjevic, S. P., V. Ramachandran, K. A. Bettelheim, B. A. Vanselow, P. Holst, G. Bailey, and M. A. Hornitzky. 2004. Serotypes and virulence gene profiles of Shiga toxin-producing Escherichia coli strains isolated from feces of pasture-fed and lot-fed sheep. Appl. Environ. Microbiol. 70:3910-3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fagan, P. K., M. A. Hornitzky, K. A. Bettelheim, and S. P. Djoredjevic. 1999. Detection of Shiga-like toxin (stx1 and stx2), intimin (eaeA), and enterohemorrhagic Escherichia coli (EHEC) hemolysin (EHEC hlyA) genes in animal feces by multiplex PCR. Appl. Environ. Microbiol. 65:868-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fegan, N., P. Vanderlinde, G. Higgs, and P. Desmarchelier. 2004. The prevalence and concentration of Escherichia coli O157 in faeces of cattle from different production systems at slaughter. J. Appl. Microbiol. 97:362-370. [DOI] [PubMed] [Google Scholar]
- 14.Friedrich, A., M. Bielaszewska, W. L. Zhang, M. Pulz, T. Kuczius, A. Ammon, and H. Karch. 2002. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185:74-84. [DOI] [PubMed] [Google Scholar]
- 15.Galland, G. J., D. R. Hyatt, S. S. Crupper, and D. W. Acheson. 2001. Prevalence, antibiotic susceptibility, and diversity of Escherichia coli O157:H7 isolates from a longitudinal study of beef cattle feedlots. Appl. Environ. Microbiol. 67:1619-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffin, P. M., B. P. Bell, P. R. Cieslak, J. Tuttle, T. J. Barrett, M. P. Doyle, A. M. McNamara, A. M. Shefer, and J. G. Wells. 1994. Large outbreak of Escherichia coli O157:H7 infections in the western United States, p. 7-12. In M. A. Karmali and A. G. Goglio (ed.), Recent advances in verocytotoxin-producing Escherichia coli infections. Elsevier Science B.V., Amsterdam, The Netherlands.
- 17.Hancock, D. D., T. E. Besser, D. H. Rice, D. E. Herriott, and P. I. Tarr. 1997. Longitudinal study of Escherichia coli O157 in fourteen cattle herds. Epidemiol. Infect. 118:193-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janda, J. M., and S. L. Abbott. 1998. Escherichia coli, p. 13-60. In J. M. Janda and S. L. Abbott (ed.), The enterobacteria. Lippincott-Raven Publishers, Philadelphia, Pa.
- 19.Jo, M. J., J. H. Kim, J. H. Lim, M. Y. Kang, H. B. Koh, Y. H. Park, D. Y. Yoon, J. S. Chae, S. K. Eo, and J. H. Lee. 2004. Prevalence and characteristics of Escherichia coli O157 from major food animals in Korea. Int. J. Food Microbiol. 95:41-49. [DOI] [PubMed] [Google Scholar]
- 20.Kaper, J. B., and A. D. O'Brien. 1998. Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains, p. 1-11. American Society for Microbiology, Washington, D.C.
- 21.Karmali, M. A. 1989. Infection by verotoxin-producing Escherichia coli. Clin. Microbiol. Rev. 2:15-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, J., J. Nietfeldt, J. Ju, J. Wise, N. Fegan, P. Desmarchelier, and A. K. Benson. 2001. Ancestral divergence, genome diversification, and phylogeographic variation in subpopulations of sorbitol-negative, β-glucuronidase-negative enterohemorrhagic Escherichia coli O157. J. Bacteriol. 183:6885-6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LeJeune, J. T., T. E. Besser, D. H. Rice, J. H. Berg, R. P. Stilborn, and D. D. Hancock. 2004. Longitudinal study of fecal shedding of Escherichia coli O157:H7 in feedlot cattle: predominance and persistence of specific clonal types despite massive cattle population turnover. Appl. Environ. Microbiol. 70:377-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leyer, G. J., L. L. Wang, and E. A. Johnson. 1995. Acid adaptation of Escherichia coli O157:H7 increases survival in acidic foods. Appl. Environ. Microbiol. 61:3752-3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin, J., M. P. Smith, K. C. Chapin, H. S. Baik, G. N. Bennett, and J. W. Foster. 1996. Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl. Environ. Microbiol. 62:3094-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maguire, A. J., D. F. J. Brown, J. J. Gray, and U. Desselberger. 2001. Rapid screening technique for class 1 integrons in Enterobacteriaceae and nonfermenting gram-negative bacteria and its use in molecular epidemiology. Antimicrob. Agents Chemother. 45:1022-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makino, S., Y. Okada, T. Maruyama, S. Kaneko, and C. Saskawa. 1994. PCR-based random amplified polymorphic DNA fingerprinting of Yersinia pseuodotuberculosis and its practical applications. J. Clin. Microbiol. 32:65-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maldonado, Y., J. C. Fiser, H. C. Nakatsu, and A. K. Bhunia. 2005. Cytotoxicity potential and genotypic characterization of Escherichia coli isolates from environmental and food sources. Appl. Environ. Microbiol. 71:1890-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.National Committee for Clinical Laboratory Standards. 2002. Performance standards for antimicrobial susceptibility testing. Twelfth informational supplement M100-S12. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 32.National Committee for Clinical Laboratory Standards. 2002. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. Approved standard M31-A2. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 33.O'Brien, A. D., V. L. Tesh, R. A. Donohue, M. P. Jackson, S. Olsnes, K. Sandving, A. A. Lindberg, and G. T. Keusch. 1992. Shiga toxin: biochemistry, genetics, mode of action and role in pathogenesis. Curr. Top. Microbiol. Immunol. 180:65-94. [DOI] [PubMed] [Google Scholar]
- 34.Pacheco, A. B. F., B. E. C. Guth, K. C. C. Soares, L. Nishimura, D. F. De Almeida, and L. C. S. Ferreira. 1997. Random amplification of polymorphic DNA reveals serotype-specific clonal clusters among enterotoxigenic Escherichia coli strains isolated from humans. J. Clin. Microbiol. 35:1521-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paton, A. W., and J. C. Paton. 1998. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbo111, and rfbo157. J. Clin. Microbiol. 36:598-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paton, A. W., R. Ratcliff, R. M. Doyle, J. Seymour-Murray, D. Davos, J. A. Lanser, and J. C. Paton. 1996. Molecular microbiological investigation of an outbreak of hemolytic uremic syndrome caused by dry fermented sausage contaminated with Shiga-like toxin-producing Escherichia coli. J. Clin. Microbiol. 34:1622-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paton, J. C., and A. W. Paton. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11:450-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pearce, M. C., D. Fenlon, J. C. Low, A. W. Smith, H. I. Knight, J. Evans, G. Foster, B. A. Synge, and G. J. Gunn. 2004. Distribution of Escherichia coli O157 in bovine fecal pats and its impact on estimates of the prevalence of fecal shedding. Appl. Environ. Microbiol. 70:5737-5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Radu, S., W. O. Ling, G. Rusul, A. I. M. Karim, and M. Nishibuchi. 2001. Detection of Escherichia coli O157:H7 by PCR and their characterization by plasmid profiling, antimicrobial resistance, RAPD and PFGE analyses. J. Microbiol. Methods 46:131-139. [DOI] [PubMed] [Google Scholar]
- 40.Renwick, S. A., J. B. Wilson, and R. C. Clarke. 1993. Evidence of direct transmission of Escherichia coli O157:H7 infection between calves and a human. J. Infect. Dis. 168:792-793. [DOI] [PubMed] [Google Scholar]
- 41.Riley, L. W., R. S. Remis, S. D. Helgerson, H. B. McGee, J. G. Wells, B. R. Davis, R. J. Hebert, E. S. Olcott, L. M. Johnson, N. T. Hargrett, P. A. Blake, and M. L. Cohen. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 308:681-685. [DOI] [PubMed] [Google Scholar]
- 42.Roberts, H. P., K. C. Davis, W. R. Garstka, and A. K. Bhunia. 2001. Lactate dehydrogenase release assay from Vero cells to distinguish verotoxin producing Escherichia coli from non-verotoxin producing strains. J. Microbiol. Methods 43:171-181. [DOI] [PubMed] [Google Scholar]
- 43.Sanderson, M. W., J. M. Gay, D. D. Hancock, C. C. Gay, L. K. Fox, and T. E. Besser. 1995. Sensitivity of bacteriologic culture for detection of Escherichia coli O157:H7 in bovine feces. J. Clin. Microbiol. 33:2616-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schroeder, C. M., C. Zhao, C. DebRoy, J. Torcolini, S. Zhao, D. G. White, D. D. Wagner, P. F. McDermott, R. D. Walker, and J. Meng. 2002. Antimicrobial resistance of Escherichia coli O157 isolated from humans, cattle, swine, and food. Appl. Environ. Microbiol. 68:576-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shere, J. A., K. J. Bartlett, and C. W. Kaspar. 1998. Longitudinal study of Escherichia coli O157:H7 dissemination on four dairy farms in Wisconsin. Appl. Environ. Microbiol. 64:1390-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sherman, P., R. Soni, and M. Karmali. 1988. Attaching and effacing adherence of Vero cytotoxin-producing Escherichia coli to rabbit intestinal epithelium in vivo. Infect. Immun. 56:756-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stewart, P. J., W. Desormeaux, and J. Chene. 1983. Hemorrhagic colitis in a home for the aged: Ontario. Can. Dis. Wkly. Rep. 9:29-32. [Google Scholar]
- 48.Su, C., and L. J. Brandt. 1995. Escherichia coli O157:H7 infection in humans. Ann. Intern. Med. 123:698-714. [DOI] [PubMed] [Google Scholar]
- 49.Witte, W. 1998. Medical consequences of antibiotic use in agriculture. Science 279:996-997. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, X., A. D. McDaniel, L. E. Wolf, G. T. Keusch, M. K. Waldor, and D. W. K. Acheson. 2000. Quinolone antibiotics induce Shiga toxin-encoding bacteriophages, toxin production, and death in mice. J. Infect. Dis. 181:664-670. [DOI] [PubMed] [Google Scholar]