Abstract
This paper compares five commercially available DNA extraction methods with respect to DNA extraction efficiency of Salmonella enterica serovar Enteritidis from soil, manure, and compost and uses an Escherichia coli strain harboring a plasmid expressing green fluorescent protein as a general internal procedural control. Inclusion of this general internal procedural control permitted more accurate quantification of extraction and amplification of S. enterica serovar Enteritidis in these samples and reduced the possibility of false negatives. With this protocol it was found that the optimal extraction method differed for soil (Mobio soil DNA extraction kit), manure (Bio101 soil DNA extraction kit), and compost (Mobio fecal DNA extraction kit). With each method, as little as 1.2 × 103 to 1.8 × 103 CFU of added serovar Enteritidis per 100 mg of substrate could be detected by direct DNA extraction and subsequent S. enterica-specific TaqMan PCR. After bacterial enrichment, as little as 1 CFU/100 mg of original substrate was detected. Finally, the study presents a more accurate molecular analysis for quantification of serovar Enteritidis initially present in soil or manure using DNA extraction and TaqMan PCR.
Environmental substrates like manure and soil have become a major concern with respect to food safety, since these substrates are suspected to play a major role in the introduction of human pathogens in the food chain (23, 29, 34). For example, salmonellae are frequently found in association with animal manure (11, 18, 23, 24), which is often applied as fertilizer to soil prior to vegetable production, thereby introducing a potential risk of contamination of vegetables grown in the manure-amended soil. This threat is evident from the fact that during recent years, the consumption of raw vegetables has been related to food-borne outbreaks (4, 7, 9, 14, 20, 21, 27). For example, each year 3.5 million cases of salmonellosis occur in the United States and Canada, leading to economic losses of up to 3.4 billion dollars a year (32). Therefore, the detection and quantification of pathogens like Salmonella enterica that are present in environmental substrates like soil, manure, and compost are of high importance. It will enable risk assessment and pathogen monitoring at different stages in the plant production chain and ensure food health and safety in the food industry.
Standardized diagnostic procedures to detect the presence of S. enterica in food samples (ISO 6579:2002) are mainly based on microbiological culturing methods, which in general require up to 5 days until results are obtained (30). In order to reduce the time demand, alternative techniques like immunological assays (1, 6) and molecular methods (5, 8, 12) have been applied to detect S. enterica in various samples. Especially real-time PCR methods, such as 5′ nuclease TaqMan PCR (15), have shown promising results due to the rapid, sensitive, and specific detection of S. enterica (3, 13, 16, 17).
However, molecular methods like (TaqMan) PCR are limited by the fact that they are dependent on the suitability of the extracted DNA for PCR (36). DNA extracted from soil, manure, or compost, in particular, can have coextracted contaminants, like humic and fulvic acids, known to cause problems during PCR amplification (2, 10). Other components besides humic and fulvic acids that are also commonly present in soil have been related to PCR inhibition (35). Moreover, the large variation in biochemical components between different substrates (2, 36) usually leads to variable efficiencies of DNA extraction methods (19, 31). Due to these deficiencies, the accurate quantification of pathogens present in different environmental substrates has not yet been accomplished using molecular techniques such as PCR.
To control the effects of inhibiting agents on PCR amplification efficiency, TaqMan PCR was improved recently by the introduction of a general internal amplification control to prevent the occurrence of false-negative results (16, 17, 26). Although this improvement provided progress in the analysis of extracted DNA from environmental substrates, a comparison between the DNA extraction efficiencies of different DNA extraction methods has been described only to a minor extent. Zhou et al. (37) investigated DNA recovery from different soils, but only one DNA extraction method was used. Another paper described a comparison of three different DNA extraction methods, evaluating the quality and quantity of DNA recovered from four soils with widely differing characteristics but not from manure or compost (19).
The objectives of this study were to evaluate five commercial DNA extraction methods with respect to DNA extraction efficiency from soil, manure, and compost. In addition, the development and application of a general internal procedural control was investigated with respect to the efficiency of the DNA extraction and TaqMan PCR amplification procedure. Moreover, the possibility of a more accurate quantification of S. enterica serovar Enteritidis from different substrates by using a general internal procedural control (GIPC) was evaluated.
MATERIALS AND METHODS
Bacterial strains and environmental substrates.
A liquid culture of S. enterica serovar Enteritidis ATCC 13076, grown overnight at 30°C in tryptic soy broth, was kindly provided by H. Aarts (RIKILT, Wageningen, The Netherlands). A bacterial culture of the genetically modified Escherichia coli strain 99507gfp, containing plasmid pVSP61TIR (22) carrying the green fluorescent protein gene (gfp) (25), was kindly provided on solid LB medium by R. Sayler (28). Each E. coli strain 99507gfp CFU contains approximately 30 plasmids with a coding sequence for GFP expression. A subculture of E. coli strain 99507gfp was grown overnight at 37°C in liquid broth medium containing 50 μg/ml ampicillin. The bacterial suspension was diluted in 50% glycerol, divided into aliquots, and stored at −80°C for further use.
Soil samples (S4O, S4C, S5O, S5C, S7O, S7C, S9O, and S9C), manure samples (M1, M2, M3, M4, M5, and M6), and compost samples (CA and CB) were obtained from A. van Diepeningen (Wageningen University and Research Centre, Biological Farming Systems, Wageningen, The Netherlands), who characterized and described the soil and manure samples extensively (11, 33). The selection of environmental samples was primarily based on a wide difference in microbial community and the presence of compounds that are suspected to inhibit or influence DNA extraction and/or PCR amplification. This was taken into account in order to cover the most common problems that arise with DNA extraction and amplification from environmental samples. Soils S4 and S9 were sandy soils with 3 to 3.5% clay, 10 to 33% silt, and 64 to 87% sand. Soils S5 and S7 were loamy soils with 8 to 11% clay, 40 to 55% silt, and 37 to 51% sand. Each soil was represented by two composite samples from neighboring organic (O) and conventional (C) farms. Manures were collected from individual Friesian Holstein cows with different diets (11). Composts originated from green garden waste (sample CA) and green household waste (sample CB) and were obtained from two large composting facilities. Each type of substrate used in this study tested negative for naturally present S. enterica in a test with bacterial enrichment of each substrate type, followed by both plating on selective Hektoen enteric agar and DNA extraction and subsequent S. enterica-specific TaqMan PCR detection.
DNA extraction.
The following commercial DNA extraction methods were used: Ultraclean soil DNA isolation kit (Mobiosoil) (MoBio Laboratories, Solana Beach, Calif.), Ultraclean fecal DNA kit (Mobiofecal) (MoBio), Bio101 extraction kit (Bio101) (Q-Biogene, Carlsbad, Calif.), Soilmaster DNA extraction kit (Episoil) (Epicenter, Madison, Wis.), plant DNeasy DNA extraction kit (Qiadneasy) (QIAGEN, Westburg, The Netherlands), and a combination of the microbial DNA extraction kit (Mobiomicro) (MoBio) with bacterial isolation using Optiprep (60% [wt/vol] solution of iodixanol; Axis-Shield, Oslo, Norway) at a density of 1.320 g/ml. All DNA extraction methods were performed following the manufacturers' instructions, including bead beating of the samples on a flatbed shaker at 250 rpm. To separate bacteria from soil using Optiprep, 400 μl of buffered peptone water (BPW) was added to 100 mg of soil sample and mixed by vortexing. Subsequently, a layer of 250 μl of Optiprep was pipetted underneath the soil suspension. The tubes were centrifuged at maximum speed (14,000 rpm) for 5 min. All supernatant on top of the Optiprep and the Optiprep solution itself were transferred to a clean tube. The suspension was mixed with 750 μl of BPW and centrifuged again at maximum speed for 5 min. The supernatant was discarded prior to further DNA extraction with the Mobiomicro method.
Primers and probes.
Sequences of the primers and probe for detection of S. enterica using TaqMan PCR were derived from Hoorfar et al. (16). To reduce false-negative results and provide a more accurate quantification of serovar Enteritidis in substrate samples, a GIPC was used. Detection of the GIPC was based on a gfp gene present in an E. coli strain harboring a multicopy plasmid containing the gfp gene. For detection of the gfp gene of the internal extraction and amplification control using TaqMan PCR, the sequences of the primers and probe were obtained from Klerks et al. (17). To allow the simultaneous detection of both targets, the S. enterica-specific detection probe was labeled at the 5′ end with 6-carboxyfluorescein, whereas the gfp gene-specific probe was labeled at the 5′ end with Yakima Yellow (Eurogentec, Maastricht, The Netherlands). Both detection probes were labeled at the 3′ end with Eclipse Dark Quencher (Eurogentec).
Preparation of bacterial dilution series and plate counting.
A fresh liquid culture of Salmonella serovar Enteritidis ATCC 13076 was maintained by daily picking two colonies from selective Hektoen enteric agar (Biotec Laboratories Ltd., United Kingdom) and growing the colonies separately in BPW overnight at 37°C and 250 rpm.
A dilution series of serovar Enteritidis was prepared each time by diluting fresh liquid culture 10-fold, up to a dilution of 108-fold (nine different dilutions in total and one negative control).
To determine the number of serovar Enteritidis CFU present in each dilution series, 40 μl of each dilution was plated on selective xylose lysine deoxycholate agar (Biotec Laboratories Ltd., United Kingdom) and selective Hektoen enteric agar in duplicate. The selective plates were incubated overnight at 37°C, and the number of colonies was determined for each plate.
Development of an internal extraction and amplification control.
As a procedural control, whole cells of E. coli strain 99507gfp were added to a substrate sample prior to DNA extraction and amplification. First, to determine the optimal amount of E. coli strain 99507gfp to be added to a sample prior to extraction, a 10-fold dilution series (up to 108-fold) of a liquid culture of E. coli strain 99507gfp (previously stored at −80°C in 50% glycerol) was prepared in BPW. Ten microliters of the 10-fold dilution series and a negative control were added to soil S4O (100 mg of soil per sample) prior to DNA extraction using the Mobiosoil method. The extracted DNA was diluted 10-fold before analysis by gfp-specific TaqMan PCR. The optimal amount of E. coli strain 99507gfp was defined by that dilution factor resulting in a cycle threshold (CT) value close to 31.5 (17). The CT value is defined by the number of cycles resulting in a detectable fluorescence signal above the threshold, defined by the mean plus four times the standard deviation (SD) of the fluorescence signal of the control samples.
The 10-fold dilution series (40 μl of each dilution) was also plated onto LB agar containing 50 μg/ml ampicillin and incubated at 37°C overnight. The number of colonies on each plate was counted, and the number of serovar Enteritidis CFU was calculated for each dilution of the dilution series added prior to DNA extraction.
Real-time PCR and internal control amplification.
The improved real-time TaqMan PCR method to simultaneously detect S. enterica and an internal amplification control (IAC) (17) were used throughout all experiments. Prior to DNA extraction, E. coli strain 99507gfp (2.5 × 104 CFU) was added to each substrate sample. Subsequent to DNA extraction, 2.5 μl of a 10-fold-diluted extracted DNA sample was used for PCR amplification (total volume of PCR mixture, 30 μl), as previously described (17). TaqMan PCR was performed using a quantitative PCR core kit (Eurogentec), and amplification was measured using the ABI Prism 7700 (Perkin Elmer, Norwalk, CT). Each DNA sample was tested by TaqMan PCR in triplicate.
Comparison of different DNA extraction methods with respect to extraction efficiency.
To determine the most favorable method to use for DNA extraction from soil, manure, or compost, the different methods were compared with respect to extraction efficiency. The extraction methods Mobiosoil, Episoil, Qiadneasy, Bio101, and Mobiomicro (with prior Optiprep treatment) were compared using a subset of soils (soils S4O, S5O, and S9O), since most extraction difficulties were expected for organically managed soils. The methods Mobiosoil, Mobiofecal, Episoil, Qiadneasy, and Bio101 were compared using a subset of manures (M1, M2, and M3) and one compost (CA). The DNA extraction efficiency was defined by the CT values obtained with TaqMan PCR, which indicated the suitability of the extracted DNA for PCR amplification.
First, 10 μl of a dilution series (the nondiluted sample and dilutions of 10-, 100-, and 1,000-fold) of serovar Enteritidis liquid culture was added to 100 mg of soils S4O, S5O, and S9O; manures M1, M2, and M3; and compost CA, in duplicate. Subsequently, DNA was extracted using the different DNA extraction methods and diluted 10-fold prior to downstream analyses.
Finally, each diluted DNA sample was subjected to TaqMan PCR including an IAC (10 fg of E. coli strain 99507gfp DNA) (17). The simultaneous S. enterica amplification and IAC coamplification (in one tube) were followed in real time using the ABI Prism 7700.
Consistency of the internal procedural control with different substrates.
To evaluate whether the previously (see section above) determined optimal amount of E. coli strain 99507gfp (2.5 × 104 CFU, resulting in a CT value of 31.5) showed consistent results in TaqMan PCR, the amount was tested with all soils, manures, and composts present. From each substrate 100 mg was used for sample preparation and DNA extraction. To each sample of soil, manure, and compost, 2.5 × 104 CFU of E. coli strain 99507gfp was added, and DNA was extracted using the Mobiosoil, the Bio101, and the Mobiofecal method, respectively, for soil, manure, and compost samples. After DNA extraction all purified DNA samples were diluted 10-fold, and 2.5 μl of each diluted sample was tested using the gfp-specific TaqMan PCR.
Evaluation of quantitative detection of S. enterica extracted from soil, manure, or compost.
First, the precision of DNA extraction and the extraction efficiency (defined by the recovery of added serovar Enteritidis based on the CT values obtained from PCR) of the optimal extraction methods (for soil, Mobiosoil; for manure, Bio101; and for compost, Mobiofecal) were tested in a large-scale evaluation. To accomplish this, the substrates were divided into separate groups, since DNA extraction and further analysis from only one group per day appeared feasible. Group 1 consisted of soils S4O, S4C, S5O, and S5C; group 2 consisted of soils S7O, S7C, S9O, and S9C; group 3 consisted of manures M1, M2, M3, and M4; and group 4 consisted of manures M5 and M6, compost CA, and compost CB. Each group was treated in a similar manner using fresh bacterial cultures each day.
A 10-fold dilution series of serovar Enteritidis was prepared. In duplicate, 40 μl of each of the five largest dilutions (105-, 106-, 107-, and 108-fold and the negative control) was plated on selective xylose lysine deoxycholate agar and Hektoen enteric agar and incubated at 37°C overnight prior to colony counting. In addition to plating, 10 μl of each dilution was added to 10-ml tubes containing 100 mg of substrate. BPW (2 ml) was added to each tube and incubated overnight at 37°C and 250 rpm. To extract DNA from enrichment samples, 1 ml of the enrichment culture was transferred to a clean tube, and 10 μl of GIPC (2.5 × 106 CFU of E. coli strain 99507gfp/ml) was added. DNA was extracted using the Mobiomicro DNA extraction method, diluted 10-fold, and stored at −20°C.
Then, 10 μl of each serovar Enteritidis dilution was added to 100 mg of each substrate, followed by the addition of 10 μl of 2.5 × 106 CFU/ml GIPC to each substrate sample. DNA was extracted from the soil, manure, and compost samples using, respectively, the Mobiosoil, the Bio101, and the Mobiofecal method. The purified DNA was diluted 10-fold and stored at −20°C. All stored DNA samples were finally analyzed by performing TaqMan PCR to detect serovar Enteritidis and the GIPC simultaneously, in triplicate.
Statistical analysis.
The most efficient method to use for DNA extraction per soil, manure, or compost was determined based on the extraction efficiency, i.e., the CT value (the number of cycles resulting in a detectable fluorescence signal above the threshold, defined by the mean plus four times the SD of the fluorescence signal of the control samples) obtained from TaqMan PCR. The methods were compared by performing a univariate analysis of variance (ANOVA). The mean CT values were calculated from the different substrates per substrate type and per dilution factor, and a post hoc Tukey's test with a 95% mean confidence interval was performed.
The applicability of using a set number (2.5 × 104 CFU) of whole cells of E. coli strain 99507gfp (GIPC) to control DNA extraction and amplification was determined per substrate (eight soils, six manures, and two composts, in triplicate) by calculating the mean CT value and its corresponding precision, i.e., the SD. The GIPC was considered applicable if the obtained mean CT value was 31.5 ± 1.
The selected methods were evaluated for their precision and extraction efficiency of GIPC DNA from different soils, manures, or composts that had been amended with a dilution series of serovar Enteritidis and the GIPC. ANOVA was performed on the CT values of the GIPC from the samples that were not positive for serovar Enteritidis when TaqMan PCR (five per substrate, tested in triplicate with PCR) was used. The mean CT values of the GIPC from the different substrates per substrate type were compared by performing a post hoc Tukey's test. The extraction precision of each DNA extraction method was estimated as the coefficient of determination for linear regression of the mean CT values versus the log(number of serovar Enteritidis CFU/100 mg of substrate) per substrate.
The effect of time of sampling (four sampling days) was assessed by multivariate ANOVA including a post hoc Tukey's test. As there were no significant differences among extraction dates for the different substrates, a regression line of all CT values versus CFU per 100 mg of soil, manure, or compost was calculated for each substrate, including 95% confidence intervals.
RESULTS
Comparison of methods to extract DNA from soil, manure, or compost.
The efficiency of DNA extraction from the different soils was optimal if the Mobiomicro method was used (mean CT value of 28.8) (Table 1). Except for the Mobiomicro method, the DNA extraction efficiency of each of the five methods was very low for soil S9O (high CT values) (Table 1). After this soil was omitted from the statistical analysis, the Mobiomicro method was not significantly different from the Mobiosoil (P = 0.962) and the Episoil (P = 0.087) methods, but the latter two methods were significantly different from each other (P = 0.016). Omitting soil S9O and the Mobiomicro method from the analysis, the Mobiosoil method was most efficient for subsequent PCR amplification (Table 1, adjusted mean CT value of 28.4).
TABLE 1.
Mean CT values of TaqMan PCR on DNA from a dilution series of S. enterica serovar Enteritidis added to 100 mg of soil, manure, or compost prior to DNA extraction and univariate analysis of variance of DNA extraction efficiency with a post hoc Tukey's test between DNA extraction methods
| Substrate and sample | Mean CT value by extraction method
|
|||||
|---|---|---|---|---|---|---|
| Mobiofecal | Mobiosoila | Episoil | Qiadneasy | Bio101 | Mobiomicro | |
| Soil | ||||||
| S4O | 28.69 | 31.11 | 40.00 | 32.70 | 29.21 | |
| S5O | 28.12 | 30.80 | 34.90 | 33.95 | 28.65 | |
| S9O | 38.75 | 35.57 | 40.00 | 40.00 | 28.56 | |
| Meanb,c | 31.85 B | 32.49 B | 38.30 D | 35.55 C | 28.81 A | |
| Adjusted meanc,d | 28.40 A | 30.95 B | 37.45 D | 33.33 C | ||
| Manure | ||||||
| M1 | 28.64 | 29.05 | 35.87 | 26.18 | 25.10 | |
| M2 | 30.16 | 38.19 | 36.73 | 31.60 | 25.49 | |
| M3 | 28.86 | 27.78 | 35.09 | 25.69 | 26.15 | |
| Meanc | 29.22 C | 31.67 D | 35.89 E | 27.82 B | 25.58 A | |
| Compost, CA | 28.63 A | 29.80 A | 36.33 B | 40.00 C | 40.00 C | |
The mean CT value obtained per substrate.
The mean CT value of the substrate obtained with each DNA extraction method.
Homogeneous subsets obtained from the post hoc Tukey's test, using a harmonic mean sample size of 24 and alpha of 0.05, separating the methods from high to low (from A to E) DNA extraction efficiency, are indicated.
The adjusted mean is based on a post hoc Tukey's test performed without the data from soil S9O and without the Mobiomicro method, with a harmonic mean sample size of 16 and alpha of 0.05.
The Bio101 method (mean CT value of 25.58) resulted in the optimal DNA extraction efficiency for each manure. Each method tested appeared significantly different with respect to DNA extraction efficiency, irrespective of the manure tested (Table 1).
DNA extraction from compost samples appeared most efficient using the Mobiofecal method (mean CT value of 28.63), although no significant difference (P = 0.863) between the Mobiosoil method and the Mobiofecal method was found (Table 1). However, a clear significant difference was observed between these two methods and the other methods, with a mean CT value of 36.33 for the Episoil method and a CT value of 40 (no PCR amplification) for the Bio101 method or the Qiadneasy method (Table 1).
GIPC for DNA extraction and amplification.
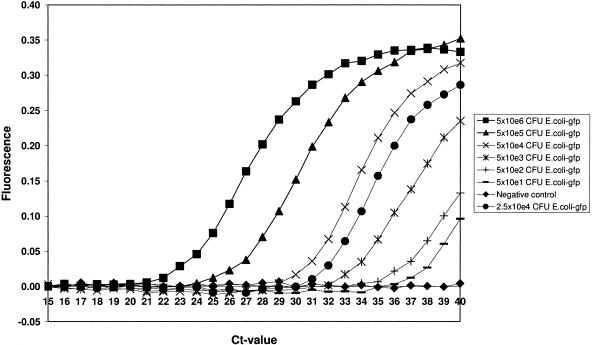
When 5 × 104 CFU or 5 × 103 CFU of the GIPC (E. coli strain 99507gfp) was added to 100 mg of substrate prior to DNA extraction, PCR amplification resulted in a CT value of 30 or 33, respectively (Fig. 1). The optimal amount resulting in a CT value of 31.5 (17) was obtained by adding 2.5 × 104 CFU of GIPC to 100 mg of each substrate prior to DNA extraction. The CT values for the GIPC varied only slightly when 2.5 × 104 CFU of GIPC was added to 100 mg of the various substrates, ranging from 31.32 ± 0.33 (mean CT ± SD) for soil extracted with Mobiosoil and 30.72 ± 0.83 for manure extracted with Bio101 to 32.38 ± 0.26 for compost extracted with Mobiofecal.
FIG. 1.
Real-time amplification and detection of a 10-fold dilution series of E. coli strain 99507gfp ranging from nondiluted bacterial culture to 105-fold diluted bacterial culture added to soil prior to DNA extraction (equal to 5 × 106 CFU/DNA extraction down to 50 CFU/DNA extraction), including the optimal amount of E. coli strain 99507gfp (giving a CT value of 31). The fluorescence increase is plotted versus the cycle number of PCR.
Evaluation of the selected S. enterica detection procedure from soil samples.
DNA was extracted from 100 mg of soil (using Mobiosoil) including the GIPC (2.5 × 104 CFU/DNA extraction) and a dilution series of serovar Enteritidis. GIPC DNA extracted from eight different soils was similar for all soils (P = 0.056) except for S4O. The mean GIPC CT value of the subset of seven soils was 31.48 ± 0.94, while that of S4O was 29.21 ± 0.93 (Table 2).
TABLE 2.
Mean CT values of TaqMan PCR for the GIPC and analysis of variance with a post hoc Tukey's test between substratesa
| Substrate | Nb | Mean CT valuec | SD |
|---|---|---|---|
| Soil | |||
| S4O | 5 | 29.21 A | 0.93 |
| S4C | 5 | 31.31 B | 1.52 |
| S5O | 5 | 30.87 B | 0.79 |
| S5C | 5 | 30.70 B | 0.80 |
| S7O | 5 | 31.47 B | 0.14 |
| S7C | 5 | 31.92 B | 0.50 |
| S9O | 5 | 32.36 B | 1.03 |
| S9C | 5 | 31.74 B | 0.26 |
| Manure | |||
| M1 | 5 | 28.52 A | 0.70 |
| M2 | 5 | 31.64 B | 1.08 |
| M3 | 5 | 29.36 A | 0.30 |
| M4 | 5 | 29.02 A | 0.53 |
| M5 | 5 | 33.85 C | 2.05 |
| M6 | 5 | 29.01 A | 0.73 |
The GIPC was added at a concentration of 2.5 × 104 CFU/100 mg of substrate prior to DNA extraction. DNA from the soil samples was extracted using the Mobiosoil method, and DNA from the manure samples was extracted with the Bio101 method.
The total number of samples where no serovar Enteritidis (negative controls and samples below detection limit) was detected by TaqMan PCR.
Mean CT value obtained from TaqMan PCR for the GIPC. The subgroups (A, B, and C) calculated from the CT values of the GIPC using Tukey's test are indicated. The mean CT value of the soils, excluding S4O, was 31.48 ± 0.94. With soil S4O, too much (10-fold) GIPC was added to the sample prior to DNA extraction, resulting in a CT value significantly lower than that of the other soils. The mean CT value of a subset of manures (M1, M3, M4, and M6) was 28.98 ± 0.63.
Salmonella serovar Enteritidis extracted directly from 100 mg of soil was detected by TaqMan PCR at a range from 1.6 × 107 down to 1.6 × 103 CFU/100 mg soil (equal to 7.8 CFU/PCR) (Table 3). After enrichment of the soil samples inoculated with serovar Enteritidis, serovar Enteritidis was detected even when only 1 CFU (calculated amount) was originally added to 100 mg of soil (Table 3).
TABLE 3.
CT values obtained from TaqMan PCR on DNA extracted directly and after enrichment from substrates that were amended with a dilution series of S. enterica serovar Enteritidis
| Method | No. of CFUa |
CT value per amount of serovar Enteritidis inb:
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Soil
|
Manure
|
Compost
|
|||||||||||||||
| S4Oc | S4C | S5O | S5C | S7O | S7C | S9O | S9C | M1 | M2 | M3 | M4 | M5 | M6 | CA | CB | ||
| Direct | Z × 107 | 25.18 | 27.09 | 25.79 | 24.24 | 24.41 | 23.70 | 26.48 | 25.52 | 21.01 | Nc | 20.20 | 20.05 | 23.04 | 21.13 | N | 24.39 |
| Z × 106 | 28.40 | 30.15 | 27.53 | 27.08 | 26.08 | 26.68 | 26.42 | 24.70 | 23.78 | N | 24.36 | 24.16 | 33.76 | 25.26 | 26.48 | 25.72 | |
| Z × 105 | 32.88 | 34.50 | 30.30 | 31.56 | 30.61 | 28.85 | 28.63 | 29.51 | 27.57 | N | 27.27 | 27.40 | N | 28.43 | 32.69 | 29.54 | |
| Z × 104 | 34.24 | 35.11 | 33.10 | 33.33 | 33.63 | 31.87 | 33.31 | 35.20 | 31.50 | N | 30.90 | 31.65 | N | 34.69 | 33.98 | 32.77 | |
| Z × 103 | 37.15 | 39.12 | 36.17 | N | 38.24 | 34.44 | 36.17 | 34.92 | 36.40 | N | 36.52 | 34.83 | N | N | N | 37.37 | |
| Z × 102 | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | |
| Neg | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | |
| Enrichment | Z × 102 | 17.38 | 15.73 | 15.74 | 16.16 | 17.47 | 15.75 | 16.06 | 15.86 | 16.93 | 15.62 | 16.80 | 16.96 | 16.09 | 16.15 | 20.71 | 24.63 |
| Z × 101 | 18.69 | 16.76 | 16.80 | 16.28 | 19.68 | 18.09 | 17.64 | 16.59 | 19.46 | 16.26 | 17.28 | 19.32 | 16.23 | 16.11 | 22.27 | 24.20 | |
| Z × 100 | 20.90 | 18.63 | 17.58 | 17.17 | 21.51 | 34.51 | 19.39 | 18.38 | 22.16 | 15.98 | 34.81 | 21.19 | 16.20 | 16.99 | 23.88 | 33.31 | |
| Z × 10−1 | N | N | N | N | N | N | N | 19.84 | 22.42 | N | N | N | N | N | N | N | |
| Neg | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | |
Calculated number of CFU of S. enterica serovar Enteritidis added to 100 mg of each substrate prior to DNA extraction. Z was different for each time-related group: Z = 1.2 for group 1 (S4O and S4C); Z = 1.3 for group 2 (S5O and S5C); Z = 1.4 for group 3 (S7O and S7C); Z = 1.6 for group 4 (S9O and S9C); Z = 1.7 for group 5 (M1 and M2); Z = 0.8 for group 6 (M3 and M4); Z = 1.8 for group 7 (M5 and M6); and Z = 1.2 for group 8 (CA and CB). Each group was treated in 1 day and in a similar manner using fresh bacterial cultures each time. Neg, negative control.
The amount was calculated from plating. A sample negative for serovar Enteritidis is indicated by N.
N represents the substrate tested for DNA extraction and subsequent TaqMan PCR.
Evaluation of the selected S. enterica detection procedure from manure samples.
The amount of GIPC DNA extracted from six manures using Bio101 was quite consistent for a subset of those manures (M1, M3, M4, and M6), with a mean CT value of 28.98 and an SD of 0.62 (P = 0.805). The mean CT values of this subset differed significantly (P = 0.001 to 0.026) from those of M2 and M5, with mean CT values of 31.64 ± 1.08 and 33.85 ± 2.05, respectively (Table 2).
Serovar Enteritidis was detected by TaqMan PCR at a range from 1.8 × 107 down to 1.8 × 103 CFU/100 mg of manure (equal to 9.2 CFU/PCR) when extracted directly from manure (Table 3). Detection of Salmonella DNA extracted from manure M2 was not possible. Manure M5 was positive for serovar Enteritidis only when large amounts (106 to 107 CFU) of serovar Enteritidis cells were added to 100 mg of substrate prior to DNA extraction (Table 3). Enrichment of manures inoculated with serovar Enteritidis enabled the detection of even 1 CFU of serovar Enteritidis originally added per 100 mg of each manure (Table 3).
Evaluation of the selected S. enterica detection procedure from compost samples.
GIPC DNA extraction from compost with Mobiofecal resulted in significantly different (P = 0.03) CT values for compost CA and CB, with mean CT values of 32.88 ± 1.84 and 31.41 ± 0.78, respectively. Serovar Enteritidis extracted from compost CB was detected by TaqMan PCR in a range of 1.2 × 107 CFU down to 1.2 × 103 CFU/100 mg of compost (equal to 6.2 CFU/PCR) (Table 2). DNA extraction from compost CA was less efficient than that from CB (Table 3). Enrichment of serovar Enteritidis inoculated in the compost samples enabled the detection of even 1 CFU originally added to 100 mg of both compost samples (Table 3).
Quantification of serovar Enteritidis present in various environmental substrates.
Regression of S. enterica CT values on the log(amount of target CFU) for each soil, manure, and compost separately resulted in a good fit for the Salmonella dilution series tested (R2 = 0.90 to 0.99), except for manures M2 and M5 (no regression analysis possible) and compost CA (R2 value of 0.87).
Slope and intercept of the regression lines for soil samples analyzed on four subsequent days (groups 1 to 4) were not significantly different in a multivariate ANOVA test (P = 0.863 and P = 0.624 for slope and intercept, respectively). These results were confirmed by a post hoc Tukey's test on the slope (P = 0.832) and intercept (P = 0.582) for each group. Similar slopes and intercepts were found when dilution series of serovar Enteritidis with three different soils, S4O, S5C, and S9O, were tested simultaneously in 1 day (data not shown).
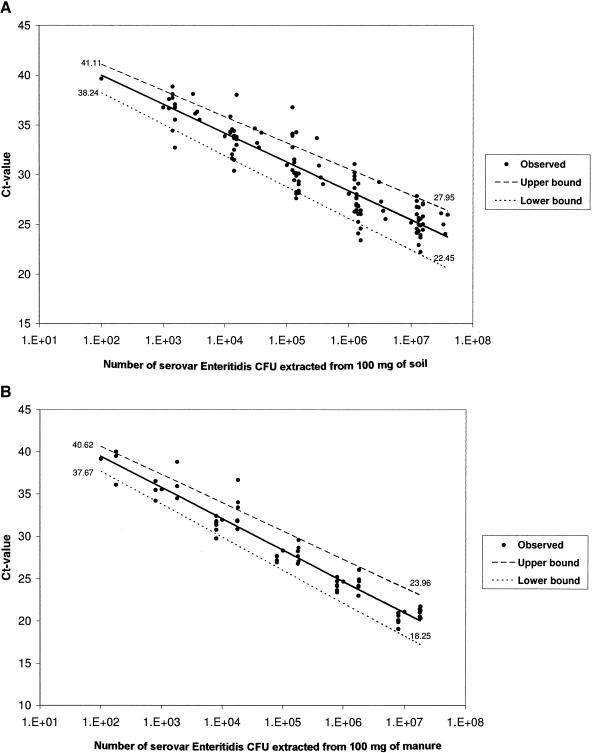
Subsequently, a regression line was estimated for all data per substrate (soil or manure), and 95% confidence intervals were calculated. The regression equations for soil (intercept = 42.571 ± 1.17; slope = −2.896 ± 0.264; R2 = 0.841) and manure (intercept = 42.756 ± 1.199; slope = −3.608 ± 0.277; R2 = 0.923) were not significantly different (Fig. 2). The variation in CT values around the means (within the 95% confidence limits) ranged from 1.43 to 2.75 (equal to 2.70 to 6.73 times the difference in initial number of CFU/100 mg of soil) for the different concentrations of serovar Enteritidis added to soil (Fig. 2A). For manure, the variations in CT values around the means were remarkably similar to those of soil, namely, 1.48 to 2.86 (equal to 2.78 to 7.25 times the difference in initial number of CFU/100 mg of manure) for the different concentrations of serovar Enteritidis added to manure (Fig. 2B).
FIG. 2.
Linear regression with 95% confidence intervals from 10-fold dilution series of Salmonella serovar Enteritidis added to soil (A) and manure (B) prior to DNA extraction and amplification. The CT value is plotted versus the log(number of serovar Enteritidis CFU/100 mg of substrate).
DISCUSSION
Until now, many different DNA extraction methods allowing subsequent PCR have been described. However, only a few papers describe a comparison of commercial DNA extraction methods with respect to extraction efficiency from soil, manure, or compost. Zhou et al. (37) described the development of a method to extract DNA from soil and evaluated the DNA recovery from eight soils with diverse composition using only this method. Lloyd-Jones and Hunter (19) compared three different DNA extraction methods with respect to DNA recovery from four soils with different composition.
In this study, six commercial DNA extraction methods were compared with respect to DNA extraction efficiency and quantification accuracy of serovar Enteritidis initially present in the substrate sample. Mobiosoil, Bio101, and Mobiofecal were found to be most efficient for DNA extraction from, respectively, soil (eight different substrates), manure (six substrates), and compost (two substrates). A sensitivity of approximately 10 CFU of serovar Enteritidis per PCR (2 × 103 CFU/100 mg of substrate) was obtained using DNA extraction followed by S. enterica-specific TaqMan PCR. In addition, even 1 CFU of serovar Enteritidis per 100 mg of substrate was clearly detected by TaqMan PCR after enrichment.
For soil, the Mobiomicro method was initially found most efficient. However, this method is based on density separation of bacteria from soil instead of chemical lysis of the soil sample. Due to the experimental setup (addition of serovar Enteritidis to the substrate prior to DNA extraction), this method would prevent a proper comparison of DNA extraction efficiencies. Therefore, this method was omitted from further experiments.
Some substrates (such as soil S9O and manures M2 and M5) did not allow the detection of serovar Enteritidis to the same extent as the other substrates tested, irrespective of the DNA extraction method used. It is likely that either little DNA was amplified from the extracted DNA from these substrates or the substrates gave a strong inhibition of DNA extraction and amplification. Both manures M2 and M5 were derived from cattle fed with low-digestible grass silage, resulting in manure of high dry matter. As the feed of cattle has a direct influence on the composition of their manure, the inhibiting components might have originated from the preserved grass. Also, due to the high dry-matter content of these manures (11), it is likely that per 100 mg of manure a higher concentration of inhibiting agents is included in the DNA extract, leading to a reduction in extracted DNA yield and/or leading to inhibited amplification. Unfortunately, the chemical and/or organic components present in manures M2 and M5, which were responsible for the reduction in DNA extraction efficiency, could not be identified by gas chromatography with mass spectrometry. The inhibiting components should be identified to allow the development of more generic DNA extraction methods for extraction of DNA from complex substrates. Nevertheless, from these data it is evident that in some cases alternative approaches for DNA extraction are required to prevent poor DNA recovery or the presence of coextracted amplification inhibitors. Recognition of the exceptions that lead to insufficient DNA extraction efficiency was previously not possible without extensively studying the efficiency. This major drawback is now countered by application of the GIPC with the substrate prior to DNA extraction.
Despite the clear differences among test kits in extraction efficiency, testing of DNA extraction from substrates inoculated with a serovar Enteritidis suspension does not completely reflect real environmental samples containing naturally present S. enterica. In fact, naturally present S. enterica might be aggregated on or between substrate particles, which could make the extraction of DNA from all cells present even more complicated. The extent to which spatial distribution affects the S. enterica DNA extraction efficiency has not been evaluated. Nevertheless, based on the rigorous lysis and homogenization during the DNA extraction procedures, it is expected to have only a minor influence on the recovery of Salmonella serovar Enteritidis DNA present in the tested substrates.
Direct evaluation of the efficiency of the complete DNA extraction and amplification procedure was enabled by implementation of a GIPC. This improvement enabled the identification of false-negative results introduced by procedural failures or mistakes. A major advantage of the developed GIPC is the fact that it is absent in natural environments (except in the cnidarian jellyfish Aequorea victoria), since its detection is based on the gfp gene of the GIPC. Therefore, independent of the substrate tested, the gfp gene can be detected simultaneously with the target (in our case serovar Enteritidis) after DNA extraction using TaqMan PCR. This is an advantage over other previously published approaches, which use housekeeping genes to control the DNA extraction and amplification efficiency. The use of such housekeeping genes is not sufficient, since the exact initial amount of control material (DNA) is not known, thus allowing only qualitative validation of a sample tested. Moreover, these controls are applicable only if the corresponding housekeeping genes are indeed present in the environmental sample (plant or animal cells) tested. Finally, the amount of housekeeping genes often exceeds the amount of target DNA, resulting in a competitive PCR amplification strongly affecting the sensitivity, precision, and accuracy of the assay. The coamplification of a different target will in each case affect the sensitivity of the primary target amplification. To reduce any influence of coamplification, the amount of GIPC is limited to a set level to ensure that the target DNA is always present in excess. Thus, the amplification of the target is influenced only at a very minor level (17).
In general, a more accurate quantification of S. enterica (or any other pathogen) from soil, manure, or compost can be obtained using the GIPC. By doing so, the presence of inhibitory factors is first determined by calculating the mean and SD of the GIPC CT values. Samples that present a statistically different CT value (of GIPC) than the water controls and the negative substrate control samples are not valid for quantification. From all other samples the amount of target CFU can then be calculated.
Applying this approach allowed a more accurate quantification of the target initially present in the substrate tested using a dilution series of serovar Enteritidis added to soil and manure. However, these data also indicated that the applied molecular approach (DNA extraction followed by TaqMan PCR) still leads to quantification errors in detecting an organism like S. enterica in environmental substrates. Until a (nearly) perfect quantification of organisms (or a specific DNA target from environmental substrates) is feasible using molecular techniques, the approach described in this paper might provide a more accurate quantification of target organisms or DNA than other currently used molecular quantification methods. Finally, the method presented in this paper might be a good addition to the standardized methods for identification and detection of S. enterica in environmental substrates, especially since preenrichment of samples is currently still obligatory to enable the detection of even 1 CFU of S. enterica in 25 g of substrate.
In conclusion, the optimized procedure provides an improved, sensitive, and precise method, eliminating a false-negative diagnosis by the introduction of a GIPC. The approach of adding a fixed amount of GIPC prior to DNA extraction provides an efficient and reliable way for evaluating and validating DNA extraction and amplification of each individual sample in one tube, independent of the substrate tested. The method is applicable for high-throughput analysis and routine diagnosis and allows a more accurate quantification of S. enterica present in soil, manure, or compost.
Acknowledgments
This research was supported by the Horticultural Product Board (Produktschap Tuinbouw), The Netherlands, Technology Foundation STW, the Applied Science Division of NWO, the Technology Program of the Ministry of Economic Affairs, and NWO Russia collaborative grant 047.014.001.
We thank H. Aarts from RIKILT, The Netherlands, for providing S. enterica serovar Enteritidis ATCC 13076 and R. J. Sayler (Department of Plant Pathology, University of Arkansas, Fayetteville, Ark.) for providing E. coli strain 99507gfp. We also thank E. Franz and A. Termorshuizen for the constructive discussions and their valuable statistical expertise.
REFERENCES
- 1.Acheson, D. W. K., S. DeBreucker, A. Donohue-Rolfe, K. Kozak, A. Yi, and G. T. Keusch. 1994. Development of a clinically useful diagnostic enzyme immunoassay for enterohemorrhagic Escherichia coli infection, p. 109-112. In M. A. Karmali and A. D. Goglio (ed.), Recent advances in verocytotoxin producing Escherichia coli infections. Elsevier Science BV, Amsterdam, The Netherlands.
- 2.Al-Soud, W. A., and P. Rådström. 1998. Capacity of nine thermostable DNA polymerases to mediate DNA amplification in the presence of PCR-inhibiting samples. Appl. Environ. Microbiol. 64:3748-3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bassler, H. A., S. J. A. Flood, K. J. Livak, J. Marmaro, R. Knorr, and C. A. Batt. 1995. Use of a fluorogenic probe in a PCR-based assay for the detection of Listeria monocytogenes. Appl. Environ. Microbiol. 61:3724-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behrsing, J., B. Tomkins, and R. Premier. 2000. The micro story on vegetables. In Proceedings of the 2nd National On-Farm Food Safety and Quality Assurance Conference. Department of Natural Resources and Environment, Victoria, Australia.
- 5.Bej, A. K., M. H. Mahbubani, M. J. Boyce, and R. M. Atlas. 1994. Detection of Salmonella spp. in oysters by PCR. Appl. Environ. Microbiol. 60:368-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolton, F. J., E. Fritz, S. Poynton, and T. Jensen. 2000. Rapid enzyme-linked immunoassay for detection of Salmonella in food and feed products: performance testing program. J. AOAC Int. 83:299-303. [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 1999. Outbreak of Escherichia coli O157:H7 and Campylobacter among attendees of the Washington County Fair-New York. Morb. Mortal. Wkly. Rep. 48:803-804. [PubMed] [Google Scholar]
- 8.De Medici, D., L. Vroci, E. Delibato, S. Di Pasquale, E. Filetici, and L. Toti. 2003. Evaluation of DNA extraction methods for use in combination with SYBR Green I real-time PCR to detect Salmonella enterica serotype Enteritidis in poultry. Appl. Environ. Microbiol. 69:3456-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fisher, I. S. 2004. Dramatic shift in the epidemiology of Salmonella enterica serotype Enteritidis phage types in Western Europe, 1998-2003: results from the Enter-net international salmonella database. Euro. Surveill. 1:9-11. [DOI] [PubMed] [Google Scholar]
- 10.Fortin, N., D. Beaumier, K. Lee, and C. W. Greer. 2004. Soil washing improves the recovery of total community DNA from polluted and high organic content sediments. J. Microbiol. Methods 56:181-191. [DOI] [PubMed] [Google Scholar]
- 11.Franz., E., A. D. van Diepeningen, O. J. de Vos, and A. H. C. van Bruggen. 2005. Effects of cattle feeding regime and soil management type on the fate of Escherichia coli O157:H7 and Salmonella enterica serovar Typhimurium in manure, manure-amended soil, and lettuce. Appl. Environ. Microbiol. 71:6165-6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heller, L. C., C. R. Davis, K. K. Peak, D. Wingfield, A. C. Cannons, P. T. Amuso, and J. Cattani. 2003. Comparison of methods for DNA isolation from food samples for detection of Shiga toxin-producing Escherichia coli by real-time PCR. Appl. Environ. Microbiol. 69:1844-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higgins, J. A., J. Ezzel, B. J. Hinnebusch, M. Shipley, E. A. Henchal, and S. S. Ibrahim. 1998. 5′ nuclease PCR assay to detect Yersinia pestis. J. Clin. Microbiol. 36:2284-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hilborn, E. D., J. H. Mermin, P. A. Mshar, J. L. Hadler, A. Voetsch, C. Wojtkunski, M. Swartz, R. Mshar, M. Lambert-Fair, J. A. Farrar, M. K. Glynn, and L. Slutsker. 1999. A multistate outbreak of Escherichia coli O157:H7 infections associated with consumption of mesclun lettuce. Arch. Intern. Med. 159:1758-1764. [DOI] [PubMed] [Google Scholar]
- 15.Holland, P. M., R. D. Abramson, R. Watson, and D. H. Gelfand. 1991. Detection of specific polymerase chain reaction product by utilizing the 5′ 3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. USA 88:7276-7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoorfar, J., P. Ahrens, and P. Rådström. 2000. Automated 5′ nuclease PCR assay for identification of Salmonella enterica. J. Clin. Microbiol. 38:3429-3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klerks, M. M., C. Zijlstra, and A. H. C. van Bruggen. 2004. Comparison of real-time PCR methods for detection of Salmonella enterica and Escherichia coli O157:H7, and quantification using a general internal amplification control. J. Microbiol. Methods 59:337-349. [DOI] [PubMed] [Google Scholar]
- 18.Lettelier, A. 1999. Distribution of Salmonella in swine herds in Quebec. Vet. Microbiol. 67:229-306. [DOI] [PubMed] [Google Scholar]
- 19.Lloyd-Jones, G., and D. W. F. Hunter. 2001. Comparison of rapid DNA extraction methods applied to contrasting New Zealand soils. Soil Biol. Biochem. 33:2053-2059. [Google Scholar]
- 20.Lyytikainen, O., J. Koort, L. Ward, R. Schildt, P. Ruutu, E. Japisson, M. Timonen, and A. Siitonen. 2004. Molecular epidemiology of an outbreak caused by Salmonella enterica serovar Newport in Finland and the United Kingdom. Epidemiol. Infect. 124:185-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michino, H., K. Araki, S. Minami, S. Takaya, N. Sakai, M. Miyazaki, A. Ono, and H. Yanagawa. 1999. Massive outbreak of Escherichia coli O157:H7 infection in schoolchildren in Sakai City, Japan, associated with consumption of white radish sprouts. Am. J. Epidemiol. 150:787-796. [DOI] [PubMed] [Google Scholar]
- 22.Miller, W. G., and S. E. Lindow. 1997. An improved GFP cloning cassette designed for prokaryotic transcriptional fusions. Gene 191:149-153. [DOI] [PubMed] [Google Scholar]
- 23.Natvig, E. E., S. C. Ingham, B. H. Ingham, L. R. Cooperband, and T. R. Roper. 2002. Salmonella enterica serovar Typhimurium and Escherichia coli contamination of root and leaf vegetables grown in soil with incorporated bovine manure. Appl. Environ. Microbiol. 68:2737-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pell, A. N. 1997. Manure and microbes: public and animal health problem? J. Dairy Sci. 80:2673-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prasher, D. C., V. K. Eckenrode, W. W. Ward, F. G. Prendergast, and M. J. Cormier. 1992. Primary structure of the Aequorea victoria green-fluorescent protein. Gene 111:229-233. [DOI] [PubMed] [Google Scholar]
- 26.Raggam, R. B., E. Leitner, G. Mühlbauer, J. Berg, M. Stöcher, A. J. Grisold, E. Marth, and H. H. Kessler. 2002. Qualitative detection of Legionella species in bronchoalveolar lavages and induced sputa by automated DNA extraction and real-time polymerase chain reaction. Med. Microbiol. Immunol. 191:119-125. [DOI] [PubMed] [Google Scholar]
- 27.Robertson, L. J., G. S. Johannessen, B. K. Gjerde, and S. Loncarevi. 2002. Microbiological analysis of seed sprouts in Norway. Int. J. Food Microbiol. 75:119-126. [DOI] [PubMed] [Google Scholar]
- 28.Semenov, A. M., A. H. C. Van Bruggen, N. N. Kunenkova, E. Franz, E. V. Semenova, V. M. Mikhailovich, A. Y. Sobolev, S. A. Lapa, Y. P. Solodovnikov, R. J. Sayler, U. Romling, and A. I. Netrusov. 2004. Experiments for the risk analysis of the spread of enteropathogens in the environment within the microbial cycle, p. 163-168. In Proceedings of the Second International Conference “Biotechnology—to Environment Preservation, Part II.” Sport and Culture, Moscow, Russia.
- 29.Solomon, B. E., S. Yaron, and K. R. Matthews. 2002. Transmission of Escherichia coli O157:H7 from contaminated manure and irrigation water to lettuce plant tissue and its subsequent internalization. Appl. Environ. Microbiol. 68:397-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stewart, D. S., M. L. Tortorell, and S. M. Gendel. 1998. Evaluation of DNA preparation techniques for detection of the SLT-1 gene of Escherichia coli O157:H7 in bovine faeces using the polymerase chain reaction. Lett. Appl. Microbiol. 26:93-97. [DOI] [PubMed] [Google Scholar]
- 31.Theron, J., and T. E. Cloete. 2000. Molecular techniques for determining microbial diversity and community structure in natural environments. Crit. Rev. Microbiol. 26:37-57. [DOI] [PubMed] [Google Scholar]
- 32.Todd, E. C. D. 1989. Costs of acute bacterial foodborne disease in Canada and the United States. Int. J. Food Microbiol. 9:313-326. [DOI] [PubMed] [Google Scholar]
- 33.Van Diepeningen, A. D., O. J. de Vos, G. W. Korthals, and A. H. C. van Bruggen. 2005. Effects of organic versus conventional management on chemical and biological parameters in agricultural soils. Appl. Soil Ecol. 31:120-135. [Google Scholar]
- 34.Wang, G., and M. P. Doyle. 1998. Survival of enterohemorrhagic Escherichia coli O157:H7 in water. J. Food Prot. 61:662-667. [DOI] [PubMed] [Google Scholar]
- 35.Watson, R. J., and B. Blackwell. 2000. Purification and characterization of a common soil component which inhibits the polymerase chain reaction. Can. J. Microbiol. 46:633-642. [DOI] [PubMed] [Google Scholar]
- 36.Wilson, I. G. 1997. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 63:3741-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou, J., M. Bruns, and J. M. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]