Abstract
Several PCR methods have recently been developed to identify fecal contamination in surface waters. In all cases, researchers have relied on one gene or one microorganism for selection of host-specific markers. Here we describe the application of a genome fragment enrichment (GFE) method to identify host-specific genetic markers from fecal microbial community DNA. As a proof of concept, bovine fecal DNA was challenged against a porcine fecal DNA background to select for bovine-specific DNA sequences. Bioinformatic analyses of 380 bovine enriched metagenomic sequences indicated a preponderance of Bacteroidales-like regions predicted to encode membrane-associated and secreted proteins. Oligonucleotide primers capable of annealing to select Bacteroidales-like bovine GFE sequences exhibited extremely high specificity (>99%) in PCR assays with total fecal DNAs from 279 different animal sources. These primers also demonstrated a broad distribution of corresponding genetic markers (81% positive) among 148 different bovine sources. These data demonstrate that direct metagenomic DNA analysis by the competitive solution hybridization approach described is an efficient method for identifying potentially useful fecal genetic markers and for characterizing differences between environmental microbial communities.
Nearly 13% of the natural waters in the United States are, at any time, designated unsafe for fishing and swimming because of high densities of fecal indicator bacteria (37). Most of this fecal contamination is attributable to either discharge of human waste treatment effluents or to agricultural practices, including open-range grazing and confined-animal feeding operations. The cattle industry is a focus of primary concern because of its relatively high volume of waste production. There are currently 104.5 million cattle in the United States (36), and the average adult bovine produces 59 to 80 pounds of feces per day (23). Bovine fecal pollution can become a significant human health risk when impacting surface waters, as bovine feces can contain human pathogens, including Escherichia coli O157:H7, Campylobacter jejuni, Salmonella spp., Leptospira interrogans, and Cryptosporidium parvum (5, 8, 13, 24, 25). Identification of bovine fecal contamination is typically confounded by the presence of fecal material and bacterial indicator species from other sources (e.g., pigs, chickens, wildlife, humans, etc.). Determining the sources of fecal pollution is essential for the accurate evaluation of human health risks, for monitoring emerging zoonotic infectious diseases, and in developing management plans to make waters safe for human use.
Methods to detect pollution from bovine feces have been previously described (26, 29). One of the most widely used approaches is based on the use of the PCR to detect Bacteroides 16S rRNA gene (rRNA) sequences (7). This bacterial group constitutes a large proportion of the normal gut microbiota of most animals, including ruminants (38). Bacterial 16S rRNA genes are useful markers because they have relatively low mutation rates (15) and are typically present in multiple operons, increasing template DNA levels for PCR (1, 19, 22, 39). While several studies have demonstrated the value of Bacteroides 16S rRNA gene-based assays, currently available Bacteroides-based PCR methods can only discriminate between ruminant and nonruminant sources (7). Alternative genetic markers capable of species level discrimination among hosts (e.g., different types of ruminants) are needed to complement existing PCR-based approaches used to identify sources of fecal pollution.
The goal of this study was to determine if a novel competitive DNA hybridization approach previously used to obtain chromosomal regions present in one bacterial genome but not another (28) could be used on a metagenomic scale to identify bovine-specific fecal DNA sequences. These experiments directly identified 380 bovine feces-specific DNA sequences while avoiding the limitations associated with any requirement for existing genetic information or growth of corresponding organisms in laboratory cultures. Three enriched DNA sequences were randomly selected and used to develop bovine-specific PCR assays. The potential utility of the latter assays for diagnosing bovine fecal pollution in natural waters was demonstrated.
MATERIALS AND METHODS
Sample collection and DNA extraction.
Fecal and septic tank samples were collected in sterile containers, and approximately 200 mg (wet weight) of feces or 200 μl of septic tank slurry was mixed with 3 ml of GITC buffer (5 M guanidine isothiocyanate, 100 mM EDTA [pH 8.0], 0.5% Sarkosyl) and stored at −80°C. Three hundred eighty-four individual fecal samples and nine septic tank samples were collected for comparison. These included 212 samples from farm animals, 99 from wildlife, 32 from birds, 25 from pets, and 16 from humans. Bovine fecal samples originated from five separate geographic locations and were collected at different times, including Delaware (in 2003), Nebraska (in 2004), West Virginia (in 2002), Georgia (in 2004), and Texas (in 2004). These specimens represented a total of 28 different animal species that likely impact watersheds nationwide. In addition to fecal and septic tank samples, 36 water samples were collected from freshwaters (Burnet Woods Pond, OH; Shepards Creek, OH; Sea Graves, GA) and marine waters (Jobos Bay, PR) known to be impacted by different sources of fecal pollution. One hundred milliliters of water was filtered through 0.2-μm-pore-size Supor-200 filters (Whatman), and each filter was placed in a sterile 15-ml tube containing 500 μl of GITC buffer and stored at −80°C.
All DNA extractions were performed with either the FastDNA Kit for Soils (Q-Biogene, Carlsbad, CA) or the UltraClean Fecal DNA Kit (MO BIO Laboratories, Inc., Carlsbad, CA). DNA extract yields were quantified with a NanoDrop ND-1000 UV spectrophotometer (NanoDrop Technologies, Wilmington, DE). The potential for amplification and presence of sufficient DNA was confirmed for each sample DNA extract by a previously described Bacteroidales-specific PCR assay with primers Bac32F and Bac708R (6).
GFE.
Genome fragment enrichment (GFE) was used to select potential fecal community-specific markers (Fig. 1) (28). This method is a modification of a nucleic acid sorting approach originally developed for bacterial RNA analysis (14) that has recently been applied to define regions of chromosomal variation between two enterococcal species (28). Briefly, biotin-labeled, sheared fecal DNA from an individual cow (source A) was first prehybridized with sheared DNA fragments of fecal DNA from an individual pig (source B) for 20 min. This “blocked” biotin-labeled DNA was then hybridized to equilibrium in solution with additional DNA fragments from the original source (bovine fecal DNA or source A) that contained defined terminal sequence tags. These terminal tags were incorporated by a prior single-round primer extension mediated by excess Klenow polymerase and DNA oligonucleotides having both a common 5′ sequence and nine random 3′ residues. DNA hybrids were then isolated by streptavidin binding, and the captured tagged genomic fragments were amplified by lone-linker PCR (16). The required specificity of lone-linker PCR with either the K9-PCR or the F9-PCR primer (14) was verified with reference sheared bovine and porcine fecal DNAs as templates. Between one and three rounds of blocked-capture enrichment and amplification were performed prior to cloning into a plasmid vector as described below. All PCRs were performed with either low-retention reaction tubes (0.2 ml) or 96-well polypropylene plates and an MJ Research DNA Engine Tetrad 2 thermal cycler (Bio-Rad, Hercules, CA).
FIG. 1.
Schematic of the GFE method used to identify bovine fecal community DNA sequences that are absent or divergent in a porcine fecal community. Biotin-labeled, sheared total metagenomic DNA from a bovine fecal sample was prehybridized with metagenomic DNA fragments from a porcine fecal sample prior to being self-hybridized with PCR-amplified bovine fecal metagenomic DNA fragments containing defined terminal sequence tags. DNA hybrids were then isolated by streptavidin binding, and the captured genomic fragments containing defined terminal sequence tags were selectively amplified by PCR.
DNA sequencing.
PCR products from triplicate parallel reaction mixtures for each round of GFE were pooled and incorporated into plasmid vector pCR4-TOPO as described by the manufacturer (Invitrogen, Carlsbad, CA). Individual E. coli clones were then subcultured in 300 μl of Luria broth containing ampicillin (10 μg/ml) and screened for inserts by PCR with primers M13F and M13R. Prior to sequencing, PCR products were purified with a QiaQuick 96 Plate (QIAGEN, Valencia, CA). Sequencing was performed on both strands at the Cincinnati Children's Hospital Medical Center Genomics Core Facility (Cincinnati, OH) by the dye terminator method on an ABI PRISM 3730XL DNA analyzer (Applied Biosystems, Foster City, CA).
Dot blot hybridizations.
Dot blot hybridizations with cloned GFE sequences and a porcine fecal DNA (GFE blocker) probe were used to identify any plasmid clone inserts obtained by GFE that were not unique to the original bovine fecal DNA source. Probe preparation, hybridization conditions, and detection were performed as previously described (28). Briefly, purified PCR products from each plasmid clone insert sequence were spotted onto nylon membranes (Li-Cor Biosciences, Lincoln, NE) with a 96-well manifold (Bio-Rad Laboratories, Hercules, CA) and hybridized with a biotin-labeled porcine fecal community DNA probe. The hybridized probe was visualized with an Odyssey Infrared Imaging System (Li-Cor Biosciences, Lincoln, NE) following detection with the streptavidin conjugate Alex Fluor 680 as described by the manufacturer (Invitrogen, Carlsbad, CA).
Data analysis.
DNA sequence reads were assembled with SeqMan II (DNAstar, Inc., Madison, WI) and used to search the National Center for Biotechnology Information (NCBI) RefSeq and Expert Protein Analysis System (ExPASy) Swiss-Prot databases with BLASTx software (3; http://www.ncbi.nlm.nih.gov/BLAST/). BLASTx hits with expectation values of ≤1e−3 were designated homologous. To determine the best BLASTx hit between the two database searches, a score density (match length/bit score) was calculated for each homolog. The highest score density value was designated as the overall best BLASTx hit. Bovine-specific GFE sequences with top BLASTx hits to genes encoded in the Bacteroides thetaiotaomicron (39), Bacteroides fragilis (22), and Porphyromonas gingivalis strain W83 (16) genomes were designated Bacteroidales-like sequences and were selected for further bioinformatic characterization. Gene function attributes for Bacteroidales-like DNA sequences were assigned on the basis of annotations of the B. thetaiotaomicron genome (39) available in the Comprehensive Microbial Resource genome database at The Institute for Genomic Research (http://www.tigr.org/tigr-scripts/CMR2/CMRGenomes.spl).
Bovine-specific PCR primer design.
Three Bacteroidales-like DNA sequences were randomly selected to develop PCR assays for detecting bovine fecal material. PCR primers were designed with PrimerSelect (DNAstar, Inc., Madison, WI) and default settings. Candidate primer sequences were aligned with homologous sequences (e value, ≤ 1e−3) from three complete Bacteroidales genome sequences, including B. thetaiotaomicron (39), B. fragilis (19), and P. gingivalis strain W83 (22), with ClustalW (35) and default settings (MegAlign; DNAstar, Inc., Madison, WI). Primer sets that align with variable DNA regions among Bacteroidales sequences were selected for optimization, host specificity, and limit of detection assays.
Primer optimization, host specificity, and limit of detection.
Optimal annealing temperatures were measured for each primer pair by thermal gradient PCR on an MJ Research DNA Engine Tetrad 2 thermal cycler (Bio-Rad, Hercules, CA). Host-specific PCR assay mixtures (25 μl) contained 1× ExTaq PCR buffer (Takara Maris Bio, Madison, WI); 2.5 mM (each) dATP, dCTP, dGTP, and dTTP; each primer at 0.2 μM; 0.04% acetamide (Sigma); 0.625 U of ExTaq (Takara Maris Bio, Madison, WI); and 2 ng of template DNA. Incubation temperatures included an initial 94°C (3 min) lysis step, followed by 35 cycles of 94°C (1 min), a gradient range of 55°C to 65°C (1 min), and 72°C (1 min). Optimized primers were then tested for host distribution among individual bovine fecal samples (n = 148) and cross-reactivity with nontarget individual animal fecal specimens (n = 292) and human septic tank samples (n = 9). Reference fecal samples represented 28 species of animals, including Bos taurus (bovine), Gallus gallus (chicken), Loragyps atratus (black vulture), Anser sp. (Canadian goose), Pavo sp. (peacock), Treron sp. (pigeon), Canis familiaris (dog), Felis catus (cat), Cavia porcellus (guinea pig), Capra aegagrus (domestic goat), Sus scrofa (pig), Ovis aries (sheep), Equus caballus (horse), Lama pacos (alpaca), Lama glama (llama), Armadillo officinalis (armadillo), Lynx rufus (bobcat), Canis latrans (coyote), Sciurus carolinensis (gray squirrel), Lepus sp. (rabbit and jackrabbit), Didelphis virginiana (opossum), Procyon lotor (raccoon), Odocoileus virginianus (whitetail deer), Meleagris sp. (wild turkey), Erinaceus sp. (hedgehog), Cynomys sp. (prairie dog), and Homo sapiens (human). To test for nonspecific amplification of DNA from representative environmental microorganisms, PCR assays were performed with 5 ng of DNA extract from freshwater (Burnet Woods Pond and Shepards Creek) and marine water (Jobos Bay) filtrates not impacted by bovine fecal pollution. DNA from water samples (Sea Graves) impacted by bovine feces was also tested. All validation PCR assays were performed in duplicate. The lower limit of detection for each primer set was estimated with serial dilutions of bovine fecal DNA starting with a concentration of 10 ng/μl. No-template, extraction blank, and water filtration blank PCR control assays were performed to test for the presence of extraneous DNA molecules introduced during laboratory experiments.
RESULTS
Identification of unique marker sequences for bovine fecal bacteria.
Four hundred sixty-eight randomly selected clones containing DNA fragments between 163 and 973 bp in size were initially characterized from plasmid clone libraries obtained by one, two, or three rounds of metagenomic GFE (Fig. 1 and Table 1). Sequence analyses showed that this subset contained a total of 380 nonredundant sequences (Table S1 in the supplemental material) and that sequences from each round of GFE contained similar numbers of redundant clones (Table 1). Dot blot hybridizations with the original porcine fecal DNA as probes were then used to identify any false-positive GFE clones obtained (Fig. 2). These analyses showed only 10 plasmid insert DNA sequences capable of hybridizing with porcine fecal DNA probes, demonstrating a very low false-positivity rate (2.6%) among GFE clones. A breakdown of false-positivity rates by GFE round (Table 1) showed that 90% (n = 9) false-positive clones were obtained following the third round of enrichment, indicating that one or two rounds of GFE is sufficient to obtain a highly enriched metagenomic clone library.
TABLE 1.
Summary of sequenced DNA clones obtained over three rounds of GFE
| GFE round | No. of clones | Avg length (bp) | No. of redundant sequences | No. of false positivesa |
|---|---|---|---|---|
| 1 | 158 | 380 | 34 | 1 |
| 2 | 157 | 448 | 35 | 0 |
| 3 | 153 | 446 | 19 | 9 |
| Total | 468 | 391 | 88 | 10 |
Number of false-positive sequences identified by dot blot hybridization.
FIG. 2.
Dot blot hybridization analysis of putative host-specific DNA fragments. PCR amplicons from all nonredundant clone sequences (88 are shown) were transferred to nylon membranes and hybridized to a biotin-labeled bovine (panel A) or porcine (panel B) fecal metagenomic DNA probe. Positive controls are indicated by boxes and included 800 ng of bovine fecal metagenomic DNA (panel A, row H, column 12) and 100 ng (panel B, row C, column 11), 200 ng (panel B, row C, column 12), 400 ng (panel B, row D, column 10), and 800 ng (panel B, row H, column 12) of porcine fecal metagenomic DNA. The porcine fecal metagenomic DNA probe cross-hybridized with 200 ng (panel B, row F, column 2) of bovine fecal metagenomic DNA. None of the no-DNA controls (panel A, row B, column 4; row C, columns 11 and 12; row D, columns 3 and 10; row F, column 2; row H, column 10; panel B, row B, column 4; row D, column 3; row H, column 10) hybridized to the probe.
Characterization of bovine fecal DNA sequences obtained by GFE.
BLASTx sequence similarity searches of the 380 nonredundant GFE clone insert sequences against the NCBI RefSeq and ExPASy Swiss-Prot protein databases identified homologous sequences for 256 clones on the basis of an expectation value cutoff of ≤1e−3 (Table S1 in the supplemental material). A best BLASTx hit for each GFE sequence was established on the basis of a comparison of score density values (bit score/match length) calculated from each database BLASTx search result (Table S1 in the supplemental material). Best BLASTx hits averaged only 51.4% sequence identity to nonredundant GFE sequences. GFE sequences were then labeled Bacteroidales-like as if the respective GFE sequence top BLASTx hit originated from a previously described Bacteroidales genome (16, 22, 39). One hundred twenty-four (32.6%) of these 380 bovine fecal community DNA sequences showed no homology to any previously reported gene sequences.
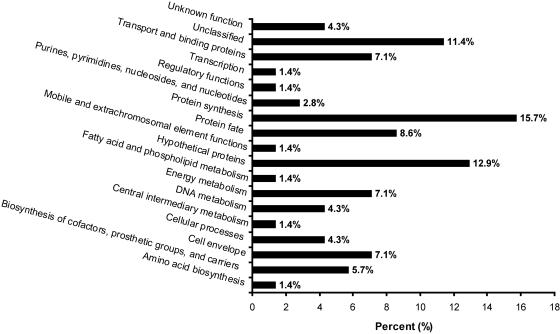
Bacteroidales-like sequences were of particular interest because they are potentially related to Bacteroides species, which are both abundant in feces and restricted in their host ranges (2, 7, 9, 11, 12, 17, 18, 20, 30). All Bacteroidales-like host-specific GFE clone sequences were then assigned to 1 of 18 functional groups (Fig. 3) based on previous annotation of the B. thetaiotaomicron VPI-5482 genome (39; Table S1 in the supplemental material). The functional groups most frequently assigned to these sequences were protein synthesis (15.7%) and hypothetical proteins (12.9%). Eleven (15.7%) of the Bacteroidales-like GFE clone sequences are predicted to encode proteins involved in protein synthesis and are potentially related to tRNA metabolism, including seven tRNA synthetases, three tRNA transferases, and an RNA methyltransferase. In contrast to our previous results obtained by direct GFE analyses of the genomes of two enterococcal species (28), no rRNA-encoding or intercistronic spacer regions were obtained by fecal metagenomic GFE assays. On the basis of additional annotations from the B. thetaiotaomicron study (39), 42 Bacteroidales-like sequences are predicted to encode membrane-associated or putative extracellular proteins, of which a surprising 80.9% are predicted secretory proteins according to SignalP analyses (4).
FIG. 3.
Functional group assignments for host-specific Bacteroidales-like GFE sequences based on a previous B. thetaiotaomicron genome study (39). Functional groups are listed along the x axis, and the percentage of Bacteroidales-like sequences (total number = 70) for each group assignment is shown along the y axis.
Five distinct B. thetaiotaomicron VPI-5482 DNA regions were indicated by two or more nonredundant (i.e., not plasmid clone sibling) Bacteroidales-like clone sequences obtained by GFE. These included variable DNA regions potentially encoding an AcrB/AcrD/AcrF family protein homologous to BT2686, a methionyl-tRNA synthetase (BT2933), a preprotein translocase (SecA subunit, BT4362), a reticulocyte binding protein (BT0749), and a conserved hypothetical protein BT0921 (39). Interestingly, all of these DNA regions are predicted to encode secretory proteins by SignalP analyses (4, 39), with the exception of the SecA-like preprotein translocase, which is potentially involved in protein export.
PCR optimization and specificity validation for bovine fecal DNA detection.
Three host-specific Bacteroidales-like GFE clone sequences were randomly selected for continued development of bovine feces-specific PCR assays (Table 2). PCR assay 1 targeted a 368-bp DNA fragment potentially encoding a conserved hypothetical secreted protein. The best BLASTx hit (8.00e−11) for this sequence shows 25% sequence identity to a B. fragilis hypothetical protein (locus BF2432). PCR assay 1 routinely detected 1 fg of bovine fecal DNA (Fig. 4) under optimized PCR conditions (62°C annealing, 30 cycles). Primers for PCR assay 2 targeted a 437-bp fragment annotated as encoding an HDIG domain protein involved in energy metabolism and electron transport (B. thetaiotaomicron genome locus BT2749). The best BLASTx hit for this sequence (32% identical; 1.00e−8) was a B. fragilis YCH46 putative membrane-associated HD superfamily hydrolase. Optimal conditions for PCR assay 2 include a 62°C annealing temperature and 35 cycles, which allow a detection limit of 10 fg of bovine fecal DNA (Fig. 4). PCR assay 3 was developed from a 569-bp DNA fragment predicted to encode a sialic acid-specific 9-O-acetylesterase secretory protein homolog (B. thetaiotaomicron genome locus BT0457) with a predicted role in cell envelope biosynthesis and degradation of surface polysaccharides and lipopolysaccharides. The best BLASTx hit for this sequence (75% identical; 8.00e−80) was a sialate O-acetylesterase protein from B. fragilis YCH46. PCR assay 3 exhibited the lowest limit of detection under optimal conditions (60°C, 35 cycles) and consistently amplified 0.1 fg of bovine fecal DNA (Fig. 4).
TABLE 2.
Host-specific PCR assay primers and optimal reaction conditions
| PCR assay no. | Primer set | Sequence (5′ to 3′) | Amplicon length (bp) | Optimal annealing temp (°C) | Optimal no. of cycles |
|---|---|---|---|---|---|
| 1 | Bac1F | TGCAATGTATCAGCCTCTTC | 196 | 62 | 30 |
| Bac1R | AGGGCAAACTCACGACAG | ||||
| 2 | Bac2F | GCTTGTTGCGTTCCTTGAGATAAT | 274 | 62 | 35 |
| Bac2R | ACAAGCCAGGTGATACAGAAAG | ||||
| 3 | Bac3F | CTAATGGAAAATGGATGGTATCT | 166 | 60 | 35 |
| Bac3R | GCCGCCCAGCTCAAATAG |
FIG. 4.
Limit of detection of host-specific primer sets with serial dilutions (1:10) of bovine fecal metagenomic DNA starting with 10 ng of bovine fecal DNA (lane 1). Panel A, 1 fg of DNA was detected by PCR assay 1 (Bac1F and Bac1R). Panel B, 10 fg of DNA was detected by PCR assay 2 (Bac2F and Bac2R). Panel C, 0.1 fg of DNA was detected by PCR assay 3 (Bac3F and Bac3R).
DNA prepared from different bovine fecal specimens was used as a template to test the host distribution and temporal stability of putative bovine feces-specific PCR assays. All three PCR assays amplified DNA of the expected size from the original target GFE bovine fecal sample, and between 72% and 100% of 148 bovine fecal samples from five different geographical locations collected over a 24-month period (Table 3). PCR assay 3 showed both the broadest target host distribution and the greatest temporal stability by successfully amplifying DNA from 91% of all 148 bovine fecal samples. Each primer set was then tested in assays with template DNA extracted from 245 individual fecal and septic tank samples representing 29 different animal species. PCR assays 1 and 2 exclusively amplified bovine fecal DNA molecules (Table 4). PCR assay 3 exhibited specificity for 99.2% of the fecal samples but did amplify DNA from two alpaca fecal samples. Each PCR assay also demonstrated high levels of specificity in feces-impacted freshwater and marine water samples (Table 4). All water samples not impacted by bovine fecal pollution tested negative in PCR assays, while bovine feces-impacted water samples tested positive in PCR assays 2 and 3. (Fig. 5).
TABLE 3.
Host distribution PCR assays
| Location | No. of cows | Collection yr | % of samples positive by:
|
||
|---|---|---|---|---|---|
| PCR assay 1 | PCR assay 2 | PCR assay 3 | |||
| Nebraska | 101 | 2004 | 70.3 | 77.2 | 87.1 |
| West Virginia | 26 | 2002 | 69.2 | 76.9 | 100 |
| Georgia | 10 | 2004 | 100 | 100 | 100 |
| Texas | 1 | 2004 | 100 | 100 | 100 |
| Delaware | 11 | 2003 | 63.6 | 100 | 100 |
| Total | 148 | 72 | 80 | 91 | |
TABLE 4.
Summary of host-specific PCR assay cross-specificity tests
| Group | No. of samples | No. of species | % of samples positive by:
|
||
|---|---|---|---|---|---|
| PCR assay 1 | PCR assay 2 | PCR assay 3 | |||
| Birds | 32 | 5 | 0 | 0 | 0 |
| Human or septic tank | 25 | 1 | 0 | 0 | 0 |
| Domestic | 64 | 6 | 0 | 0 | 2 |
| Wildlife | 99 | 11 | 0 | 0 | 0 |
| Pets | 25 | 3 | 0 | 0 | 0 |
| Water not impacted by cattle | 34 | 0 | 0 | 0 | |
| Total | 279 | 27 | 0 | 0 | 0.8 |
FIG. 5.
Gel electrophoresis of PCR products from reactions with bovine-specific PCR assays 1, 2, and 3 (panels A, B, and C, respectively) and a Bacteroidales-specific PCR assay (panel D) with primers Bac32F and Bac708R (6). Each PCR assay was tested against DNA extracts from feces-impacted fresh and marine water samples (five are shown). Lanes: Jobos Bay, PR, lane 1; Shepards Creek, OH, lane 2; Burnet Woods Pond, OH, lane 3; Chandler Farms, GA, lanes 4 and 5; no-template control, lane 6; bovine fecal DNA, lane 7.
Experimental controls.
Each fecal, water, and septic tank reference sample yielded the expected PCR products when amplified with Bacteroidales 16S rRNA-specific reference primers Bac32F and Bac708R (6), indicating a lack of potential PCR inhibitors. The presence of extraneous DNA molecules introduced during laboratory manipulations by performing no-template (n = 276), extraction blank (n = 27), and water filtration blank (n = 18) control PCR assays was also tested for. In all cases, the results were negative. Because of the requirement for the lone primer amplification step (16) to selectively amplify DNA with terminal tag sequences and because of the complex nature of the metagenomic templates used in these experiments, we also tested the specificity of the K9-PCR and F9-PCR primers. No amplification was observed when the K9-PCR primer was tested against sheared bovine metagenomic DNA or when the F9-PCR primer was tested against sheared porcine metagenomic DNA (data not shown).
DISCUSSION
Several hundred candidate bovine fecal community-specific DNA sequences were obtained by the application of GFE to total DNAs extracted from individual bovine and porcine fecal specimens. Enrichment for the desired sequences with regard to the two original sources was confirmed for 97.4% of these fragments by dot blot hybridizations, indicating that GFE is an efficient approach to obtain unique or highly divergent DNA sequences between metagenomic nucleic acid pools and is a valuable targeted alternative approach to large-scale sequencing for researchers whose diverse goals include the need to characterize genetic variation among complex microbial communities. Although sharing some aspects of other DNA-sorting methods, GFE is neither subtractive hybridization (31) nor PCR-mediated selective amplification (10, 21) of target molecules. Rather, GFE obtains a sampling of the difference between two nucleic acid pools by competitive solution hybridization and physical separation, followed by amplification of all of the target molecules obtained. In contrast, subtractive hybridization relies on the inherently difficult process of complete physical removal of all shared sequences as heterologous nucleic acid hybrids prior to amplification of all of the remaining hybrid DNA strands. Although target molecules captured by GFE as hybrids are amplified by a PCR targeting terminal tag sequences, it does not rely on the PCR process itself to select unique nucleic acids from pools, as do suppression subtractive hybridization (10) and representational difference analysis (21).
Bovine feces-specific GFE sequences exhibited an average of only 51% sequence identity with NCBI RefSeq and ExPASy Swiss-Prot database sequences, reflecting the fact that many conserved but quite divergent bacterial chromosomal regions were obtained. Approximately one-third of the GFE clone sequences showed no similarity to any previously described genes, suggesting that many of the DNA fragments originate from previously uncharacterized prokaryotic genes. It is important to note that in order to generate a complete assessment of the genetic diversity between the bovine and porcine fecal metagenomes, hundreds of thousands of GFE fragments must be sequenced. The intent of this study was not to characterize every genetic difference between these microbial communities but to determine if GFE could isolate DNA sequences unique to one bacterial community and absent in another that could be used to develop host-specific PCR assays.
Classification of Bacteroidales-like GFE clone sequences based on B. thetaiotaomicron genome annotations (39) indicates an abundance of genes that encode bacterial surface-associated and secreted proteins, suggesting that a potential major difference between bovine and porcine fecal Bacteroidales-like genes is in their capacity for producing secreted factors. This is perhaps not surprising considering that genome studies have shown that the majority of the B. fragilis (19) and B. thetaiotaomicron (39) proteomes are dedicated to metabolizing dietary polysaccharides harvested in the gastrointestinal tract. Large sequence variations in genes involved in membrane-associated activities have also been reported among closely related microorganisms, including E. coli O55 and O157 (32), enterococci (28), and other opportunistic and obligate pathogens (27, 33, 34).
The central objective of this study was to determine if GFE could be used to enrich for DNA fragments from a bovine fecal metagenome that were divergent or absent in a porcine fecal metagenome. After many such sequences were identified, three putative bovine community-specific PCR assays were designed and optimized on the basis of randomly selected GFE Bacteroidales-like DNA sequences. These assays consistently amplified target DNAs from more than 80% of 148 bovine fecal samples regardless of their geographic origin. Markers also showed remarkable stability over a 24-month period at these sample locations. Considering that DNA sequences for these PCR assays were obtained from an initial comparison limited to just two individual fecal samples, it seems likely that GFE will be a useful approach to identify additional discriminatory genetic markers for other fecal microbial communities.
The stability and broad distribution of the PCR markers in bovine populations encouraged us to test target specificity against other animal groups. All three PCR assays showed extremely high levels of specificity for bovine fecal DNA (Table 4). These results corroborate the notion that genes encoding bacterial proteins directly involved in interactions with hosts exhibit increased levels of specificity relative to bacterial 16S rRNA gene sequences (7, 11). Current 16S rRNA gene-based PCR assays can only discriminate between ruminant and nonruminant fecal sources (7). The bovine-specific PCR assays described here that target nonribosomal sequences were able to differentiate among cattle and five other ruminant or pseudoruminant species, including goats, sheep, alpacas, llamas, and whitetail deer, with the exception of bovine-specific PCR assay 3 (Table 4), which produced positive signals for two alpaca fecal samples.
To explore the potential of our bovine-specific PCR assays for fecal source identification in natural waters, each PCR assay was challenged against water samples collected from a stream situated in central Georgia that is directly impacted by a local dairy farm; PCR assays 2 and 3 tested positive. These initial experiments suggest that our novel PCR assays may have utility in environmental monitoring. However, in order to realize the full potential of these PCR assays for fecal source tracking applications, several issues need to be addressed that go beyond the scope of this study, such as the survival of target DNA molecules in water, the relevance of each PCR assay to current regulatory fecal indicator methods used to monitor water quality (such as E. coli and enterococci), and linking of the prevalence of a specific DNA target sequence in the environment to relevant public health risks.
Supplementary Material
Acknowledgments
This research was funded in part by a New Start Award from the National Center for Computational Toxicology of the U.S. Environmental Protection Agency Office of Research and Development.
We are grateful to Sam Myoda, Don Stoeckel, George DiGiovanni, Jason Vogel, and Michelle Bonkosky for providing fecal and water samples. Donald Reasoner, Rich Haugland, and Marirosa Molina are recognized for early discussions.
Any opinions expressed in this paper are ours and do not necessarily reflect the official positions and policies of the U.S. Environmental Protection Agency. Any mention of products or trade names does not constitute recommendation for use by the U.S. Environmental Protection Agency.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Acinas, S. G., L. A. Marcelino, V. Klepac-Ceraj, and M. F. Polz. 2004. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J. Bacteriol. 186:2629-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allsop, K., and J. D. Stickler. 1985. An assessment of Bacteroides fragilis group organisms as indicators of human faecal pollution. J. Appl. Bacteriol. 58:95-99. [DOI] [PubMed] [Google Scholar]
- 3.Althschul, S. F., F. Thomas, L. Madden, A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 5.Benenson, A. S. 1995. Control of communicable diseases manual. American Public Health Association, Washington, D.C.
- 6.Bernhard, A. E., and K. G. Field. 2000. Identification of nonpoint sources of fecal pollution in coastal waters by using host-specific 16S ribosomal DNA genetic markers from fecal anaerobes. Appl. Environ. Microbiol. 66:1587-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernhard, A. E., and K. G. Field. 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl. Environ. Microbiol. 66:4571-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Covert, T. C. 1999. Salmonella, waterborne pathogens, AWWA manual M48. American Water Works Association, Denver, Colo.
- 9.Daly, K., C. S. Stewart, H. J. Flint, and S. P. Shirazi-Beechey. 2001. Bacterial diversity within the equine large intestine as revealed by molecular analysis of cloned 16S rRNA genes. FEMS Microbiol. Ecol. 38:141-151. [Google Scholar]
- 10.Diatchenko, L., Y. C. Lau, A. P. Cambell, A. Chenchik, F. MoQadam, B. Huang, S. Lukyanov, K. Lukynov, N. Gurskaya, E. D. Sverdlov, and P. D. Siebert. 1996. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl. Acad. Sci. USA 93:6025-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dick, L. K., A. E. Bernhard, T. J. Brodeur, J. W. Santo Domingo, J. M. Simpson, S. P. Walters, and K. G. Field. 2005. Host distributions of uncultivated fecal Bacteroidales bacteria reveal genetic markers for fecal source identification. Appl. Environ. Microbiol. 71:3184-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fiksdal, L., J. S. Make, S. J. LaCroix, and J. T. Staley. 1985. Survival and detection of Bacteroides spp., prospective indicator bacteria. Appl. Environ. Microbiol. 49:148-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fricker, C. 1999. Campylobacter, waterborne pathogens, AWWA manual M48. American Water Works Association, Denver, Colo.
- 14.Graham, J. E., and J. E. Clark-Curtiss. 1999. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc. Natl. Acad. Sci. USA 96:11554-11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grimont, F., and P. A. Grimont. 1986. Ribosomal ribonucleic acid gene restriction patterns as potential taxonomic tools. Ann. Inst. Pasteur Microbiol. 137B:165-175. [DOI] [PubMed] [Google Scholar]
- 16.Grothues, D., C. R. Cantor, and C. L. Smith. 1993. PCR amplification of megabase DNA with tagged random primers (T-PCR). Nucleic Acids Res. 21:1321-1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hold, G. L., S. E. Pryde, V. J. Russell, E. Furrie, and H. J. Flint. 2002. Assessment of microbial diversity in human colonic samples by 16S rDNA sequence analysis. FEMS Microbiol. Ecol. 39:33-39. [DOI] [PubMed] [Google Scholar]
- 18.Kreader, C. A. 1995. Design and evaluation of Bacteroides DNA probes for the specific detection of human fecal pollution. Appl. Environ. Microbiol. 61:1171-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuwahra, T., A. Yamashita, H. Hirakawa, H. Nakayama, H. Toh, N. Okada, S. Kuhara, M. Hattori, T. Hayashi, and Y. Ohnishi. 2004. Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation. Proc. Natl. Acad. Sci. USA 101:14919-14924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leser, T. D., J. Z. Amenuvor, T. K. Jensen, R. H. Lindecrona, M. Boye, and K. Moller. 2001. Culture-independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl. Environ. Microbiol. 68:673-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lisitsyn, N., N. Lisitsyn, and M. Wigler. 1993. Cloning the differences between two complex genomes. Science 259:946-951. [DOI] [PubMed] [Google Scholar]
- 22.Nelson, K. E., R. D. Fleischmann, R. DeBoy, I. T. Paulsen, D. E. Fouts, J. A. Eisen, S. C. Daugherty, R. J. Dodson, A. S. Durkin, M. Gwinn, D. H. Haft, J. Kolonay, W. C. Nelson, T. Mason, L. Tallon, J. Gray, D. Granger, H. Tettelin, H. Dong, J. L. Galvin, M. J. Duncan, F. E. Dewhirst, and C. M. Fraser. 2003. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J. Bacteriol. 185:5591-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohio State University. 1992. Ohio livestock manure and wastewater management guide bulletin 604. Ohio State University, Columbus.
- 24.Okhuysen, P. C., J. H. Chappell, J. H. Crabb, C. R. Sterling, and H. L. DuPont. 1999. Virulence of three distinct Cryptosporidium parvum isolates for healthy adults. J. Infect. Dis. 180:1275-1278. [DOI] [PubMed] [Google Scholar]
- 25.Pruimboom-Brees, I. M., T. W. Morgan, M. R. Ackermann, E. D. Nystrom, J. E. Samuel, N. A. Cornick, and H. W. Moon. 2000. Cattle lack vascular receptors for Escherichia coli O157:H7 Shiga toxins. Proc. Natl. Acad. Sci. USA 97:10325-10329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scott, T. M., J. B. Rose, T. M. Jenkins, S. R. Farrah, and J. Lukasik. 2002. Microbial source tracking: current methodology and future directions. Appl. Environ. Microbiol. 68:5796-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Selander, R. K. 1997. DNA sequence analysis of the genetic structure and evolution of Salmonella enterica, p. 191-213. In B. A. M. van der Zeijst, W. P. M. Hoekstra, and J. D. A. van Embden (ed.), Ecology of pathogenic bacteria: molecular and evolutionary aspects. Royal Netherlands Academy of Arts and Sciences, Amsterdam, The Netherlands.
- 28.Shanks, O. C., J. W. Santo Domingo, and J. E. Graham. 6 February 2006. Use of competitive DNA hybridization to identify differences in the genomes of bacteria. J. Microbiol. Methods. [Online.] doi: 10.1016/j.mimet.2005.12.006. [DOI] [PubMed]
- 29.Simpson, J. M., D. J. Reasoner, and J. W. Santo Domingo. 2002. Microbial source tracking: state of the science. Environ. Sci. Technol. 36:5279-5288. [DOI] [PubMed] [Google Scholar]
- 30.Simpson, J. M., J. W. Santo Domingo, and D. J. Reasoner. 2004. Assessment of equine fecal contamination: the search for alternative bacterial source-tracking targets. FEMS Microbiol. Ecol. 47:65-75. [DOI] [PubMed] [Google Scholar]
- 31.Straus, D., and F. M. Ausubel. 1990. Genomic subtraction for cloning DNA corresponding to deletion mutations. Proc. Natl. Acad. Sci. USA 87:1889-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tarr, P. I., L. M. Schoening, Y. L. Yea, T. R. Ward, S. Jelacic, and T. S. Whittam. 2000. Acquisition of the rfb-gnd cluster in evolution of Escherichia coli O55 and O157. J. Bacteriol. 182:6183-6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 34.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain BC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 35.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.U.S. Department of Agriculture. 2005. Cattle inventory report. U.S. Department of Agriculture, Washington, D.C.
- 37.U.S. Environmental Protection Agency. 2002. National water quality inventory report EPA-841-R-02-001. U.S. Environmental Protection Agency, Washington, D.C.
- 38.Wood, J., K. P. Scott, G. Avgustin, C. J. Newbold, and H. J. Flint. 1998. Estimation of the relative abundance of different Bacteroides and Prevotella ribotypes in gut samples by restriction enzyme profiling of PCR-amplified 16S rRNA gene sequences. Appl. Environ. Microbiol. 64:3683-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu, J., M. K. Bjursell, J. Himrod, S. Deng, L. K. Carmichael, H. C. Chiang, L. V. Hooper, and J. I. Gordon. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074-2076. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.