Abstract
Helicobacter pylori BabA is the ABO blood group antigen binding adhesin, which has a closely related paralogue (BabB) whose function is unknown. PCR and DNA sequence analysis showed extensive genotypic diversity in babA and babB across different strains, as well as within a strain colonizing an individual patient. We hypothesize that diverse profiles of babA and babB reflect selective pressures for adhesion, which may differ across different hosts and within an individual over time.
Approximately 4% of the Helicobacter pylori genome encodes a diverse repertoire of outer membrane proteins (OMPs), the largest of which is the 21-gene Hop family (18). Several of the H. pylori Hop proteins have been identified as adhesins, the best studied of which is the ABO blood group binding antigen, BabA (4, 6, 7, 9). The predicted H. pylori OMPs all have one region of similarity at the amino-terminal end and seven regions of similarity at the carboxy-terminal end (1, 18). This has led to the suggestion that recombination events might lead to a mosaic organization of OMPs that could be the basis for antigenic variation (18). This suggestion is supported by the observation that H. pylori J99 and 26695 have babA and babB in complementary loci (2, 18) and by evidence of RecA-dependent recombination in vitro (11). Many of the OMPs also have dinucleotide CT repeats in the 5′ coding region, which have been postulated to regulate their expression by phase variation through slipped-strand mispairing (10, 14, 20). This plasticity in the profile of H. pylori OMPs may provide a mechanism for adaptation to the different niches and microenvironments within the stomach, either by immune evasion, adhesion, or both, or perhaps by some other mechanism.
We recently observed an apparent gene conversion after experimental infection of rhesus macaques with H. pylori strain J166 (17). The inoculated J166 strain had babA and babB in the same chromosomal loci as J99, but most of the strains recovered after infection had replaced babA with a second copy of babB. The few recovered strains that retained babA did not express it, as a result of changes in the number of CT dinucleotide repeats preceding the 5′ signal peptide sequence. This result, combined with evidence that babA is absent in some human strains (13, 21), led us to hypothesize that the recombination events we observed in monkeys might reflect a response to selective pressure that may also be apparent in human clinical isolates of H. pylori. In this report we describe the composition of babA and babB at two loci in 44 human H. pylori isolates and provide evidence that the genotypes of these two OMPs are highly variable.
Determination of the babA and babB genotypes.
In H. pylori J99, babA (JHP0833) is downstream of hypD (JHP0835) and babB (JHP1164) is downstream of s18 (JHP1165). In strain 26695, the locations of babA (HP1243) and babB (HP0317) are reversed (2). For simplicity, the chromosomal locations downstream of hypD and s18 will be referred to as the babA and babB loci, respectively. We performed four anchored PCRs with primer pairs designed to query whether the gene at the babA locus was babA (primer pair HypDF1-BabAR1 [Table 1]) or babB (HypDF1-BabBR1) and whether the gene at the babB locus was babA (S18F1-BabAR1) or babB (S18F1-BabBR1). Each of the four PCRs (94°C for 5 min; 40 cycles of 94°C for 30 s, 56.5°C for 30 s, 72°C for 2 min; 72°C for 5 min) was performed on chromosomal DNA from low-passage-number, single-colony isolates (n = 44) by using standard concentrations of reagents. Primers (Table 1) were designed from H. pylori J99 (JHP0835 and JHP1165) or from multiple sequence alignments used to identify conserved regions that reliably discriminate babA and babB (12). PCR products were electrophoresed in 1% agarose (Gibco BRL) using J99 and 26695 as controls. All PCR products of the predicted size (babA locus, 2.1 to 2.6 kb; babB locus, 1.0 to 1.5 kb) from each of these four reactions were sequenced over a mean (standard deviation [SD]) of 676 (85) bp and compared to the GenBank nucleotide database using FASTA. PCR products for which it appeared that both babA and babB were present at the same locus (e.g., both S18F1-BabAR1 and S18F1-BabBR1 PCRs positive) were also sequenced. The presence of babA or babB at a given locus was defined in all cases as a positive PCR product of the predicted size whose DNA sequence was most closely related to babA or babB. To determine if a gene could be assigned correctly to babA or babB based on a mean of 676 bp, we performed the analysis on 10 complete sequences of babA and 10 of babB available in GenBank. In all 20 instances, the assignment was made correctly (data not shown). Mean (SD) percent similarities of the identified babA and babB genes to their orthologues in strain J99 were 90.4% (3.3%) and 89.2% (4.3%), respectively.
TABLE 1.
Primers used for amplification, cloning, and real-time PCR
| Primer designation | Genea | Position | Sequence (5′-3′) |
|---|---|---|---|
| S18F1 | JHP1165 | 197-218 | CTTTAATCCCCTACATTGTGGA |
| S18F2 | JHP1165 | 137-157 | GCAATAGCAAAAAGTGGC AAG |
| S18F3 | JHP1165 | 190-211 | CACATGGCTTTAATCCCCTACA |
| HypDF1 | JHP0835 | 675-697 | TTTTGAGCCGGTGGATATATTAG |
| HypDF2 | JHP0835 | 732-754 | CAAAGAAGCCAAGCTAGAAATCC |
| BabAR1 | JHP0833 | 723-705 | TTTGCCGTCTATGGTTTGG |
| BabAR2 | JHP0833 | 444-424 | ATACCCTGGCTCGTTGTTGAA |
| BabAR3 | JHP0833 | 255-236 | ATCGTTACGCACCCCATTGA |
| BabAR4 | JHP0833 | 363-341 | GCCTAAGACATTCCAAAACCCTA |
| BabBR1 | JHP1164 | 772-750 | TCGCTTGTTTTAAAAGCTCTTGA |
| BabBR2 | JHP1164 | 481-459 | CATGTCCTGGCTCATAATACGAA |
| BabBR3 | JHP1164 | 389-370 | TCATTGCTACCAGGACCACA |
| BabBR4 | JHP1164 | 285-261 | GGTTTTGACATCAAGCAAATTCCTA |
H. pylori strain J99 designation.
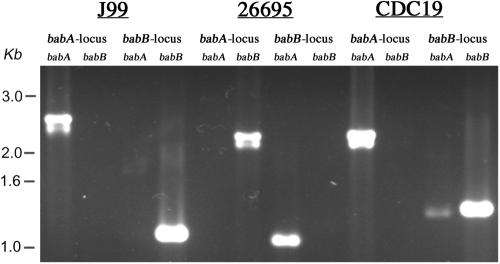
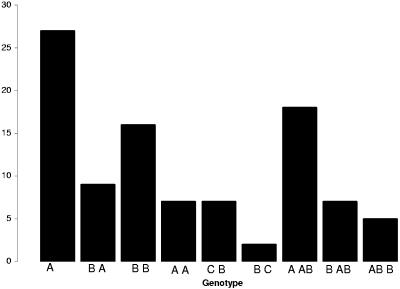
A representative agarose gel for the two sequenced strains, J99 and 26695, and a clinical isolate is shown in Fig. 1. In total, 27% (12/44) of isolates were J99-like, with babA in the babA locus and babB in the babB locus (designated AB), and 9% (4/44) were 26695-like, with babA in the babB locus and babB in the babA locus (designated BA) (Table 2; Fig. 2). In 23% (10/44) of the isolates, either babA or babB alone was present at both loci (AA or BB). In 30% (13/44) of the isolates, there was a mixed genotype, where the population of cells (derived from a single colony) contained both babA and babB at the same locus, a phenomenon previously observed for strain J166 (17). In all but 2 of these 15 isolates, the mixed genotype was at the babB locus (e.g., A AB). To determine the limit of detection for babA at the babB locus in a mixed-genotype strain, PCR was performed (primers S18F1 and BabAR1) using a mixture of DNA from 26695 (AB) and serial dilutions of strain H45 (BB) in ratios from 1 to 10−6. The results showed that babA could be detected as a minority population in the babB locus when it was present at a fraction of 10−5 or greater (data not shown). Interestingly, 9% (4/44) of the PCR products yielded sequences that were most closely related (mean homology, 85%; SD, 1.9%) to HP0317 (hopU) in strain 26695. The function of HP0317 is unknown, but it is closely related to babA and babB and is here designated babC. One strain (D5131) contained babB at the babB locus but did not contain an identified OMP at the babA locus. These results suggest that the babAB genotype of H. pylori is complex and that some strains may have undergone a gene conversion event much like that observed in experimentally infected rhesus monkeys (17), with deletion of babA and duplication of babB.
FIG. 1.
PCR products stained with ethidium bromide and electrophoresed in a 1% agarose gel. Results are shown for each of the four PCRs (babA in the babA and babB loci; babB in the babA and babB loci [see Materials and Methods for details]) performed on control strains J99 (genotype AB) and 26695 (genotype BA) and on a sample clinical isolate (CDC19 [genotype A AB]). A kilobase ladder is shown on the left.
TABLE 2.
H. pylori strain genotype and Leb attachment
| Strain | Genotypea
|
Leb attachmentb | |
|---|---|---|---|
| babA locus | babB locus | ||
| CDC2 | A | A | + |
| TR86-1 | A | A | + |
| H135-1 | A | A | + |
| CDC12 | A | AB | − |
| CDC22 | A | AB | − |
| D5135 | A | AB | − |
| H106-1 | A | AB | − |
| 95-66 | A | AB | + |
| CDC19 | A | AB | + |
| CDC3 | A | AB | + |
| H28-2 | A | AB | + |
| CDC16 | A | B | − |
| AR20 | A | B | − |
| AR3-1 | A | B | − |
| D5129 | A | B | − |
| H116B | A | B | − |
| 878787B6 | A | B | + |
| CDC21 | A | B | + |
| CDC24 | A | B | + |
| D5127 | A | B | + |
| J99 | A | B | + |
| D5132 | A | B | + |
| CDC1 | A | B | − |
| CDC10 | AB | B | + |
| J166 | AB | B | + |
| 26695 | B | A | − |
| CDC11 | B | A | − |
| D2371 | B | A | + |
| TR106-3 | B | A | + |
| AR19 | B | AB | − |
| TR56-5 | B | AB | + |
| H13-1 | B | AB | + |
| 88-23 | B | B | − |
| CDC25 | B | B | − |
| H34-JP | B | B | − |
| H11 | B | B | ND |
| H40-2 | B | B | ND |
| H45-1 | B | B | − |
| H48-1 | B | B | ND |
| AR18 | B | C | − |
| CDC8 | C | B | − |
| H12 | C | B | − |
| H100 | C | B | − |
| D5131 | B | − | |
A, babA; B, babB; AB, babA and babB; C, babC.
+, positive; −, negative; ND, not determined.
FIG. 2.
Percentage of strains (n = 44) that showed each of the genotypes indicated. For example, strains with genotype AB have babA at the babA locus and babB at the babB locus. A AB, B AB, and AB B indicate the genotypes of strains that had more than one bab gene present at a given locus within the population derived from a single colony.
Intrastrain genotype variation.
Since H. pylori shows marked genomic diversity, we next examined multiple single-colony isolates from each of three patients to determine if the diversity we found across patients could also be detected within a patient at a single point in time. Multiple (9 to 12) additional single colonies were isolated from the original cultures of a strain (H106) with genotype A AB (babA at the babA locus and both babA and babB at the babB locus), a J99-like strain (H116) with genotype AB, and a strain with genotype AA (H135). Each of the four PCRs and DNA sequencing of the products were performed as before on each isolate. For strain H106, the babAB genotype was A AB in all 12 isolates. However, although most isolates of strain H135 and H116 were AA and AB, respectively, like the original isolate (Table 2), 4 of 12 (33%) and 4 of 9 (44%), respectively, were A AB. Therefore, all patients from whom we examined multiple isolates were colonized with strains that were least partially AB at the babB locus. Repetitive extragenic palindromic PCR fingerprints (16) were identical for all isolates from each of the three patients (data not shown), indicating that patients are commonly colonized with a single strain but that there is considerable heterogeneity in the babAB genotype within the population, particularly at the babB locus.
Rates of recombination.
Relative abundances of babA and babB at the babB locus were determined within the population for four strains (H106, D5135, CDC12, and CDC3) with genotype A AB. In each case babB appeared predominant by standard PCR (for example, strain CDC19 in Fig. 1). Copy numbers of babA and babB at the babB locus were determined using quantitative real-time DNA PCR as previously described (17). The results showed that for each strain, the frequency of babA at the babB locus was between 10−3 and 10−4, a result similar to the finding we reported previously for strain J166 (17). The relative frequency of babA at the babB locus appeared to be stable for a given strain, since assays performed on 10 independent colonies of H106 were highly reproducible (mean, 2.2 × 10−4; SD, 1.9 × 10−4).
Site of recombination.
DNA sequences from 10 strains (8 with genotype A AB, 1 with genotype B AB, and 1 with genotype AB B) were inspected to determine the upstream location at which the recombination event occurred for the second copy of babA or babB. The approximate site of recombination was identified as the position at which the second copy of babA, for example (in a strain with genotype A AB), showed homology to babA and not babB. In five strains (all genotype A AB), the recombination event was identified from approximately 50 to 200 bp downstream of the ATG, while in the other five strains, it was upstream of the ATG. Therefore, recombination of a second copy of babA or babB occurs before the unique region of the gene that most distinguishes babA from babB.
CT Repeats in babA and babB.
H. pylori strains 26695 and J99 each contain CT repeats in the 5′ coding region of babB, which likely serve to make it phase variable, while in other strains, CT repeats are absent (12). BabA has not been reported to be phase variable (14) until recently (17). We therefore analyzed the babA and babB sequences from each of the 44 strains for the presence of 5′ CT repeats. The results show that CT repeats (minimum, 5; maximum, 11) are more common in babB (43/53 [81%]) than in babA (13/43 [30%]; χ2 = 25.5; P < 0.001) and also more common at the babB locus (42/54 [78%]) than at the babA locus (14/42 [33%]; χ2 = 19.2; P < 0.001). In strains that had duplicate copies of babA (n = 11) or babB (n = 12), both copies were in frame more often for babB (33%) than for babA (9%), but the results were not statistically significant.
Leb attachment.
We used an enzyme-linked immunosorbent assay as described previously (17) to measure attachment to Leb, which serves as an assay for functional binding of babA to blood group antigens. Of 41 strains tested, 19 (46%) showed binding to Leb (Table 2). All strains that bound Leb had at least one copy of babA, though many strains had one or more copies of babA and did not bind Leb. In one case, a strain (AR19) with babA did not bind Leb due to a frameshift, which produced a stop codon; in other cases, one or both copies of babA were in frame but no binding was seen (e.g., AR20, CDC1, D5129). This suggests that there is heterogeneity in binding of BabA to Leb, which is consistent with recent work (8) as well as with the observation that H. pylori J99 binds Leb while 26695 does not (17), even though babA is transcribed and translated in both strains.
Perspective.
The primary ecological niche for H. pylori is the mucus that overlies human gastric mucosa. Elegant studies in the gerbil model suggest that of the approximately 100 μm total thickness of this mucus layer, H. pylori is found only in the 25 μm closest to the epithelium, and only a small minority of bacteria are actually attached to host cells (15). The precise localization of bacteria within the mucus layer may represent a compromise between selective factors that promote adherence, such as nutrient acquisition (19) from disruption of epithelial-cell tight junctions (3), and those that oppose it, such as evasion of reactive oxygen species and other aspects of the host immune response. This hypothesis suggests that fine control and modulation of adhesion may be a critical feature that permits H. pylori to persistently colonize its host despite an active and multifaceted innate and adaptive immune response. Here we show that while most H. pylori strains have babA and babB, which are typically located at the same genomic loci as in strain J99, in 27% of strains (12/44) babA appeared to be absent. This result is in close agreement with results obtained using PCR with Western blotting (21) and a whole-genome DNA microarray (13). Most strains that did not contain babA had a second copy of babB in its place, like strains recovered from experimentally or naturally infected macaques (17). All strains without evidence of babA also tested negative for attachment to Leb (Table 2). The babB gene was identified in 95% of our isolates (41/44) and in 100% (15/15) of isolates studied by DNA microarray (13), suggesting that the presence of babB confers a stronger selective advantage than does the presence of babA. BabB may itself serve as a lectin to bind an unidentified receptor on the host epithelium, or perhaps to modulate binding of other adhesins.
Diversity at the babAB loci is not limited simply to selection for a particular combination of babA, babB, or babC. Examination of multiple colonies from individual patients with babA at the babA locus showed that in all cases both babA and babB were expressed at the babB locus, though in different proportions. This may in fact be true for all such strains. We have, for example, occasionally found strain J99 to have genotype A AB, which may suggest that the mixed genotype of this strain at the babB locus is near the limit of our detection (10−5). CT dinucleotide repeats that permit phase variation were common in the 5′ coding region, particularly in babB, but also in babA when it was found in the babB locus. This evidence for marked phase variation (CT repeats) and antigenic variation (presence of babA and B in different proportions) is consistent with the recent suggestion that babB constitutes an expression or contingency locus (5). We hypothesize that the diversity in babABC reflects both the glycophenotype of the host and selective pressures for adhesion, which may differ across different hosts, across gastric regions within a host, and also within an individual over time.
Nucleotide sequence accession numbers.
All DNA sequences from this study were deposited in the NCBI GenBank database with accession numbers AY743975 to AY744069.
Acknowledgments
We thank Martin Blaser, Benjamin Gold, David Graham, Guillermo Perez-Perez, Julie Parsonnet, and Yoshio Yamaoka for contributing H. pylori strains and Nina Salama for helpful comments on the manuscript.
This work was supported by Public Health Service grants AI42081 and RR14298 from the National Institutes of Health.
Editor: V. J. DiRita
REFERENCES
- 1.Alm, R. A., J. Bina, B. M. Andrews, P. Doig, R. E. Hancock, and T. J. Trust. 2000. Comparative genomics of Helicobacter pylori: analysis of the outer membrane protein families. Infect. Immun. 68:4155-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alm, R. A., L. L. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 3.Amieva, M. R., R. Vogelmann, A. Covacci, L. S. Tompkins, W. J. Nelson, and S. Falkow. 2003. Disruption of the epithelial apical-junctional complex by Helicobacter pylori CagA. Science 300:1430-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aspholm-Hurtig, M., G. Dailide, M. Lahmann, A. Kalia, D. Ilver, N. Roche, S. Vikstrom, R. Sjostrom, S. Linden, A. Backstrom, C. Lundberg, A. Arnqvist, J. Mahdavi, U. J. Nilsson, B. Velapatino, R. H. Gilman, M. Gerhard, T. Alarcon, M. Lopez-Brea, T. Nakazawa, J. G. Fox, P. Correa, M. G. Dominguez-Bello, G. I. Perez-Perez, M. J. Blaser, S. Normark, I. Carlstedt, S. Oscarson, S. Teneberg, D. E. Berg, and T. Boren. 2004. Functional adaptation of BabA, the H. pylori ABO blood group antigen binding adhesin. Science 305:519-522. [DOI] [PubMed] [Google Scholar]
- 5.Backstrom, A., C. Lundberg, D. Kersulyte, D. E. Berg, T. Boren, and A. Arnqvist. 2004. Metastability of Helicobacter pylori bab adhesin genes and dynamics in Lewis b antigen binding. Proc. Natl. Acad. Sci. USA 101:16923-16928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boren, T., P. Falk, K. A. Roth, G. Larson, and S. Normark. 1993. Attachment of Helicobacter pylori to human gastric epithelium mediated by blood group antigens. Science 262:1892-1895. [DOI] [PubMed] [Google Scholar]
- 7.Falk, P., K. A. Roth, T. Boren, T. U. Westblom, J. I. Gordon, and S. Normark. 1993. An in vitro adherence assay reveals that Helicobacter pylori exhibits cell lineage-specific tropism in the human gastric epithelium. Proc. Natl. Acad. Sci. USA 90:2035-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hennig, E. E., R. Mernaugh, J. Edl, P. Cao, and T. L. Cover. 2004. Heterogeneity among Helicobacter pylori strains in expression of the outer membrane protein BabA. Infect. Immun. 72:3429-3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ilver, D., A. Arnqvist, J. Ogren, I. M. Frick, D. Kersulyte, E. T. Incecik, D. E. Berg, A. Covacci, L. Engstrand, and T. Boren. 1998. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science 279:373-377. [DOI] [PubMed] [Google Scholar]
- 10.Mahdavi, J., B. Sonden, M. Hurtig, F. O. Olfat, L. Forsberg, N. Roche, J. Angstrom, T. Larsson, S. Teneberg, K. A. Karlsson, S. Altraja, T. Wadstrom, D. Kersulyte, D. E. Berg, A. Dubois, C. Petersson, K. E. Magnusson, T. Norberg, F. Lindh, B. B. Lundskog, A. Arnqvist, L. Hammarstrom, and T. Boren. 2002. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science 297:573-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pride, D. T., and M. J. Blaser. 2002. Concerted evolution between duplicated genetic elements in Helicobacter pylori. J. Mol. Biol. 316:629-642. [DOI] [PubMed] [Google Scholar]
- 12.Pride, D. T., R. J. Meinersmann, and M. J. Blaser. 2001. Allelic variation within Helicobacter pylori babA and babB. Infect. Immun. 69:1160-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. S. Tompkins, and S. Falkow. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 97:14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salaun, L., B. Linz, S. Suerbaum, and N. J. Saunders. 2004. The diversity within an expanded and redefined repertoire of phase-variable genes in Helicobacter pylori. Microbiology 150:817-830. [DOI] [PubMed] [Google Scholar]
- 15.Schreiber, S., M. Konradt, C. Groll, P. Scheid, G. Hanauer, H. O. Werling, C. Josenhans, and S. Suerbaum. 2004. The spatial orientation of Helicobacter pylori in the gastric mucus. Proc. Natl. Acad. Sci. USA 101:5024-5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Solnick, J. V., L. M. Hansen, D. R. Canfield, and J. Parsonnet. 2001. Determination of the infectious dose of Helicobacter pylori during primary and secondary infection in rhesus monkeys (Macaca mulatta). Infect. Immun. 69:6887-6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Solnick, J. V., L. M. Hansen, N. Salama, J. K. Boonjakuakul, and M. Syvanen. 2004. Modification of Helicobacter pylori outer membrane protein expression during experimental infection of rhesus macaques. Proc. Natl. Acad. Sci. USA 101:2106-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, J. C. Venter, et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 19.van der Ende, A., and K. van Amsterdam. 2004. Nutrients released by gastric epithelial cells enhance Helicobacter pylori growth. Helicobacter 9:614-621. [DOI] [PubMed] [Google Scholar]
- 20.Yamaoka, Y., H. M. Malaty, M. S. Osato, and D. Y. Graham. 2000. Conservation of Helicobacter pylori genotypes in different ethnic groups in Houston, Texas. J. Infect. Dis. 181:2083-2086. [DOI] [PubMed] [Google Scholar]
- 21.Yamaoka, Y., J. Souchek, S. Odenbreit, R. Haas, A. Arnqvist, T. Boren, T. Kodama, M. S. Osato, O. Gutierrez, J. G. Kim, and D. Y. Graham. 2002. Discrimination between cases of duodenal ulcer and gastritis on the basis of putative virulence factors of Helicobacter pylori. J. Clin. Microbiol. 40:2244-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]