Abstract
A safer and more effective vaccine than the previously developed live attenuated vaccine is needed for combating Francisella tularensis, a highly infectious bacterial pathogen. To search for potential candidates for inclusion in a new vaccine, we characterized the proteins present in the culture filtrates of a virulent recent clinical isolate and the attenuated live vaccine strain of F. tularensis using a proteomic approach. We identified a total of 12 proteins; among these, catalase-peroxidase was much more abundant in the culture filtrate of the virulent clinical isolate, whereas bacterioferritin was more abundant in the culture filtrate of the live vaccine strain. Streptolysin O treatment of infected human macrophages indicated that catalase-peroxidase and the heat shock protein GroEL are released intracellularly by actively growing F. tularensis. Mice immunized with F. tularensis developed significant cell-mediated immune responses to catalase-peroxidase, the heat shock protein GroEL, and bacterioferritin as measured by splenic lymphocyte proliferation and gamma interferon production. Finally, we expressed the major culture filtrate proteins that are promising vaccine candidates in Escherichia coli at high levels in soluble form to facilitate study of their immunobiology and potential role in vaccines.
The gram-negative bacterium Francisella tularensis is the causative agent of the zoonotic disease tularemia. Humans acquire tularemia from contact with infected tissues or materials, insect bites, consumption of contaminated food or water, or inhalation of aerosols (35). F. tularensis consists of three subspecies—tularensis, holarctica, and mediasiatica—which differ in their geographic distributions and in their virulence in humans (9). F. tularensis subsp. tularensis (found almost exclusively in North America) is highly virulent for humans. As few as 10 organisms subcutaneously or 25 organisms by inhalation can lead to a severe infection (31, 32). F. tularensis subsp. holarctica (found in North America and in Europe) and subsp. mediasiatica (found in Asia) are of lower virulence. Because of its high infectivity and capacity to cause severe morbidity and mortality, F. tularensis subsp. tularensis is considered a potential agent of bioterrorism.
An attenuated live vaccine strain (LVS) derived from a colony variant of F. tularensis subsp. holarctica, developed in the former Soviet Union in the 1930s, confers substantial protective immunity against virulent F. tularensis in humans. However, this vaccine suffers major drawbacks. First, the vaccine does not provide full protection against aerosol challenge with relatively low doses (10 to 50 organisms) of virulent F. tularensis (32). Second, the vaccine retains significant virulence raising concerns regarding its safety, especially in children and immunocompromised individuals. In addition, the immunoprotective moieties in the vaccine have not been determined. In view of these drawbacks, a safer and more effective vaccine against tularemia is needed.
As a first step toward designing a better vaccine, we sought to identify protein candidates for inclusion in a vaccine. F. tularensis is a facultative intracellular bacterium that retards the maturation of its phagosome and subsequently escapes into and multiplies within the cytoplasm of host macrophages (8, 13). In the case of the intracellular pathogens Legionella pneumophila and Mycobacterium tuberculosis, we have previously shown that proteins that are secreted or abundantly released by these bacteria can be formulated into vaccines that confer immunoprotection in animal models of the diseases caused by the pathogens (5, 15, 26). With this concept in mind, we have sought in this study to identify the major proteins exported by F. tularensis. To achieve this, we have employed 35S metabolic radiolabeling, one- and two-dimensional polyacrylamide gel electrophoresis, and N-terminal amino acid sequencing to evaluate the proteins released by F. tularensis growing in a defined medium. We have compared the protein profile obtained from a virulent recent clinical isolate (RCI) of F. tularensis subsp. tularensis with that from the attenuated F. tularensis subsp. holarctica (LVS). Moreover, we have cloned and expressed genes coding for five of the F. tularensis major extracellular proteins that we postulate are promising vaccine candidates as soluble recombinant proteins. One of these, catalase-peroxidase (KatG), one of the most abundant proteins in the culture filtrate of the virulent clinical isolate of F. tularensis, exhibits a functional signal peptide and is exported by the recombinant Escherichia coli host cells.
MATERIALS AND METHODS
Bacteria.
F. tularensis live vaccine strain and a virulent recent clinical isolate (NY 96-3369) were obtained from the Centers for Disease Control and Prevention (Atlanta, GA). Bacteria were cultivated on chocolate II agar (BD BBL, Sparks, MD), passaged through THP-1 cells, a human monocytic cell line (American Type Culture Collection), and stored at −80°C (8).
Culture media.
Chemically defined Chamberlain medium was prepared as described previously (7). All ingredients used in preparation of the medium were purchased from Sigma (St. Louis, MO). The medium was stirred overnight and sterilized by passing through a 0.2-μm filtration unit. The final medium had a pH of 6.3 to 6.5. Mueller-Hinton broth (BD BBL) was supplemented with 0.025% ferric pyrophosphate and 2% IsoVitaleX (BD BBL). Brain heart infusion broth (BD BBL) was supplemented with 1% β-cyclodextrin (Sigma). Tryptic soy broth (BD BBL) was supplemented with 0.1% cysteine hydrochloride (Sigma).
Bacterial cultures.
Overnight cultures of F. tularensis on chocolate II agar plates were scraped and washed twice in normal saline and resuspended in Chamberlain medium to an optical density at 540 nm (OD540) of 0.1. Bacterial cultures were grown at 37°C in Erlenmeyer flasks equipped with 0.2-μm filter caps (Corning Incorporated, Corning, NY), rotated at 200 rpm, and harvested at selected time points by centrifugation at 3,500 rpm and 4°C for 30 min. Culture supernatant was passed sequentially through 0.45-μm and 0.2-μm filters. Culture supernatants with a volume of 200 ml or less were concentrated using 5-kDa-cutoff Centricon Plus-20 centrifugal filter devices (Millipore Corporation, Bedford, MA). Culture supernatants with a volume of greater than 200 ml were first concentrated using an Amicon ultrafiltration cell with a YM10 membrane (Millipore) and then further concentrated using a Centricon Plus-20 centrifugal filter device. Bacterial pellets were resuspended in Dulbecco's phosphate-buffered saline (PBS) and subjected to sonication on ice with a W-375 sonication Ultrasonic processor (Heat Systems-Ultrasonics, Inc., Farmingdale, N.Y.) at 50% duty cycle with a pulse and strength setting of 5 for three 1-min sessions. Insoluble material and unbroken bacteria were removed by centrifugation. Protein concentration in the culture filtrate and in the clear lysate obtained from bacterial sonication was determined by bicinchonic acid protein assay (Pierce Biotechnology, Rockford, IL) with albumin as the standard.
Radiolabeling of culture filtrate proteins.
F. tularensis RCI was cultured in Chamberlain medium for 4 h at 37°C to an OD540 of 0.4 to 0.5. The bacteria were pelleted by centrifugation, washed once in normal saline, and suspended in methionine-free Chamberlain medium. [35S]methionine (Amersham Pharmacia Biotech, Little Chalfont, England) was added to the culture to a final concentration of 100 μCi/ml, the culture was incubated for 1 h, and the radiolabeled culture filtrate was obtained and processed as described above for nonradiolabeled bacterial cultures.
LDH assay.
The activity of lactate dehydrogenase (LDH) was measured by a modification of the method of Reeves and Fimognari (29). Concentrated protein samples from 20-ml equivalent of culture filtrate or 1-ml equivalent of bacterial lysate were added to 0.85 ml of PBS, 50 μl of NADH (2.5 mg/ml), and 50 μl of sodium pyruvate (2.5 mg/ml). The decrease in optical density at 340 nm was measured over a 2-min period at room temperature in an optical cuvette with 1.0-cm path length. One unit of LDH activity is defined as the amount of enzyme required to produce 1.0 μmol of lactate within a minute.
Protease assays.
Protease activity was examined both by zymogram analysis and a quantitative colorimetric protease assay. Proteins concentrated from 40 ml of culture filtrate were examined by zymogram analysis using polyacrylamide gels embedded with 10% casein or 12% gelatin (Bio-Rad Laboratories, Hercules, CA) according to the manufacturer's instructions. Proteinase K (Epicentric, Madison, WI) (1 μg) was loaded onto the same zymogram gel as a positive control. Culture filtrate proteins harvested from 200-ml bacterial cultures were desalted by passing through an Excellulose GF-5 column (Pierce). Protein effluent was monitored at 280 nm, combined, and concentrated for use in the assay. Protease was quantified using the QuantiCleave protease assay kit (Pierce). A standard curve was prepared using tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK) trypsin at concentrations of 0.5 mg/ml to 0.5 ng/ml.
Two-dimensional polyacrylamide gel electrophoresis (2-DE).
Culture filtrate proteins (200 μg) were dissolved in sample buffer containing 8 M urea, 2% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 50 mM dithiothreitol (DTT), and 0.2% (wt/vol) Bio-Lyte 3/10 ampholytes and separated using an isoelectric focusing (IEF) tube gel (16 cm × 1.5 mm) (ratio of Biolyte 5/7 ampholytes to Biolyte 3/10 ampholytes of 4:1) at 200 V for 2 h, 500 V for 2 h, and 800 V for 14 h. The IEF gel was equilibrated for 15 min each time with buffer I containing 6 M urea, 2% sodium dodecyl sulfate (SDS), 0.375 M Tris-HCl (pH 8.8), 20% glycerol, and 2% (wt/vol) DTT and buffer II containing 6 M urea, 2% SDS, 0.375 M Tris-HCl (pH 8.8), 20% glycerol, and 2.5% (wt/vol) iodoacetamide. Each equilibrated IEF gel was loaded onto a 12.5% SDS-polyacrylamide gel (16 cm × 16 cm × 1 mm). The second-dimension gel system was run at 20 mA for the stacking gel and 30 mA for the separating gel. The gels were fixed in 40% ethanol and 10% acetic acid for 1 h, stained with 0.1% (wt/vol) Coomassie brilliant blue G, 10% ethanol, 2% phosphoric acid, and 1% (wt/vol) ammonium sulfate for 4 h, and destained with three or four changes of 10% acetic acid.
N-terminal amino acid sequencing.
Proteins separated by one- or two-dimensional polyacrylamide gel electrophoresis were transferred to a polyvinylidene difluoride (PVDF) membrane in 10 mM CAPS (3-[cyclohexylamino]-1-propane sulfonic acid), pH 11, and stained with Coomassie brilliant blue R-250. Protein spots cut from the membrane were subjected to N-terminal amino acid sequencing by Edman degradation at the Molecular Structure Facility of the University of California, Davis.
Protein homology search.
F. tularensis (Schu 4) genome was downloaded from the website http://artedi.ebc.uu.se/Projects/Francisella/data. A collection of open reading frames encoding ≥50 amino acids were translated using the computer program Artemis (30), and N-terminal amino acid sequences obtained for individual protein spots were searched. We assigned an open reading frame to a particular protein spot only if it met all three of the following criteria. (i) The open reading frame encoded a polypeptide whose N terminus (within the first three methionines of the open reading frame) matched the sequence of the first seven (or more) amino acid residues of the protein spot. (ii) This matched sequence was the only one found in the entire collection of open reading frames. (iii) The calculated molecular mass of the polypeptide predicted by the open reading frame approximated the size of the protein spot observed on SDS-polyacrylamide gels. We then conducted a BLAST search in the GenBank database using the entire protein sequence. We assigned a potential identity to an open reading frame on the basis of its high percentage of identity or similarity in primary sequence to a protein of a known physiological function and/or by virtue of its possessing one or more conserved domain(s) of such a protein.
Amplification of F. tularensis genes.
Genomic DNA of F. tularensis RCI was isolated by the previously published method (28). F. tularensis RCI cultured in Chamberlain medium was pelleted by centrifugation and incubated at 50°C for 12 h in a lysis solution containing 10 mM NaCl, 20 mM Tris-HCl, pH 8, 1 mM EDTA, 0.5% SDS, and 0.7 mg/ml protease. Bacterial lysate was treated with sodium perchlorate at room temperature for 1 h and extracted three times with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1). Nucleic acids were precipitated from the aqueous phase, resuspended, and treated with RNase at 37°C for 1 h. After another round of extraction with phenol-chloroform-isoamyl alcohol, genomic DNA was precipitated and dissolved in buffer containing 10 mM Tris-HCl, pH 8, and 1 mM EDTA. Primer pairs used for amplification of individual genes from the isolated F. tularensis RCI genomic DNA were as follows: 5′-CCGCTCGAGCATATGCTAAAGAAAATTGTAACTGCTTTAGGAATGTCTGGAATGCTACTAGC-3′ and 5′-CCGCTCGAGTTAACAAATTTATTGTTGAACATCAAATCTGCCAAGCATCATAACTTTATGCC-3′ for katG; 5′-CGGGATCCCATATGGCTGCAAAACAAGTTTTATTTTCAGATGAAGCTC-3′ and 5′-CGGGATCCCTATTACATCATGCCAGGCATACCGCCCATGCCACCGCC-3′ for groEL; 5′-CGGGATCCCATATGTCAAAAACAGCTGTAGTTTTTCCTGGTCAAGGTTC-3′ and 5′-CGGGATCCTTAAATATTTTCTAAACTATCAATACTGTTTGTATCTTTT-3′ for fabD; 5′-CGGGATCCGGCTTACCACATGATCTAATTGCCGTTGCTGCATGCGC-3′ and 5′-CGGGATCCAGCTATCAAGATCACAACCACTATTGATAAAACCCCTA-3′ for sodB; and 5′-CCGGATCCCCATCTTTAAAT GTACAGGTTGTATCTAGACTTTCTGC-3′ and 5′-CGGGATCCGTCACTGAATATCTCG ATAGCGCATTCTAGTGAATCCAAG-3′ for bfr. DNA polymerase from the PfuUltra Hotstart (Stratagene, La Jolla, CA) or FailSafe (Epicentric, Madison, WI) system was used in the amplification reactions under a typical condition of 1 cycle at 95°C for 1 min; 30 cycles at 95°C for 1 min, 50°C for 1 min, and 72°C for 1 to 2 min depending on the length of the target gene; and 1 cycle at 72°C for 10 min. The amplified product was gel purified and ligated with pZero (Invitrogen, Carlsbad, CA). The identity of each cloned gene was confirmed by nucleotide sequencing. Nucleotide sequencing was carried out by the sequencing core facility at the University of California, Los Angeles.
Cloning and expression of F. tularensis genes.
The groEL and fabD genes were subcloned directly from pZero into pET15b (Novagen, Madison, WI) between the NdeI and BamHI restriction sites. The sodB and bfr genes were amplified from the corresponding pZero constructs with proper restriction sites and inserted between the NdeI and BamHI restriction sites on pET15b. For expression of the full-length katG gene, the gene was released from pZero with NdeI and XhoI and ligated with pET22b (Novagen) treated with the same enzymes. F. tularensis genes cloned into the expression vector pET15b produce recombinant proteins with a thrombin-cleavable histidine tag at the N terminus, and the katG gene cloned into pET22b produces a recombinant protein with the histidine tag fused to its C terminus. E. coli BL21 CodonPlus(DE3)-RIL (Stratagene) transformed with the pET15b or pET22b construct was first grown in LB medium to an absorbance at 540 nm of 0.4 and then induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 5 h before harvesting. The culture filtrate and the soluble and insoluble fractions from the bacterial sonicate were analyzed on 12.5% SDS-polyacrylamide gels to assess the expression and solubility of the recombinant proteins.
Protein purification and antibody production.
Recombinant bacterioferritin (Bfr), FabD, SodB, and GroEL were purified by Ni-nitrilotriacetic acid (Ni-NTA) metal affinity chromatography and by elution with imidazole. The N-terminal histidine tag was cleaved off from the recombinant proteins by digestion with biotinylated thrombin, which was subsequently removed with streptavidin agarose (Novagen). Residual recombinant proteins still bearing the histidine tag were removed by a second round of Ni-NTA chromatography. Purification of recombinant KatG was carried out with a combination of Ni-NTA and Q-Sepharose chromatography. The purity of the recombinant proteins was assessed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE). Purified recombinant proteins were used to immunize rabbits for antibody production (Covance, Denver, PA).
Western blot analysis.
Proteins separated by 10% or 12.5% SDS-PAGE were transblotted in Tris-glycine buffer at 45 V for 5 h onto a 0.45-μm nitrocellulose membrane (Bio-Rad). For colorimetric detection of recombinant proteins, the membrane blot was probed first with an anti-histidine tag monoclonal antibody (Amersham Pharmacia) at a dilution of 1:3,000 and then with alkaline phosphatase-conjugated goat anti-mouse antibodies (Sigma). Immunoreactive protein bands were visualized with developing reagent containing 5-bromo-4-chloro-3-indolylphosphate p-toluidine salt and p-nitroblue tetrazolium chloride. For chemiluminescence detection, the membrane blot was reacted with antibodies specific to KatG, GroEL, or Bfr at a dilution of 1:5,000 and subsequently with a 1:25,000 dilution of horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (Bio-Rad). Signals were developed by incubating with chemiluminescent substrate (Pierce) and detected by exposing X-ray film. The anti-GroEL antibody was preabsorbed with acetone extracts of THP-1 cells to remove antigen cross-reactivity (14).
Analysis of bacterial proteins released during infection of macrophages.
Differentiated THP-1 cells (107) were infected with F. tularensis RCI (108) in RPMI medium containing 10% human serum type AB for 90 min at 37°C and 5% CO2, washed three times with PBS, and incubated in RPMI medium containing gentamicin (2 μg/ml) and 10% heat-inactivated fetal bovine serum (HI-FBS). The monolayer was washed after 10 h of incubation, harvested by scraping into homogenization buffer containing 250 mM sucrose, 20 mM HEPES-KOH, pH 7.3, and 0.5 mM EGTA, and centrifuged at 600 × g and 4°C for 15 min. The pelleted cells were resuspended in ice-cold homogenization buffer containing DTT-activated streptolysin O (25 U/ml). The cells were incubated on ice for 15 min and subsequently at 37°C for another 15 min to effect permeabilization and then centrifuged at 14,000 × g for 10 min. The supernatant was passed through a protein desalting spin column (Pierce) and analyzed by SDS-PAGE and Western immunoblotting.
Lymphocyte proliferation assay.
Female BALB/c mice purchased from Charles River Laboratories were injected intradermally with 5 × 104 live or heat-killed F. tularensis LVS or with PBS alone. Mice were euthanized 5, 9, and 15 weeks later. Splenocytes were prepared as described previously (26) and cultured in 96-well U-bottom plates at a concentration of 2 × 106 cells per 0.1 ml per well in RPMI medium containing 10% HI-FBS, polymyxin B (100 U/ml), and purified recombinant proteins (10 μg/ml). After 72 h of incubation, splenocytes were pulsed with 0.25 μCi of methyl-[3H]thymidine (Amersham Pharmacia Biotech) per well and harvested 16 h later with a cell harvester (Skatron). The amount of incorporated [3H]thymidine was determined by counting in a liquid scintillation counter. The stimulation index was defined as the ratio of the counts per minute obtained from cells incubated with the purified protein antigen divided by the counts per minute obtained from cells incubated without antigen.
Intracellular cytokine staining.
BALB/c mice were immunized intradermally with 5 × 104 live or heat-killed F. tularensis LVS or with PBS alone. Spleens were harvested 9 weeks later from the immunized mice. Red blood cells were lysed with PharmLyse (BD Pharmingen, San Diego, CA). Splenocytes were washed and seeded at a density of 2 × 106 cells per 0.1 ml per well in RPMI medium containing 10% HI-FBS and interleukin 2 (50 U/ml) and incubated with or without purified protein antigen (10 μg/ml) for 24 h at 37°C and 5% CO2. Golgi-Plug (BD Pharmingen) was added, and the cells were incubated for an additional 12 h. Cells were pelleted at 250 × g and 4°C for 5 min and resuspended in PBS containing 3% FBS, 0.09% sodium azide (staining buffer), and Fc-Block (BD Pharmingen). After incubation at 4°C for 15 min, cells were stained with fluorescein isothiocyanate (FITC)-labeled anti-CD4 or CyChrome (CyC)-labeled anti-CD8 antibody at a 1:100 dilution on ice for 30 min. Cells were washed twice in staining buffer, fixed with Cytofix solution for 20 min at 4°C, and washed twice with Perm/Wash solution. Cells were then stained for intracellular gamma interferon (IFN-γ) with phycoerythrin-labeled rat anti-mouse IFN-γ or a phycoerythrin-labeled isotypic control immunoglobulin G at a dilution of 1:100. Stained cells were washed, resuspended in staining buffer, and analyzed on a FACSCalibur flow cytometer using CellQuest software.
Nucleotide sequence accession numbers.
The nucleotide sequences for the genes of F. tularensis RCI have been submitted to GenBank under the following accession numbers: AY662299 (katG), AY662300 (groEL), AY662301 (fabD), AY662302 (sodB), and AY662303 (bfr).
RESULTS
Growth of F. tularensis in Chamberlain medium and profiles of culture filtrate proteins.
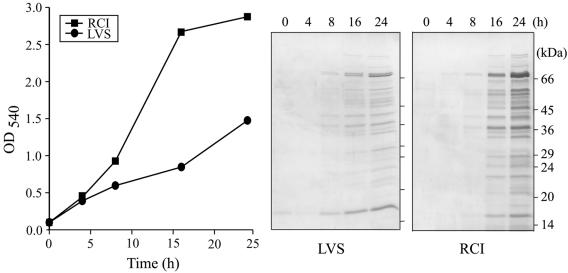
We inoculated the partially attenuated live vaccine strain and a virulent recent clinical isolate of F. tularensis into Chamberlain medium at an initial OD540 of 0.1 and monitored bacterial growth over a 24-h period (Fig. 1). For the first 4 h of incubation, F. tularensis LVS and RCI grew at comparable rates. Thereafter, the growth of LVS markedly lagged behind that of RCI. At 24 h, F. tularensis RCI had attained an optical density of 3.0, whereas F. tularensis LVS had attained an optical density of only 1.5. The slower growth of LVS in Chamberlain medium was not attributable to a lower percentage of viable bacteria in the culture, since fluorescence staining of the LVS culture at 5 h and 15 h using the Molecular Probes Dead-Live reagents (ethidium bromide and Sytox-9) revealed that at both time points nearly 100% of the bacteria were viable. F. tularensis LVS bacteria that had been heated to 60°C for 1 h showed less than 1% viability when stained with the same reagents.
FIG. 1.
Growth and culture filtrate protein profiles of F. tularensis in Chamberlain medium. The growth of F. tularensis LVS and RCI strains was monitored by measurement of culture density (absorbance at 540 nm) over a 24-h period. Culture filtrates harvested from bacterial cultures at the indicated time points were concentrated, and the concentrate derived from 10 ml of original culture for each sample was analyzed by SDS-PAGE on a 12.5% gel and stained with Coomassie blue. This experiment was performed twice with similar results.
We concentrated the culture filtrates of F. tularensis LVS and RCI harvested at various time points over a 24-h period and analyzed sample volumes corresponding to 10 ml of original culture filtrate at each time point by SDS-PAGE (Fig. 1). At 8 h, protein bands of 76, 42, 38, and 16 kDa were visible in the culture filtrates of both F. tularensis LVS and RCI. With longer incubation, the number of protein bands found in the culture filtrates of both strains increased. At 24 h, the most prominent protein bands in the culture filtrate of F. tularensis LVS had molecular masses of 76, 74, and 16 kDa, and the most prominent protein bands in the culture filtrate of F. tularensis RCI had molecular masses of 76, 74, 60, 57, 47, 42, 38, and 16 kDa. The proteins expressed by the two strains were similar, but their intensities differed. At 16 h and 24 h, more proteins were observed in equal volumes of culture filtrate from RCI than from LVS, a difference which may result from the superior growth of RCI in the Chamberlain medium.
Comparison of the growth of F. tularensis RCI and LVS in chemically defined and complex media.
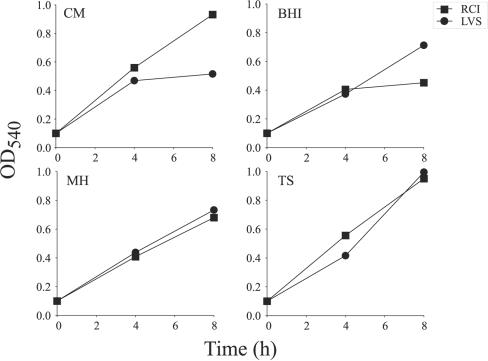
To investigate further the differences in growth between F. tularensis LVS and RCI in liquid culture media, we examined the growth of the two strains in the chemically defined Chamberlain medium and in three different complex media—brain heart infusion broth, Mueller-Hinton broth, and tryptic soy broth. Whereas F. tularensis LVS grew slower than RCI in Chamberlain medium as previously noted, it grew at a comparable rate, if not faster in the three complex media tested (Fig. 2). Hence, the lower growth rate of F. tularensis LVS in Chamberlain medium is primarily due to the constituents of the medium rather than an inability of F. tularensis LVS to achieve a growth rate or culture density that is comparable to that of F. tularensis RCI. Chamberlain medium and tryptic soy broth supported the growth of F. tularensis RCI equally well; the strain attained an OD540 of 1.0 at 8 h in these media. In contrast, Mueller-Hinton broth and brain heart infusion broth were inferior to Chamberlain medium in supporting the growth of F. tularensis RCI.
FIG. 2.
Growth of F. tularensis in defined and complex media. The growth of F. tularensis LVS and RCI in four different media was monitored by assaying the culture density over an 8-h period. The media are abbreviated as follows: CM, Chamberlain medium; BHI, brain heart infusion broth; MH, Mueller-Hinton broth; TS, tryptic soy broth. This experiment was performed twice with similar results.
Comparison of protein profiles of the culture filtrate and cell lysate of F. tularensis.
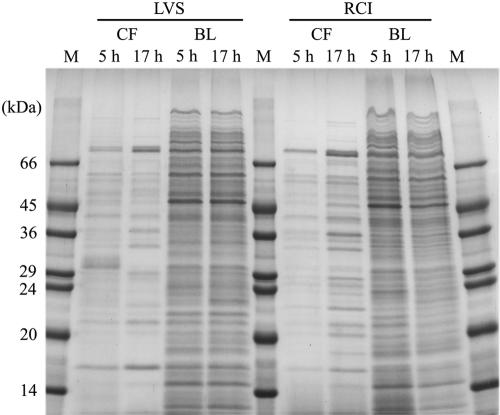
Bacterial proteins can be released into the culture medium through active secretion, leakage during active bacterial growth and division, or autolysis. The profile of culture filtrate proteins from a bacterial culture that is undergoing autolysis would be expected to resemble the profile of cell lysate proteins, i.e., soluble proteins extracted from the bacterial cells. To determine whether culture filtrate proteins of F. tularensis are released by bacterial autolysis, we grew F. tularensis LVS and RCI in Chamberlain medium, harvested the cultures at 5 h or 17 h, and compared culture filtrate and bacterial lysate protein profiles by SDS-PAGE. We observed marked differences between the protein profiles of the culture filtrate and cell lysate for both F. tularensis LVS and RCI at both 5 h and 17 h (Fig. 3). These results argued against the culture filtrate proteins being released by autolysis.
FIG. 3.
Protein profiles of the culture filtrates and the corresponding bacterial lysates of F. tularensis LVS and RCI. An equal amount (30 μg) of culture filtrate (CF) and sonicated bacterial lysate (BL) obtained from F. tularensis LVS and RCI grown in Chamberlain medium for 5 h or 17 h was analyzed by SDS-PAGE on a 12.5% gel and stained with Coomassie blue. Lanes M contain molecular mass markers (in kilodaltons). This experiment was performed twice with similar results.
LDH activity in the culture filtrate of F. tularensis.
Lactate dehydrogenase is a stable cytoplasmic enzyme, and the measurement of its enzymatic activity in the culture supernatant serves as an indicator of bacterial leakage or autolysis. To confirm that culture filtrate proteins were not released to any great extent by autolysis, we assayed the LDH activity in the culture filtrate and bacterial lysate prepared from F. tularensis LVS and RCI grown in Chamberlain medium for 5 or 14 h. Whereas abundant LDH activity was present in the cell lysates of F. tularensis LVS and RCI at both time points, LDH activity in the culture filtrates of the two strains was undetectable or minimally above the background level even after 14 h of cultivation (Table 1).
TABLE 1.
Lactate dehydrogenase activity in F. tularensis culture filtrate and cell lysate
| F. tularensis strain | Growth time (h)a | OD540 | Amt of protein (μg/ml)
|
LDH activity (mU/ml) (mean ± SE)d
|
||
|---|---|---|---|---|---|---|
| CFb | BLc | CF | BL | |||
| LVS | 5 | 0.503 | 0.49 | 112 | 0.000 ± 0.000 | 10.008 ± 0.284 |
| 14 | 0.718 | 0.40 | 120 | 0.008 ± 0.006 | 8.360 ± 0.455 | |
| RCI | 5 | 0.544 | 0.39 | 100 | 0.004 ± 0.000 | 12.098 ± 0.171 |
| 14 | 1.984 | 0.84 | 333 | 0.002 ± 0.003 | 37.259 ± 0.171 | |
F. tularensis LVS and RCI were grown in Chamberlain medium for 5 or 14 h.
CF, culture filtrate.
BL, sonicated bacterial lysate.
LDH assay was performed as described in Materials and Methods. The experiment was performed twice with similar results.
Protease activity in the culture filtrate of F. tularensis.
Proteases are released by many bacteria either for nutritional purposes (to metabolize complex nutrients) or as part of a pathogenic mechanism to degrade particular host proteins (12). To investigate whether F. tularensis LVS or RCI bacteria release proteases into their extracellular milieu, we harvested the culture supernatant from bacteria grown in Chamberlain medium and screened it for protease activity by zymogram analysis on nondenaturing polyacrylamide gels and by a quantitative colorimetric assay for proteolytic activity. We observed no proteolytic activity in zymograms using the culture filtrate from F. tularensis LVS or RCI grown for 5 h or 17 h with either casein or gelatin as the substrate. As a positive control, proteinase K was loaded on the same gels; it produced a broad clear band indicative of proteolytic activity (data not shown). We also did not detect any protease activity in the culture filtrate of either strain by a quantitative colorimetric protease assay, which is sensitive to trypsin proteolytic activity at enzyme concentrations as low as 50 ng/ml (data not shown).
Metabolic labeling and identification of culture filtrate proteins of F. tularensis.
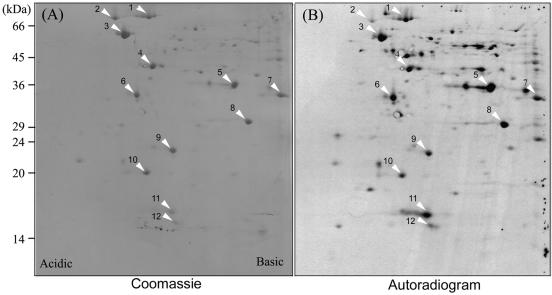
To label culture filtrate proteins, we grew F. tularensis RCI for 4 h in Chamberlain medium, pelleted the bacteria by centrifugation, washed them, and inoculated them into methionine-free Chamberlain medium at an OD540 of around 0.4. We then incubated the bacterial culture for 1 h at 37°C in the presence of [35S]methionine to allow metabolic labeling of proteins that are actively released by F. tularensis. Culture filtrate proteins harvested from the radiolabeled bacterial culture were mixed with culture filtrate proteins from a nonradiolabeled culture and separated by 2-DE (Fig. 4). The separated proteins were transferred to a PVDF membrane, stained with Coomassie blue, and exposed to an X-ray film to determine the profile of radiolabeled proteins. For each major protein spot on the Coomassie blue-stained PVDF membrane, we measured the intensity of the spot on the PVDF membrane and on X-ray film and calculated an intensity ratio (radiolabel intensity/Coomassie blue stain intensity). A total of 12 distinct major protein spots with high intensity ratios were subjected to N-terminal amino acid sequencing. In all cases, the N-terminal sequence was found to match exactly to a peptide sequence from an open reading frame in the F. tularensis Schu 4 genomic database. The N-terminal sequences obtained for the 12 protein spots, the potential identities (determined by sequence homology), and calculated masses and pIs are listed in Table 2.
FIG. 4.
2-DE separation of radiolabeled F. tularensis culture filtrate proteins. Early culture filtrate proteins of F. tularensis RCI radiolabeled with [35S]methionine were mixed with nonradiolabeled early culture filtrate proteins of RCI and separated by 2-DE. Coomassie blue-stained protein spots on PVDF membrane (A) and the corresponding radiolabeled spots on X-ray film (B) are indicated by the white arrowheads. This experiment was performed twice with similar results.
TABLE 2.
N-terminal amino acid sequence of protein spots from 2-DE of F. tularensis culture filtrate proteins
| Spota | N-terminal sequence | Best homologyb | Protein name or abbreviation | Calculated mass (kDa)c | Calculated pIc |
|---|---|---|---|---|---|
| 1 | EDTTTKNDNLSPQSVDLS | Catalase-peroxidase | KatG | 80 | 5.4 |
| 2 | GKIIGIDLGTTNS | 70-kDa chaperone | DnaK | 69 | 4.8 |
| 3 | AAKQVLFSDEAR | 60-kDa chaperone | GroEL | 57 | 4.9 |
| 4 | MNLHEYQAKDLL | Succinyl-CoA synthetase, β subunit | SucC | 42 | 5.2 |
| 5 | MRVVINGFG | Glyceraldehyde 3-phosphate dehydrogenase | GAPDH | 35 | 5.7 |
| 6 | SKTAVVFPGQGS | (Acyl carrier protein) S-melanoyltransferase | FabD | 33 | 5.0 |
| 7 | SVLVDKNT | Succinyl-CoA synthetase, α subunit | SucC | 30 | 6.4 |
| 8 | SLPTPFI | Purine-nucleoside phosphorylase | DeoD | 27 | 7.8 |
| 9 | MKFELPKLPYAV | Superoxide dismutase | SodB | 22 | 5.6 |
| 10 | TKKVPNVTFKTR | Peroxiredoxin | Ahp1 | 20 | 5.1 |
| 11 | MELQLEN-QEIId | Bacterioferritin | Bfr | 17 | 5.6 |
| 12 | MNIRPLQ | 10-kDa chaperone | GroES | 10 | 5.4 |
The spot numbers correspond to those indicated in Fig. 4.
The protein with which the N-terminal sequence showed the best homology. Best homology is assigned on the basis of BLAST search results using the entire protein sequence. CoA, coenzyme A.
Mass and pI were calculated from the entire protein sequence.
The unidentified amino acid residue in the N-terminal sequence is indicated by a hyphen.
Comparison of 2-DE patterns of the culture filtrate proteins of F. tularensis LVS and RCI.
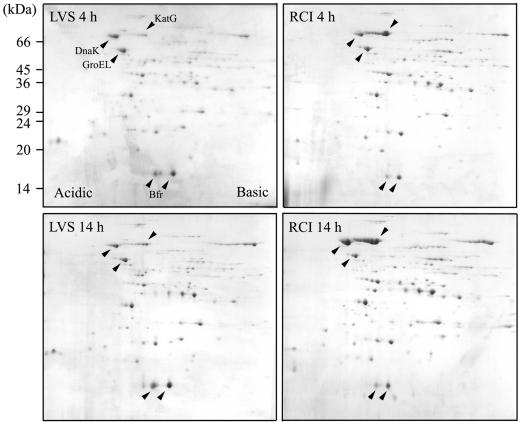
We employed 2-DE to compare the profiles of culture filtrate proteins released by F. tularensis LVS and RCI growing in Chamberlain medium. Culture filtrate proteins (200 μg) were separated on the first dimension by nonlinear IEF gels using a mixture of pH 5 to 7 and pH 3 to 10 ampholytes at a ratio of 4:1 to obtain a better separation of individual protein spots than that obtained on linear broad-range (pH 3 to 10) IEF gels. The proteins were then separated on the second dimension by SDS-PAGE on 12.5% gels. We observed on average 90 distinct protein spots on Coomassie blue-stained 2-D gels from the culture filtrates of F. tularensis harvested at 4 h and 14 h. While the majority of the protein spots were present in the culture filtrates of both F. tularensis LVS and RCI, the relative abundance of several individual protein spots was different (Fig. 5).
FIG. 5.
Comparative 2-DE of F. tularensis LVS and RCI culture filtrate proteins. Equal amounts of culture filtrate proteins (200 μg) harvested from F. tularensis LVS and RCI grown in Chamberlain medium for 4 h or 14 h were separated by 2-DE and stained with Coomassie blue. The spots corresponding to KatG, DnaK, GroEL, and Bfr are indicated by the black arrowheads. Three independent experiments for LVS and RCI at each time point were performed with similar results.
KatG, DnaK, and GroEL are among the most abundant proteins found in the culture filtrate of F. tularensis. Since GroEL is present at about the same level in the culture filtrates of both strains at both time points, we designated it the reference spot to measure the staining intensity ratios of these three proteins in the culture filtrate of F. tularensis RCI harvested at 4 h (KatG/DnaK/GroEL ratio of 3:1:1) and 14 h (11:6:1), and in the culture filtrate of F. tularensis LVS harvested at 4 h (0.2:1:1) and 14 h (0.8:1:1). KatG is ∼14-fold more abundant in the RCI culture filtrate than in the LVS culture filtrate at both time points; it increases ∼4-fold between 4 h and 14 h in the cultures of both strains such that the relative amount in the cultures of the two strains remains about the same at both time points. DnaK is present at about the same level in the LVS and RCI culture filtrates at 4 h; however, at 14 h, it is significantly increased in RCI but not LVS culture filtrate such that it is approximately sixfold more abundant. Bacterioferritin was also identified as an abundant extracellular protein. Typically, Bfr appears on 2-D gels as one major and one minor isoform in the RCI culture filtrate, whereas three or four Bfr spots of the same molecular mass but different pIs are present in the LVS culture filtrate. N-terminal sequences of the two most abundant Bfr isoforms in the LVS are identical to each other (MELKLENKQ) and differ from that of the RCI by a single amino acid (i.e., Lys for LVS and Gln for RCI at residue 4). Interestingly, the most prominent Bfr spot in the LVS culture filtrate has the highest pI among various Bfr isoforms and is totally absent from the RCI culture filtrate.
Cloning and recombinant expression of genes encoding F. tularensis culture filtrate proteins.
We amplified genes coding for KatG, GroEL, FabD, SodB, and Bfr, the five abundant proteins identified in culture filtrate, from genomic DNA of F. tularensis RCI and determined their nucleotide sequences, which were found to be identical to the corresponding gene sequence of the F. tularensis Schu 4 strain (21). We cloned and expressed each of the five genes in E. coli at high levels as soluble recombinant proteins (Fig. 6). KatG, which contained a putative 23-amino-acid signal peptide (25), was released by the recombinant E. coli into its culture medium. N-terminal amino acid sequencing of the recombinant KatG protein released by E. coli revealed a sequence of the mature protein identical to that of KatG from the culture filtrate of F. tularensis RCI. This result strongly indicated that the 23 amino acids preceding the mature protein constituted a functional signal peptide. We purified the recombinant proteins for use in antibody production and in assays measuring host cellular immune response to these proteins in the F. tularensis-infected mice.
FIG. 6.
Recombinant expression in E. coli of genes corresponding to F. tularensis culture filtrate proteins. E. coli BL21 CodonPlus(DE3)-RIL expressing F. tularensis culture filtrate proteins with a N-terminal histidine tag were disrupted by sonication. (A) The released soluble proteins were analyzed by subjecting them to SDS-PAGE, staining with Coomassie blue and by immunoblotting using an anti-histidine tag monoclonal antibody. Lanes 1, vector alone; lanes 2, Bfr; lanes 3, SodB; lanes 4, FabD; lanes 5, GroEL; lane M, molecular mass markers. (B) Culture filtrate proteins from the recombinant E. coli expressing F. tularensis KatG without (−) or with (+) IPTG induction were analyzed by subjecting them to SDS-PAGE and staining with Coomassie blue and by immunoblotting using an anti-histidine tag monoclonal antibody. The protein band indicated by the small black arrowhead was subjected to N-terminal amino acid sequencing. The first seven amino acids of the protein band corresponded to those in the N-terminal sequence of the mature KatG of F. tularensis. (C) The sequence of the first 40 amino acid residues of the full-length KatG protein is shown. The cleavage site of the KatG signal peptide is indicated by the black arrow pointing down.
Release of culture filtrate proteins by intracellular F. tularensis.
To investigate whether the major culture filtrate proteins identified in this study are also expressed and released by the bacterium intracellularly in host macrophages, we infected THP-1 cells with F. tularensis RCI for 10 h, by which time the bacteria have escaped into and multiplied within the cytoplasm of the host cell (8). We then permeabilized the infected cells with streptolysin O and analyzed the soluble cytosolic proteins released from the permeabilized cells by immunoblotting. KatG was detected in the cytosol of the infected host cells but was not detected in equal numbers of uninfected THP-1 cells or bacteria that were subjected to streptolysin O permeabilization (Fig. 7). GroEL was detected in both the cytosol of the infected THP-1 cells and in the bacteria alone. Nevertheless, the GroEL signal detected in the infected cytosol was 9.5-fold greater than that detected in the bacteria alone. We reasoned that the GroEL signal detected in the bacteria alone was most likely from the spontaneous release of the protein by the bacteria, since the same intensity of GroEL signal was also detected in the bacteria without streptolysin O treatment (data not shown). Unlike KatG and GroEL, Bfr was not detected in the cytosol of the infected THP-1 cells. Results from these experiments indicated that KatG and GroEL are both released by F. tularensis intracellularly in human macrophages. The absence of a Bfr-immunoreactive band in the streptolysin lysate of F. tularensis-infected THP-1 cells may indicate absence of release of this protein by bacteria growing intracellularly as opposed to bacteria growing extracellularly in synthetic culture medium. Alternatively, the absence of Bfr immunoreactivity in the F. tularensis-infected cells might reflect degradation of Bfr subsequent to its extracellular release.
FIG. 7.
Immunodetection of the intracellular release of culture filtrate proteins by F. tularensis. F. tularensis RCI (108), THP-1 cells (107), or RCI-infected THP-1 cells were treated with streptolysin O (+), and the soluble supernatant released from each sample was divided equally for analysis by immunoblotting with antibodies specific to KatG, GroEL, or Bfr.
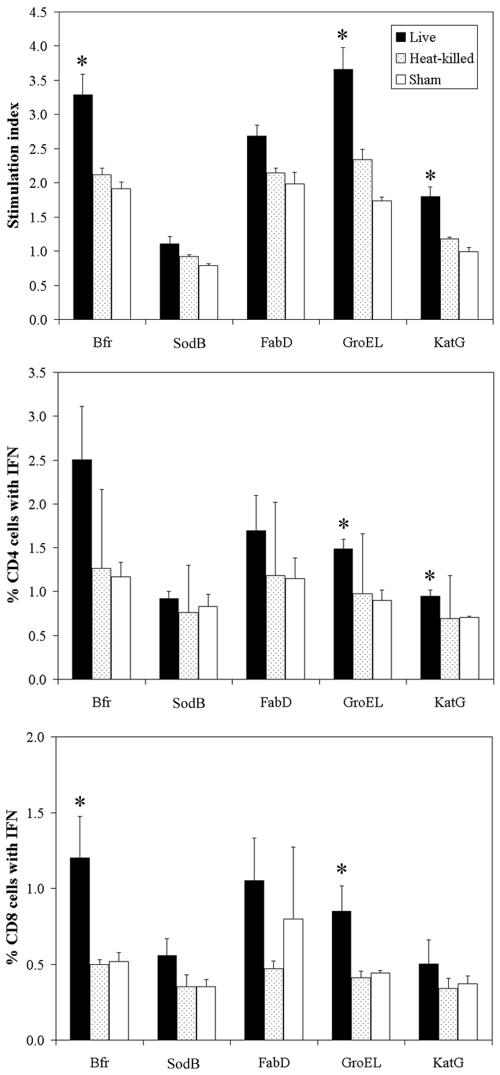
Culture filtrate proteins of F. tularensis are targets of cellular immune response in the infected mice.
Cellular immunity plays a dominant role in protection from intracellular bacterial infections. Both the CD4+ and CD8+ T cells are implicated in controlling F. tularensis infection (39). To assess whether culture filtrate proteins identified in this study are targets of the host immune system during a natural infection, we infected mice with F. tularensis LVS by the intradermal route, and at 5, 9, and 15 weeks postinfection, we assayed T-cell proliferation against F. tularensis culture filtrate proteins that were purified to homogeneity from recombinant E. coli. Control mice were immunized with heat-killed LVS or sham immunized (injected with saline only). At each of the three postinfection time points, T cells from mice infected with live LVS or injected with heat-killed LVS proliferated in response to individual F. tularensis culture filtrate proteins in a remarkably similar fashion. Hence, we present in Fig. 8 the combined T-cell proliferation from the three time points postinfection. In comparison with T cells from sham-immunized mice, T cells from mice infected with live LVS proliferated strongly in response to the culture filtrate proteins of F. tularensis. Stimulation indices for T-cell proliferation reached statistical significance for Bfr, GroEL, and KatG. In contrast, T cells from mice immunized with heat-killed LVS proliferated marginally in response to culture filtrate proteins. Moreover, Bfr, GroEL, and KatG stimulated IFN-γ secretion from both CD4 and CD8 T cells in mice immunized with live LVS but stimulated little IFN-γ secretion in mice immunized with the heat-killed bacteria. These results indicate that during an infection, the culture filtrate proteins are actively produced by the live bacterium and are processed and presented by the infected cells to the host immune system. The fact that Bfr, GroEL, and KatG stimulate functional CD4 and CD8 T-cell responses suggests that these protein antigens are promising candidates for inclusion in a vaccine.
FIG. 8.
T-cell responses to culture filtrate proteins in mice immunized with F. tularensis. BALB/c mice were injected with PBS or with heat-killed or live F. tularensis LVS. Proliferation of T cells to individual protein antigens was measured, and the stimulation index (SI) was calculated as described in Materials and Methods. Data are the mean SI values ± standard errors (SE) (error bars) for nine animals in a group. The frequency of IFN-γ-secreting CD4+ and CD8+ T cells specific for individual protein antigens was determined by flow cytometry analysis. Data are presented as means ± SE for three mice per group. Values for animals immunized with live LVS and sham-immunized animals that were significantly different (P value of less than 0.05 by Student's t test) are indicated by an asterisk.
DISCUSSION
We have identified 12 major extracellular proteins released by the attenuated LVS and fully virulent F. tularensis RCI. Among the major extracellular proteins identified, we observed important differences between LVS and RCI in 2-DE profiles for three of these proteins—KatG, DnaK, and Bfr. KatG and DnaK were more abundant in the RCI culture filtrate, whereas Bfr was more abundant in the LVS culture filtrate.
In this study, we cultivated F. tularensis in a chemically defined medium and observed that F. tularensis RCI grows more rapidly and to a higher culture density than F. tularensis LVS in a 24-h period. This difference in growth rate is highly reproducible and cannot be explained by a lower number of viable F. tularensis LVS in the initial inoculum. However, this divergence in growth rates is absent for the first 4 h during which time both strains grow from an initial OD of 0.1 to an OD of ∼0.4. It is conceivable that the nutrients stored in F. tularensis from growth on solid chocolate agar are sufficient to support two generations of bacterial division. After the initial growth phase, the growth rate of F. tularensis LVS lags behind that of F. tularensis RCI by at least twofold. Although growing significantly slower in the defined medium, with an extended cultivation time, F. tularensis LVS can still achieve the same ultimate cell density as that of F. tularensis RCI. In one experiment, for example, the F. tularensis RCI culture reached an OD of 3.5 in 20 h, whereas it took 45 h for F. tularensis LVS to reach a similar culture density. The lower growth rate of F. tularensis LVS in Chamberlain medium could be due to slower adaptation or to less efficient transport or utilization of certain nutrient(s) in the defined medium. F. tularensis LVS is a strain used widely in tularemia research and presumably has gone through many passages in laboratories over the years. It is possible that the slower growth of LVS in Chamberlain medium is merely the result of extensive passages on enriched agars and media. Alternatively, the differential growth rates of F. tularensis LVS and RCI in Chamberlain medium may reflect a fundamental difference between F. tularensis subsp. holarctica and the more virulent subspecies tularensis.
KatG is the most abundant protein released by F. tularensis RCI. Both the amount of KatG released into culture filtrate and the abundance relative to other proteins is significantly less for F. tularensis LVS than for F. tularensis RCI. Interestingly, for both strains, there was an approximately fourfold increase in the abundance of KatG in the culture filtrate as the cultures grew from 4 h to 14 h. The more abundant KatG in the culture filtrate of F. tularensis RCI observed at these two time points may be due to the combination of a higher production and a higher rate of export of the protein. In one experiment, we assayed catalase activity and total bacterial protein in the culture filtrate and the pellet of F. tularensis cultures harvested at the 14-h time point (optical densities of the culture were 2.4 for RCI and 0.7 for LVS). We found that catalase enzymatic activity in the bacterial pellet was 8-fold higher for RCI (i.e., RCI/LVS ratio of 8:1), while the total amount of bacterial protein in the pellet was only 3.2-fold higher for RCI. At the same time, we found that the catalase activity in the culture filtrate was 14-fold higher for RCI than for LVS, whereas the total amount of bacterial protein in the culture filtrate was only 1.5-fold higher for RCI than for LVS. Normalizing for total bacterial protein, the RCI strain had 2.5-fold-more catalase activity in the bacterial pellet and 9.3-fold-more catalase activity in the culture filtrate than the LVS strain did.
Catalase-peroxidase is a bifunctional enzyme exhibiting both catalase and broad-spectrum peroxidase activities (40). It contains a heme prosthetic group and catalyzes a multistep oxidation reaction involving hydrogen peroxide as the electron acceptor. KatG of F. tularensis is phylogenetically closest to that of Yersinia pestis (GenBank accession number AF135170) and L. pneumophila (GenBank accession number AF276752) both with 58% identity and 72% similarity and to that encoded on a plasmid in E. coli (O157:H7) (GenBank accession number X89017) with 56% identity and 72% similarity. In addition to their apparent sequence homology, the catalase-peroxidase of each of these four bacterial species includes a signal peptide as a unique feature, which is absent from other known members of the catalase-peroxidase subfamily (40) and are probably exported via a Sec-dependent pathway (34). Our analysis of the cytosol extract from streptolysin O-permeabilized macrophages demonstrates that KatG is secreted by F. tularensis growing intracellularly in macrophages. Using cryoimmunoelectron microscopy, we have found that in addition to being secreted away from the bacterium, the majority of immunogold staining for KatG is located at the periphery of the bacteria, consistent with a periplasmic distribution (unpublished results). L. pneumophila produces two catalase-peroxidases, KatA and KatB. Whereas KatB is located exclusively in the bacterial cytosol, KatA possesses a signal peptide and is present in the periplasm of the bacterium. Amemura-Maekawa and coworkers proposed that the periplasmic catalase-peroxidase (KatA) protects L. pneumophila from oxidative stresses by inactivating the hydrogen peroxide converted from superoxide radicals by the action of SodC, a Cu-Zn-superoxide dismutase which is usually located in bacterial periplasm (1). Gene mutation experiments demonstrated that phagosomes containing L. pneumophila katA or katB mutant exhibited a higher frequency of colocalization with LAMP-1, and the bacterial mutants showed defective intracellular multiplication in primary macrophages and amoebae (2). F. tularensis and L. pneumophila are both intracellular pathogens that must defend against host oxidative stress. It is conceivable that KatG of F. tularensis plays a role equivalent to the role of KatA in L. pneumophila and contributes to the pathogenesis and intracellular life style of F. tularensis. At present, KatG is the only catalase-peroxidase identified in the F. tularensis Schu 4 genome.
The remaining 11 proteins that we have identified in the early culture filtrates possess no predictable signal peptide, and based on their potential physiological function, these proteins are likely to have a major presence in the bacterial cytosol. Nevertheless, our metabolic radiolabeling studies show that these proteins are released early into the culture filtrate by actively growing bacteria. Whether these proteins are nonspecifically released by the bacterium or exported through a specific, yet unidentified, secretion mechanism remains to be determined. The different protein profiles between proteins from the culture filtrate and bacterial fraction combined with the rapid metabolic labeling and export of these proteins argue against autolysis as the main mechanism. We cannot exclude the possibility that these proteins are passively released during the active growth and division of the bacterium. ABC transporters are responsible for the export of a number of proteins that do not have the typical signal sequence (20). Mechanosensitive channel MscL has been shown to release DnaK and EF-Tu in an osmolarity-sensitive manner in E. coli (3). Another likely mechanism of export for these proteins is membrane shedding or membrane blebbing, a well-described phenomenon in gram-negative bacteria (4, 16, 19, 37). Proteins on membranes and in the periplasmic space can be enclosed in membrane vesicles and blebbed away from the bacterium.
Accumulated evidence has shown that in addition to their intracellular role, some cytosolic proteins are released and function on the bacterial surface as receptors. Glyceraldehyde-3-phosphate dehydrogenase is a protein of multiple functions. It is the major transferrin-binding protein in the cell walls of staphylococci (23). It is also identified as a major surface protein in group A streptococci that is capable of binding fibronectin, plasminogen, lysozyme, and actin (11, 27). In addition, glyceraldehyde-3-phosphate dehydrogenase and DnaK of Listeria monocytogenes are present in the cell wall and show strong binding affinity to plasminogen (33). Export of GroEL has been reported previously for Bartonella bacilliformis (22), Helicobacter pylori (36), and L. pneumophila (10). In the case of H. pylori and L. pneumophila, the exported GroEL may mediate gastric colonization and adherence to HeLa cells, respectively (10, 18). Whether the 11 proteins that we have identified in the F. tularensis culture filtrate have physiological roles in this extracellular site remains to be elucidated.
Bacterioferritin of F. tularensis RCI exhibits a moderate homology of approximately 21% identity and 44% similarity to that of Brucella melitensis (Swiss-Prot accession number P49944) and a strict anaerobe, Desulfovibrio desulfuricans (Swiss-Prot accession number Q93PP9). In the cases studied, bacterioferritin assembles as a spherical protein shell of 24 subunits with 12 heme b prosthetic groups between subunit pairs surrounding a central nonheme iron storage cavity (24, 38). Bacterioferritin acts as an iron storage protein and may play a role in protection against cell-damaging free radicals generated from oxygen in the presence of free iron (6). F. tularensis LVS and RCI each have multiple Bfr isoforms. Among these various Bfr isoforms, the most abundant LVS isoform is absent from the RCI culture filtrate. Similarly, a Bfr protein spot unique to F. tularensis subsp. holarctica was recently reported in a comparative proteome analysis of the bacterial cell lysates from F. tularensis subspecies holarctica and tularensis (17).
Analysis of the cytosol of F. tularensis-infected macrophages demonstrated that KatG and GroEL were released into the host cell cytosol. These proteins would thus be expected to be available for processing and presentation by the host cell. Consistent with this, T cells from LVS-infected mice proliferated strongly and released IFN-γ in response to these proteins. Although the presence of Bfr in macrophage cytosol was not confirmed in this study, T cells also responded strongly to Bfr.
In summary, we have identified F. tularensis proteins that are present in abundance in the culture filtrate and demonstrated that mice infected with F. tularensis LVS develop a significant cell-mediated immune response to these culture filtrate proteins. Our successful expression and purification of these proteins in soluble form will enable us to continue studies of the host immune response to these proteins and their vaccine potential. It is particularly noteworthy that we have found that KatG is secreted in abundance by the fully virulent F. tularensis but is secreted at a lower level by the attenuated LVS. It is possible that a vaccine utilizing this protein will have advantages over LVS in conferring protection against virulent F. tularensis.
Acknowledgments
We are grateful to the F. tularensis (Schu 4) genome sequencing consortium for sharing unpublished sequence data.
This work was supported by grant DAMD17-03-1-0052 from the U.S. Army Medical Research and Material Command and by grants AI065359 and HL077000 from the National Institutes of Health.
Editor: D. L. Burns
REFERENCES
- 1.Amemura-Maekawa, J., S. Mishima-Abe, F. Kura, T. Takahashi, and H. Watanabe. 1999. Identification of a novel periplasmic catalase-peroxidase KatA of Legionella pneumophila. FEMS Microbiol. Immunol. 176:339-344. [DOI] [PubMed] [Google Scholar]
- 2.Bandyopadhyay, P., B. Byrne, Y. Chan, M. S. Swanson, and H. M. Steinman. 2003. The Legionella pneumophila catalase-peroxidases are required for proper trafficking and growth in primary macrophages. Infect. Immun. 71:4526-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berrier, C., A. Garrigues, G. Richarme, and A. Ghazi. 2000. Elongation factor Tu and DnaK are transferred from the cytoplasm to the periplasm of Escherichia coli during osmotic downshock presumably via the mechanosensitive channel MscL. J. Bacteriol. 182:248-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beveridge, T. J. 1999. Structures of gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 181:4725-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blander, S. J., and M. A. Horwitz. 1989. Vaccination with the major secretory protein of Legionella pneumophila induces cell-mediated and protective immunity in a guinea pig model of Legionnaires' disease. J. Exp. Med. 169:691-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carrondo, M. A. 2003. Ferritins, iron uptake and storage from the bacterioferritin viewpoint. EMBO J. 22:1959-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chamberlain, R. E. 1965. Evaluation of a live tularemia vaccine prepared in a chemically defined medium. Appl. Microbiol. 13:232-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clemens, D. L., B. Lee, and M. A. Horwitz. 2004. Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect. Immun. 72:3204-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis, J., P. C. F. Oyston, M. Green, and R. W. Titball. 2002. Tularemia. Clin. Microbiol. Rev. 15:631-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garduno, R. A., G. Faulkner, M. A. Trevors, N. Vats, and P. S. Hoffman. 1998. Immunolocalization of Hsp60 in Legionella pneumophila. J. Bacteriol. 180:505-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gase, K., A. Gase, H. Schirmer, and H. Malke. 1996. Cloning, sequencing and functional overexpression of the Streptococcus equisimilis H46A gapC gene encoding a glyceraldehyde-3-phosphate dehydrogenase that also functions as a plasmin(ogen)-binding protein. Purification and biochemical characterization of the protein. Eur. J. Biochem. 239:42-51. [DOI] [PubMed] [Google Scholar]
- 12.Goguen, J. D., N. P. Hoe, and Y. V. Subrahmanyam. 1995. Proteases and bacterial virulence: a view from the trenches. Infect. Agents Dis. 4:47-54. [PubMed] [Google Scholar]
- 13.Golovliov, I., V. Baranov, Z. Krocova, H. Kovarova, and A. Sjöstedt. 2003. An attenuated strain of the facultative intracellular bacterium Francisella tularensis can escape the phagosome of monocytic cells. Infect. Immun. 71:5940-5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harlowe, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 15.Horwitz, M. A., B.-W. E. Lee, B. J. Dillon, and G. Harth. 1995. Protective immunity against tuberculosis induced by vaccination with major extracellular proteins of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 92:1530-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hozbor, D., M. E. Rodriguez, J. Fernandez, A. Lagares, N. Guiso, and O. Yantorno. 1999. Release of outer membrane vesicles from Bordetella pertussis. Curr. Microbiol. 38:273-278. [DOI] [PubMed] [Google Scholar]
- 17.Hubálek, M., L. Hernychová, J. Havlasová, I. Kasalová, V. Neubauerová, J. Stulík, A. Macela, M. Lundqvist, and P. Larsson. 2003. Towards proteome database of F. tularensis. J. Chromatogr. B 787:149-177. [DOI] [PubMed] [Google Scholar]
- 18.Huesca, M., S. Borgia, P. Hoffman, and C. A. Lingwood. 1996. Acidic pH changes receptor binding specificity of Helicobacter pylori: a binary adhesion model in which surface heat shock (stress) proteins mediate sulfatide recognition in gastric colonization. Infect. Immun. 64:2643-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keenan, J., T. Day, S. Neal, B. Cook, G. Perez-Perez, R. Allardyce, and P. Bagshaw. 2000. A role for the bacterial outer membrane in the pathogenesis of Helicobacter pylori infection. FEMS Microbiol. Lett. 182:259-264. [DOI] [PubMed] [Google Scholar]
- 20.Kuchler, K. 1993. Unusual routes of protein secretion: the easy way out. Trends Cell Biol. 3:421-426. [DOI] [PubMed] [Google Scholar]
- 21.Larsson, P., P. C. F. Oyston, P. Chain, M. C. Chu, M. Duffield, H. H. Fuxelius, E. Garcia, G. Halltorp, D. Johansson, K. E. Isherwood, P. D. Karp, E. Larsson, Y. Liu, S. Michell, J. Prior, R. Prior, S. Malfatti, A. Sjostedt, K. Svensson, N. Thompson, L. Vergez, J. K. Wagg, B. W. Wren, L. E. Lindler, S. G. Andersson, M. Forsman, and R. W. Titball. 2005. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat. Genet. 37:153-159. [DOI] [PubMed] [Google Scholar]
- 22.Minnick, M. F., L. S. Smitherman, and D. S. Samuels. 2003. Mitogenic effect of Bartonella bacilliformis on human vascular endothelial cells and involvement of GroEL. Infect. Immun. 71:6933-6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Modun, B., and P. Williams. 1999. The staphylococcal transferrin-binding protein is a cell wall glyceraldehyde-3-phosphate dehydrogenase. Infect. Immun. 67:1086-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore, G. R., S. Mann, and J. V. Bannister. 1986. Isolation and properties of the complex nonheme-iron containing cytochrome b-557 (bacterioferritin) from Pseudomonas aeruginosa. J. Inorg. Biochem. 28:329-336. [DOI] [PubMed] [Google Scholar]
- 25.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 26.Pal, P. G., and M. A. Horwitz. 1992. Immunization with extracellular proteins of Mycobacterium tuberculosis induces cell-mediated immune response and substantial protective immunity in a guinea pig model of pulmonary tuberculosis. Infect. Immun. 60:4781-4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pancholi, V., and V. A. Fischetti. 1992. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J. Exp. Med. 176:415-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prior, R. G., L. Klasson, P. Larsson, K. Williams, L. Lindler, A. Sjöstedt, T. Svensson, I. Tamas, B. W. Wren, P. C. Oyston, S. G. E. Andersson, and R. W. Titball. 2001. Preliminary analysis and annotation of the partial genome sequence of Francisella tularensis strain Schu 4. J. Appl. Microbiol. 91:614-620. [DOI] [PubMed] [Google Scholar]
- 29.Reeves, W. J., and G. M. Fimogarni. 1966. L-lactate dehydrogenase: Heart (H4). Methods Enzymol. 9:288-290. [Google Scholar]
- 30.Rutherford, K., J. Parkhill, J. Crook, T. Hornsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 31.Saslaw, S., H. T. Eigelsbach, J. Prior, H. Wilson, and S. Carhart. 1961. Tularemia vaccine study. I. Intracutaneous challenge. Arch. Intern. Med. 107:121-133. [DOI] [PubMed] [Google Scholar]
- 32.Saslaw, S., H. T. Eigelsbach, J. Prior, H. Wilson, and S. Carhart. 1961. Tularemia vaccine study. II. Respiratory challenge. Arch. Intern. Med. 107:702-714. [DOI] [PubMed] [Google Scholar]
- 33.Schaumburg, J., O. Diekmann, P. Hagendorff, S. Bergmann, M. Rohde, S. Hammerschmidt, L. Jänsch, J. Wehland, and U. Kärst. 2004. The cell wall subproteome of Listeria monocytogenes. Proteomics 4:2991-3006. [DOI] [PubMed] [Google Scholar]
- 34.Strathopoulos, C., D. R. Hendrixson, D. G. Thanassi, S. J. Hultgren, J. W. St. Geme III, and R. Curtiss III. 2000. Secretion of virulence determinants by the general secretory pathway in Gram-negative pathogens: an evolving story. Microbes Infect. 2:1061-1072. [DOI] [PubMed] [Google Scholar]
- 35.Titball, R. W., and A. Sjöstedt. 2003. Francisella tularensis: an overview. ASM News 69:558-563. [Google Scholar]
- 36.Vanet, A., and A. Labigne. 1998. Evidence for specific secretion rather than autolysis in the release of some Helicobacter pylori proteins. Infect. Immun. 66:1023-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wai, S. N., B. Lindmark, T. Soderblom, A. Takade, M. Westermark, J. Oscarsson, J. Jass, A. Richter-Dahlfors, Y. Mizunoe, and B. E. Uhlin. 2003. Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial ClyA cytotoxin. Cell 115:25-35. [DOI] [PubMed] [Google Scholar]
- 38.Yariv, J., A. J. Kalb, R. Sperling, E. R. Bauminger, S. G. Cohen, and S. Ofer. 1981. The composition and the structure of bacterioferritin of Escherichia coli. Biochem. J. 197:171-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yee, D., T. R. Rhinehart-Jones, and K. L. Elkins. 1996. Loss of either CD4+ or CD8+ T cells does not affect the magnitude of protective immunity to an intracellular pathogen, Francisella tularensis strain LVS. J. Immunol. 11:5042-5048. [PubMed] [Google Scholar]
- 40.Zámocky, M., Š. Janecek, and F. Koller. 2000. Common phylogeny of catalase-peroxidases and ascorbate peroxidases. Gene 256:169-182. [DOI] [PubMed] [Google Scholar]